Abstract
Genome-wide association studies (GWAS) have identified over 40 genetic loci associated with colorectal cancer (CRC) risk. The association of single nucleotide polymorphisms (SNPs) at these loci with CRC risk and survival has not been adequately evaluated in East Asians. GWAS-identified CRC risk variants were used to construct weighted genetic risk scores (GRSs). We evaluated these GRSs in association with CRC risk in 3,303 CRC cases and 3,553 controls using logistic regression models. Associations with overall and CRC-specific survival were assessed in 731 CRC patients using Cox regression models. The association between the GRSs (overall and Asian-specific) and CRC risk was approximately 2-fold (highest versus lowest quintile), and the shape of the dose-response was linear (Ptrend =1.24×10−13 and 3.02×10−14 for overall GRS and Asian-specific GRS, respectively). The association of the GRS with CRC risk was stronger among those with a family history of CRC (Pinteraction =0.007). Asian-specific GRS using previously reported survival SNPs increased risk for mortality and the shape of the dose-response was linear for CRC-specific and all-cause mortality (Ptrend =0.01 and 0.006, respectively). Furthermore, the minor alleles of rs6983267 and rs1957636 were associated with worse CRC-specific and overall survival. We show that GRSs constructed using GWAS-identified common variants are strongly associated with CRC risk in Asians. We confirm previous findings for the possible association between some SNPs with survival, and provide evidence for two additional CRC risk variants that may be related to CRC survival.
Keywords: GWAS, single nucleotide polymorphism, colorectal cancer, survival, genetics, review
Introduction
Colorectal cancer (CRC) is the third most common cancer and the fourth leading cause of cancer-related mortality worldwide, resulting in over 1.6 million new cases and 771,000 deaths each year.1 Recently, genome-wide association studies (GWAS) have identified multiple single nucleotide polymorphisms (SNPs) at approximately 40 loci associated with CRC susceptibility.2–25 Of these, 18 loci were identified in previous studies conducted in the Asia Colorectal Cancer Consortium.9,11,12,17–19,23 Many of the risk variants initially identified in GWAS conducted among European descendants have not yet been investigated in Asian populations. In addition, no study has investigated the association of all these known risk variants collectively with CRC risk in Asians. Moreover, only a few studies have investigated the association between GWAS-identified risk variants and CRC prognosis, and the findings from these studies were inconsistent.26–33 In the present study, we provided a summary of all CRC risk loci identified by GWAS and examined their associations with CRC risk and survival among an Asian population.
Materials and Methods
Study population
Data used for the current project came from studies conducted in Shanghai. Details of the study design are described elsewhere.9,11,17,23 Briefly, the project includes 3,303 CRC cases from three resources: the Shanghai Women’s Health Study (SWHS) (N = 489), the Shanghai Men’s Health Study (SMHS) (N = 239) and the Shanghai Cancer Registry (N = 2,575). Cancer-free controls for the study (N = 3,553) were randomly selected from the SWHS (N = 956), the SMHS (N = 692), and the Shanghai GWAS of breast cancer (N = 1,905).
The cases from the population-based Shanghai Cancer Registry were recruited between January 2009 and February 2011. The SWHS and the SMHS are two on-going, population-based, prospective cohort studies, conducted in Shanghai, China. Details of the study designs and baseline questionnaires were published previously.34,35 Briefly, permanent residents of the study communities in Shanghai were approached for the study by trained interviewers. At baseline recruitment, anthropometric measurements and information on socio-demographic characteristics, lifestyle factors, and medical history were collected through in-person interviews. The SWHS recruited 74,942 women from 1997 to 2000 (participation rate 92.7%), and the SMHS recruited 61,480 men from 2002 to 2006 (participation rate 74.0%). These two cohorts have been followed up through a combination of in-person surveys every 2 to 4 years and annual record linkage with the population-based Shanghai Cancer Registry and the Shanghai Vital Statistics Registry to identify incident cancer cases and cause-specific mortality. CRC diagnosis and staging were assessed according to World Health Organization classifications and TNM classifications from the 7th edition of the American Joint Committee on Cancer. For cancer cases, clinical information was obtained by reviewing medical records and pathological slides. Participants with known hereditary CRC (e.g., Lynch syndrome) were excluded from the case populations. All participants provided informed consent for genetic analysis, and all studies were approved by the relevant institutional review boards for human research. The present analysis was restricted to study participants who were diagnosed with incident invasive colorectal adenocarcinoma during the study follow-up period and after the date of blood or buccal collection for DNA testing. For survival analysis, inclusion was restricted to the 731 CRC patients from the SWHS and SMHS for whom survival outcome data were available. For the analysis of genetic variants with CRC risk, however, all cases from the SWHS, SMHS, and those recruited through Shanghai Cancer Registry were included, along with their controls.
Genotyping, quality control and genotype imputation
Genomic DNA was extracted from blood samples or buccal cells using conventional methods. The details of genotyping and quality control for samples from the SWHS and SMHS were reported previously.9,11,17,23 The samples from the Shanghai Cancer Registry were genotyped with Infinium OncoArray-500K Bead Chip (Illumina, San Diego, CA) in accordance with the manufacturer’s protocol. Samples were excluded if the call rate was < 95%, heterozygosity was < 5% or > 40%, were from close relatives, or contained discrepant gender information between self-reported and genetically determined. SNPs that met any of the following criteria were excluded: call rate was < 95%, P value was < 10−7 in the Hardy-Weinberg equilibrium test among the controls, a call rate of <95%, or a poor genotyping cluster plot. All the samples meeting the quality-control criteria were imputed with the 1000 Genome Project Phase 3 data as reference using the program Minimac3 (University of Michigan, Ann Arbor, MI).
SNP selection
We compiled a list of all the loci reported to be associated with colorectal cancer risk at P < 5×10−8 through literature review, which identified 57 SNPs at 42 loci associated with CRC (Table 1). SNPs were excluded for the following reasons: a minor allele frequency of ≤ 5% in our study population (N = 8), a low imputation quality (R2 < 0.8) (N = 13), SNPs in high linkage disequilibrium (LD) with each other (N = 4), or SNP on the X chromosome (N = 1). We excluded the SNP on the X chromosome, given this SNP is located in the non-pseudoautosomal region of the X chromosome (X: 9,751,474, Table 1). If SNPs were in linkage disequilibrium (LD; r2 ≥ 0.4, based on the 1000 Genome Project Phase 3 Asian population), the SNP derived from the largest sample size was included. After these exclusions, 38 SNPs were retained for analysis (Supplementary Table 1).
Table 1.
