SUMMARY
Intrahepatic cholangiocarcinoma (ICC) and hepatocellular carcinoma (HCC) are clinically disparate primary liver cancers with etiological and biological heterogeneity. We identified common molecular subtypes linked to similar prognosis among 199 Thai ICC and HCC patients through systems integration of genomics, transcriptomics, and metabolomics. While ICC and HCC share recurrently mutated genes, including TP53, ARID1A, and ARID2, mitotic checkpoint anomalies distinguish the C1 subtype with key drivers PLK1 and ECT2, whereas the C2 subtype is linked to obesity, T-cell infiltration and bile acid metabolism. These molecular subtypes are found in 582 Asian, but less so in 265 Caucasian patients. Thus, Asian ICC and HCC, while clinically treated as separate entities, share common molecular subtypes with similar actionable drivers to improve precision therapy.

INTRODUCTION
Primary liver cancer consists of two main histologically-distinct subtypes, i.e., hepatocellular carcinoma (HCC) and intrahepatic cholangiocarcinoma (ICC) confined within the liver, whose diagnoses and treatment decisions are uniquely based on their baseline clinical features. Diagnosis of HCC and ICC is traditionally based on radiologic, serologic and/or pathologic evaluations. Various HCC staging guidelines have been introduced to better triage patients for appropriate treatments, such as molecularly targeted therapies, but their effectiveness is limited, as evident by the recent setbacks of multiple failures of Phase III studies (Worns and Galle, 2014). While ICC is morphologically distinct from HCC, its classification has been of intense debate in recent years due to its complex histology and biology (Banales et al., 2016; Bridgewater et al., 2014). It should be noted that these varying classification models do not distinguish tumors with unique tumor biology, which is necessary for applying effective therapies. Moreover, like many other solid cancers, both HCC and ICC are genetically and biologically heterogeneous, which makes them highly resistant to treatment, ranking them as the second most lethal malignancies worldwide (Theise, 2014; Wang and Thorrgeirsson, 2014).
The extensive inter-tumor genomic heterogeneity of HCC and ICC is attributed to the presence of complex, multifactorial etiologies, including environmental factors such as hepatitis B virus (HBV), hepatitis C virus (HCV), parasitic infections and chemical carcinogens. Other risk factors include an unhealthy lifestyle, such as cigarette smoking, excess alcohol intake and dietary factors (El-Serag, 2011), in addition to sex and race/ethnic disparities wherein liver cancer mainly affects men and is highly prevalent in Asian populations (http://globocan.iarc.fr/). HBV and HCV are the major causative etiological factors for HCC, accounting for up to 90% of liver cancer globally, while ICC is uncommon, except in South-East Asia, such as northeastern Thailand, where infection with liver fluke (Opisthorchis viverrini) is endemic and approximately 70% of liver cancers are ICC (Sripa et al., 2007). These global disparities may be attributed to the presence of different etiological factors among different ethnic groups. One hypothesis is that various causative factors can evoke distinct molecular mechanisms to independently initiate malignant transformation, which results in vast genomic heterogeneity among patients. Such unique risk factor patterns of HCC and ICC provide an opportunity to study cancer heterogeneity and associated distinct tumor biology. In the current study, we sought to define stable molecular subtypes of ICC and HCC.
RESULTS
Tumor molecular subtypes defined by transcriptome and consensus clustering
To improve our understanding of disease susceptibility and progression as well as patient outcomes, we established the Thailand Initiative in Genomics and Expression Research for Liver Cancer (TIGER-LC) consortium to create a comprehensive biorepository with biospecimens linked to etiologies and clinical features from 3,000 patients with liver cancer, and 3,000 high risk and healthy individuals who reside in Thailand. A systematic integration of transcriptomic, genomic, somatic copy number alteration (SCNA), and metabolomic profiles of biospecimens provides a comprehensive approach to better classify molecular subtypes and related drivers in cancer. In this study, we utilized this experimental strategy to define molecular subtypes of the first sequential 199 enrolled Thai patients. Common molecular subtypes were further validated in 847 liver cancer patients from Asia, Europe and North America. The experimental strategy is outlined in Figure S1A – B.
Genomic analyses of ICC and HCC by whole genome or exome sequencing has shown a complex mutational landscape with vast inter-tumor heterogeneity (Guichard et al., 2012; Nakamura et al., 2015; Totoki et al., 2014). In contrast, transcriptome profiling of ICC and HCC have revealed stable molecular subtypes linked to tumor biology and patient outcomes (Hoshida et al., 2009; Lee et al., 2004; Roessler et al., 2015; Ye et al., 2003). To define molecular patterns in Thai ICC and HCC patients, Affymetrix Human Transcriptome Array 2.0 was performed on paired tumor and non-tumor specimens derived from 199 patients. The clinical features of these patients are summarized in Table S1. Among them, 153 tumor and 151 non-tumor samples passed the quality control tests and were used for transcriptome analyses. We first performed unsupervised hierarchical clustering and principle component analyses of all primary liver tumors to determine transcriptomic patterns using genes based on variance among tumor specimens (Figure1A–B). ICC and HCC are considered as two distinct clinical/histologic tumor types and consistently, we found that these primary liver tumors have some distinct transcriptomic patterns, however several specimens showed an overlapping pattern, indicating that some ICC and HCC share similar molecular features (Figure 1A – B). To compare these molecular features, we first identified subtypes in ICC or HCC utilizing a consensus clustering (cCluster) method, which was proven to be effective in defining stable tumor subtypes as previously described (Monti et al., 2005). cCluster of tumor samples revealed 3 major subtypes of HCC and 4 major subtypes of ICC based on consensus distributions and the corresponding consensus matrices (Figure S1C–D). The relationship among the HCC or ICC subtypes defined by cCluster can be visualized through unsupervised hierarchical clustering following a ranking method of the transcriptome used by TCGA (The Cancer Genome Atlas Research, 2011) (Figure 1C – D). This analysis revealed unique subtypes within HCC or ICC cases with distinct gene expression patterns that were independent of known diagnostic factors, cirrhosis, staging or etiology (Figure 1C – 1D).
Figure 1. Identification of ICC and HCC molecular-based tumor subtypes.
(A) A heatmap of ICC and HCC samples is shown by unsupervised hierarchical clustering of the most variable genes (+/− 2SD; n=587) among tumor specimens. (B) A principal components (PC) analysis of ICC and HCC tumor specimens is shown. (C) A heatmap of HCC subtypes is shown based on consensus clustering. The x-axis represents HCC subtype consensus clusters. HCC samples are represented in columns, grouped by the dendrogram into 3 main clusters and genes (n=370) are represented in rows. Z-scored gene expression are shown from −4 to 4. Clinical data of the samples are included below the heatmap. (D) A heatmap of ICC subtypes is shown as in (C). The x-axis represents ICC subtype consensus clusters and genes (n=1115) are represented in rows. See also Figure S1, Table S1.
