Abstract
Many viruses deliver their genomes into the nucleoplasm for viral transcription and replication. Here, we describe a novel cell-free system to elucidate specific interactions between viruses and nuclear pore complexes (NPCs). Nuclei reconstituted in vitro from egg extracts of Xenopus laevis, an established biochemical system to decipher nuclear functions, were incubated with GFP-tagged capsids of herpes simplex virus, an alphaherpesvirus replicating in the nucleus. Capsid binding to NPCs was analyzed using fluorescence and field emission scanning electron microscopy. Tegument-free capsids or viral capsids exposing inner tegument proteins on their surface bound to nuclei, while capsids inactivated by a high-salt treatment or covered by inner and outer tegument showed less binding. There was little binding of the four different capsid types to nuclei lacking functional NPCs. This novel approach provides a powerful system to elucidate the molecular mechanisms that enable viral structures to engage with NPCs. Furthermore, this assay could be expanded to identify molecular cues triggering viral genome uncoating and nuclear import of viral genomes.
Keywords: herpesviruses, Herpes Simplex Virus, Xenopus nuclei, reconstitution, nuclear pore
Introduction
More than 30 proteins termed nucleoporins (Nups) assemble into NPCs that serve as specialized gateways in the nuclear envelope to allow active and passive bidirectional molecular traffic between the cytosol and the nucleoplasm (1–3). The structural organization of NPCs is similar in humans and amphibians; both possess a central channel with an inner diameter of about 50 nm at their center surrounded by an elaborate scaffold structure with 8-fold rotational symmetry (4, 5). Their nucleoplasmic face is capped by the nuclear basket, while filaments of up to 50 nm in length protrude freely from the cytosolic face. NPCs allow the passage of molecules up to about 5 nm in diameter by diffusion, while shuttling nuclear transport receptors, called importins and exportins, are required for active translocation of cargoes (6–8). A RanGTP/RanGDP gradient across the nuclear envelope and members of the importin β superfamily control the directionality of transport, but the translocation process itself does not require GTP hydrolysis (8–12).
Herpesviruses comprise a large group of DNA viruses that have been identified from humans to mollusks (13). Herpes simplex virus type 1 (HSV1) is a human alphaherpesvirus with a genome of 152 kb that is packaged into an icosahedral capsid of 125 nm in diameter (14). The major capsid protein VP5 forms 150 hexons on the capsid faces and edges and 11 pentons at vertices, while the 12th vertex is occupied by a dodecamer of pUL6 that operates as a portal (15). The capsids are enclosed by an eccentric tegument of about 25 distinct proteins that have been classified into a more ordered inner and a less structured outer layer (16–22). The outer tegument binds to cytosolic domains of viral glycoproteins embedded into the viral envelope, overall forming a virion of about 225 nm in diameter (16, 22). Three types of capsids can be readily purified from the nuclei of infected cells (c.f. Fig. 1, top left): C capsids, which are the heaviest on sedimentation gradients, contain a viral genome; B capsids contain the scaffold proteins but no DNA; and A capsids, which are empty, lack both scaffold proteins and DNA. The packaging of progeny genomes into nuclear, rather spherical pro-capsids results in the formation of C capsids that are then predominantly exported across the nuclear envelope, while the nuclear A and B capsids are considered dead-end products (14, 23). The cytosolic capsids recruit inner tegument proteins such as pUL36 and pUL37 and are transported to cytoplasmic membranes to acquire the outer tegument and their envelopes (22–26). The virions are transported within secretory vesicles to the plasma membrane for release by exocytosis (22–24).
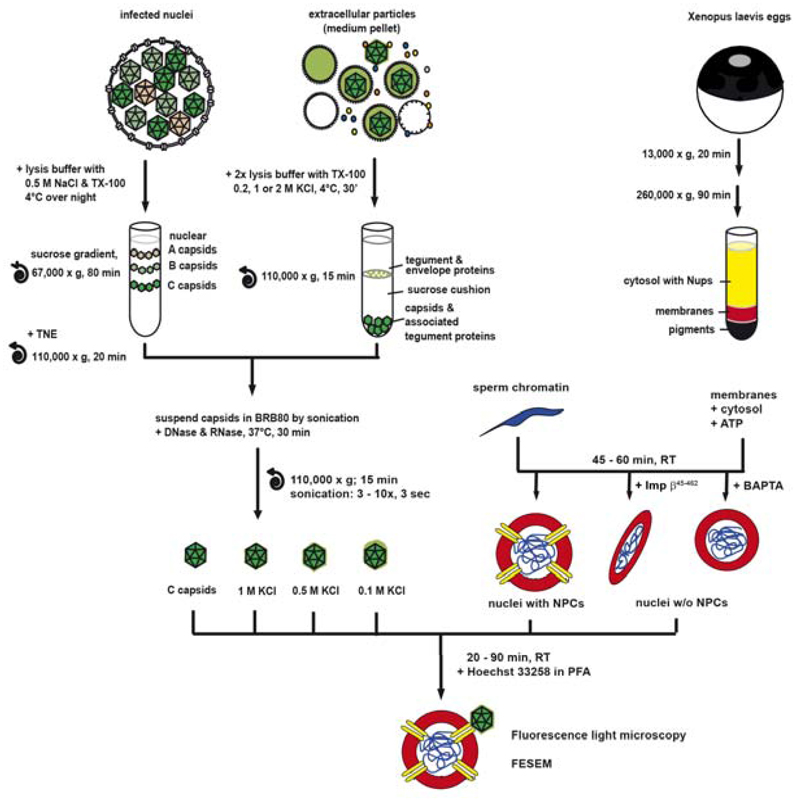
Figure 1. An in vitro assay to analyze the binding of viral capsids to NPCs.
Nuclear capsids (dark green) were isolated from the nuclei of HSV1 infected cells by gradient sedimentation (top left). Viral capsids were generated from mature extracellular particles released from HSV1 infected cells by lysis with 1% TX-100 in the presence of 1 M, 0.5 M or 0.1 M KCl (light and dark green), and purified through sucrose cushions (top middle). The four different capsid types were re-suspended in BRB80 buffer before tip sonication and DNase/RNase treatment. Nuclei harboring functional NPCs were generated in vitro from Xenopus egg extracts around a chromatin template. The formation of NPCs was inhibited in some reactions by the addition of Imp β45-462 or BAPTA to the assembly reaction to create nuclei enclosed by continuous membranes lacking functional NPCs (top right). After nuclear assembly and re-sedimentation the nuclear or viral 1 M, 0.5 M or 0.1 M capsids were incubated with different nuclei for 20 to 90 min at room temperature. Nuclear assembly and capsid binding were analyzed by fluorescence light microscopy or field emission scanning electron microscopy (FESEM). Modified from Figure 1 of (19).
After HSV1 cell entry by fusion with host membranes, inner tegument proteins such as pUL36 and pUL37 remain capsid-associated, while most or all outer tegument proteins detach (25–31). The capsids recruit the motor protein dynein and its cofactor dynactin for transport along microtubules towards the nucleus (32, 33). Electron microscopy images show parental capsids interacting with the cytosolic filaments emanating from NPCs, with one vertex facing the central channel (33–39). While most of these capsids appear empty, fortuitous cross sections seem to visualize the viral DNA genome as it is ejected from one vertex through the NPC (33, 39). Similarly, after a short adsorption to a solid surface, the DNA is also ejected in vitro as a single double helix from one vertex, most likely the portal (40). These observations are consistent with the notion that HSV1 utilizes energy from ATP hydrolysis to package genomes into capsids, while a pressure-driven mechanism pushes the genomes out of the capsids and through the permeability barrier of NPCs (41–43). The capsids are able to withstand this internal pressure by further stabilization of the capsid shell that seems to be initiated by DNA packaging after protein scaffold expulsion (44).
A biochemical cell-free system based on Xenopus laevis extracts has greatly aided studies of NPC assembly and nuclear transport (c.f. Fig. 1, right). Adding chromatin to fractionated egg extracts triggers a complex series of events including membrane vesicle recruitment, vesicle fusion, and assembly of NPCs into the newly forming nuclear envelopes (45–48). NPC assembly can be blocked by the addition of Imp β45-462, a truncated form of importin β, or by the Ca2+-chelator BAPTA [1,2-bis(o-aminophenoxy)-ethane-N,N,N',N'-tetraacetic acid], resulting in the formation of apparently pore-free nuclei in which the chromatin is fully enclosed by membranes lacking any signs of NPCs (49). These and other biochemical interventions make this in vitro system a powerful tool for the analysis of various nuclear functions (50).
Much remains to be learned about capsid docking at NPCs, viral genome uncoating, and viral genome translocation into the nucleoplasm. Since NPCs are essential for cell survival, it is a challenge to elucidate their function in intact cells. Our first in vitro study has shown that isolated HSV1 capsids derived from extracellular virions by TX-100 lysis in the presence of 0.5 M NaCl can bind to nuclei isolated from rat liver, and that importin β, the RanGTP/GDP cycle, and capsid-NPC interactions are required to trigger the release of the viral genomes (37). Similarly, capsids isolated from virions by TX-100 lysis in the presence of 0.5 M KCl interact in vitro with dynein, its cofactor dynactin, kinesin-1 and kinesin-2 from both, pig brain cytosolic extracts or from Xenopus egg extracts, and assemble functional capsid-motor complexes that can translocate along microtubules in vitro (summarized in Table 1; (19, 20, 51)).
Table 1. Protein Characterization of isolated HSV1 Capsids.