Summary of genetic variants identified to date by genome-wide association studies in relation to colorectal cancer risk at the significant level of P< 5×10−8
| Locus | SNP | Position (hg19) | Allelesa | Nearby genes (putative gene target)b | RAFc | Study populationd | Author (reference #e) |
|---|---|---|---|---|---|---|---|
| 1p36.12 | rs72647484 | 22,587,728 | T/C | MIR4418 (CDC42 or WNT4) | 0.92/1.00 | European | Al-Tassan et al. (2) |
| 1q25.3 | rs10911251 | 183,081,194 | A/C | LAMC1 (LAMC1) | 0.55/0.48 | European | Whiffin et al. (3) |
| 1q41 | rs6691170 | 222,045,446 | T/G | RP11-191N8.2 (DUSP10) | 0.40/0.00 | European | Houlston et al. (4) |
| 1q41 | rs6687758 | 222,164,948 | G/A | RP11-400N13.2 (DUSP10) | 0.22/0.20 | European | Houlston et al. (4) |
| 2q32.3 | rs11903757 | 192,587,204 | C/T | NABP1 (MYO1B) | 0.17/0.07 | European | Peters et al. (5) |
| 2q35 | rs992157 | 219,154,781 | A/G | PNKD, TMBIM1 (PNKD or TMBIM1) | 0.58/0.62 | European | Orlando et al. (6) |
| 3p22.1 | rs35360328 | 40,924,962 | A/T | RP11-761N21.1 (CTNNB1) | 0.16/0.08 | European and Asian | Schumacher et al. (7) |
| 3p14.1 | rs812481 | 66,442,435 | G/C | LRIG1 (LRIG1) | 0.56/0.79 | European and Asian | Schumacher et al. (7) |
| 3q26.2 | rs10936599 | 169,492,101 | C/T | MYNN (TERC) | 0.76/0.42 | European | Houlston et al. (4) |
| 4q32.2 | rs35509282 | 163,333,405 | A/T | FSTL5 (NAF1) | 0.11/0.41 | European | Schmit et al. (8) |
| 5q31.1 | rs647161 | 134,499,092 | A/C | PITX1 (PITX1) | 0.66/0.30 | Asian and European | Jia et al. (9) |
| 6p21.2 | rs1321311 | 36,622,900 | A/C | CDKN1A (CDKN1A) | 0.22/0.17 | European | Dunlop et al. (10) |
| 6p21.1 | rs4711689 | 41,692,812 | A/G | TFEB (TFEB) | 0.54/0.80 | Asian | Zeng et al.(11) |
| 6q25.3 | rs7758229 | 160,840,252 | T/G | SLC22A3 (SLC22A3) | 0.32/0.25 | Asian | Cui et al. (12) |
| 8q23.3 | rs2450115 | 117,624,093 | T/C | EIF3H (EIF3H) | 0.81/0.54 | Asian | Zeng et al.(11) |
| 8q23.3 | rs16892766 | 117,630,683 | C/A | EIF3H (EIF3H) | 0.09/0.00 | European | Tomlinson et al. (13) |
| 8q23.3 | rs6469656 | 117,647,788 | A/G | EIF3H (EIF3H) | 0.89/0.67 | Asian | Zeng et al.(11) |
| 8q24.21 | rs6983267 | 128,413,305 | G/T | RP11-382A18.2 (MYC) | 0.50/0.39 | European | Tomlinson et al. (14) |
| 8q24.21 | rs7014346 | 128,424,792 | A/G | POU5F1B (MYC) | 0.33/0.29 | European | Tenesa et al. (16) |
| 10p14 | rs10795668 | 8,701,219 | G/A | RNA5SP299 (GATA3) | 0.68/0.63 | European | Tomlinson et al. (13) |
| 10q22.3 | rs704017 | 80,819,132 | G/A | ZMIZ1 (ZMIZ1 or AS1) | 0.56/0.28 | Asian | Zhang et al. (17) |
| 10q24.2 | rs1035209 | 101,345,366 | T/C | snoU13 (NKX2-3) | 0.19/0.18 | European | Whiffin et al. (3) |
| 10q24.2 | rs11190164 | 101,351,704 | G/A | snoU13 (NKX2-3) | 0.28/0.23 | European and Asian | Schumacher et al. (7) |
| 10q24.32 | rs4919687 | 104,595,248 | G/A | CYP17A1 (CYP17A1) | 0.70/0.75 | Asian | Zeng et al.(11) |
| 10q25.2 | rs12241008 | 114,280,702 | C/T | VTI1A (VTI1A) | 0.10/0.30 | Asian and African | Wang et al. (18) |
| 10q25.2 | rs10506868 | 114,319,380 | T/C | VTI1A (VTI1A) | 0.03/0.29 | Asian | Zeng et al.(11) |
| 10q25.2 | rs11196172 | 114,726,843 | A/G | TCF7L2 (TCF7L2) | 0.12/0.65 | Asian | Zhang et al. (17) |
| 11q12.2 | rs174537 | 61,552,680 | G/T | MYRF (FEN1, FADS1 or FADS2) | 0.65/0.43 | Asian | Zhang et al. (17) |
| 11q13.4 | rs3824999 | 74,345,550 | G/T | POLD3 (POLD3) | 0.52/0.42 | European | Dunlop et al. (10) |
| 11q23.1 | rs3802842 | 111,171,709 | C/A | COLCA2 (COLCA2) | 0.27/0.40 | European | Tenesa et al. (16) |
| 12p13.32 | rs10774214 | 4,368,352 | T/C | CCND2 (CCND2) | 0.38/0.32 | Asian and European | Jia et al. (9) |
| 12p13.32 | rs3217810 | 4,388,271 | T/C | CCND2 (CCND2) | 0.12/0.01 | European | Whiffin et al. (3) |
| 12p13.31 | rs10849432 | 6,385,727 | T/C | PLEKHG6 (CD9) | 0.90/0.81 | Asian | Zhang et al. (17) |
| 12p13.31 | rs11064437 | 6,982,162 | C/T | SPSB2 (SPSB2) | 0.99/0.72 | Asian | Zeng et al.(11) |
| 12p13.2 | rs2238126 | 12,009,741 | G/A | ETV6 (ETV6) | 0.19/0.47 | Asian | Wang et al. (19) |
| 12q13.12 | rs7136702 | 50,880,216 | T/C | LARP4 (DIP2B or ATF1) | 0.34/0.44 | European | Houlston et al. (4) |
| 12q13.12 | rs11169552 | 51,155,663 | C/T | ATF1 (DIP2B or ATF1) | 0.75/0.60 | European | Houlston et al. (4) |
| 12q24.12 | rs3184504 | 111,884,608 | C/T | SH2B3 (SH2B3) | 0.54/1.00 | European and Asian | Schumacher et al. (7) |
| 12q24.12 | rs73208120 | 117,747,590 | G/T | NOS1 (NOS1) | 0.08/0.00 | European and Asian | Schumacher et al. (7) |
| 14q22.2 | rs4444235 | 54,410,919 | C/T | MIR5580 (BMP4) | 0.49/0.47 | European | Houlston et al. (20) |
| 14q22.2 | rs1957636 | 54,560,018 | T/C | BMP4 (BMP4) | 0.41/0.66 | European | Tomlinson et al. (21) |
| 15q13.3 | rs16969681 | 32,993,111 | T/C | SCG5 (GREM1) | 0.07/0.37 | European | Tomlinson et al. (21) |