Several recent studies have described transcriptomic similarity between ICC and HCC (Oishi et al., 2012; Woo et al., 2010), which is consistent with the overlapping patterns observed in the Thai specimens of this study (Figure 1A – B). In order to compare the subtypes between ICC and HCC, we used an unsupervised Subclass Mapping method (SubMap) (Hoshida et al., 2007). The SubMap method performs a pairwise comparison of the molecular features between each of the predetermined ICC and HCC subtypes, identified in Figure 1C–D, outputting a statistical likelihood that two subclasses share the same or similar underlying transcriptomic patterns, represented by a Bonferroni adjusted p value <0.05. Accordingly, this analysis revealed that the molecular features of the ICC-C1 and HCC-C1 subtypes are significantly similar (p=0.01), as are the ICC-C2 and HCC-C2/C3 subtypes (p=0.01) (Figure 2A). However, this relationship was not observed in non-tumor tissues, suggesting that subtype-related genes are tumor-specific (Figure S1E). Moreover, an unsupervised hierarchical clustering of the common subtypes found in ICC and HCC, namely C1 and C2, reveal a striking similarity in the molecular patterns of each subtype among primary liver tumors, independent of known diagnostic factors, cirrhosis, staging or etiology (Figure 2B). Gene Set Enrichment Analysis (GSEA) of subtype-specific gene signatures revealed that both ICC-C1 and HCC-C1 subtypes were enriched for mitotic checkpoint signaling pathways, suggesting that this subtype contains high chromosomal instability (Figure 2C). In contrast, ICC-C2 and HCC-C2 subtypes were enriched for cell immunity-related pathways, suggesting that inflammatory responses are linked to the C2 subtype (Figure 2C, Table S2). Interestingly, cases defined as HCC-C1 had poor survival while those in HCC-C2 had better survival, with a similar trend observed for ICC-C1 or ICC-C2 patients (Figure 2D). These results indicate that despite the distinct histological differences between ICC and HCC, common subtypes are evident between these primary liver cancers with similar transcriptome patterns, tumor biology and outcome.
Figure 2. Identification of common C1 and C2 molecular subtypes of ICC and HCC.
(A) Subclass Mapping of ICC and HCC subtypes is shown. Significant relationships between subtypes are represented by Bonferroni adjusted p values. Significant associations showing similarity between subtypes are shown in red with p<0.05 while differences between subtypes (Bonferroni adjusted p=1) are shown in blue. (B) A heatmap of ICC and HCC C1 and C2 samples is shown by unsupervised hierarchical clustering of genes (n=1378) differentiating the C1 and C2 groups among tumor specimens. (C) Significant pathways, identified by GSEA analysis, of HCC or ICC-C1 or C2 subtypes is shown, represented by log10 p values from 2 to 0 (p value from 0.01 to 1). (D) Kaplan-Meier survival analysis of HCC subtypes (top panel) or ICC subtypes (bottom panel) is shown. See also Figure S1, Tables S2 and S3.
Several gene signatures have been linked to ICC/HCC prognostic subtypes, cancer stem cell features and tumor metastasis (Andersen et al., 2012; Hoshida et al., 2009; Lee et al., 2004; Sia et al., 2013; Yamashita et al., 2008; Ye et al., 2003). We thus examined the relationship between the Thai HCC subtypes and known signatures using a nearest template prediction algorithm (Hoshida, 2010). We found that the HCC-C1 and ICC-C1 subtypes are significantly enriched for S1–2-related genes (p<0.05) (Hoshida study) and stem cell genes (Lee study), whereas ICC-C2 and HCC-C2 are significantly enriched for cases that are negative for these signatures (Figure S1F–G, Table S3). HCC-C1 is also significantly enriched in EpCAM genes (Yamashita study). Likewise, CCA-like signatures are also enriched in the above subtypes while C3/4 subtypes contain mixed cases. In contrast, metastasis signature, the class 2 signature (Andersen study) and the proliferation signature (Sia study) did not separate common subtypes well.
Race/ethnicity-related common tumor subtypes
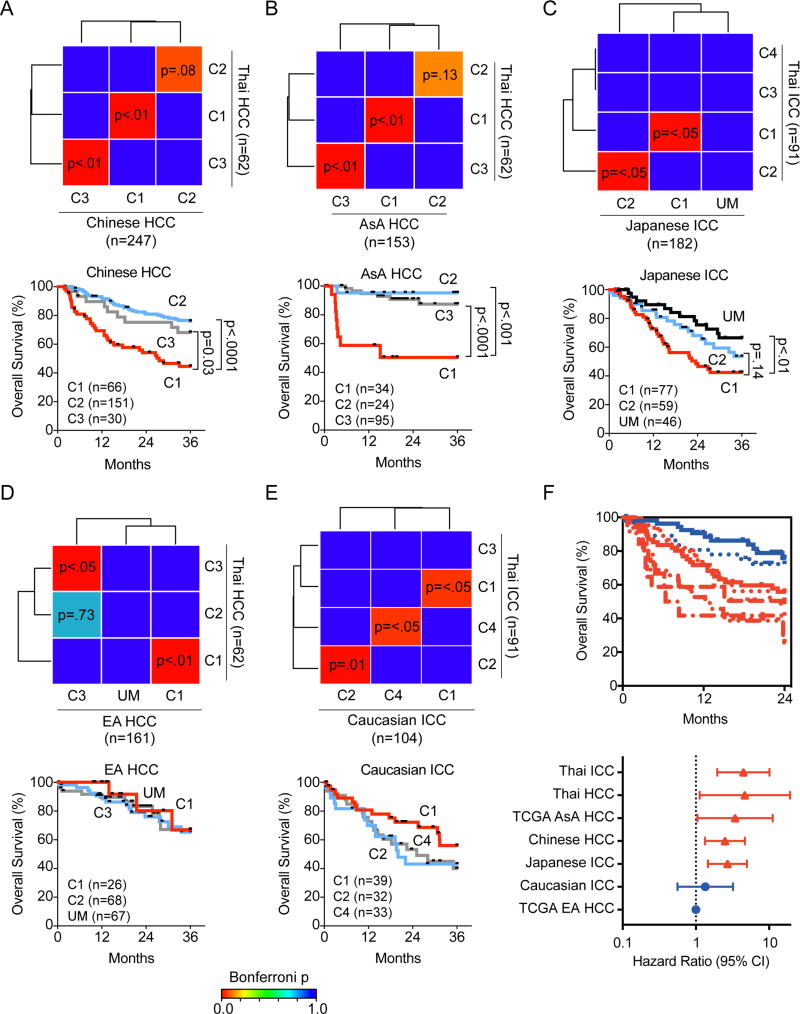
To further determine whether the common molecular subtypes of ICC and HCC observed in the Thai samples are universal, we examined several independent cohorts with available transcriptome data from patients who resided in Asia, Europe and North America. These include 247 HCC patients from China, 314 HCC patients from the U.S. (TCGA data), 104 ICC patients from Europe and 182 ICC patients from Japan (Table S4). SubMap was used to determine similarities among various subtypes identified by the transcriptome. We found that the C2 molecular subtype was observed in Chinese HCC patients (Bonferroni p=0.08; unadjusted p=0.003), Asian American (AsA, n=153) HCC patients (Bonferroni p=0.13; unadjusted p=0.005), Japanese ICC patients (Bonferroni p<0.05), and Caucasian ICC patients (Bonferroni p=0.01), but not in European American (EA, n=161) HCC patients (Figure 3A – E). In contrast, we found a molecularly similar C1 subtype in all ICC and HCC patients regardless of their race/ethnicity and histologic types. Interestingly, the C2 subtype of Thai ICC, Thai HCC, Chinese HCC, Japanese ICC had a better prognosis than Caucasian ICC, whereas the C1 subtype from all race/ethnicity had a poor prognosis. To further examine the relation between the potential overlapping molecular/prognostic subtypes of HCC and ICC, we compared the C1 and C2 subtypes of HCC or ICC among several Asian cohorts by unsupervised hierarchical clustering. This analysis reveals two clusters, whereby the C1 subtypes of Asian ICC and HCC cluster together and are distinct from the C2 subtypes of Asian ICC and HCC which also cluster together (Figure S2). There is a statistically significant enrichment in C1- and C2 in each cluster (fisher’s exact test two-sided p<2.2 × 10e-16). In contrast, a statistical enrichment of ICC and HCC in the clusters was not found (p=0.086). These data thus confirm and display the distinct transcriptome profile of C1 and C2 subtypes and their common presence among ICC and HCC. Since the C1 molecular subtype can be found in both Asians and Caucasians (Figure 3A – E), we performed Kaplan-Meier survival analyses to compare the C1 subtypes between Asian and Caucasian individuals among various cohorts (Figure 3F) Interestingly, these data show that although Caucasian ICC and HCC patients among the C1 subtype have similar tumor transcriptome patterns with Asian patients (Figure 3A – E), they have a different outcome