Abstract
Induced pluripotent stem cell (iPSC) technology has enormous potential to provide improved cellular models of human disease. However, variable genetic and phenotypic characterisation of many existing iPSC lines limits their potential use for research and therapy. Here, we describe the systematic generation, genotyping and phenotyping of 711 iPSC lines derived from 301 healthy individuals by the Human Induced Pluripotent Stem Cells Initiative (HipSci: http://www.hipsci.org). Our study outlines the major sources of genetic and phenotypic variation in iPSCs and establishes their suitability as models of complex human traits and cancer. Through genome-wide profiling we find that 5-46% of the variation in different iPSC phenotypes, including differentiation capacity and cellular morphology, arises from differences between individuals. Additionally, we assess the phenotypic consequences of rare, genomic copy number mutations that are repeatedly observed in iPSC reprogramming and present a comprehensive map of common regulatory variants affecting the transcriptome of human pluripotent cells.
Introduction
Induced pluripotent stem cells (iPSCs) are important model systems for human disease1. A major open question is whether iPSCs can be used to study the functions of genetic variants associated with complex traits and normal human phenotypic variation. Previous work has suggested that individual iPSC lines are highly heterogeneous2–5, although some of these differences may arise due to genetic background of the donor6,7. Nonetheless, high variability could make iPSCs unsuitable cellular models for genetic variants with small effects. Existing iPSC lines also frequently have limited genetic and phenotypic data of variable quality, or are derived from individuals with severe genetic disorders, limiting their utility for studying other phenotypes.
The Human Induced Pluripotent Stem Cells Initiative (HipSci: www.hipsci.org) was established to generate a large, high-quality, open-access reference panel of human iPSC lines. A major focus of the initiative is the systematic derivation of iPSCs from hundreds of healthy volunteers using a standardised and well-defined experimental pipeline. The lines are extensively characterised and available to the wider research community along with the accompanying genetic and phenotypic data. Here, we report initial results from the characterization of the first 711 iPSC lines derived from 301 healthy individuals. We provide a high-resolution map of recurrent copy number aberrations in iPSCs, identify putative candidate genes under selection in these regions, and assess the functional consequences of these changes. We show that common genetic variants produce readily detectable effects in iPSCs and provide the most comprehensive map of regulatory variation in human iPSCs to date. We also demonstrate that differences between donor individuals have pervasive effects at all phenotypic levels in iPSCs, from the epigenome, transcriptome and proteome to cell differentiation and morphology.
Sample collection and iPSC derivation
Samples were collected from healthy, unrelated research volunteers via the NIHR Cambridge BioResource (Methods). We established 711 lines from 301 donors (>1 line for 82% of donors, >2 lines for 50%), which were profiled using an initial set of ‘Tier 1‘ assays (Fig. 1a). These included array-based genotyping and gene expression profiling of the iPSCs and their fibroblast progenitors, as well as an assessment of the pluripotency and differentiation properties of the iPSCs. Using immunohistochemistry followed by quantitative image analysis (hereafter ‘Cellomics’), we measured protein expression of pluripotency markers in 307 lines and differentiated 372 lines into neuroectoderm, mesoderm, and endoderm8 measuring expression of three lineage-specific markers in each germ layer (Fig. 1a; Extended Data Fig. 1). We then selected 1-2 lines per donor to minimise the number of genetic abnormalities and performed further phenotyping (hereafter ‘Tier 2’) using RNA-seq, DNA methylation arrays, quantitative proteomics and cell morphological imaging in 239, 27, 16 and 24 lines, respectively (Supplementary Table 1).
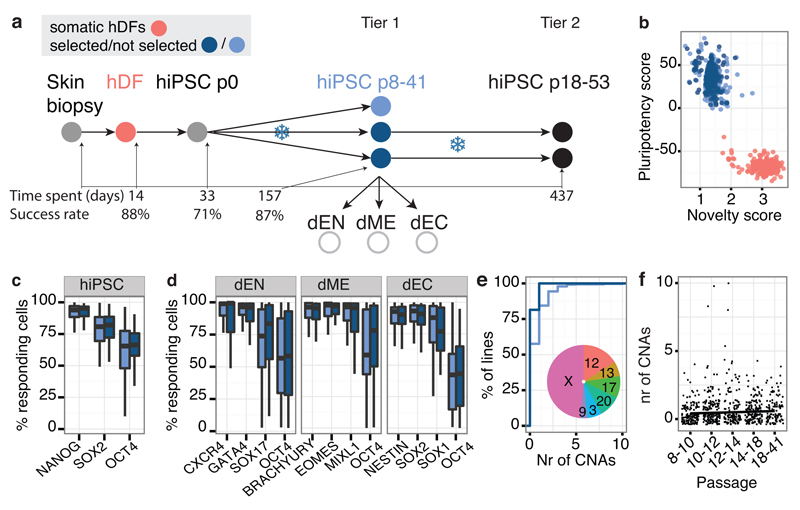
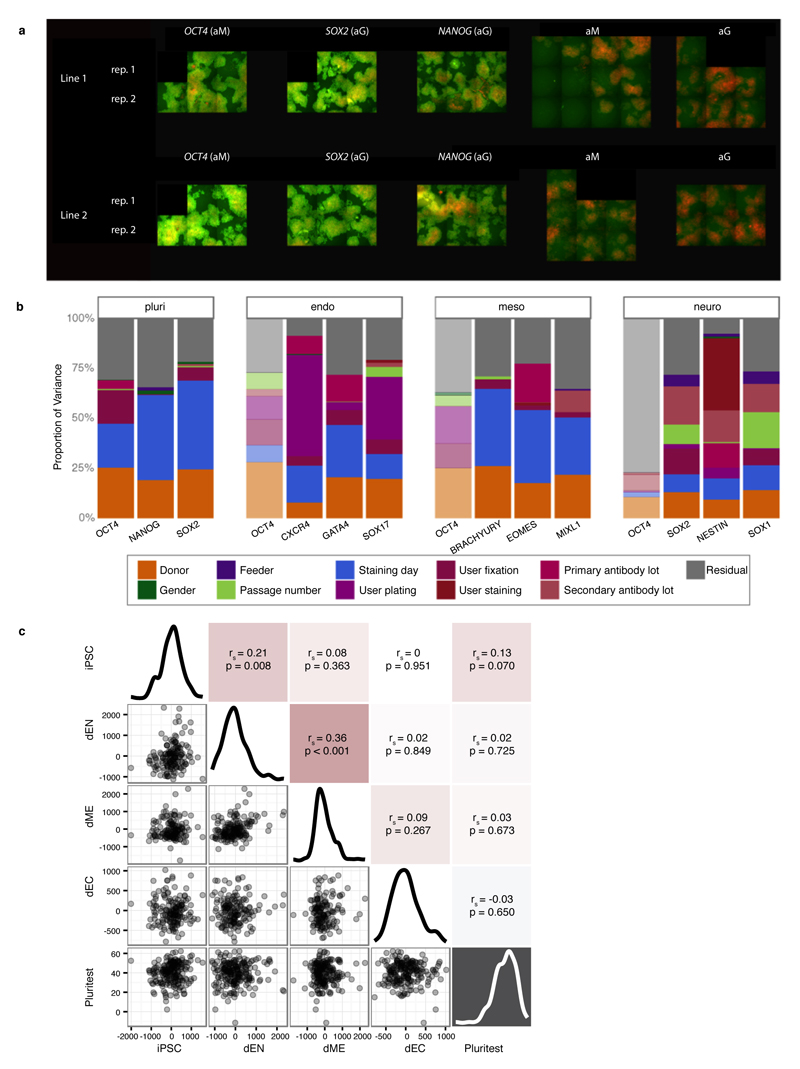
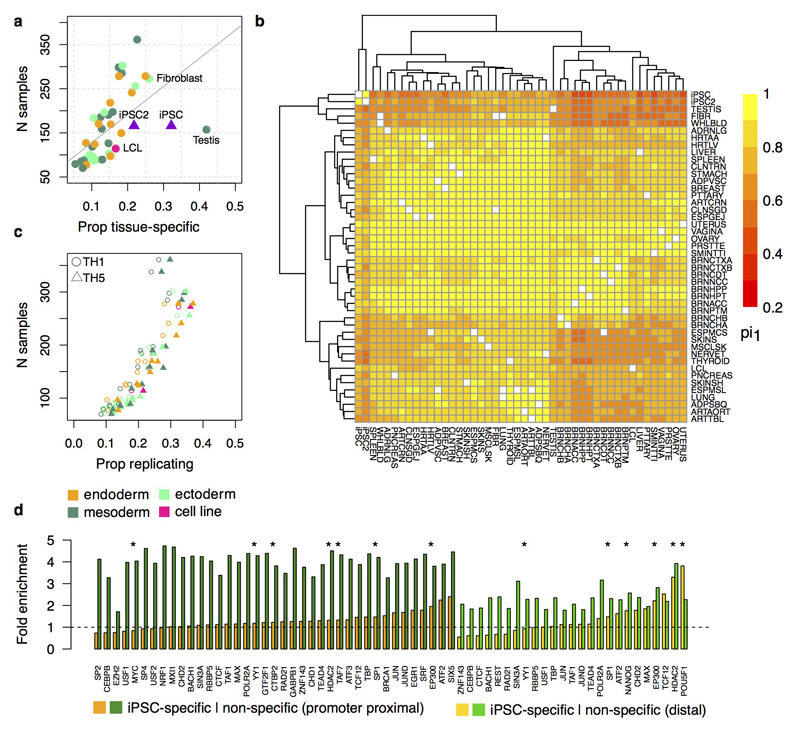
Figure 1. iPSC line generation and quality control.
Throughout light blue = not selected, dark blue = selected lines. (a) hDF: human dermal fibroblasts; dEN: differentiated endoderm; dME: differentiated mesoderm; dEC: differentiated neuroectoderm. The x-axis shows the median number of days, including freeze/thaw cycles (snowflakes), at each pipeline stage, with stage-specific success rates. (b) PluriTest pluripotency versus novelty score9. (c,d) Percentage of cells expressing pluripotency and differentiation markers. (e) Cumulative distribution of number of CNAs, fraction of trisomies per chromosome (inset). (f) Relationship between CNA counts and line passage number.
Pluripotency and genetic stability
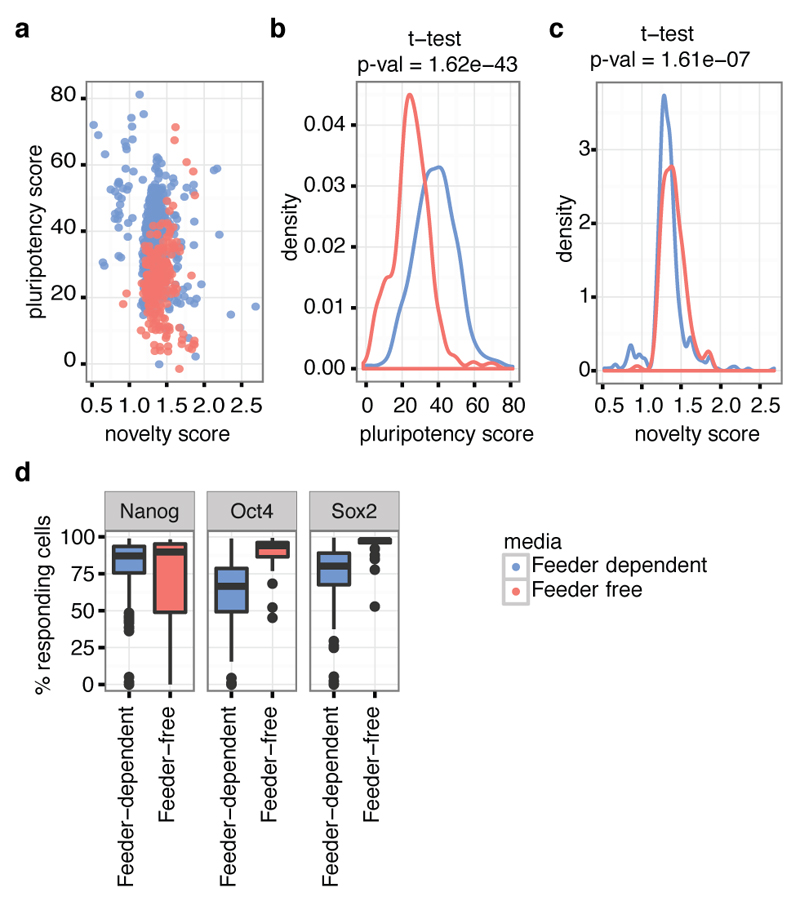
Using Tier 1 expression data, 84% of our lines were classified as pluripotent by PluriTest9 (score > 20) and 97% had a pluripotency score of >10, which yields almost identical sensitivity and specificity in the PluriTest training set (Fig. 1b). Most lines with a pluripotency score <20 (69%) had been cultured on feeder free Essential 8 media (OR 5.4, P < 8x10-13, Fisher’s exact test), which likely reflects that PluriTest was primarily trained using lines grown in feeder-dependent conditions (Extended Data Fig. 2). Using the Cellomics imaging data we quantified the fraction of cells expressing each pluripotency marker individually and estimated that, on average, between 18% and 62% of cells in the iPSC lines co-expressed all three markers NANOG, POU5F1 (OCT4) and SOX2 (Fig. 1c). Almost all lines (>99%) successfully produced cells from all three germ layers during directed differentiation with the average line producing up to 70%, 84% and 77% of cells expressing all three markers of dEN, dME and dEC, respectively (Fig. 1d). We assessed correlations of differentiation capacity between different germ layers and found a positive correlation between endoderm and mesoderm marker expression (Spearman r = 0.36, P < 0.001), and between endoderm and pluripotency marker expression (Spearman r = 0.21, P < 0.008) (Extended Data Fig. 1c). Taken together, our data indicate that virtually all of the iPSC lines we have derived are pluripotent, although we observed some variability in differentiation between lines.
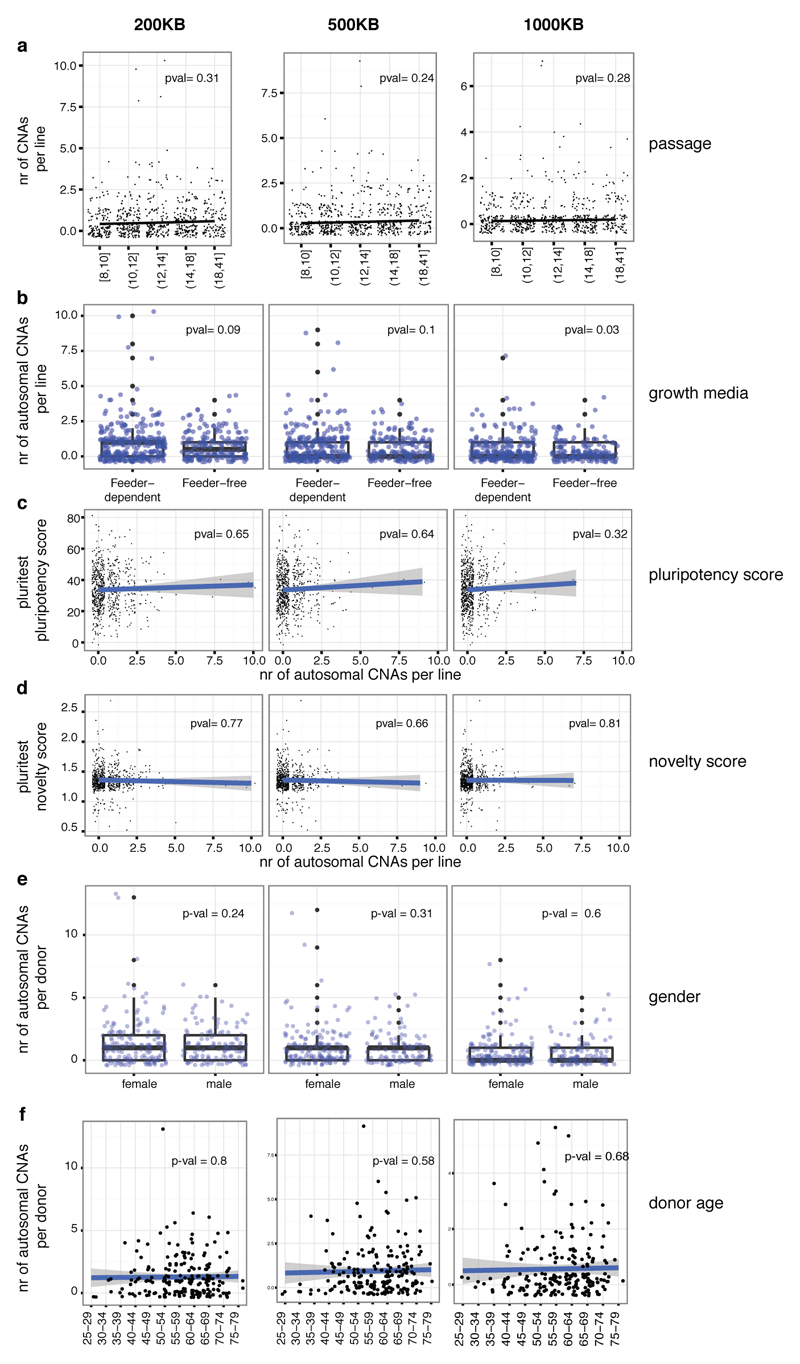
Next, we used genotyping arrays to detect copy number alterations (CNAs) between the iPSC lines and their progenitor fibroblasts. For this purpose, we developed a computational approach10 that can detect genetic abnormalities of >200 Kb occurring in 20% or more cells. We identified trisomies in 4% of lines (none of the selected lines), and 41% of lines (18% of the selected lines) harboured one or more CNAs of, on average, 7.15 Mb in length with duplications outnumbering deletions by 2.8 to 1 (Fig. 1e, Supplementary Table 2). Although the majority of CNAs were unique to single iPSC lines, 22% were also observed in at least one replicate line from the same donor (at least one base pair overlap), and 15% were identified in all replicates (Fig. 1f). We found no significant association between the number of CNAs and either passage number, donor age, gender or PluriTest score of a line (P > 0.09, Fig. 1g, Extended Data Fig. 3).