| 15q13.3 | rs4779584 | 32,994,756 | T/C | SCG5 (GREM1) | 0.20/0.81 | European | Jaeger et al. (22) |
| 15q13.3 | rs11632715 | 33,004,247 | A/G | SCG5 (GREM1) | 0.46/0.77 | European | Tomlinson et al. (21) |
| 16q22.1 | rs9929218 | 68,820,946 | G/A | CDH1 (CDH1) | 0.71/0.77 | European | Houlston et al. (20) |
| 17p13.3 | rs12603526 | 800,593 | C/T | NXN (NXN) | 0.01/0.20 | Asian | Zhang et al. (17) |
| 18q21.1 | rs7229639 | 46,450,976 | A/G | SMAD7 (SMAD7) | 0.10/0.13 | Asian | Zhang et al. (23) |
| 18q21.1 | rs4939827 | 46,453,463 | T/C | SMAD7 (SMAD7) | 0.53/0.31 | European | Broderick et al. (24) |
| 19q13.11 | rs10411210 | 33,532,300 | C/T | RHPN2 (RHPN2) | 0.90/0.81 | European | Houlston et al. (20) |
| 19q13.2 | rs1800469 | 41,860,296 | G/A | TMEM91 (TGFB1) | 0.69/0.45 | Asian | Zhang et al. (17) |
| 20p12.3 | rs961253 | 6,404,281 | A/C | CASC20 (BMP2) | 0.36/0.11 | European | Houlston et al. (20) |
| 20p12.3 | rs4813802 | 6,699,595 | G/T | RP5-859D4.3 (BMP2) | 0.32/0.23 | European | Tomlinson et al. (21) |
| 20p12.3 | rs2423279 | 7,812,350 | C/T | HAO1 (BMP2) | 0.27/0.33 | Asian and European | Jia et al. (9) |
| 20q13.13 | rs6066825 | 47,340,117 | A/G | PREX1 (PREX1) | 0.62/0.71 | European and Asian | Schumacher et al. (7) |
| 20q13.33 | rs4925386 | 60,921,044 | C/T | LAMA5 (LAMA5) | 0.67/0.74 | European | Houlston et al. (4) |
| 20q13.33 | rs6061231 | 60,956,917 | C/A | RPS21 (LAMA5) | 0.70/0.84 | Asian | Zeng et al.(11) |
| Xp22.2 | rs5934683 | 9,751,474 | T/C | GPR143 (SHROOM2) | 0.39/0.82 | European | Dunlop et al. (10) |
Abbreviations: RAF – risk allele frequency
risk/reference allele;
based on the information from GENCODE 19;
RAF in Europeans/East Asians based on 1000 Genome Project Phase 3 allele frequencies;
The association for the six SNPs reported by Schumacher et al. (7) was mainly driven by the meta-analysis in European populations and thus they were classified as “first reported in European population”.
Reference serial number.
Statistical analysis
Using an additive genetic model, we calculated weighted genetic risk scores (GRSs) using the natural log-odds ratios (ORs or β) as the SNP-specific weight which was obtained from previous studies having the largest combined sample sizes and showing statistically significant associations (Supplementary Table 1). Two GRSs were calculated as the sum of the product of the weight (i.e., β) and the number of risk alleles (i.e., dosage) for k SNPs included in the GRS (i.e., ) per individual. The overall GRS was calculated as the sum of weighted risk alleles across all 38 SNPs. Asian-specific GRS was calculated based on 25 SNPs that were identified or replicated in Asian populations. The GRSs were categorized into quintiles (Q) using the following cut-points for the overall GRS: Q1≤4.14, Q2: 4.15 to 4.45, Q3: 4.46 to 4.72, Q4: 4.73 to 5.02, Q5≥5.03; and the following cut-points for the Asian-specific GRS: Q1≤ 2.57, Q2: 2.58–2.79, Q3: 2.80 to 2.99, Q4: 3.00 to 3.21, Q5≥3.22. Differences in socio-demographic characteristics and lifestyle risk factors between controls and cases were evaluated using a t-test for continuous variables or a chi-square test for categorical variables. ORs and 95% confidence intervals (95% CIs) of weighted GRSs for CRC risk were derived from logistic regression models adjusted for age at diagnosis, sex, study sets, and principal components for ancestry, which were estimated using GWAS data. Stratified analyses were performed to assess whether the associations differed by age at diagnosis (< 60, ≥ 60 years), sex, BMI (< 25.0, 25.0–29.9, or > 30.0 kg/m2), or family history of CRC.
For survival analysis, we calculated the time from CRC diagnosis to death from CRC or any cause, or December 31, 2013 (the end of the follow-up period), whichever came first. Hazard ratios (HRs) and 95% CIs for CRC-specific and all-cause mortality were estimated using Cox proportional hazard models to evaluate the associations of CRC survival with weighted GRSs. All models were adjusted for age, sex, tumor location, and stage. In addition, we conducted analyses of the GRSs in relation to survival stratified by age at diagnosis, sex, BMI (< 25.0, 25.0–29.9, or > 30.0 kg/m2), tumor location (colon/rectum), and stage at diagnosis. We also used Cox proportional hazard regression models to estimate HRs and 95% CIs for individual SNPs. Kaplan-Meier survival curves were used to visualize the differences in unadjusted survival curves, and the log-rank test was performed to test the differences between groups. The proportional hazard assumption was evaluated by using the likelihood ratio test for the multiplicative interaction term between weighted GRS and time in nested models; no violation was observed. All analyses were performed using SAS, version 9.4 (SAS Institute, Inc., Cary, NC), and all tests of statistical significance were set at P < 0.05 for two-sided analyses.
