Abstract
Heparanase (HPSE) is the only mammalian endoglycosidase which has been widely implicated in cancer because of its capability to degrade heparan sulfate chains (HS) of HS proteoglycans (HSPGs). Specifically, the cell surface HSPG syndecans 1 and 4 (SDC1 and SDC4) are modulators of growth factor action and SDC4 is implicated in cell adhesion as a key member of focal adhesion complexes. We hypothesized that extracellular heparanase modulates brain metastatic breast cancer (BMBC) cell invasiveness by affecting cytoskeletal dynamics, SDC4 carboxy terminal-associated proteins, and downstream targets. We used two independently-derived human BMBC cell systems (MB-231BR, MB-231BR3), which possess distinct cellular morphologies and properties. Highly aggressive spindle-shaped 231BR3 cells changed to a round-cell morphology associated with expression of the small GTPase guanine nucleotide exchange factor-H1 (GEF-H1). We demonstrated that GEF-H1 is a new component of the SDC4 signaling complex in BMBC cells. Treatment with HPSE resulted in regulation of the SDC4/protein kinase Cα axis while maintaining a constitutive GEF-H1 level. Third, GEF-H1 knockdown followed by cell exposure to HPSE caused a significant regulation of activities of Rac1 and RhoA, which are GEF-H1 targets and fundamental effectors in cell plasticity control. Fourth, L-HPSE augmented expression of β1 Integrin in BMBC cells and of vascular cell adhesion molecule 1 (VCAM1; the major β1 Integrin receptor) in human brain microvascular endothelial cells. Finally, using a newly developed blood-brain barrier in vitro model, we show that BMBC cell transmigration was significantly reduced in GEF-H1 knockdown cells. These findings implicate heparanase in mechanisms of cytoskeletal dynamics and in the cross-talk between tumor cells and vascular brain endothelium. They are of relevance because they elucidate molecular events in the initial steps leading to BMBC onset and capturing distinct roles of latent and active HPSE in the brain microenvironment.
Keywords: heparanase, GEF-H1, Rac/Rho, cytoskeletal dynamics, brain metastatic breast cancer
Introduction
Patients with brain metastatic breast cancer (BMBC) have an exceptionally poor prognosis, making BMBC the most devastating and feared consequence of breast cancer. Despite increasing incidence of BMBC and its recognition as a high clinical priority, mechanisms causing brain metastasis of breast cancer are understudied and remain largely unknown, limiting the effectiveness of therapeutic interventions (1–3). Heparan sulfate proteoglycans (HSPGs) are ubiquitous macromolecules located in the extracellular matrix and on the cell surface, consisting of a core protein and covalently-attached HS. Syndecans and glypicans are families of cell surface HSPGs whose members regulate the cross-talk between tumor and host cells by acting as co-receptors for HS-binding factors. The syndecans are expressed by a family of four genes, SDC1–4 (4). Syndecans are type I transmembrane proteins oriented with an extracellular amino terminus and an intracellular carboxy terminus (4). Each of the SDC proteins has a unique extracellular and intracellular domain, thus allowing for distinct co-receptor phenotypes and intracellular signal transduction (4). Their functions place them at the center of cell signaling integration and are thus linked to tumor progression (5–8). Importantly, HSPGs are targets of heparanase (HPSE), the only mammalian endoglycosidase (endo-β-D-glucuronidase), whose activity has been widely implicated in cancer metastasis (9, 10). HPSE cleaves HS at specific intrachain sites resulting in fragments (10–20 sugar subunits). These fragments are biologically active and can bind potent growth factors and angiogenic factors (9, 10). Of interest, HPSE also has non-enzymatic functions in its unprocessed, latent 65 kDa form (L-HPSE), independent of its known endoglycosidase activity, which has been ascribed to the fully processed, active 58 kDa form of HPSE (a heterodimer consisting of 50kDa and 8kDa subunits or A-HPSE) (11–13). Further, our laboratory has recently provided evidence of microRNA mechanisms suppressing BMBC by directly targeting heparanase (14).
As tumor cells invade, they establish, abolish, and/or relocate transient focal adhesions. The invasive cell phenotype requires altered cytoskeletal dynamics, which are driven by the small GTPases: Rac1 and RhoA (15–20). The Rho guanine nucleotide exchange factor GEF-H1 is crucial in coupling microtubule dynamics to Rho GTPase activation in a variety of biological settings. Importantly, GEF-H1 is a microtubule-binding protein which influences the dynamics of the actin cytoskeleton by inhibiting Rac1 and facilitating RhoA activity (21). Tumor cell extravasation requires a dynamically changing cell shape, which is associated with extensive cytoskeletal alterations. These, in turn, are driven by the small GTPases, Rac1 and RhoA, whose regulation is involved in tumor cell plasticity (19). Focal adhesion dynamics require Rac1 and RhoA activities with roles played by cell surface HSPGs as recent evidence indicate (19). Further, we have reported that GEF-H1 differentially associates with SDC4 following cell exposure to purified preparations of human latent (L-HPSE) or active heparanase (A-HPSE) (11). In order to negotiate with the blood-brain barrier (BBB), tumor cells often change their morphology inside the vascular lumen and form cytoplasmic protrusions which expand, but not disrupt, the vessel wall. These observations are of relevance in the pathology of brain metastasis (22), and require molecular deciphering.
We hypothesized that heparanase modulates HSPG-associated signaling in BMBC cells through mechanisms unrelated to its endoglycosidase activity. To this end, we utilized two human BMBC cell systems consisting of isogenic MDA-MB-231 variants which were independently selected in vivo. Both variants have increased ability to form brain metastasis, and possess distinct cellular phenotypic characteristics. We demonstrate a differential HPSE-induced modulation of BMBC cell morphology, adhesion, invasion and Rac1/RhoA GTPase activities depending upon treatment with the active versus the latent form of heparanase. Second, we demonstrate that heparanase actions on Rac1 and RhoA are mediated by GEF-H1, suggesting roles for heparanase in the initial events of BMBC pathogenesis, e.g., cell adhesion, cytoskeletal dynamics, and cell extravasation, which are independent of its enzymatic activity. These findings establish heparanase as a critical modulator of the brain metastatic potential of breast cancer.
Materials and Methods
Cell culture
Brain metastatic breast cancer cells (BMBC) cells from two MDA-MB-231 cell systems independently derived from the in vivo selection of parental counterparts (231P and the brain metastatic variants 231BR and 231BR3 for brevity) were provided by Drs. Patricia Steeg (The National Cancer Institute, Bethesda, MD) and Janet Price (MD Anderson Cancer Center, Houston, TX). Cell lines were obtained between late 2009 – beginning 2010, and freshly recovered (< than 6 months) from liquid nitrogen before they were used for indicated experiments. The 231BR clones were obtained at early passage and tested (2011) for consistent in vivo abilities to metastasize to brain. Cells were grown in DMEM/F12 plus FBS (10%) and penicillin/streptomycin (1%), as published (14, 23). Human brain microvascular endothelial cells (isolate # 4 or HBMEC-4) were obtained following isolation of brain capillaries and their culturing as previously described (24, 25), and used at early passages. Human astrocytes were purchased from Lonza (Lonza Group Ltd., Basel, Switzerland), and grown in media supplemented with AGM Bulletkit for astrocytes (astrocyte growth media, cat # CC-1423).
RT-PCR
Total RNA was isolated from the cells using the RNeasy Plus mini-kit (QIAGEN, Valencia, CA) according to manufacturer instructions. RNA yield was determined using a NanoDrop ND1000 spectrophotometer (NanoDrop products, Wilmington, DE). To ensure lack of genomic DNA contamination, 1 μg of total RNA was digested with DNase I (Invitrogen, Carlsbad, CA) prior to first-strand synthesis. Two μl of the inactivated DNase I digestion was used as template with a First Strand Synthesis kit utilizing SuperScript II reverse transcriptase according to manufacturer instructions (Invitrogen, Carlsbad, CA). The first strand synthesis reaction was then diluted 1:1 with ddH2O to be utilized as single strand cDNA template. PCR amplification was performed in 20 μl reactions consisting of 1X AmpliTaq Gold buffer (Applied Biosystems, Foster City, CA), 2 mM MgCl2, 300 μM dNTP mix, 400 nM primer pair, 2 μl single strand cDNA template, and 2 u AmpliTaq Gold Taq polymerase (Applied Biosystems, Foster City, CA). PCR conditions were: 94°C, 2 min.; 40 cycles of 94°C, 20 sec.; 58°C, 15 sec.; 72°C, 42 sec.; 72°C, 30 sec. Gene accession numbers and DNA sequences for the oligonucleotide primer pairs utilized are shown in supplemental table 3. The PCR reactions were conducted using in a Mastercycler epgradient thermocycler (Eppendorf North America, Westbury, NY).
Western blotting
Proteins were resolved by SDS–PAGE, transferred to nitrocellulose membranes (Bio-Rad, Hercules, CA), and blocked with 5% (w/v) non-fat dry milk (for most antibodies), or 3% (w/v) bovine serum albumin (for anti-Rac1 or anti-RhoA antibodies) in TBS with 0.5% (v/v) TWEEN 20 before being probed with appropriate antibodies. The antibodies and dilutions used in experiments were: SDC1 (1:1,000, clone B-A38), Grb2 (1:1,000, 3972), purchased from Cell Sciences (Canton, MA), SDC4 (1:1,000, ab24511) was from Abcam (Cambridge, MA), full-length PKCα (1:1,000, #2506), GEF-H1 (1:1,000, #4076) and GAPDH (1:1,000, 14C10) were from Cell Signaling (Danvers, MA), β1Integrin (1:1,000, clone 656) was from BD Biosciences (Sparks, MD), Rac1 (1:1,000, clone 23A8) and RhoA (1:500, clone 55) were from Millipore (Danvers, MA), Tiam1 (1:1,000, sc872) and β-actin (1:1,000, sc-69879) were from Santa Cruz Biotechnology (Santa Cruz, CA). Blots were washed with TBS containing 0.5% (v/v) TWEEN®20 in TBS (pH 7.4), before probing with horseradish peroxidase conjugated secondary antibodies which were diluted in blocking solution (1:6,000, sc-2030 and sc-2031 from Santa Cruz Biotechnology). Blots were then exposed to film using SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific, Pittsburgh, PA).
GST-pulldown analyses
Affinity chromatography by glutathione-S-transferase pulldowns (GST-PD) were performed as previously described (26). Briefly, GST-SDC1 or GST-SDC4 fusion constructs in a pGEX-4T-3 vector (GE Healthcare, Piscataway, NJ) were transformed into the Escherichia coli strain BL21. Fusion protein expression was induced by the addition of isopropyl β-D-1-thiogalactopyranoside (IPTG) to a concentration of 200μM to the LB culture media. Induced fusion proteins from bacteria were obtained by lysis using the B-PER reagent (Thermo), and the fusion proteins were purified by passing cell lysates over columns containing Immobilized Glutathione beads (Thermo). GST fusion proteins were eluted from the columns with reduced glutathione. Thirty micrograms of purified GST fusion proteins were bound to glutathione bead columns and whole cell lysates (500mg) from BMBC cell lines were passed over the columns. These were then washed thrice and bound proteins eluted with 2X Laemmli buffer with subsequent heating at 95°C for 5 min. Eluted proteins were resolved by SDS-PAGE, transferred to nitrocellulose membranes (Bio-Rad Inc., Hercules, CA), and proteins were identified by Western blotting.
