Abstract
Macrophage stimulation with interferon-γ (IFN-γ) and interleukin 4 (IL-4) triggers distinct and opposing activation programs. During mixed infections or cancer macrophages are often exposed to both cytokines, but how these two programs influence each other remains unclear. We found that IFN-γ and IL-4 mutually inhibited epigenomic and transcriptional changes induced by each cytokine alone. Computational and functional analyses revealed the genomic bases for gene-specific cross-repression. For instance, while STAT1 and IRF1 motifs were associated with robust and IL-4-resistant responses to IFN-γ their coexistence with binding sites for auxiliary transcription factors such as AP-1, generated vulnerability to IL-4-mediated inhibition. These data provide a core mechanistic framework for the integration of signals that control macrophage activation in complex environmental conditions.
When exposed to micro-environmental stimuli, macrophages acquire new functional properties in a dynamic and reversible fashion 1. M1 and M2 programs, instigated by the exposure to interferon-γ (IFN-γ and interleukin 4 (IL-4), respectively, represent two extreme poles of a broad spectrum of macrophage activation states 2 and were named after the cytokines specifically produced by TH1 and TH2 lymphocyte subsets 3, 4. While the M1 program is typically associated with a pro-inflammatory phenotype and high microbicidal activity, M2 macrophages are linked to resistance to helminths and tissue repair 5, 6. Although the characterization of these two functional extremes has greatly contributed to advance our understanding of macrophage biology, several elements of complexity must be taken into consideration. First, macrophage activation states are in principle as diverse as the variety of stimuli these cells can be exposed to 2. Second, such activation states, although associated with some forms of cellular memory 7, 8, 9, do not drive terminal and irreversible differentiation programs, implying the possibility of inter-conversion between macrophage states when micro-environmental conditions change 10. Third, macrophages are exposed to multiple and often conflicting micro-environmental stimuli that may impact their biology in a complex fashion 11.
Co-existence of antagonistic signals impacting on macrophage function is frequently observed in vivo. Upon co-infection with pathogens eliciting type 1 and type 2 immunity (such as viruses and helminths, respectively), the pathways leading to IL-4 and IFN-γ release are concomitantly activated and the interplay between these conflicting programs has important biological consequences. The activation of an IL-4-dependent type 2 immune response to helminths results in a broad spectrum of immune-regulatory and suppressive effects 12, 13 that also depend on myeloid cells 14, 15. Consistently, epidemiological data indicate that helminths interfere with the ability of the immune system to control co-infections by microbial pathogens 16 and macrophages activated by IL-4 favor Mycobacterium tuberculosis replication 17, 18. Similar occurrences are observed in conditions whereby a single pathogen, such as Leishmania major, concomitantly triggers M1 and M2 responses 19. Furthermore, both in tumors and during the resolution phase of inflammation, macrophages are co-exposed to pro-and anti-inflammatory signals driving concomitant and frequently co-existing M1 and M2 profiles 20. Collectively, these observations highlight the need to dissect the relationships between multiple environmental signals to understand their impact on macrophage biology.
Here, we set out to address some critical and basic aspects of macrophage polarization, namely whether the M1 and M2 polarization programs are mutually exclusive; whether one of the two programs is dominant over the other and finally what are the mechanisms underlying the cross-talk between M1 and M2 polarizing stimuli. Transcriptional and epigenomic profiling of macrophages treated with IL-4, IFN-γ or their combination showed that either cytokine exerted inhibitory effects on the opposite activation program. These effects were of broad amplitude but in most cases of limited magnitude, indicating lack of mutual exclusivity of the M1 and M2 programs and a high degree of macrophage plasticity. However, induction of selected genes with critical biological roles as well as hundreds of enhancers characteristic of either program was strongly suppressed upon co-stimulation. While STAT1 and IRF1 binding were associated with resistance of IFN-γ-responsive enhancers to IL-4-mediated inhibition, the involvement of auxiliary transcription factors, such as AP-1 and C/EBPβ, generated vulnerability to the inhibitory effects of IL-4. In the context of IL-4-dependent macrophage activation, induction of Myc was instead required to trigger a component of the response that was resistant to IFN-γ-mediated inhibition. These data provide a mechanistic framework for the interpretation of cross-regulatory effects of stimuli driving opposing macrophage activation programs.
Results
Cross-regulation between macrophage polarization programs
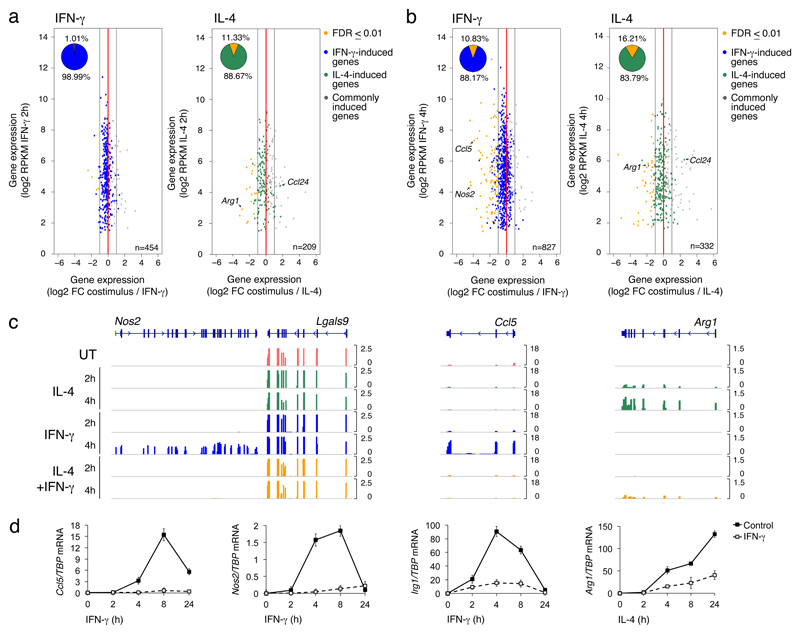
We used RNA-seq in mouse bone marrow derived macrophages (BMDMs) to determine the effects of the co-stimulation with IL-4 and IFN-γ on the gene expression programs induced by individually administered cytokines. In pilot studies, we determined the optimal stimulus concentrations, namely the minimal dose required for IL-4 and IFN-γ to stimulate phosphorylation of the transcription factors STAT6 and STAT1, respectively, and maximal induction of canonical M2 and M1 genes such as Arg1 and Nos2 (Supplementary Fig. 1a). Flow cytometry and immunofluorescence analysis (Supplementary Fig. 2) with antibodies recognizing the phosphorylated forms of the transcription factors STAT1 (Tyr701) and STAT6 (Tyr641), showed that these critical mediators of the IL-4 and IFN-γ induced programs respectively, can be activated within the same cells in the analyzed population. BMDMs were stimulated for 2h or 4h and poly-adenylated RNA from biological triplicates was extracted and sequenced at high depth (Supplementary Table 1). The overall correlation among samples was very high (R-squared range: 0.974-0.99) (Supplementary Fig. 1b). We first analyzed the effects of the co-stimulation on the genes induced at 2h or 4h by IFN-γ (454 and 827, respectively) or IL-4 (209 and 332). For both IFN-γ and IL-4-induced