HSV1 capsids were isolated from extracellular virions using TX-100 in the presence of 0.1 M, 0.5 M or 1 M KCl or from infected nuclei (nuclear) and the association of microtubule-associated factors was analyzed by immunoblot and quantitative electron microscopy (19). Furthermore, the composition of structural HSV1 proteins was analyzed by immunoblot, quantitative mass spectrometry and quantitative immunoelectron microscopy (summarized in (51). The signal intensities of the immunoblots for each protein were evaluated qualitatively (+++ for the strongest band, ++, +, and – if there was no or only a negligible band) (19). For mass spectrometry data, the amount of the respective protein in gradient-purified extracellular virions was set to 100%, and the amount of the proteins present on the respective viral capsids were normalized accordingly (19). For the immunoelectron microscopy, the labelling intensity of the capsid type with the highest amount of surface labelling was set to 100%, and the labelling intensities on the other capsids were normalized accordingly. The information on the estimated copy number per virion (copies / virion) was compiled from several reports (17, 21, 68, 87–89).
| Association of Host Factors | Detection Method | viral 0.5 M KCl |
nuclear IB on B and C IEM on B and C |
viral 1 M KCl |
viral 0.1 M KCl |
|---|---|---|---|---|---|
| Dynein | IB IEM [%] |
+++ 100 |
- 0 |
+++ 98 |
- 33 |
| Dynactin | IB IEM [%] |
+++ 100 |
- 0 |
++ 66 |
- 3 |
| Kinesin-1 | IB IEM [%] |
+++ 100 |
- 0 |
- 19 |
- 4 |
| Kinesin-2 | IB IEM [%] |
+++ 100 |
- 35 |
++ 43 |
+ 0 |
|
HSV1 proteins (copies/virion) |
Detection Method |
viral 0.5 M KCl |
nuclear IB on B and C MS on B IEM on C |
viral 1M KCl |
viral 0.1 M KCl |
| Capsid Proteins | +++ | +++ | +++ | +++ | |
| VP5 (955) | MS [%] | 100 | 100 | 100 | 100 |
| VP19c (320) | MS [%] | 100 | 100 | 100 | 100 |
| VP23 (640) | MS [%] | 100 | 100 | 100 | 100 |
| VP24 (150) | MS [%] | 100 | 100 | 100 | 100 |
| VP26 (600) | MS [%] | 100 | 100 | 100 | 100 |
| pUL17 (60) | IB MS [%] |
+++ 100 |
+++ 100 |
+++ 100 |
+++ 100 |
| pUL25 (60) | IB MS [%] IEM [%] |
+++ +++ ++ |
+++ +++ ++ |
+++ +++ +++ |
+++ +++ ++ |
| Inner Tegument Proteins | +++ | - | +++/- | +++ | |
| pUS3 (?) | IB IEM [%] |
+++ 90 |
+ 19 |
+++ 75 |
+++ 100 |
| pUL36 (150) | IB middle | ++ | -/+ | ++ | +++ |
| MS [%] | 28 | - | 18 | 20 | |
| MS – Nterm [%] | 189 | - | 183 | 161 | |
| IEM - middle Ab [%] | 100 | 1 | 81 | 76 | |
| IEM - Cterm Ab [%] | 100 | 36 | 2 | 58 | |
| pUL37 (150) | MS [%] IEM (GFP) [%] |
49 94 |
- 1 |
21 32 |
48 100 |
| ICP0 (?) | IB | +++ | - | ++ | +++ |
| pUL14 (?) | IB | +++ | + | +++ | +++ |
| pUL16 (?) | MS [%] | 45 | - | 21 | 42 |
| pUL21 (?) | MS [%] | 55 | - | 19 | 50 |
| Outer Tegument Proteins | ++ | - | + | +++ | |
| pUL41 - vhs (?) | IB | ++ | - | + | +++ |
| pUL11 (?) | IB | + | - | + | +++ |
| ICP4 (?) | IB | ++ | - | + | +++ |
| ICP34.5 (?) | IB | ++ | - | ++ | +++ |
| VP13/14 (1300-1800) | MS [%] IEM [%] |
65 71 |
- -/+ |
12 56 |
124 100 |
| VP16 (700-2000) | MS [%] IEM [%] |
41 89 |
- -/+ |
19 65 |
72 100 |
| VP22 (700-1500) | MS [%] IEM [%] |
55 72 |
- -/+ |
23 24 |
143 100 |
These studies show that key virus-host interactions are conserved from amphibians to mammals, and that capsids derived from extracellular virions by detergent lysis at 0.5 M NaCl or 0.5 M KCl maintain their affinity for specific host factors. In contrast, capsids isolated from the nuclei of infected cells, or capsids prepared by detergent lysis of extracellular virions in the presence of 0.1 M KCl do not bind to microtubule motors, while viral capsids isolated in the presence of 1 M KCl maintain their ability to interact with dynein, but no longer bind to kinesins (summarized in Table 1; c.f. Fig. 1 left; (19, 20)). Our subsequent immunoblot analysis, quantitative mass spectrometry and quantitative immunoelectron microscopy studies have shown that viral capsids treated with 0.5 M KCl expose the inner tegument proteins pUL36, pUL37 and pUS3 and only low amounts of outer tegument proteins such as VP13/14, VP16, VP22, while capsids treated with 0.1 M KCl harbor in addition to inner tegument high amounts of outer tegument (summarized in Table 1; (19, 20)).
In this report, we describe a novel cell-free assay that measures the binding of HSV1 capsids to reconstituted Xenopus nuclei (c.f. Fig. 1, bottom). Our results show that nuclear capsids that display only outer capsid proteins on their surfaces and 0.5 M KCl viral capsids exposing outer capsid proteins in addition to inner tegument proteins, but not the other viral capsid types bind specifically to nuclei harboring functional NPCs. This new biochemical assay has broad potential for the study of interactions between viral structures with distinct components of the NPC as well as the nuclear import of viral proteins and genomes for transcription and replication.
Results
HSV1 capsids with defined surface features
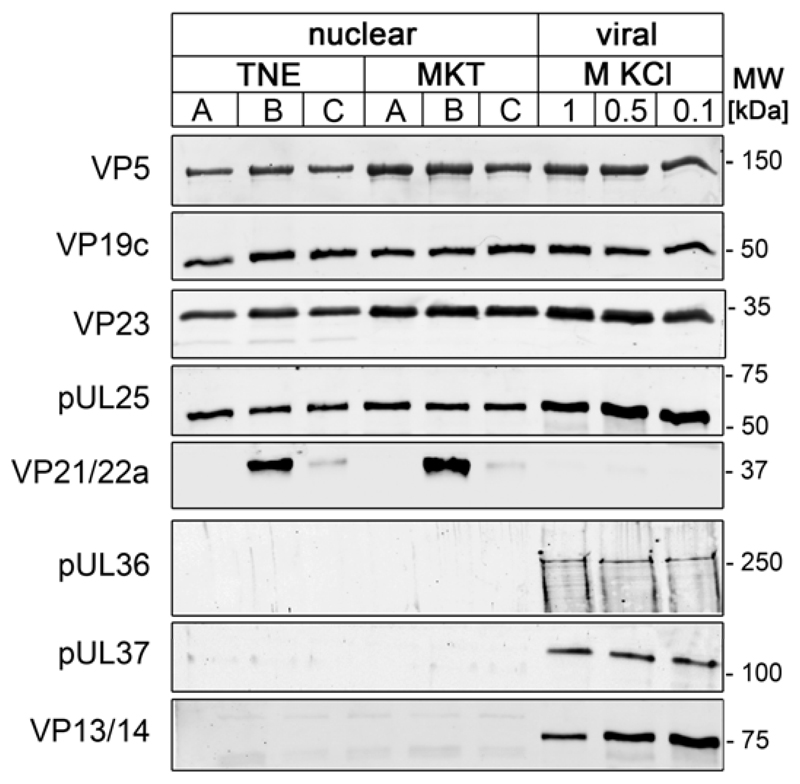
To identify the role of HSV1 structures in binding to NPCs, we prepared capsids characterized by different surface features. To isolate nuclear capsids (c.f. Fig. 1, left), we treated infected cells prior to lysis with two different buffers: with TNE containing 0.5 M NaCl as reported by Newcomb and Brown (52, 53), or with MKT containing a physiological concentration of 0.1 M KCl before cell lysis as described in our previous studies (19, 20, 51). Viral capsids were prepared from extracellular virions using TX-100 in the presence of 1 M, 0.5 M or 0.1 M KCl as described in our previous studies (19, 20); c.f. Fig. 1, middle). Immunoblot analysis did not reveal any differences in the protein composition of the different nuclear capsid preparations irrespective of the buffer used prior to cell lysis (Fig. 2). Nuclear capsids contained the capsid proteins VP5, VP19c and VP23 in similar amounts to viral capsids, although there were some minor variations between different preparations. B capsids also contained the scaffold proteins VP21 and VP22a. The amount of pUL25 was similar on A, B and C capsids, but appeared higher on viral capsids than on nuclear capsids. When compared to such nuclear capsids, the three types of viral capsids also contained similar amounts of the inner tegument proteins pUL36 and pUL37. By contrast, the outer tegument protein VP13/14 was most prominent on viral 0.1 M KCl capsids, but not present on nuclear capsids, consistent with our previous analysis (summarized in Table 1; (19, 20, 51)).
Figure 2. Protein composition of different HSV1 capsids types.
Nuclear capsids were isolated from BHK cells infected with HSV1-GFPVP26 using TNE- or MKT-buffer for cell suspension, and their protein composition was compared to that of viral capsids treated with TX-100 and 1.0 M, 0.5 or 0.1 M KCl by immunoblotting with antibodies raised against the capsid proteins VP5 (mAb H1.4), pUL25 (mAb #166), VP19c (pAb NC-2), VP21/VP22a (pAb NC3/4), or VP23 (pAb5) or the tegument proteins pUL36 (pAb #147), pUL37 (pAb anti-pUL37), or VP13/14 (pAb R220).