CNAs observed in pluripotent stem cells (PSCs) are known to recur at certain genomic locations11–13. We observed 35 regions where CNAs occurred significantly more often than expected under a uniform genomic distribution, including whole chromosome duplication of the X chromosome (P = 1.5x10-9), 20 sub-chromosomal duplications, 11 deletions and three regions with both duplications and deletions (Fig. 2a, Supplementary Table 2). The three most frequent CNAs (X trisomy, chromosome 17 and 20) have been previously observed in PSCs12,14,15, but others are newly identified, to our knowledge.
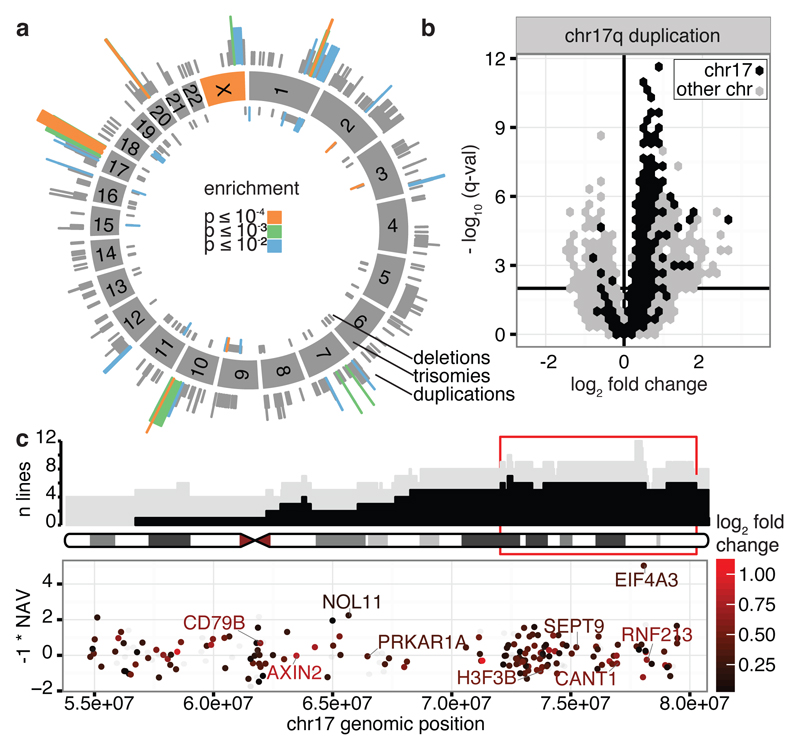
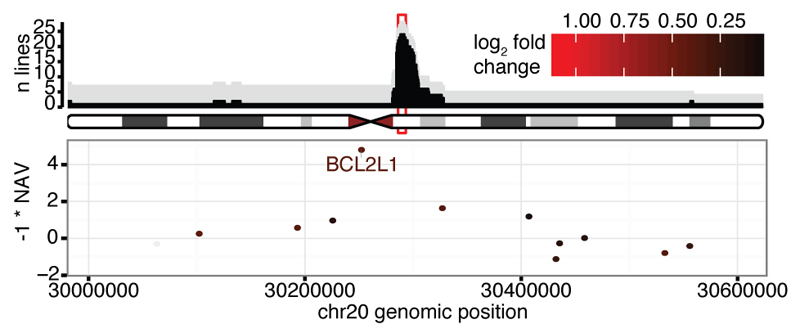
Figure 2. Locations and consequences of recurrent CNA regions.
(a) Genomic locations of CNAs. Colours denote the significance level of recurrence. (b) Genes differentially expressed between lines with CN 2 and 3 for the recurrent chr17 CNA. Horizontal bar denotes 1% FDR threshold (Benjamini-Hochberg). (c) Top panel shows genomic location versus number of lines with CN 3 (grey) and with a CNA (black). Bottom panel shows the NAV gene score from ref22 and log2 gene expression fold change between the iPSC lines with CN 2 and 3 (color scale), in the region highlighted in red in the top panel. Highlighted genes are up-regulated when copy number increases, known onco/tumour-suppressor genes and/or genes with NAV score in the top 2%.
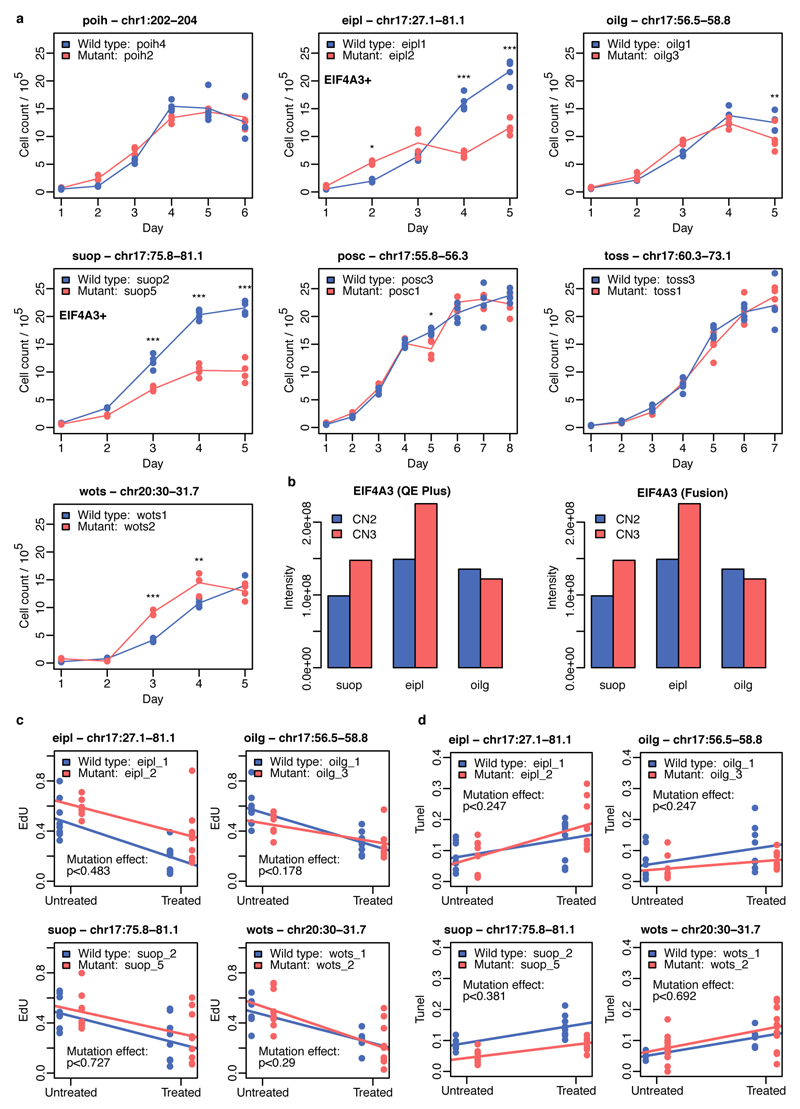
Although recurrent CNAs could be due to mutational hotspots we did not find a significant overlap between our recurrent CNA set and annotated chromosome fragile sites16 (17% overlap, P = 0.075). Recurrent CNAs could also arise if duplication or deletion of specific genes led to a selective advantage. To identify potential targets of selection, we defined peak regions of amplification (regions of maximum recurrence e.g. Fig. 2c and Extended Data Fig. 4) within each CNA and identified expressed genes (FPKM>0 in >10% of lines). Fourteen candidate regions contained fewer than six expressed genes including genes with establised roles in cancer progression (DOCK1, FATS, WWOX, STAG2 and XIAP)17–21. In regions with larger numbers of genes we searched for: (i) significant differential expression between lines with copy number 2 and 3 (ii) reported oncogenes from COSMIC20 and (iii) high scoring genes (top 2%) in a genome-wide siRNA screen for hESC cell proliferation22 (Fig. 2c, Extended Data Fig. 4; Supplementary Table 2). This approach identified BCL2L112 on chr20q11.21, EIF4A3, NOL11 and seven other genes on chr17q and UTP6 and SUZ12 on chr17q11.2. One candidate, EIF4A3, scored more highly than BCL2L1 in reducing ESC proliferation (Fig. 2c, top 0.1% of genes), was highly expressed in iPSCs, and over-expressed in lines with increased copy number, at both the mRNA (Q = 2x10-5) and protein level (Extended Data Fig. 5). Finally, we compared lines from the same donor with and without CNAs to test for genome-wide effects on gene expression levels and, in a subset of cases, for effects on cell growth, proliferation and apoptosis (Fig. 2b, Extended Data Fig. 5). The recurrent duplication on chromosome 17 was associated with the largest number of changes in gene expression, including 1,098 genes (FDR < 1%) in trans located on other chromosomes, which were enriched for ‘Neural Crest Differentiation’ and ‘DNA strand elongation’ pathways (PathCards23, Supplementary Table 2). We also detected significant increases and decreases in cell growth rate associated with CNAs on chromosome 17 and 20 (Extended Data Fig. 5).
Sources of iPSC heterogeneity
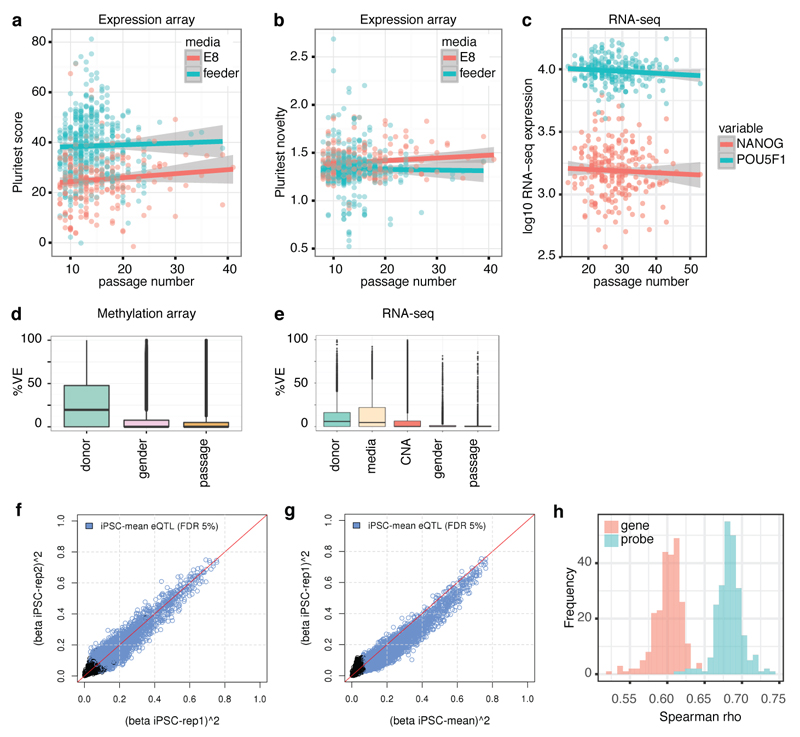
Characterisation of multiple lines per donor enabled us to quantify the variance contributed by between-individual differences (hereafter, ‘donor effects’) and systematically compare this with variance from other factors, substantially extending previous analyses in smaller cohorts6,7 (Fig. 3a-c). We identified consistent donor effects for most measured iPSC phenotypes, ranging from DNA methylation, through mRNA and protein abundance to pluripotency, differentiation, and cell morphology (Fig. 3b,c). After accounting for assay-specific batch factors (full list in Methods), donor effects explained 5.2-26.3% of the variance in the genome-wide assays (Fig. 3a), 21.4-45.8% in protein immunostaining (Fig. 3b), and 7.8-22.8% in cellular morphology (Fig. 3c). Collectively, these results indicate that differences between donor individuals affect most iPSC cellular traits.
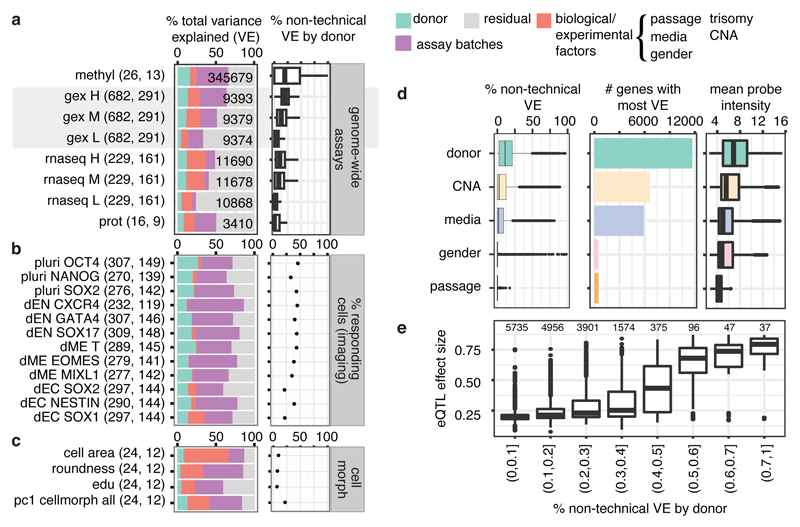
Figure 3. Variance component analysis of HipSci assays.
(a-c) Partitioning of variance in genomic and proteomic assays (a), differentiation and pluripotency markers (b) and cell morphology (c). Panels show total variance (left) and proportion of variance explained by donor, accounting for technical covariates (right), with numbers of lines and donors in parenthesis. For genomic assays, genes are divided into low (L), medium (M) and high (H) expression. (d) Partitioning of variance in microarray gene expression into donor, media, CNA, gender or passage number at the time of the expression assay. Left: the distribution of variance components. Middle: the number of genes where each factor explains the most variance. Right: mean expression of genes with most variance explained by a factor. (e) Donor variance component versus expression array eQTL effect sizes. Numbers denote the number of array probes in each bin.
We further partitioned iPSC gene expression variation using the Tier 1 expression array data, the assay with the largest number of donors and lines. Of the 25,434 probes analysed (16,829 genes) (Supplementary Table 3), donor effects explained the largest proportion of variation in 46.4% of probes (53.3% of genes), substantially more than any other factor, including copy number status (23.4%), culture conditions (26.2%), passage (2%) and gender (1.9%, Fig. 3d). Donor effects were common, and consistent across large numbers of genes, while others such as CNA status had larger effects on a smaller number of genes (Fig. 3d). We observed minor effects of gender and line passage number on RNA-seq, methylation and protein immunofluorescence (Fig. 3d, Extended Data Fig. 6). Likewise, we did not observe substantial changes in PluriTest scores, or pluripotency marker expression across passages (P > 0.3, Extended Data Fig. 6), reflecting that pluripotency was maintained during culture. In principle, the estimated donor variation could arise due to shared reprogramming environment because lines were derived from the same population of fibroblast cells. However, expression quantitative trait locus (eQTL) effect sizes mapped using Tier 1 expression data (Supplementary Table 4, Extended Data Fig. 6) revealed that higher donor variation was associated with larger effect sizes of lead eQTL variants (Fig. 3e), suggesting that donor variance primarily reflects genetic differences.