genes, co-administration of the other cytokine caused a transcriptional cross-inhibition of the respective program that was both broad in amplitude and overall limited in magnitude, at both 2h (Fig. 1a) and 4 hours (Fig. 1b) post stimulation. Down-regulation of IFN-γ-induced genes by IL-4 co-stimulation became more evident at 4h (Fig. 1b), suggesting that genes induced by IFN-γ with slower kinetics were more sensitive to IL-4 inhibition. A relatively small fraction of IFN-γ- and IL-4-induced genes was especially sensitive to co-stimulation (Fig. 1b, Supplemental Table 2), showing almost complete inhibition by the antagonistic stimulus. Notably, canonical markers of M1 or M2 polarization were included in this category. For instance, induction of Nos2 (encoding the inducible nitric oxide synthase) and Ccl5 were strongly inhibited by IL-4 at the mRNA (Fig. 1c) and protein levels (Supplementary Fig. 1c). Similarly, the M2 signature genes Arg1, encoding for arginase 1 and Retnla (Resistin-like alpha) were induced by IL-4 and repressed by co-stimulation with IFN-γ (Fig. 1c, Supplementary Fig. 1d, Supplementary Table 2). The transcriptional antagonism between IFN-γ and IL-4 at the genes analyzed persisted over longer times of co-stimulation (Fig. 1d), and occurred through a broad range of concentrations, being in some cases influenced by the cytokine dose (Supplementary Fig. 1d). Moreover, consistent with the increased susceptibility to co-stimulation of late IFN-γ-induced genes, transcription of Nos2 and Ccl5 by IFN-γ which peaked at >4 hours post-stimulation, could be inhibited by the addition of IL-4 even after IFN-γ stimulation (Supplementary Fig. 1e). In some cases, as for Ccl24, co-stimulation increased gene expression above the levels obtained with individual stimuli (Fig. 1a, b), but this occurrence was uncommon. Finally, IL-4-mediated inhibition of IFN-γ-induced gene expression was abrogated in STAT6-/- BMDMs (Supplementary Fig. 3), suggesting it required STAT6. Overall, these data indicate that IFN-γ and IL-4 exert a mutual transcriptional antagonism in BMDMs.
Fig. 1. Effects of the co-stimulation on the transcriptional programs induced by IFN-γ and IL-4.
(a,b) Scatter plots showing inhibition of IFN-γ- (left panel, blue) or IL-4-stimulated genes (right panel, green) upon co-stimulation for 2 hours (a) or 4 hours (b). Genes selectively induced by IFN-γ or IL-4 were identified based on an FDR ≤ 0.01 and a fold change ≥ 2 versus untreated. Orange dots indicate genes whose inhibition by co-stimulation is statistically significant (FDR ≤ 0.01) and with a fold change ≥ 2 versus IFN-γ or IL-4 alone, respectively. (c) Representative RNA-seq snapshots of genes induced by IFN-γ (Nos2, Ccl5) or IL-4 (Arg1) and sensitive to co-stimulation. (d) Expression kinetics by RT-qPCR of selected genes induced by IFN-γ (Ccl5, Nos2 and Irg1) or IL-4 (Arg1) and sensitive to co-stimulation.
Lack of mutual IFN-γ vs. IL-4 antagonism on signaling pathways
We next investigated the mechanistic bases of the interplay between IFN-γ and IL-4 in BMDMs. IFN-γ-induced phosphorylation of STAT1, ERK1/2 or AKT was unaffected by co- or pre-administration of IL-4, and induction of STAT6 phosphorylation occurred normally in the presence of IFN-γ (Supplementary Fig. 4a). Moreover, the conditioned supernatant from IL-4-treated BMDM, in which IL-4 was depleted with a neutralizing antibody, was unable to inhibit IFN-γ-stimulated induction of Nos2 and Ccl5 (Supplementary Fig. 4b), indicating that the inhibitory effects of the co-stimulation were not mediated by the autocrine activities of soluble molecules released in the culture medium. Because in vivo macrophages may be exposed to different cytokines in sequential or temporally distinct waves, we determined whether the IL-4-mediated inhibition persisted after its removal from the cell culture medium. BMDMs were conditioned with IL-4 for 4h, washed and stimulated with IFN-γ 24h after the addition of fresh medium. In all tested cases, transient pre-conditioning with IL-4 reduced the expression of IFN-γ-induced genes (Supplementary Fig. 4c), indicating a short-term memory effect of macrophage conditioning by IL-4. Collectively, these data indicate a cell-intrinsic regulatory mechanism controlling the IL-4-IFN-γ crosstalk downstream of cytokine-induced signaling pathways.
Mutual repression of cytokine-induced histone acetylation
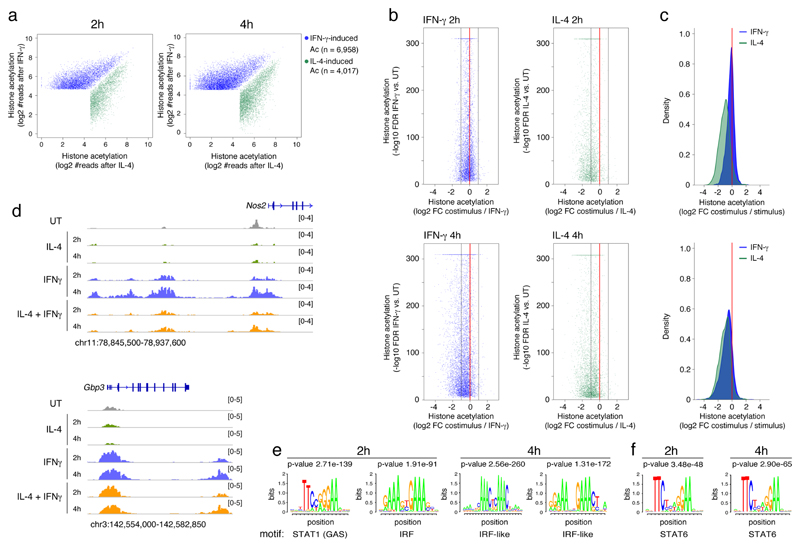
Because individual genes showed differential sensitivity to the inhibitory effects of the co-stimulation, we investigated if the cis-regulatory elements (enhancers and promoters) controlling the transcription of co-stimulation-sensitive genes had a different composition and organization than those controlling co-stimulation-resistant genes. We used chromatin immunoprecipitation coupled to next-generation sequencing (ChIP-seq) to map genome-wide changes in histone acetylation (H3K27Ac) in BMDMs stimulated with IFN-γ, IL-4 or their combination for 2 and 4 hours. Histone acetylation is associated with active cis-regulatory elements 21, 22 and is dynamically regulated in response to acute stimulation 7, 23, reflecting changes in the binding or activity of sequence-specific transcription factors. We focused on the inducible acetylation events at 2h and 4h that were selectively activated by either IFN-γ or IL-4 (Fig. 2a, Supplementary Table 3). The following main trends were evident. First, co-administration of IFN-γ and IL-4 caused a global reduction of inducible H3K27Ac triggered by either IFN-γ or IL-4 alone (Fig. 2b). Second, the repressive effects caused by IFN-γ on the IL-4 program were of higher magnitude than those exerted by IL-4 on the IFN-γ program, particularly at 2h (Fig. 2c), suggesting a dominance of IFN-γ over IL-4. Third, IFN-γ and IL-4 appeared to repress deposition of H3K27Ac by the antagonistic stimulus in a temporally different manner (Fig. 2b, c). While repression of IL-4-induced acetylation by IFN-γ was already maximal at 2h post-stimulation, IL-4-dependent repression of IFN-γ responses was only detectable at 4h, suggesting the involvement of transcription factors acting late in the response, such as those whose expression is induced by IFN-γ. As representative examples, acetylation of several enhancers upstream of the Nos2 gene was induced by IFN-γ and suppressed by IL-4 co-stimulation, while acetylation induced by IFN-γ in the Gbp3 locus was not significantly impacted by IL-4 (Fig. 2d).