Results
There are 57 SNPs at 42 genetic loci reported to date in association with CRC risk at genome-wide significance (P < 5×10−8) (Table 1). Of these, 18 loci were first reported in GWAS conducted in populations of East Asian ancestry or in pooled analyses of data from East-Asian descendants and data from other populations (Table 1).9,11,12,17–19,23 An additional 6 SNPs were replicated in Asian-ancestry populations.9,12,17 We excluded 19 SNPs from this analysis because they have a minor allele frequency < 0.05 in Asians, had a low imputation quality (R2 < 0.8) in our study, had high LD with other nearby SNPs selected for the study, or was on the X chromosome. After these exclusions, 38 SNPs remain for the present study (Supplementary Tables 1 and 2).
Association of Genetic Risk Scores with CRC Risk
Socio-demographic characteristics and lifestyle risk factors of controls and CRC cases are shown in Supplementary Table 3. Because of the study design, more female controls than male controls were included in the analysis. CRC cases were more likely to be overweight, to have more education and a family history of CRC than controls.
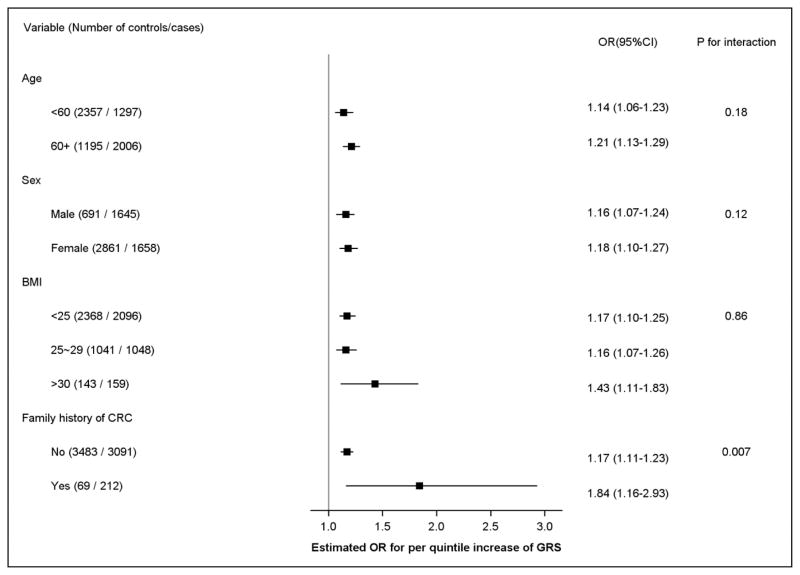
Both overall GRS (based on 38 SNPs) and Asian-specific GRS (based on 25 SNPs) were statistically significantly associated with CRC risk after adjusting for age, sex, study, and principal components following a dose-response pattern (P for trend = 1.24×10−13 and 3.02×10−14 for overall GRS and Asian-specific GRS, respectively; Table 2). The ORs (95% CIs) comparing the fifth to the first quintile were 2.00 (95% CI: 1.60 – 2.50) and 2.05 (95% CI: 1.65 – 2.56) for overall GRS and Asian-specific GRS, respectively. Because of similar results for the overall and Asian-specific GRSs, additional analyses were performed for Asian-specific GRS only. We additionally examined the association of the overall GRS and Asian-specific GRS with CRC risk after further adjusting for potential CRC risk factors including smoking, alcohol use, physical activity, and red meat intake. However, our results did not change appreciably after adjustment (data not shown). Stratified analyses showed a statistically significant interaction between the GRS and family history of CRC (P for interaction = 0.007) with a stronger association of GRS with CRC risk among individuals with a family history of CRC (per quintile: OR = 1.84, 95% CI = 1.16 – 2.93) than those without a family history of CRC (per quintile: OR = 1.17, 95% CI = 1.11 – 1.23) (Figure 1). No apparent multiplicative interaction was observed for the association between the Asian-specific GRS and colorectal cancer risk when stratified by sex (p for interaction = 0.12), age (<60 vs. 60+; p for interaction=0.18), and across BMI categories (<25 kg/m2, 25–29, and >30; p for interaction=0.86).
Table 2.
Association of weighted genetic risk scores (GRSs) of GWAS-identified CRC risk variants with CRC risk in Asians.
| GRS Quintilesa | Overall GRSb | Asian-specific GRSc | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Controls (N = 3,553) | Cases (N = 3,303) | OR | 95% CI | Controls (N = 3,553) | Cases (N = 3,303) | OR | 95% CI | |
| Q1(low) | 711 | 460 | 1.00 | reference | 711 | 477 | 1.00 | reference |
| Q2 | 711 | 563 | 1.25 | 0.99–1.59 | 711 | 573 | 1.15 | 0.91–1.46 |
| Q3 | 709 | 678 | 1.47 | 1.17–1.86 | 709 | 631 | 1.41 | 1.12–1.78 |
| Q4 | 712 | 697 | 1.76 | 1.40–2.22 | 712 | 677 | 1.37 | 1.09–1.72 |
| Q5 | 710 | 905 | 2.00 | 1.60–2.50 | 710 | 945 | 2.05 | 1.65–2.56 |
| P for trend | 1.24×10−13 | 3.02×10−14 | ||||||
Adjusted for age, sex, study, and the global genomic principal components.
Quintile cut-points for (1) Overall GRS: Q1≤4.14, Q2: 4.15 to 4.45, Q3: 4.46 to 4.72, Q4: 4.73 to 5.02, Q5≥5.03; and (2) Asian-specific GRS: Q1≤ 2.57, Q2: 2.58–2.79, Q3: 2.80 to 2.99, Q4: 3.00 to 3.21, Q5≥3.22.
Overall GRS was constructed using all CRC risk variants identified to date and available to this study.
Asian-specific GRS was constructed using CRC risk variants identified or replicated in Asian populations to date and available to this study.
Figure 1.
Colorectal cancer risk associated with weighted Asian-specific genetic risk score (GRS) stratified by age, sex, BMI and family history of CRC.
ORs: Adjusted by age, sex, study, and the global genomic principal components.
Association of Genetic Risk Score with survival
Demographic, clinical, and pathologic characteristics of the 731 patients included in the survival analysis are shown in Supplementary Table 4. The median age at diagnosis was 67 years (range 42 – 83 years); 241 (33%) patients were male; and 442 (60.5%) patients were diagnosed with colon cancer. The clinical stages were as follows: 147 patients at stage I (23.6%), 168 patients at stage II (27.0%), 228 patients at stage III (36.6%), and 80 patients at stage IV (12.8%). The majority of the patients received surgery (N = 688, 94.1%) and/or chemotherapy (N = 586, 80.2%). By December 31, 2013, a total of 359 deaths had been documented, of which 319 were attributed to CRC.