Identification and characterization of iPSC-specific regulatory variants
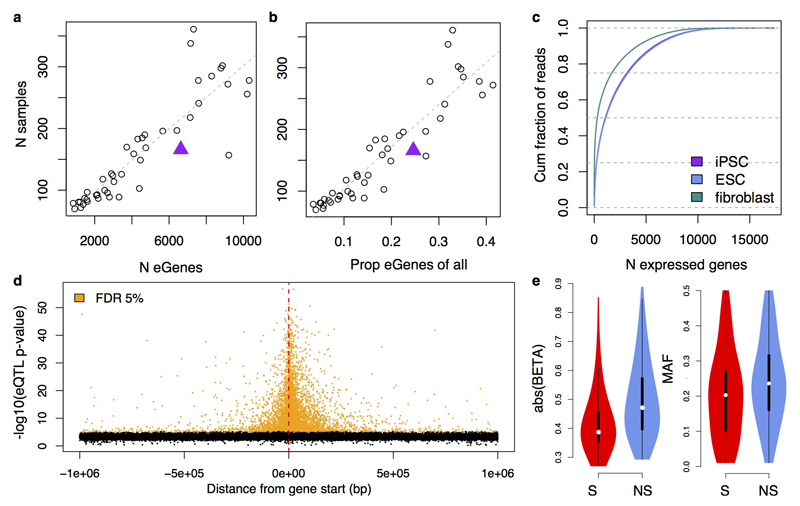
Using RNA-seq data from 166 unrelated donors (median sequencing depth 38M reads), we next mapped eQTLs in a 1 Mb cis-window from the gene start. We identified 6,631 genes with an eQTL (FDR 5%, hereafter ‘eGenes’), 598 of which had a significant secondary eQTL (Supplementary Table 4). Power to discover eGenes in iPSCs was comparable to that in somatic tissues24 given our sample size, and iPSC eQTLs showed similar genomic properties to eQTLs in cell lines and tissues (Extended Data Fig. 7; Supplementary Table 5).
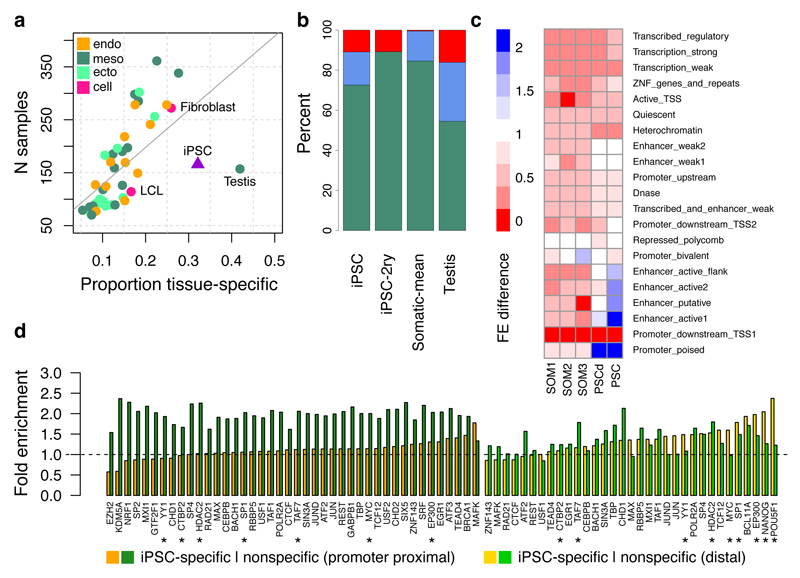
As many eQTLs are shared among tissues9,25,26, we sought to place iPSC eQTLs in the broader context of somatic tissues. We assessed iPSC eQTL replication across 44 GTEx tissues (lead eQTLs and proxy variants, r2 > 0.8, defining replication as P < 0.01/45; Methods), revealing 2,131 eQTLs that were specific to iPSCs (Fig. 4a). We also considered secondary eQTLs, identifying a similar proportion of iPSC-specific genetic effects (both 32%). Most tissue-specific signals (72%) occurred in genes with at least one GTEx eQTL that was not in high linkage disequilibrium (LD) with the lead iPSC eQTL variant, suggesting that iPSC-specific eQTLs are frequently driven by alternative regulatory variants. Only 11% of the iPSC-specific eQTLs could be attributed to tissue-specific gene expression (Fig. 4b), despite greater numbers of expressed genes in iPSCs compared with somatic tissues (Extended Data Fig. 7). Similarly, most somatic tissue-specific eQTLs were also driven by alternative regulatory variants, with only testis showing a substantial fraction (16%) of eQTLs attributable to tissue-specific gene expression (Fig. 4b). Using alternative methods for eQTL detection and assessing the extent of sharing between eQTLs in iPSCs and GTEx tissues, we confirmed that our conclusions were robust to methodological differences between GTEx and our study (Extended Data Fig. 8). However, due to variation in sample size, we cannot rule out that a fraction of the iPSC-specific eQTLs may have a weak effect on gene expression in some somatic tissues.
Figure 4. Comparison of iPSC and somatic tissue eQTLs.
(a) Proportion of tissue-specific eQTLs in iPSCs and 44 GTEX tissues24. (b) Most likely source of tissue-specific eQTLs in iPSCs (lead and secondary), testis and somatic tissues in GTEx (averaged; including cell lines, excluding testis). Breakdown: gene not expressed (red); gene expressed but no eQTL (blue); eQTL effect is driven by distinct lead variants (r2 < 0.8; green). (c) Heatmap of the fold enrichment (FE) difference between iPSC-specific and non-specific eQTLs at chromatin states from the Roadmap Epigenomics Project27, shown for five aggregated clusters representing 127 cell types (SOM, somatic; PSCd, PSC-derived). Colouring: enriched for iPSC-specific eQTLs (blue), enriched for non-specific eQTLs (red). (d) Enrichment of iPSC eQTLs at promoter proximal and distal transcription factor binding sites in H1-hES cells from the ENCODE Project49. Fold enrichments per factor are shown for iPSC-specific and non-specific eQTLs. Pluripotency-associated factors are indicated with an asterisk.
The transcriptional regulatory networks that maintain pluripotency are unique to stem cells. We next investigated how common genetic variants modulate these networks to produce iPSC-specific genetic effects on expression. We used chromatin state annotations from 127 reference epigenomes from the Roadmap Epigenomics Project27 to quantify the fold enrichment of iPSC-specific and nonspecific eQTL sets across 25 chromatin states (using matched null variants; Methods). iPSC-specific eQTLs were enriched in active enhancers and poised promoters in PSCs and PSC-derived cell types, while shared eQTLs were enriched for active promoters and transcribed regions in somatic tissues (Fig. 4c). iPSC-specific eQTLs were also enriched for binding sites of NANOG, POU5F1 (OCT4), and multiple other pluripotency factors9,28 (Fig. 4d; Extended Data Fig. 8). Our results suggest that common genetic differences between individuals may affect expression regulation during early stages of development.
iPSC eQTLs tag common disease variants
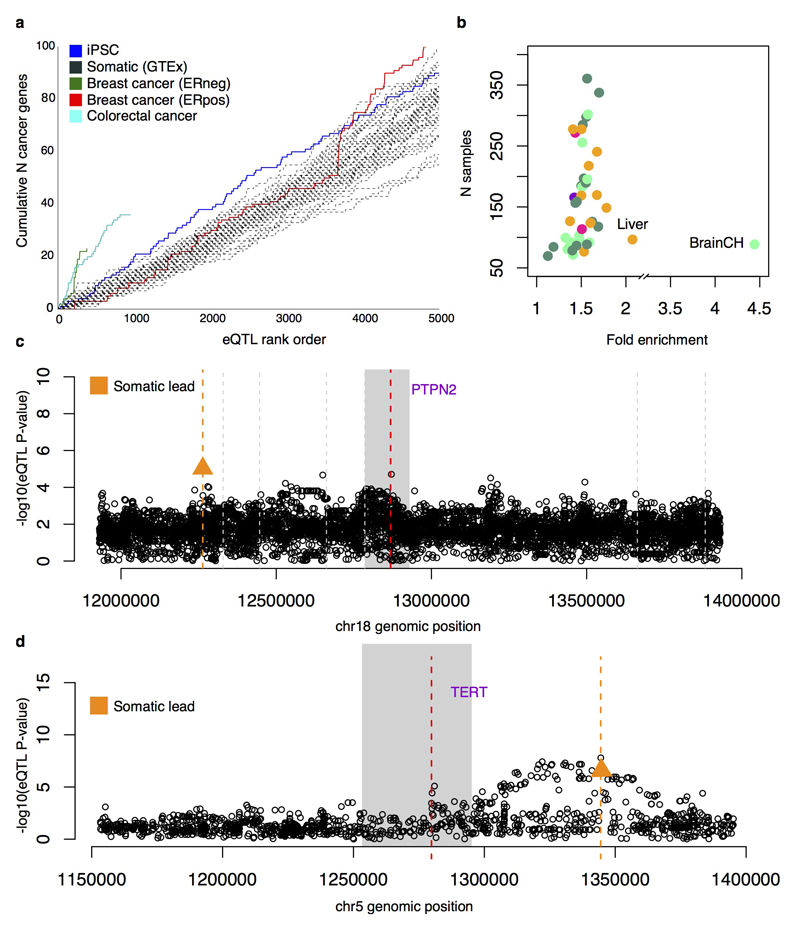
We next identified iPSC eQTLs that may be associated with disease. iPSC eQTLs tagged 322 variants associated in genome-wide association studies with 145 different disease traits, corresponding to a 1.4-fold global enrichment over control variants (Fisher’s exact P = 1.4x10-6), and trait-specific enrichments for seven traits (Supplementary Table 6), a comparable level of enrichment to eQTLs from most somatic tissues (Extended Data Fig. 9). We also observed that iPSC eQTLs tagged a larger number of known cancer genes (COSMIC cancer census 27/04/201620) than somatic tissue eQTLs, with only cancer eQTLs tagging more (Extended Data Fig. 9).
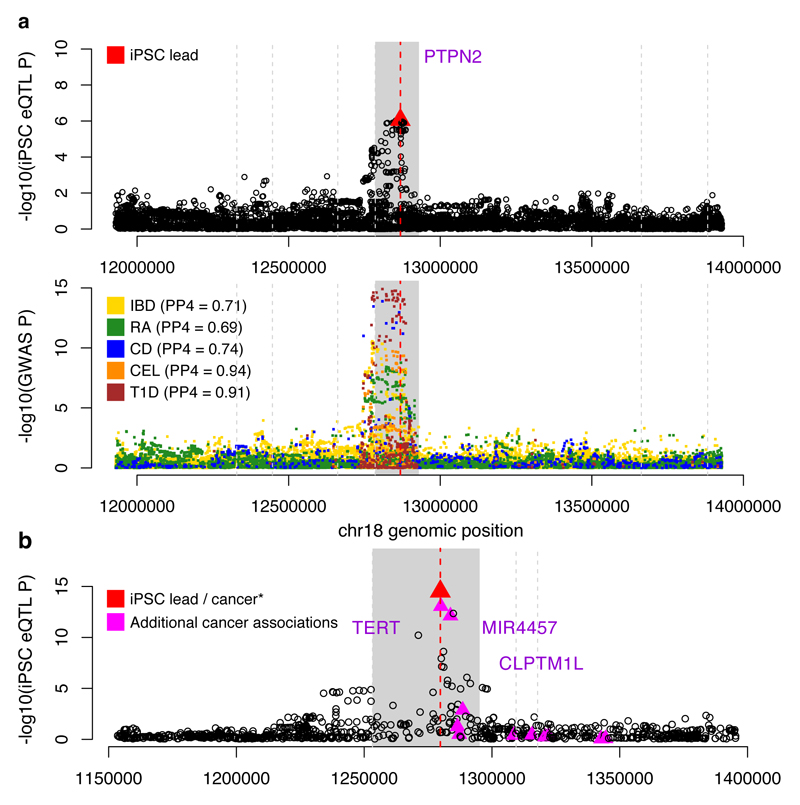
Next, we used statistical colocalisation29 to identify loci where the same causal variant appeared to be driving both an iPSC eQTL and an association with one of 14 complex traits, identifying 233 loci where the posterior probability of a joint association exceeded 0.5 (Supplementary Table 6). Of these, 45 were iPSC-specific, including PTPN2, an iPSC-specific eQTL that strongly colocalised with risk variants for four autoimmune disorders (Fig. 5a). Previous eQTL studies in both immune cells30–32 and GTEx tissues have not identified a PTPN2 eQTL (Extended Data Fig. 9), suggesting that disease risk variants at PTPN2 may function in stem cells, or early development.
Figure 5. iPSC eQTLs tag disease-associated variation.
(a) Colocalised association signal for iPSC expression of PTPN2 (top) and five common diseases (bottom; inflammatory bowel disease, IBD; rheumatoid arthritis, RA; Crohn’s disease, CD; celiac disease, CEL; and type 1 diabetes, T1D). PP4 is the posterior probability that the disease and gene expression associations are driven by the same causal variant29. (b) An iPSC-specific eQTL for TERT (rs10069690) that is associated with risk for breast, ovarian and other cancers.33,34 The lead variant is indicated with a red triangle, the focal gene region in solid grey, and other protein-coding gene start positions by vertical grey lines.
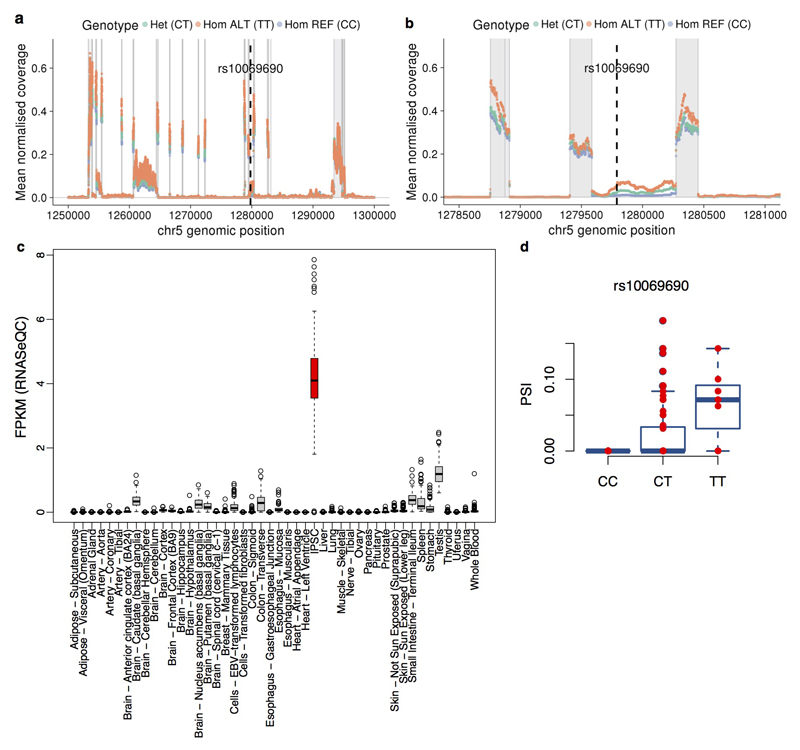
Statistical colocalisation analysis is limited to instances where full summary statistics are available for both traits. For other disease traits available in the GWAS catalogue, we searched for sharing of lead iPSC-specific eQTL and GWAS SNPs. We found six variants where the lead eQTL variant was identical to a catalogued GWAS variant, with no other common variants in LD (r2 < 0.8). One example was rs10069690, the lead eQTL variant for the TERT (Telomerase Reverse Transcriptase) gene (Fig. 5b). Although this variant is associated with germline predisposition to seven cancers33–35, this eQTL is not reported in cancer eQTL studies36–38 nor in any GTEx tissue. Previous studies have reported aberrant splicing of TERT caused by rs1006969039. We quantified TERT intron retention rates and found that the minor allele of rs10069690 increased the fraction of TERT transcripts in which intron four is retained (P = 1.7x10-9, Bonferroni adjusted) (Fig. 5d, Extended Data Fig. 10). Somatic TERT promoter mutations only manifest in differentiated cells, resulting in increased telomerase activity40. We speculate that the germline TERT eQTL we identified in iPSCs results in genotype-dependent variability in telomerase activity in somatic cell types, leading to differential cancer susceptibility.