Fig. 2. Cross-regulation of histone acetylation induced by IFN-γ and IL-4 by co-stimulation.
(a) Scatter plots showing H3K27 histone acetylation events (identified using SICER) (Xu et al., 2014) specifically induced by stimulation with IFN-γ (blued dots) or IL-4 (green dots) at 2h or 4h.
(b) Scatter plots showing inhibition of IFN-γ- (left panels, blue) or IL-4-stimulated (right panels, green) histone acetylation upon co-stimulation at 2 (top panels) or 4 hours (bottom panels). The Y-axis reports the -log10 of the FDR obtained by SICER when comparing acetylation in cells stimulated with IFN-γ or IL-4 with acetylation in unstimulated cells. The X-axis shows the log2 of the fold change in acetylation measured in stimulated vs. co-stimulated cells. (c) Density plots showing the effects of the co-stimulation on histone acetylation induced by IFN-γ (blue) or IL-4 (green) after 2 hours (top) or 4 hours (bottom). (d) ChIP-Seq snapshots showing representative genomic regions in which acetylation induced by one stimulus is affected (Nos2 locus, left) or not affected (Gpb3 locus, right) by co-stimulation. (e-f) Over-representation analysis of consensus TF motifs within regulatory elements induced by IFN-γ (e) or IL-4 (f) at the indicated time points.
The transcription factors associated with regions acetylated in response to IFN-γ partially differed at 2h and 4h (Fig. 2e and Supplementary Table 4). At 2h, the most over-represented motif was the canonical STAT1 binding site (GAS, gamma-activated sequence), which enables direct binding of STAT1 homodimers, followed by an IRF motif to which STAT1 is recruited via complexes with an IRF protein that provides DNA binding specificity 24 (Fig. 2e). At 4h, the most overrepresented motifs in regions acetylated in response to IFN-γ were variants of canonical IRF sites, while STAT1 motifs were not retrieved anymore (Fig. 2e). To determine the molecular bases of this finding, we generated STAT1 ChIP-Seq datasets in BMDMs that were either untreated or stimulated with IFN-γ for 2h and 4h. Consistent with the motif discovery analysis, STAT1 binding was increased genome-wide at 2h post-stimulation and returned to baseline level at 4h along with a reduction in phospho-STAT1 (Supplementary Fig. 5a-c). At the genomic regions where H3K27Ac was induced by IL-4 stimulation, a canonical STAT6 was the most over-represented motif at both time points (Fig. 2f).
To obtain additional insight into the cross-regulation between IFN-γ and IL-4, we analyzed the genomic distribution of STAT1 and STAT6 in BMDMs. 14,576 STAT1 peaks were detected 2h after IFN-γ stimulation (Fig. 3a) and co-stimulation with IL-4 had a marginal impact (Fig. 3b, Supplementary Table 5), indicating that the antagonistic effect of IL-4 on IFN-γ-induced histone acetylation cannot be ascribed to a reduced association of STAT1 with its genomic targets. Consistently, STAT1 and STAT6 binding events were similarly frequent in proximity of IL-4-resistant and IL-4-sensitive genes and they were unaffected by co-stimulation. 23,306 STAT6 peaks were detected above threshold in IL-4 stimulated BMDMs (Fig. 3a). Co-treatment with IFN-γ caused a trend towards a reduction of STAT6 association with chromatin, although IL-4-induced binding was in general maintained (Fig. 3c, d). As examples, the reduction of acetylation induced by IL-4 at the Ccl5 and the Rsad2/Cmpk2 locus was not associated with detectable changes in STAT1 occupancy (Fig. 3e). The Arg1 locus showed mild but detectable attenuation of STAT6 recruitment upon co-stimulation with IFN-γ (Fig. 3e). The DNA sequences enriched in STAT1 peaks were both GAS and IRF motifs, suggesting that its recruitment occurs both via direct and IRF-mediated DNA binding, while those associated with STAT6 peaks included canonical STAT6 motifs and IRF/PU.1-like motifs. Overall, these data indicate that both at the transcriptome and epigenome level, co-stimulation induced a continuum of inhibitory effects over the changes caused by individually administered IFN-γ or IL-4.
Fig. 3. STAT1 and STAT6 binding in response to IFN-γ, IL-4 or co-stimulation.
(a) Scatter plot showing STAT1 peaks induced by IFN-γ and not overlapping with STAT6 peaks (blue dots), and STAT6 peaks induced by IL-4 and not overlapping STAT1 (green dots). Overlapping STAT1/STAT6 peaks are represented by red dots. (b) STAT1 binding in cells stimulated with IFN-γ +/- IL-4 for 2h. (c) STAT6 binding in cells stimulated with IL-4 +/- IFN-γ for 2h. (d) Density plot showing the overall effects of the co-stimulation on STAT1 (blue) or STAT6 (green) genomic binding induced by IFN-γ or IL-4. (e) Representative snapshots showing binding of STAT6 and STAT1 and H3K27Ac signals after single or combined stimulation at cross-regulated genomic loci.