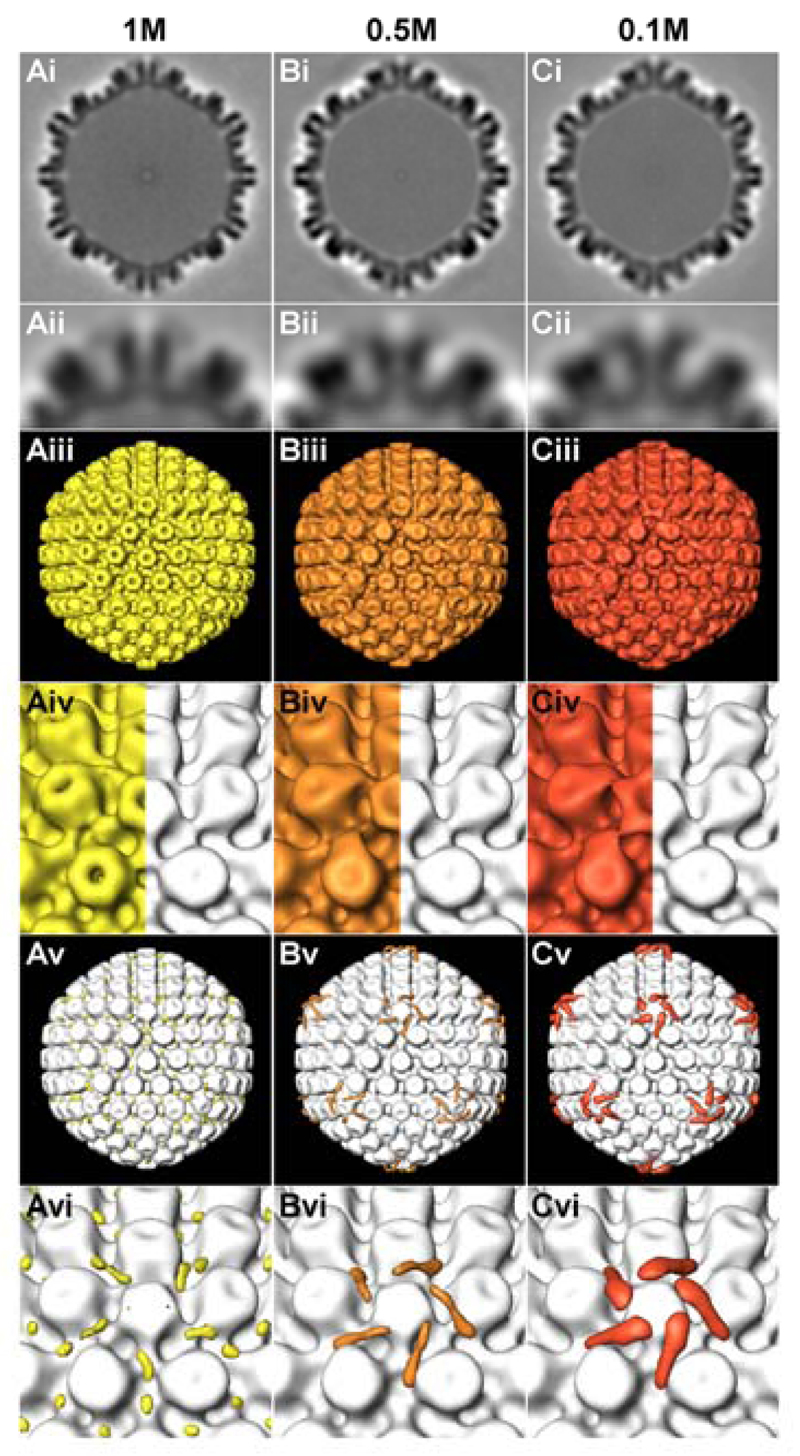
We next analyzed the capsid surfaces using electron cryo tomography and sub-volume averaging, and compared the degree of tegumentation of viral capsids to that of nuclear C capsids (Fig. 3). Since viral capsids had a tendency to aggregate, we selected single capsids present in holes of the carbon support film for analysis. In total, we averaged 183, 246 or 198 capsids isolated in the presence of 1 M, 0.5 M or 0.1 M KCl, respectively. The capsid averages were filtered to a resolution of 5.6 nm, and the differences in densities were calculated by subtracting the gray values of nuclear capsids from those of KCl-treated capsids. We detected little differences between viral capsids treated with 1 M KCl and nuclear capsids (yellow in Fig. 3Av, 3Avi). By contrast, there was a clear extra density on viral capsids prepared with 0.5 M KCl (orange in Fig. 3Bv, 3Bvi) and even more pronounced on 0.1 M KCl capsids (red in Fig. 3Cv, 3Cvi). These densities were located at the vertices protruding from the top of the pentons and from one side of the neighboring hexons, but not on the other hexons. Thus, viral and nuclear HSV1 capsids vary substantially in their protein composition and in their surface structure, and may therefore differ in their interactions with shuttling nuclear transport receptors and NPCs.
Figure 3. Electron cryo tomography reveals surface features of different HSV1 capsids types.
Results from sub-volume averaging are presented for capsids isolated from intact virions (HSV1 strain F) and extracted with TX-100 and 1 M (A), 0.5 M (B) or 0.1 M (C) KCl. Row (i): cross sections of the averages from 183 (1 M), 246 (0.5 M) or 217 (0.1 M) capsid sub-volumes. Row (ii): close-up view of the top vertex in row (i). Row (iii): iso-surface representation of the averages. Row (iv): close-up view of a salt treated capsid vertex (left), compared to nuclear C-capsids (66). Row (v): difference map between the respective salt treated capsid group and the nuclear C-capsids average, superimposed onto the nuclear C-capsids average. Row (vi): close-up view of a vertex from row (v). The capsids have a diameter of 125 nm.
Nuclei harboring NPCs reconstituted from Xenopus laevis egg extracts
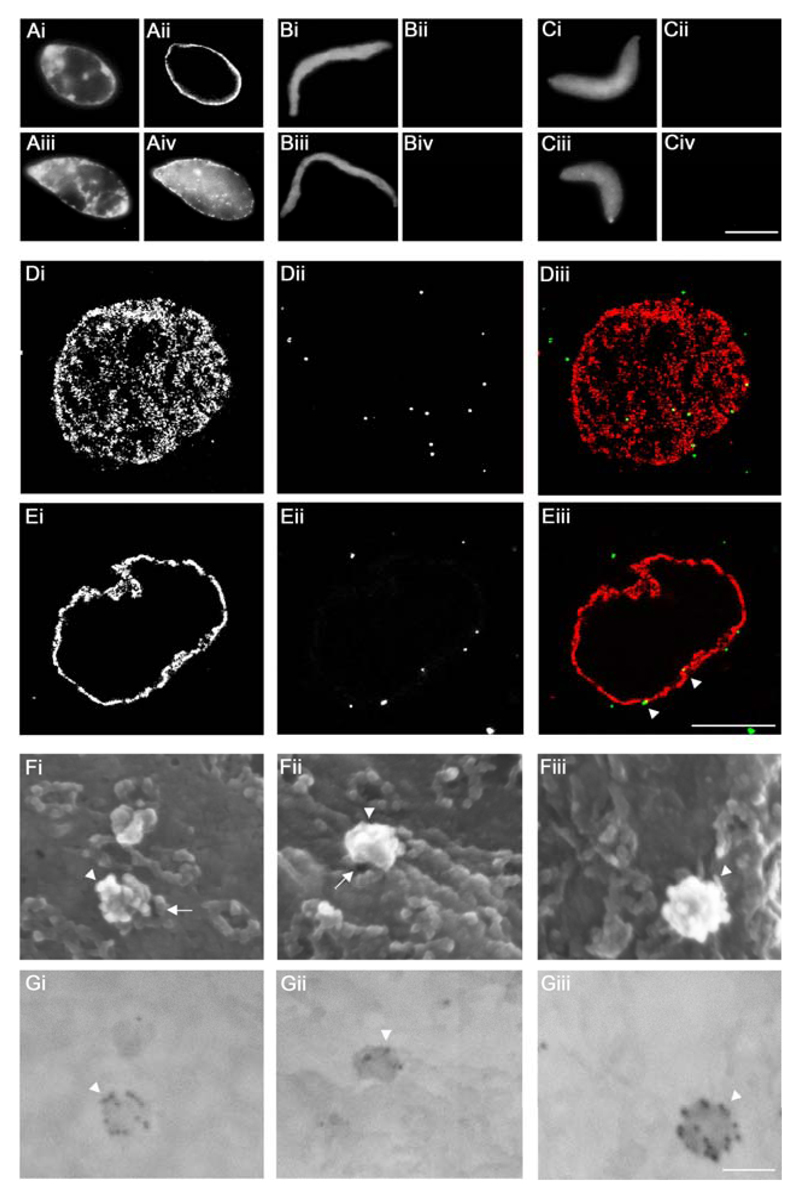
To develop a novel method to study capsid interactions with NPCs, we assembled nuclei in vitro from Xenopus laevis egg extracts. De-membranated sperm chromatin was incubated with membrane vesicles as a source for nuclear membranes and a cytosolic fraction as a source for Nups (Fig. 1, top left; (46, 54)). Nuclear assembly was validated for each experiment by labeling with the monoclonal antibody mAb414 recognizing FG-repeat Nups in mature NPCs (55). A rim-like signal around the chromatin indicated the formation of nuclei surrounded by nuclear envelopes harboring NPCs (Fig. 4Aii). No NPC staining was observed when either Imp β45-462 (Fig. 4Bii) or BAPTA (Fig. 4Cii) had been added to the reconstitution mixture. Furthermore, TRITC-NLS-BSA, a reporter cargo of the classical importin α/β pathway, was imported into functional control nuclei (Fig. 4Aiv), but not into pore-free nuclei (Figs. 4Biv and 4Civ).
Figure 4. Targeting of HSV1 capsids to NPCs on Xenopus nuclei in vitro.
A to C. Functional (A), Imp β45-462 (B) or BAPTA (C) nuclei were reconstituted in vitro. Completion of nuclear assembly was monitored by epi-fluorescence microscopy by DNA staining with Hoechst 33258 (i and iii), NPC staining with fluorescently labeled mAb414 (ii) and the transport capacity of the assembled NPCs was confirmed by the nuclear import of TRITC-NLS-BSA (iv). Scale bar: 10 µm. D, E. Binding of viral 0.5 M KCl capsids extracted from extracellular particles of HSV1-GFPVP26 (ii and iii, green, white arrowheads) to Xenopus nuclei was analyzed using confocal fluorescence laser scanning microscopy and a serial z-slicing of 0.37 μm optical sections. A surface view (D) and one midsection (E) of a representative nucleus are shown. NPCs were labeled with Imp β45-462-TRITC (i and iii, red). Scale bar: 10 µm. F, G. HSV1-GFPVP26 viral 0.5 M capsids were incubated with Xenopus nuclei harboring functional NPCs for 45 min, labeled with a polyclonal rabbit antiserum raised against intact HSV1 capsids (Remus, bleed V) and colloidal gold with a diameter of 12 nm coated with anti-rabbit antibodies. The specimens were analyzed by field emission scanning electron microscopy. (F) 3D surface topography of reconstituted nuclei from different views in the in-lens images (Fig. 4Fi-iii). The same areas were imaged through a backscatter electron detector revealing the positions of gold-conjugated antibodies (Fig. 4Gi-iii; inverted color mode). HSV1 capsids (white arrowheads) are bound to the cytoplasmic face of the NPCs (white arrows). Scale bar: 100 nm.
