ABSTRACT
Human epidermal growth factor receptor 3 (HER3, also known as ErbB3) has emerged as relevant target for antibody-mediated tumor therapy. Here, we describe a novel human antibody, IgG 3–43, recognizing a unique epitope formed by domain III and parts of domain IV of the extracellular region of HER3, conserved between HER3 and mouse ErbB3. An affinity of 11 nM was determined for the monovalent interaction. In the IgG format, the antibody bound recombinant bivalent HER3 with subnanomolar affinity (KD = 220 pM) and HER3-expressing tumor cells with EC50 values in the low picomolar range (27 - 83 pM). The antibody competed with binding of heregulin to HER3-expressing cells, efficiently inhibited phosphorylation of HER3 as well as downstream signaling, and induced receptor internalization and degradation. Furthermore, IgG 3–43 inhibited heregulin-dependent proliferation of several HER3-positive cancer cell lines and heregulin-independent colony formation of HER2-overexpressing tumor cell lines. Importantly, inhibition of tumor growth and prolonged survival was demonstrated in a FaDu xenograft tumor model in SCID mice. These findings demonstrate that by binding to the membrane-proximal domains III and IV involved in ligand binding and receptor dimerization, IgG 3–43 efficiently inhibits activation of HER3, thereby blocking tumor cell growth both in vitro and in vivo.
KEYWORDS: Cancer therapy, ErbB3, HER3, heregulin, internalization, phage display, receptor tyrosine kinase, therapeutic antibody
Abbreviations
- ADCC
antibody-dependent cell-mediated cytotoxicity
- AUC
area under the curve
- HRG
heregulin
- scFv
single-chain fragment variable
- SCID
severe combined immunodeficiency
Introduction
Human epidermal growth factor receptor 3 (HER3, also known as ErbB3) is a member of the ErbB receptor family further comprising epidermal growth factor receptor (EGFR or ErbB1), HER2 (ErbB2), and HER4 (ErbB4).1,2 Although previously neglected as a suitable target for cancer therapy, accumulating evidence now supports its role in tumorigenesis, cancer progression and the evolvement of resistance to therapy.3,4 A meta-analysis showed that expression of HER3 in solid tumors is associated with worse survival, especially in tumors in which HER2 is overexpressed.5 Moreover, acquired resistance to anti-EGFR and anti-HER2 treatment is frequently coupled to compensatory signaling of HER3, which can be a result of increased HER3 expression, decreased HER3 dephosphorylation, increased membrane localization, or upregulation of the HER3 ligands.6,7
The extracellular region of HER3 is composed of 4 domains. Domains I and III are involved in ligand binding, domains II and IV in receptor dimerization.8 Two natural ligands exist for HER3, heregulin-1 (HRG-1) and heregulin-2 (HRG-2), also known as neuregulin-1 (NRG-1) and neuregulin-2 (NRG-2), which comprise various isoforms also capable of binding to HER4.9 HER3, similar to EGFR and HER4, can adopt either an inactive tethered or an extended form.10 The extended form is stabilized by ligand binding, which liberates the dimerization domains II and IV, allowing the dimerization with other members of the HER family.11 Phosphorylation of the intracellular receptor domains results in activation of 2 major signaling pathways, the PI3K/AKT pathway and the MAPK/ERK pathway, regulating a variety of cellular processes, including survival, proliferation, migration and invasion.12,13 In contrast to the other family members, the protein tyrosine kinase homology domain of HER3 was found to be catalytically inactive.14 Because of its impaired kinase domain, phosphorylation of HER3 requires heterodimerization with other members of the receptor family. The prime partner is HER2, but EGFR and other receptor tyrosine kinases such as IGF-1R and c-Met can also form heterodimers with HER3.2,15,16 The presence of 6 binding sites for p85, the regulatory subunit of PI3K, within the HER3 cytoplasmic tail provides an explanation for the strong activation of this pathway by HER3-containing receptor heterodimers and the contribution of HER3 to survival signaling and therapy resistance.17,18
Although some weak autophosphorylation of HER3 was recently reported,19,20 the lack of substantial kinase activity makes HER3 rather unsuitable as a therapeutic target for small molecule inhibitors. Furthermore, oncogenic HER3 mutations have been identified in a subset of colon and gastric cancers, but these HER3 mutants still required the activity of HER2 to transform cells.21 Antibodies and various scaffold proteins binding to the extracellular region of HER3 have, therefore, been developed to interfere with HER3 activation.22 Mechanistically, neutralizing molecules such as antibodies can affect various steps of HER3 activation.11 Antibodies can bind to the ligand-binding sites formed by domain I and III, thereby directly inhibiting ligand binding. Alternatively, binding outside the ligand-binding site may sterically prevent the receptor from adopting a conformation required for high affinity ligand binding and dimerization, thus stabilizing the inactive tethered conformation. Furthermore, antibodies can bind to the dimerization domains II or IV directly, inhibiting receptor dimerization. Many of the anti-HER3 antibodies developed for clinical use are capable of inhibiting ligand binding.13,22,23 Others, such as LJM716 and KTN3379, lock HER3 in an inactive conformation, and thus inhibit ligand-dependent and independent receptor activation.24,25
Here, we describe a novel human anti-HER3 antibody, IgG 3–43, derived from a naïve antibody library by phage display. Using fragments of the extracellular region of HER3, we show that the antibody recognizes a unique epitope formed by domain III and IV. IgG 3–43 was further analyzed for binding to HER3-expressing cell lines, inhibition of heregulin binding and receptor phosphorylation, establishing that the antibody is capable of efficiently inhibiting HER3 activation and cell proliferation in both ligand-dependent and ligand-independent manner. Finally, anti-tumor activity was demonstrated in a mouse xenograft tumor model.
Results
Selection of anti-HER3 scFv
A panel of anti-HER3 scFv was isolated from the human antibody phage library HAL7/8 by selection against a human HER3-Fc fusion protein (amino acids (aa) 20–643). A total of 8 clones showing binding to HER3-Fc by ELISA was further analyzed for binding to HER3-expressing MCF-7 cells by flow cytometry. Here, only 3 clones (3–39, 3–43, 3–44) showed binding, with the strongest binding seen for scFv 3–43 (Fig. S1a, b). ScFv 3–39 and 3–43 were further tested for inhibition of HER3 phosphorylation using MCF-7 cells in the absence or presence of heregulin (Fig. S1c). Potent inhibition of heregulin-induced HER3 phosphorylation was seen after 15 and 60 min of pre-incubation with scFv 3–43 (10 µg/ml), whereas scFv 3–39 showed only partial inhibition.
Specificity and epitope mapping of IgG 3–43
ScFv 3–43 was chosen for further analysis and was converted into a human IgG1 (IgG 3–43) carrying 2 mutations (S239D/I332E) in the Fc region demonstrated to increase Fc receptor binding and to enhance antibody-dependent cell-mediated cytotoxicity (ADCC).26 The antibody was produced in HEK293–6E cells and purified by protein A chromatography. Approximately 43 mg protein could be purified from 1 L of supernatant. Integrity of the protein was confirmed by SDS-PAGE and size-exclusion chromatography (SEC) (Fig. 1a, b). The antibody retained the antigen specificity as shown by ELISA. Here, strong binding was detected for HER3-Fc, whereas no binding was found for EGFR-Fc and HER2-Fc (Fig. 1c). By ELISA, IgG 3–43 bound with an EC50 of 90 pM to immobilized HER3-Fc (Fig. 1d). IgG 3–43 also recognized mouse ErbB3-Fc although with a higher EC50 value of 2.7 nM, indicative of binding to a conserved epitope, (Fig. 1d). Immunoblotting analysis with scFv 3–43 revealed binding to non-reduced HER3-Fc, but not to reduced antigen, suggesting that the antibody binds to a conformational epitope (Fig. S2). Binding was further analyzed by quartz crystal microbalance (QCM) measurements using immobilized IgG 3–43 and either a dimeric HER3-Fc or a monomeric His-tagged HER3 fragment (aa 27–643). Strong binding with a KD of 220 pM was determined for HER3-Fc binding (Fig. 1e). By contrast, monomeric HER3 bound with a KD of 11.2 nM, especially due to a faster off-rate (Fig. 1f; Table 1). We next analyzed whether IgG 3–43 competes with ligand binding. To this end, MCF-7 cells were incubated with his-tagged heregulin (Fig. 1g). Here, IgG 3–43 strongly reduced heregulin binding, while cetuximab, included as a negative control, showed only marginal effects.
Figure 1.