Discussion
Here we present the most comprehensive analysis yet of genetic and phenotypic data from human iPSC lines. Our study substantially extends on previous work6,7 by demonstrating widespread functional consequences of genetic variation for many molecular and cellular phenotypes in human pluripotent stem cell lines, including in the efficiency with which iPS cells differentiate41–43. This is potentially a consequence of variation in core components of the regulatory networks controlling cellular differentiation and responses to external environmental stimuli, as observed previously in hematopoietic cells and mouse and fly embryos44–46.
We have also created a high-resolution map of recurrent genetic abnormalities in hiPSCs and identified plausible candidate targets of selection. The majority of these recurrent loci are rare and were not reliably identified in previous studies with smaller sample sizes. Compared to previous work11,12, we observed substantially lower levels of genetic aberrations. One possible explanation is that access to donor-matched reference samples helped us more accurately identify germline CNAs that would otherwise have inflated our estimates, while previous studies in ESCs were unable to perform similar comparisons.
Our study provides the highest resolution map to date of common regulatory variation in human PSCs. We show that variation in local gene regulation in iPSCs is similar to that in somatic tissues, with eQTLs driving cell-type specific expression profiles through distal tissue-specific regulatory elements. We have identified eQTLs that function primarily in pluripotent cells, a subset of which tag loci associated with disease. These loci may drive disease-susceptibility through molecular changes early in development or, more generally, in cells with ‘stem-like’ characteristics, which are not well captured by studies of differentiated primary tissues from adult individuals. A compelling example of this is the iPSC-specific eQTL regulating TERT expression. In human tissues, telomerase activity is mainly restricted to stem cells, with most somatic tissues silencing TERT expression. However, cancer cells bypass this tumour suppressive mechanism by reactivating telomerase activity47. This result highlights how iPSCs could be used to study the genetic effects of diseases that manifest in transient states during cellular growth and differentiation, including in cancer48.
The analysis of the recurrence of CNAs and the eQTL map we present are based on a large sample, providing a high-confidence map of molecular associations in iPSCs. We have presented preliminary experimental characterisation of some of the CNAs and eQTLs we detected, however our results are inconclusive. An important next step will be to perform more extensive functional characterisation to understand how iPSC cellular phenotypes are influenced by CNAs and iPSC eQTLs. We anticipate that the lines and data we have generated here will be a valuable starting point for future studies to understand how germline and somatic genetic variation influences iPSC growth and differentiation.
In summary, our study provides a detailed picture of the genetic and phenotypic variability in human pluripotent stem cells, including the major drivers of this variation. Data and cell lines from this study are being made available through www.hipsci.org, the European Collection of Authenticated Cell Cultures (ECACC) and the European Bank for Induced Pluripotent Stem Cells (EBiSC). As the HipSci resource continues to expand in sample size and assays, it will enable the study of subtler genetic effects, under a wider range of conditions, in an increasing range of disease-relevant differentiated cell types.
Methods
Generation of iPSC lines
All samples for the HipSci resource were collected from consented research volunteers recruited from the NIHR Cambridge BioResource (http://www.cambridgebioresource.org.uk). Samples were collected initially under ethics for iPSC derivation (REC Ref: 09/H0304/77, V2 04/01/2013), with later samples collected under a revised consent (REC Ref: 09/H0304/77, V3 15/03/2013).
Fibroblast isolation
Primary fibroblasts were derived from 2 mm skin punch biopsies from each donor. Biopsies were collected in fibroblast growth medium (Advanced DMEM, 10% FBS, 1% L-Glutamine, 0.007% 2-mercaptoethanol and 1% Pen/Strep) in falcon tubes at room temperature. Biopsies were manually dissected using a microscope under a drop of fibroblast medium using sterile scalpels. The biopsy fragments were transferred onto a 60 mm Petri dish containing several drops of fibroblast growth medium. Sterile cover slips were placed onto the dissected pieces of tissue to hold them in place against the bottom of the plate. The explants were cultured for five days and the spent media was removed and replaced with a few drops of media (1 ml) to prevent dehydration. The explants were fed every five days with 1 ml fibroblast media until fibroblast outgrowths appeared. The explants were screened for presence of mycoplasma using a standard PCR kit (EZ-PCR Kit, Gene flow (41106313-001)). On average outgrowths appeared within 14 days, with a small fraction of samples failing to produce outgrowths (12% of cases). Failures were due to contamination (0.5%) or lack of observed outgrowths after 30 days (11%). Approximately 30 days post dissection, when the fibroblasts had reached confluence, the culture was trypsinized and passaged into a 25 cm2 tissue culture flask. When 80-90% confluent, the fibroblasts were further passaged into a 75 cm2 flask. Cells were then expanded to confluency in 225 cm2 flasks (at a split ratio of 1:3) and either cryopreserved at 1-2 million cells per vial in FBS and 10% DMSO or seeded immediately for reprogramming.
iPSC derivation
Fibroblasts were transduced in one well of a six-well plate using Sendai vectors expressing hOCT3/4, hSOX2, hKLF4, and hc-MYC 50 (CytoTune™, Life Technologies, Cat. no. A1377801). The transduced cells were cultured on an irradiated mouse embryonic fibroblast (MEF-CF1) feeder layer on a 10 cm2 tissue culture dish in iPSC medium consisting of Advanced DMEM (Life technologies, UK) supplemented with 10% Knockout Serum Replacement (KOSR, Life technologies, UK), 2 mM L-glutamine (Life technologies, UK) 0.007% 2-mercaptoethanol (Sigma-Aldrich, UK), 4 ng/mL of recombinant Zebrafish Fibroblast Growth Factor-2 (CSCR, University of Cambridge), and 1% Pen/Strep (Life technologies, UK). Cells with an iPSC morphology appeared approximately 25 to 30 days post-transduction. The undifferentiated colonies (six per donor) were picked between days 30-40, transferred onto 12-well MEF-CF1 feeder plates and cultured in iPSC medium with daily media change until ready to passage. Cells were passaged every five to seven days, depending on the confluence and morphology of the cells, at a maximum 1:3 split ratio until established – usually at passage five or six. Once the iPSC lines were established in culture, three of the six lines were selected based on morphological qualities (undifferentiated, roundness and compactness of colonies) and expanded for banking and characterisation.
Transfer to feeder-free culture
Between passages four to eight, selected feeder-dependent iPSC lines were transferred to feeder-free culture. The feeder-dependent iPSC lines were split and passaged onto both feeder-dependent and feeder-free conditions. The feeder dependent lines continued to be cultured on MEF-CF1 feeder plates in iPSC medium, whilst the feeder free lines were cultured in Essential 8 (E8) medium on tissue culture dishes coated with 10 µg/ml Vitronectin XF (StemCell Technologies, UK, 07180). E8 complete medium consists of basal medium DMEM/F-12(HAM) 1:1(Life technologies, UK, A1517001) supplemented with E8 supplement (50X) (Life technologies, UK, A1517001) and 1% Pen/Strep (Life technologies, UK, 15140122). Media was changed daily. To passage feeder-free iPSC lines, cells were washed with PBS and incubated with PBS-EDTA solution (0.5 mM) for 5-8 minutes. PBS-EDTA solution was removed, and cells were resuspended in E8 medium and seeded at split ratios ranging from 1:3 to 1:6 onto Vitronectin coated tissue culture dishes. Cells were passaged every four to seven days (depending on the confluence and morphology of the cells). Once the feeder-free iPSC lines were established in culture, the cells were expanded for banking and characterisation.
iPSC line selection and molecular assays
Each iPSC line was passaged on average 16 times before being expanded for the collection of initial molecular data for quality control (‘Tier 1 assays’). These included genotyping (‘gtarray’), gene expression data (‘gexarray’), and an assessment of the pluripotency and differentiation potential of each line (‘Cellomics’). Pluripotency of the lines was additionally verified in silico, using the PluriTest assay 9. Following Tier 1 assays, one or two lines were selected from a subset of donors (hereafter ‘selected lines’) and further expanded to enable collection of a richer set of molecular data (‘Tier 2 assays’). The criteria for line selection were: (i) level of pluripotency, as determined by the PluriTest assay (ii) number of copy number abnormalities and (iii) ability to differentiate into each of the three germ layers. These included proteomics, DNA methylation (‘mtarray’), RNA-sequencing and high-content cellular imaging. Once Tier 1 genotyping data were collected (see below) cell lines originating from the same donor were checked for possible sample swaps using BCFtools (bcftools gtcheck -G1).
Nucleic acid extraction
DNA and RNA from iPSC lines were extracted using Qiagen Chemistry on a QIAcube automated extraction platform. Sample volume was checked using a BioMicroLab automated volume check system. The PicoGreen assay was used to measure the concentration of the samples, using both Beckman FX liquid handling platforms and Molecular Devices plate readers. Invitrogen E-Gels were run to check sample integrity; the loading of these gels was automated using Beckman FX/NX liquid handling platforms. A standard Fluidigm genotyping assay containing 24 SNPs (22 autosomal and 2 gender markers) was performed to produce a fingerprint of the samples, which was used to confirm sample identity after sequencing or genotyping. The gender markers also allowed for sample swaps and plate orientation issues to be identified prior to downstream analysis. Samples that passed quality control were quantified to 50 ng/µl by the onsite sample management team prior to submission for sequencing.
Genotyping (‘gtarray’)
Experimental processing of arrays
Samples were hybridised to the Illumina HumanCoreExome-12 Beadchip according to the manufacturer's guidelines. Four microlitres (200 ng) of DNA is required for the pre-amplification reaction using a Tecan Freedom Evo. The process is automated except for manual agitation/centrifugation step midway through and at the end of the process. Post-amplification processes (fragmentation, precipitation, resuspension, hybridization to beadchip and xStaining) were completed over three days as per Illumina protocol. Following the staining process, beadchips were coated for protection and dried completely under vacuum before scanning on the Illumina iScan paired with Illumina Autoloader 2.x. Prior to downstream analysis, all samples were subjected to initial quality control to establish that the assay was successful. Sample call-rates below 92.5% were flagged before loading samples into Illumina’s GenomeStudio software. Using Illumina’s QC dashboard, sample performance was assessed by measuring dependant and non-dependent controls that are manufactured onto each beadchip during production.
Genotype calling and imputation
After primary quality control, the Genotyping (GT) module of the GenomeStudio software (Illumina, CA, USA) was used to call the genotypes. For each probe, the GT module estimates the Log R ratio and B-allele frequency for each sample using a clustering model applied to the distribution of signal intensities. These statistics are used internally by GenomeStudio to assign the sample genotypes for each marker. Variant coverage was further increased using statistical imputation and phasing. We constructed a reference panel of haplotypes from a combination of SNPs and small insertions and deletions (indels) in the UK10K cohorts and 1000 Genomes Phase 1 data 51,52. Samples were independently imputed using IMPUTE2 v2.3.1 53 and subsequently phased using SHAPEIT v2.r790 54. This analysis was done in chunks of average 5 Mb, with 300 Kb buffer regions on each side. IMPUTE2 was used with its default MCMC options (-Ne 20000 -k 80) for autosomes and -Ne 15000 -k 100 for X chromosome. SHAPEIT was run without MCMC iteration (-no-mcmc) so that each sample was phased independently using the reference panel as the haplotype scaffold regardless of the phasing of the other samples. Single-sample VCFs were merged together and INFO scores were re-calculated from genotype posterior probabilities (GPs). Variants with INFO score less than 0.4 were excluded from further analysis. Cell lines originating from the same donor were checked for possible sample swaps using BCFtools (bcftools gtcheck -G1). Swapped samples typically had large number of discordant genotypes (>20%), whereas in samples from the same donor the number of discordant genotypes was low (<0.3%), even in the presence of large copy number variation.
Gene expression arrays (‘gexarray’)
Experimental processing of arrays
500 ng of total RNA for each sample was amplified and purified using the Illumina TotalPrep-96 RNA Amplification kit (Life Technologies, UK), according to the manufacturer’s instructions. Biotin-Labelled cRNA was then normalized to a concentration of 150 ng/ul and 750 ng was hybridised to Illumina Human-12 v4 BeadChips (Illumina, CA, USA) for 16 hours (overnight) at 58°C. Following hybridisation, BeadChips were washed and stained with streptavidin-Cy3 (GE Healthcare, UK). BeadChips were then scanned using the BeadArray reader and image data was then processed using GenomeStudio software (Illumina, CA, USA).
Re-mapping of array probes
Prior to analysis, array probe sequences (Nprobes = 47,230; length 50 bp) were re-mapped against the human genome build 37 using BWA version 0.7.5 55.
We first mapped the sequences allowing no mismatches (-n 0, seeding disabled) and kept uniquely mapping probes with a minimum mapping quality (MAPQ) of 10 (–q 10). These sequences were then mapped again, this time allowing one mismatch (-n 1). Again, only uniquely mapping probes with MAPQ > 10 were retained, resulting in a total of N = 37,740 probes. We further removed all probes that overlapped with any variant with a minor allele frequency greater than 0.05 in the main imputed dataset (Nlines = 858). Remaining probes were annotated with Gencode version 19 gene annotations 56 and only probes mapping uniquely to a single gene were kept (final probeset Nprobes = 25,604, representing 17,116 unique genes of which 14,569 are protein coding.
Pre-processing and normalization of data
Gene expression profiles were measured with Illumina HumanHT-12 v4 Expression BeadChips. After limiting the dataset to iPSC lines derived from fibroblast of healthy donors, we obtained data from 711 iPSC lines and 301 somatic fibroblast lines. Probe intensity estimates were normalised separately for the two cell types using the variance-stabilizing transformation implemented in the R/Bioconductor vsn package 57. After normalization, the datasets were limited to the final remapped set of probes (Nprobes= 25,604). We refer to this version of the “gexarray” data by vsn log2 (iPSC/somatic).
Cellular differentiation assay (‘Cellomics’)
Differentiation potential assay
Selected iPSC lines were assessed for their pluripotency and differentiation properties by culturing the cells under conditions favouring the formation of the three embryonic germ layers, and subsequent immunostaining with markers specific for pluripotency and differentiation. Differentiation was performed as described previously 58. Briefly, iPSCs grown in feeder-dependent or feeder-free conditions were harvested using either collagenase and dispase or EDTA, respectively. Colonies were collected, washed in media and mechanically broken up before being re-plated onto 24-well mouse embryonic fibroblast (MEF) feeder plates or pre-coated gelatine/FBS plates. For pluripotency assays, feeder-dependent colonies were seeded on MEF feeder plates and feeder-free colonies onto Vitronectin plates. For the differentiation assay, colonies were grown on gelatine/FBS plates. Prior to differentiation to mesoderm (dME), endoderm (dEN), and neuroectoderm (dEC), cells were cultured overnight in pre-differentiation media CDM-PVA supplemented with recombinant Activin-A (10 ng/ml; CSCR, University of Cambridge) and zebrafish FGF2 (12 ng/ml; CSCR, University of Cambridge).
For differentiation into mesoderm following culture in pre-differentiation media, spent media was removed and replaced with fresh CDM-PVA media containing bone morphogenic protein 4 (BMP4, 10 ng/ml, R&D Systems Inc.), FGF2 (20 ng/ml; CSCR, University of Cambridge), recombinant Activin-A (10 ng/ml; CSCR, University of Cambridge), LY29004 (10 mM, Promega, UK.), CHIR99021 (5 mM, Selleckchem) and subsequently cultured for three days. Media was changed daily.