An in vitro assay to decipher the interactions of viral capsids with NPCs
To detect capsids, we used an HSV1-GFPVP26 strain in which the outer capsid protein VP26 has been replaced by a GFPVP26 fusion protein that is incorporated into virions during capsid assembly (56). Although a fraction of GFPVP26 seems to dissociate during cytosolic passage in epithelial cells, sufficient amounts remain associated with and are detected on HSV1 capsids docked at the nuclei (57). Furthermore, HSV1-GFPVP26 capsids utilize microtubule transport and, like wild-type capsids, recruit dynein from cytosolic extracts of Xenopus laevis eggs (20, 57). Thus, the GFP tag on VP26 does not interfere with crucial cytosolic capsid-host interactions. Of the capsids tested here, viral capsids treated with 0.5 M KCl are most similar in terms of protein composition to the ones that bind to NPCs of rat liver nuclei or to microtubule motors (19, 20, 37). To investigate whether such capsids can also bind to Xenopus NPCs, we incubated 0.5 M KCl viral capsids of HSV1-GFPVP26 with reconstituted nuclei. To delineate the nuclear envelopes, fluorescent Imp β45-462 was added during the last 10 min of the respective binding reaction. Initial time course experiments revealed that the number of bound capsids varied between the different capsid types and preparations, but remained rather constant during a time window of 30 to 90 min (data not shown). While surface views in top and bottom confocal laser microscopy sections showed punctate labelling typical of NPCs (Fig. 4Di, red in Fig. 4Diii) and HSV1 capsids scattered over the nuclear surface (Fig. 4Dii, green in Fig. 4Diii), mid sections revealed capsids (Fig 4Eii, green in Fig. 4Eiii, white arrowheads) closely juxtaposed to the rim labelling of the nuclear envelopes (Fig. 4Ei, red in Fig. 4Eiii).
To obtain further information on the association of capsids with Xenopus nuclei, we used high resolution field emission scanning electron microscopy (FESEM) to image the nuclear surface. Samples were labeled with a polyclonal antiserum raised against viral capsids, followed by anti-rabbit-antibodies conjugated to 12 nm colloidal gold particles. The three-dimensional surface topography of reconstituted nuclei, including the cytoplasmic surface of NPCs is seen in the in-lens images (Fig. 4F), while the corresponding backscatter electron detector images reveal the position of gold-conjugated antibodies (Fig. 4G). Thus, HSV1 capsids could be unequivocally identified by multiple gold particles, which often lined up along the capsid edges and vertices (Fig. 4G; white arrowheads). Each capsid blocked a considerable area of the nuclear envelope from view, but depending on the imaging angle, it was possible to obtain partial views of NPCs underneath the capsids (Fig. 4Fi, 4Fii; white arrows). Controls in which either the capsids or the primary antibodies had been omitted revealed little background labeling (not shown). These experiments demonstrated that HSV1 capsids are capable of docking onto NPCs of in vitro reconstituted Xenopus nuclei.
Specific capsid features and functional NPCs are required for HSV1 capsid binding to Xenopus nuclei
To determine which HSV1 capsid types were targeted to NPCs, we tested nuclear C capsids, as well as viral capsids extracted at 1 M, 0.5 M or 0.1 M KCl, for binding to functional nuclei harboring NPCs or to pore-free nuclei assembled in the presence of Imp β45-462 or BAPTA. Several images of HSV1-GFPVP26 capsids were acquired by epi-fluorescence microscopy at different focal planes, merged and superimposed with a middle focal plane image of the DNA-stained nucleus. Only immobile capsids colocalizing with the DNA were scored as bound to nuclei, and their number was determined for 5 to 20 nuclei in each condition. This setup allowed a comparison of several conditions on the same day. However, no binding of HSV1 capsids to Xenopus nuclei was observed if either component had been stored for several hours at room temperature, on ice or frozen. Instead, it was mandatory to isolate HSV1 capsids for each experiment and to reconstitute Xenopus nuclei just prior to their co-incubation, and only a few parameters could be compared within one given experiment. Therefore, we integrated all experiments into one analysis, although some conditions had been evaluated more often than others (Table 2).
Table 2. Binding of HSV1 capsids to NPCs of Xenopus nuclei reconstituted in vitro.
HSV1 capsids were isolated from extracellular virions using TX-100 in the presence of 0.1 M, 0.5 M or 1 M KCl (viral capsids) or from infected nuclei (nuclear capsids) and incubated with functional Xenopus nuclei or with nuclei assembled in the presence of Imp β45-462 or BAPTA. The samples were fixed, and bound capsids were counted. The number of experiments and the number of capsids bound per nucleus with the standard error of the mean have been tabulated for the different conditions.
| HSV1 capsid | Functional | Imp β45-462 | BAPTA | |||
|---|---|---|---|---|---|---|
| # of experiments | # of capsids per nucleus | # of experiments | # of capsids per nucleus | # of experiments | # of capsids per nucleus | |
| 0.1 M | 22 | 7.2 ± 1.3 | 4 | 4.6 ± 0.6 | 2 | 5.5 ± 1.5 |
| 0.5 M | 24 | 14.8 ± 1.6 | 4 | 4.2 ± 0.4 | 2 | 6.5 ± 0.5 |
| 1 M | 22 | 7.5 ± 1.3 | 3 | 6.3 ± 3.0 | 10 | 3.7 ± 0.6 |
| nuclear | 25 | 13.1 ± 1.7 | 3 | 6.7 ± 3.2 | 11 | 6.7 ± 2.1 |
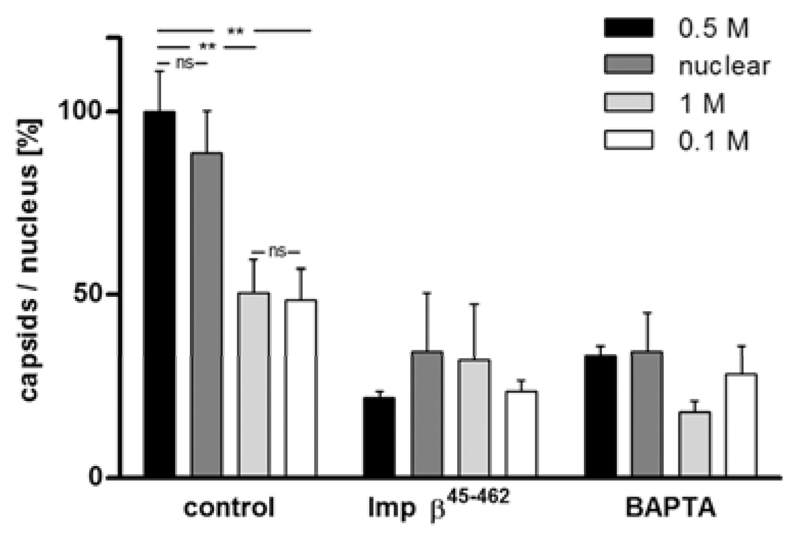
To summarize the data of many experiments in a comparative manner, we set the mean number of the 0.5 M KCl viral capsids (14.8 capsids) bound per functional nucleus to 100% (Table 2, Fig. 5). All types of viral and nuclear C capsids were targeted to Xenopus nuclei to some extent, with the viral 0.5 M KCl capsids and the nuclear capsids showing the highest degree of binding. While the number of viral 0.5 M KCl capsids was comparable to that of nuclear capsids, 0.1 M and 1 M capsids bound significantly less efficiently (p = 0.001). Furthermore, the binding of all capsid types was substantially reduced for nuclei that had been reconstituted in the presence of Imp β45-462 or BAPTA. Since the binding of 1 M viral capsids to BAPTA-treated nuclei was the lowest of all conditions tested, it may be considered as nonspecific background. These results show that HSV1 capsids docked specifically onto NPCs of in vitro reconstituted Xenopus nuclei, and that this interaction was mediated by outer capsid and/or inner tegument proteins exposed in a functional conformation on the surface of 0.5 M KCl and nuclear C capsids.
Figure 5. HSV1 capsid binding to nuclei requires specific capsid surface features and NPCs.
Viral capsids isolated from viral particles with TX-100 and 0.1, 0.5 or 1.0 M KCl or nuclear C capsids were incubated with Xenopus nuclei reconstituted in vitro. The number of HSV1 capsids bound to nuclei harboring functional NPCs (control), or to nuclei assembled in the presence of Imp β45-462 or BAPTA was determined by epi-fluorescence microscopy. Images were acquired at different focal planes, merged, bound capsids were counted, and the data normalized to 100% for 0.5 M viral capsids binding to functional nuclei. The error bars indicate the standard error of the mean. Asterisks indicate p<0.0015 and “ns” p>0.05 as determined in two-tailed student’s t-test.
Discussion
A cell-free assay to analyze viral interactions with NPCs
Very few biochemical studies have been able to address an association of viral structures with NPCs (37, 58–61). To study the nuclear targeting of mammalian viruses, we have harnessed Xenopus nuclei reconstituted in vitro that have been used extensively to study nuclear assembly and nuclear transport (48, 54, 62, 63). HSV1 viral capsids, isolated from virions with TX-100 in the presence of 0.5 M KCl, as well as nuclear C capsids interacted specifically with nuclei harboring functional NPCs. High resolution FESEM imaging demonstrated that capsids bound specifically to the cytoplasmic surface of NPCs. In cells, incoming HSV1 capsids require microtubule motors for their transport towards NPCs, and indeed viral 0.5 M KCl capsids, but not nuclear C capsids recruit microtubule motors from Xenopus egg and pig brain cytosol (19, 20). Notably, both 0.5 M KCl and nuclear C capsids were shown here to bind specifically to NPCs.