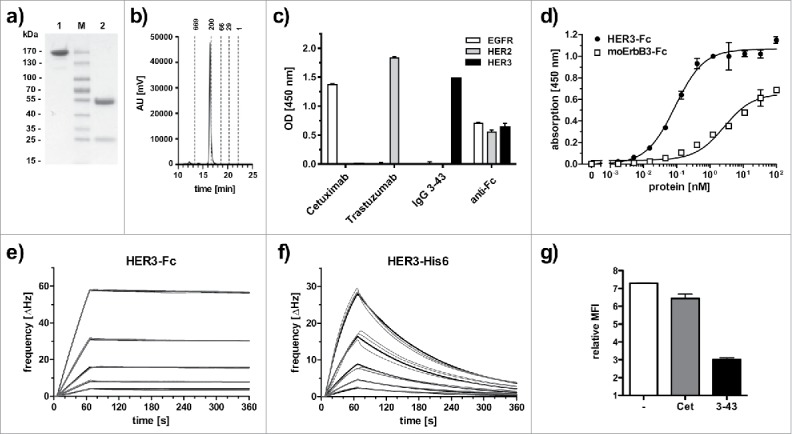
Biochemical characterization and antigen binding of IgG 3–43. (a) SDS-PAGE analysis (Coomassie stained) under non-reducing (1) and reducing (2) conditions. (b) Size-exclusion chromatography of IgG 3–43. (c) Selectivity for HER3 analyzed by ELISA with immobilized human EGFR-Fc, HER2-Fc, and HER3-Fc. Cetuximab (anti-EGFR) and trastuzumab (anti-HER2) were included as positive controls. An anti-Fc antibody was included as a coating control. (d) Binding of IgG 3–43 to HER3 and mouse ErbB3-Fc in ELISA. Bound protein was detected with HRP-conjugated anti-human Fab antibody. (e, f) Quartz crystal microbalance measurements with IgG 3–43 immobilized on a carboxyl chip and incubation with either dimeric HER3-Fc (0.625 - 10 nM) (e) or a monomeric his-tagged extracellular region of HER3 (1.25 - 20 nM) (f). Curve fits are shown as bold lines. (g) Inhibition of binding of his-tagged recombinant human heregulin to MCF-7 cells by preincubation with a 60-fold molar excess of IgG 3–43 was analyzed by flow cytometry. Cetuximab (Cet) was included as negative control.
Table 1.
Monovalent and bivalent antigen binding of IgG 3–43 determined by QCM with immobilized IgG 3–43.
| analyte | Bmax | ka (M−1s−1) | kd (s−1) | KD (nM) |
|---|---|---|---|---|
| HER3-Fc | 237.32 | 4.7 × 105 | 1.0 × 10−4 | 0.22 |
| HER3-His6 | 63.55 | 6.2 × 105 | 7.0 × 10−3 | 11.2 |
The epitope of IgG 3–43 was determined by ELISA using fragments of human HER3 expressed as Fc fusion proteins (Fig. S3). Binding was seen for full-length HER3, and fragments containing domains II-IV and III-IV, whereas domain IV alone showed no binding. Fragments containing only a part of domain III with deletions from the N-terminal end and the entire domain IV (fragments 5 and 6) showed no binding, indicating the importance of the N-terminal region of domain III for antibody binding. Surprisingly, fragment 7 lacking domain IV, i.e., covering domains I - III (aa 20 - 531), showed significantly reduced binding (by ∼50%). Fragments consisting of domains I-II (fragment 9) or domain I-II plus the first 29 residues of domain III were not recognized by IgG 3–43. These findings indicate that domain III and IV both contribute to the formation of the epitope. C-terminal shortening of domain IV (fragments 10 and 11) showed that a fragment comprising aa 329 - 587, i.e., containing the entire domain III and 56 amino acids of the N-terminal region of domain IV, was the minimal region required for the formation of the IgG 3–43 epitope.
Binding to HER3-expressing tumor cells
Next, different cancer cell lines were analyzed for binding of IgG 3–43. These cell lines (MCF-7, FaDu, BT474, A431, NCI-N87, A549, SKBR3) were first quantified by indirect immunofluorescence staining for expression of EGFR, HER2, and HER3 (Table 2). This analysis showed that all cell lines were HER3-positive, expressing low to moderate levels of the receptor. IgG 3–43 bound to these HER3-expressing cell lines with low EC50 values in a range between 27 to 83 pM (Fig. 2, Table 2).
Table 2.
Expression levels (receptors per cell) of EGFR, HER2, and HER3 on various cancer cell lines and binding of IgG-43 as analyzed by flow cytometry.
| IgG 3–43 binding | ||||
|---|---|---|---|---|
| EGFR | HER2 | HER3 | (EC50 in pM) | |
| MCF7 | 1,000 | 21,230 | 17,280 | 37 |
| FaDu | 143,250 | 15,800 | 2,880 | 30 |
| BT474 | 7,220 | > 572,000 | 11,240 | 59 |
| A431 | > 572,000 | 8,740 | 4,370 | 44 |
| NCI-N87 | 16,180 | > 572,000 | 3,320 | 27 |
| A549 | 64,130 | 6,390 | 1,470 | 83 |
| SKBR-3 | 29,800 | > 572,000 | 14,080 | 42 |
Figure 2.
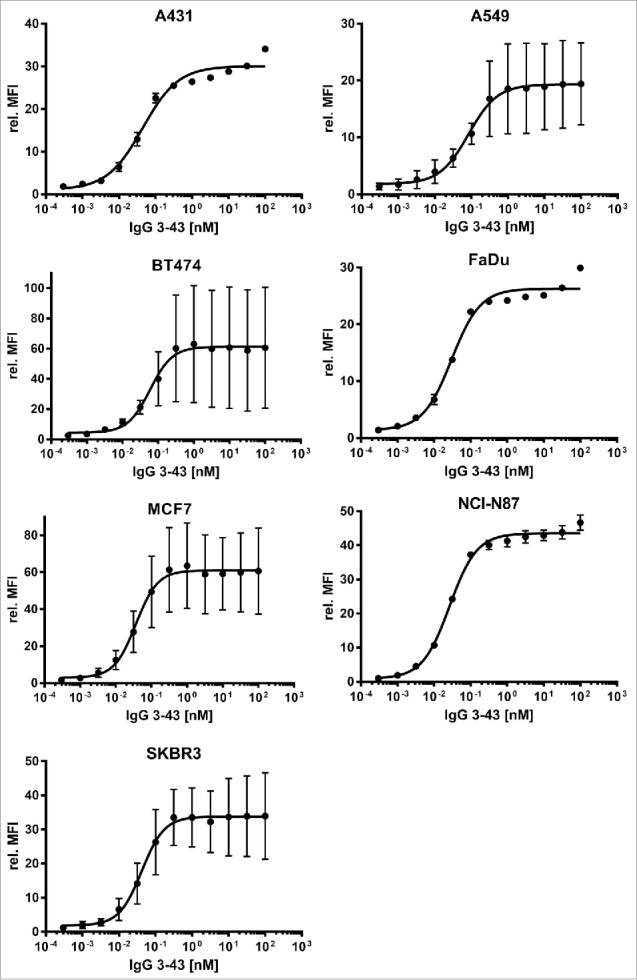
Binding of IgG 3–43 to HER3-expressing tumor cell lines. Various tumor cell lines (as indicated) were incubated with varying concentrations of IgG 3–43 and bound antibody was detected with a PE-labeled secondary anti-human Fc antibody and analyzed by flow cytometry (n = 3, ± SD).
Antibody-dependent cell-mediated cytotoxicity
SKBR3 cells were incubated with natural killer (NK) cell-containing human PBMCs at a ratio of 1:2 in the presence of varying concentrations of IgG 3–43 for 1 day. Efficient lysis of tumor cells was observed with an EC50 value of 2.4 pM. Compared to IgG 3M6 (also comprising the 2 mutations (S239D/I332E) in the Fc region to enhance ADCC), which is derived from the anti-HER3 antibody MM-121 (seribantumab) currently in Phase 2 clinical trials,27 an ∼4-fold stronger ADCC was determined (Fig. S4). This might be due to a weaker binding of 3M6 to HER3-Fc in ELISA and HER3-expressing cells (Fig. S4).
Inhibition of HER3 phosphorylation and downstream signaling proteins
IgG 3–43 was analyzed for its capacity to interfere with phosphorylation of HER3 in different cell lines (MCF-7, NCI-N87, A431, A549, FaDu) (Fig. 3). Semi-confluent cells were serum-starved overnight and incubated with IgG 3–43 for one hour, and then either left unstimulated or were stimulated with heregulin (50 ng/ml) for 15 minutes. Western blot analyses of the cell lysates showed basal HER3 phosphorylation and heregulin-mediated upregulation of HER3 phosphorylation for some of the cell lines. In all 5 cell lines, HER3 phosphorylation could be efficiently blocked by IgG 3–43, whereas a control antibody showed no or only marginal effects. In addition, decreased levels of pAkt and pErk (MCF-7, A549) were observed after incubation of heregulin-stimulated cells with IgG 3–43. Using MCF-7 cells, a titration of IgG 3–43 further revealed an IC50 value in the picomolar range (109 ± 35 pM, n = 3) for blockade of heregulin-induced HER3 phosphorylation (Fig. S5). Increased inhibition of HER3 phosphorylation was observed compared with IgG 3M6 (IC50 value of 640 pM) (Fig. S5). Importantly, IgG 3–43 alone, i.e., without the addition of heregulin, did not induce HER3 phosphorylation, demonstrating the absence of any agonistic activity. Instead, the basal HER3 phosphorylation detectable in FaDu, A549 and A431 cells and basal Akt and Erk phosphorylation in FaDu cells were also reduced by IgG 3–43. These experiments also showed that the levels of HER3 receptor decreased after incubation with IgG 3–43. This was studied in more detail in MCF-7 cells over an incubation period of 6 h (Fig. 4a). A sustained downregulation of HER3 was observed, with strongly reduced levels seen by 60 minutes of incubation, indicating antibody-mediated internalization and degradation of HER3 (Fig. 4b). This was confirmed by confocal microcopy studies using Cy5-labeled IgG 3–43. After 1 to 2 hours, a strong intracellular accumulation of IgG 3–43 was detectable (Fig. 4c).