For differentiation into endoderm, following culture in pre-differentiation media cells were further cultured in differentiation media for three days. Briefly, day one media was removed and replaced with fresh CDM-PVA media supplemented with recombinant Activin-A (100 ng/ml; CSCR, University of Cambridge), zebrafish FGF2 (80 ng/ml; CSCR, University of Cambridge), BMP4 (10 ng/ml R&D Systems Inc.), LY29004 (10 mM), and CHIR99021 (3 mM). Day two media was removed and replaced with fresh CDM-PVA supplemented with recombinant Activin-A (100 ng/ml), zebrafish FGF2 (80 ng/ml), BMP4 (10 ng/ml), and LY29004 (10 mM). Day three media was removed and replaced with RPMI media supplemented with B27 (1x, Life Technologies UK), recombinant Activin-A (100 ng/ml), zebrafish FGF2 (80 ng/ml), and Non-Essential Amino Acids (1x, Life Technologies UK).
For differentiation to neuroectoderm, iPSCs were grown for 12 days in CDM-PVA supplemented with SB431542 (10mM; Tocris Bioscience), FGF2 (12 ng/ml, CSCR University of Cambridge), and Noggin (150 ng/ml, R&D Systems Inc.). Media was changed daily.
Immunostaining for pluripotency and differentiation markers
For the detection of pluripotency and differentiation markers, cells grown in 24-well plates were fixed with 4% paraformaldehyde for 20 minutes. Cells were permeabilized and blocked with 10% donkey serum and 0.1% Triton X-100 in PBS. Subsequently, cells were stained with primary antibodies overnight at 4°C and finally incubated with fluorochrome-labeled secondary antibodies (Invitrogen, UK). The primary antibodies used for detecting pluripotency markers were: anti-OCT4 (SC-5279, Santa Cruz Biotech, USA), anti-SOX2 (AF2018, R&D, UK), anti-NANOG (AF1997, R&D, UK). The primary antibodies used for detecting endoderm markers were: anti-SOX17 (AF1924, R&D, UK), anti-CXCR4 (MAB173-100, R&D, UK and anti-GATA4 (SC-25310, Santa Cruz Biotech, USA). The primary antibodies used for detecting mesoderm markers were: anti-Brachyury (AF2085, R&D, UK), anti-EOMES (Ab23345, Abcam, UK) and anti-MIXL1 (SC-98664, Santa Cruz Biotech, USA). The primary antibodies used for detecting neuroectoderm markers were: anti-NESTIN (AB22035, Abcam, UK), and anti-SOX1 (AF3369, R&D, UK) anti-SOX2 (AF2018, R&D, UK). The secondary antibodies used were: Donkey anti-goat AF488 (Invitrogen, UK), Donkey anti-mouse AF488 (Invitrogen, UK), Donkey anti-rabbit AF488 (Invitrogen, UK). Additionally, DAPI staining was used to label cell nucleus in order to facilitate cell segmentation.
Images were captured and quantified using a Cellomics Array Scan imaging system. Briefly, images were taken in 24-well plates. Individual plates were used to either measure pluripotency markers or markers to assess differentiation for one of the germ layers. Each plate contained cells from one or two cell lines, as well as technical replicates for each measurement. Three types of plate layouts were considered throughout the project: Two-channel, three-channel, and three-channel with single staining. For all layouts, the signal from the DAPI staining was read in the first channel. The first columns of each plate were used for marker staining; subsequent columns (one or two) were stained with the secondary antibody to measure background signal (Extended Data Fig. 1).
Processing of images on the Cellomics instrument
Individual wells in the plate were imaged consecutively, either until the whole plate was imaged or until 10,000 individual cells were detected. Cell detection was performed based on nucleus segmentation from DAPI staining. All considered markers except for CXCR4 are nuclear markers, so their signal intensities were measured in the segmented nucleus area. The cell surface marker CXCR4 was quantified in a circle around the segmented nucleus. For each cell and marker, we used the average intensity within the respective quantification area as final readout. Each batch of lines for staining included the reference line (‘CTRL0214pf-iely’). This reference line was used to determine parameter values for cell size (usually around 30-400) and an approximate intensity threshold for detecting responding cells.
To quantify Cellomics phenotypes, we fit a Gamma mixture model to the Cellomics raw intensities (Supplementary Information). Briefly, this model was fit to primary wells as well as background wells (Extended Data Fig. 1), thereby estimating both the proportion of responding cells as well as the overall intensity (expression) of the corresponding cells.
For downstream analyses, technical replicates on each plate were aggregated using average values. Analogously to the processing steps for gene expression arrays, we regressed out batch (derived from the date of staining), media type, gender, passage number, plating technician, fixation technician, and the technician in charge of the staining. Analyses of proportions of responding cells are based on estimates of the proportion of responding cells (Fig. 1c,d). Analyses that consider intensities were based on quantitative expression estimates, averaged across individual markers for a given layer (Extended Data Fig. 1c).
Proteomics
Sample preparation
Frozen iPSC pellets were thawed and washed with PBS twice prior to lysis. The protein content of the cells was extracted by re-dissolving the pellets in 8 M urea, 100 mM TEAB, pH 8.5 and mixing at room temperature for 15 minutes. Next, the DNA content of the cells was sheared using ultrasonication. The protein amount was determined using a fluorescence based assay (EZQ, Life Technologies) prior to double digestion using mass spectrometry grade lysyl endopeptidase (Wako, Japan) and trypsin (Pierce) in a substrate-to-enzyme ratio of 1:50; w:w, at a final urea concentrations of 2 M and 0.8 M, respectively. The digested proteins were desalted using sepak vacuum cartridges (waters) and dried in vacuo. The desalted peptides were redissolved in (10 mM borate at pH 9.3 : acetonitrile; 80:20; v:v) for hSAX fractionation using a 40 minute gradient. A total of 16 fractions were collected, desalted and dried. The hSAX fractions were redissolved in 5% formic acid for label-free LC-MS analysis. In addition to individual samples, a composite reference sample (‘HPSI_composite_1503’) was constructed by pooling together protein lysates from 43 iPSC lines. 2 mg of protein was used for each. All samples in this reference were of fibroblast origin and reprogrammed with sendai virus.
For Tandem Mass Tag (TMT)-based quantification, the dried peptides were re-dissolved in 100mM TEAB (50 µL) and their concentration was measured using a fluorescent assay (CBQCA) (Life Technologies). 100 µg of peptides from each cell line to be compared, in 100 µL of TEAB, were labelled with a different TMT tag (20 µg ml-1 in 40 µL acetonitrile) (Thermo Scientific), for two hours at room temperature. After incubation, the labelling reaction was quenched using 8 µl of 5% hydroxylamine (Pierce) for 30 minutes and the different cell lines/tags were mixed and dried in vacuo.
The TMT samples were fractionated using off-line high pH reverse phase chromatography: samples were loaded onto a 4.6 x 250 mm Xbridge™ BEH130 C18 column with 3.5 µm particles (Waters). Using a Dionex bioRS system, the samples were separated using a 25-minute multistep gradient of solvents A (10 mM formate at pH 9) and B (10 mM ammonium formate pH 9 in 80% acetonitrile), at a flow rate of 1 ml/min. Peptides were separated into 48 fractions, which were consolidated into 24 fractions. The fractions were subsequently dried and the peptides re-dissolved in 5% formic acid and analysed by LC-MS.
LC-MS/MS
Label-free analysis
RP-LC was performed using a Dionex RSLC nano HPLC (Thermo Scientific). Peptides were injected onto a 75 μm × 2 cm PepMap-C18 pre-column and resolved on a 75 μm × 50 cm RP- C18 EASY-Spray temperature controlled integrated column-emitter (Thermo) using a four-hour multistep gradient from 5% B to 35% B with a constant flow of 200 nL min-1 as described previously 59,60. The mobile phases were: 2% ACN incorporating 0.1% FA (Solvent A) and 80% ACN incorporating 0.1% FA (Solvent B). The spray was initiated by applying 2.5 kV to the EASY-Spray emitter and the data were acquired on a Q-Exactive Orbitrap (Thermo Scientific) under the control of Xcalibur software in a data dependent mode selecting the 15 most intense ions for HCD-MS/MS.
TMT-based analysis
5% of the material was analysed using an orbitrap fusion tribrid mass spectrometer (Thermo Scientific), equipped with a Dionex ultra high-pressure liquid chromatography system (nano RSLC). RP-LC was performed using a Dionex RSLC nano HPLC (Thermo Scientific). Peptides were injected onto a 75 μm × 2 cm PepMap-C18 pre-column and resolved on a 75 μm × 50 cm RP- C18 EASY-Spray temperature controlled integrated column-emitter (Thermo), using a four-hour multistep gradient from 5% B to 35% B with a constant flow of 200 nL min-1. The mobile phases were: 2% ACN incorporating 0.1% FA (Solvent A) and 80% ACN incorporating 0.1% FA (Solvent B). The spray was initiated by applying 2.5 kV to the EASY-Spray emitter and the data were acquired under the control of Xcalibur software in a data dependent mode using top speed and 4 s duration per cycle. The survey scan is acquired in the orbitrap covering the m/z range from 400 to 1400 Th, with a mass resolution of 120,000 and an automatic gain control (AGC) target of 2.0 e5 ions. The most intense ions were selected for fragmentation using CID in the ion trap with 30 % CID collision energy and an isolation window of 1.6 Th. The AGC target was set to 1.0 e4 with a maximum injection time of 70 ms and a dynamic exclusion of 80 s.
During the MS3 analysis for more accurate TMT quantifications, 5 fragment ions were co-isolated using synchronous precursor selection using a window of 2 Th and further fragmented using HCD collision energy of 55%. The fragments were then analysed in the orbitrap with a resolution of 60,000. The AGC target was set to 1.0 e5 and the maximum injection time was set to 105 ms.
Quantification
Label-free proteomics samples were analysed with MaxQuant v. 1.3.0.5 software 61 as a single batch against a Uniprot reference database, constructed from all Swissprot entries (N = 20,043) and their isoforms (N = 21,914). Run parameters have been deposited to PRIDE along with the data and the full MaxQuant quantification output (PXD003903).
For the selected TMT-based experiments (Extended Data Fig. 5b), the TMT-labelled samples were analysed using Maxquant v. 1.5.3.30. Proteins and peptides were identified using the UniProt human reference proteome database (Swiss Prot). Run parameters have been deposited to PRIDE along with the full MaxQuant quantification output (PXD005506).
Pre-processing and normalization of data
Data for analysis was obtained from the “ProteinGroups.txt” output of MaxQuant. Contaminant and reverse hits (N = 3,419) were excluded from analysis. For each sample, the total protein abundance was calculated by summing up protein intensity (‘Intensity’) values across all proteins and protein groups. This value was then used to scale all quantification values (‘iBAQ’) per sample. For a protein or a protein group to be considered, we required at least one unique peptide mapping to it. Overall, we quantified 10,097 protein groups (4,877 unique proteins) in at least one of the samples. Only unique protein entries quantified in at least half of the samples were used in the subsequent analyses (3,435 proteins). The mean pair-wise correlation of samples was 0.87 for unique proteins (Spearman rank correlation). Based on the clustering of samples (principal component analysis and pairwise correlation of protein quantification; data not shown), one sample appeared as an outlier (‘HPSI0713i-darw_1’) and was excluded from further analyses.
DNA methylation (‘mtarray’)
Sample preparation and experimental processing of arrays
500 ng of DNA was used for bisulfite conversion using the Zymo Research EZ-96 DNA Methylation Kit. The bisulfite converted DNA extracts were hybridized to Infinium 450K BeadChips (Illumina). Due to the differences in sample plates between the completed Zymo assay and the Illumina assay, pre-amplification was performed manually by following the Illumina MSA4 SOP. Once complete, sample and reagent barcodes were simmed through the Illumina LIMS tracking software. Four microlitres (200 ng) of sample is required (Illumina guidelines) for the pre-amplification reaction using the Tecan Freedom Evo. No further quantification step was performed after the completion of the Zymo assay. Labelling was performed automatically during the post-amplification xStain process with Biotin and DNP labelled antibodies. The HumanMethylation450_15017482_v.1.1 arrays were scanned with iScan (Illumina) in accordance with the manufacturer’s protocol.
Quantification and normalization of data
Methylation profiles were measured with HumanMethylation450_15017482_v.1.1 arrays. GenomeStudio v2011.1 (Methylation Module 1.9.0; Illumina) was used to export the raw data as. idat files, which were then processed with the minfi Bioconductor package 62. Samples were normalised using the stratified quantile normalisation implemented in the ‘preprocessQuantile’ function and probes were annotated to genomic locations using the IlluminaHumanMethylation450kanno.ilmn12.hg19 Bioconductor annotation package. Subsequently, genotyping probes or probes overlapping any dbSNP or 1000 Genomes variant loci were discarded. M-values, defined as the logarithm of the fraction of the methylated and unmethylated channels M = log( Meth / Unmeth ), were used in the variance component and other downstream analysis.
RNA-sequencing
Library preparation and sequencing
mRNA in total RNA was isolated and converted into non-stranded or stranded libraries. Non-stranded libraries were produced manually using reagents provided in the Illumina TruSeq RNA Sample Preparation Kit v2 in accordance with manufacturer’s recommendations. The protocol was modified to produce size-selected libraries by modifying the fragmentation conditions and using a Caliper LabChip XT instrument. Stranded libraries were prepared using a NeoPrep Library Prep System and the reagents provided in the Illumina TrueSeq Stranded mRNA Library Preparation kit. The stranded library prep workflow is similar to the non-stranded workflow, except that it involves additional ribosomal reduction chemistry to maximise the percentage of uniquely mapped reads. Following purification, the RNA was fragmented and synthesised into cDNA using a reverse transcriptase process. The products were then enriched with PCR (maximum 10 cycles) to create the final cDNA library. Enriched libraries were subjected to 75 base paired-end sequencing using Illumina HiSeq 2000 v3 kits following manufacturer’s instructions.
Pre-processing of sequence data
Raw RNA-seq reads were aligned using STAR v2.4.0 63 against the 1000 Genomes Phase2 reference genome assembly that integrates the GRCh37 primary assembly with the human decoy sequence 37d5. Exon-intron junctions derived from Gencode v19 transcript annotations 56 were used to improve the alignments.
The same approach was taken to re-align data for two tissues from the GTEx Project (Extended Data Fig. 8). Raw fastq-files were obtained from dbGap (accession phs0004242.v6.p1.c1) for Adrenal Gland (N=126 samples) and Esophagus Gastroesophageal Junction (N=127 samples; limited to 126 unique samples used in the GTEx V6p map).
Quantification and normalization of data
Mapped reads were quantified on the level of genes using HTSeq version 0.6.1p1 64 and annotations from Gencode v19 56. We used the ‘union’ method of ‘htseq-count’ for unstranded libraries (-s no) and considered only uniquely mapping reads (-a 255; with 255 indicating uniquely mapped reads from the STAR aligner). Of note, STAR only outputs properly paired reads. Raw gene counts were scaled across individuals with scaling factors obtained with DESeq 65. The same approach was used to generate ‘probe-level’ counts using the final re-mapped set of gexarray probes. These were used to filter expressed probes in the CNA analysis. Finally, an alternative set of gene-level quantifications was generated for quality control purposes using RNA-SeQC v1.1.8 66. To match the original GTEx v6p quantifications as closely as possible, we ran RNA-SeQC with the -strictMode flag and used custom exon annotations generated and used by GTEx (gencode.v19.genes.v6p_model.patched_contigs.gtf.gz) to obtain gene RPKMs. The same RNA-SeQC quantification pipeline was applied to the two re-mapped GTEx tissues.