Immunofluorescence microscopy
Adherent cells (1 × 105 cells per well) were grown on Lab-Tek II Chamber Slide w/Cover, 2 well size (Nalge Nunc Intl., Roskilde, Denmark). Cells were gently washed twice with 1X PBS, and fixed with 4% paraformaldehyde in 1X PBS for 10 min. Then, cells were gently washed once with 1X PBS, extracted with chilled extraction buffer consisting of: 0.5% Triton X-100, 20mM HEPES (pH 7.4), 50mM NaCl, 3mM MgCl2, and 300mM sucrose on ice for 20 min. This was followed by two gentle 5 min. washes with wash buffer consisting of 0.1% NP-40 in 1X PBS, cells were blocked with blocking buffer consisting of 5% horse serum and 0.1% NP-40 in 1X PBS for 1 hr. at room temperature (25°C). Cells were exposed to primary antibodies in blocking solution for 16 hr. at 4°C, and then three gentle 5 min. washes with wash buffer were performed. The cells were exposed to secondary antibodies in blocking solution containing Hoechst 33258 for 1 hr. at room temperature (25°C). This was followed by three gentle 5 min. washes with wash buffer. The chambers were then removed from slides and mounting medium was placed directly upon stained cells, and cover slips were applied. The slides were cured 16 hrs. at 25°C, the cover slips were then sealed to the slides with nail polish. Antibodies and dilutions were: 1:500 VCAM1(BBIG-V1 (4B2), BBA5, R&D Systems, Minneapolis, MN, USA), 1:100 GEF-H1 (Cl B4/7, HM2152, Hycult Biotech, Plymouth Meeting, PA, USA), 1:400 tubulin (EP1332Y, ab52866, Abcam, Cambridge, MA, USA), 1:150 SDC4 (ab24511, Abcam), 1:50 PKCα (#2056, Cell Signaling Technology, Danvers, MA, USA), 1:40 phalloidin (594, Invitrogen, Carlsbad, CA, USA), 2.5ug/ml Hoechst 33258 mounting medium with antifade: ProLong (Invitrogen). Immunofluorescence (IF) images were acquired using a confocal microscope: laser source: X-Cite series 120, microscope: Zeiss Axio Imager.Z1, objectives: Plan-APOCHROMAT 63X or 100X 1.4 oil DIC, software version: AxioVision 4.6.3. A consistent calibration of the confocal microscope, the presence of controls which were performed in parallel, and the choice of fluorophore combination, resulted in only a minimal overlap in wavelength which was readily compensated by the microscope.
In vitro blood-brain barrier transmigration assay
Blood-brain barrier extravasation assays were performed based upon a protocol already described (27). Briefly, 6.5mm transwell membrane inserts with 8μM pores (Corning, New York, NY) were treated on the underside with 100μl of gelatin for 30 minutes, which was then gently removed. Astrocytes were then seeded to the underside of the inserts at a density of 1 × 105 cells/well, and allowed to grow in astrocyte growth media for 4 hr. The growth media was changed, and 1 × 104 HBMEC cells were added to the top of each insert. The astrocyte-HBMEC coated inserts were allowed to grow in astrocyte growth media for 3 days with fresh media daily. After 3 days, BMBC cells were treated with recombinant human heparanase (100ng/ml) (none, latent, or active) in DMEM/F12, 5% FBS, 10mM HEPES (pH 6.5) with penicillin/streptomycin (8) for 1 hr. at 37°C, then trypsinized and counted. Cells were suspended at a density of 3 × 105 cells: 300μl DMEM/F12, 0.1% BSA, then 300μl of cell suspension was added to the tops of the inserts. Lower chambers contained HBMEC - conditioned media with 5μM N-Formyl-Met-Leu-Phe (FMLP, Sigma, St. Louis, MO). The cells were then left to invade for appropriate time points, then inserts were gently cleaned with Q-tips, and membranes removed and placed on slides for visualization (28). Red fluorescent protein expressing HBMEC cells were produced by stably transfecting cells with pmCherry-C1 (cat # 632524, Clontech, Mountain View, CA) using FuGene 6 Reagent (Roche Diagnostics, Indianapolis, IN). Procedures were repeated under similar conditions using GFP-labeled 231BR3 GEF-H1 shRNA knockdown clones. An equal number (1 × 104 post-FACS cells) of viable GFP-labeled 231BR3 GEF-H1 shRNA knockdown cells was added to the top chamber at the start of invasion assays. Astrocytes and HBMEC cells were subtracted from quantification because we included control Transwell inserts with these two cell lines only, but no BMBC cells. An equivalent number of astrocytes and HBMEC cells was added to each assay, allowing for standardization/normalization of results. Further, HBMEC cells were tagged (Texas red fluorescence) to efficiently identify them for ease of analyses.
Rac1 and RhoA activity assays
BMBC cells were cultured in 0.5% DMEM/F12 with penicillin/streptomycin for 16 hr. The following day, the BMBC cells were treated with DMEM/F12, 0.5% serum, 10mM HEPES (pH 6.5) with recombinant human HPSE (100ng/ml) (none, latent, or active) and penicillin/streptomycin for 1 hr. at 37°C. The 1 hour time was chosen because it aligns well with standardized HPSE activity determinations provided by the HS-degrading enzyme assay (Takara Inc., Takarazuka, Japan; 11, 28). Rac1 and RhoA small GTPase activity assays were performed using the GLISA kits (BK124, BK125) (Cytoskeleton, Denver, CO) according to manufacturer’s instructions. Second, Rac1 and RhoA activities were normalized to total Rac1/RhoA protein content by performing corresponding Western blotting analyses. We determined the cell number to cell lysis buffer volume ratio that would reproducibly result in protein concentrations adjusted to 0.5mg/ml. Cell lysates were then snap-frozen in liquid nitrogen to preserve GTP-bound GTPase (Rac1 or RhoA) activity. GTP-bound Rac1 GTPase was bound by Rac1-GTP binding protein linked wells of 96-well plates. Bound active Rac1 was detected using a Rac1 specific antibody. The degree of Rac1 activation was determined by comparison to activity values obtained from lysates derived from untreated cells. Similarly, GTP bound RhoA GTPase was bound by RhoA-GTP-binding protein linked wells of 96-well plates. Bound active RhoA was detected with a RhoA specific antibody. Absorbance readings were obtained at 490nm for both Rac1 and RhoA activities using a SpectraMax Plus384 spectrophotometer (Molecular Devices, Sunnyvale, CA). Both Rac1 and RhoA activity assays were performed in quadruplicate and normalized to total Rac1 and RhoA within the same sample, as measured by Western blotting.
Knockdown of protein expression
The expression plasmids encoding shRNA to GEF-H1 and scrambled control were purchased from GeneCopoeia (Rockville, MD) and transfected into 293Ta cells using manufacturer’s protocol. Supernatant containing lentiviral particles were then transduced into 231BR3 cells using 2μM polybrene (Millipore) and selected with 2.5μg/ml puromycin (Sigma). Transduction efficiency was determined by GFP expression, and knockdown of GEF-H1 protein expression was confirmed by Western blotting analysis. The precise target sequences can be found in supplemental table 1. The oligos encoding siRNA to HPSE and scramble control were purchased from Dharmacon (Thermo) and transfected into BMBC cells using manufacturer’s protocol. Knockdown of HPSE protein expression was confirmed by Western blotting analysis. The precise target sequences can be found in supplemental table 2.
FACS
Fluorescently labeled (green fluorescent protein or GFP) cell samples (scrambled, GEF-H1 shRNA 1/4 clones) were analyzed and sorted using the BD FACS Aria II 3 Laser High-speed Sorting Flow Cytometer equipped with 12 independent fluorescent channel capabilities and DIVA acquisition software. Each sample staining set included single-color controls to facilitate rigorous instrument set-up and compensation. For each sorted sample, events were collected per list mode data file, gating and sorting for GFP-negative and GFP-positive cells (suppl. Fig. 3). Cells were collected in 0.5 ml RPMI media (Invitrogen), and then used for culturing and visualization.
Cell adhesion assays
Fibronectin solution (Sigma) (0.5μg/ml, 100μl) was added to wells of a 96-well plate and allowed coat the plate for 16 hr. Subsequently, the excess fibronectin solution was removed and 100μl of HBMEC suspended at a concentration of 2×105 cells/ml in serum-free DMEM were added to the wells and allowed to adhere for 16 hr. HBMEC cells were then treated with recombinant human heparanase (100ng/ml) (none, latent, or active) for 1 hr. at 37°C in DMEM supplemented with: 10mM HEPES (pH 6.5), 5% FBS, and penicillin/streptomycin. The cells were then washed with PBS. BMBC cells (2 × 105 cells/ml, 100μl) suspended a in serum-free DMEM/F12 were added to the wells and allowed to adhere for 2 hr. at 37°C. Wells were washed with 1X PBS three times, and then 0.5% crystal violet solution was added (200μl) to wells for 10 min. Subsequently, wells were washed with ddH2O 4 times, and extracted with 200μl 10% acetic acid for 10 min. with gentle shaking. The extracted sample (150μl) was then transferred to new wells, and absorbance readings were obtained (560nm). Cell adhesion was determined by subtracting values for cells without BMBC cells from wells with BMBC cells.
Statistical analyses
Data were represented as mean + standard deviation. Significance values were obtained by Student’s paired t-test (*p < 0.05, **p < 0.01) when compared to untreated controls, unless otherwise indicated. All figures were representative of at least three independent experiments. Error bars in figures signify standard deviation. P values of < 0.05 were considered statistically significant.
Results
Differential regulation of BMBC cell adhesion, invasion, and GTPase activities by latent vs. active HPSE
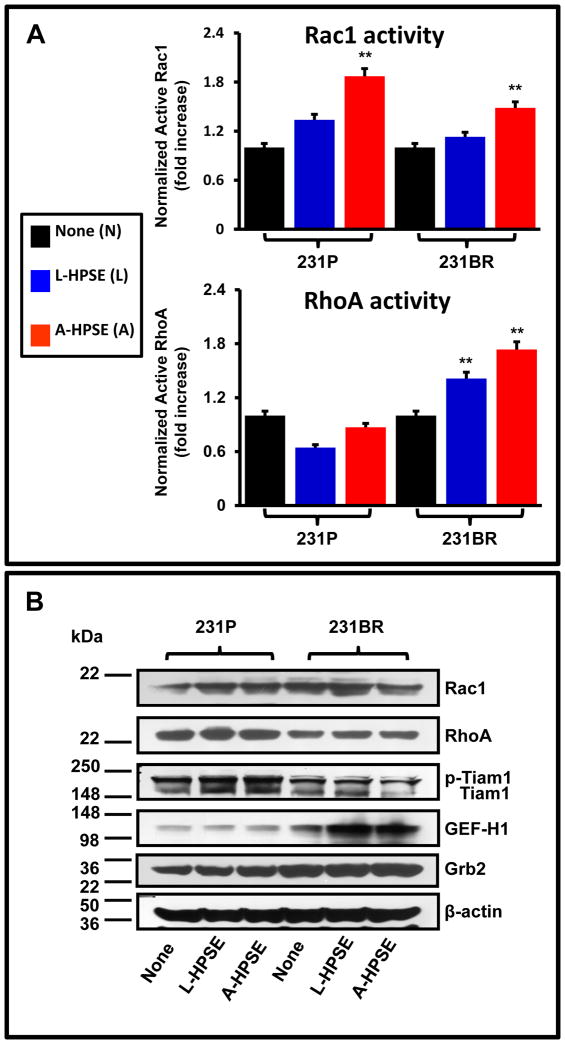
To test the hypothesis that GEF-H1 expression and the activities of Rac1 and RhoA are relevant in BMBC cells, we incubated 231P and the brain metastatic variant 231BR with human latent or active heparanase (L-HPSE or A-HPSE, respectively). Human latent and active HPSE properties, e.g., molecular weight and HPSE enzymatic activity of forms, were verified (supplemental figure 1). Figure 1 shows a significant (p < 0.05) increase of Rac1 GTPase activity following exposure to HPSE in both 231P and 231BR relative to untreated control (none), particularly with the exogenous A-HPSE treatment (p < 0.01). The Rac1 activity response of the two cell lines to HPSE was similar. However, the effect of HPSE on RhoA activity was quite different when comparing 231P and 231BR cells (Fig. 1A), as only the 231BR cells had significantly increased RhoA activity in response to exogenous treatment with either L- or A-HPSE (p < 0.01). Interestingly, HPSE (both latent and active) increased GEF-H1 expression in 231BR cells, but not in parental 231P cells (Fig. 1B). We also investigated the expression of Rac1 guanine exchange factor Tiam1, known to be present in BMBC cells and regulated by c-src and FAK (29), and the adapter Grb2, a scaffold protein associated with these tyrosine kinases (30) (Fig. 1B).