Figure 3.
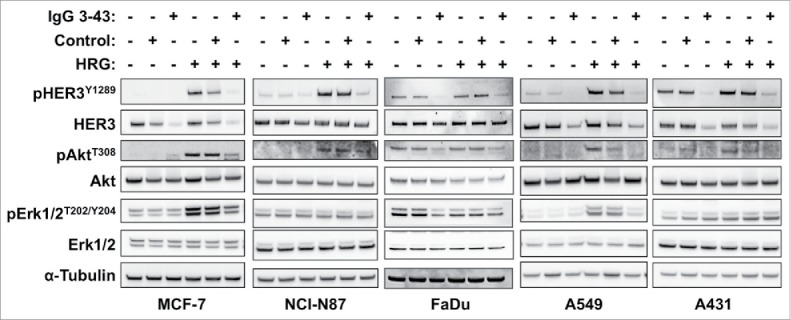
IgG 3–43 inhibits HRG-induced phosphorylation of HER3 and downstream targets. Indicated cell lines were serum-starved overnight and incubated for one hour with 100 nM IgG 3–43 or control IgG (rituximab) in the absence or presence of 50 ng/ml human heregulin-β1 (HRG). Subsequently, cells lysates were analyzed by western blotting.
Figure 4.
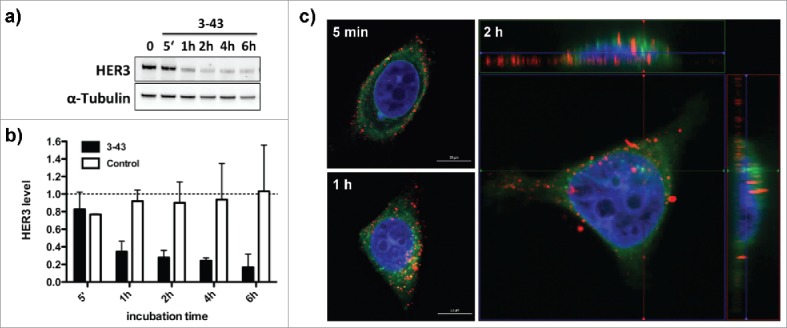
Internalization of IgG 3–43. (a) Cellular HER3 expression levels after incubation of MCF-7 cells with IgG 3–43 (100 nM) analyzed by immunoblotting. (b) Quantification of HER3 levels after incubation with IgG 3–43 or an irrelevant control antibody for the indicated time points. HER3 levels were analyzed by western blot. (c) Confocal fluorescence analysis of MCF-7 cells incubated with Cy5-labeled IgG 3–43 (red) for the indicated time points. Cells were counter-stained with DAPI (blue) and Con-A (green).
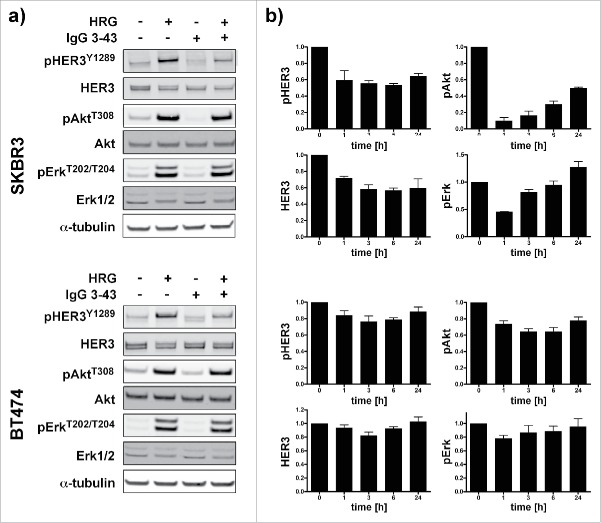
Furthermore, we analyzed SKBR3 and BT474 cells, which can activate HER3 in a ligand-independent manner due to HER2 overexpression, for inhibition of HER3 phosphorylation and cell proliferation. To specifically address basal phosphorylation, these experiments were performed in growth medium containing 10% serum. Under these conditions, inhibition of HER3 phosphorylation was observed in unstimulated and heregulin-stimulated cells (Fig. 5a). Consistent with the previous results, analysis of basal pHER3, HER3, pAkt and pErk levels over a period of 24 h showed downregulation within the first hour of incubation followed by a recovery after 24 h (Fig. 5b).
Figure 5.
Inhibition of HER3 phosphorylation and downstream signaling in SKBR3 and BT474 cells. (a) Cells were serum starved overnight and then incubated for 1 h with IgG 3–43 (50 nM) followed by stimulation with or without heregulin (50 ng/ml) for 15 min. (b) SKBR3 and BT474 cells were grown without serum starvation and incubated with IgG 3–43 for up to 24 h without the addition of heregulin. Expression of pHER3, HER3, pAkt, and pErk levels were quantified by immunoblotting (n = 2 for SKBR3, n = 3 for BT474).
Inhibition of ligand-dependent and ligand-independent cell proliferation
Next, IgG 3–43 was evaluated for its ability to reduce tumor cell proliferation in vitro. MCF-7, NCI-N87, A549 cell lines were grown in low serum in the presence of heregulin (10 ng/ml) and IgG 3–43 or an irrelevant control antibody (10 µg/ml). Proliferation was determined after 1 week of incubation. For all 3 cell lines, a significant reduction of proliferation compared with the control antibody was observed (Fig. 6a). In addition, we analyzed FaDu cells known to endogenously express heregulin30 for inhibition of proliferation by IgG 3–43. Here, an IC50 value of 273 pM was determined for IgG 3–43, whereas a control antibody showed no effects (Fig. 6b). These findings demonstrate that IgG 3–43 efficiently inhibits heregulin-mediated proliferation of different tumor cell lines. In further experiments we analyzed whether IgG 3–43 is also capable of inhibiting ligand-independent proliferation of SKBR3 and BT474. Using a colony formation assay, we could show that IgG 3–43 inhibited proliferation of both cell lines, similar to trastuzumab included as a positive control (Fig. 6c).
Figure 6.
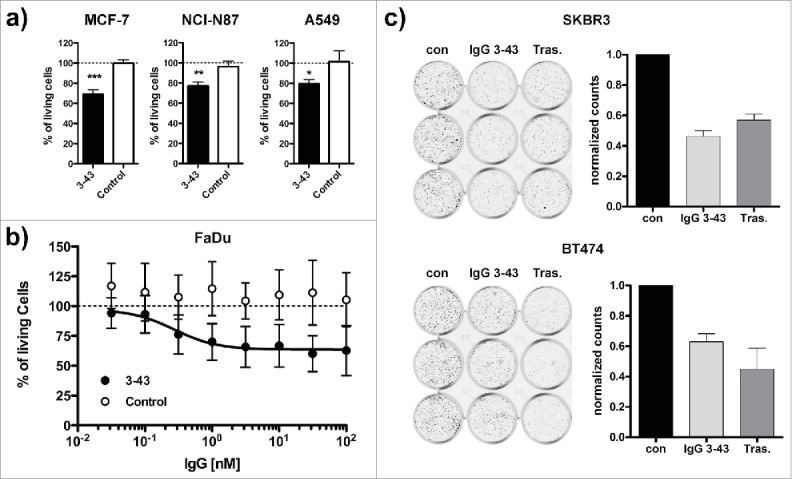
Inhibition of ligand-dependent and ligand-independent proliferation. (a) Inhibition of proliferation of MCF-7, NCI-N87, and A549 cells incubated for one week under low (0.2%) serum concentrations in the presence of 10 ng/ml heregulin and either IgG 3–43 (3–43) or rituximab (Control) as a negative control antibody. (b) Inhibition of proliferation of FaDu cells without the addition of heregulin under the same conditions as in (a). (c) Colony formation assay. SKBR3 or BT474 were grown in 12-well plates (1,000 cells per well) and incubated with IgG 3–43 (50 nM) or trastuzumab (Tras.) for 12 d. Medium and antibodies were exchanged after 7 d. Cells incubated without antibody were included as a control (con). Shown are crystal violet-stained wells and the quantification of 2 independent experiments performed with triplicate wells.