than Asian HCC and ICC C1 patients. This pattern was observed at either a 2-year time point (Figure 3F) or a 5-year time point (data not shown). In forest plots based on the survival data, using TCGA Caucasian HCC as the reference (since this group has the best outcome), a statistically increased hazard ratio (p<0.05) is observed when comparing TCGA Caucasian HCC C1 patients to any of the Asian ICC or HCC C1 patients, however a similar hazard ratio was found when comparing the TCGA Caucasian HCC C1 patients to the Caucasian ICC C1 patients (Figure 3F). To avoid platform bias when comparing different cohorts using different molecular profiling techniques, we performed unsupervised hierarchical clustering using only the TCGA cohort (Figure S2B) which shows that the C1 subtype (red) clusters together among Asian or Caucasian individuals (yellow or white bars), however the C2 subtype (blue bars) is only present in Asian TCGA patients. In addition, we performed SubMap analysis on the TCGA cohort between Asians and Caucasians which shows the presence and similarity among the C1 or C3 subtypes of HCC patients (Figure S2C). However, the C2 subtype is not observed in Caucasian individuals in TCGA. It seems that C1 and C3 subtypes are observed in ICC and HCC patients of Asian or European descent, however the association of the C1 subtype with poor outcome is more readily observed in Asian individuals. Taken together, these results suggest that common molecular subtypes of ICC and HCC are also related to race/ethnicity.
Figure 3. Similar HCC and ICC tumor subtypes are found in Asians from China, Japan and U.S.
(A, B) Subclass Mapping of Thai HCC versus Chinese (A) or Asian American (AsA, B) HCC subtypes. (C) Subclass Mapping of Thai ICC versus Japanese ICC subtypes. (D) Subclass Mapping of Thai HCC versus Caucasian American (EA) HCC subtypes. (E) Subclass Mapping of Thai ICC versus Caucasian ICC subtypes. For A-E, significantly similar relationships between clusters are represented by Bonferroni-adjusted p values from 0 (similar) to 1 (different). Significant associations between clusters are shown in red with p<0.05 while differences between subtypes with p=1 are shown in blue. Subtypes in cohorts with no matching subtype to the Thai cohort is indicated with an UM (unmatched). The lower panels show Kaplan-Meier survival analysis of the subtypes indicated in the corresponding upper panel, with the number of samples in each subtype indicated. (F) Kaplan-Meier survival analysis of the ICC or HCC C1 subtype among various cohorts indicated (top panels) at a 2-year survival time cutoff with log-rank p value. Forest plots (bottom panels) show the hazard ratio with 95% confidence interval (CI) of the C1 subtype among various cohorts with the European American HCC patients from TCGA as the referent group since these individuals have the best overall survival. See also Figure S2, Table S4.
Subtype-related cancer drivers
Next, to define subtype-related driver genes, we performed targeted exome sequencing of Thai ICC and HCC. The exons and surrounding noncoding genomic regions of 562 protein-coding genes most frequently recurrent and mutated across diverse solid tumor types defined by the COSMIC database (Table S5) were captured in 197 pairs of tumor and paired non-tumor tissues and sequenced at an average of 125x coverage. We found that mutation types are similar between ICC and HCC (Figure 4A). The average mutation rate was 4.4 mutations per megabase for ICC and 3.7 mutations per megabase for HCC with no significant difference between the two tumor types (Figure 4B). Both HCC and ICC included a subset of 12 hypermutated samples [greater than 10 somatic single nucleotide variants (snvs)/Mb], which may be candidates for immune checkpoint inhibitor trials. These hypermutated tumors included several cases with mutations in mismatch repair genes as well as one case with a somatic mutation signature consistent with aristolochic acid exposure (Figure 4B, Table S6). We noticed that ICC has more C>T transition mutations, while HCC has more A>C, A>T and C>A transversion mutations (Figure 4C–F, Table S6).
Figure 4. Mutation profiles and their functional consequences in Thai ICC and HCC.
(A) Overview distribution of mutation types in ICC (n=129) and HCC (n=68). (B) Plot of ranked nucleotide substitutions presented as single nucleotide variant (SNV) density per Mb for ICC and HCC. (C) Proportions (mean ±SEM with 95% confidence interval) of SNVs identified in ICC and HCC. Differences calculated by Students t-test (*p<0.05). (D) Proportions (percentage) of SNVs in ICC and HCC as separated by either transitions or transversions. (E) Number (mean ±SEM) of transition (C>T) or transversions (A>C, C>T, and C>A) in ICC and HCC according to subtypes. (F) Overview of SNV per sample (number of variant changes per sample) according to different subtypes. See also Tables S5 and S6.
Among 22 candidate driver genes in ICC and 32 candidate driver genes in HCC, eight genes (TP53, ARID1A, ARID2, CSMD3, RYR2, NF1, PRKDC, PSIP1) were common in both ICC and HCC (Figure 5). A majority of these genes have been found by other genome sequencing studies of liver cancer with some variations of their frequencies (Guichard et al., 2012; Nakamura et al., 2015; Ong et al., 2012; Totoki et al., 2014). A comparison of the candidate drivers in ICC or HCC Thai patients show consistent mutation frequencies when compared to Japanese ICC, COSMIC ICC or COSMIC HCC patients (Table S7). Consistent with other published studies, we observed a complex mutational landscape of ICC and HCC with a vast inter-tumor heterogeneity without evident dominant driver genes as most of these genes are mutated in a low frequency in these tumor types (Figure 5). It should also be noted that genes such as RYR2, CSMD3 and SYNE1 are no longer considered cancer genes even their mutation recurrence is still relatively high.
Figure 5. The landscape of driver genes in Thai ICC and HCC.
An overview of driver genes in ICC (top panel) and HCC (bottom panel). Shown are genes with nonsynonymous and indel mutations of >5% frequencies. Genes were sorted by frequencies (right bar) and their alterations in each sample classified by cCluster-defined subtypes. Genes in bold are common between ICC and HCC. The status of hepatitis virus or Opisthorchis viverrini (OV) infection and variant frequency per sample are noted. Hypermutated samples are indicated as those with variant frequency above 10, indicated by the dotted horizontal line. See also Table S7.