DNA sequence features of inducibly acetylated genomic regions
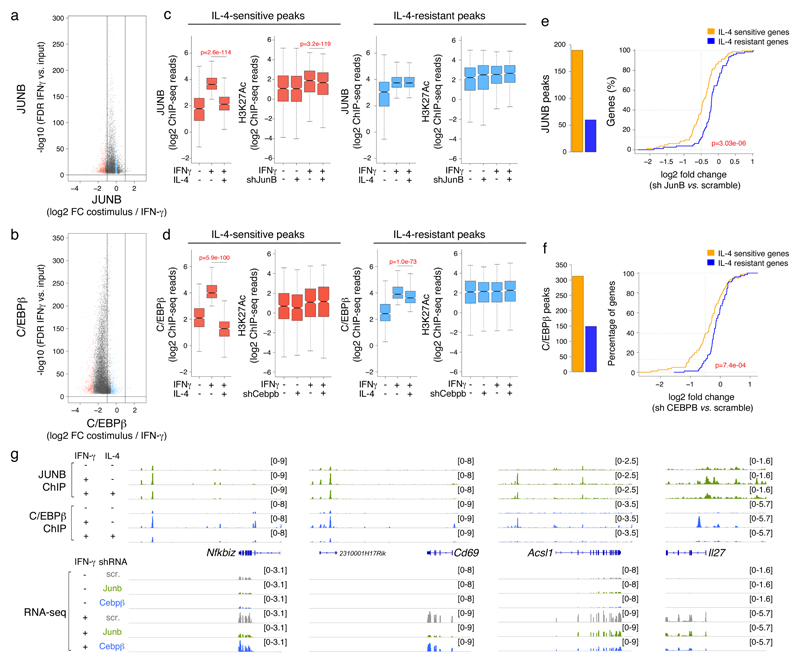
To dissect the genomic bases of the different sensitivity of cis-regulatory elements to the inhibitory effects of the co-stimulation, we defined discrete sub-groups of enhancers with clearly differing behaviors and compared them with each other. We first focused on IFN-γ-activated elements. At one extreme, we identified 773 genomic regions whose acetylation was induced by IFN-γ and was highly sensitive to the inhibitory effects of the co-stimulation (Fig. 4a, b). At the other end, we identified 736 IFN-γ-inducible cis-regulatory elements that were completely resistant to the co-stimulation (Fig. 4a-b, Supplementary Table 6). We next used motif enrichment analyses to identify over-represented transcription factor motifs in the groups of IL-4-sensitive vs. IL-4 resistant enhancers. When compared with the FANTOM5 collection of active enhancers25 (Fig. 4c, Supplementary Table 7), both the IL-4-sensitive and the IL-4-resistant subsets of IFN-γ-inducible enhancers showed a similarly significant over-representation of STAT and IRF motifs. Therefore, the molecular bases for the responsiveness of both classes of elements to IFN-γ stimulation appear to be similar and linked to the activation of STAT1-IRF124. However, when directly comparing IL-4-sensitive with IL-4-resistant enhancers, we noticed the over-representation of AP-1, ATF, C/EBP and NF-κB motifs in the IL-4-sensitive group (Fig. 4c, Supplementary Table 8). IL-4-resistant elements did not show any strongly significant enrichment relative to the IL-4-sensitive ones. Overall, the main difference between IL-4-resistant and IL-4-sensitive elements activated by IFN-γ appeared to be the presence in the IL-4-sensitive group of motifs recognized by transcription factors other than STAT1 and IRF1.
Fig. 4. Characterization of IL-4 sensitive and IL-4-resistant groups of enhancers induced by IFN-γ.
(a) Scatter plot showing the identified groups of IFN-γ-induced acetylated regions highly sensitive (orange dots, n=773) or completely resistant (blue dots, n=736) to IL-4 inhibition. (b) Box plots showing the quantification of histone acetylation changes in the IL-4-sensitive and resistant groups defined in a). *** p = 3.53e-128. (c) Over-representation analyses of transcription factor motifs within IL-4-sensitive and resistant regions either relative to the FANTOM5 collection of active enhancers or in the cross-comparison. The complete analyses are reported in Supplementary Tables 7 and 8. (d) mRNA expression (from RNA-seq data) of selected TFs belonging to IRF, AP-1 and CEBP families. Bars indicate means +/- SE. (e) Genomic snapshots of the JunB and Cebpb loci showing binding of both STAT6 and STAT1 as well as RNA levels. Adjacent genes are shown for comparison.
Transcription factors mediating the inhibitory effects of IL-4
These data led us to investigate whether a genomic response mediated exclusively by STAT1 and IRF1 may by default be resistant to the inhibitory effects of IL-4, and conversely, whether the dependence of a cis-regulatory element on additional transcription factors other than STAT1-IRF1 (such as AP-1 and C/EBP), determined its vulnerability to IL-4. First, we analyzed in BMDMs the genomic distribution of IRF1, whose mRNA was induced by IFN-γ in a sustained and IL-4-resistant manner (Supplementary Fig. 4a, Fig. 4d) and found that, similarly to STAT1, it was largely unaffected by IL-4 co-stimulation (Supplementary Fig. 6). We next analyzed the expression or activity in BMDMs stimulated with IFN-γ without or with IL-4, of auxiliary transcription factors whose consensus DNA binding sites were over-represented in the IL-4-sensitive enhancers. Among them, Cebpb and JunB were induced by IFN-γ and repressed by IL-4 costimulation (Fig. 4d, Supplementary Table 7). Moreover, the Junb and Cebpb loci were bound by both STAT1 and STAT6 in response to co-stimulation (Fig. 4e), suggesting direct cross-regulation. These observations prompted us to determine the role of C/EBPβ and JUNB as mediators of the transcriptional program induced by IFN-γ and of its crosstalk with IL-4. We used ChIP-Seq to analyze the genomic distribution of JUNB and C/EBPβ in BMDMs stimulated for 4 hours with IFN-γ in the absence or presence of IL-4, and found that the binding of both transcription factors to DNA was significantly reduced in co-stimulated BMDMs as compared to those stimulated with IFN-γ alone (Fig. 5a, b). We next analyzed the IFN-γ-induced histone acetylation and gene expression in BMDMs depleted of Junb or Cebpb through lentivirus-mediated delivery of interfering short hairpin RNAs (Supplementary Fig. 7). The IFN-γ-inducible JUNB and C/EBPβ ChIP-seq peaks were sorted into discrete subgroups, namely those most inhibited and those least affected by IL-4 (Fig. 5a-d); then, we analyzed the effects of Junb or Cebpb depletion on H3K27Ac levels in response to IFN-γ at the two groups of regions. Knock down of Junb resulted in a significant, albeit moderate reduction in H3K27Ac at genomic regions where JUNB binding was strongly inhibited by IL-4, but not at those where JUNB binding was IL-4-resistant (Fig. 5c), indicating that JUNB binding contributes to the deposition of H3K27Ac in response to IFN-γ and that its depletion partially phenocopies the inhibition by IL-4 co-stimulation. Instead, Cebpb knock down had no clear effect on IFN-γ-induced histone acetylation at sites of IL-4-sensitive or IL-4-resistant C/EBPβ occupancy (Fig. 5d). JUNB and, to a lesser extent, C/EBPβ peaks were found more commonly in the proximity of IL-4-sensitive than IL-4-resistant genes that were induced by IFN-γ (Fig. 5e-f, Supplementary Table 9). Notably, depletion of Junb and Cebpb preferentially impaired the activation of IFN-γ-inducible genes that were sensitive to IL-4 inhibition (Fig. 5e-f, Supplementary Table 10). As an example, Nfkbiz and Cd69 were dependent on JUNB for their inducibility by IFN-γ, while Acsl1 and Il27 were dependent on CEBPβ and to a lesser extent on JUNB (Fig. 5g). Taken together, these data indicate that IFN-γ-inducible genes whose activation requires auxiliary transcription factors (such as JUNB and C/EBPβ) in addition to STAT1 and IRFs, were vulnerable to the inhibitory effects of IL-4.
Figure 5. Analysis of the role of JUNB and C/EBPβ in the IFN-γ response and IL-4-mediated antagonism.