Xenopus reconstitution reactions can be fine-tuned to block nuclear assembly at specific stages (64, 65). The addition of Imp β45-462 or BAPTA results in the formation of membrane-sealed assembly intermediates that lack any architectural signs of NPCs and are thus considered to be pore-free (49). We observed a residual affinity of HSV1 capsids to such nuclei, which most likely indicates the level of background binding to nuclear envelopes. The preferential binding of 0.5 M KCl and nuclear C capsids cannot be solely attributed to the fact that nuclei assembled in the presence of Imp β45-462 or BAPTA do not expand as much as in the controls (Fig. 4; (49, 54)). While they present a reduced surface area, there was little specificity among the four capsid types in binding to Imp β45-462 or BAPTA nuclei. In contrast, 0.5 M KCl and nuclear capsids had a significantly higher affinity for nuclei harboring functional NPCs.
Surface features on HSV1 capsids
Using electron cryo tomography, we have identified substantial differences on the surface of the different capsid types. There were pronounced extra densities on 0.5 M KCl capsids and even more prominent densities on 0.1 M capsids when compared to nuclear C capsids. The shape of these densities is similar to those on cytosolic capsids imaged in vivo in the axons of neurons (66), and on viral capsids extracted with detergent and 0.5 M NaCl (67). Previous protein analyses have also revealed differences: whereas nuclear capsids lack tegument proteins, viral 0.5 M KCl capsids contain mainly inner tegument proteins, and viral 0.1 M KCl capsids maintain both inner and outer tegument proteins (summarized in Table 1; (19, 20)). Similar to viral 0.5 M KCl capsids, viral 0.5 M NaCl capsids also contain pUL36 and pUL37 and only low amounts of outer tegument proteins (68).
These data are consistent with the notion that viral capsids extracted with TX-100 and 0.5 M salt maintain additional proteins around their pentons, which most likely include inner tegument proteins. By contrast, we detected only minor structural differences between 1 M KCl viral and nuclear capsids, although the former contain more tegument proteins than the latter (c.f. Fig. 2; (19, 20)). Apparently these proteins do not form an ordered density on the capsid surface and were therefore not revealed in the icosahedrally symmetrized averages.
HSV1 proteins interacting with Nups
The outer capsid protein pUL25, and the inner tegument protein pUL36, are likely candidates to target incoming capsids to NPCs.
pUL17 and pUL25 form complexes that connect the pentons and possibly also the portal with the surrounding hexons (69, 70). Furthermore, capsid-associated pUL25 is required for proper genome uncoating at the NPC, and biochemical assays have shown that pUL25 can interact with Nup214 and hCG1, two Nups facing the cytosol (39, 71–73). pUL25 epitopes are accessible on the surface of the four capsid types tested here, as shown by immunoelectron microscopy of isolated capsids (c.f. Table 1; (19)).
pUL36 and pUL37 form complexes that are presumably located at the pentons, and that remain capsid-associated during transport to the nucleus (25, 26, 28, 67, 68, 71). Antibodies directed against pUL36 reduce nuclear targeting, incoming capsids lacking pUL36 or lacking the N-terminal nuclear localization sequence in pUL36 are not targeted to NPCs, capsids with a point-mutation in pUL36 fail to release their genomes, and proteolysis of pUL36 seems to be a prerequisite for the nuclear import of viral DNA (34, 74–77). Polyclonal antibodies have access to more pUL36 epitopes on the surface of 0.5 M KCl capsids than on 1 M or 0.1 M capsids, whereas pUL36 cannot be detected on nuclear capsids by these antibodies or by mass spectrometry (c.f. Table 1; (19)).
Thus, surface exposed pUL25 could mediate the interaction of both viral 0.5 M KCl and nuclear capsids with NPCs, while surface exposed pUL36 could only be involved in the association of viral 0.5 M KCl capsids with NPCs. However, additional pUL36 on the 0.5 M KCl capsids when compared to nuclear capsids did not stimulate NPC association. Furthermore, the treatment with 0.1 M or 1 M KCl apparently impaired HSV1 factors contributing to NPC binding. The treatment with 1 M salt might perturb the conformation of pUL36, pUL25 or another viral cofactor, while the treatment with 0.1 M KCl might prevent the removal of a putative inhibitor involved in NPC binding. Thus, we were able to demonstrate that the surface of the four HSV1 capsid types harbor structural differences that modulate their capability to interact with Xenopus NPCs as well as their affinity to antibodies directed against structural HSV1 proteins. However, so far the molecular and structural details of these changes remain elusive, and could not yet be used to unequivocally pinpoint to specific proteins mediating the interaction of HSV1 capsids with NPCs.
Clearly, more studies are required to characterize the interactions of viral capsids with NPCs. With this new approach, protein domains and peptide motifs suggested by protein-protein interaction assays using isolated proteins could be tested in the context of the complex capsid and NPC structures. Furthermore, this cell-free system may be used to determine the requirements for genome release from the capsids and their import into the nucleoplasm. It provides many advantages: nuclei harboring NPCs but lacking a particular Nup can be generated (62, 63), and even for mutants that do not form virions, cell entry can be bypassed if sufficient amounts of viral particles can be generated in heterologous systems. Further insight into capsid-docking and viral genome uncoating may be obtained by modulating other host factors such as kinases and using nuclei lacking specific Nups or capsids harboring subtle mutations. Such studies can identify molecular interactions essential for nuclear targeting, which should be maintained in viral vectors but could be targeted in antiviral therapy.
Materials and Methods
HSV1 virion preparation
Extracellular virions of HSV1 strain F (ATCC VR-733) or HSV1-GFPVP26 strain KOS (K26GFP; (56)) were prepared as before (19, 33, 57). BHK cells (ATCC CCL-10) grown to a density of 1 to 2 x 107 cells/175 cm2 flask in MEM (Cytogen, Sinn, Germany) containing 10% fetal calf serum (PAA, Pasching, Austria) were infected with an MOI of 0.01 to 0.02 pfu/cell for 2 to 3 days. The media containing extracellular virions and infected cells were centrifuged at 3,000 g for 10 min at 4°C. The cell pellet was suspended in an equal volume of TNE (0.5 M NaCl, 20 mM Tris-HCl, pH 7.5, 1 mM EDTA) to prepare TNE-nuclear capsids, or of MKT (0.1 M KCl, 30 mM MES, 20 mM Tris, pH 7.4) to prepare MKT-nuclear capsids. The cells were aliquoted, frozen in liquid N2, and stored at -80°C. Extracellular viral particles were sedimented from the supernatant at 12,000 rpm for 90 min (Type19 rotor, Beckman Coulter, Fullerton, USA), and the pellet was suspended in MKT buffer, aliquoted, frozen in liquid N2, and stored at -80°C.
HSV1 capsid preparation
Nuclear capsids were isolated from infected cells that were homogenized in H2O with 10 mM DTT and protease inhibitors (PI; 10 μg/ml antipain, 2 μg/ml bestatin, 2 μg/ml pepstatin, 2 μg/ml aprotinin, 10 μg/ml E-64, 2 μg/ml leupeptin, 160 μg/ml PMSF; Sigma-Aldrich), and the nuclei were sedimented at 230 g and 4°C for 7 min and lysed in 1% Triton X-100 in TNE supplemented with 10 mM DTT and PIs. These lysates were layered onto linear sucrose gradients (20% to 50% sucrose in TNE), and the capsids were banded at 67,000 g for 80 min at 4°C (cf. Fig. 1, top left; (19, 20, 51, 53)). The fractions containing the A, B or C capsids were diluted 1 to 4 with TNE buffer with DTT and PIs, and sedimented at 50,000 rpm (TLA 100.2 rotor, Beckman) for 20 min and at 4°C.
Viral capsids were prepared as described before (19, 20, 37, 51). To isolated viral capsids, we mixed the sediment of the medium of infected cells with an equal volume of a two-fold capsid lysis buffer (2% [w/v] Triton X-100 in 0.2 M, 1 M or 2 M KCl respectively, 20 mM MES, 30 mM Tris, pH 7.4, 20 mM DTT, PIs), incubated for 30 min on ice, and layered onto sucrose cushions (20% [w/v] sucrose in 0.1 M, 0.5 M or 1 M KCl respectively, 20 mM MES, 30 mM Tris pH 7.4, 10 mM DTT, PIs). The capsids were separated from solubilized envelope and tegument proteins by ultracentrifugation at 110,000 g for 15 min at 4°C (TLA 100.2 rotor), suspended in BRB80 (80 mM PIPES, pH 6.8, 12 mM MgCl2, 1 mM EGTA, 10 mM DTT, PIs), and treated with 0.1 to 0.2 U/μl DNase I (Promega, Madison, USA or Roche, Basel, Switzerland) and 100 μg/ml RNase (Roth GmbH, Karlsruhe, Germany) for 30 min at 37°C to facilitate suspension. After an overnight incubation at 4°C, the capsids were sedimented at 110,000 g for 15 min at 4°C (TLA 100.2 rotor) and suspended in ELB-S buffer (250 mM sucrose, 50 mM KCl, 10 mM HEPES, pH 7.6, 10 mM DTT, 2.5 mM MgCl2, 1 mg/ml BSA, PIs) using a tip sonifier (Sonic B-12; Branson, Danbury, USA) at 40 W for initially 3 and up to 10 pulses of 1 to 2 seconds. If the capsids still appeared clumped by fluorescence microscopy (BX61TRF, 20x objective, Olympus, Melville, USA), further pulses were applied. The extent of sonication required for dispersion increased from C-capsids to viral capsids isolated at 1 M, 0.5 M or 0.1 M KCl.
Antibodies
VP5 was detected by mouse monoclonal antibodies (mAb) H1.4 (Biodesign International - Meridian Life Science, Memphis, USA), VP19c by rabbit polyclonal antibodies (pAb) NC2 (78, 79), VP21/22a by pAb NC3/4 (78), VP23 by pAb NC5 (78), pUL25 by mAb #166 (80), pUL36 by pAb #147 (25), pUL37 by pAb pUL37 (26, 81), and VP13/14 by pAb R220 (82). NPCs were visualized with fluorescently labeled mAb 414 (A488-120L; Covance, Princeton, NJ, USA; (55)), and DNA with Hoechst 33258 (2 μg/ml; Sigma-Aldrich, St. Louis, MO). HSV1 structural proteins were detected by pAb Remus that has been raised against tegumented capsids (37).