Figure 1.
The modulation of Rac1 and RhoA GTPases by cell treatment with latent or active heparanase (L-HPSE and A-HPSE, respectively). Parental (231P) or brain metastatic (231BR) breast cancer cells were exposed to recombinant human L- or A-HPSE (100ng/ml for 1 hr. at 37°C), cell lysates were then prepared and analyzed accordingly. A. Rac1 and RhoA activities by G-LISA assays normalized to total Rac1/RhoA protein content (Western blotting), respectively. G-LISA assays were performed in triplicate. Statistical comparisons via Student’s paired t-test were made from 231P to 231BR exposed to identical treatments, and differences were statistically significant; **p < 0.01. B. Western blotting analyses of total Rac1 and RhoA, GEF-H1, and associated signaling molecules following L- and A-HPSE cell treatments. Expression of Rac1 guanine exchange factor Tiam1, known to be expressed in BMBC cells and regulated by c-src and FAK (29), and the adapter Grb2, which is a scaffold protein associated with these tyrosine kinases (30), are shown. Blots were probed for β-actin as a loading control. Refer to the “Materials and Methods” section for additional details.
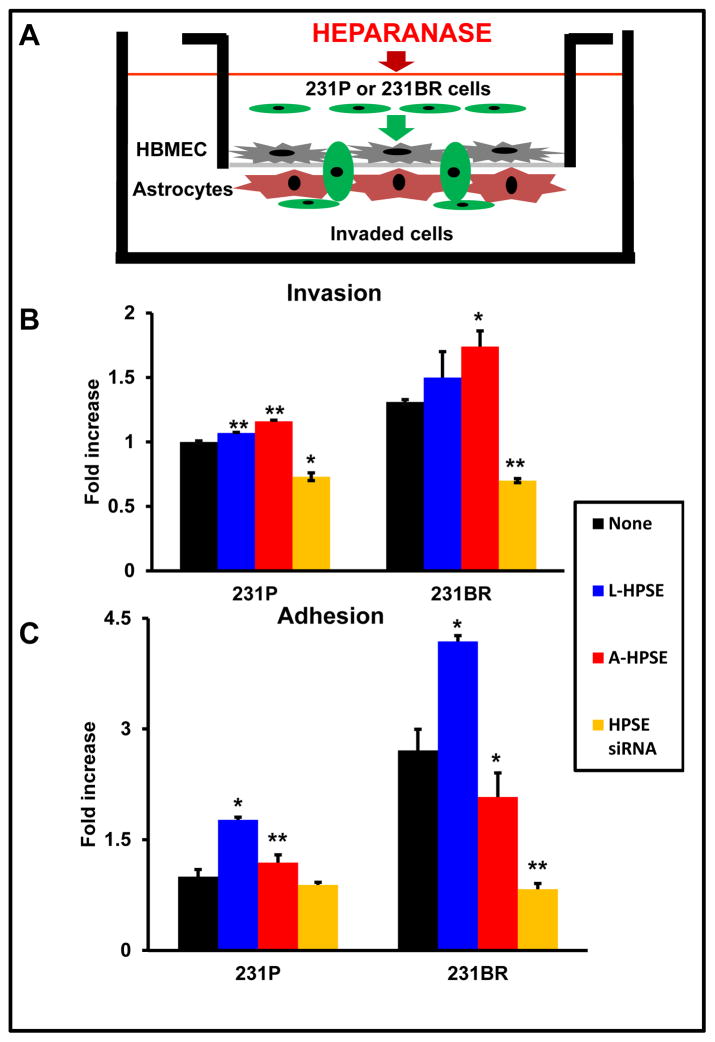
To brain metastasis to occur, cancer cells must first adhere to the vascular endothelium, then cross the endothelial layer of the blood-brain barrier (BBB), and finally invade and proliferate within the brain parenchyma. To assess heparanase involvement in cytoskeletal dynamics and cell extravasation, we developed a model to mimic the BBB, in which human brain endothelial cells (HBMEC-4) and astrocytes were cultured on opposite sides of the porous membrane of transwells (Fig. 2A)(13). Next, we aimed to analyze roles of Rac1/RhoA activities and expression, and BMBC cell transmigration using this in vitro co-culture model (27). As initial investigations, we found differential effects on cell invasion and adhesion upon exposure of 231BR cells to recombinant HPSE (Fig. 2B, 2C). Exogenous L- or A-HPSE caused increased invasiveness in both 231P and 231BR cells (Fig. 2B) and knockdown of endogenous HPSE with siRNA reduced invasion of both 231P and 231BR cells (Fig. 2B). Both 231P and 231BR cells were also more adhesive in response to L- or A-HPSE (Fig. 2C). Of note, differential effects on cell adhesion vs. invasion were detected in 231BR cells compared to 231P counterpart: While cell invasion was upregulated by both L- and A-HPSE (14), cell adhesion was predominantly mediated by the latent form of the molecule (L-HPSE) (Fig. 2C). Interestingly, HPSE siRNA reduced adhesion below baseline only in 231BR, but not the 231P cells (Fig. 2C). These data indicate a differential role for L- vs. A-HPSE in adhesion and invasion of 231P and 231BR cells.
Figure 2.
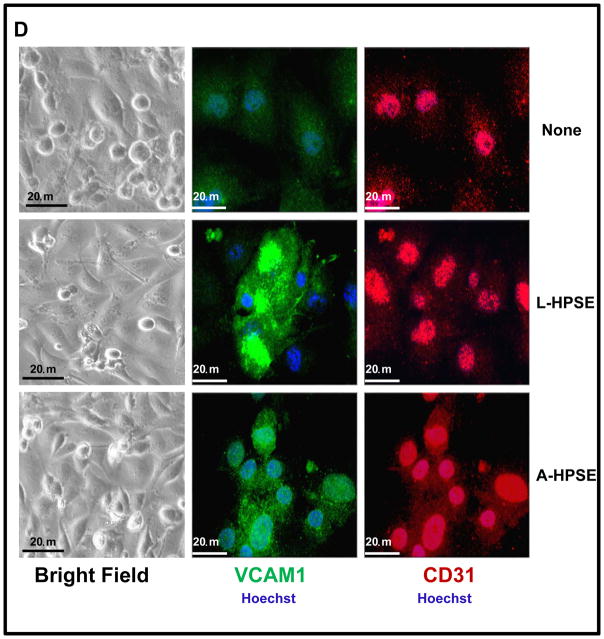
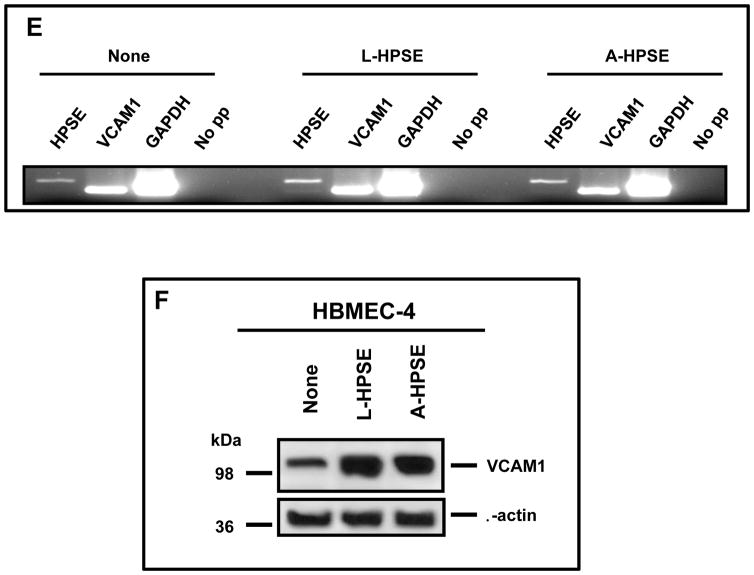
Heparanase treatment increases the invasiveness and adhesiveness of BMBC cells while inducing VCAM1 expression in HBMEC. A. An in vitro blood-brain barrier (BBB) transmigration assay model to study 231P and 231BR cells. Human astrocytes were seeded onto the bottom of MatrigelTM - coated transwell membrane inserts (8 mM pores) and allowed to adhere. Human brain endothelial cells (HBMEC-4) were then seeded onto membranes (top chambers), allowed to adhere, and cultures were examined microscopically to assess monolayer integrity. BMBC (231BR) cells were treated with A-HPSE, L-HPSE, SmartPool HPSE siRNA (23), or untreated, added to inserts, and examined. Bottom chambers contained HBMEC-4 conditioned medium as chemoattractant. B. Effects of L- and A-HPSE on cell invasion. Cells (231P and 231BR) were treated with L-HPSE, A-HPSE, or HPSE siRNA SmartPool (22), and allowed to invade for 12 hr, then stained with crystal violet, extracted, and quantified by reading OD values (560 nm). C. Effects of L- and A-HPSE on cell adhesion. Cells were treated as in B., then seeded onto fibronectin - coated wells and allowed to adhere for 1 hr. Cells were then stained and extracted, and quantified by measuring O.D. values (560 nm). Data are representative of quadruplicate replicates of BBB model invasion chambers for each cell line, as previously described (11). Statistical comparisons were made between treated vs. untreated of the indicated cell line (Fig. 2B, 2C); *p < 0.05. D. Heparanase treatment induces VCAM1 expression in CD31 - positive human brain endothelial cells. Human brain endothelial cells (HBMEC-4) were grown in chamber slides and treated with human recombinant latent (L-HPSE) or active (A-HPSE) heparanase at 100 ng/ml for 1 hr at 37°C. Cells were then stained with antibodies raised against CD31 (red) or VCAM1 (green). Nuclei were stained with Hoechst 33258 (blue). Immunofluorescence and confocal microscopy were subsequently performed and images taken using a 60X objective. E. Analysis of HPSE and VCAM1 gene expression from human brain endothelial cells (HBMEC-4) by RT-PCR. Samples were analyzed for GAPDH gene expression control. Representative RT-PCR products: No exogenous heparanase treatment (None), treatment with L-HPSE (100ng/ml for 1 hr. at 37°C), treatment with A-HPSE (100ng/ml for 1 hr. at 37°C). Each set of RT-PCR reactions was performed in triplicate. Refer to the ”Materials and Methods” section for additional details.