(a-b) Scatter plots reporting ChIP-seq data for JUNB (a) and C/EBPβ (b) in macrophages stimulated for 4h with IFN-γ in the presence or absence of IL-4. Red and light blue dots indicate peaks that were highly sensitive or relatively resistant to IL-4-mediated inhibition, respectively. (c-d) For each TF, data were divided in two groups based on sensitivity or resistance of transcription factor binding to IL-4-mediated inhibition. In each group, the box plot on the left shows transcription factor ChIP-seq read counts in the three indicated conditions. The box plot on the right shows histone acetylation read counts in untreated or IFN-γ-treated cells transduced with scrambled or TF-specific lentiviral shRNA. (e-f) Left: JUNB and C/EBPβ peaks in proximity of IFN-γ-inducible genes that are either sensitive or resistant to IL-4-mediated inhibition. Right: empiric cumulative distribution functions (ECDFs) showing the effects of Junb (e) or Cebpb (f) depletion on the induction by IFN-γ of IL-4-sensitive and resistant genes.
Genomic roles of Myc in the interplay between IL-4 and IFN-γ
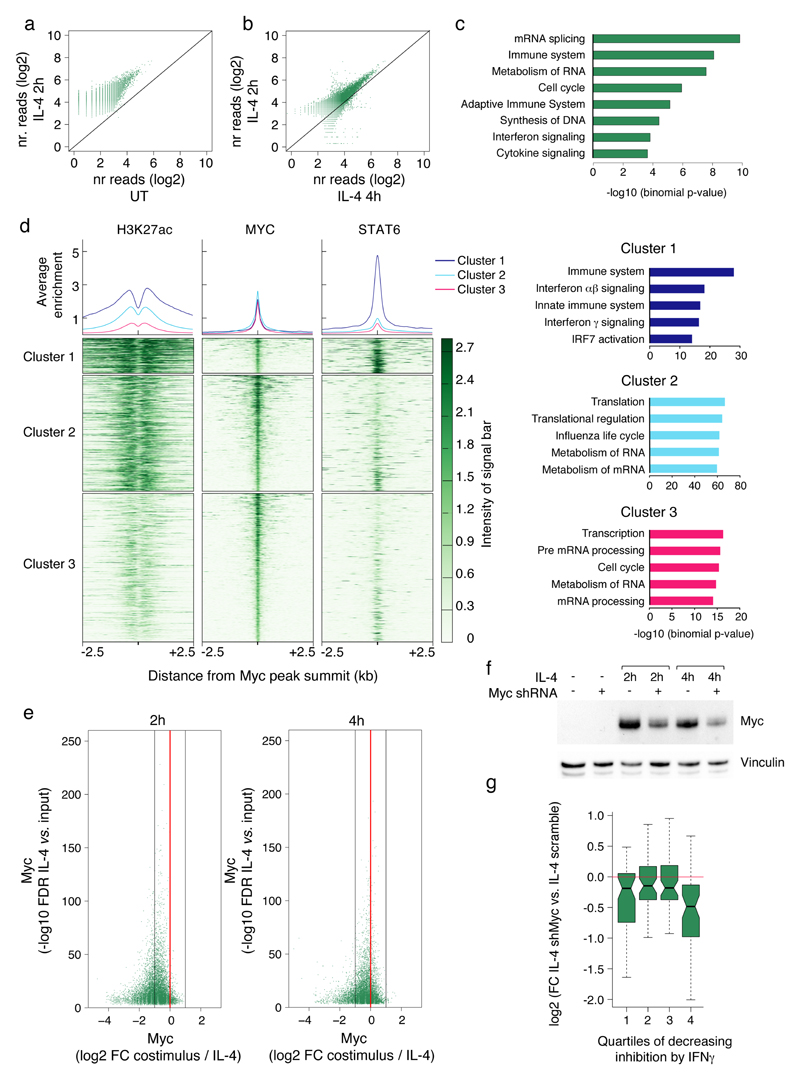
We next analyzed over-represented transcription factor motifs in the sets of IL-4-activated cis-regulatory regions whose acetylation was most or least sensitive to IFN-γ-mediated inhibition (Fig. 6a, b). Because of the strong inhibitory effect of IFN-γ on the genomic response induced by IL-4, we could only analyze a small set of 317 acetylated regions resistant to cross-inhibition by IFNγ. When analyzed against the FANTOM 5 collection of active enhancers, the IFN-γ-sensitive group was strongly enriched for the STAT6 motif (Fig. 6c, Supplementary Table 11), consistent with the genome-wide attenuation of STAT6 binding in BMDMs co-stimulated with IFN-γ. When compared directly with the set of IFN-γ-resistant regulatory elements, the IFN-γ-sensitive enhancers were enriched for MAF motifs (Fig. 6c), which may relate to the inhibitory effect of IFN-γ on IL-4-mediated induction of MafB (Supplementary Table 2). Conversely, the most enriched motif in the IFN-γ-resistant enhancers was the canonical E-box recognized by Myc (Fig. 6c), which is induced by IL-4 26. The STAT6 DNA binding site was also over-represented in this subset of genomic regions but its enrichment was less statistically significant than that of Myc (Supplementary Table 11). Consistently, E-boxes were also over-represented in IFN-γ-resistant compared to IFN-γ-sensitive elements, together with motifs with high G+C content, which relates to the high frequency of E-boxes in CpG islands (Fig. 6c, Supplementary Table 12). Myc mRNA and protein expression were induced by IL-4 (Fig. 6d-e), probably via direct STAT6 binding to the Myc promoter (Fig. 6d), and this response was largely preserved in BMDMs co-stimulated with IFN-γ. These data suggest that Myc, a transcription factor mainly associated with control of proliferation, may participate in the IL-4 response downstream of STAT6 and that the Myc-dependent genomic regulation of IL-4-activated enhancers may confer robustness to antagonistic signals.
Figure 6. Analysis of the cis-regulatory bases for inhibition of IL-4-mediated macrophage activation by IFN-γ.
(a) Scatter plot showing a group of IL-4-induced acetylated regions highly sensitive (orange dots) or resistant (green dots) to IFN-γ-dependent inhibition. (b) Box plots showing the quantification of histone acetylation changes in the IFN-γ-sensitive and resistant groups. *** p = 2.004e-60. (c) Over-representation of transcription factor motifs in IFN-γ-sensitive and resistant regions relative to the FANTOM5 collection of active enhancers or in the cross-comparison. The top ranked motif in each comparison is reported. The complete analysis is reported in Supplementary Table 11. (d) Snapshot showing the epigenomic and transcriptional organization of the Myc locus in cells treated with IL-4 +/- IFN-γ. (e) Anti-Myc western blot in macrophages treated with IL-4 +/- IFN-γ as indicated.