Immunoblot analysis
Nuclear or viral capsids suspended in sample buffer [1% (w/v) SDS, 50 mM Tris-HCl, pH 6.8, 5% (v/v) glycerol, 1% (v/v) β-mercaptoethanol, 0.001% (w/v) bromphenol blue] were loaded onto 6 to 16% linear gradient SDS-PAGE gels, and the proteins were transferred to nitrocellulose membranes (Pall Corporation, NY). After blocking with 5% (w/v) fat-free milk in PBS-T (0.1% [v/v] Tween-20, 2.7 mM KCl, 137.9 mM NaCl, 1.5 mM KH2PO4, 8.1 mM Na2HPO4, pH 7.4), the membranes were probed with primary antibodies and IRDye anti-rabbit or anti-mouse antibodies (LI-COR Biosciences, Bad Homburg, Germany), and imaged using an Odyssey® Infrared Scanner (LI-COR Biosciences).
Electron cryo tomography
Viral capsids were sonicated for 3 times 10 seconds (BB6 cup horn, Sonoplus HD3200, 60% maximal output, Bandelin, Berlin, Germany) directly before adding them onto the grids, and processed for electron cryo tomography as described before (66). 5 μl of a capsid preparation were added together with 2 μl of a homemade colloidal gold suspension (10 nm gold coated with BSA in PBS) onto the holey carbon support film of electron microscopy grids (R2-1, Cu 200 mesh, Quantifoil, Jena, Germany), and excess liquid was removed by blotting with filter paper. Specimens were vitrified by plunge-freezing into liquid ethane and stored in liquid N2.
Data were collected using transmission electron microscopy; some of the 1 M KCl viral capsid tilt series were collected with a CM300 (43 of the 183 subvolumes; FEI-Philips, Eindhoven, The Netherlands), while the rest including the tilt series from other capsid types were collected with a Tecnai Polara (FEI, Eindhoven, The Netherlands). The microscopes were operated at 300 kV; the pixel size was 0.68 nm (CM300) or 0.81 nm (Polara) at the specimen level, and tilt series were collected with an angular increment of 2° or 3° from -60° to 60°. Defocus was measured along the tilt axis after each tilt and automatically maintained at -8 μm +/- 0.5μm to gain phase contrast. The total electron dose received at the specimen level was kept between 60 and 90 electrons/Å2. The applied electron dose was kept proportional to 1/cosα of the tilt angle (α). Images were acquired on a 2Kx2K Multiscan CCD camera (GIF 2002 post-column energy filter, Gatan, Pleasanton, CA).
Image processing and averaging of tomographic sub-volumes
Alignment of tilted projection image series was performed using 10 nm gold beads as fiducial markers. Three-dimensional reconstructions were calculated using the software IMOD (83), and subsequent processing using Bsoft as described before (66, 84). Capsids were located in unbinned tomograms, and sub-volumes with a size of 180x180x180 pixels were extracted. The 3D orientation of all sub-volumes was determined using a 22 Å resolution structure of the HSV1 capsid (85) as a template. The oriented sub-volumes were averaged and icosahedral symmetry was applied. Symmetrized averages were used as templates for the next iteration of orientation refinement. Three iterations were performed.
The resolution of the averages was determined by Fourier shell correlation using the 0.5 criterion, after splitting the data in two halves, calculating two separate averages and imposing icosahedral symmetry. To calculate the difference map of the two averages gray values were scaled to the same radial density maximum within the capsid and minimum just outside of the capsid. The difference of densities was then calculated by subtracting the capsids without tegument from the capsids with tegument. All capsid reconstructions were first scaled against the nuclear C capsid reconstruction. Magnification differences up to 3.5% were detected and these were compensated for by creating up-scaled or down-scaled maps.
Reconstitution of nuclei
Xenopus laevis egg extracts, cytosolic and membrane fractions, demembranated sperm chromatin, and an ATP regeneration system were prepared, and nuclei reconstituted within 50 to 60 min at RT (47, 64). To assemble pore-free nuclei, 8 μM of recombinant Imp β45-462 or 5 mM BAPTA (Calbiochem, La Jolla, CA, USA) were added to the mixture and incubated for 60 min (48). In each experiment the functionality of the reconstituted nuclei was tested by removing a fraction of the nuclei for staining with mAb 414 and Hoechst 33258, and nuclear import analysis with bovine serum albumin coupled to a nuclear localization sequence and to tetramethyl-rhodaminyl-isothiocyanate (TRITC-NLS-BSA; (48, 50)).
Binding of HSV1 capsids to Xenopus nuclei
The reconstitution reactions were diluted threefold with 1x ELB-S-buffer (pH 7.6, supplemented with 2.5 mM MgCl2), 10 μl of nuclear or viral HSV1 capsids were added to 10 μl of the nuclei, and the reactions were agitated at 7 rpm for 20 to 90 min at room temperature (Fig. 1, bottom). Small aliquots were stained with Hoechst 33258 in 12% [w/v] para-formaldehyde, 250 mM sucrose, 10 mM HEPES, pH 7.5, 0.4% [w/v] N-propylgalate for 5 min. Capsid binding to nuclei was analyzed by epifluorescence microscopy (UPlanSApo 100x NA 1.4 oil immersion, Olympus BX61TRF equipped with a DP70 digital camera), or by laser scanning confocal fluorescence microscopy (63x NA 1.4 oil immersion, ZEISS LSM 510 META, Oberkochen, Germany). 10 to 25 nuclei of each experimental condition were randomly chosen, and the number of capsids bound to each nucleus was quantified over all focal planes.
Field Emission Scanning Electron Microscopy (FESEM)
In vitro reconstituted intact Xenopus nuclei incubated with HSV1 capsids for 60 min were labeled with Remus serum. Its dilution was optimized by fluorescence microscopy and adjusted for immunogold labeling. Colloidal gold of 12 nm diameter and coated with goat-anti-rabbit antibodies (Jackson Immuno-Research, West Grove, USA) was titrated to obtain a minimal background signal in the absence of primary antibodies (64, 65). To facilitate the capture of nuclei, they were sedimented at 1,000 g for 10 min onto silicon chips (Ted Pella, Irvine, USA) that had been coated with 0.2 mg/ml poly-lysine for 15 min. The chips were transferred to 24-well plates, washed in 1x ELB-K (100 mM KCl, 10 mM HEPES, pH 7.6, 2.5 mM MgCl2, 1 mM DTT, 5 μg/ml cycloheximide, 5 μg/ml cytochalasin B, 10 μg/ml aprotinin, 10 μg/ml leupeptin), and fixed in 3.7% para-formaldehyde with 0.2% glutaraldehyde (Electron Microscopy Sciences; Hatfield, PA, USA) in 150 mM sucrose, 80 mM PIPES, pH 6.8, 1 mM MgCl2 for 30 min at RT. All subsequent steps were as described in (86), with post-fixation in aqueous 0.5% osmium tetroxide, critical-point drying on a CPD030 apparatus (Bal-Tec AG, Liechtenstein), and sputter coating with 2 nm of chromium (K575X coater, Emitech, London, England). The samples were imaged using a FESEM (Ultra plus, ZEISS, Oberkochen, Germany) with an in-lens detector for secondary electrons to reveal surface structures, and an energy selective backscatter electron detector to localize gold particles.
Synopsis.
A novel assay based on nuclei reconstituted in vitro from Xenopus egg extracts was used for studying the interactions between viruses and nuclear pore complexes (NPCs). The targeting of GFP-tagged herpes simplex virus capsids to NPCs was followed by fluorescence and scanning electron microscopy. Surface features of different capsid types were analyzed by electron cryo tomography and compared to their targeting efficiency in the in vitro assay. Capsids exposing outer capsid and inner tegument proteins are targeted preferentially to NPCs.
Acknowledgement
We thank Katinka Döhner and Anna Buch (Virology, Hannover Medical School) for constructive discussions throughout the study. We are grateful to Prashant Desai for providing HSV1(KOS)-GFPVP26 (Johns Hopkins University, Baltimore, MD) and to Ari Helenius (ETH, Zürich, Switzerland), David M. Meredith (University of Leeds, UK), Gary H. Cohen and Roselyn J. Eisenberg (University of Pennsylvania, PA, USA), Thomas C. Mettenleiter (Friedrich-Loeffler-Institut, Greifswald, Germany), and Valerie G. Preston (University of Glasgow, UK) for generously donating essential antibodies. This study was supported by funds from the Niedersächsisches Ministerium für Wissenschaft und Kultur and the Technion Society to AH and BS, by the German Research Council (DFG, SFB 900, TP2 to BS), and by the EraNet NanoSci+ Initiative to AH (ISF 2045/09), BS (DFG, So403/4), and KG (DFG GR1990/3).