As integrins are important in regulating migration and invasion and may be engaged in BMBC-brain endothelial cell interactions as initial steps leading to brain metastasis, and because the vascular cell adhesion molecule 1 (VCAM1; α4β1 integrin is the major receptor of VCAM1)(47) can function as a physical attachment site for a cell surface β1 Integrin/SDC4 complex, we hypothesized that extracellular HPSE may affect cell surface expression of VCAM1 in brain microvascular endothelial cells (HBMEC-4). Therefore, we analyzed HBMEC-4 for expression of VCAM1 in conjunction with the endothelial cell marker CD31 (control). We observed an overall increased staining for VCAM1 in response to exogenous L-HPSE and A-HPSE treatments which was most pronounced in a sub-population of HBMEC cells (Fig. 2D, 2F). Importantly, we observed no change in morphology of HBMEC-4 following treatment with exogenous L- or A-HPSE (Fig. 2D). We analyzed HPSE and VCAM1 gene expression by RT-PCR: comparable levels of HPSE and VCAM1 gene expression were observed among cell lines. These levels were not significantly altered by exogenous L- or A-HPSE exposure (Fig. 2E). While mRNA of VCAM1 was not altered, VCAM1 protein expression was significantly up-regulated by HPSE exposure (Fig. 2F).
These experiments suggest that the altered invasion of the breast cancer cells induced by HPSE may be at least in part due to post-transcriptional effects of HPSE to increase VCAM1 presence in the endothelial cell barrier of the brain.
Heparanase alters the cell morphology in distinct BMBC 231BR cell systems
We sought to assess HPSE-mediated changes in cell morphology in the two isogenic BMBC cell systems associated with cytoskeletal dynamics, e.g., alterations in the extent of polymerized actin and tubulin, the components forming actin stress fibers and microtubules, respectively
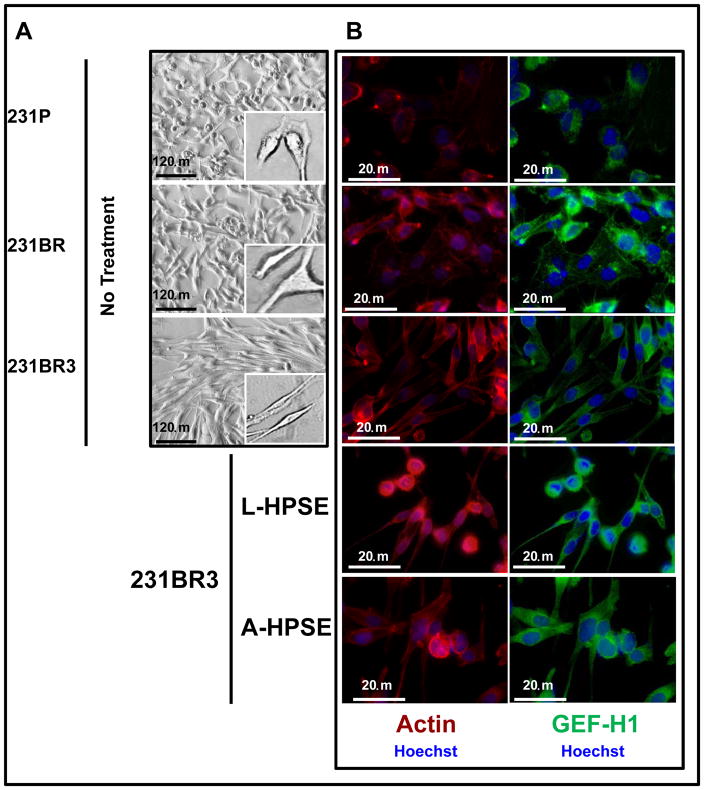
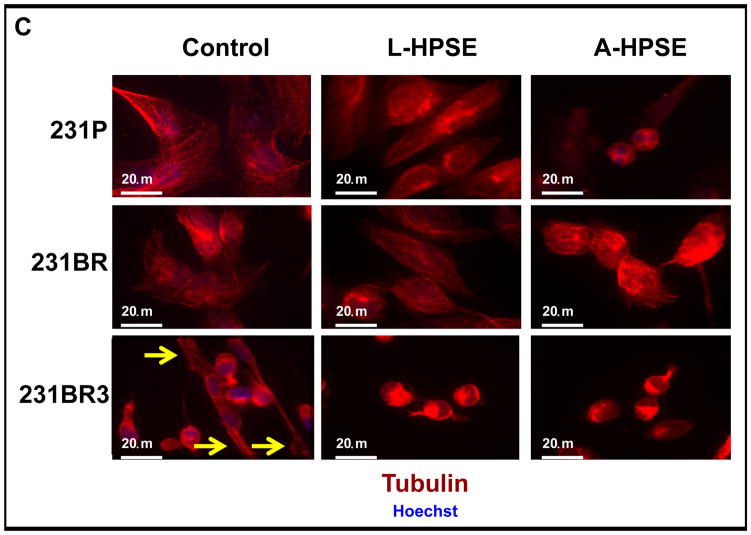
The actin stress fibers among the MDA-MB-231 derived cell lines were strikingly different, as the fibers were thicker, longer, and more pronounced in 231BR3 cells, which also possessed a more spindle-shaped morphology (Fig. 3A, 3B). This contrasted with the 231P and 231BR cells which had a mixture of rhomboidal and spherical cell shapes (Fig. 3A, 3B). Of note, the distinctive cell morphologies by the two BMBC cell lines did not change during continuous passages (almost 20) in culture. Second, high basal levels of GEF-H1 in 231BR cells were observed to correlate with round/rhomboidal cell morphology. Interestingly, the exposure of 231BR cells to recombinant L- and/or A-HPSE resulted in 231BR cells changing morphology to a round-cell shape associated with higher GEF-H1 expression (Fig. 3B). Conversely, networks of tubulin, an important cytoskeleton component (16–20), in these cell lines were mostly comparable; however, we detected tubulin which extended to the tips of the spindle-shaped 231BR3 cells (Fig. 3C). These results show that GEF-H1 is an important regulator of BMBC cell morphology that correlates with tubulin arrangement.
Figure 3.
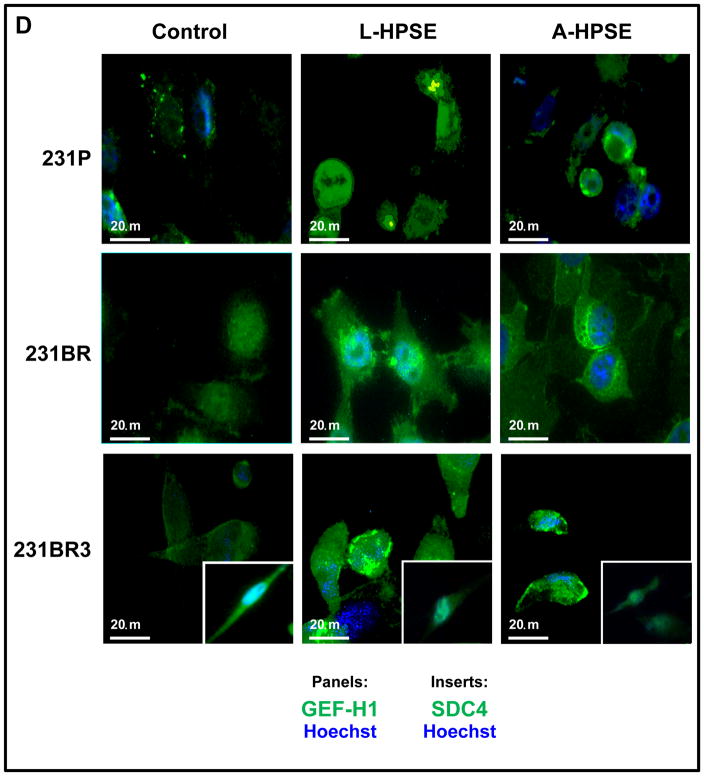
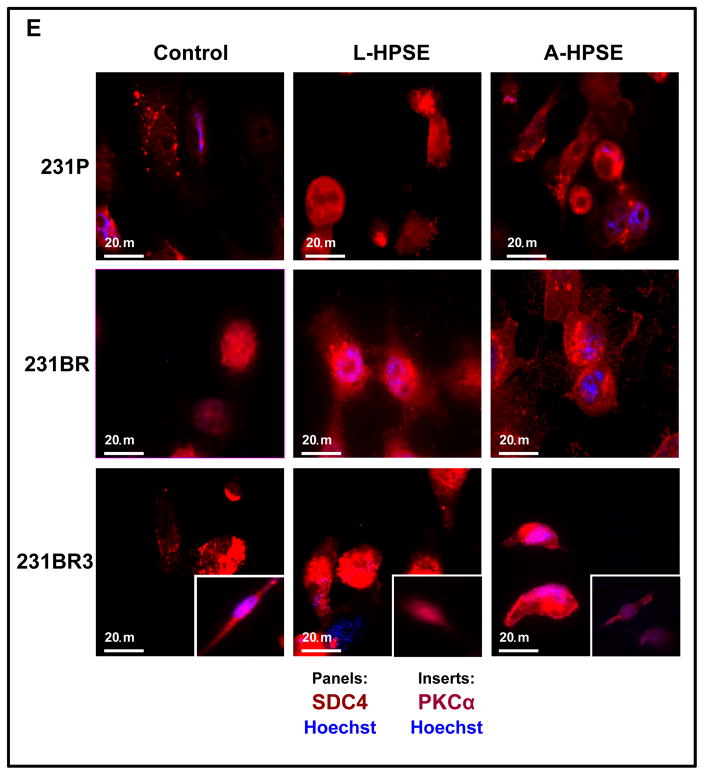
231BR3 cell morphology is distinct from that of 231P and 231BR. A. Brightfield images demonstrating morphologies of three BMBC cell lines. Upper panel: BMBC 231P cells. Middle panel: BMBC 231BR cells. Lower panel: BMBC 231BR3 cells. Inserts show representative images of cells at higher magnification. B. Immunofluorescence staining for actin (red), GEF-H1 (green), and nucleus (blue) of 231P, 231BR (panel image mostly shows GEF-H1 overexpressor cells), and 231BR3 breast cancer cells. Cells were treated with human recombinant heparanase either in its latent or active form (100 ng/ml for 1hr. at 37°C). C. Immunofluorescence staining for tubulin (red) and nucleus (blue). Arrows indicate tips of spindle in 231P, 231BR, and 231BR3 breast cancer cells. Controls refer to no HPSE treatment. Refer to the “Materials and Methods” section for additional details. D. Immunofluorescence staining for GEF-H1 (green, panels), SDC4 (green, inserts), and nucleus (blue) for 231P, 231BR, and 231BR3 breast cancer cells. Cells were treated with human recombinant heparanase either in its latent or active form (100ng/ml for 1 hr. at 37°C). Control refers to no HPSE treatment. E. Immunofluorescence staining for SDC4 (red, panels) PKCα (red, inserts), and nucleus (blue) for 231P, 231BR, and 231BR3 breast cancer cell lines. Cells were treated with human recombinant heparanase either in the latent or active form (100ng/ml for 1 hr. at 37°C). Refer to the “Materials and Methods” section for additional details.