Since gene mutation data were available for 182 Japanese ICC tumors, we determined whether any relationships existed between the common subtypes identified in this study and the top 32 mutated genes described by Nakamura et al (Nakamura et al., 2015). We found that while mutation data do not clearly discriminate common subtypes, and mutation frequencies range from 1–43% for each subtype, mutations in TP53, KRAS, MYC, and GNAS (>10% mutations) showed enrichment in the C1 subtype with poor prognosis, and mutations in BAP1 and IDH1 were more frequent in UM (unmatched), a subtype that did not match the C1/C2 subtypes but was associated with a good prognosis (Figure S3A). Noticeably, 15% of the UM subtype, but none of C1 and C2 subtypes, carries IDH1 mutations, suggesting that this unique subtype of ICC has a distinct gene mutation pattern and better prognosis. Interestingly, we found that a statistically significant number of genes (3 of 51 driver genes, i.e., NRAS, PRKCI and ECT2), based on SCNA and the transcriptome (Figure 6) overlap with these 32 mutated genes (p=0.0004; hypergeometric test). It is noted that mutation frequencies vary among different cohorts. It is possible that these heterogeneities could be contributed by differences in populations or to clinical tumor heterogeneity in ICC and HCC with differences in underlying etiological factors. However, a clear relationship between major etiological factors and mutation profiles was not observed. As evident by our further analysis of IDH1, IDH2, BAP1 mutations among various cohorts, which were discovered by recent exome sequencing projects to have a relative high mutation frequency in ICC (Jiao et al., 2013), we found that mutations of these 3 genes vary widely among different cohorts (Figure S3B). It fact, the data indicate that BAP1 mutations occur with higher frequency, and IDH1 and IDH2 mutations occur solely, in those of European descent versus Asian or African descent ICC patients (Figure S3C). Consistently, we only found one case of Thai ICC with IDH1 mutations (Table S6). In contrast, among the Thai cohort, the C1 common subtypes contain more p53 mutations than C2 subtypes (Figure 5). We also found that p53 R249S mutation, an aflatoxin signature mutation, is only associated with the HCC-C3 subtype (data not shown), suggesting a unique environmental exposure associated with this subtype. Thus, targeted exome sequencing based on the Oncovar design can identify driver mutations with relatively high frequencies, similar to whole exome sequencing.
Figure 6. Integration of somatic copy number alterations (SCNA) and gene expression to define subtype-related driver genes in ICC and HCC C1 and C2 subtypes.
(A) The frequency of chromosomal aberrations is shown for the ICC-C1 subtype (top panel) or ICC-C2 subtype (bottom panel). Copy number gain or loss is shown in red or blue, respectively. (B) The frequency of chromosomal aberrations is shown for the HCC-C1 subtype (top panel) or HCC C2 subtype (bottom panel). (C) A VENN diagram showing a comparison between the number of driving events (high concordance of SCNA and gene expression) in the C1 subtype of ICC or HCC with onesided Fisher’s exact p=0.001 for the overlapping genes. (D) The relationship between high concordant genes and tumor subtypes is shown along with the frequency of samples with copy number variation (CNV). In the upper panel, red bars represent copy number gain (Gain), blue bars represent copy number loss (Loss), while dark red or black bars represent chromosomal amplification with increased gene expression (Gain + Up) or chromosomal deletion with increased gene expression (Loss + Up). Gene expression is represented by the pink and light blue bars to indicate increased (Up) or decreased (Down) expression in tumors. In the lower panel, the ratio of samples showing copy number changes for each subtype is shown. The color of the column bars indicates the tumor subtypes shown on the y-axis in the upper panel. (E) Ingenuity Pathway Analysis of the 51 driver genes indicating a relationship with the PLK signaling network. (F) Representative images of ICC and HCC cases are shown based on immunohistochemical staining for ECT2 or PLK1. Scale bars represent 10 mm. (G) Kaplan-Meier survival analysis of all ICC (top panel) and HCC (lower panel) cases based on the ratiometric combination of protein expression (ECT2/PLK1) is shown with log rank p value. Low and high cutoff is defined by ECT2/PLK1 ratio ≤1 for the low and >1 for the high group. See also Figure S3, Tables S8 and S9.
We also determined somatic copy number alterations (SCNA) among Thai tumor specimens and paired non-tumor tissues as controls using Affymetrix Genome-Wide Human SNP Array 6.0. Consistent with previously published studies (Roessler et al., 2012), a typical SCNA profile with recurrent gains and losses on 1q, 6p, 8q and 4q, 8p, 13q, 16, 17p, respectively, was evident in HCC specimens (Figure S3D). A similar pattern of SCNA was found in TCGA HCC patients of Asian origin as indicated by common gain of 1q, 6p and 8q or loss of heterozygosity at 4q, 8p, 13q, 16 and 17p, but not in TCGA HCC patients of Caucasian origin (Figure S3E–F, Table S8). We found that SCNA profiles between ICC and HCC differ considerably (Figure S3D and S3G, Table S8). However, when we analyzed SCNA profiles based on C1 and C2 subtypes, there was a significantly higher degree of recurrent gains and losses found in both ICC-C1 and HCC-C1, when compared to ICC-C2 and HCC-C2 (Figure 6A – B). This result is consistent with the transcriptome and GSEA analyses and suggests that ICC-C1 and HCC-C1 contain mitotic checkpoint defects, which may result in higher degrees of aneuploidy.
To further determine potential subtype-related driver genes based on the notion that a driver gene should have high concordant alteration among SCNA and gene expression (Roessler et al., 2012; Woo et al., 2009), we first performed Pearson’s correlation between SCNA and the transcriptome. This analysis revealed that there is a significant positive correlation among a subset of genes (Figure S3H), i.e., 239 genes for ICC-C1 and 89 genes for HCC-C1 tumor specimens. Among them, 51 genes overlapped between ICC-C1 and HCC-C1, suggesting the existence of common subtype-specific drivers in ICC and HCC (Figure 6C, Table S9). Consistently, more copy number gain and elevated expression were associated with the C1 subtype than the C2 subtype (Figure 6D). Consistent with our findings above, a network analysis of these 51 genes revealed enrichment of mitotic checkpoint signaling pathways linked to PLK1 signaling (Figure 6E, Table S9). In addition, ECT2 was the top ranking differentially expressed gene between the C1 and C2 subtypes (Table S9). Similar results were observed using the TCGA HCC dataset (Figure S3I). Consistent with Thai ICC and HCC, we found that an association of 51 common subtype-related genes and race/ethnicity-related prognosis were also observed in HCC patients from the U.S. (Figure S3J). Noticeably, 6% of the Japanese ICC-C1 subtype also showed mutations in ECT2, consistent with our finding that ECT2 is a functional driver for the common C1 subtype (Figure 6).