Multivariable-adjusted HRs and 95% CIs for CRC-specific and all-cause mortality by quintiles of Asian-specific GRS were shown in Table 3. No significant association was found between Asian-specific GRS and CRC-specific or all-cause mortality in the multivariable Cox regression analyses (Table 3) or in the Kaplan-Meier survival analysis (data not shown). We also constructed an Asian-specific GRS utilizing SNPs that have previously shown to be associated with survival among Asians (i.e., rs1321311, rs6983267, rs4939827, rs10411210, rs961253)29,30,32 and found a statistically significant linear trend with both CRC-specific and all-cause mortality (Table 3). Similar associations were observed for overall GRS with both CRC-specific and all-cause mortality (data not shown). We also evaluated whether the association of Asian-specific GRS with CRC-specific mortality may differ across the strata of potential predictors of survival, including age at diagnosis, sex, BMI, tumor location and stage (Supplementary Figure 1). There was no evidence of significant effect modification by any of these variables (P for heterogeneity > 0.05). Finally, similar to the CRC risk analysis, we also examined the association between the overall GRS and Asian-specific GRS with mortality after adjusting for potential CRC risk factors including smoking, alcohol use, physical activity, and red meat intake. However, results from these sensitivity analyses did not alter our conclusions (data not shown).
Table 3.
Association between Asian-specific GRS and mortality among CRC patients
| GRS Quintilesa | No. of patients | CRC-specific mortality | All-cause mortality | ||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| No. of events | HR | 95% CI | No. of events | HR | 95% CI | ||
| GRS using All SNPs | |||||||
| Q1(low) | 146 | 66 | 1.00 | reference | 73 | 1.00 | reference |
| Q2 | 146 | 65 | 1.12 | 0.79–1.59 | 70 | 1.08 | 0.77–1.50 |
| Q3 | 146 | 64 | 1.04 | 0.73–1.47 | 71 | 1.03 | 0.74–1.44 |
| Q4 | 147 | 64 | 1.26 | 0.89–1.79 | 76 | 1.34 | 0.96–1.86 |
| Q5 | 146 | 60 | 1.07 | 0.75–1.52 | 69 | 1.08 | 0.77–1.50 |
| P for trend | 0.55 | 0.35 | |||||
| GRS using survival SNPs | |||||||
| Q1(low) | 146 | 68 | 1.00 | reference | 74 | 1.00 | reference |
| Q2 | 146 | 34 | 1.15 | 0.76–1.75 | 38 | 1.15 | 0.77–1.70 |
| Q3 | 146 | 81 | 1.21 | 0.88–1.68 | 94 | 1.29 | 0.95–1.75 |
| Q4 | 147 | 74 | 1.17 | 0.84–1.63 | 84 | 1.23 | 0.89–1.68 |
| Q5 | 146 | 62 | 1.50 | 1.06–2.12 | 69 | 1.51 | 1.09–2.10 |
| P for trend | 0.01 | 0.006 | |||||
Adjusted by age, sex, tumor location, and stage.
GRS was constructed using CRC risk variants (n=25) identified or replicated in Asian populations to date and available to this study (All SNPs). GRS using survival SNPs incorporates 5 SNPs previously identified in the literature 29,30,32. Quintile cut-points for GRS using All 25 Asian-specific SNPs : Q1≤ 2.67, Q2: 2.68 to 2.88, Q3: 2.89 to 3.08, Q4: 3.09 to 3.35, Q5≥3.36. Quintile cut-points for GRS using 5 Asian-specific survival SNPs: Q1≤ 0.31, Q2: 0.32 to 0.36, Q3: 0.37 to 0.45, Q4: 0.46 to 0.56, Q5≥0.56.
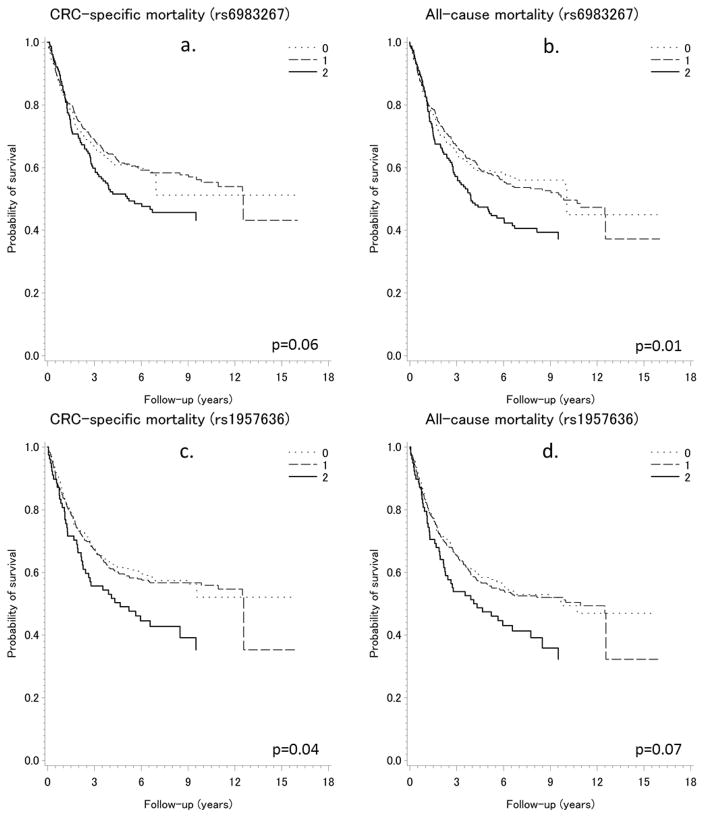
We also investigated whether any individual SNP may be associated with CRC survival and found two SNPs (rs6983267 near gene MYC and rs1957636 near gene BMP4) showing suggestive associations. In Kaplan-Meier curves showing survival probability according to the genotypes of rs6983267 and rs1957636, the minor alleles of both SNPs (GG for rs6983267; CC for rs1957636) were associated with higher risk of CRC-specific death (rs6983267: log-rank P = 0.06; rs1957636: log-rank P = 0.04) and death due to any cause (rs6983267: log-rank P = 0.01; rs1957636: log-rank P = 0.07) (Figure 2). Multivariate Cox regression analyses also showed that these two SNPs were associated with CRC survival (Table 4). When we calculated a risk score limited to these two SNPs, we observed significantly poorer CRC-specific survival (HR = 2.22, 95% CI = 1.11–4.47) and overall survival (HR = 2.05, 95% CI = 1.03–4.07) for patients homozygous for the minor allele in both SNPs. There was a dose-response relationship between the number of minor alleles of these two SNPs and risk of death due to CRC (P for trend = 0.01) or any cause (P for trend = 0.003) (Table 4). None of the remaining 36 SNPs was significantly associated with survival.