To directly address these possibilities, we analyzed Myc genomic distribution in BMDMs left untreated or treated with IL-4 for 2h or 4h. While Myc bound to a limited number of sites (433 peaks over background) in unstimulated BMDMs, it generated 11,357 peaks at 2h and 8,747 at 4h post-stimulation (Fig. 7a, Supplementary Table 13). Myc binding intensity decreased over time (Fig. 7b), along with the mild reduction in protein levels (Fig. 6e). Myc binding occurred in the vicinity of both genes associated with housekeeping functions, such as mRNA metabolism and cell cycle, and genes linked to immune system-related GO terms, such as immune response and interferon signaling (Fig. 7c). Clustering analysis showed that Myc-bound regions could be divided into a small cluster (cluster 1, 11.6% of peaks) associated with strong STAT6 recruitment, high H3K27Ac, the STAT6 motif and the E-box motif; and two larger groups with limited or negligible STAT6 binding, high (cluster 2) or low (cluster 3) acetylation, and a strong over-representation of the E-box motif (Fig. 7d, Supplementary Table 14). GO terms associated with immune responses were selectively enriched in cluster 1, while cluster 2 and 3 were both enriched for canonical Myc-associated GO terms, such as RNA metabolism and cell cycle (Fig. 7d, Supplementary Table 15). BMDMs co-stimulated with IL-4 and IFN-γ showed a reduction of Myc genomic occupancy compared to IL-4-stimulated BMDMs (Fig. 7e), consistent with the partial reduction of its mRNA and protein levels upon co-stimulation.
Figure 7. Involvement of Myc in the IL-4-dependent response.
(a) Myc genomic occupancy in untreated vs. IL-4-treated (2h) macrophages. (b) Myc genomic occupancy in macrophages treated with IL-4 for 2h and 4h. (c) Selected gene-ontology (GO) categories retrieved from GREAT analysis of the Myc ChIP-seq data set (IL-4 2h). (d) Myc peaks (IL-4 2h) were clustered into three groups together with histone acetylation and STAT6 ChIP-seq data. The average enrichment plot (top panels) and the heatmaps were centered on the summit of Myc peaks and extended for + 2.5kb. Gene ontology categories associated with the three Myc clusters (shown on the right) were retrieved using GREAT (supplemental methods). (e) Effects of IFN-γ co-treatment on Myc genomic occupancy in IL-4-treated macrophages at 2h (left) and 4h (right). (f) Anti-Myc western blot in cells transduced with a Myc-specific shRNA and treated with IL-4 as indicated. (g) Effects of Myc depletion on IL-4-induced gene expression. IL-4-inducible genes were divided into quartiles based on the inhibitory effect of IFN-γ co-stimulation (the first quartile representing the one with the strongest inhibitory effect and the fourth the one including the least inhibited genes).
To directly address the role of Myc in the IL-4 response, we used lentiviral-mediated delivery of a Myc-specific shRNA or a scramble control (Fig. 7f) and used RNA-seq to analyze the macrophage response to IL-4 at 2h and 4h. The IL-4-induced genes were assigned to four quartiles on the basis of their sensitivity to the inhibitory effects of IFN-γ (determined in the experiments described before), the first quartile including the most IFN-γ-inhibited genes and the fourth quartile the least IFN-γ-inhibited ones. Although Myc knock down showed broad effects of low magnitude, it impacted more evidently on the most IFN-γ-resistant IL-4-induced genes (Fig. 7g). Overall, Myc depletion reduced the expression of 54 IL-4-inducible genes (Supplementary Table 16), which included mainly IFN-γ-resistant genes, such as Ch25h, Cd274, Ccl2, Ccl7, Ccl12, Flt1, Serpina3f.
Discussion
In this study, we identified key principles governing the interplay between stimuli that drive macrophage function towards contrasting programs. This framework may provide the basis for a more systematic mechanistic analysis of the interplay among macrophage activators in physiology and disease.
The IL-4- and IFN-γ-driven polarization states entail distinct and non-overlapping gene expression changes that reflect the dynamic evolution of the transcriptional regulatory circuits activated by polarizing cytokines. Upon co-stimulation, IFN-γ and IL-4 could still induce their specific gene expression programs and the associated epigenomic changes. However, IL-4 and IFN-γ co-stimulation extensively, yet only partially attenuated the responses elicited by each stimulus alone. Overall, the IFN-γ- and the IL-4-induced programs could co-exist to a large extent, but in conditions of IL-4 and IFN-γ co-stimulation macrophages are stretched between two opposite functional poles, resulting in the reciprocal attenuation of both of them. In a sense, the co-delivery of the two polarizing cytokines showed the remarkable extent, and at the same time the maxima to which macrophage plasticity can be driven.
Both the transcriptional and epigenomic impact of co-stimulation was not homogeneous, but instead more evident at subsets of genes and cis-regulatory elements. Specifically, the induction of a small number of M1 or M2 signature genes, such as Nos2 and Arg1, respectively was almost completely abolished in the presence of the antagonistic cytokine, while many others, such as typical IFN-stimulated antiviral genes were unaffected. Robustness of the IFN-γ-elicited program is essential to prevent an extreme increase in susceptibility to infections when type 2 immunity is concomitantly activated. At the same time, the inhibitory activity of IL-4 on the IFN-γ-activated program and the suppression of some genes involved in microbial killing, such as Nos2, can explain the negative impact of type 2 immunity on resistance to some infections.
Although the cross-inhibition between IL-4 and IFN-γ likely entails multiple regulatory circuits, we identified some basic mechanistic principles. Simple cis-regulatory elements containing only the minimal motifs responsive to IFN-γ, namely GAS and IRF sites, were resistant to the antagonistic effects of IL-4. Conversely, susceptibility to IL-4 inhibition was limited to complex cis-regulatory elements containing, in addition to GAS and IRF sites, motifs for auxiliary transcription factors such as JUNB, whose expression was induced by IFN-γ and antagonized by IL-4 co-stimulation. Such different genomic organization of cis-regulatory elements controlling IFN-γ-activated genes reflects an optimal solution to two opposing needs that become evident when macrophages are exposed to multiple stimuli: maintaining the robustness of IFN-γ-stimulated responses critical for survival to infections, while at the same time enabling the plasticity required to modulate macrophage properties in response to complex inputs12.
Antagonistic regulation of IL-4-induced gene expression and chromatin changes by IFN-γ follows different rules, as STAT6 binding to cognate sites was globally attenuated by IFN-γ. While this finding is consistent with an overall dominance of the IFN-γ- over the IL-4-induced program, the mechanisms underlying this effect are still unclear. Our data also demonstrate the involvement of Myc, a hub in the control of cell proliferation27, in the IFN-γ-resistant component of the IL-4 response. Myc was induced by IL-4, bound a set of genomic regions largely distinct from those occupied by STAT6, and contributed to the activation of a large panel of IL-4-inducible genes. Because IL-4 is required for macrophage proliferation during parasitic infections 28, 29, it is likely that Myc contributes to this proliferative response. However, the requirement for Myc in IL-4-induced activation of many immune response genes, such as CCL-family chemokines, hints at a broader role in the IL-4 response.
An additional intriguing aspect is the possibility that STAT1 and STAT6 may exert a direct mutual control on each other's activity at a large number of genomic regulatory elements. In response to IFN-γ, STAT1 also bound many regulatory elements associated with IL-4-inducible genes. Analogously, IL-4-activated STAT6 bound a sizeable fraction of the regulatory elements of IFN-γ-activated genes. Overall, a large fraction of STAT1 and STAT6 binding events occurred at overlapping or nearby genomic regions. These data hint at a genomic organization that enables mutual inhibitory interactions between STAT1 and STAT6. Whether different combinations of signals with pro- and anti-inflammatory functions will produce similar outcomes will require additional investigations. In this context, IFN-γ and lipopolysaccharide have been shown to act synergistically to enhance induction of cytokine production in macrophages through a process dependent on chromatin priming 30.