The data above suggested that ECT2 and PLK1 could be clinically relevant functional biomarkers useful to detect ICC and HCC subtypes since both have been previously linked to tumor progression (Cook et al., 2014; Strebhardt, 2010; Vigil et al., 2010). We thus evaluated ECT2 and PLK1 by immunohistochemistry (IHC) on tissue microarrays (TMAs) of ICC and HCC that were constructed from 199 Thai patients. We found that PLK1 is detected in the cytoplasm, but is not expressed in the normal hepatocytes (Figure 6F, Figure S3K, and data not shown). Meanwhile, ECT2 is expressed in the nucleus, but is absent in normal hepatocytes (Figure 6F, Figure S3K, and data not shown). The expression levels of these proteins detected by IHC correlated with mRNA data (Figure S3L). Further analysis demonstrated correlation of PLK1 and ECT2 expression at the RNA level and protein expression level (Figure S3M). PLK1 and ECT2 were subject to survival analysis individually (Figure S3N), showing that patients with a high TMA score of PLK1 or ECT2 had a poor outcome trend. Given the substantial correlation in expression between PLK1 and ECT2 and the fact that PLK1 has been demonstrated to phosphorylate ECT2 in vitro (Niiya et al., 2006), we evaluated the combination of the two biomarkers to predict outcome. Based on prior examples of ratiometric combination of related biomarkers (Chung et al., 2009; Kitano et al., 2014), we generated a PLK1/ECT2 ratio from the TMA scores, and performed survival analysis (Figure 6G), which demonstrated that the PLK1/ECT2 ratio was robust to discriminate outcomes. To determine whether the combination of PLK1 and ECT2 expression was a universal predictor of outcome, we assessed high/low mRNA combination groups based on the median expression of the two genes in various cohorts. We found that a combined high PLK1 and ECT2 expression is associated with poor survival compared to a combined low PLK1 and ECT2 expression in HCC (Figure S3O) patients among those who are Asian, however an association with prognosis is not observed in Caucasian patients. A similar trend is observed in Asian ICC and Caucasian ICC patients (Figure S3P). These data indicate that protein levels of PLK1 and ECT2 may be more indicative of patient outcome and suitable for diagnostic purposes rather than mRNA based levels.
Obesity and tumor subtypes
To determine whether any of the etiological/demographic/clinical features are linked to the identified tumor subtypes, we compared C1 and C2 subtypes based on available clinical variables that include age, gender, tobacco and alcohol consumption, body mass index (BMI), and tumor characteristics. Only age, BMI status and tumor size appeared to be different between the C1 and C2 subtypes of Thai ICC and HCC (Table S1). It should be noted that alcohol consumption, HBV and HCV status, cirrhosis by Child-Pugh score, levels of alkaline phosphatase (ALP), CA19-9, alpha-fetoprotein (AFP), and tumor staging differed significantly between ICC and HCC (Table S1). It is interesting that these etiological factors are not associated with common molecular subtypes, but they are linked to histological subtypes.
Since higher BMI was found among Thai C2 patients when compared to C1, we further determined BMI profiles in Thai, AsA and EA patients with available BMI data. We found that EA patients tend to have higher BMI than Asian patients regardless of whether they live in Asia or in the U.S. (Figure S4A). C2 subtypes tend to have a higher BMI than C1 subtypes and the difference is statistically significant (Figure S4B). BMI is measure of obesity, a disease associated with alterations in metabolism. We thus determined whether metabolic alterations were associated with the prognostic subtypes we identified in HCC and ICC, by performing untargeted metabolomic profiling among 188 ICC and HCC tumor tissue specimens from Thai patients (Table S10). Since integrating metabolite and gene expression profiles are a powerful method of reducing false positivity and increasing the chances of defining functional metabolites (Budhu et al., 2013), we performed a Pearson correlation analysis between metabolite and gene expression profiles (Figure S4C–D). A total of 77 metabolites for ICC and 81 metabolites for HCC from a total of 178 most variable metabolites showed a high correlation with gene expression (R> − 0.5 or 0.5; p <0 .05; >20% of the genes). We found a statistically significant number of overlapping metabolites (n=46) between ICC and HCC (hypergeometric p=0.0007). Moreover, metabolites that showed a high concordance with gene expression discriminated the C1 and C2 subtypes (Figure 7A, Figure S4E). The top networks of the most significant metabolites from ICC and HCC were strikingly similar (Figure 7B, Figure S4F, Table S10). We found that bile acid-related metabolites, such as taurochenodeoxycholate and tauroursodeoxychoate (TUDCA), are significantly more abundant in both ICC-C2 and HCC-C2 than in C1 subtypes (Figure 7C, Figure S4G).
Figure 7. Bile-acid metabolism and inflammation are altered in the HCC C1 and C2 subtype.
(A) Hierarchical clustering of HCC (n=29) based on 81 metabolites. Samples are represented in columns, and metabolites are represented in rows. Metabolite abundance is represented in log2. (B) Ingenuity Pathway Analysis of the highly concordant metabolite/gene network is shown. Upregulated metabolites in the C1 subtype or the C2 subtype are noted in pink or green, respectively. (C) Box-plots of the abundance of three representative bile-acid-related metabolites in C1 (n=15) and C2 (n=14) HCC samples are shown as first quartile, median and third quartile (bottom box, middle line and top box, respectively) with Student’s t-test p values. Whiskers represent minimum and maximum values. The number of cases in each subtype is indicated in parentheses. (D) CIBERSORT analysis of the HCC C1 versus the HCC C2 subtype is shown. High or low associations between cell types are shown on a scale from red to blue (1 to −1). The size of circles indicates the significance of the association, with larger circles representing higher significance. (E) The relative fraction of leukocyte types associated with C1 and C2 are shown. (F) Box-plots of the abundance of three leukocyte types in C1 and C2 HCC samples are shown as first quartile, median and third quartile (bottom box, middle line and top box, respectively) with Student’s t-test p value. Whiskers represent minimum and maximum values. See also Figure S4, Table S10.
The C2 subtype is associated with an increased BMI, which is known to be linked to metabolic disease and cellular inflammation (Calle et al., 2003; Park et al., 2010). In addition, the C2 subtype-related bile acid metabolites are known to be linked to inflammation and immunity (Yoshimoto et al., 2013). In fact, the gene expression profile of the C2 subtype was also linked to immune and inflammation pathways (Figure 2C). We thus examined infiltrating immune cells in Thai ICC and HCC using CIBERSORT (Newman et al., 2015). We found that the activity of leukocyte infiltrates is much higher in the C2 than in the C1 subtype (Figure 7D, Figure S4H). Noticeably, elevated CD4+ memory T cells, along with γδ T cells, but reduced Treg cells, are associated with the C2 subtype (Figure 7E–F, Figure S4I–J). Taken together, the results indicate that while the C1 subtype contains mitotic checkpoint defects with altered PLK1 and ECT2, the C2 subtype has elevated BMI, immune cell abnormality and abnormal bile acid metabolism.
DISCUSSION
A major hallmark of liver cancer is its association with various types of etiological factors and its high heterogeneity in clinical presentation and underlying tumor biology. Consequently, most patients with liver cancer are refractory to treatment and have a dismal outcome. One of the essential requirements needed to improve their outcome is to provide a diagnostic tool kit that is capable of accurately defining homogenous molecular subtypes, each displaying unique tumor biology linked to potentially druggable driver genes in order to implement rational treatment choices based on molecular subtypes. Accordingly, the development of well-annotated biobanks of cancer patients, such as the efforts by TCGA and ICGC, are key resources to further develop the goals of precision medicine. Currently, TCGA and ICGC include HCC specimens largely derived from North America, Europe and Japan. Considering that ICC and HCC are much more prevalent in Asian populations, we established a TIGER-LC consortium to generate a biorepository and conduct a case-control study in Thailand where liver cancer, especially ICC, is endemic.