We performed immunofluorescence (IF) staining of the breast cancer cells (231P, 231BR, and 231BR3) for GEF-H1, SDC4, and PKCa. First, we performed co-immunostaining for GEF-H1, SDC4, and nuclei (Hoechst). We observed a robust GEF-H1 signal that co-localized with the SDC4 signal from the 231BR and the 231BR3 cells (Fig. 3D, 3E). Syndecan-4 was not only detected as a punctate pattern throughout the cell, but also localized to small intracellular vesicles by a particularly intense immunodetection (Fig. 3D, 3E; 231BR center panel). This represents a similar staining pattern that others have observed using the BT549 (for SDC4) and 231P (for SDC1) breast cancer cell lines (31). Next, IF co-immunostaining were performed for expression of PKCα, SDC4, and nuclei (Hoechst). Low PKCα expression was detected among 231P cells not treated with heparanase while high constitutive PKCα was particularly present in 231BR3 cells (Fig. 3E, insert); however, while PKCα and SDC4 were generally co-expressed, treatment of BMBC cells with L- or A-HPSE significantly reduced PKCα expression, notably in the 231BR3 line (Fig. 3E, inserts).
GEF-H1 is a critical regulator of BMBC cell GTPase activity
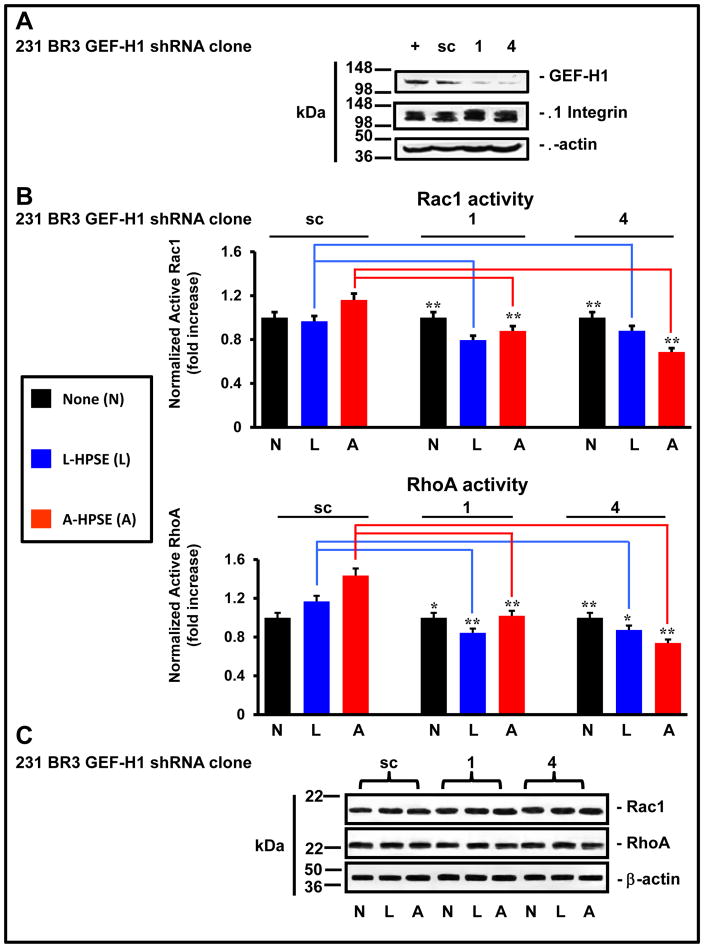
Because we identified a heparanase–induced modulation of the cytoskeleton which is closely associated with the morphology of human 231BR3 cells, we interrogated effects of knocking down GEF-H1 on Rac1 and RhoA activities. Cells (231BR3) were stably transduced with scramble control or one of four shRNA lentiviral constructs (clones 1–4; GeneCopoeia) (supplemental tables 1 and 2), then expression of GEF-H1 and β1 Integrin were investigated by Western blotting. We detected a significant decrease in GEF-H1 protein expression with a concomitant β1 Integrin expression, particularly for clones 1 and 4, which were used thereafter (Fig. 4A and supplemental Fig. 2).
Figure 4.
Reduced GEF-H1 expression results in increased Rac1 activity and decreased RhoA activity. A. GEF-H1 expression is reduced by lentiviral delivered shRNA. BMBC (231BR3) cells were stably transduced with an shRNA scrambled control (sc) or 1 of 4 distinct shRNA expressing lentivirus. Whole cell lysates were analyzed for GEF-H1 and b1 Integrin expression by Western blotting. Blots were probed for b-actin as a loading control. B. The modulation of Rac1 and RhoA small GTPase activity. BMBC (231BR3) cells were stably transduced by a shRNA scrambled control (sc) or 1 of 4 distinct shRNA expressing lentivirus. Cells were plated on fibronectin-coated dishes, subsequently treated with HPSE (none, latent, or active) (100 ng/ml for 1 hr. at 37°C). Whole cell lysates were analyzed for Rac1 and RhoA activity by G-LISA assays normalized to total Rac1/RhoA protein content (Western blotting), respectively. *p < 0.05; **p < 0.01. Asterisks refer to p values comparing HPSE treatment of the GEF-H1 shRNA clones to the scrambled control (no treatment). G-LISA assays were performed in triplicate. C. Effects of HPSE on Rac1 and RhoA expression. 231BR3 cells were treated with or without A-HPSE or L-HPSE, and whole cell lysates were analyzed for Rac1 and RhoA expression levels by Western blotting. Blots were probed for β-actin as loading control. Refer to the “Materials and Methods” section for additional details.
Next, to measure the effect of exogenous HPSE treatment upon the activities and expression of Rac1 and RhoA small GTPases, we treated stably transduced 231BR3 GEF-H1 shRNA cells with recombinant human HPSE (none, latent, or active). We measured significant increases of basal Rac1 activity in 231BR3 GEF-H1 shRNA clones 1 and 4 (12% and 15%; p = 0.0004 and p = 0.0003, respectively) compared to scramble control (Fig. 4B). Notably, we observed little change in Rac1 or RhoA protein expression concomitant with shRNA knockdown of GEF-H1 expression (Fig. 4C). Treatment of the scramble control with A-HPSE increased Rac1 activity above basal Rac1 activity without changing Rac1 protein expression (Fig. 4B, 4C). Treatment with L-HPSE did not appreciably increase Rac1 activities in GEF-H1 knockdown clones above corresponding basal levels (Fig. 4B). Importantly, when 231BR3 GEF-H1 knockdown clones were exposed to A-HPSE, Rac1 activity was significantly reduced (−4% and −12%; p = 4 × 10−4 and 1 × 10−3, respectively) which was not associated with decreased Rac1 expression (Fig. 4B, 4C).
As for RhoA, we measured reduced basal activity (p = 0.021 and 3.1 × 10−3, respectively) when comparing the RhoA activities from scramble control versus the basal RhoA activity of the GEF-H1 shRNA knockdown clones, which was not associated with decreased RhoA protein expression (Fig. 4B, 4C). The RhoA activity of both GEF-H1 shRNA knockdown clones were further reduced subsequent to treatment with exogenous L-HPSE (p = 2 × 10−4 and 1.4 × 10−4, respectively) compared to the identical treatment of scramble control, again, not associated with decreased RhoA protein expression (Fig. 4B, 4C). Treatment of the scramble control with L- or A-HPSE maintained or slightly increased RhoA activity compared to untreated control (Fig. 4B). However, the RhoA activity was significantly decreased in response to A-HPSE treatment (32% and 59%; p = 2.04 × 10−4 and 4.26 × 10−5, respectively) in 231BR3 cells stably transduced to knockdown GEF-H1 expression, when compared to the RhoA activity from identical treatment of the scrambled control, which was not accompanied by reduced RhoA protein expression (Fig. 4B, 4C).
GEF-H1 knockdown reduces cell invasion
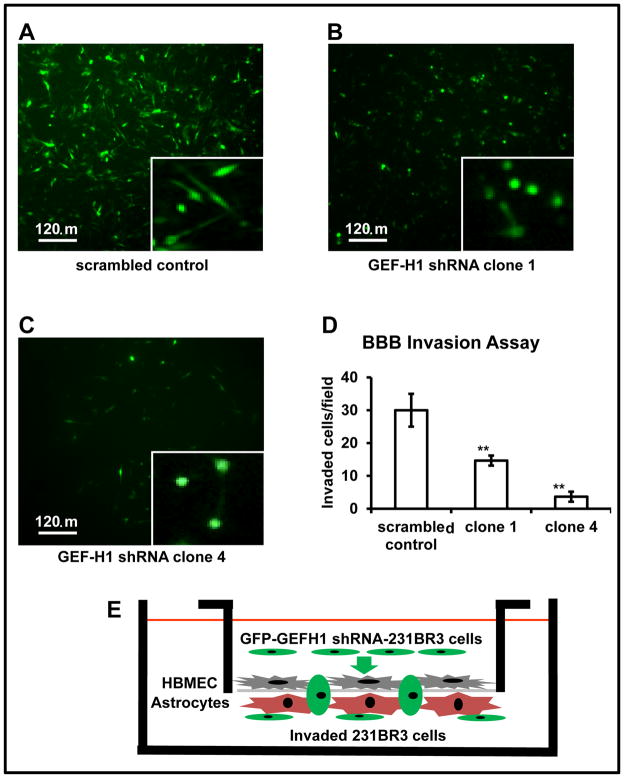
To form brain metastases, cancer cells must first negotiate with the blood-brain barrier (BBB), implement mechanisms to cross this barrier, and extravasate. We devised an in vitro assay to assess the invasive phenotypic changes that may occur as a result of knocking-down GEF-H1 expression in the highly aggressive 231BR3 cell line. To aid in the visualization of the morphological changes, we utilized green fluorescent protein (GFP) expression in conjunction with lentiviral GEF-H1 shRNA knockdowns and IF microscopy (Fig. 5A–5C, and supplemental Fig. 3). GFP expression by knockdown cells does not equate to shRNA expression due to stochastic factors. While shRNA is driven by the U6 promoter, GFP expression is driven by the CMV promoter. Additionally, all GFP tagged cells are puromycin resistance - selected cells. Using our in vitro BBB transmigration assay, we found that the 231BR3 scrambled control cell line was highly invasive while both GEF-H1 shRNA 231BR3 clones were poorly invasive. Figures 5A–C shows images of underside of invasion chambers, visualizing the morphologies of invaded GFP - expressing 231BR3 cells whose number was quantified (Fig. 5D; see also suppl. Fig. 4). Of note, the 231BR3 GEF-H1 shRNA knockdown clones had strikingly fewer GFP - expressing cells that had invaded through this in vitro BBB model (Fig. 5D). Also, while the morphology of the invaded shRNA scrambled control 231BR3 cells was primarily spindle-shaped, poorly invasive GEF-H1 shRNA clones were mostly rounded (Fig. 5A–C). The in vitro BBB transmigration assays were performed in triplicate.
Figure 5.