The interplay between IFN-γ and IL-4 has important biological implications, since these cytokines frequently coexist in vivo. The crosstalk between these cytokines may be involved in the transition from pro- to anti-inflammatory programs during resolution of inflammation; it may underlie some of the pathogenic effects observed during co-infections with pathogens eliciting type 1 or type 2 immune responses; and it may explain complex and diverging phenotypes of macrophages associated with different tumors.
Online methods
Mice and Cell cultures
Animal experiments were performed in accordance with the Italian Laws (D.L.vo 116/92), which enforce the EU 86/609 Directive. Bone marrow-derived macrophage (BMDM) were prepared ex vivo from mouse bone marrow cells cultured in L929-conditioned medium containing M-CSF as described 31. After 6 days in culture, adherent cells were >99% Cd11b+ F4/80+ as assessed by flow cytometry. Infections were carried out as described 31. Junb and Cebpb lentiviral shRNA vectors were gifts of Ido Amit (Weizman Institute). BM cells from Stat6-deficient mice were a kind gift of Dr. David Voehringer (University Hospital Erlangen, Germany).
Cytokines and antibodies
Recombinant mouse IFN-γ and IL-4 were from R&D Systems. Unless otherwise stated, cytokines were used at a concentration of 100ng/ml and 10ng/ml, respectively. Anti-IL4 blocking antibody (clone 11B11) was from eBioscience. CD11B (clone M1/70), F4/80 (BM8), NOS2 (CXNFT) antibodies for flow cytometry were from eBioscience. Antibodies used in ChIP experiments against STAT1 (sc-592), STAT6 (sc-981), MYC (sc-764), JUNB (sc-46x), C/EBPβ (sc-150X) were all from Santa Cruz. The anti-H3K27Ac (ab4729) antibody was from Abcam. The anti-Pu.1 rabbit polyclonal antibody was generated in-house against the N-terminus of Pu.1 (aa. 1-100; NP_035485.1) 32. Antibodies used in western blot experiments: Stat1 (#9172), phospo-Stat1 (Tyr701, #9171), Stat6 (#9362), phospho-Stat6 (Tyr641, #9361), Erk1/2 (#9102), phospho-Erk1/2 (Thr202/Tyr204, #9101), phospho-Akt (Ser473, #9271); anti-actin (A4700) from Sigma-Aldrich. The following antibodies were used for flow cytometry: CD11b (clone M1/70), F4/80 (BM8), Nos2 (CXNFT) from eBioscience; phospho-STAT1-AlexaFluor 647 (Cat. 612597) and phospho-STAT6-PE (Cat. 558252) from BD Biosciences. The following antibodies were used for immunofluorescence: Alexafluor 488 goat anti-rabbit secondary antibody (Cat. R37116) and DAPI (Cat. 62247) from Thermo Scientific; phospho-STAT1-AlexaFluor 647 (Cat. 612597) and phospho-STAT6-PE (Cat. 558252) from BD Biosciences.
Flow cytometry intracellular staining
After cytokine stimulation for the indicated time points, BMDM were collected and stained with LYVEDEAD Fixable Yellow (L34959, Molecular Probes) for dead-cell exclusion and with anti-Cd11b. For Nos2 intracellular staining cell fixation and permeabilization were performed using buffers and protocols from eBiosciences (00-8222-49/56). For phospho-STAT1 and phospho-STAT6 analyses, cells were fixed with BD PhosflowTM Fix Buffer I (Cat. No. 557870) at 37°C for 10 minutes, washed and permeabilized with BD PhosflowTM Perm Buffer III (Cat. No. 558050). Cells were resuspended in FACS buffer and analyzed on a FACS Canto II (BD Biosciences). The specificity of the staining was verified with isotype, unstained, and fluorescence minus one (FMO) controls. Data were analyzed with FACS DIVA Software 6.1.2 (BD Biosciences) and FlowJo Software 9.3.2.
Immunofluorescence analyses
Bone marrow cells were plated on coverslips within a six-wells plate at 105 cells/well and differentiated for 6 days. BMDM were then stimulated with IFNγ, IL-4, or both cytokines, washed with cold PBS and fixed with 1% Paraformaldehyde (PFA) in PBS at room temperature (RT) for 10 minutes. Fixed cells were then permeabilized with cold methanol (100%) for 10 minutes at -20°C, washed three times with 0.3% Triton X-100/PBS, and blocked for 1 hour with blocking buffer (5% BSA in 0.3% Triton-X100/PBS). For the analysis of phospho-STAT1 and phospho-STAT6, cells were stained with conjugated antibodies (1:20) overnight at 4°C. Cells were counterstained with 4',6-diamidino-2-phenylindole, dihydrochloride (DAPI, 100µg/mL) for 10 minutes at RT, and mounted with Aqua/Poly mount (Polysciences, Cat. 18606) on slides. Images were taken on a Nikon Eclipse E600 microscope (1024x1024, 40hex) or on a Leica TCS SP2 confocal microscope (1024x1024, 40hex, 2x optical zoom) and analysed with Fiji Image J software (v 2.0.0-rc-43).
ChIP-seq
ChIP was carried out as previously described 7. Briefly, 1-3x106 (for H3K27ac ChIP-Seq) or 5-10x107 (for TFs) macrophages were fixed with 1% formaldehyde and nuclear fractions isolated and lysed. After chromatin shearing by sonication, lysed nuclei were incubated overnight at 4°C with protein G Dynabeads (Life Technologies) previously coupled with 1-5 μg of antibody. Beads were recovered using a 96-wells magnet, washed extensively, and DNA eluted and decrosslinked overnight at 65°C. DNA was purified with Solid-phase reversible immobilization (SPRI) beads (Agencourt AMPure XP, Beckman Coulter) and quantified with PicoGreen (Life Technologies). 1-5 ng of ChIP DNA was used for ChIP-Seq library preparation, using a previously described protocol 33 with minor modifications 7 and sequenced on a HiSeq2000 (Illumina).
RNA analyses
Total RNA was isolated using RNEasy Mini Kit (Qiagen) with in-column DNase treatment, quantified with Nanodrop (ThermoScientific) and reverse transcribed using ImProm-II RT (Promega) according to manufacturers’ instructions. 10ng of cDNA were used for amplification. mRNA-Seq library preparation from 1 μg of total RNA was performed with the TruSeq RNA Sample Prep kit (Illumina) and sequenced on a HiSeq2000 (Illumina).