Figure 2.
Kaplan-Meier estimates of survival functions for CRC-specific and all-cause mortality by genotypes of SNPs rs6983267 and rs1957636 among patients with colorectal cancer (for rs6983267: 0=TT, 1=TG, 2=GG; for rs1957636: 0=TT, 1=CT, 2=CC)
Table 4.
Association of overall and colorectal cancer-specific mortality with SNP rs6983267 and rs1957636.
| SNPs | No. of cases (N=731) | CRC-specific mortality | Overall mortality | ||
|---|---|---|---|---|---|
|
|
|
||||
| No. of events | HR(95% CI) | No. of events | HR(95% CI) | ||
| rs6983267 | |||||
| TT | 208 | 85 | 1.00 (ref) | 91 | 1.00 (ref) |
| TG | 369 | 154 | 1.10 (0.84–1.43) | 176 | 1.17 (0.91–1.51) |
| GG | 154 | 80 | 1.39 (1.02–1.89) | 92 | 1.52 (1.13–2.03) |
| Trend tests | P = 0.041 | P = 6.02×10−3 | |||
| rs1957636 | |||||
| TT | 308 | 127 | 1.00 (ref) | 145 | 1.00 (ref) |
| CT | 345 | 147 | 1.01 (0.79–1.28) | 166 | 1.01 (0.81–1.26) |
| CC | 78 | 45 | 1.43 (1.01–2.01) | 48 | 1.37 (0.98–1.91) |
| Trend tests | P = 0.127 | P = 0.160 | |||
| No. of risk allelesa | |||||
| 0 | 81 | 24 | 1.00 (ref) | 27 | 1.00 (ref) |
| 1 | 271 | 122 | 1.54(0.99–2.40) | 136 | 1.56(1.03–2.36) |
| 2 | 248 | 102 | 1.50(0.96–2.34) | 114 | 1.50(0.98–2.28) |
| 3 | 113 | 59 | 1.93(1.20–3.12) | 70 | 2.10(1.34–3.28) |
| 4 | 18 | 12 | 2.22(1.11–4.47) | 12 | 2.05(1.03–4.07) |
| Trend tests | P = 0.010 | P = 0.003 | |||
Adjusted by age, sex, tumor location and stage.
Risk allele: G for rs6983267; C for rs1957636.
Bonferroni-adjusted α = 0.05/152 = 3.29×10−4 based on 38 SNPs, 2 comparisons per SNP, and 2 survival outcomes per SNP. None of the associations reached statistical significance after correction for multiple comparisons.
Discussion
In this study, we found that combinations of GWAS-identified risk variants for CRC, as measured by both overall GRS and Asian-specific GRS, were strongly associated with CRC risk following a dose-response pattern in a Chinese population. However, neither overall GRS nor Asian-specific GRS was associated with survival among CRC patients. Two SNPs, rs6983267 on chromosome 8q24.21 and rs1957636 on 14q22.2, were significantly associated with CRC survival, providing some evidence that certain genetic variants identified for CRC risk may also be related to CRC survival.
Since 2008, genetic variants at 42 loci have been found to be associated with the risk of CRC. Although the association of CRC risk with each of these genetic variants is weak, typically with an OR of 1.2 or lower, the GRS, an aggregate measure of the effect of multiple risk variants, showed a strong association with CRC risk. In both overall and Asian-specific GRS, individuals in the highest quintile of the weighted GRS had a 2-fold increased risk of CRC compared with those in the lowest quintile, providing evidence that the GRS might be useful, in combination with other predictors, to identify high-risk individuals for primary prevention and cancer screening. A family history of CRC is one of the most important risk factors for CRC, and in our study the association of CRC risk with the GRS was stronger among individuals with a family history of CRC than for those without a family history of CRC. It has been shown previously that GRS improved the prediction of CRC risk when family history of CRC was considered.36,37 Our results, along with those from these previous studies, provide support for using GRS to further classify CRC patients with a family history of CRC into different risk groups for personalized prevention of this common cancer.
Several studies have examined the relationship between GWAS-identified CRC risk variants and CRC survival, most of which were conducted among European descendants and focused on the evaluation of individual SNPs; their results were inconsistent. There were two null association reports.27,28 However, positive findings were also reported. For example, Phipps et al. found the minor allele in rs4939827 (SMAD7) was associated with reduced overall and CRC-specific survival.29 Dai et al. reported five SNPs (rs961253, rs355527, rs4464148, rs6983267 and rs10505477) were associated with survival for patients with stage III disease.30 Morris et al. reported that rs4444235 was significantly associated with survival in CRC patients.31 Smith et al. evaluated the influence of 20 GWAS-identified CRC risk SNPs in 7,635 cases, and found that patients who were homozygous for the minor allele (AA genotype) of rs9929218 had a poorer overall survival rate.33 To date, only two studies have been conducted among East Asians. The study of Xing et al. found an association between rs4779584 and a reduced risk of CRC mortality in 380 Chinese CRC patients.26 Kang et al. reported that rs1321311 (CDKN1A) and rs10411210 (RHPN2) were associated with survival for Korean patients with surgically resected CRC.32
In our study, we found that the GG genotype of rs6983267 and the CC genotype of rs1957636 were significantly associated with poorer survival outcomes. The SNP rs6983267 was first identified in GWAS as a CRC susceptibility loci mapping to 8q24.1 in European descendants,14 and was later replicated in East Asians.17 This SNP has previously been reported to be significantly associated with survival outcomes for CRC patients30 in the same direction as observed in the present study. The G allele of rs6983267 confers an increased CRC risk through the mechanism of Wnt signaling by disrupting an enhancer element and interacting with the promoter of the MYC oncogene.38 MYC is a well-known oncogene and is overexpressed in many tumors, including CRC.39 It lies 116 kb telomeric to rs6983267, and some reports have shown that rs6983267 has a long-range physical interaction with MYC in CRC cell lines.38,40 The variant rs6983267 that is associated with increased risk of colorectal adenomas 14 could also be associated with increased risk of CRC recurrence and initiation. Thus, it is possible that SNPs in MYC involved in CRC risk could also be related to CRC pathological severity and prognosis.