Abbreviations
- BAPTA
1,2-bis(o-aminophenoxy)ethane-N,N,N',N'-tetraacetic acid
- FESEM
field emission scanning electron microscopy
- GFP
green fluorescence protein
- HSV1
herpes simplex virus type 1
- NLS
nuclear localization signal
- NPC
nuclear pore complex
- Nup
nucleoporin
- PI
protease inhibitors
- pULX
viral protein encoded by the gene ULX
- RT
room temperature
- VPX
viral protein X
References
- 1.Brohawn SG, Partridge JR, Whittle JR, Schwartz TU. The nuclear pore complex has entered the atomic age. Structure. 2009;17:1156–1168. doi: 10.1016/j.str.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hetzer MW, Wente SR. Border control at the nucleus: biogenesis and organization of the nuclear membrane and pore complexes. Dev Cell. 2009;17:606–616. doi: 10.1016/j.devcel.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frenkiel-Krispin D, Maco B, Aebi U, Medalia O. Structural analysis of a metazoan nuclear pore complex reveals a fused concentric ring architecture. J Mol Biol. 2010;395:578–586. doi: 10.1016/j.jmb.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 5.Maimon T, Elad N, Dahan I, Medalia O. The human nuclear pore complex as revealed by cryo-electron tomography. Structure. 2012;20:998–1006. doi: 10.1016/j.str.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 6.Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- 7.Pante N, Kann M. Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol Biol Cell. 2002;13:425–434. doi: 10.1091/mbc.01-06-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wente SR, Rout MP. The nuclear pore complex and nuclear transport. Cold Spring Harb Perspect Biol. 2010;2:a000562. doi: 10.1101/cshperspect.a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strom AC, Weis K. Importin-beta-like nuclear transport receptors. Genome Biol. 2001;2:3008. doi: 10.1186/gb-2001-2-6-reviews3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fried H, Kutay U. Nucleocytoplasmic transport: taking an inventory. Cell Mol Life Sci. 2003;60:1659–1688. doi: 10.1007/s00018-003-3070-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Görlich D, Pante N, Kutay U, Aebi U, Bischoff FR. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- 12.Lee SJ, Matsuura Y, Liu SM, Stewart M. Structural basis for nuclear import complex dissociation by RanGTP. Nature. 2005;435:693–696. doi: 10.1038/nature03578. [DOI] [PubMed] [Google Scholar]
- 13.Davison AJ, Eberle R, Ehlers B, Hayward GS, McGeoch DJ, Minson AC, Pellett PE, Roizman B, Studdert MJ, Thiry E. The order Herpesvirales. Arch Virol. 2009;154:171–177. doi: 10.1007/s00705-008-0278-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown JC, Newcomb WW. Herpesvirus capsid assembly: insights from structural analysis. Curr Opin Virol. 2011;1:142–149. doi: 10.1016/j.coviro.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rochat RH, Liu X, Murata K, Nagayama K, Rixon FJ, Chiu W. Seeing the portal in herpes simplex virus type 1 B capsids. J Virol. 2011;85:1871–1874. doi: 10.1128/JVI.01663-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grünewald K, Desai P, Winkler DC, Heymann JB, Belnap DM, Baumeister W, Steven AC. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science. 2003;302:1396–1398. doi: 10.1126/science.1090284. [DOI] [PubMed] [Google Scholar]
- 17.Loret S, Guay G, Lippe R. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J Virol. 2008;82:8605–8618. doi: 10.1128/JVI.00904-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou ZH, Chen DH, Jakana J, Rixon FJ, Chiu W. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J Virol. 1999;73:3210–3218. doi: 10.1128/jvi.73.4.3210-3218.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radtke K, Kieneke D, Wolfstein A, Michael K, Steffen W, Scholz T, Karger A, Sodeik B. Plus- and minus-end directed microtubule motors bind simultaneously to herpes simplex virus capsids using different inner tegument structures. PLoS Pathog. 2010;6:e1000991. doi: 10.1371/journal.ppat.1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolfstein A, Nagel CH, Radtke K, Dohner K, Allan VJ, Sodeik B. The inner tegument promotes herpes simplex virus capsid motility along microtubules in vitro. Traffic. 2006;7:227–237. doi: 10.1111/j.1600-0854.2005.00379.x. [DOI] [PubMed] [Google Scholar]
- 21.Kelly BJ, Fraefel C, Cunningham AL, Diefenbach RJ. Functional roles of the tegument proteins of herpes simplex virus type 1. Virus Res. 2009;145:173–186. doi: 10.1016/j.virusres.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 22.Mettenleiter TC, Klupp BG, Granzow H. Herpesvirus assembly: an update. Virus Res. 2009;143:222–234. doi: 10.1016/j.virusres.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 23.Johnson DC, Baines JD. Herpesviruses remodel host membranes for virus egress. Nat Rev Microbiol. 2011;9:382–394. doi: 10.1038/nrmicro2559. [DOI] [PubMed] [Google Scholar]
- 24.Henaff D, Radtke K, Lippe R. Herpesviruses exploit several host compartments for envelopment. Traffic. 2012;13:1443–1449. doi: 10.1111/j.1600-0854.2012.01399.x. [DOI] [PubMed] [Google Scholar]
- 25.Schipke J, Pohlmann A, Diestel R, Binz A, Rudolph K, Nagel CH, Bauerfeind R, Sodeik B. The C terminus of the large tegument protein pUL36 contains multiple capsid binding sites that function differently during assembly and cell entry of herpes simplex virus. J Virol. 2012;86:3682–3700. doi: 10.1128/JVI.06432-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sandbaumhüter M, Dohner K, Schipke J, Binz A, Pohlmann A, Sodeik B, Bauerfeind R. Cytosolic herpes simplex virus capsids not only require binding inner tegument protein pUL36 but also pUL37 for active transport prior to secondary envelopment. Cell Microbiol. 2013;15:248–269. doi: 10.1111/cmi.12075. [DOI] [PubMed] [Google Scholar]
- 27.Antinone SE, Smith GA. Retrograde axon transport of herpes simplex virus and pseudorabies virus: a live-cell comparative analysis. J Virol. 2010;84:1504–1512. doi: 10.1128/JVI.02029-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coller KE, Lee JI, Ueda A, Smith GA. The capsid and tegument of the alphaherpesviruses are linked by an interaction between the UL25 and VP1/2 proteins. J Virol. 2007;81:11790–11797. doi: 10.1128/JVI.01113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Granzow H, Klupp BG, Mettenleiter TC. Entry of pseudorabies virus: an immunogold-labeling study. J Virol. 2005;79:3200–3205. doi: 10.1128/JVI.79.5.3200-3205.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luxton GW, Haverlock S, Coller KE, Antinone SE, Pincetic A, Smith GA. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc Natl Acad Sci U S A. 2005;102:5832–5837. doi: 10.1073/pnas.0500803102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maurer UE, Sodeik B, Grunewald K. Native 3D intermediates of membrane fusion in herpes simplex virus 1 entry. Proc Natl Acad Sci U S A. 2008;105:10559–10564. doi: 10.1073/pnas.0801674105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Döhner K, Wolfstein A, Prank U, Echeverri C, Dujardin D, Vallee R, Sodeik B. Function of dynein and dynactin in herpes simplex virus capsid transport. Mol Biol Cell. 2002;13:2795–2809. doi: 10.1091/mbc.01-07-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sodeik B, Ebersold MW, Helenius A. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J Cell Biol. 1997;136:1007–1021. doi: 10.1083/jcb.136.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Batterson W, Furlong D, Roizman B. Molecular genetics of herpes simplex virus. VIII. further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J Virol. 1983;45:397–407. doi: 10.1128/jvi.45.1.397-407.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Granzow H, Weiland F, Jons A, Klupp BG, Karger A, Mettenleiter TC. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J Virol. 1997;71:2072–2082. doi: 10.1128/jvi.71.3.2072-2082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyamoto K, Morgan C. Structure and development of viruses as observed in the electron microscope: XI Entry and uncoating of herpes simplex virus. J Virol. 1971;8:910–918. doi: 10.1128/jvi.8.6.910-918.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ojala PM, Sodeik B, Ebersold MW, Kutay U, Helenius A. Herpes simplex virus type 1 entry into host cells: reconstitution of capsid binding and uncoating at the nuclear pore complex in vitro. Mol Cell Biol. 2000;20:4922–4931. doi: 10.1128/mcb.20.13.4922-4931.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peng L, Ryazantsev S, Sun R, Zhou ZH. Three-dimensional visualization of gammaherpesvirus life cycle in host cells by electron tomography. Structure. 2010;18:47–58. doi: 10.1016/j.str.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rode K, Dohner K, Binz A, Glass M, Strive T, Bauerfeind R, Sodeik B. Uncoupling uncoating of herpes simplex virus genomes from their nuclear import and gene expression. J Virol. 2011;85:4271–4283. doi: 10.1128/JVI.02067-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newcomb WW, Booy FP, Brown JC. Uncoating the herpes simplex virus genome. J Mol Biol. 2007;370:633–642. doi: 10.1016/j.jmb.2007.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Labokha AA, Gradmann S, Frey S, Hulsmann BB, Urlaub H, Baldus M, Gorlich D. Systematic analysis of barrier-forming FG hydrogels from Xenopus nuclear pore complexes. EMBO J. 2013;32:204–218. doi: 10.1038/emboj.2012.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roos WH, Ivanovska IL, Evilevitch A, Wuite GJ. Viral capsids: mechanical characteristics, genome packaging and delivery mechanisms. Cell Mol Life Sci. 2007;64:1484–1497. doi: 10.1007/s00018-007-6451-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang K, Homa F, Baines JD. Putative terminase subunits of herpes simplex virus 1 form a complex in the cytoplasm and interact with portal protein in the nucleus. J Virol. 2007;81:6419–6433. doi: 10.1128/JVI.00047-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roos WH, Radtke K, Kniesmeijer E, Geertsema H, Sodeik B, Wuite GJ. Scaffold expulsion and genome packaging trigger stabilization of herpes simplex virus capsids. Proc Natl Acad Sci U S A. 2009;106:9673–9678. doi: 10.1073/pnas.0901514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Newmeyer DD, Forbes DJ. Nuclear import can be separated into distinct steps in vitro: nuclear pore binding and translocation. Cell. 1988;52:641–653. doi: 10.1016/0092-8674(88)90402-3. [DOI] [PubMed] [Google Scholar]
- 46.Newport J. Nuclear reconstitution in vitro: stages of assembly around protein-free DNA. Cell. 1987;48:205–217. doi: 10.1016/0092-8674(87)90424-7. [DOI] [PubMed] [Google Scholar]
- 47.Harel A, Orjalo AV, Vincent T, Lachish-Zalait A, Vasu S, Shah S, Zimmerman E, Elbaum M, Forbes DJ. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol Cell. 2003;11:853–864. doi: 10.1016/s1097-2765(03)00116-3. [DOI] [PubMed] [Google Scholar]