Inhibition of tumor growth in a xenograft tumor model
Finally, the antitumor activity of IgG 3–43 was tested in a subcutaneous FaDu xenograft model in SCID mice. Cells were injected into both flanks of the mice and treatment started when tumors reached a volume of ∼80 mm3 (14 d after tumor cell inoculation). Mice received twice weekly intravenous injections for 3 weeks (q2wx3) at doses of 30, 100, and 300 µg, including phosphate-buffered saline (PBS) as negative control. Antitumor effects were observed for all 3 dosing regimens of IgG 3–43, with increased survival and a tumor growth inhibition (Fig. 7). A significantly increased median survival was determined for the 2 higher doses (59 d and 71.5 d for the 100 and 300 µg dose, respectively, versus 51.5 d for untreated animals) and significantly reduced tumor volumes for all 3 doses after completion of treatment (day 42) (Fig. 7e, f). Serum levels of IgG 3–43 were determined after the first and last injection (Fig. 7g). The AUC0–72h values increased approximately 4- to 6-fold between the first to the last injection, indicating an accumulating dose of antibody. This resulted also in increased terminal half-lives for all 3 doses, while initial half-lives were not affected (Table 3). Prolonged half-lives seen for the highest dose (∼10 mg/kg) indicates saturation of HER3 and, thus a target-mediated drug disposition, described also for other anti-HER3 antibodies, e.g., RG7116.28 No effects on body weights were observed during treatment with the 3 dosing regimens. Taken together, the anti-proliferative effects observed in vitro translated into inhibition of tumor growth in vivo.
Figure 7.
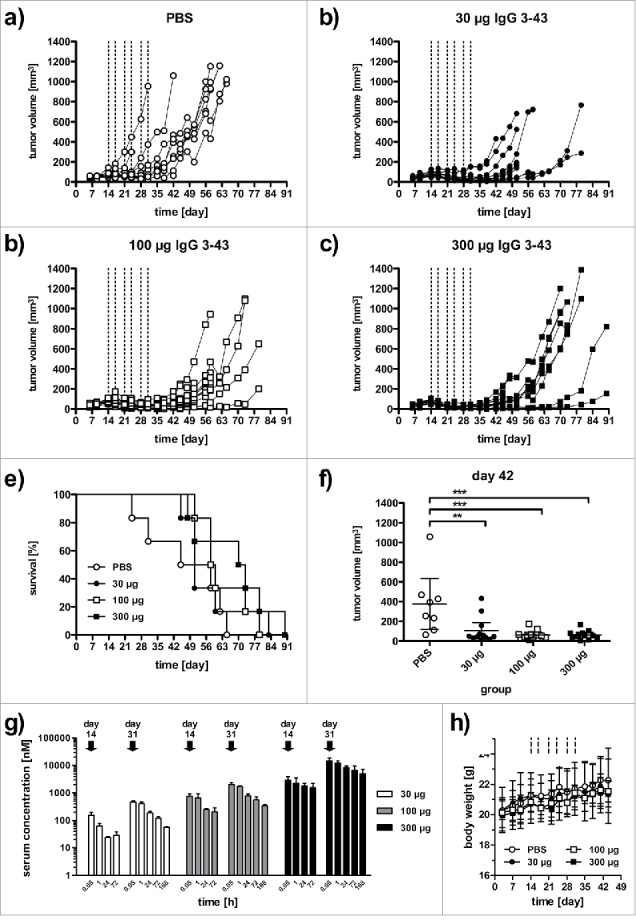
IgG 3–43 inhibits growth of a subcutaneous xenograft FaDu tumor model in SCID mice. Mice were treated when tumors reached a size of ∼100 mm3 (2 weekly injections for 3 weeks, see lines) with the indicated doses. (a-d) Growths of individual tumors in mice treated with PBS (a), 30 µg IgG 3–43 (b), 100 µg IgG 3–43 (c), or 300 µg IgG 3–43 (d). (e) Kaplan-Meier plot of survival. (f) Tumor volumes at day 42. (g) Pharmacokinetics of IgG 3–43. Serum concentrations of IgG 3–43 were determined by ELISA after the first intravenous injection (day 14) and last intravenous injection (day 31) of treatment at the indicated doses. (h) Body weight of animals during treatment.
Table 3.
PK properties of IgG 3–43 after the first (day 14) and last (day 32) injection into tumor-bearing mice.
| dose [µg] | injection | t½α [h] | t½β [h] | AUC0–72h [nM·h] | Cmax [nM] |
|---|---|---|---|---|---|
| 30 | first | 11.7 ± 0.8 | 59.1 ± 28.4 | 2,365 ± 84 | 157 ± 35 |
| last | 19.7 ± 4.0 | 87.1 ± 9.9 | 14,498 ± 873 | 480 ± 32 | |
| 100 | first | 17.3 ± 5.3 | 47.8 ± 7.2 | 22,101 ± 5,882 | 717 ± 147 |
| last | 18.7 ± 3.0 | 126.5 ± 10.1 | 62,605 ± 5,046 | 2,172 ± 345 | |
| 300 | first | 32.4 ± 9.5 | 127.9 ± 9.6 | 151,558 ± 38,462 | 4,453 ± 1,569 |
| last | 37.1 ± 9.4 | 224.2 ± 116.6 | 610,044 ± 137,488 | 22,582 ± 5,472 |
Discussion
A HER3-binding scFv (3–43) cross-reactive with mouse ErbB3 was isolated from a naïve human antibody phage display library. The affinity of the scFv for monomeric HER3 was determined to be 11.2 nM, whereas IgG 3–43 showed binding in the picomolar range both to a bivalent HER3-Fc fusion protein and to HER3-expressing cells. This indicates that avidity effects cause an approximate 10- to 100-fold increase in binding, in accordance with data obtained for anti-EGFR antibodies with affinities in the nanomolar range.29
The epitope of IgG 3–43, which is sensitive to reduction, was mapped to a region comprising domain III and the N-terminal region of domain IV (aa 329–587). Surprisingly, a smaller fragment (aa 20–550) was not able to bind IgG 3–43 and a fragment comprising domains I-III showed strongly reduced binding, indicating that the antibody recognizes a unique, complex epitope spanning domains III and IV, which discriminates IgG 3–43 from other known anti-HER3 antibodies (Fig. S6). Supported by the observation that IgG 3–43 inhibits ligand binding and ligand-dependent HER3 activation, but is also able to inhibit ligand-independent activation of HER3 as shown for HER2-overexpressing cell lines, these findings indicate that, mechanistically, IgG 3–43 interferes with both receptor dimerization and ligand binding, e.g., either by trapping the receptor in an inactive conformation or sterical interference with the transition of the receptor from the tethered into the open configuration. Of note, this inhibitory effect is also seen for scFv 3–43, indicating that even the small and monovalently binding antibody fragment is sufficient to inhibit HER3. Trapping of HER3 in its inactive conformation was described for 2 antibodies (KTN3379, LJM716), both recognizing an epitope formed by 2 different HER3 domains. Whereas KTN3379 binds to a region spanning domains II and III, LJM716 simultaneously binds to domains II and IV of tethered HER3.24,25 IgG 3–43 differs from these antibodies by binding to domain III and IV, indicating that structurally restraining these domains also stabilizes the tethered conformation. Upon ligand binding, major structural changes occur between domain II and III, resulting in a rotation of ∼130° with respect to domains III and IV.30 Aligning the tethered structure of HER3 with a model of the open conformation build on HER2 (pdb entry 3wlw)31 indicates also some minor movements of domain III (not shown). Further studies, e.g., by mutational and structural analyses, are necessary to identify the exact epitope of IgG 3–43 and to shed light on the molecular mechanism of HER3 inhibition.
Inhibition of HER3 activation by IgG 3–43 reduced cell proliferation and colony formation in vitro both in ligand-dependent and ligand-independent settings. In the analyzed cell lines, the antibody inhibited phosphorylation of HER3 and, furthermore, reduced the cellular levels of HER3. Quantification of receptor levels in SKBR3 and BT474 cells incubated with IgG 3–43 for up to 24 h showed that reduced levels of pHER3 correlate with reduced HER3 levels. This indicates that internalization and degradation of HER3 might be the primary effect leading to reduced pHER3 levels in these cell lines. This basal HER3 turnover is controlled by the ubiquitin ligases Nrdp1 and Nedd4.16,32 A recent study with antibody 9F7-F11, which is directed against domain I of HER3 and also induces rapid internalization of the receptor, showed that JNK1/2-dependent ITCH/AIP4 activation is responsible for receptor ubiquitinylation and degradation.33 Associated with the inhibition of HER3 phosphorylation and receptor downregulation, IgG 3–43 reduced basal and heregulin-stimulated Akt phosphorylation. Since the PI3K/Akt pathway is one of the most important cancer-associated pathways mediating survival, proliferation and apoptosis resistance of tumor cells, the efficient targeting of the PI3K-Akt signaling axis is a primary goal in anti-cancer therapy.