SNP rs1957636 was reported to be associated with increased risk of colorectal adenoma41 suggesting that this SNP might play a role in the initiation of CRC. We did not find any significant association of this SNP with CRC risk in our study. However, we observed a significant association of the C allele in rs1957636 with poorer survival after CRC diagnosis. One would expect that genetic factors involved in colorectal tumor progression but not initiation might be more strongly associated with risk of CRC mortality. The reasons for our findings are unclear. SNP rs1957636 is upstream of the transcriptional start site of BMP4 (136 kb upstream) and 150 kb downstream to CRC susceptibility SNP rs4444235; however, rs1957636 and rs4444235 are not in strong LD.21 Members of the BMP signaling pathway have been shown to interact with transforming growth factor-β (TGF-β)42, which behaves as a tumor suppressor by inhibiting cell proliferation in normal tissue but promotes metastasis by enhancing angiogenesis and extracellular matrix disruption in the tumor.29 The interaction between BMP4 and TGF-β and the pleiotropic functions of the TGF-β pathway may explain the seemingly opposite association of rs1957636 with null association for CRC but poor survival after CRC diagnosis.
The primary limitation of our study is the small sample size in the survival analysis. The statistical power is limited in the detection of a weak association with individual SNPs, particularly in stratified analyses by cancer stages. The significant association observed in this study for SNPs rs6983267 and rs1957636 was no longer statistically significant after taking into consideration multiple comparisons. Therefore, these associations should be further evaluated in future studies. Furthermore, the GRS included 17 SNPs that were initially identified in relation to CRC risk in the Asia Colorectal Cancer Consortium that include samples from the two Shanghai studies used in this analysis. However, the two Shanghai studies only account for ~23% of samples in the Asia consortium. Furthermore, the current analysis includes data from Shanghai that was not included in the Asia Colorectal Cancer Consortium study that identified these risk variants. The strength of the association across the three studies was similar for all quintiles and the estimates were not significantly different from one another (i.e., p for heterogeneity for Quintiles 2, 3, 4, 5, versus Quintile 1 = 0.71, 0.92, 0.97, 0.53, respectively). Thus, a pooled analysis of the individual-level data was conducted and adjusted for each individual study in the models. It is also possible that the alleles utilized in this analysis that were identified among European Americans may not be the purported risk allele among Asians. Thus, in the future the GRS could be further refined to include newly identified Asian-specific risk alleles.
In conclusion, our study indicated that GRSs constructed from risk SNPs identified in previous GWAS were significantly associated with CRC risk following a dose-response pattern, but they were not associated with CRC survival. However, common germline variants in GWAS-identified loci near the MYC and BMP4 genes may be associated with CRC survival. Further studies to independently evaluate these associations are warranted.
Supplementary Material
Novelty and Impact.
We systematically evaluated genetic risk variants identified by genome-wide association studies for colorectal cancer (CRC) in relation to CRC risk and survival in East Asians. We found that these variants combined were strongly associated with CRC risk, and the association was modified by CRC family history. We also provided evidence for a suggestive association of two risk variants (rs6983267 and rs1957636) in relation to CRC survival.
Acknowledgments
This work was supported by grants from the United States National Institutes of Health (R37 CA070867, R01 CA188214, UM1 CA182910, and UM1 CA173640). The authors thank the study participants and research staff for their contributions and support to this project, Regina Courtney and Jie Wu for DNA preparation, and Kim Kreth for editing the manuscript. Sample preparation was conducted at the Survey and Biospecimen Shared Resources, which are supported in part by the Vanderbilt-Ingram Cancer Center (P30 CA68485).
Abbreviations
- BMI
body mass index
- CI
confidence interval
- CRC
colorectal cancer
- GRS
genetic risk scores
- GWAS
Genome-wide association studies
- HR
hazard ratio
- LD
linkage disequilibrium
- OR
odds ratio
- SNP
single nucleotide polymorphisms
- SMHS
Shanghai Men’s Health Study
- SWHS
Shanghai Women’s Health Study
- TGF-β
transforming growth factor-β
Footnotes
Conflict of Interest: The authors declare no conflicts of interest or financial disclosures.
References
- 1.Global Burden of Disease Cancer Collaboration. Fitzmaurice C, Dicker D, et al. The Global Burden of Cancer 2013. JAMA Oncol. 2015;1:505–27. doi: 10.1001/jamaoncol.2015.0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Tassan NA, Whiffin N, Hosking FJ, et al. A new GWAS and meta-analysis with 1000Genomes imputation identifies novel risk variants for colorectal cancer. Sci Rep. 2015;5:10442. doi: 10.1038/srep10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whiffin N, Hosking FJ, Farrington SM, et al. Identification of susceptibility loci for colorectal cancer in a genome-wide meta-analysis. Hum Mol Genet. 2014;23:4729–37. doi: 10.1093/hmg/ddu177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Houlston RS, Cheadle J, Dobbins SE, et al. Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nat Genet. 2010;42:973–7. doi: 10.1038/ng.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peters U, Jiao S, Schumacher FR, et al. Identification of Genetic Susceptibility Loci for Colorectal Tumors in a Genome-Wide Meta-analysis. Gastroenterology. 2013;144:799–807. e24. doi: 10.1053/j.gastro.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orlando G, Law PJ, Palin K, et al. Variation at 2q35 (PNKD and TMBIM1) influences colorectal cancer risk and identifies a pleiotropic effect with inflammatory bowel disease. Hum Mol Genet. 2016 doi: 10.1093/hmg/ddw087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schumacher FR, Schmit SL, Jiao S, et al. Genome-wide association study of colorectal cancer identifies six new susceptibility loci. Nat Commun. 2015;6:7138. doi: 10.1038/ncomms8138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmit SL, Schumacher FR, Edlund CK, et al. A novel colorectal cancer risk locus at 4q32.2 identified from an international genome-wide association study. Carcinogenesis. 2014;35:2512–9. doi: 10.1093/carcin/bgu148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jia W-H, Zhang B, Matsuo K, et al. Genome-wide association analyses in East Asians identify new susceptibility loci for colorectal cancer. Nat Genet. 2013;45:191–6. doi: 10.1038/ng.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunlop MG, Dobbins SE, Farrington SM, et al. Common variation near CDKN1A, POLD3 and SHROOM2 influences colorectal cancer risk. Nat Genet. 2012;44:770–6. doi: 10.1038/ng.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeng C, Matsuda K, Jia W-H, et al. Identification of Susceptibility Loci and Genes for Colorectal Cancer Risk. Gastroenterology. 2016 doi: 10.1053/j.gastro.2016.02.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui R, Okada Y, Jang SG, et al. Common variant in 6q26–q27 is associated with distal colon cancer in an Asian population. Gut. 2011;60:799–805. doi: 10.1136/gut.2010.215947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomlinson IPM, Webb E, Carvajal-Carmona L, et al. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet. 2008;40:623–30. doi: 10.1038/ng.111. [DOI] [PubMed] [Google Scholar]