In the current study, we found that both ICC and HCC, regardless of their difference in histology and associated etiologies, consist of several common molecular subtypes shared mainly among Asian patients. Interestingly, certain common ICC and HCC subtypes share distinct gene expression matrices and have similar aggressiveness, suggesting the presence of common molecular types beyond the traditionally viewed histological tumor subtypes. While exomic sequencing reveals 8 common driver genes among ICC and HCC in the midst of a complex mutational landscape with considerable inter-tumor heterogeneity, systematic integration of the cancer transcriptome, SCNA and metabolome revealed additional key oncogenic drivers linked to an aggressive molecular subtype. Specifically, the C1 subtype is enriched for p53 mutations and contains mitotic checkpoint defects, while the C2 subtype is linked to inflammation, obesity and bile acid biogenesis. These results suggest that treatment stratification should not be based on histological types, but rather on molecular types with a unified perspective of the disease (Dotto and Rustgi, 2016).
Hepatocarcinogenesis is a complex process resulting from an accumulation of genetic and epigenetic alterations of various cancer drivers over many decades. Tumor evolution is expected to vary significantly between different tumor types due to the fact that each tumor cell needs to adapt to the microenvironment’s stress induced by different etiological factors. Consequently, liver cancer is especially prone to genetic heterogeneity. While whole genome/exome sequencing approaches are powerful in identifying potential cancer drivers, candidate ICC or HCC-related driver mutations are extremely heterogeneous, as each tumor carries a large number of low frequency mutated genes in various combinations (Guichard et al., 2012; Nakamura et al., 2015; Totoki et al., 2014). Furthermore, most of these genetic abnormalities characterized by whole exome sequencing have been considered as either passenger or histological mutations that do not have any functional impact on the tumors being diagnosed (Helleday et al., 2014). For those genes with a relatively higher mutation frequency that have been characterized as candidate drivers, vast inter-tumor heterogeneity among different tumor lesions that carry varying combinations of these candidate drivers is clearly evident (Guichard et al., 2012; Totoki et al., 2014). The results of our exomic sequencing analysis are consistent with this observation. It is conceivable that a combination of different cancer drivers may emerge as new convergent adaptive pathways are rewired for cancer cell survival that are unique to different subtypes. It should be noted that cancer drivers can not only be acquired by somatic mutations, but also via epigenetic mechanisms, such as DNA methylation of oncogenes or tumor suppressor genes. This may explain why finding stable tumor subtypes with unique tumor biology by whole exome sequencing is very challenging as it only captures a fraction of tumor characteristics. In contrast, transcriptomic profiling has been successful in defining stable HCC subtypes that may reflect their unique tumor biology (Hoshida et al., 2009; Lee et al., 2006; Nault et al., 2013; Roessler et al., 2010; Yamashita et al., 2008), although defining key driver genes among different tumor subtypes by transcriptome-based approaches has been challenging. We suggest that an integrated omics approach, correlating genomics, epigenomics, transcriptomics, proteomics, and metabolomics data, is key to addressing tumor heterogeneity and identifying cancer drivers.
Our study revealed that the C1 subtype is linked to mitotic checkpoint defects and p53 mutations, and that PLK1 and ECT2 are two key, clinically relevant genes for C1. A link between p53 and the mitotic checkpoint is well documented (Cross et al., 1995). Loss of p53 function by mutations found in the C1 subtype may explain the increased aneuploidy observed in this subtype. In addition, we found that both PLK1 and ECT2 expressions are highly expressed in the C1 subtype and are robust in defining tumor subtypes using IHC. This is clinically meaningful since IHC is a preferable method for pathological diagnosis. The ECT2 gene is also preferentially amplified or mutated in the C1 subtype, consistent with the hypothesis that it is a driver gene for this subtype. Consistently, both PLK1 and ECT2 have been functionally linked to cancer. PLK1 is a mitotic serine/threonine protein kinase and is required to initiate mitosis and regulate spindle assembly. Overreactive PLK1 signaling has been found in human tumors, including HCC, and has been proposed to serve as a potential target for the treatment of cancer because of its crucial functional node in the oncogenic network (Strebhardt, 2010). Many PLK1 inhibitors are currently being tested in solid tumors (Yim, 2013). ECT2 belongs to the Ras superfamily GEFs and GAPs (Vigil et al., 2010) and is a classical oncogene originally identified in 1991 based on its ability to transform NIH 3T3 cells (Cook et al., 2014). Its role as a tractable target for cancer therapy, including HCC, has been suggested since it is responsible for promoting early recurrence of HCC (Chen et al., 2015; Vigil et al., 2010). Our results suggest that PLK1 and ECT2 expression could serve as biomarkers for patient stratification and molecular targets for the C1 subtype of Asian ICC and HCC.
Metabolic liver diseases and obesity have been linked to liver inflammation and cancer (Calle et al., 2003; Cohen et al., 2011; Park et al., 2010; Welzel et al., 2011). However, the molecular mechanisms underlying these associations are unclear. We found that a common clinical feature linked to the C2 subtype is an increased BMI, suggesting a possible association with liver-related metabolic diseases. As BMI is linked to inflammatory responses and metabolic disorders, we also examined tumor-associated leukocytes among the molecularly defined subtypes. We found that lymphoid cells such as CD4+ memory T cells and γδ T cells, but not myeloid cells, are significantly elevated in the C2 subtype. These results are consistent with the gene expression data of the C2 subtype, which was enriched for cell immunity-related pathways, reaffirming the idea that an inflammatory response is linked to the C2 subtype. It is interesting that several bile acid metabolites such as TUDCA, taurocholic acid and glycochenodeoxycholate are also consistently much more abundant in the C2 subtype than the C1 subtype in both ICC and HCC. Bile acids have recently emerged as versatile signaling molecules to regulate cholesterol metabolism, energy and glucose homeostasis, which forms the basis for developing drug targets to treat common metabolic and hepatic diseases (Thomas et al., 2008). It is conceivable that some of these drugs may be applicable to treat the C2 subtype of liver cancer. Our results are also consistent with recent studies indicating that the obesity-induced gut microbial metabolite deoxycholic acid promotes liver carcinogenesis through the senescence secretome and that diet can alter the human gut microbiome, facilitating diet-related diseases, such as obesity (David et al., 2014; Yoshimoto et al., 2013). Moreover, dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in IL10-deficient mice, revealing a plausible mechanistic basis for diets with certain saturated fats for induction of immune-mediated diseases in genetically susceptible hosts (Devkota et al., 2012).