Antitumor activity, including tumor regression during the treatment phase and a prolonged survival, was demonstrated in a subcutaneous FaDu xenograft tumor model in SCID mice. The antiproliferative activity of IgG 3–43 seen in vitro might result in tumor cell death and tumor regression in vivo through induction of apoptosis. Induction of apoptosis has been described for some anti-HER3 antibodies, whereas others did not induce apoptosis on their own but were able to enhance apoptosis induced by chemotherapeutic drugs.34,35 Furthermore, immune effector cells might contribute to antitumor activity, shown for example for the glycoengineered anti-HER3 antibody RG7116 in an orthotopic lung xenograft model of A549 cells in SCID mice.36 Here, a contribution of murine monocytes and macrophages expressing Fc RIV was discussed. This is consistent with results from a comparative study of anti-HER2 IgG and F(ab')2 fragments demonstrating the importance of an Fc region for regression of large xenograft tumors in SCID mice.37 In addition, antiangiogenic activities might contribute to antitumor effects in vivo. This was shown for an anti-EGFR antibody (h-R3/nimotuzumab) capable of inhibiting angiogenesis through inhibition of VEGF production, resulting in tumor regression in a subcutaneous tumor model in SCID mice associated with a reduced micro-vascular density and an elevated apoptosis index.38 The contribution of HER3 to tumor angiogenesis was demonstrated with a micro-RNA (miR148a), which directly targets and inhibits HER3 expression.39 The identification of the exact mechanisms leading to tumor regression in vivo, besides the reduced proliferation of tumor cells, necessitates further studies, e.g., through the use of Fc-modified IgG molecules and histological analysis of treated tumors.
In summary, we developed a novel human anti-HER3 antibody, directed against a complex epitope formed by domain III and IV, capable of inhibiting heregulin-dependent and ligand-independent receptor activation, downstream signaling and cell proliferation. Thus, IgG 3–43 may be suitable as a therapeutic agent to treat various types of cancer dependent on HER3 signaling, including HER2-overexpressing tumors. In addition to monotherapy, the antibody might be especially useful in combination treatment with antibodies targeting other members of the HER family or other epitopes of HER3, or with small molecule inhibitors of receptors or downstream signaling molecules to potentiate antitumor activities and circumvent resistance mechanisms of established therapies.
Materials and methods
Materials
CCK-8 Cell Counting Kit-8 and recombinant heregulin-β were purchased from Sigma-Aldrich (St. Louis, MO, USA), his-tagged heregulin (Ser20-Lys241 NRG1, c-Terminal 6-His-tag) and mouse HER3-Fc fusion protein were purchased from R&D systems (Minneapolis, MN, USA). Dako QIFIKIT for expression analysis was purchased from Biozol (Eching, Germany). Antibodies used for immunoblotting experiments were purchased from Cell Signaling (Danvers, MA, USA: rabbit anti-phospho-HER3 (Tyr1289) (#4791), mouse anti-Akt (pan) (#2920), rabbit anti-phospho-Akt (Thr308) XP® (#13038), mouse anti-phospho-Akt (Thr308) (#5106), mouse anti-Erk1/2 (#9107), polyclonal rabbit anti-phospho-Erk1/2 (Thr202/Tyr204) (#9101)), Thermo Fischer Scientific (Waltham, MA, USA: mouse anti-HER3 (#MS201-P1)) and Sigma-Aldrich (St. Louis, MO, USA: monoclonal mouse anti-α-tubulin (#T6793), polyclonal anti-mouse IgG (Fc specific)-HRP (#A2554) and goat polyclonal anti-rabbit-IgG peroxidase (#A0545)). Antibodies used for receptor expression analysis (anti-human EGFR (clone AY13) (#352902), anti-human erbB2/HER-2 (clone 24D2) (#324402), and anti-human erbB3/HER-3 (clone1B4C3) (#324702)) were purchased from BioLegend (San Diego, CA, USA). Polyclonal anti-human IgG (Fc specific)-HRP (#A0170) and polyclonal anti-human IgG (whole molecule)-HRP (#A8792) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Mouse monoclonal anti His6-HRP (#sc8036) was from Santa Cruz Biotechnology (Dallas, TX, USA). R-phycoerythrin conjugated γ-chain specific anti-human IgG (#9170) was purchased from Sigma-Aldrich (St. Louis, MO, USA).Cetuximab and rituximab were kindly provided by Thomas Mürdter und Jens Schmidt (IKP, Stuttgart, Germany). An anti-TNFR1 antibody (Atrosab) used as negative control was kindly provided by Andreas Herrmann (Baliopharm, Basel, Switzerland). Antibody 3M6 was derived from MM-121 (seribantumab)40 mutating Cys89 in CDRL1 to serine. A549 cells were obtained from CLS Cell Lines Services (Eppelheim, Germany). Caco-2 cells and HCT-116 cells were originally obtained from Interlab Cell Line Collection (Genova, Italy). FaDu cells were obtained from DSMZ German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany). SKBR-3 cells were obtained from CLS (Eppelheim, Germany). BT-474 cells were originally obtained from Nancy Hynes (Friedrich Miescher Institute, Basel, Switzerland). MCF-7 cells were originally obtained from Cornelius Knabbe (Institute of Clinical Pharmacology, Stuttgart, Germany). NCI-N87 cells were kindly provided by TRON (Mainz, Germany). HEK293 cells were cultured in RPMI 1640 containing 5% fetal calf serum (FCS). A549, A431, BT-474, NCI-N87 and MCF-7 cells were cultured in RPMI 1640 supplemented with 10% FCS. FaDu and SKBR-3 were cultured in DMEM supplemented with 10% FCS. HEK293–6E were cultured under agitation in F17 Freestyle medium supplemented with L-glutamine, Kolliphor P-188 and 25 µg/ml G418. All cell lines were incubated in a humidified atmosphere of 5% CO2 at 37 °C. Human peripheral blood mononuclear cells (PBMCs) were obtained from the blood bank of the Klinikum Stuttgart.
Production of recombinant proteins
ScFv were produced in E. coli and purified by immobilized-metal affinity chromatography from periplasmic preparations as described previously.41 All receptor Fc fusion proteins were cloned into pSecTagA and produced with stably transfected HEK293T cells grown in Opti-MEM (Invitrogen, Karlsruhe, Germany). DNA encoding the heavy and light chain of IgG 3–43 was cloned into pEE14.4 and pEE6.4, respectively. Plasmids were then combined and transfected into HEK293–6E cells and grown in F17 Freestyle medium. IgG and Fc fusion proteins were purified from the supernatant by protein A chromatography. The protein concentration was determined with a spectrophotometer (NanoDrop Products, Wilmington, USA) using the calculated molecular mass and molar extinction coefficient. Aliquots were stored at −80 °C.
Phage selection
Antibody panning was performed in 96-well microtiter plates (Costar High Binding, Corning, Amsterdam, The Netherlands).42 A well was coated with the HER3-Fc fusion in PBS pH 7.4 for 1 h at RT. In parallel, one well for each panning was coated with a non-related Fc fusion. Both wells were blocked with 2% bovine serum albumin (BSA) (PAA, Cölbe, Germany) in PBS for 1 h or overnight. 2.5 × 1011 phage particles of Hyperphage43 packaged human naïve antibody gene libraries HAL7 and HAL844 were diluted in PBST with 1% skim milk and 1% BSA and preincubated for 1 h in the well with the non-related Fc fusion to avoid Fc-specific binders and sticky binders. The supernatant, containing the depleted library, was transferred to an HER3-Fc-coated well and incubated at RT for 2 hr, followed by 10 washing steps with PBST. Afterwards, bound scFv phage particles were eluted with 200 µl trypsin solution (10 µg/ml trypsin in PBS) at 37 °C for 30 min. The supernatant containing eluted scFv phage particles was transferred into a new tube. Ten microliter of eluted scFv phage were used for titration as described.428 Twenty ml E. coli XL1-Blue MRF' (Agilent, Böblingen, Germany) culture in logarithmic growth phase (OD600 = 0.4 - 0.5) were infected with the remaining scFv-phage at 37 °C for 30 min without shaking. The infected cells were harvested by centrifugation for 10 min at 3,220xg, and the pellet was resuspended in 250 µl 2xTY medium, supplemented with 100 mM glucose and 100 µg/ml ampicillin (2xTY-GA), plated on a 15 cm 2xTY agar plate supplemented with 100 mM glucose and 100 µg/ml ampicillin, and incubated overnight at 37 °C. Colonies were harvested with 5 ml 2xTY-GA. Thirty milliliter of 2xTY-GA were inoculated with 100 µl of the harvested colony suspension, and grown to an OD600 of 0.4 to 0.5 at 37 °C and 250 rpm. Five ml bacterial suspension (∼2.5 × 109 bacteria) were infected with 5 × 1010 helper phage M13K07 (Stratagene), incubated at 37 °C for 30 min without shaking, followed by 30 min at 250 rpm. Infected cells were harvested by centrifugation for 10 min at 3,220xg and the pellet was resuspended in 30 ml 2xTY, supplemented with 100 µg/ml ampicillin and 50 µg/ml kanamycin (2xTY-AK). Antibody phage were produced at 30 °C and 250 rpm for 16 h. Cells were harvested by centrifugation for 10 min at 3220xg. The supernatant containing the antibody phage (∼1 × 1012 cfu/ml) were directly used for the next panning round or stored at 4 °C for a few days.