231BR3 high GFP expressing cells in culture subsequent to FACS selection of the scrambled control and the GEF-H1 shRNA clones demonstrate spindle-shaped morphology in culture while GEF-H1 knockdown clones have reduced invasiveness with a spheroid cell morphology. A. Invaded cells stably transduced with the lentivirus scrambled control expressing GFP. B. Invaded 231BR3 GEF-H1 shRNA (clone 1) cells expressing GFP. C. Invaded 231BR3 GEF-H1 shRNA (clone 4) cells expressing GFP. The presence and morphologies of GFP-expressing cells from the underside of chemoinvasion chambers were visualized. Inserts to figures 5A–C indicate cells at higher magnification with apparent morphological changes. D. The in vitro BBB transmigration assay. Quantification of invaded 231BR3 cells, scrambled control, GEF-H1 shRNA (clone 1), or GEF-H1 shRNA (clone 4). Asterisks refer to p values (*p < 0.05, ** p < 0.01), comparing the GEF-H1 shRNA clones to the scrambled control when statistically compared via Student’s paired t-test. In vitro BBB transmigration assays were performed in triplicate. Images were acquired by microscopy using a 10X objective. Cells were identified by eye and manually counted for multiple quadrants within the chamber fields, and their number statistically quantified. Images shown in A–C, although representative, are not directly related to quadrants (fields) for the quantitative assessment of cell invasion by in vitro BBB transmigration assays. Refer to the “Materials and Methods” section for additional details. E. Model of the BBB transmigration assay studying 231BR3 cells.
GEF-H1 interactions with HSPG signaling components
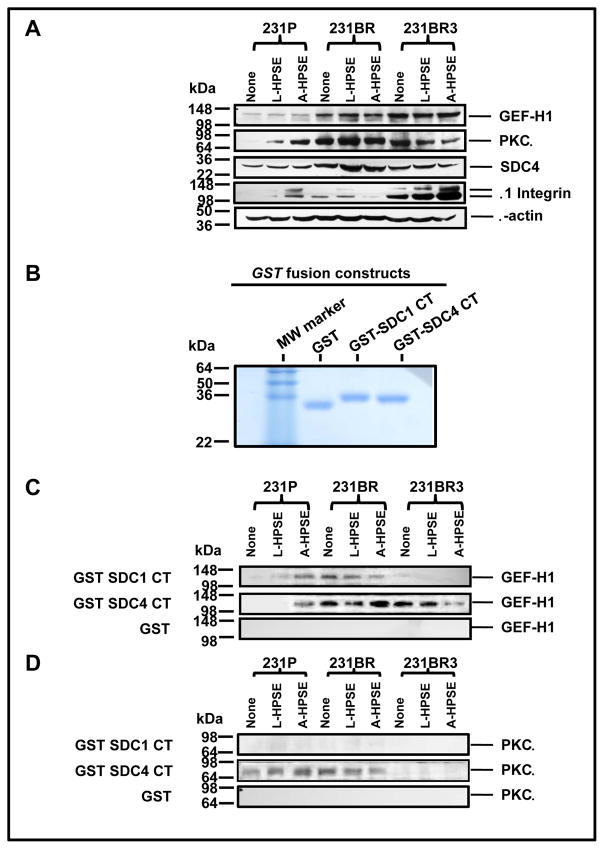
Because signal transduction emanating from the SDC4 intracellular CT domain of this HSPG component of focal adhesion is of relevance, we analyzed cell lysates from 231P, 231BR, and 231BR3 cells to determine whether similar associations could be identified in these cells. We identified a gradient of GEF-H1 expression (231P < 231BR < 231BR3) (Fig. 6A). Additionally, we determined the expression of GEF-H1 protein under normal conditions as well as following treatment with L- or A-HPSE. The expression of GEF-H1 was not modulated by heparanase (Fig. 6A). Conversely, heparanase treatment regulated the expression of PKCα differentially among the three cell lines. For example, we observed that treatment of 231P cells with heparanase induced PKCα expression, particularly in the presence of A-HPSE, while the opposite was true analyzing 231BR3 cells (Fig. 6A). The 231BR cell line had the highest detected PKCα expression while 231BR3 cells displayed highest levels of β1 Integrin which was upregulated in the presence of heparanase cell exposure (Fig. 6A). Syndecan 4 was detected from each of the cell lines tested, however its expression was not regulated by HPSE (Fig. 6A). Therefore, we investigated whether an association of GEF-H1 with SDC4 could occur in BMBC cells and, if so, whether the association would be modulated by treatment with exogenous heparanase. We employed the intracellular carboxy-terminal (CT) region of SDC4 (Fig. 6B) in GST pulldown (GST-PD) analyses and detected a HPSE-regulated association of GEF-H1 with the intracellular SDC4 CT domain. The 231BR cells displayed an augmented SDC4 CT association subsequent to treatment with A-HPSE unlike 231BR3 cells, whose GEF-H1 association with the SDC4 CT decreased subsequent to treatment with A-HPSE (Fig. 6C). Analyses for PKCα interactions with the intracellular CT of SDC4 by GST-PD revealed that 231P and 231BR cells did demonstrate an association between PKCα and SDC4 CT, while 231BR3 cells did not (Fig. 6D). Conversely, SDC1 CT did not possess the same association with GEF-H1 and PKCα as SDC4 CT in all 3 cell lines (Fig. 6C, 6D). These results demonstrate that GEF-H1 intimately associates with HSPG signaling components, and is a target for regulation by HPSE.
Figure 6.
BMBC demonstrate differential GEF-H1 and PKCα protein expression and HPSE treatment modulated SDC4 CT association. A. GEF-H1 and PKCa are SDC CT associated proteins in HPSE-treated BMBC cells. Whole cell lysates from BMBC cells treated with L- or A-HPSE were immunoblotted for GEF-H1, PKCa, SDC4 or β1Integrin to determine protein expression levels in different BMBC cell lines. Blots were probed for β-actin as a loading control. Data presented are representative of four reproducible experiments. B. The production of GST fusion proteins. Affinity glutathione-S-transferase pulldowns for SDC1 and SDC4 were performed as previously described (11). Figure indicates eluted lysates (1 mg/lane) run on a 15% SDS-PAGE gel under reducing conditions. C. GST pulldown of GEF-H1 by SDC1/4CT. D. GST pulldowns of PKCa by SDC1/4CT. BMBC cells were treated with or without latent or active (L- or A-HPSE) (100ng/ml for 1 hr. at 37°C), then whole cell lysates were generated, passed over GST-SDC1/4 CT fusion protein affinity columns. The same lysates were loaded onto GST protein affinity columns followed by immunoblotting for GEF-H1 or PKCa as controls. Refer to the “Materials and Methods” section for additional details.
Discussion
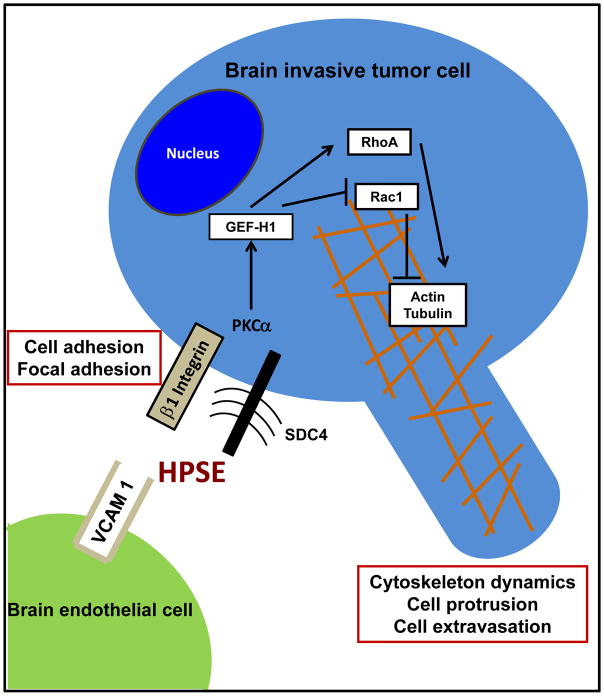
The work presented provides first-time evidence demonstrating that: 1) exogenous heparanase treatment of GEF-H1 – expressing isogenic human BMBC cells altered cell adhesion and invasion; 2) these phenotypic changes are dependent upon the type of exogenous heparanase treatment, e.g., employing the active vs. the latent form of HPSE; 3) heparanase – induced, GEF-H1 - mediated effects altering Rac1 and RhoA activities and cytoskeletal dynamics; 4) an association of SDC4/PKCα and GEF-H1 may be relevant to govern these events (see also Fig. 7).
Figure 7.
A model for the proposed mechanisms involving heparanase – induced, GEF-H1 – mediated, regulation of Rac1 and RhoA activities in the cytoskeletal dynamics; and HPSE activities on SDC4/β1 integrin as members of focal adhesion altering cell adhesion of brain metastatic breast cancer cells.
Notions that cell surface syndecans are important to cell invasion and focal adhesion function, and that Rac1 and RhoA are involved in tumor cell migration and phenotypic plasticity, have been well established (17, 18, 32). GEF-H1 is uniquely positioned to participate in this interplay since it binds and regulates these GTPases (33–35). Equally relevant, heparanase is long known to act as a potent pro-tumorigenic, pro-angiogenic, and pro-metastatic enzyme (9, 10), and recent evidence implicate HPSE in cancer cell signaling independent of its enzymatic activity (11–13, 36). Here, we demonstrate that exogenous active heparanase (58 kDa), and even more the latent form of heparanase (65 kDa), can alter the cell morphology status and phenotypic characteristics in human isogenic BMBC cell systems, and expression of GEF-H1 in its abilities to affect Rac1/RhoA GTPases differentially. We noted the 231BR3 cell line to have a spindle-shaped cell morphology and high GEF-H1 expression compared to 231P cells. Because cytoskeletal dynamics is a fundamental process for cell invasiveness, we hypothesized that upon reducing GEF-H1 expression in the brain metastatic 231BR3 breast cancer variant, the efficiency of invasion would be decreased, thereby reducing a core event of the metastatic process. Indeed, knocking-down GEF-H1 expression reduced 231BR3 cell invasiveness, however it also changed the morphology from a characteristic spindle-shape to a spheroid-type. The morphological change of these GEF-H1 knockdown cell lines was revealed by visualizing invaded cells in our BBB transmigration assay. We consider this an indication that GEF-H1 may be involved maintaining the spindle-shape morphology of the 231BR3 cells during invasive events. While the two MDA-MB-231 derived cell lines employed here are isogenic, 231BR3 are distinct from 231BR cells from a morphological perspective. Further, 231BR3 differ from 231BR cells not only in the number of selection steps applied in vivo (3 vs. 6), in the cell injection route into animals (intracarotid vs. intracardiac), and the expression of EGFR and HER2, two important biomarkers and hallmarks of BMBC; with 231BR3 cells expressing EGFR and HER2 (23) while 231BR cells having EGFR but very low HER2 expression (37).
We postulate that the spindle-shaped morphology presented by the 231BR3 cells is a result of an underlying cytoskeletal organization. We showed the spindles that are in polar opposition to each other are supported by actin stress fibers, known to be maintained by small GTPases Rac1 and RhoA whose activities are regulated by GAPs and GEFs (17, 21). Similarly, investigating HPSE-mediated alterations of tubulin, a key component of microtubules (16–20), was considered of relevance. Accordingly, we analyzed the expression of one specific GEF, GEF-H1. We discovered that GEF-H1 was expressed in each of the three MDA-MB-231 derived cell lines studied. Of note, GEF-H1 was highly expressed in MDA-231BR variants possessing high propensities to metastasize to brain.
The HBMEC-4 cells were tested for the expression of VCAM1, an important component for the formation of focal adhesions involving endothelial cells (39). Treatment with exogenous HPSE, particularly L-HPSE, induced VCAM1 expression in HBMEC-4 cells and that such increased expression was regulated post-transcriptionally. Interestingly, unlike the breast cancer cells we tested, the HBMEC-4 cells did not change to a spheroid morphology in response to heparanase treatment.