While our results indicate that the common C1-C2 subtypes are mainly associated with Asian patients, the reason for this race/ethnicity-related association is unclear. It is plausible that Western-type diets may induce dysbiosis differently between Asians and Caucasians, and that race/ethnicity-related gut microbiome differences may cooperate with bile acid metabolism to induce distinct carcinogenesis processes observed in the Asian-related common C2 subtype. Increased infiltrating T cells in the C2 subtype also suggest a plausible scenario whereby these tumors may be sensitive to immune checkpoint inhibitors. In summary, our integrated omics approach has defined common molecular subtypes of ICC and HCC across several Asian populations and identified potential driver genes and metabolic processes linked to the specific subtypes. Further efforts of the TIGER-LC Consortium are underway to more fully understand ICC and HCC biology, and to improve outcomes for liver cancer patients. Thus, TIGER-LC serves as a rich resource from which numerous investigations can follow, spanning a breadth of disciplines, including genomics, epidemiology, functional and clinical studies, which, when amalgamated, may significantly enhance the scope of our liver cancer knowledge-base and allow us to more successfully manage patient care with an outlook towards precision medicine.
STAR METHODS
Contact For Reagent And Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Xin Wei Wang (xw3u@nih.gov).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cohorts and Clinical Specimens
A set of 398 surgical paired tumors and nontumor specimens derived from 199 sequential patients of the TIGER-LC cohort (130 ICC patients and 69 HCC patients) were used in this study. Tumor diagnosis was independently confirmed by pathological assessment by resident pathologists at each participating center in Thailand as well as a surgical pathologist in the U.S. Mixed HCC-ICC cases were excluded from this study. Clinical, demographic, socioeconomic and morbidity data were abstracted from comprehensive questionnaires and medical chart records. A list of clinical variables assessed in this study is provided in Table S1. The characteristics of 153 Asian HCC patients and 161 Caucasian HCC patients from TCGA were also used in this study (TCGA Research Network: http://cancergenome.nih.gov/). The characteristics of 104 Caucasian ICC patients and 182 Japanese patients from independent cohorts were described recently (Andersen et al., 2012; Nakamura et al., 2015). The HCC cohort of 247 Chinese patients from LCI was previously described (Roessler et al., 2010). Informed consent was obtained from all patients included in this study and approved by the Institutional Review Boards of the respective institutions (NCI protocol number 13CN089; CRI protocol number 18/2555; Chulabhorn Hospital protocol number 11/2553; Thai NCI protocol number EC163/2010; Chiang Mai University protocol number TIGER-LC; Khon Kaen University protocol number HE541099). A study design diagram representing the use of cohorts in this study is shown in Figure S1.
METHOD DETAILS
RNA Isolation and Transcriptomics
Total RNA was extracted from frozen tissue using TRIzol (Invitrogen) according to the manufacturer’s protocol. Only RNA samples with good RNA quality as confirmed with the Agilent 2100 Bioanalyzer (Agilent Technologies) were included for array studies. The Affymetrix Human Transcriptome Array 2.0 was used to measure transcripts among paired tumor and nontumor tissue specimens. Raw gene expression data were normalized using the Robust Multi-array Average (RMA) method (Irizarry et al., 2003) and sketch quantile normalization method. For genes with more than one probe set, the mean gene expression was calculated. The microarray platform and data were submitted to the Gene Expression Omnibus (GEO) public database at NCBI following MIAME guidelines (GEO Series GSE76297). Gene expression data of the three independent cohorts are accessible through GEO Series (GSE14520 and GSE26566), TCGA and European Genome-phenome Archive (EGA) database (EGA00001000950). All expression data were log2 transformed and normalized using R statistical packages (https://cran.r-project.org/doc/FAQ/R-FAQ.html) based on mixed model ANOVA to overcome differences between the platforms. Unsupervised hierarchical clustering of the most variable genes (+/− 2SD) among tumor specimens of ICC or HCC was performed. Consensus clustering (cCluster; hierarchical clustering; Pearson distance; complete linkage; 1000 resampling iteration) was used to define subtypes among HCC or ICC (Monti et al., 2005; Wilkerson and Hayes, 2010). Heatmaps were generated using the Complex Heatmap package in R (Gu et al., 2016) to determine the relationship among samples or cCluster-defined subgroups. In subtype analyses, genes were selected based on ANOVA between subtypes of HCC or ICC (Bonferroni corrected p=0.05) An unsupervised subclass mapping method (SubMap) was used to identify common subgroups between independent cohorts using the default setting found in the SubMap module (http://genepattern.broadinstitute.org/) (Hoshida et al., 2007). Significant similarity between clusters found by the SubMap method are defined as those with p<0.05 and denoted by red colored boxes, while borderline significance (p approaching 0.05) is denoted by an orange box and lack of significance (statistical difference, p~1.0) is denoted by a blue box.
DNA Isolation and Somatic Copy Number Alterations (SCNA)
Total DNA was extracted from frozen tissue using a Phenol/Chloroform extraction protocol. Samples with sufficient amount of double-stranded DNA as confirmed by Quant-iT PicoGreen dsDNA Assay Kit (Thermo Fisher Scientific) were included for array studies. The Affymetrix Genome-Wide Human SNP Array 6.0 was used to determined somatic copy number alterations (SCNA) among paired tumor and nontumor tissue specimens. The raw SCNA data is accessible through GEO Series GSE76213. SCNA of ICC and HCC was analyzed using Partek Genomics Suite 7.5 using the paired non-tumor tissue as the reference for each patient. The segmented regions in ICC and HCC were found using the genomic segmentation algorithm in Partek. To identify genes that are concordantly regulated with SCNA, the Pearson correlation value was calculated between the copy number segmented region and the transcriptome data from either ICC or HCC tissues. To define the C1 and C2 subtype-specific concordant genes, the genes located in segmented regions with a positive correlation value and p values ≤0.005 were considered. To define the copy number concordant genes for the C1 and C2 subtype in ICC and HCC, the p value from a Student’s t-test and fold change of the log2 transformed expression value among the most variable genes used for the class prediction between C1 and C2 ICC or HCC subtypes were used. To identify C1 subtype-specific copy number concordant genes in ICC and HCC, we selected the genes with a Student’s t-test p value ≤0.005 when C1 and C2 subtypes were compared.
Exome Sequencing and Data Processing
Simultaneous fragmentation and adaptor ligation was performed on input gDNA (50 ng) by tagmentation using the Nextera DNA Library Preparation kit, according to manufacturer’s protocol (Illumina). Products with a mean size of 350 bp +/− 20% were purified using the Agencourt AmpureXP Purification System (Beckman Coulter). Amplification and dual indexing of purified samples was performed using Illumina PCR primers InPE1.0 and InPE2.0 and primer indices (8bp). Hybridization capture of pooled, indexed libraries was performed according to the manufacturer’s protocol using NCI OncoVar V4, an Agilent SureSelect Custom DNA kit (Agilent Technologies) targeting 2.93 Mb of sequence in 562 genes found to be mutated in diverse solid tumors (Tablele S6). In addition, xGen® Blocking Oligos (Integrated DNA Technologies Inc., Coralville, IA) specific to Nextera library adaptor sequences were used during hybridization according to manufacturer’s recommendations. The libraries were sequenced on an Illumina NextSeq 500 or HiSeq 2500 instrument by paired end 2×75bp to an average target region depth of 125x. Alignments to the hg19 human reference genome assembly were performed with BWA mem 0.7.10-r789 (Li and Durbin, 2009), indel realignment by GATK IndelRealigner 3.4-0-g7e26428 (McKenna et al., 2010) and duplicates were marked with picard MarkDuplicates 1.129 (Li et al., 2009). Somatic single nucleotide variants and small insertions and deletions were called with strelka 2.0.17 (Saunders et al., 2012).