High-content cellular imaging
Sample preparation and cellular imaging
Each line was cultured and plated as previously described 67. Briefly, 96-well plates were coated with three concentrations of Fibronectin in alternated columns in a randomised fashion. Cell lines were seeded also in rows in a randomised fashion. 3,000 cells were plated and fixed after 24 hours. EdU was incorporated 30 minutes before fixation. Plates were then fixed and stained with DAPI and cell mask and EdU staining. Images were acquired using the Operetta (Perkin Elmer) high content device. Using the Harmony software, measurements were derived for each cell. Measurements included intensity features (DAPI, EdU), morphology features (cell area, cell roundness, cell width to length ratio, nucleus area, nucleus roundness, nucleus width to length ratio) and context features related with cell adhesion properties (number of cells per clump). Processing quantification and normalization of data was performed as previously described 67. The Cellomics fluorescence imaging data was used to quantify the fraction of cells expressing each protein marker independently. In the absence of co-staining information we sought to use the marginal fractions of cells expressing each marker to calculate lower and upper bounds for the fractions of cell estimating all markers simultaneously. Let be the fraction of cells expressing protein marker and the fraction of cells expressing all n markers simultaneously, then this value is bounded by:
Variance component analysis
Feature (gene, protein, or probe) intensity estimates for each of the assays were pre-processed and normalised as described in the individual assay sections and subsequently transformed into a standard normal distribution across lines. For each feature in each assay, variance was partitioned using a linear mixed model (implemented in the lme4 R package) fitted with all metadata variables as random effects. Only lines with complete metadata information were included in each one of the analyses, these numbers are shown in parenthesis in Fig. 3a-c. The variance components were normalized to sum to one and subsequently averaged across the different sets of features considered (Fig. 3). The fraction of non-technical variance explained by each biological or experimental factor refers to the variance explained by the factor, divided by the total variance minus the variance explained by assay batches (see below for a definition of experimental and assay batch factors). Confidence intervals for the Cellomics variance components were obtained with the ‘profile’ method implemented in the ‘confint’ function of the lme4 package. The following random effects were included for each assay:
Methylation - Donor. Experimental factors (summed up to produce Fig. 3a): gender, passage interval at time of assay. Assay batches (summed up to produce Fig. 3a): sentrix id, sentrix array (within sentrix id), bisulfite conversion plate, year and month of assay, year and month of Tier 1 assays.
Expression microarrays - Donor. Experimental factors: gender, passage interval at the time of assay, culture system at time of assay, trisomy status, recurrent CNA status. Assay batches: array batch, year and month of assay, beachip id, beadchip array (within beadchip id), technician id, assay performed before or after April 2014.
RNA-seq - Donor. Experimental factors: gender, passage interval at time of assay, trisomy status, recurrent CNA status, culture system at time of assay. Assay batches: year and month of assay, year and month of Tier 1 assays.
Proteomics (uniquely identified proteins) - Donor. Experimental factors: gender. Assay batches: year and month of QC assays, instrument, year and month of analysis.
Cellomics - Donor. Experimental factors: gender, culture system at the time of assay, passage interval at time of assay. Assay batches: date of staining, plating technician id, fixation technician id, staining technician id, primary antibody lot and secondary antibody lot.
Cellular morphology assays (cell area, roundness, EDU, PC1 cellmorph) - Donor. Experimental factors: gender, cell line, fibronectin concentration. Assay batches: plate, row.
CNA analysis
Pairwise fibroblast-iPSC CNA detection
Copy number differences between fibroblast and iPSC lines from the same donor were checked using a HMM algorithm implemented in BCFtools/cnv for this purpose 10. In order to distinguish between normal and novel copy number variation as well as to reduce the number of false calls, the program was run in the pairwise mode (bcftools cnv -c <donor> -s <derived>) with default parameters. The CNA calls were filtered to exclude calls with quality score smaller than 2, deletions with fewer than 10 markers, and duplications with fewer than 10 heterozygous markers. Three sets of CNA calls were generated: a more lenient set containing all calls >= 0.2 Mb in length, a set with all calls >= 0.5 Mb and a stricter subset of the previous with calls >= 1 Mb.
Statistical significance of recurrent CNAs was estimated from the complementary cumulative distribution function of the binomial distribution, and the significance of sub-chromosomal events was estimated using a permutation test (Supplementary Information).
Overlap with annotated regions
To assess the significance of the overlap between CNAs and annotated regions (namely chromatin fragile sites 16 and recurrent somatic copy number altered regions in cancer 68 we randomly generated a set of 2,000 matched control regions for each CNA. Control regions were generated so that they had the same size as the CNA, did not overlap with telomeres or centromeres, and did not overlap the original CNA. Overlaps were determined between the CNA and the annotated regions and between the matched control regions and the annotated regions to calculate an empirical P-value.
Association between CNAs and gene expression
To determine the functional consequence of CNAs with regards to gene expression we first selected CNAs for which five (~1%) or more of the cell lines had a copy number different from two (regardless of copy number in the corresponding somatic cells). For each of these CNAs we took the copy number at the region of peak coverage for each cell line (see CNA coverage plots in Fig. 2c and Extended Data Fig. 4). We then defined the set of expression array probes to test by choosing only expressed probes. Here a probe was defined as expressed if the number of RNA-seq fragment counts (normalised between samples for sequencing depth) mapping to the genomic regions targeted by the probe is greater than 0 in 10% or more of the lines. Finally, we used a linear mixed model to independently test for association between copy number of each CNA and the intensity of each probe. We included culture condition, gender and an interaction between copy number and culture condition as fixed effects; and used donor and assay batch as random effects. Q-values were obtained for each CNA using Benjamini Hochberg to adjust for multiple testing. The same approach was employed to test for association between X chromosome copy number and gene expression, but we limited the tests to female samples and to probes on the X chromosome.
Gene set enrichment analysis
Pathway enrichment analysis of the genes regulated by chr17 was performed with GSEA 69 on the full list of genes ordered by effects size of association between gene expression with copy number and with a custom set of pathways. The custom set of pathways considered comprised 1,156 super pathways from the PathCards database 23 filtered to exclude pathways relating to infectious diseases and pharmacokinetics. Multiple testing correction was performed as described in 69.
Expression quantitative trait loci (eQTL)
eQTL mapping
Pre-processing of genotype data
Variants in the original VCF files were renamed to format “type_chr_pos” (e.g. ‘snp_1_236887241’ or ‘indel:2D_1_18847945’), and filtered for polymorphic and bi-allelic sites with VCFtools v.0.1.12b 70 (vcftools –gzvcf IN --mac 1 --min-alleles 2 --max-alleles 2 --recode --recode-INFO-all –out OUT). The resulting VCF files were then converted to ‘012’ format (vcftools --gzvcf IN --012 --out OUT.012), where 0, 1, and 2 represent the number of non-reference alleles, and further to HDF5 format using a converter function (-g012) from LIMIX (). This resulted in a set of N = 14,644,791 variant sites. To obtain allele dosages, genotype likelihoods (GL) in the original VCF were converted to genotype probabilities (GP) with BCFtools (bcftools +tag2tag IN -O z -o OUT -- --gp-to-gl) and used to define allele dosage as follows: Dosage of alternative allele = GP(REF/ALT) + 2*GP(ALT/ALT). Genotype dosage information was converted to HDF5 format using LIMIX converter (LIMIX converter, --g012_dosage). For eQTL mapping, we included autosomal variants with a minimum minor allele frequency of 1% in our samples and maximum 10% missing values across individuals. Variant sites were further required to have a minimum IMPUTE2 INFO score of 0.4 to assure good imputation quality. Missing genotypes were mean imputed and the dosage of the alternative allele used for mapping.
Pre-processing of expression data
Scaled gene counts were filtered for missing values (maximum 90% missing values, i.e. zero counts, allowed per gene). Zero values were offset by 1, after which the data was log10 transformed and quantile normalized across individuals using R limma normalizeQuantiles function 71. We then ran PEER 72 with full pre-normalized dataset with the following parameters: K=30; covariates = gender, iPSC growth condition (feeder-dependent/E8), mean expression (‘addMean=True’ in PEER); maximum iterations = 10,000. Residuals for each gene were gaussianised, i.e. converted to the quantiles of a standard normal distribution and finally mean centered and standardized prior to mapping. In total, we had 26,936 and 17,116 genes available for mapping (RNA-seq and ‘gexarray’, respectively).
Linear mixed model
Expression quantitative trait loci (eQTL) were identified using a linear mixed model implemented in LIMIX 73,74. eQTLs were mapped in cis, considering a window of 1Mb around the gene start (as defined by Gencode v19 annotations). We modelled the genotype as a fixed effect, with population structure included as random effect. Population structure was modelled with a kinship matrix, calculated as the dot product of the genotypes in trans for each cis window (realized relationship).
eQTL mapping was performed with the following datasets (Supplementary Table 4):
-
1)
iPSC, 166 donors (239 lines), RNA-seq data (hereon the ‘main’ eQTL map)
-
2)
iPSC, 301 donors (711 lines), ‘gexarray’ data
When multiple lines were available for a donor, the mean expression value of lines was used. In the array-based iPSC dataset, which had the largest number of replicate lines available per donor (246 donors with multiple lines), we additionally mapped the eQTLs using two randomly drawn sets of individual lines per donor to assess the replicability of the iPSC eQTLs (Extended Data Fig. 6f-h). In the main RNA-seq based map, we identified both primary and secondary eQTL effects. To identify secondary effects, we repeated the mapping with the genotypes of the lead eQTL variant included as a covariate in the model. Finally, an alternative version of the main eQTL map was generated using a pipeline matched with GTEx V6p eQTLs for quality control purposes (See Supplementary Information).
Multiple testing correction
For cis eQTLs (primary and secondary), to adjust for multiple testing, we permuted genotype sample labels in each cis window 10,000 times, keeping everything else in the model constant. To derive an empirical P-value distribution, the test statistic of the most significant variant in each permutation round was stored. A region-wise adjusted cis P-value was derived from the proportion of permuted test statistics that were larger than the most significant observed test statistic in the region. These threshold P-values were further adjusted for genome-wide analysis using the Benjamini-Hochberg (BH) correction. A gene was considered an eGene if its final BH-adjusted P-value was less than 0.05.
Identification of tissue-specific cis eQTLs
In this study, tissue-specific eQTLs were defined as eQTL effects not replicating in any of the 44 tissues analysed by the GTEx Project 24. Replication was tested on the level of individual eQTL variants between all pairs of tissues. For this analysis, we considered full cis eQTL output of iPSC eQTLs from HipSci and 44 tissues from GTEx (V6p results; 2,025 tissue pairs). The included tissues and cell lines are detailed in Supplementary Table 5a. For each discovery tissue, we tested for the replication of all lead eQTL effects (lead eQTL variant - target eGene; hereon referred to as ‘ePair’) originally reported (FDR 5% in both HipSci and GTEx; Supplementary Table 5a). Ideally, the exact same ePair would have been tested across all tissues. However, due to differences in genotyping methods and allele-frequency due to sample size it was not always possible to query the exact same ePair in all tissues. To account for this, if the original lead variant was not available to test in the query tissue, a proxy variant was tested instead. We note that the selected proxy may differ across the replication tissues. The approach to define the proxy variant is summarized in the Supplementary Information.
Replication was defined as the query variant (original lead or proxy) having a nominal eQTL P < 2.2x10-04 for the same eGene (corresponding to P = 0.01 / 45, where 45 refers to the total number of tissues tested). A lenient threshold was chosen in order to rule out any evidence of replication. High-LD proxies for a lead were defined as having r2 > 0.8 in the UK10K European reference panel and located in the same cis window. All available LD-proxies were tested for replication and the variant with the most significant eQTL P-value in the discovery tissue was stored as the proxy. If no LD-proxies could be tested, the cis variant with the most significant eQTL P-value in the discovery tissue overall was selected. Overall, the same lead variant was available to test in 95% of tests across tissues (median of discovery tissues, with each discovery tissue represented as the median of tests across all replication tissues; 90% for iPSC eQTLs). A high-LD proxy for the lead was tested 3.6% of the time (7.3% in iPSC), while the best available cis variant was tested only 1.1% of the time (2.7% in iPSC). Of the rare cases when the best available variant was tested, the selected variant was independent of the original eQTL effects (r2 < 0.1) 0.2% of the time (1.2% in iPSC), indicating that in the vast majority of cases, the same eQTL effect was tested for replication and the choice of variant is unlikely to have a marked effect on the results. Statistics of the replication of the iPSC eQTLs, including numbers of selected proxy variants, are provided in Supplementary Table 5b. If a gene was not tested for eQTLs in the query tissue (for e.g. due to gene not expressed), replication information was defined as missing. A replication profile was derived for each eGene in the discovery tissue, indicating whether the lead eQTL effect replicated (yes | no) or could not be tested (‘NA’). We then extracted eGenes, for which the lead eQTL effect did not show evidence of replication in any other tissue (P > 2.2x10-04) or could not be tested (hereon referred to as ’tissue-specific eQTLs’). Of note, in this analysis, for an eQTL effect to replicate, it has to affect the expression of the same gene in both tissues. It the same variant is an eQTL for two different genes in the two tissues, it is not considered replicating. We also investigated the impact of the specific replication threshold, considering a threshold of P<0.01 and P<0.05 (Extended Data Fig. 8c). This analysis showed that the ability to replicate an eQTL signal in a second tissue is primarily determined by the sample size of the replication tissue and not the specific choice of threshold. This dependency needs to be taken into account when interpreting tissue-specific eQTL effects.
As an alternative strategy for comparing eQTLs among different tissues, we also calculated the π1 statistic (π1 = 1 - π0; 75) for all pairs of tissues (Extended Data Fig. 8b) using the qvalue package in R. The π1 statistic provides a global measure of similarity between a pair of tissues by estimating the proportion of eQTL signal discovered in one tissue that shows evidence of replication in a second tissue. For this analysis, for each discovery tissue, we queried all significant variants per eGene in the full cis output of all other tissues and estimated q0 with the ‘bootstrap’ method of the qvalue package.
Functional annotation of eQTLs
Matched sets of variants
For all functional enrichment analyses, 100 matched sets of variants (hereon ‘control variants’) were used as the null. These sets were generated with SNPsnap 76 using unique lead eQTL variants from different datasets as the input. Variants were matched for minor allele frequency, number of SNPs in LD (‘LD buddies’; r2 > 0.5), distance to the nearest gene, and gene density, allowing for maximum deviation of +/- 50% for each criterion. HLA SNPs (defined as falling between positions 25,000,000 and 35,000,000 on chromosome 6) were excluded from the analysis and all matched sets were non-overlapping with the input variants. 1000 Genomes Phase 3 European population was used as the genotype reference panel. Both target and control sets of variants were further expanded with their high-LD proxies (r2 > 0.8) derived from the European UK10k reference panel. These expanded sets form the basis for all subsequent enrichment analyses.
Overlap with chromatin state annotations
Functional enrichment of eQTLs was assessed using chromatin state data from the Roadmap Epigenomics Project 27. The data comprised of 25 chromatin states derived from reference epigenomes from 127 cell types. We overlapped target eQTL variants separately with each chromatin state and cell type. We also overlapped 100 sets of control variants with the same annotations and derived an empirical P-value for each enrichment. This was defined as the number of control variant sets (N) that showed a higher overlap with the target annotation than the eQTL lead variants (P = N/100). The empirical P-values were further adjusted for the number of tests (25 states x 127 cell types) within each eQTL set using the Q-value 75 package in R. Annotations with a Q-value < 0.05 were considered significantly enriched. We tested two sets of eQTLs for enrichment: iPSC-specific eQTLs (N = 2,131) and non-specific eQTLs (N = 4,500).
For visualization purposes we aggregated the 127 cell types into five clusters, using k-means clustering. The number of clusters was chosen based on the number of different sample types annotated by Roadmap (primary cell, primary culture, primary tissue, cell line, ESC derived). A heatmap of the difference in fold enrichment between iPSC-specific and non-specific eQTLs (DIFF = FEspecific - FEnonspecific) was generated with the pheatmap package in R (Fig. 4d) in order to assess how well our definition of iPSC-specific enriches for functional elements active in stem cells.
Overlap with transcription factor binding sites
Functional enrichment of eQTLs was additionally assessed using ChIP-seq based transcription factor (TF) binding sites (TFBS) from the ENCODE Project 49. Specifically, we used a set of proximal and distal TFBSs, where proximal is defined as a 2,000 bp window centered on Gencode v19 annotated transcription start sites (TSS). These sets were constructed by overlaying TF ChIP-seq peaks from all available cell types with DNase peaks. We limited our analysis to those peaks that overlapped with H1-ESC derived TF peaks. As with chromatin state annotations, we overlapped target and control eQTL variants with binding sites for each individual factor (all sites where the given factor is bound or co-bound). An empirical P-value for the enrichment was derived based on the control sets and adjusted for multiple testing using the Benjamini-Hochberg method. Factors with an adjusted P < 0.05 were considered enriched. Plotted in Fig. 4 are factors with at least 10 observed overlaps.