Computational methods
Single-end reads (51 nt) were quality-filtered according to the Illumina pipeline and mapped to the mm10 reference genome. For ChIP-Seq, reads were mapped using Bowtie2 v 2.2.6 34. We used default parameters with the options --very-sensitive, --no-unal and with the pre-built bowtie2 index. Only uniquely mapping reads were retained. Peak calling for acetylation was performed using SICER v1.1 35 using a redundancy threshold of 1, a window size of 200bp, a gap size of 600 bp and a False Discovery Rate (FDR) cutoff of 1 × 10−3. Fragment size was set to 150 and the effective genome fraction to 0.80. Peak calling for TFs was performed using MACS2 v.2.1.0.20150731 36, using default parameters with the exception of the bandwidth (–bw 100), the minimum q-value cutoff for peaks detection (-q 0.01) and the effective genome size (-g 1.87e9). For RNA-Seq, single-reads were mapped using TopHat v2.1.0 using the option --b2-very-sensitive 37. Differential expression was evaluated with an exact test for the negative binomially distributed counts using edgeR v3.10.5 with limma v3.24.15 Bioconductor package38, 39, 40, 41. Detailed computational methods are described in the Supplementary note.
Supplementary Material
Acknowledgements
This project was supported by the European Research Council (Advanced ERC grant to GN); the Italian Ministry of University and Research (grant FIRB RBAP11H2R9 to GN); the Telethon Foundation (SR-Tiget Core Grant TGT16F04 to RO); the Cariplo Foundation (Giovani Ricercatori, 2015-0990 to RO) and the Italian Association for Research on Cancer (AIRC) to BA. We thank Silvia Monticelli (IRB) for critical feedback on the manuscript; L. Rotta, T. Capra, and S. Bianchi (IEO and IIT@SEMM) for the preparation and processing of the sequencing libraries; Francesco Cilenti (SR-TIGET) for help with immunofluorescence experiments; Ido Amit (Weizmann Institute) for the JunB and Cebpb shRNA vectors; D. Voheringer for the STAT6-/- bone marrow cells.
Footnotes
Author contributions
Conceptualization: VP, RO and GN. RO and AC designed and carried out most of the experiments. MG, SG, MS generated a subset of the data. VP analyzed all data and generated figure panels. AS and BA provided critical reagents. Supervision, GN. Funding acquisition, GN and RO. GN wrote the manuscript with inputs from all authors.
Competing financial interest
The authors declare no competing financial interests
Accession numbers. Raw data sets are available for download at the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/gds/) under the accession number GSE84520.
References
- 1.Glass CK, Natoli G. Molecular control of activation and priming in macrophages. Nature immunology. 2016;17:26–33. doi: 10.1038/ni.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murray PJ, et al. Macrophage Activation and Polarization: Nomenclature and Experimental Guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. The Journal of clinical investigation. 2012;122:787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lawrence T, Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nature reviews Immunology. 2011;11:750–761. doi: 10.1038/nri3088. [DOI] [PubMed] [Google Scholar]
- 5.Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nature reviews Immunology. 2011;11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ostuni R, et al. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 8.Monticelli S, Natoli G. Short-term memory of danger signals and environmental stimuli in immune cells. Nature immunology. 2013;14:777–784. doi: 10.1038/ni.2636. [DOI] [PubMed] [Google Scholar]
- 9.Netea MG, et al. Trained immunity: A program of innate immune memory in health and disease. Science (New York, N.Y.) 2016;352 doi: 10.1126/science.aaf1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lavin Y, et al. Tissue-Resident Macrophage Enhancer Landscapes Are Shaped by the Local Microenvironment. Cell. 2014;159 doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xue J, et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. 2014;40:274–288. doi: 10.1016/j.immuni.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allen JE, Maizels RM. Diversity and dialogue in immunity to helminths. Nature reviews Immunology. 2011;11:375–388. doi: 10.1038/nri2992. [DOI] [PubMed] [Google Scholar]
- 13.Elliott DE, Weinstock JV. Helminth-host immunological interactions: prevention and control of immune-mediated diseases. Annals of the New York Academy of Sciences. 2012;1247:83–96. doi: 10.1111/j.1749-6632.2011.06292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osborne LC, et al. Virus-helminth coinfection reveals a microbiota-independent mechanism of immunomodulation. Science. 2014;345:578–582. doi: 10.1126/science.1256942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reese TA, et al. Helminth infection reactivates latent  -herpesvirus via cytokine competition at a viral promoter. Science. 2014 doi: 10.1126/science.1254517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salgame P, Yap GS, Gause WC. Effect of helminth-induced immunity on infections with microbial pathogens. Nature immunology. 2013 doi: 10.1038/ni.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris J, et al. T helper 2 cytokines inhibit autophagic control of intracellular Mycobacterium tuberculosis. Immunity. 2007;27:505–517. doi: 10.1016/j.immuni.2007.07.022. [DOI] [PubMed] [Google Scholar]
- 18.Potian JA, et al. Preexisting helminth infection induces inhibition of innate pulmonary anti-tuberculosis defense by engaging the IL-4 receptor pathway. The Journal of experimental medicine. 2011;208:1863–1874. doi: 10.1084/jem.20091473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schleicher U, et al. TNF-Mediated Restriction of Arginase 1 Expression in Myeloid Cells Triggers Type 2 NO Synthase Activity at the Site of Infection. Cell Reports. 2016;15:1062–1075. doi: 10.1016/j.celrep.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ostuni R, Kratochvill F, Murray PJ, Natoli G. Macrophages and cancer: from mechanisms to therapeutic implications. Trends Immunol. 2015;36:229–239. doi: 10.1016/j.it.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 21.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaikkonen MU, et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Molecular cell. 2013;51:310–325. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nature reviews Immunology. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andersson R, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pello OM, et al. Role of c-MYC in alternative activation of human macrophages and tumor-associated macrophage biology. Blood. 2012;119:411–421. doi: 10.1182/blood-2011-02-339911. [DOI] [PubMed] [Google Scholar]
- 27.Kress TR, Sabo A, Amati B. MYC: connecting selective transcriptional control to global RNA production. Nat Rev Cancer. 2015;15:593–607. doi: 10.1038/nrc3984. [DOI] [PubMed] [Google Scholar]
- 28.Jenkins SJ, et al. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science. 2011;332:1284–1288. doi: 10.1126/science.1204351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sieweke MH, Allen JE. Beyond stem cells: self-renewal of differentiated macrophages. Science. 2013;342:1242974. doi: 10.1126/science.1242974. [DOI] [PubMed] [Google Scholar]
- 30.Qiao Y, et al. Synergistic activation of inflammatory cytokine genes by interferon-γ-induced chromatin remodeling and toll-like receptor signaling. Immunity. 2013;39:454–469. doi: 10.1016/j.immuni.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Austenaa LM, et al. Transcription of Mammalian cis-Regulatory Elements Is Restrained by Actively Enforced Early Termination. Molecular cell. 2015;60:460–474. doi: 10.1016/j.molcel.2015.09.018. [DOI] [PubMed] [Google Scholar]
- 32.Mancino A, et al. A dual cis-regulatory code links IRF8 to constitutive and inducible gene expression in macrophages. Genes & development. 2015;29:394–408. doi: 10.1101/gad.257592.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garber M, et al. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Molecular cell. 2012;47:810–822. doi: 10.1016/j.molcel.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zang C, et al. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson MD, Smyth GK. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics. 2008;9:321–332. doi: 10.1093/biostatistics/kxm030. [DOI] [PubMed] [Google Scholar]
- 41.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.