Screening of scFv
Polypropylene microtiter plates (96-well) (U96 PP 0.5 ml, Greiner, Frickenhausen, Germany) containing 150 µl phosphate-buffered 2xTY-GA (2xTY-GA supplemented with 10% (v/v) potassium phosphate buffer (0.17 M KH2PO4, 0.72 M K2HPO4)) were inoculated with colonies from the titration plate of the third panning round. Plates were incubated overnight at 37 °C at 1,000 rpm in a shaker (Thermoshaker PST-60HL-4, Lab4You, Berlin, Germany). A volume of 180 µl phosphate-buffered 2xTY-GA in each PP-MTP well was inoculated with 10 µl of the overnight culture and grown at 37 °C and 800 rpm for 2 h. Bacteria were harvested by centrifugation for 10 min at 3,220xg, and 180 µl supernatant was removed. The pellets were resuspended in 180 µl buffered 2xTY supplemented with 100 µg/ml ampicillin, 100 mM sucrose, 50 µM isopropyl-β-D-thiogalactopyranoside and incubated at 30 °C and 800 rpm overnight. Bacteria were pelleted by centrifugation for 10 min at 3,220xg at 4 °C. The scFv-containing supernatant was transferred to a new microtiter plate and stored at 4 °C before analysis. For identification of binders, supernatants containing monoclonal scFv were incubated with the antigen-coated plates (coating as described above) for 1.5 h at RT followed by 3 PBST washing steps. Bound scFv were detected using murine anti-c-Myc tag mAb 9E10 and a goat anti-mouse Ig serum, conjugated with horseradish peroxidase (HRP) (Sigma; 1:10,000). The visualization was performed with TMB (3,3´,5,5´-tetramethylbenzidine) substrate. The staining reaction was stopped by adding 100 µl 1 N sulfuric acid. The absorbance at 450 nm and scattered light at 620 nm were measured and the 620 nm values were subtracted using a SUNRISE microtiter plate reader (Tecan, Crailsheim, Germany).
Biochemical characterization
Purified proteins were analyzed by SDS-PAGE under reducing and non-reducing conditions and stained with Coomassie brilliant blue G-250. HPLC SEC was performed using a Phenomenex Yarra SEC-2000 column (Phenomenex, Aschaffenburg, Germany) and a Waters 2695 HPLC (Waters Corporation, Milford, USA) at a flow rate of 0.5 ml/min. The mobile phase was 0.1 M Na2HPO4/NaH2PO4, 0.1 M Na2SO4, pH 6.7. As standard proteins, thyroglobulin (669 kDa, 8.5 nm), β-amylase (200 kDa, 5.4 nM), BSA (67 kDa, 3.55 nm), carbonic anhydrase (29 kDa, 2.35 nm), and FLAG peptide (1 kDa) were used.
Quartz crystal microbalance
Affinities of IgG 3–43 for the monomeric receptor extracellular domain of HER3 and dimeric HER3-Fc fusion were determined via QCM measurements using an Attana Cell A200 instrument. IgG 3–43 was immobilized on the surface of a low nonspecific-binding carboxyl chip using the amine coupling kit (EDC + sNHS, Attana AB, Stockholm, Sweden) in a density that resulted in a frequency change of about 90 Hz. The measurement was performed at 25 °C with a flow-rate of 25 µl/min of PBST (0.1% Tween) pH7.4. Regeneration of the binding was performed twice with 3 M MgCl2 for 15 s. After every second measurement a buffer injection was performed to determine the baseline, which was subsequently subtracted from the measurements. Soluble his-tagged HER3 was injected in a 2-fold dilution series in PBST in random order, with concentrations between 1.25 to 20 nM. Dimeric HER3-Fc was injected in a 2-fold dilution series in PBST in random order, with concentrations between 0.625 to 10 nM. Data were analyzed with the Attana evaluation software and TraceDrawer.
ELISA
Fc fusion proteins (300 ng/well) were coated overnight at 4 °C and the remaining binding sites were blocked with 3% (w/v) non-fat dry milk/PBS (MPBS). Purified proteins were diluted in MPBS and titrated in duplicates. Bound proteins were detected with an HRP-conjugated anti-human IgG-Fab antibody or an anti-His-tag antibody for detection of scFv using 100 µl of 3,3′,5,5′-tetramethylbenzidine (TMB) substrate per well (0.1 mg/ml TMB, 100 mM sodium acetate buffer, pH 6.0, 0.006% H2O2). The reaction was stopped with 1 M H2SO4 (50 µl/well) and optical density was measured at 450 nm in an ELISA reader.
Flow cytometry
Flow cytometry studies were performed with HER3-expressing MCF-7, FaDu, BT474, A431, NCI-N87, A549, and SKBR3 cells. Cells were briefly trypsinized at 37 °C, trypsin was quenched with FCS-containing medium and removed by centrifugation. Cells (200,000 cells per sample) were seeded and incubated with varying concentrations of IgG 3–43 for at least one hour at 4 °C. Cells were washed twice with PBA (2% (v/v) FCS, 0.02% (w/v) NaN3 in 1 × PBS). Cells were incubated with PE-labeled murine anti-human Fc antibody for another hour to visualize bound antibody molecules. After 2 further washing steps, fluorescence was measured with a MACSQuant® Analyzer 10 and median fluorescence intensities relative to unstained cells were calculated using the FlowJo software. Quantification of receptor expression was performed by indirect immunofluorescence staining using the QIFIKIT (Dako) according to the manufacturer's protocol and anti-EGFR antibody, anti-HER2 antibody, and anti-HER3 antibody (diluted 1:10). Data analysis was performed using the FlowJo software, Excel and GraphPad Prism.
Inhibition of heregulin binding
MCF-7 cells were briefly trypsinized at 37 °C, trypsin was quenched with FCS-containing medium and removed by centrifugation. Cells (200,000 cells per sample) were seeded and incubated with 3 µM of IgG 3–43 or control antibody (cetuximab) at 4 °C. After 30 minutes, 50 nM of recombinant 6His-tagged heregulin was added and the cells were mixed and incubated for at least another hour at 4°C. Washing was performed twice with PBA (2% (v/v) FCS, 0.02% (w/v) NaN3 in 1 × PBS). Cells were incubated with PE-labeled anti-His antibody for another hour to visualize bound heregulin molecules. After 2 further washing steps, fluorescence was measured with a MACSQuant® Analyzer 10 and median fluorescence intensities relative to unstained cells were calculated using the FlowJo software.
Internalization
IgG 3–43 was conjugated with Amersham Cy5 malemide mono-reactive dye (Amersham) according to the manufacturer's instructions. MCF-7 cells were seeded on 8-well glass chamber slides (BD) one day before the experiment to be semi-confluent on the next day. Cy5-labeled IgG 3–43 was incubated in growth medium at 37°C for the indicated time points. The cells were fixed with 4% paraformaldehyde in PBS (200 µl/well) for 15 minutes at 37 °C. Fixed cells were washed with PBS and counterstained with Concanavalin A and DAPI for 10 minutes. After 3 more washing steps with PBS, the cells were covered with Fluoromount and coverslips. Dried slides were stored at 4 °C and analyzed using a spinning disk microscope.
Immunoblot analysis of protein phosphorylation
Cells were grown in 6-well plates (2 × 105 cells per well) overnight and then starved in medium containing 0.2% FCS for another day. Next, cells were incubated with indicated concentrations of antibodies in starvation medium at 37 °C for 1 h or the indicated time. Subsequently, cells were stimulated with heregulin (50 ng/ml) for 15 min, before being lysed using protease and phosphatase inhibitor containing RIPA buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1% Triton-X 100, 0.5 sodium deoxycholate, 0.1% SDS, 1 mM sodium orthovanadate, 10 mM sodium fluoride and 20 mM β-glycerophosphate plus complete protease inhibitors (Roche, Basel, Switzerland)) at 4 °C. Lysates were centrifuged (13,200 rpm, 30 min, 4 °C) and supernatants were collected. Protein concentrations in each lysate were assessed using the Bio-Rad DC™ Protein Assay. Lysates were fractionated by SDS-PAGE and transferred onto nitrocellulose membranes (iBlot®Gel Transfer Stacks; Invitrogen) using the iBlot® 2 Dry Blotting System. Membranes were blocked with 0.5% blocking reagent (Roche) in PBS containing 0.1% Tween-20 and incubated with indicated primary antibodies, diluted in 0.5% blocking reagent, 0.05% NaN3, overnight at 4 °C, followed by 5 washing steps with 0.5% PBST and incubation with HRP-conjugated secondary antibody for 1 h at RT. After washing, activity of HRP was detected with ECL substrate and visualized by the FUSION SOLO Imager (Vilber Lourmat, Eberhardzell, Germany).
Cell proliferation
Cells (103 per well) were grown in 100 µl medium containing 10% FCS in 96-well plates at 37 °C for one day. Next, cells were incubated with the indicated concentrations of antibodies in reduced serum medium (0.2% FCS) at 37 °C for 6 d in the presence or absence of 10 ng/ml heregulin. The number of viable cells was determined using the Cell-Counting-Kit-8. The absorbance at 450 nm was measured in an ELISA reader.
Colony formation assay
Cells (SKBR3, BT474) were seeded into 12-well plates (1,000 cells/well) in medium containing 10% FCS. The next day, antibody was added in 1 ml medium containing 2% FCS. After 7 days, 0.2 ml medium with antibody was added and incubated for additional 5 d. Cells were then fixed with Histofix and stained with crystal violet.