Overlap with the GWAS catalogue
The NHGRI-EBI GWAS catalogue was downloaded on 2016-04-18 (release 2016-04-10). Entries with missing positional or P-value information were removed and the positions of the remaining entries were converted to hg19 with the UCSC liftOver function in the “rtracklayer” R/Bioconductor package 77 and the ‘hg38ToHg19’ chain file. This resulted in 24,861 catalogue entries, corresponding to 18,446 unique variants of which 6,681 were significantly associated to a trait (P < 5x10-08). However, many studies included in the complete catalogue are not reliable, so we parsed the sample size information in the catalogue and excluded studies that did not report effect sizes (odds ratio or regression coefficient), had a sample size below 1,000, or assayed fewer than 100,000 variants. Then, following the approach taken in 78, we further filtered the set of remaining associations to retain only traits that had at least six significantly associated variants, and kept all associations with P < 1x10-06 for these traits. This approach yielded a filtered set of 9,562 associations for 6,059 variants with 358 diseases and traits. Each variant position was then parsed for trait overlap (all traits associated to the given variant). If a variant - trait association was reported multiple times (e.g. by different studies), the most significant association was kept.
We first tested whether eQTLs in iPSCs and somatic tissues (lead variants and their high-LD proxies, control variants as before) showed global enrichment in this final set of disease-associated variants compared with matched sets of variants (Extended Data Fig. 9b). Fold enrichments were derived from a comparison to 100 sets of matched controls (mean of control overlaps per tissue). For iPSCs, we additionally tested enrichment for individual traits, deriving an empirical P-value. Traits with an adjusted P < 0.05 (Benjamini-Hochberg) and minimum five observed overlaps were considered enriched (Supplementary Table 6c).
Lastly, we parsed each disease variant position for overlap with iPSC eQTLs, again considering lead variants and their high-LD proxies for each tissue as follows. We report all disease variants that were tagged by iPSC eQTLs (lead or proxy) and the eGene(s) regulated by the eQTL lead/proxy (Supplementary Table 6a). We additionally report a subset of these variants that are exact matches with iPSC lead eQTL variants and highlight the number of high-LD proxies (r2 > 0.8) each variant has (Supplementary Table 6b).
Colocalisation analysis
We used coloc v2.3-1 29 to test for colocalisation between molecular QTLs and GWAS hits from Alzheimer’s disease 79, celiac disease 80, inflammatory bowel disease, ulcerative colitis, and Crohn’s disease 81, multiple sclerosis 82, 83, narcolepsy 83, primary biliary cirrhosis 84, psoriasis 85, rheumatoid arthritis 86, schizophrenia 87, systemic lupus erythematosus 88, type 1 diabetes 89 and type 2 diabetes 90. We ran coloc on a 250 Kb region centered on the eQTL gene for all eQTL variants that were less than 100 Kb away from at least one GWAS variant with nominal P < 1x 10-5. We then applied a set of filtering steps to identify a stringent set of eQTLs that colocalised with GWAS hits. We removed all cases with <50 SNPs in the cis region and selected only loci where PP3 + PP4> 0.5 and PP4/(PP3+PP4) > 0.5 to only keep loci where coloc strongly preferred a model of QTL for both traits, of a single shared causal variant driving both association signals over a model of two distinct causal variants. We excluded all colocalisation results from the MHC region (GRCh37: 6:28477897-33448354) because these could exhibit an elevated false positives rate due to the complicated LD patterns in this region. We kept only results where the minimal GWAS P-value was < 5x10-8.
Overlap with the COSMIC genes
To assess the overlap of iPSC eQTLs with known cancer genes in the context of somatic and cancerous tissues, we calculated the cumulative number of cancer genes (COSMIC cancer census 27/04/2016; Ngenes = 571 20) regulated by eQTLs in iPSCs, somatic tissues (GTEx V6p), and three different cancers (ER positive and negative breast cancer, colorectal cancer) 36,37 (Extended Data Fig. 9a).
Splicing of TERT
Alternative splicing of the TERT gene was analysed using Leafcutter 91, which focuses on introns and quantifies both known and novel alternative splicing events by quantifying reads mapping to exon-exon junctions. Annotations were derived from Gencode v19. Introns supported by fewer than 30 reads (-m 30; default) across all samples were removed. We obtained quantifications for eight intron clusters within TERT. After removing individual introns with a mean intron usage of zero, we had a total of 22 introns to test. We used the intron excision proportions to assess genotype-dependent effect of rs10069690 on TERT splicing (linear model between genotype and excision proportion, Bonferroni correction of P-values for the total number of introns tested). One intron showed evidence of a splicing-QTL effect (P < 0.05, Bonferroni adjusted; Extended Data Fig. 10).
Data availability
The assay data used in this publication are listed in the Biostudies archive (https://www.ebi.ac.uk/biostudies/studies) with accession identifier S-BSMS5. All data can be accessed via the HipSci data portal (http://www.hipsci.org), which references to EMBL-EBI archives that are used to store the HipSci data. Managed access data from all assays are accessible via EGA under the study EGAS00001001465. Open access genotyping array data and RNA-seq data are available from ENA under the studies PRJEB11752 and PRJEB7388. Open access gene expression array data are available in the ArrayExpress database under accession number E-MTAB-4057. The mass spectrometry proteomics data have been deposited to ProteomeXchange via the PRIDE repository with the dataset identifiers PXD003903 and PXD005506. Data types from specialized assays for which none of the existing archives are appropriate are available from the HipSci FTP site (ftp://ftp.hipsci.ebi.ac.uk/vol1/ftp). Intermediate result files for this study, such as processed gene expression levels, can be found at: ftp://ftp.hipsci.ebi.ac.uk/vol1/ftp/data. For full details see Supplementary Information.
Code availability
Scripts that were used to process the raw data and for implementing the statistical analyses presented are available from https://github.com/hipsci/Nature2017.
Extended Data
Extended Data Figure 1. Overview of the Cellomics assay.
(a) Example plate layout for the cellular differentiation assay. Images are shown for the pluripotency markers (Oct4, Sox2, and Nanog) as they are measured in the Cellomics imaging device. Each line is measured in two rows of the same plate as technical replicates. The secondary antibody used for each marker is shown in parenthesis. Each plate also has measurements for staining with the secondary antibody only, which serves as a means to assess background fluorescence. The red channel shows the signal from the DAPI staining, the green channel the marker signal. As expected, there is only little signal from the green channel in the wells stained only for the secondary antibody. Image acquisition stops as soon as 10,000 cells have been detected. (b) Detailed variance components of the Cellomics markers (Methods). Substantial proportions of the marker variance could be attributed to batch factors, including staining, technician effects and antibody lots. These effects mean that the fraction of cells expressing particular markers need to be interpreted with caution (Fig. 1c,d). (c) Pairwise correlation between quantitative expression scores derived from immunostaining for pluripotency and differentiation and the PluriTest score.
Extended Data Figure 2. Pluritest scores in the two culture conditions.
(a-c) Comparison of PluriTest novelty score versus pluripotency score for the 711 lines generated. Lines grown on feeder-free conditions (E8 media) scored systematically lower than Feeder-dependent lines (P = 1.62x10-43 t-test, for pluripotency score). We note that, while we cannot rule out that Feeder-free lines are less pluripotent, Feeder-free conditions are not well represented in the PluriTest training dataset, which may explain this result (of the 204 ESC/IPSC lines in the pluriTest paper that have media metadata available, none were on E8 and only 37 were on a variety of other feeder free formulations such as MTSER). (d) Despite lower pluripotency scores, lines grown on Feeder-free conditions have higher fractions of cells expressing canonical protein markers of pluripotency.
Extended Data Figure 3. Extended CNA analysis.
Relationship between the number of CNAs using three CNA minimum length thresholds for calling CNAs: 200 Kb, 500 Kb and 1,000 Kb and other experimental factors. Values on the x-axis have been ‘jittered’ (i.e. small random ‘noise’ has been added to the true values) to enhance the visualisation. Data points underlying the boxplots are shown as semi-transparent blue dots. (a) Number of CNAs per line versus passage number. P-values shown are from a generalized linear mixed model (Poisson regression) with donor random effect. (b) Boxplot of the number of autosomal CNAs per line versus growth media. P-values are for a Poisson regression on culture condition. (c-d) Number of autosomal CNAs per line versus PluriTest pluripotency and novelty scores. P-values are for a linear mixed model on the number of autosomal CNAs per line with a donor random effect. (e-f) Number of CNA counts per donor versus gender and donor age. CNA counts refer to the total number of unique CNAs across all lines derived from the same donor. CNAs that are shared between lines of the same donor (overlap by at least one base) are counted only once. P-values shown are for a Poisson regression on either gender or age.
Extended Data Figure 4. Location and consequence of the recurrent CNA on chr20 (related to Fig. 2).
Top panel shows genomic location versus number of lines with CN three (grey) and with a CNA (black). Bottom panel shows the NAV gene score from ref22 and log2 gene expression fold change between the iPSC lines with CN two and three (color scale), in the region highlighted in red in the top panel. Highlighted genes are up-regulated when copy number increases, known onco/tumour-suppressor genes and/or genes with NAV score in the top 2%.
Extended Data Figure 5. Functional assessment of CNAs using growth assays.
Cell growth rate (a), proliferation (b) and apoptosis (c) in cell lines with copy number two (“wild type”, blue dots) or copy number three (“mutant”, red dots) in a recurrently duplicated region in iPSCs on chromosome 1, 17 or 20. Plot titles show the donor name and the genomic coordinates of the CNA. (a) Shown are cell counts taken on successive days in culture, for pairs of lines (one mutant, one wild type) grown on the same 24-well plates. Star symbols denote significance levels for statistical interactions between day and copy number in a linear mixed model, using fixed effects to fit day and copy number, and random effects to account for culture plate effects. “EIF4A3” denotes whether a copy number variant overlaps one of the suspected candidate genes on chromosome 17. * - P < 0.05; ** - P < 0.01; *** - P < 0.001. (b) Protein expression level measured using Tandem Mass Tag (TMT)-based quantitation on the Q-exactive plus (labelled “QE Plus”) orbitrap and a fusion (labelled “Fusion”) orbitrap MS platforms. (c) Estimated fraction of fluorescing nuclei following EdU assay in mutant and wild type lines, following exposure to mitomycin ("Treated"), or in a control sample ("Untreated"). (d) Estimated fraction of fluorescing nuclei following Terminal deoxynucleotidyl transferase dUTP nick end labelling assay (TUNEL) in mutant and wild type lines, following exposure to mitomycin ("Treated"), or in a control sample ("Untreated"). Solid trend lines are least squares regression fits. P-values in b and c denote the significance of statistical interactions between copy number and mitomycin treatment condition (“Treated” or “Untreated”).
Extended Data Figure 6. Effect of passage on Tier 1 and Tier 2 data and overview of iPSC cis eQTLs mapped with ‘Tier 1’ gene expression array data.
(a,b) Passage number versus PluriTest pluripotency and novelty scores shows no significant association between passage number and pluripotency. Trend lines shown are fit using linear regression of PluriTest scores on passage number (score P = 0.66, novelty P = 0.21). Association was also not deemed significant when including gender and media as fixed effects and batch variables and donor as random effects (score P = 0.3, novelty P = 0.14). (c) Passage number versus log10 RNA-seq expression of pluripotency factors Nanog and Pou5f1 (Oct4) shows no significant association between passage number and pluripotency. Trend lines are fit using linear regression of log10 expression on passage number (Nanog P = 0.5, Pou5f1 P = 0.15). Association was also not deemed significant when considering the two genes together and when including gender and media as fixed effects and batch variables and donor as random effects (passage P = 0.28, passage-gene interaction P = 0.96). (d,e) Variance component analysis for Tier 2 assays, showing that for the majority of genes gender and passage explained little of the total variance. (f,g) Comparison of eQTL effect sizes (squared beta) at lead variants of the main gexarray eQTL map (derived using mean expression levels per donor). Plotted are the effect sizes for all tested genes (FDR < 5% eGenes indicated in blue) derived from (f) iPSC line replicate sets 1 and 2, one per donor, drawn randomly (rho = 0.47 genome-wide, rho = 0.80, FDR < 5% eGenes, P < 2.2e-16; Spearman rank correlation) and (g) replicate set 1 and the main map (rho = 0.57 genome-wide, rho = 0.88, FDR < 5% eGenes, P < 2.2e-16). Panel (g) shows that the effect sizes obtained using the mean expression values per donor are higher than when using individual lines. (h) Pairwise correlation between gene expression levels in iPSCs measured with RNA-seq and gexarray. Plotted are the Spearman rank correlation coefficients of either gene (pink) or gexarray probe (blue) region based read counts, demonstrating higher correlation of probe-based counts.
Extended Data Figure 7. Properties of iPSC cis eQTLs in comparison to somatic eQTLs.
Plotted is the power to detect eQTLs, comparing 44 somatic tissues from GTEx 24 (V6p) and the HipSci RNA-seq -based eQTL map (purple triangle), considering either the absolute (a) or relative (b) number of eQTLs identified (eGenes, FDR < 5%). The major determinant of eQTL detection power is sample size. (c) Cumulative fraction of RNA-seq reads relative to the number of protein coding genes expressed. Plotted is the mean read count derived from 20 iPSC lines (10 donors, two lines each), five fibroblast lines, and two embryonic stem cell (ESC) lines. In iPSCs, half of the reads are explained by the expression of 1,071 genes, while 75% and 90% of the reads are explained by the expression of 3,159 and 5,814 genes, respectively (total protein coding genes with non-zero counts N = 17,332). (d) Distribution of iPSC eQTLs around the annotated gene start position. Plotted is the -log10 (eQTL P-value) against the distance (bp) from the gene start for lead eQTL variants genome-wide, highlighting significant eQTLs (FDR < 5%) in orange. (e) Comparison of the magnitude of eQTL effect size (absolute beta; left panel) and minor allele frequency (MAF; right panel) between iPSC-specific (N = 2,131; labelled as ‘S’) and non-specific eQTLs (N = 4,500; labelled as ‘NS’), demonstrating that overall, iPSC-specific eQTLs have smaller effects on the transcriptome than eQTLs shared among multiple tissues (P = 9.97x10-161; Wilcox test) and have a lower minor allele frequency (P = 1.08x10-35, Wilcox test).
Extended Data Figure 8. Comparison of eQTL mapping pipelines between HipSci and GTEx (V6p).
(a) Proportion of tissue-specific eQTLs as a function of the discovery sample size. For iPSC, shown are the two sets of tissue-specific eQTLs obtained with the two different mapping pipelines (Methods), namely the standard HipSci pipeline (‘iPSC’; purple triangle) and the alternative ‘GTEx-like’ pipeline (‘iPSC2’; purple triangle). Points other than iPSC are from the GTEx Consortium (44 somatic tissues and cell lines) 24. (b) Heatmap of pairwise π1 values (π1 = 1 - π0) between iPSCs and GTEx tissues, with rows representing the discovery tissue and columns the replication tissue. Clustering of tissues is based on euclidean distance (R hclust, method=average). (c) Effect of eQTL replication threshold on the definition of tissue-specific effects. Shown is the replication profile of iPSC eQTLs across GTEx tissues relative to discovery sample size in each replication tissue. Plotted is the proportion of iPSC lead eQTLs that replicate in each tissue, with replication defined using two different replication thresholds (TH1: nominal eQTL P < 0.01/N_tissues; TH5: P < 0.05/N_tissues; plotted as dots and triangles, respectively). (d) Enrichment of alternative iPSC eQTLs (‘GTEx-like”) at promoter proximal and distal (defined as less than or greater than 2 Kb from the transcription start site) transcription factor binding sites (TFBS) in H1-hES cells from the ENCODE Project 49. Fold enrichments per factor are shown for iPSC-specific and non-specific eQTLs (minimum 10 observed overlaps) (Methods). Pluripotency-associated factors are indicated with an asterisk. The profile of enrichments is comparable to that obtained with the standard HipSci pipeline (Fig. 4d).
Extended Data Figure 9. iPSC eQTLs and disease.