- 48.Savulescu AF, Shorer H, Kleifeld O, Cohen I, Gruber R, Glickman MH, Harel A. Nuclear import of an intact preassembled proteasome particle. Mol Biol Cell. 2011;22:880–891. doi: 10.1091/mbc.E10-07-0595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Macaulay C, Forbes DJ. Assembly of the nuclear pore: biochemically distinct steps revealed with NEM, GTP gamma S, and BAPTA. J Cell Biol. 1996;132:5–20. doi: 10.1083/jcb.132.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chan RC, Forbes DI. In vitro study of nuclear assembly and nuclear import using Xenopus egg extracts. Methods in molecular biology. 2006;322:289–300. doi: 10.1007/978-1-59745-000-3_20. [DOI] [PubMed] [Google Scholar]
- 51.Radtke K, Anderson F, Sodeik B. A precipitation-based assay to analyze interactions of viral particles with cytosolic host factors. Methods in molecular biology. 2014;1144:191–208. doi: 10.1007/978-1-4939-0428-0_13. [DOI] [PubMed] [Google Scholar]
- 52.Newcomb WW, Brown JC. Time-dependent transformation of the herpesvirus tegument. J Virol. 2009;83:8082–8089. doi: 10.1128/JVI.00777-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Newcomb WW, Brown JC. Structure and capsid association of the herpesvirus large tegument protein UL36. J Virol. 2010;84:9408–9414. doi: 10.1128/JVI.00361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harel A, Chan RC, Lachish-Zalait A, Zimmerman E, Elbaum M, Forbes DJ. Importin beta negatively regulates nuclear membrane fusion and nuclear pore complex assembly. Mol Biol Cell. 2003;14:4387–4396. doi: 10.1091/mbc.E03-05-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wente SR, Rout MP, Blobel G. A new family of yeast nuclear pore complex proteins. J Cell Biol. 1992;119:705–723. doi: 10.1083/jcb.119.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Desai P, Person S. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J Virol. 1998;72:7563–7568. doi: 10.1128/jvi.72.9.7563-7568.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Döhner K, Radtke K, Schmidt S, Sodeik B. Eclipse phase of herpes simplex virus type 1 infection: Efficient dynein-mediated capsid transport without the small capsid protein VP26. J Virol. 2006;80:8211–8224. doi: 10.1128/JVI.02528-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kann M, Sodeik B, Vlachou A, Gerlich WH, Helenius A. Phosphorylation-dependent binding of hepatitis B virus core particles to the nuclear pore complex. J Cell Biol. 1999;145:45–55. doi: 10.1083/jcb.145.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cohen S, Pante N. Pushing the envelope: microinjection of Minute virus of mice into Xenopus oocytes causes damage to the nuclear envelope. J Gen Virol. 2005;86:3243–3252. doi: 10.1099/vir.0.80967-0. [DOI] [PubMed] [Google Scholar]
- 60.Porwal M, Cohen S, Snoussi K, Popa-Wagner R, Anderson F, Dugot-Senant N, Wodrich H, Dinsart C, Kleinschmidt JA, Pante N, Kann M. Parvoviruses cause nuclear envelope breakdown by activating key enzymes of mitosis. PLoS Pathog. 2013;9:e1003671. doi: 10.1371/journal.ppat.1003671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmitz A, Schwarz A, Foss M, Zhou L, Rabe B, Hoellenriegel J, Stoeber M, Pante N, Kann M. Nucleoporin 153 arrests the nuclear import of hepatitis B virus capsids in the nuclear basket. PLoS Pathog. 2010;6:e1000741. doi: 10.1371/journal.ppat.1000741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Finlay DR, Forbes DJ. Reconstitution of biochemically altered nuclear pores: transport can be eliminated and restored. Cell. 1990;60:17–29. doi: 10.1016/0092-8674(90)90712-n. [DOI] [PubMed] [Google Scholar]
- 63.Walther TC, Fornerod M, Pickersgill H, Goldberg M, Allen TD, Mattaj IW. The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. EMBO J. 2001;20:5703–5714. doi: 10.1093/emboj/20.20.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rotem A, Gruber R, Shorer H, Shaulov L, Klein E, Harel A. Importin beta regulates the seeding of chromatin with initiation sites for nuclear pore assembly. Mol Biol Cell. 2009;20:4031–4042. doi: 10.1091/mbc.E09-02-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shaulov L, Harel A. Improved visualization of vertebrate nuclear pore complexes by field emission scanning electron microscopy. Structure. 2012;20:407–413. doi: 10.1016/j.str.2012.01.022. [DOI] [PubMed] [Google Scholar]
- 66.Ibiricu I, Huiskonen JT, Dohner K, Bradke F, Sodeik B, Grunewald K. Cryo electron tomography of herpes simplex virus during axonal transport and secondary envelopment in primary neurons. PLoS Pathog. 2011;7:e1002406. doi: 10.1371/journal.ppat.1002406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cardone G, Newcomb WW, Cheng N, Wingfield PT, Trus BL, Brown JC, Steven AC. The UL36 tegument protein of herpes simplex virus 1 has a composite binding site at the capsid vertices. J Virol. 2012;86:4058–4064. doi: 10.1128/JVI.00012-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Newcomb WW, Jones LM, Dee A, Chaudhry F, Brown JC. Role of a reducing environment in disassembly of the herpesvirus tegument. Virology. 2012;431:71–79. doi: 10.1016/j.virol.2012.05.017. [DOI] [PubMed] [Google Scholar]
- 69.Cockrell SK, Huffman JB, Toropova K, Conway JF, Homa FL. Residues of the UL25 protein of herpes simplex virus that are required for its stable interaction with capsids. J Virol. 2011;85:4875–4887. doi: 10.1128/JVI.00242-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Conway JF, Cockrell SK, Copeland AM, Newcomb WW, Brown JC, Homa FL. Labeling and localization of the herpes simplex virus capsid protein UL25 and its interaction with the two triplexes closest to the penton. J Mol Biol. 2010;397:575–586. doi: 10.1016/j.jmb.2010.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pasdeloup D, Blondel D, Isidro AL, Rixon FJ. Herpesvirus capsid association with the nuclear pore complex and viral DNA release involve the nucleoporin CAN/Nup214 and the capsid protein pUL25. J Virol. 2009;83:6610–6623. doi: 10.1128/JVI.02655-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Preston VG, Murray J, Preston CM, McDougall IM, Stow ND. The UL25 gene product of herpes simplex virus type 1 is involved in uncoating of the viral genome. J Virol. 2008;82:6654–6666. doi: 10.1128/JVI.00257-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Trus BL, Newcomb WW, Cheng N, Cardone G, Marekov L, Homa FL, Brown JC, Steven AC. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-Filled HSV-1 capsids. Mol Cell. 2007;26:479–489. doi: 10.1016/j.molcel.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Abaitua F, Hollinshead M, Bolstad M, Crump CM, O'Hare P. A Nuclear localization signal in herpesvirus protein VP1-2 is essential for infection via capsid routing to the nuclear pore. J Virol. 2012;86:8998–9014. doi: 10.1128/JVI.01209-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Copeland AM, Newcomb WW, Brown JC. Herpes simplex virus replication: roles of viral proteins and nucleoporins in capsid-nucleus attachment. J Virol. 2009;83:1660–1668. doi: 10.1128/JVI.01139-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jovasevic V, Liang L, Roizman B. Proteolytic cleavage of VP1-2 is required for release of herpes simplex virus 1 DNA into the nucleus. J Virol. 2008;82:3311–3319. doi: 10.1128/JVI.01919-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roberts AP, Abaitua F, O'Hare P, McNab D, Rixon FJ, Pasdeloup D. Differing roles of inner tegument proteins pUL36 and pUL37 during entry of herpes simplex virus type 1. J Virol. 2009;83:105–116. doi: 10.1128/JVI.01032-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cohen GH, Ponce de Leon M, Diggelmann H, Lawrence WC, Vernon SK, Eisenberg RJ. Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. J Virol. 1980;34:521–531. doi: 10.1128/jvi.34.2.521-531.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Eisenberg RJ, Long D, Ponce de Leon M, Matthews JT, Spear PG, Gibson MG, Lasky LA, Berman P, Golub E, Cohen GH. Localization of epitopes of herpes simplex virus type 1 glycoprotein D. J Virol. 1985;53:634–644. doi: 10.1128/jvi.53.2.634-644.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thurlow JK, Murphy M, Stow ND, Preston VG. Herpes simplex virus type 1 DNA-packaging protein UL17 is required for efficient binding of UL25 to capsids. J Virol. 2006;80:2118–2126. doi: 10.1128/JVI.80.5.2118-2126.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Leege T, Granzow H, Fuchs W, Klupp BG, Mettenleiter TC. Phenotypic similarities and differences between UL37-deleted pseudorabies virus and herpes simplex virus type 1. J Gen Virol. 2009;90:1560–1568. doi: 10.1099/vir.0.010322-0. [DOI] [PubMed] [Google Scholar]
- 82.Whittaker GR, Riggio MP, Halliburton IW, Killington RA, Allen GP, Meredith DM. Antigenic and protein sequence homology between VP13/14, a herpes simplex virus type 1 tegument protein, and gp10, a glycoprotein of equine herpesvirus 1 and 4. J Virol. 1991;65:2320–2326. doi: 10.1128/jvi.65.5.2320-2326.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 84.Heymann JB, Belnap DM. Bsoft: image processing and molecular modeling for electron microscopy. J Struct Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 85.Cheng N, Trus BL, Belnap DM, Newcomb WW, Brown JC, Steven AC. Handedness of the herpes simplex virus capsid and procapsid. J Virol. 2002;76:7855–7859. doi: 10.1128/JVI.76.15.7855-7859.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Allen TD, Rutherford SA, Murray S, Sanderson HS, Gardiner F, Kiseleva E, Goldberg MW, Drummond SP. Generation of cell-free extracts of Xenopus eggs and demembranated sperm chromatin for the assembly and isolation of in vitro-formed nuclei for Western blotting and scanning electron microscopy (SEM) Nat Protoc. 2007;2:1173–1179. doi: 10.1038/nprot.2007.138. [DOI] [PubMed] [Google Scholar]
- 87.Loret S, Lippe R. Biochemical analysis of infected cell polypeptide (ICP)0, ICP4, UL7 and UL23 incorporated into extracellular herpes simplex virus type 1 virions. The Journal of general virology. 2012;93:624–634. doi: 10.1099/vir.0.039776-0. [DOI] [PubMed] [Google Scholar]
- 88.Roizman B, Campadelli-Fiume G. Alphaherpes viral genes and their functions. In: Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K, editors. Human Herpesviruses, Biology, Therapy and Immunoprophylaxis. New York: Cambridge University Press; 2007. pp. 70–92. [PubMed] [Google Scholar]
- 89.Toropova K, Huffman JB, Homa FL, Conway JF. The herpes simplex virus 1 UL17 protein is the second constituent of the capsid vertex-specific component required for DNA packaging and retention. J Virol. 2011;85:7513–7522. doi: 10.1128/JVI.00837-11. [DOI] [PMC free article] [PubMed] [Google Scholar]