Our data show that increased Rac1 activity correlates with decreased GEF-H1 expression since shRNA knockdown of GEF-H1 in 231BR3 cells resulted in increased Rac1 activity under basal conditions. This suggests that the increased Rac1 activity is likely due to removal of sustained Rac1 inhibition by GEF-H1 in 231BR3 cells. Further, the increased Rac1 activity subsequent to exposing these cells to exogenous A-HPSE (scrambled control) was not measured from the GEF-H1 knockdown clones.
The decreased Rac1 activity we measured by comparing A-HPSE - treated scrambled control vs. the GEF-H1 shRNA knockdown clone was not expected considering that Rac1 expression was unchanged in the GEF-H1 knockdown clones and that GEF-H1 acts as inhibitor of Rac1 GTPase activity. This could be explained as knocking down GEF-H1 in 231BR3 cells results in the compensatory expression of another Rac1 inhibitor. Alternatively, this may involve a Rac1 inhibitory process that requires GEF-H1 but in a particular post-translational state. Further, GEF-H1 may inhibit Rac activities independent of Rac1, e.g., Rac2 and Rac3. However, Rac1 is the predominant form of Rac: analyses in our cells indicate that there is only very little Rac2 (which is hemapoietically specific; 48) and Rac3 (unpublished observations; see also 49), with GEF-H1 being the specific inhibitor of Rac1 (34). Second, the reduced RhoA activity detected when comparing the scrambled control to the two GEF-H1 knockdown cell lines, under all conditions tested, is likely attributed to reduced GEF-H1 expression since we observed no decrease in RhoA protein expression and GEF-H1 was the only RhoA-GEF removed from the experimental conditions. This implicates GEF-H1 as a functional regulator of basal RhoA GTPase activity in 231BR3 cells and supports the idea that exogenous heparanase can stimulate GTPase activity, but that the increased RhoA activity subsequent to HPSE treatment involves GEF-H1. Treatment with latent or active exogenous heparanase of 231BR3 cells stably transduced to constitutively express shRNA directed against GEF-H1 resulted in decreased RhoA activation, further supporting the notion that heparanase modulates RhoA activity via GEF-H1.
Third, we demonstrated a potential mechanistic link for heparanase-regulated signal transduction to the cytoskeleton. This can be of relevance since it is known that the direct binding of PKCα to SDC4 CT increases its localization to focal adhesion sites (40). Focal adhesions are involved in cell migration and SDC4 is a known component of focal adhesions (33, 34). The carboxy terminus of SDC4 interacts with protein kinase Ca (PKCα), which has also been implicated in cytoskeletal dynamics (41). Further, protein kinase Cα and SDC4 provide mechanistic links to the extracellular environment for focal adhesion formation (42–45), and we have previously demonstrated an association between the SDC4 carboxy terminal (CT) domain and both GEF-H1 and PKCα employing other cell systems (11, 46). Of note, SDC1 did not present the same association as SDC4 (Fig. 6C, 6D). Also, the combination of GEF-H1 knockdown and HPSE treatment downregulated cell invasiveness and altered Rac/Rho activity. This indicates that the two might act through a pathway originating with SDC4, and suggest that HPSE is an important regulator of cytoskeletal dynamics along with GEF-H1.
In conclusion, we have demonstrated a GEF-H1 association with SDC4 using three breast cancer cell lines representing two BMBC models. We have shown a phenotypic morphology of spindle-shaped cells changing to a round cell-shape during invasion as a result of GEF-H1 knockdown with consequent diminished cell invasiveness. Further, we have shown that treatments with exogenous latent or active heparanase differentially regulate the GEF-H1/SDC4 association and activities of small GTPases Rac1 and RhoA as effectors of cytoskeletal dynamics. These findings are of relevance because they directly relate to the pathology of BMBC in its initial events leading to brain colonization, e.g., interactions between BMBC and brain vasculature cells, and the formation of BMBC cytoplasmic cell protrusions and invadopodia stretching the brain microvessel wall for subsequent tumor cell extravasation (22). They substantiate roles for heparanase independent of its enzymatic activity, e.g., acting as a cell adhesion molecule and signal transducer, and provide added impetus for developing inhibitors targeting latent heparanase and/or the use of HPSE inhibitors in novel cancer therapies, particularly in breast cancer brain metastasis.
Supplementary Material
Supplemental Table 1. Sequences used for the GEF-H1 knockdown experiments via lentivirus expressed shRNA.
Supplemental Table 2. Sequences used for the HPSE knockdown experiments via transiently transfected siRNA.
Supplemental Table 3. Gene accession numbers and DNA sequences for the oligonucleotide primer pairs utilized for RT-PCR.
Supplemental Figure 1. A. Western blot of purified preparations of human latent and active HPSE recombinant proteins showing distinct molecular weights, β-actin was probed as a loading control. M.W. standards were run in parallel with samples (left lane). B. Dose-dependent inhibition of heparanase (A-HPSE) endoglycosidase activity by SST0001, a glycol-split, non – anticoagulant form of heparin (50, 51). The heparanase inhibitory activity and structural details of SST0001 has been described ( 50, 51). Aliquots of purified recombinant human active HPSE were treated with various concentrations of SST0001 (0, 1, 3, and 10 mg/ml, respectively) for 2 hr. at 37°C. HPSE activity (U/ml) was then determined using the heparan sulfate degrading enzyme assay kit™ (Takara, Inc.: cat # MK412) following manufacturer’s protocol.
Supplemental Figure 2. Western blot probed for GEF-H1 used to generate GEF-H1 knockdowns (fig. 4A). β-actin was probed as a loading control.
Supplemental Figure 3. Gated populations from FACS demonstrating the selection of transduced high GFP - expressing 231BR3 cells.
Supplemental Figure 4. 231BR3 GFP - expressing cells. A. 231BR3 scrambled control cells expressing GFP. B. 231BR3 GEF-H1 shRNA (clone 1) cells expressing GFP. C. 231BR3 GEF-H1 shRNA (clone 4) cells expressing GFP. Upper panel: brightfield images. Lower panel: GFP images.
Acknowledgments
We express our gratitude to Dr. Israel Vlodavsky (The Bruce Rappaport Medical Faculty, Technion, Israel) for providing preparations of human recombinant latent and active heparanase. We also thank Disha Kumar for her expert editorial assistance. This work was supported by NIH grants CA086832 and CA160335-01 to DM, and CA98568 to AEE.
Abbreviations used
- BMBC
Brain metastatic breast cancer
- CT
carboxy-terminal
- ddH2O
double-distilled water
- DMEM
Dulbecco’s Modified Eagle’s Medium
- EGFR
epidermal growth factor receptor
- HER2
epidermal growth factor receptor2
- FAK
focal adhesion kinase
- GFP
green fluorescent protein
- GAP
GTPase-activating protein
- GEF
guanine nucleotide exchange factor
- GST-PD
glutathione-S-transferase pulldowns
- HBMEC
human brain microvascular endothelial cells
- HEPES
4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid
- HPSE
heparanase
- A-HPSE
active heparanase
- L-HPSE
latent heparanase
- HS
heparan sulfate glycosaminoglycan chains
- HSPG
heparan sulfate proteoglycan
- O.D
optical density
- SDS-PAGE
sodium dodecyl sulfate - polyacrylamide gel electrophoresis
- SDC
syndecan
- TBS
tris-buffered-saline
- VCAM1
vascular cell adhesion molecule1
Footnotes
Conflict of Interest. The authors declare no competing conflicts of interest.
References
- 1.Gavrilovic IT, Posner JB. Brain metastases: epidemiology and pathophysiology. J Neurooncol. 2005;75:5–14. doi: 10.1007/s11060-004-8093-6. [DOI] [PubMed] [Google Scholar]
- 2.Steeg PS, Camphausen KA, Smith QR. Brain metastases as preventive and therapeutic targets. Nat Rev Cancer. 2011;11:352–63. doi: 10.1038/nrc3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eichler AF, Chung E, Kodack DP, Loeffler JS, Fukumura D, Jain RK. The biology of brain metastases-translation to new therapies. Nat Rev Clin Oncol. 2011;8:344–56. doi: 10.1038/nrclinonc.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernfield M, Kokenyesi R, Kato M, et al. Biology of the syndecans: a family of transmembrane heparan sulfate proteoglycans. Ann Rev Cell Biol. 1992;8:365–93. doi: 10.1146/annurev.cb.08.110192.002053. [DOI] [PubMed] [Google Scholar]
- 5.Iozzo RV. Heparan sulfate proteoglycans: intricate molecules with intriguing functions. J Clin Invest. 2001;108:165–7. doi: 10.1172/JCI13560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Connell MP, Fiori JL, Kershner EK, et al. Heparan sulfate proteoglycan modulation of Wnt5A signal transduction in metastatic melanoma cells. J Biol Chem. 2009;284:28704–12. doi: 10.1074/jbc.M109.028498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanderson RD, Yang Y. Syndecan-1: a dynamic regulator of the myeloma microenvironment. Clin Exp Metastasis. 2008;25:149–59. doi: 10.1007/s10585-007-9125-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reiland J, Kempf D, Roy M, Denkins Y, Marchetti D. FGF2 binding, signaling, and angiogenesis are modulated by heparanase in metastatic melanoma cells. Neoplasia. 2006;8:596–606. doi: 10.1593/neo.06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ilan N, Elkin M, Vlodavsky I. Regulation, function and clinical significance of heparanase in cancer metastasis and angiogenesis. Int J Biochem Cell Biol. 2006;38:2018–39. doi: 10.1016/j.biocel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 10.Vreys V, David G. Mammalian heparanase: what is the message? J Cell Mol Med. 2007;11:427–52. doi: 10.1111/j.1582-4934.2007.00039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ridgway LD, Wetzel MD, Marchetti D. Modulation of GEF-H1 induced signaling by heparanase in brain metastatic melanoma cells. J Cell Biochem. 2010;111:1299–309. doi: 10.1002/jcb.22854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zetser A, Bashenko Y, Miao HQ, Vlodavsky I, Ilan N. Heparanase affects adhesive and tumorigenic potential of human glioma cells. Cancer Res. 2003;63:7733. [PubMed] [Google Scholar]