(a) Cumulative number of cancer genes (COSMIC cancer census 27/04/2016; Ngenes = 571 20) regulated by eQTLs in iPSCs, somatic tissues (GTEx V6p), and three different cancers (ER positive and negative breast cancer, colorectal cancer) 33,34. (b) Enrichment of iPSC and somatic eQTLs (lead variants and their high-LD proxies) at disease-associated variants in the NHGRI-EBI GWAS catalogue (2016-04-10). Plotted is the fold enrichment of eQTLs over 100 random sets of matched variants for each tissue relative to eQTL discovery sample size. The tissues showing the highest fold enrichment are liver and brain (cerebellar hemisphere; ‘BrainCH’’). (c) Somatic eQTL signal for PTPN2 (Protein Tyrosine Phosphatase, Non-Receptor Type 2) locus on chromosome 18. This locus contains a colocalising association signal for PTPN2 gene expression in iPSCs and five immunological disease phenotypes (Fig. 5a). (d) Somatic eQTL signal for TERT (Telomerase Reverse Transcriptase) locus on chromosome 5 (Fig. 5b). In both (c) and (d), the lead eQTL variant locations are indicated with red and orange vertical lines for iPSC and somatic tissues, respectively. The focal gene regions are indicated in solid grey and gene start positions of other protein-coding genes on the same strand with vertical grey lines.
Extended Data Figure 10. Tissue expression and alternative splicing results at the TERT locus.
(a,b) Normalised RNA-seq per-base coverage across the TERT locus stratified by rs10069690 genotype. Plotted in the full locus (a), while (b) shows a zoomed view of the region around the lead eQTL and cancer risk variant rs10069690, indicated with a dotted line on each plot. Grey regions indicate annotated exons from Ensembl v75. Coverage was computed from indexed BAM files using the coverageBed function from the bedtools (v2.25.0) 92. Raw coverage was divided by total library size in millions (total number of mapped reads) per sample to obtain normalised coverage, which was then averaged over samples with the same rs10069690 genotype to obtain mean normalised coverage for each genotype group. (c) Profile of TERT expression in iPSCs and across somatic tissues from GTEx. Shown are gene FPKM values obtained with RNA-SeQC (GTEx V6p). (d) Splicing-QTL of TERT. We quantified TERT intron retention rates using Leafcutter {Li, 2016 #443} and identified one alternative splicing event associated with rs10069690, the lead iPSC eQTL variant for TERT (Fig. 5b). Shown is TERT intron 4 retention ratio (PSI, percent spliced in) in iPSC lines of all individual donors stratified by their genotype at rs10069690. This variant affects the splicing of the intron where it is located, with the minor allele (T) increasing the fraction of TERT transcripts in which intron 4 is retained (P = 1.7x10-9, Bonferroni adjusted linear regression).
Supplementary Information
Acknowledgements
This work was funded with a strategic award from the Wellcome Trust and Medical Research Council (WT098503). We thank the staff in the Cellular Genetics and Phenotyping and Sequencing core facilities at the Wellcome Trust Sanger Institute. Work at the Wellcome Trust Sanger Institute was further supported by Wellcome Trust grant WT090851. HK is supported by a MRC eMedLab Medical Bioinformatics career development award from the Medical Research Council [MR/L016311/1]. FMW gratefully acknowledges financial support from the Department of Health via the NIHR Biomedical Research Centre award to Guy’s & St Thomas’ National Health Service Foundation Trust in partnership with King’s College London and King’s College Hospital NHS Foundation Trust. We gratefully acknowledge the participation of all NIHR Cambridge BioResource volunteers, and thank the NIHR Cambridge BioResource centre staff for their contribution. We thank the National Institute for Health Research and NHS Blood and Transplant. The NIHR/Wellcome Trust Cambridge Clinical Research Facility supported the volunteer recruitment. We acknowledge Life Science Technologies Corporation as the provider of Cytotune. The authors thank Franz-Josef Müller (Zentrum für Integrative Psychiatrie, Kiel, Germany) for insights regarding the PluriTest method, and the GTEx consortium for making raw data and intermediate results available. The datasets used for parts of the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/gap through dbGaP accession number phs0004242.v6.p1.c1.
Footnotes
Author contributions
HK, AG, OS, DG: Wrote the paper with input from all authors.
HK, AG, DB, YM, IS, PD, DMcC, AA, MP, DD, AL, OS, DG: Contributed to the supplementary material
HK, AG, AL, FPC, PD, DMcC, DD: Analysed the data
SA, WO: Managed and supervised collection of research volunteer samples
FS, CA, AA, RN, SH, MP, CK: Generated iPSC lines, Tier 1 assay data, RNA-seq and methylation data
VA, DB: Generated and processed the proteomics data
AL, OC, RM, NM, DD: Generated and processed the high content cellular imaging data
SMcC, YM: Initial data quality control and bioinformatics processing/pipelines
AF, PH, IS, LC: Curated and managed data and project website
RH, AKK: Coordinated the project
DD, PB, WO, EB, LV, AIL, FW, RD, OS, DG Supervised and designed the research
Author information
Reprints and permissions information is available at www.nature.com/reprint.
+ Competing financial information statement.
Details of the data generated during the project, including archive accession identifiers for obtaining the data, are described in the Supplementary Information. The HipSci website (www.hipsci.org) also has full details of all publicly available data and instructions for researchers to apply for access to data in European Genome-phenome Archive (EGA). Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Sterneckert JL, Reinhardt P, Scholer HR. Investigating human disease using stem cell models. Nat Rev Genet. 2014;15:625–639. doi: 10.1038/nrg3764. [DOI] [PubMed] [Google Scholar]
- 2.Kim K, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim K, et al. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat Biotechnol. 2011;29:1117–1119. doi: 10.1038/nbt.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lister R, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nazor KL, et al. Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell Stem Cell. 2012;10:620–634. doi: 10.1016/j.stem.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rouhani F, et al. Genetic background drives transcriptional variation in human induced pluripotent stem cells. PLoS Genet. 2014;10:e1004432. doi: 10.1371/journal.pgen.1004432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burrows CK, et al. Genetic Variation, Not Cell Type of Origin, Underlies the Majority of Identifiable Regulatory Differences in iPSCs. PLoS Genet. 2016;12:e1005793. doi: 10.1371/journal.pgen.1005793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vallier L, et al. Signaling pathways controlling pluripotency and early cell fate decisions of human induced pluripotent stem cells. Stem Cells. 2009;27:2655–2666. doi: 10.1002/stem.199. [DOI] [PubMed] [Google Scholar]
- 9.Muller FJ, et al. A bioinformatic assay for pluripotency in human cells. Nat Methods. 2011;8:315–317. doi: 10.1038/nmeth.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danecek P, McCarthy SA, HipSci C, Durbin R. A Method for Checking Genomic Integrity in Cultured Cell Lines from SNP Genotyping Data. PLoS One. 2016;11:e0155014. doi: 10.1371/journal.pone.0155014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laurent LC, et al. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell. 2011;8:106–118. doi: 10.1016/j.stem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.International Stem Cell, I et al. Screening ethnically diverse human embryonic stem cells identifies a chromosome 20 minimal amplicon conferring growth advantage. Nat Biotechnol. 2011;29:1132–1144. doi: 10.1038/nbt.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abyzov A, et al. Somatic copy number mosaicism in human skin revealed by induced pluripotent stem cells. Nature. 2012;492:438–442. doi: 10.1038/nature11629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayshar Y, et al. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell. 2010;7:521–531. doi: 10.1016/j.stem.2010.07.017. [DOI] [PubMed] [Google Scholar]
- 15.Taapken SM, et al. Karotypic abnormalities in human induced pluripotent stem cells and embryonic stem cells. Nat Biotechnol. 2011;29:313–314. doi: 10.1038/nbt.1835. [DOI] [PubMed] [Google Scholar]
- 16.Hussein SM, et al. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- 17.Laurin M, Cote JF. Insights into the biological functions of Dock family guanine nucleotide exchange factors. Genes Dev. 2014;28:533–547. doi: 10.1101/gad.236349.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, et al. FATS is a transcriptional target of p53 and associated with antitumor activity. Molecular Cancer. 2010;9:244–244. doi: 10.1186/1476-4598-9-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lo JY, Chou YT, Lai FJ, Hsu LJ. Regulation of cell signaling and apoptosis by tumor suppressor WWOX. Exp Biol Med (Maywood) 2015;240:383–391. doi: 10.1177/1535370214566747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Futreal PA, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duckett CS, et al. A conserved family of cellular genes related to the baculovirus iap gene and encoding apoptosis inhibitors. The EMBO Journal. 1996;15:2685–2694. [PMC free article] [PubMed] [Google Scholar]
- 22.Chia NY, et al. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010;468:316–320. doi: 10.1038/nature09531. [DOI] [PubMed] [Google Scholar]
- 23.Belinky F, et al. PathCards: multi-source consolidation of human biological pathways. Database (Oxford) 2015;2015 doi: 10.1093/database/bav006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grundberg E, et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat Genet. 2012;44:1084–1089. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roadmap Epigenomics, C et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu H, et al. ESCAPE: database for integrating high-content published data collected from human and mouse embryonic stem cells. Database (Oxford) 2013;2013:bat045. doi: 10.1093/database/bat045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giambartolomei C, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dubois PC, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 2010;42:295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stranger BE, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeller T, et al. Genetics and beyond--the transcriptome of human monocytes and disease susceptibility. PLoS One. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purrington KS, et al. Genome-wide association study identifies 25 known breast cancer susceptibility loci as risk factors for triple-negative breast cancer. Carcinogenesis. 2014;35:1012–1019. doi: 10.1093/carcin/bgt404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia-Closas M, et al. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat Genet. 2013;45:392–398. doi: 10.1038/ng.2561. 398e391-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Z, et al. Imputation and subset-based association analysis across different cancer types identifies multiple independent risk loci in the TERT-CLPTM1L region on chromosome 5p15.33. Hum Mol Genet. 2014;23:6616–6633. doi: 10.1093/hmg/ddu363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Q, et al. Integrative eQTL-based analyses reveal the biology of breast cancer risk loci. Cell. 2013;152:633–641. doi: 10.1016/j.cell.2012.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ongen H, et al. Putative cis-regulatory drivers in colorectal cancer. Nature. 2014;512:87–90. doi: 10.1038/nature13602. [DOI] [PubMed] [Google Scholar]
- 38.Chen QR, Hu Y, Yan C, Buetow K, Meerzaman D. Systematic genetic analysis identifies Cis-eQTL target genes associated with glioblastoma patient survival. PLoS One. 2014;9:e105393. doi: 10.1371/journal.pone.0105393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bojesen SE, et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat Genet. 2013;45:371–384. doi: 10.1038/ng.2566. 384e371-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiba K, et al. Cancer-associated TERT promoter mutations abrogate telomerase silencing. Elife. 2015;4 doi: 10.7554/eLife.07918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kyttala A, et al. Genetic Variability Overrides the Impact of Parental Cell Type and Determines iPSC Differentiation Potential. Stem Cell Reports. 2016;6:200–212. doi: 10.1016/j.stemcr.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kajiwara M, et al. Donor-dependent variations in hepatic differentiation from human-induced pluripotent stem cells. Proc Natl Acad Sci U S A. 2012;109:12538–12543. doi: 10.1073/pnas.1209979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi J, et al. A comparison of genetically matched cell lines reveals the equivalence of human iPSCs and ESCs. Nat Biotechnol. 2015;33:1173–1181. doi: 10.1038/nbt.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gerrits A, et al. Expression quantitative trait loci are highly sensitive to cellular differentiation state. PLoS Genet. 2009;5:e1000692. doi: 10.1371/journal.pgen.1000692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spies N, et al. Constraint and divergence of global gene expression in the mammalian embryo. Elife. 2015;4:e05538. doi: 10.7554/eLife.05538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cannavo E, et al. Genetic variants regulating expression levels and isoform diversity during embryogenesis. Nature. 2017;541:402–406. doi: 10.1038/nature20802. [DOI] [PubMed] [Google Scholar]
- 47.Kim NW, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 48.Kelly LM, Gilliland DG. Genetics of myeloid leukemias. Annu Rev Genomics Hum Genet. 2002;3:179–198. doi: 10.1146/annurev.genom.3.032802.115046. [DOI] [PubMed] [Google Scholar]
- 49.Encode Project, C. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 51.Consortium, U. K. et al. The UK10K project identifies rare variants in health and disease. Nature. 2015;526:82–90. doi: 10.1038/nature14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Genomes Project, C et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2012;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 55.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(Suppl 1):S96–104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- 58.Vallier L, et al. Early cell fate decisions of human embryonic stem cells and mouse epiblast stem cells are controlled by the same signalling pathways. PLoS One. 2009;4:e6082. doi: 10.1371/journal.pone.0006082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ly T, et al. A proteomic chronology of gene expression through the cell cycle in human myeloid leukemia cells. Elife. 2014;3:e01630. doi: 10.7554/eLife.01630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bensaddek D, et al. Micro-proteomics with iterative data analysis: Proteome analysis in C. elegans at the single worm level. Proteomics. 2016;16:381–392. doi: 10.1002/pmic.201500264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 62.Aryee MJ, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.DeLuca DS, et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012;28:1530–1532. doi: 10.1093/bioinformatics/bts196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leha A, et al. A high-content platform to characterise human induced pluripotent stem cell lines. Methods. 2016;96:85–96. doi: 10.1016/j.ymeth.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zack TI, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stegle O, Parts L, Durbin R, Winn J. A Bayesian framework to account for complex non-genetic factors in gene expression levels greatly increases power in eQTL studies. PLoS Comput Biol. 2010;6:e1000770. doi: 10.1371/journal.pcbi.1000770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lippert C, Casale FP, Rakitsch B, Stegle O. LIMIX: genetic analysis of multiple traits. BioRxiv. 2014 003905. [Google Scholar]
- 74.Casale FP, Rakitsch B, Lippert C, Stegle O. Efficient set tests for the genetic analysis of correlated traits. Nat Methods. 2015;12:755–758. doi: 10.1038/nmeth.3439. [DOI] [PubMed] [Google Scholar]
- 75.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pers TH, Timshel P, Hirschhorn JN. SNPsnap: a Web-based tool for identification and annotation of matched SNPs. Bioinformatics. 2015;31:418–420. doi: 10.1093/bioinformatics/btu655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lawrence M, Gentleman R, Carey V. rtracklayer: an R package for interfacing with genome browsers. Bioinformatics. 2009;25:1841–1842. doi: 10.1093/bioinformatics/btp328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Farh KK, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lambert JC, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Trynka G, et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet. 2011;43:1193–1201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu JZ, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.International Multiple Sclerosis Genetics, C et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet. 2013;45:1353–1360. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Faraco J, et al. ImmunoChip study implicates antigen presentation to T cells in narcolepsy. PLoS Genet. 2013;9:e1003270. doi: 10.1371/journal.pgen.1003270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cordell HJ, et al. International genome-wide meta-analysis identifies new primary biliary cirrhosis risk loci and targetable pathogenic pathways. Nat Commun. 2015;6:8019. doi: 10.1038/ncomms9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tsoi LC, et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet. 2012;44:1341–1348. doi: 10.1038/ng.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Okada Y, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schizophrenia Working Group of the Psychiatric Genomics, C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bentham J, et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat Genet. 2015;47:1457–1464. doi: 10.1038/ng.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Onengut-Gumuscu S, et al. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat Genet. 2015;47:381–386. doi: 10.1038/ng.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Morris AP, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li YI, Knowles DA, Pritchard JK. LeafCutter: Annotation-free quantification of RNA splicing. bioRxiv. 2016 doi: 10.1038/s41588-017-0004-9. 044107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li YI, et al. RNA splicing is a primary link between genetic variation and disease. Science. 2016;352:600–604. doi: 10.1126/science.aad9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The assay data used in this publication are listed in the Biostudies archive (https://www.ebi.ac.uk/biostudies/studies) with accession identifier S-BSMS5. All data can be accessed via the HipSci data portal (http://www.hipsci.org), which references to EMBL-EBI archives that are used to store the HipSci data. Managed access data from all assays are accessible via EGA under the study EGAS00001001465. Open access genotyping array data and RNA-seq data are available from ENA under the studies PRJEB11752 and PRJEB7388. Open access gene expression array data are available in the ArrayExpress database under accession number E-MTAB-4057. The mass spectrometry proteomics data have been deposited to ProteomeXchange via the PRIDE repository with the dataset identifiers PXD003903 and PXD005506. Data types from specialized assays for which none of the existing archives are appropriate are available from the HipSci FTP site (ftp://ftp.hipsci.ebi.ac.uk/vol1/ftp). Intermediate result files for this study, such as processed gene expression levels, can be found at: ftp://ftp.hipsci.ebi.ac.uk/vol1/ftp/data. For full details see Supplementary Information.