ADCC
Human PBMC were thawed and cultivated overnight at 37 °C, 5% CO2 to allow for monocyte adherence. The next day, the percentage of NK cells (CD3−, CD56+) was determined by flow cytometry. SKBR3 cells (20,000/well) were seeded and incubated overnight at 37°C, 5% CO2. SKBR3 cells were incubated with serial dilutions of the antibodies or medium for 15 min at room temperature, before addition of monocyte-depleted PBMC containing 40,000 NK cells/well. After 24 h at 37°C, 5% CO2, supernatants were removed, followed by 3 washing steps and addition of 200 µl medium. Living SKBR3 cells were detected by MTT assay (addition of 10 µl MTT solution (5 mg/ml), incubation at 37°C, 5% CO2 for 2 h, addition of 90 µl lysis buffer (7.5% (w/v) SDS in 50% (v/v) dimethylformamide and 50% (v/v) H2O, pH 4.5–5.0 adjusted with 80% acetic acid) followed by incubation at room temperature overnight and determination of OD595nm-OD655nm).
Pharmacokinetics
Animal care and all experiments performed were in accordance with federal guidelines and have been approved by university and state authorities. Three animals of each treatment group of the pharmacodynamics experiment described in the following (twice weekly injections with either 30 µg, 100 µg or 300 µg IgG 3–43 for 3 weeks) were used to determine serum half-life of IgG 3–43. Blood samples (around 50 µl) were collected from the tail after 5 min, 1 h, 1 day, and 3 d of the first injection (day 14), and after 5 min, 1 h, 1 day, 3 d and one week of the last injection (day 31). Serum samples were incubated on ice for 10 minutes, clotted blood was centrifuged (16,100x g, 20 min, 4 °C) and serum samples were stored at 4 °C. IgG serum concentration was analyzed by ELISA as described above using 300 ng/well HER3-Fc for coating and HRP-coupled anti-human IgG (Fab-specific) antibody diluted 1:20,000 for detection by interpolation from a standard curve of purified IgG 3–43. Initial and terminal half-lives (t1/2α, t1/2β) and AUC were calculated with Excel, Cmax was calculated with PKSolver.
Pharmacodynamics
FaDu cells (5 × 106) were injected subcutaneously in the left and right flanks of female SCID beige mice (Charles River). During the injection of the cells, the mice were anesthetized with isoflurane. The treatment was initiated when the tumor volume reached ∼80 mm3. Each treatment group consisted of 6 animals. All mice received 6 intravenous injections twice weekly for 3 weeks beginning on day 14 after inoculation and ending on day 31. Treatment formulations contained either PBS (control group) or a total of 30 µg, 100 µg or 300 µg IgG 3–43 per injection dose. Tumor growth was monitored with a caliper and tumor volume was calculated as follows: tumor volume = (a × b2)/2 (a, longitudinal diameter of tumor; b, transverse diameter of tumor). The 95% confidential interval (95%CI) was used for statistical analyses of the in vivo experiment. Significances were calculated by GraphPad Prism 5.0.1 and results were compared via one-way ANOVA followed by Tukey's multiple comparison test (post-test).
Supplementary Material
Disclosure of potential conflicts of interest
L.C.S, O.S., M.H., J.Z, M.H., S.D., M.A.O, and R.E.K. are named inventor on a patent application covering the 3-43 antibody technology.
Acknowledgments
We would like to thank Sabine Münkel (Institute of Cell Biology and Immunology, University of Stuttgart) and Saskia Helmsing (Technische Universität Braunschweig) for technical assistance. We thank Jens Schmid and Thomas Mürdter (IKP, Stuttgart, Germany) for providing Cetuximab and Rituximab, BioNTech (Mainz, Germany) for providing cell lines and Andreas Herrmann (Baliopharm, Basel, Switzerland) for providing Atrosab.
Funding
This work was supported by BMBF (eBio: Predict).
References
- 1.Wieduwilt MJ, Moasser MM. The epidermal growth factor receptor family: Biology driving targeted therapeutics. Cell Mol Life Sci 2008; 65:1566-84; PMID: 18259690 PMCID; https://doi.org/ 10.1007/s00018-008-7440-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kol A, Terwisscha van Scheltinga AG, Timmer-Bosscha H, Lamberts LE, Bensch F, de Vries EG, Schröder CP. HER3, a serious partner in crime: Therapeutic approaches and potential biomarkers for effect of HER3-targeting. Pharmacol Ther 2014; 143:1-11; PMID: 24513440; https://doi.org/ 10.1016/j.pharmthera.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 3.Baselga J, Swain SM. Novel anticancer targets: Revisiting ErbB2 and discovering ErbB3. Nat Rev Cancer 2009; 9:463-75; PMID: 19536107; https://doi.org/ 10.1038/nrc2656. [DOI] [PubMed] [Google Scholar]
- 4.Aurisicchio L, Marra E, Roscilli G, Mancini R, Ciliberto G. The promise of anti-ErbB3 monoclonals as new cancer therapeutics. Oncotarget 2012; 3:744-58; PMID:22889873; https://doi.org/ 10.18632/oncotarget.550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ocana A, Vera-Badillo F, Seruga B, Templeton A, Pandiella A, Amir E. HER3 overexpression and survival in solid tumors: A meta-analysis. J Natl Cancer Inst 2012; 105:266-73; PMID: 23221996; https://doi.org/ 10.1093/jnci/djs501. [DOI] [PubMed] [Google Scholar]
- 6.Amin DN, Cambell MR, Moasser MM. The role of HER3, the unpretentious member of the HER family, in cancer biology and cancer therapeutics. Semin Cell Dev Biol 2010; 21:944-50; PMID: 20816829; https://doi.org/ 10.1016/j.semcdb.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kruser TJ, Wheeler DL. Mechanisms of resistance to HER family targeting antibodies. Exp Cell Res 2010; 316:1083-100; PMID: 20064507; https://doi.org/ 10.1016/j.yexcr.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 8.Roskoski R. The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol Res 2014; 79:34-74; PMID: 24269963; https://doi.org/ 10.1016/j.phrs.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Breuleux M. Role of heregulin in human cancer. Cell Mol Life Sci 2007; 64:2358-77; PMID: 17530167; https://doi.org/ 10.1007/s00018-007-7120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho HS, Leahy DJ. Structure of the extracellular region of HER3 reveals an interdomain tether. Science 2002; 297:1330-33; PMID: 12154198; https://doi.org/ 10.1126/science.1074611. [DOI] [PubMed] [Google Scholar]
- 11.Schmitz KR, Ferguson KM. 2009 Interaction of antibodies with ErbB receptor extracellular regions. Exp Cell Res 2007; 315:659-70; PMID: 18992239; https://doi.org/ 10.1016/j.yexcr.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yarden Y, Sliwkowski MX. Untangeling the ErbB signalling network. Nat Rev Mol Cell Biol 2001; 2:127-37; PMID: 11252954; https://doi.org/ 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 13.Gaborit N, Lindzen M, Yarden Y. Emerging anti-cancer antibodies and combination therapies targeting HER3/ErbB3. Human Vaccin Immunother 2015; 12:576-92; PMID: 26529100; https://doi.org/ 10.1080/21645515.2015.1102809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sierke SL, Cheng K, Kim HH, Koland JG. Biochemical characterization of the protein tyrosine kinase homology domain of the ErbB3 (HER3) receptor protein. Biochem J 1997; 322:757-63; PMID: 9148746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graus-Porta D, Beerli RR, Daly JM, Hynes NE. ErbB-2, the preferred heterodimerization partner of all ErbB receptors, is a mediator of lateral signaling. EMBO J 1997; 16:1647-55; PMID: 9130710; https://doi.org/ 10.1093/emboj/16.7.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanizaki J, Okamoto I, Sakai K, Nakagawa K. Differential roles of trans-phosphorylated EGFR, HER2, HER3, and RET as heterodimerisation partners of MET in lung cancer with MET amplification. B J Cancer 2011; 105:807-13; PMID: 21847121; https://doi.org/ 10.1038/bjc.2011.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mujoo K, Choi BK, Huang Z, Zhang N, An Z. Regulation of ErbB3/HER3 signaling in cancer. Oncotarget 2014; 5:10222-36; PMID: 25400118; https://doi.org/ 10.18632/oncotarget.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu X, Cheng Y, Li G, Xia L, Gu R, Wen X, Ming X, Chen H. Her3 is associated with poor survival of gastric adenocarcinoma: Her3 promotes proliferation, survival and migration of human gastric cancer mediated by PI3K/AKT signaling pathway. Med Oncol 2014; 31:903; PMID: 24623015; https://doi.org/ 10.1007/s12032-014-0903-x. [DOI] [PubMed] [Google Scholar]