- 14.Tomlinson I, Webb E, Carvajal-Carmona L, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39:984–8. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 15.Zanke BW, Greenwood CMT, Rangrej J, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39:989–94. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 16.Tenesa A, Farrington SM, Prendergast JGD, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet. 2008;40:631–7. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang B, Jia W-H, Matsuda K, et al. Large-scale genetic study in East Asians identifies six new loci associated with colorectal cancer risk. Nat Genet. 2014;46:533–42. doi: 10.1038/ng.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H, Burnett T, Kono S, et al. Trans-ethnic genome-wide association study of colorectal cancer identifies a new susceptibility locus in VTI1A. Nat Commun. 2014;5:4613. doi: 10.1038/ncomms5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang M, Gu D, Du M, et al. Common genetic variation in ETV6 is associated with colorectal cancer susceptibility. Nat Commun. 2016;7:11478. doi: 10.1038/ncomms11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.COGENT Study. Houlston RS, Webb E, et al. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat Genet. 2008;40:1426–35. doi: 10.1038/ng.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomlinson IPM, Carvajal-Carmona LG, Dobbins SE, et al. Multiple common susceptibility variants near BMP pathway loci GREM1, BMP4, and BMP2 explain part of the missing heritability of colorectal cancer. PLoS Genet. 2011;7:e1002105. doi: 10.1371/journal.pgen.1002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaeger E, Webb E, Howarth K, et al. Common genetic variants at the CRAC1 (HMPS) locus on chromosome 15q13.3 influence colorectal cancer risk. Nat Genet. 2008;40:26–8. doi: 10.1038/ng.2007.41. [DOI] [PubMed] [Google Scholar]
- 23.Zhang B, Jia W-H, Matsuo K, et al. Genome-wide association study identifies a new SMAD7 risk variant associated with colorectal cancer risk in East Asians. Int J Cancer. 2014;135:948–55. doi: 10.1002/ijc.28733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Broderick P, Carvajal-Carmona L, Pittman AM, et al. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat Genet. 2007;39:1315–7. doi: 10.1038/ng.2007.18. [DOI] [PubMed] [Google Scholar]
- 25.Peters U, Hutter CM, Hsu L, et al. Meta-analysis of new genome-wide association studies of colorectal cancer risk. Hum Genet. 2012;131:217–34. doi: 10.1007/s00439-011-1055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xing J, Myers RE, He X, et al. GWAS-identified colorectal cancer susceptibility locus associates with disease prognosis. Eur J Cancer Oxf Engl 1990. 2011;47:1699–707. doi: 10.1016/j.ejca.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 27.Tenesa A, Theodoratou E, Din FVN, et al. Ten common genetic variants associated with colorectal cancer risk are not associated with survival after diagnosis. Clin Cancer Res Off J Am Assoc Cancer Res. 2010;16:3754–9. doi: 10.1158/1078-0432.CCR-10-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoskins JM, Ong P-S, Keku TO, et al. Association of eleven common, low-penetrance colorectal cancer susceptibility genetic variants at six risk loci with clinical outcome. PloS One. 2012;7:e41954. doi: 10.1371/journal.pone.0041954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phipps AI, Newcomb PA, Garcia-Albeniz X, et al. Association between colorectal cancer susceptibility loci and survival time after diagnosis with colorectal cancer. Gastroenterology. 2012;143:51–4. e4. doi: 10.1053/j.gastro.2012.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dai J, Gu J, Huang M, et al. GWAS-identified colorectal cancer susceptibility loci associated with clinical outcomes. Carcinogenesis. 2012;33:1327–31. doi: 10.1093/carcin/bgs147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morris EJA, Penegar S, Whiffin N, et al. A retrospective observational study of the relationship between single nucleotide polymorphisms associated with the risk of developing colorectal cancer and survival. PloS One. 2015;10:e0117816. doi: 10.1371/journal.pone.0117816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang BW, Jeon H-S, Chae YS, et al. Association between GWAS-identified genetic variations and disease prognosis for patients with colorectal cancer. PloS One. 2015;10:e0119649. doi: 10.1371/journal.pone.0119649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith CG, Fisher D, Harris R, et al. Analyses of 7,635 Patients with Colorectal Cancer Using Independent Training and Validation Cohorts Show That rs9929218 in CDH1 Is a Prognostic Marker of Survival. Clin Cancer Res Off J Am Assoc Cancer Res. 2015;21:3453–61. doi: 10.1158/1078-0432.CCR-14-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng W. The Shanghai Women’s Health Study: Rationale, Study Design, and Baseline Characteristics. Am J Epidemiol. 2005;162:1123–31. doi: 10.1093/aje/kwi322. [DOI] [PubMed] [Google Scholar]
- 35.Shu X-O, Li H, Yang G, et al. Cohort Profile: The Shanghai Men’s Health Study. Int J Epidemiol. 2015;44:810–8. doi: 10.1093/ije/dyv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsu L, Jeon J, Brenner H, et al. A model to determine colorectal cancer risk using common genetic susceptibility loci. Gastroenterology. 2015;148:1330–9. e14. doi: 10.1053/j.gastro.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jo J, Nam CM, Sull JW, et al. Prediction of Colorectal Cancer Risk Using a Genetic Risk Score: The Korean Cancer Prevention Study-II (KCPS-II) Genomics Inform. 2012;10:175–83. doi: 10.5808/GI.2012.10.3.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pomerantz MM, Ahmadiyeh N, Jia L, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41:882–4. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hutter CM, Slattery ML, Duggan DJ, et al. Characterization of the association between 8q24 and colon cancer: gene-environment exploration and meta-analysis. BMC Cancer. 2010;10:670. doi: 10.1186/1471-2407-10-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tuupanen S, Turunen M, Lehtonen R, et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet. 2009;41:885–90. doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]
- 41.Carvajal-Carmona LG, Zauber AG, Jones AM, et al. Much of the genetic risk of colorectal cancer is likely to be mediated through susceptibility to adenomas. Gastroenterology. 2013;144:53–5. doi: 10.1053/j.gastro.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fernandez-Rozadilla C, Palles C, Carvajal-Carmona L, et al. BMP2/BMP4 colorectal cancer susceptibility loci in northern and southern European populations. Carcinogenesis. 2013;34:314–8. doi: 10.1093/carcin/bgs357. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.