- 13.Cohen-Kaplan V, Doweck I, Naroditsky I, Vlodavsky I, Ilan N. Heparanase augments epidermal growth factor receptor phosphorylation: correlation with head and neck tumor progression. Cancer Res. 2008;68:10077–85. doi: 10.1158/0008-5472.CAN-08-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L, Sullivan PS, Goodman JC, Gunaratne PH, Marchetti D. MicroRNA-1258 suppresses breast cancer brain metastasis by targeting heparanase. Cancer Res. 2011;71:645–54. doi: 10.1158/0008-5472.CAN-10-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nalbant P, Chang YC, Birkenfeld J, Chang ZF, Bokoch GM. Guanine nucleotide exchange factor-H1 regulates cell migration via localized activation of RhoA at the leading edge. Mol Biol Cell. 2009;20:4070–82. doi: 10.1091/mbc.E09-01-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ridley AJ, Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–99. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- 17.Sanz-Moreno V, Gadea G, Ahn J, et al. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 2008;135:510–23. doi: 10.1016/j.cell.2008.09.043. [DOI] [PubMed] [Google Scholar]
- 18.Sanz-Moreno V, Marshall CJ. Rho-GTPase signaling drives melanoma cell plasticity. Cell Cycle. 2009;8:1484–7. doi: 10.4161/cc.8.10.8490. [DOI] [PubMed] [Google Scholar]
- 19.Symons M, Segall JE. Rac and Rho driving tumor invasion: who’s at the wheel? Genome Biol. 2009;10:213. doi: 10.1186/gb-2009-10-3-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heasman SJ, Carlin LM, Cox S, Ng T, Ridley AJ. Coordinated RhoA signaling at the leading edge and uropod is required for T cell transendothelial migration. J Cell Biol. 2010;190:553–63. doi: 10.1083/jcb.201002067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Birkenfeld J, Nalbant P, Yoon SH, Bokoch GM. Cellular functions of GEF-H1, a microtubule-regulated Rho-GEF: is altered GEF-H1 activity a crucial determinant of disease pathogenesis? Trends Cell Biol. 2008;18:210–9. doi: 10.1016/j.tcb.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 22.Lorger M, Felding-Habermann B. Capturing changes in the brain micro-environment during initial steps of breast cancer brain metastasis. Am J Pathol. 2010;176:2958–71. doi: 10.2353/ajpath.2010.090838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang L, Sullivan P, Suyama J, Marchetti D. Epidermal growth factor-induced heparanase nucleolar localization augments DNA topoisomerase I activity in brain metastatic breast cancer. Mol Cancer Res. 2010;8:278–90. doi: 10.1158/1541-7786.MCR-09-0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu J, Millard M, Ren X, Cox OT, Erdreich-Epstein A. C-Abl mediates endothelial apoptosis induced by inhibition of integrins alphavbeta3 and alphavbeta5 and by disruption of actin. Blood. 2010;115:2709–18. doi: 10.1182/blood-2009-05-223776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stins MF, Shen Y, Huang SH, Gilles F, Kalra VK, Kim KS. Gp120 activates children’s brain endothelial cells via CD4. J Neurovirol. 2001;7:125–34. doi: 10.1080/13550280152058780. [DOI] [PubMed] [Google Scholar]
- 26.Ridgway LD, Kim EY, Dryer SE. MAGI-1 interacts with Slo1 channel proteins and suppresses Slo1 expression on the cell surface. Am J Physiol Cell Physiol. 2009;297:C55–65. doi: 10.1152/ajpcell.00073.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eugenin EA, Berman JW. Chemokine-dependent mechanisms of leukocyte trafficking across a model of the blood-brain barrier. Methods. 2003;29:351–61. doi: 10.1016/s1046-2023(02)00359-6. [DOI] [PubMed] [Google Scholar]
- 28.Roy M, Reiland J, Murry BP, Chouljenko V, Kousoulas KG, Marchetti D. Antisense-mediated suppression of heparanase gene inhibits melanoma cell invasion. Neoplasia. 2005;7:253–62. doi: 10.1593/neo.04493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minard ME, Kim LS, Price JE, Gallick GE. The role of the guanine nucleotide exchange factor Tiam1 in cellular migration, invasion, adhesion and tumor progression. Breast Cancer Res Treat. 2004;84:21–32. doi: 10.1023/B:BREA.0000018421.31632.e6. [DOI] [PubMed] [Google Scholar]
- 30.Batzer AG, Rotin D, Urena JM, Skolnik EY, Schlessinger J. Hierarchy of binding sites for Grb2 and Shc on the epidermal growth factor receptor. Mol Cell Biol. 1994;14:5192–201. doi: 10.1128/mcb.14.8.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burbach BJ, Friedl A, Mundhenke C, Rapraeger AC. Syndecan-1 accumulates in lysosomes of poorly differentiated breast carcinoma cells. Matrix Biol. 2003;22:163–77. doi: 10.1016/s0945-053x(03)00009-x. [DOI] [PubMed] [Google Scholar]
- 32.Dokmanovic M, Hirsch DS, Shen Y, Wu WJ. Rac1 contributes to trastuzumab resistance of breast cancer cells: Rac1 as a potential therapeutic target for the treatment of trastuzumab-resistant breast cancer. Mol Cancer Ther. 2009;8:1557–69. doi: 10.1158/1535-7163.MCT-09-0140. [DOI] [PubMed] [Google Scholar]
- 33.Birkenfeld J, Nalbant P, Bohl BP, Pertz O, Hahn KM, Bokoch GM. GEF-H1 modulates localized RhoA activation during cytokinesis under the control of mitotic kinases. Dev Cell. 2007;12:699–712. doi: 10.1016/j.devcel.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ren Y, Li R, Zheng Y, Busch H. Cloning and characterization of GEF-H1, a microtubule-associated guanine nucleotide exchange factor for Rac and Rho GTPases. J Biol Chem. 1998;273:34954–60. doi: 10.1074/jbc.273.52.34954. [DOI] [PubMed] [Google Scholar]
- 35.Guilluy C, Swaminathan V, Garcia-Mata R, O’Brien ET, Superfine R, Burridge K. The Rho GEFs LARG and GEF-H1 regulate the mechanical response to force on integrins. Nat Cell Biol. 2011;13:722–7. doi: 10.1038/ncb2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levy-Adam F, Feld S, Suss-Toby E, Vlodavsky I, Ilan N. Heparanase facilitates cell adhesion and spreading by clustering of cell surface heparan sulfate proteoglycans. PLoS One. 2008;3:e2319. doi: 10.1371/journal.pone.0002319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palmieri D, Bronder JL, Herring JM, et al. Her-2 overexpression increases the metastatic outgrowth of breast cancer cells in the brain. Cancer Res. 2007;67:4190–8. doi: 10.1158/0008-5472.CAN-06-3316. [DOI] [PubMed] [Google Scholar]
- 38.Guo F, Debidda M, Yang L, Williams DA, Zheng Y. Genetic deletion of Rac1 GTPase reveals its critical role in actin stress fiber formation and focal adhesion complex assembly. J Biol Chem. 2006;281:18652–9. doi: 10.1074/jbc.M603508200. [DOI] [PubMed] [Google Scholar]
- 39.Schwartz BR, Wayner EA, Carlos TM, Ochs HD, Harlan JM. Identification of surface proteins mediating adherence of CD11/CD18-deficient lymphoblastoid cells to cultured human endothelium. J Clin Invest. 1990;85:2019–22. doi: 10.1172/JCI114668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim ST, Longley RL, Couchman JR, Woods A. Direct binding of syndecan-4 cytoplasmic domain to the catalytic domain of protein kinase C alpha (PKC alpha) increases focal adhesion localization of PKC alpha. J Biol Chem. 2003;278:13795–802. doi: 10.1074/jbc.M208300200. [DOI] [PubMed] [Google Scholar]
- 41.Dovas A, Yoneda A, Couchman JR. PKCbeta-dependent activation of RhoA by syndecan-4 during focal adhesion formation. J Cell Sci. 2006;119:2837–46. doi: 10.1242/jcs.03020. [DOI] [PubMed] [Google Scholar]
- 42.Beauvais DM, Rapraeger AC. Syndecans in tumor cell adhesion and signaling. Reprod Biol Endocrinol. 2004;2:3. doi: 10.1186/1477-7827-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oh ES, Woods A, Couchman JR. Syndecan-4 proteoglycan regulates the distribution and activity of protein kinase C. J Biol Chem. 1997;272:8133–6. doi: 10.1074/jbc.272.13.8133. [DOI] [PubMed] [Google Scholar]
- 44.Horowitz A, Simons M. Phosphorylation of the cytoplasmic tail of syndecan-4 regulates activation of protein kinase Calpha. J Biol Chem. 1998;273:25548–51. doi: 10.1074/jbc.273.40.25548. [DOI] [PubMed] [Google Scholar]
- 45.Horowitz A, Simons M. Regulation of syndecan-4 phosphorylation in vivo. J Biol Chem. 1998;273:10914–8. doi: 10.1074/jbc.273.18.10914. [DOI] [PubMed] [Google Scholar]
- 46.Ridgway LD, Wetzel MD, Marchetti D. Heparanase Modulates Shh and Wnt3a Signaling in Human Medulloblastoma Cells. Exp Ther Med. 2011;2:229–38. doi: 10.3892/etm.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Plow EF, Hass TA, Zhang Li, et al. Ligand binding to integrins. J Biol Chem. 2000;275(29):21785–8. doi: 10.1074/jbc.R000003200. [DOI] [PubMed] [Google Scholar]
- 48.Haataja L, Groffen J, Heisterkamp N. Characterization of RAC3, a novel member of the Rho family. J Biol Chem. 1997;272(33):20384–8. doi: 10.1074/jbc.272.33.20384. [DOI] [PubMed] [Google Scholar]
- 49.Mira JP, Benard V, Groffen J, Sanders LC, Knaus UG. Endogenous, hyperactive Rac3 controls proliferation of breast cancer cells by a p21-activated kinase-dependent pathway. Proc Natl Acad Sci U S A. 2000;97(1):185–9. doi: 10.1073/pnas.97.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Naggi A, Casu B, Perez M, Torri G, Cassinelli G, Penco S, et al. Modulation of the heparanase-inhibiting activity of heparin through selective desulfation, graded N-acetylation, and gycol splitting. J Biol Chem. 2005;280:12103–13. doi: 10.1074/jbc.M414217200. [DOI] [PubMed] [Google Scholar]
- 51.Ritchie JP, Ramani VC, Ren Y, et al. SST0001, a chemically modified heparin, inhibits myeloma growth and angiogenesis via disruption of the heparanase/syndecan-1 axis. Clin Cancer Res. 2001;17(6):1382–93. doi: 10.1158/1078-0432.CCR-10-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. Sequences used for the GEF-H1 knockdown experiments via lentivirus expressed shRNA.
Supplemental Table 2. Sequences used for the HPSE knockdown experiments via transiently transfected siRNA.
Supplemental Table 3. Gene accession numbers and DNA sequences for the oligonucleotide primer pairs utilized for RT-PCR.
Supplemental Figure 1. A. Western blot of purified preparations of human latent and active HPSE recombinant proteins showing distinct molecular weights, β-actin was probed as a loading control. M.W. standards were run in parallel with samples (left lane). B. Dose-dependent inhibition of heparanase (A-HPSE) endoglycosidase activity by SST0001, a glycol-split, non – anticoagulant form of heparin (50, 51). The heparanase inhibitory activity and structural details of SST0001 has been described ( 50, 51). Aliquots of purified recombinant human active HPSE were treated with various concentrations of SST0001 (0, 1, 3, and 10 mg/ml, respectively) for 2 hr. at 37°C. HPSE activity (U/ml) was then determined using the heparan sulfate degrading enzyme assay kit™ (Takara, Inc.: cat # MK412) following manufacturer’s protocol.
Supplemental Figure 2. Western blot probed for GEF-H1 used to generate GEF-H1 knockdowns (fig. 4A). β-actin was probed as a loading control.
Supplemental Figure 3. Gated populations from FACS demonstrating the selection of transduced high GFP - expressing 231BR3 cells.
Supplemental Figure 4. 231BR3 GFP - expressing cells. A. 231BR3 scrambled control cells expressing GFP. B. 231BR3 GEF-H1 shRNA (clone 1) cells expressing GFP. C. 231BR3 GEF-H1 shRNA (clone 4) cells expressing GFP. Upper panel: brightfield images. Lower panel: GFP images.