- 19.Shi F, Telesco SE, Radhakrishnan R, Lemmon MA. ErbB3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation. Prot Natl Acad Sci USA 2010; 107:7692-97; PMID: 20351256; https://doi.org/ 10.1073/pnas.1002753107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steinkamp MP, Low-Nam ST, Yang S, Lidke KA, Lidke DS, Wilson BS. ErbB3 is an active tyrosine kinase capable of homo- and heterointeractions. Mol Cell Biol 2014; 34:965-77; PMID: 24379439; https://doi.org/ 10.1128/MCB.01605-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaiswal BS, Kljavin NM, Stawiski EW, Chan E, Parikh C, Durinck S, Chaudhuri S, Pujara K, Guillory J, Edgar KA, et al.. Oncogenic ErbB3 mutations in human cancers. Cancer Cell 2013; 23:603-17; PMID: 23680147; https://doi.org/ 10.1016/j.ccr.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 22.Malm M, Frejd FY, Stahl S, Löfblom J. Targeting HER3 using mono- and bispecific antibodies or alternative scaffolds. MAbs 2016; 8:1195-209; PMID: 27532938; https://doi.org/ 10.1080/19420862.2016.1212147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang N, Chang Y, Rios A, An Z. HER3/ErbB3, an emerging cancer therapeutic target. Acta Biochim Biophys Sin 2016; 48:39-48; PMID: 26496898; https://doi.org/ 10.1093/abbs/gmv103. [DOI] [PubMed] [Google Scholar]
- 24.Garner AP, Bialucha CU, Spraque ER, Garrett JT, Sheng Q, Li S, Sineshchekova O, Saxena P, Sutton CR, Chen D, et al.. An antibody that locks HER3 in the inactive conformation inhibits tumor growth driven by HER2 or Neuregulin. Cancer Res 2013; 73:6024-35; PMID: 23928993; https://doi.org/ 10.1158/0008-5472.CAN-13-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee S, Greenlee EB, Amick JR, Ligon GF, Lillquist JS, Natoli Jr EJ, Hadari Y, Alvarado D, Schlessinger J. Inhibition of ErbB3 by a monoclonal antibody that locks the extracellular domain in an inactive configuration. Proc Natl Acad Sci USA 2015; 112:13225-30; PMID: 26460020; https://doi.org/ 10.1073/pnas.1518361112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horton HM, Bernett MJ, Pong E, Peipp M, Karki S, Chu SY, Richards JO, Vostiar I, Joyce PF, Repp R, et al.. Potent in vitro and in vivo activity of an Fc engineered anti-CD19 monoclonal antibody against lymphoma and leukemia. Cancer Res 2008; 68:8049-57; PMID: 18829563; https://doi.org/ 10.1158/0008-5472.CAN-08-2268. [DOI] [PubMed] [Google Scholar]
- 27.Liu JF, RayCoqiard O, Selle F, Poveda AM, Cibula D, Hirte H, Hilpert F, Raspagliesi F, Gladieff L, Harter P, et al.. Randomized Phase II trial of Seribantumab in combination with paclitaxel in patients with advanced Platinum-Resistant or -Refractory ovarian cancer. J Clin Oncol 2016; 34:4345-53; PMID: 27998236; https://doi.org/ 10.1200/JCO.2016.67.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meneses-Lorente G, Friess T, Kolm I, Hölzlwimmer G, Bader S, Meille C, Thomas M, Bossenmaier B. Preclinical pharmacokinetics, pharmacodynamcis, and efficacy of RG7116: A novel humanized, glycoengineered anti-HER3 antibody. Cancer Chemother Pharmacol 2015; 75:837-50; PMID: 25702049; https://doi.org/ 10.1007/s00280-015-2697-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Y, Goenaga AL, Harms BD, Zhou H, Lou J, Conrad F, Adams GP, Schoeberl B, Nielsen UB, Marks JD. Impact of intrinsic affinity on functional binding and biological activity of EGFR antibodies. Mol Cancer Ther 2012; 11:1467-76; PMID: 22564724; https://doi.org/ 10.1158/1535-7163.MCT-11-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lemmon MA. Ligand-induced Erbb receptor dimerization. Exp Cell Res 2009; 315:638-48; PMID: 19038249; https://doi.org/ 10.1016/j.yexcr.2008.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu S, Sun Y, Meng Y, Wang X, Yang W, Fu W, Guo H, Qian W, Hou S, Li B, et al.. Molecular architecture of the ErbB2 extracellular domain homodimer. Oncotarget 2015; 6:1695-706; PMID: 25633808; https://doi.org/ 10.18632/oncotarget.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang Z, Choi BK, Mujoo K, Fan X, Fa M, Mukherjee S, Owiti N, Zhang N, An Z. The E3 ubiquitin ligase NEDD4 negatively regulates HER3/ErbB3 level and signaling. Oncogene 2015; 34:1105-15; PMID: 24662824; https://doi.org/ 10.1038/onc.2014.56. [DOI] [PubMed] [Google Scholar]
- 33.Le Clorennec C, Lazrek Y, Dubreuil O, Larbouret C, Poul M-A, Mondon P, Melino G, Pèlegrin A, Chardès T. The anti-HER (ErbB3) therapeutic antibody 9F7-F11 induces HER3 ubiquitination and degradation in tumors through JNK1/2-dependent ITCH/AIP4 activation. Oncotarget 2016; 7:37013-29; PMID: 27203743; https://doi.org/ 10.18632/oncotarget.9455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lazrek Y, Dubreuil O, Garambois V, Gaborit N, Larbouret C, Le Clorennec C, Thomas G, Leconet W, Jarlier M, Pugnière M, et al.. Anti-HER3 domain 1 and 3 antibodies reduce tumor growth by hindering HER2/HER3 dimerization and AKT-induced MDM2, XIAP, and FoxO1 phosphorylation. Neoplasia 2013; 15:335-47; PMID: 23479511 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang S, Huang J, Lyu H, Cai B, Yang X, Li F, Tan J, Edgerton SM, Thor AD, Lee CK, et al.. Therapeutic targeting of erbB3 with MM-121/SAR256212 enhances antitumor activity of paclitaxel against erbB2-overexpressing breast cancer. Breast Cancer Res 2013; 15:R101; PMID: 24168763; https://doi.org/ 10.1186/bcr3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mischberger C, Schiller CB, Schräml M, Dimoudis N, Friess T, Gerdes CA, Reiff U, Lifke V, Hoelzlwimmer G, Kolm I, et al.. RG7116, a therapeutic antibody that binds to inactive HER3 receptor and is optimized for immune effector activation. Cancer Res 2013; 73:5183-94; PMID: 23780344; https://doi.org/ 10.1158/0008-5472.CAN-13-0099. [DOI] [PubMed] [Google Scholar]
- 37.Spiridon CI, Guinn S, Vitetta ES. A comparison of the in vitro and in vivo activities of IgG and F(ab')2 fragments of a mixture of three monoclonal anti-HER-2 antibodies. Clin Cancer Res 2004; 10:3542-51; PMID: 15161714; https://doi.org/ 10.1158/1078-0432.CCR-03-0549. [DOI] [PubMed] [Google Scholar]
- 38.Crombet-Ramos T, Rak J, Pérez R, Viloria-Petit A. Antiproliferative, antiangiogenic and proapoptotic activity of h-R3: A humanized anti-EGFR antibody. Int J Cancer 2002; 101:567-75; PMID: 12237899; https://doi.org/ 10.1002/ijc.10647. [DOI] [PubMed] [Google Scholar]
- 39.Yu J, Li Q, Xu Q, Liu L, Jiang B. MiR-148a inhibits angiogenesis by targeting ErbB3. J Biomed Res 2011; 25:170-7; PMID: 23554686; https://doi.org/ 10.1016/S1674-8301(11)60022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schoeberl B, Faber AC, Li D, Liang MC, Crosby K, Onsum M, Burenkova O, Pace E, Walton Z, Nie L, et al.. An ErbB3 antibody, M121, is active in cancers with ligand-dependent activation. Cancer Res 2010; 70:2485-94; PMID: 20215504; https://doi.org/ 10.1158/0008-5472.CAN-09-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Müller D, Trunk G, Sichelstiel A, Zettlitz KA, Quintanilla M, Kontermann RE. Murine endoglin-specific single-chain Fv fragments for the analysis of vascular targeting strategies in mice. J Immunol Methods 2008; 339:90-8; PMID: 18790696; https://doi.org/ 10.1016/j.jim.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 42.Frenzel A, Kügler J, Wilke S, Schirrmann T, Hust M. Construction of human antibody gene libraries and selection of antibodies by phage display. Methods Mol Biol 2014; 1060:215-43; PMID: 24037844; https://doi.org/ 10.1007/978-1-62703-586-6_12. [DOI] [PubMed] [Google Scholar]
- 43.Rondot S, Koch J, Breitling F, Dübel S. A helper phage to improve single-chain antibody presentation in phage display. Nat Biotechnol 2001; 19:75-8; PMID: 11135557; https://doi.org/ 10.1038/83567. [DOI] [PubMed] [Google Scholar]
- 44.Hust M, Meyer T, Voedisch B, Rülker T, Thie H, El-Ghezal A, Kirsch MI, Schütte M, Helmsing S, Meier D, et al.. A human scFv antibody generation pipeline for proteome research. J Biotechnol 2011; 152:159-70; PMID: 20883731; https://doi.org/ 10.1016/j.jbiotec.2010.09.945. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.