Metabolomics
Metabolon’s Discover HD4 Platform was employed to measure small biochemical species among tumor tissue specimens. Both liquid chromatography in positive and negative modes (LC+/LC−) and gas chromatography/mass spectrometry (GC/MS) were employed. A total of 718 metabolites were measured. The missing values were imputed using the minimum value of each metabolite. The 178 most variable metabolites common in both HCC and ICC cohorts were selected (Filter: 1.5-fold change from metabolite’s median value, Log intensity variation p value>0.01). Pearson correlation values between the selected metabolites and the most variable genes were calculated for each cohort separately. Only significantly correlated metabolites and genes within the same samples (p<0 .05) were included in further analysis. Furthermore, metabolites that were significantly associated with at least 20% of the genes were selected. Metabolomics data is available in Table S10.
Immunohistochemistry
Tissue microarrays (TMAs) were constructed with 1.0 mm cores from formalin fixed, paraffin embedded tissue that had been reviewed by SH (Kononen et al., 1998) as separate TMAs for cholangiocarcinoma and hepatocellular carcinoma. Matched TMAs of normal tissue from the patients were constructed as a reference. All TMAs contained internal control tissues. Immunohistochemistry was performed on 5µm TMA sections. First, slides were deparaffinized in xylene, and rehydrated in graded alcohol. Antigen retrieval was performed in a pressure cooker for 20 min with a pH6 citrate buffer. Anti-PLK1 (mouse monoclonal, clone CN05–844, Millipore) was applied at a 1:1000 dilution in a 2% non-fat milk solution at room temperature for 2 hr. Anti-ECT2 (mouse monoclonal, “E-1” cat SC-514769, Santa Cruz Biotechnology) was applied at a dilution of 1:500 in a 2% non-fat milk solution. Antigen-Antibody complexes were detected with Envision+ (Dako) secondary, and DAB. Slides were counterstained with hematoxylin, dehydrated, cleared and coverslipped. Positive and negative controls were performed. A TMA of matched normal liver was stained concurrent with the tumor TMAs. Interpretation of immunohistochemical staining was performed at 200X magnification. Tumors were scored for percentage of tumor cells stained (0–4 in quartiles) and intensity of staining (0–4). The two values were multiplied (range 0–16). Normal tissue TMAs failed to demonstrate staining of tumor markers. ECT2/PLK1 ratios were calculated from raw data, and cut points were determined as ECT2/PLK1 ratio ≤1 for the low and >1 for the high group, similar to that previously described (Kitano et al., 2014), and plotted as Kaplan-Meier plots. High or low mRNA levels of PLK1 and ECT2 were determined using a median score. The Cox-Mantel log-rank test was used for statistical analysis.
QUANTIFICATION AND STATISTICAL ANALYSIS
Pathway analysis was performed using Gene Set Enrichment Analysis (GSEA) version 16 and Ingenuity Pathway Analysis (IPA) version 24718999. Kaplan-Meier survival analysis was used to compare patient survival using GraphPad Prism 6–7 and the statistical p value was generated by the Cox-Mantel log-rank test. All p values are two-sided and the statistical significance was defined as p<0.05 unless otherwise noted. The relationship between previously reported signatures and the Thai ICC and HCC cohorts was determined using a nearest template prediction algorithm (Hoshida, 2010) implemented in GenePattern (Subramanian et al., 2005) based on a prediction confidence false discovery rate (FDR) cut-off of 0.05. For comparisons among various cohorts and platforms, a z-score was applied to normalize each set. An estimation of the relative fractions of immune/inflammatory cell subsets from tissue expression profiles of ICC or HCC was conducted using CIBERSORT (Newman et al., 2015). The gene expression data were converted by quantile normalization of the log2 scaled expression matrix and relative fractions of leukocytes in the C1 and C2 subtype of ICC and HCC were quantified according to the website (cibersort.stanford.edu/) with implemented analyses using the built-in LM22 signature matrix (LM22). Welch’s Two Sample t-test was used to compare each of the 22 relative leucocyte fractions between the C1 and C2 subtype. Pearson correlation analysis was used to determine the correlation between leukocytes and BMI in the C1 and C2 subtypes.
Data And Software Availability
All DNA sequencing data have been deposited in the dbGaP (Accession # phs001199.v1.p1). All gene expression and SCNA data used in this study have been deposited in the NCBI Gene Expression Omnibus (GEO) under accession codes GSE76297 and GSE76213. Metabolomics data is provided Table S10. Software used in this study are noted in the Method Details section above and the Key Resources Table.
Key Resources Table
Supplementary Material
Table S7, related to Figure 5. Comparison of somatic mutation frequency in ICC and HCC cohorts.
Table S8, related to Figure 6. Comparison of somatic copy number alteration amplification or deletion in TIGER-LC and TCGA HCC patients.
Table S9, related to Figure 6. Driver genes.
Table S10, related to Figure 7. Metabolite analysis of TIGER-LC tumors.
Table S1, related to Figure 1. Demographic, clinical and laboratory characteristics of ICC and HCC patients in the TIGER-LC cohorts at the time of surgery.
Table S2, related to Figure 2. Significantly different pathways among subtypes from GSEA analysis (cut off NOM p <0.05).
Table S3, related to Figure 2. Gene signatures tested in the study.
Table S4, related to Figure 3. Demographic, clinical and laboratory characteristics of patients in the validation cohorts.
Table S5, related to Figure 4. List of genes sequenced in this study.
Table S6, related to Figure 4. OncoVar mutation summary of the TIGER-LC cohort.
Highlights.
ICC and HCC consist of several stable molecular types
ICC and HCC share common molecular subtypes and driver genes
Asian-specific ICC and HCC subtypes are linked to unique metabolic processes
Oncogenic PLK1 and ECT2 are subtype-related biomarkers
Chaisaingmongkol et al. identify common molecular subtypes linked to similar prognosis in intrahepatic cholangiocarcinoma and hepatocellular carcinoma, clinically different malignancies, among Thai patients. These molecular subtypes are also found in other Asian patients, but rarely in Caucasian patients.
Significance.
Primary liver cancers have a complex mutational landscape with vast inter-tumor heterogeneity, which poses a major challenge to define actionable drivers. Well-defined patient populations with well-annotated molecular characteristics are needed to accurately define homogenous molecular subtypes with unique tumor biology linked to potentially druggable driver genes. Here, we demonstrate that common molecular subtypes with key drivers are shared among Asian ICC and HCC patients through systematic integration of the genome, transcriptome, and metabolome. Our results indicate that ICC and HCC, while clinically treated as separate entities, share common molecular determinants, suggesting that a unified molecular landscape of liver cancer is required to improve diagnosis and therapy.
Acknowledgments
We thank other members of the TIGER-LC consortium, patients and families who contributed to this study. This work was supported in part by the intramural research program of the Center for Cancer Research, National Cancer Institute of the United States. Funding support for the establishment of the tissue/tumor biobank and collection of specimens and clinical diagnosis was received from the Chulabhorn Research Institute, Bangkok, Thailand.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
XWW, CM, MR, CCH, RHW conceived the idea, designed and implemented the consortium; JC, AB, HD, SR, BP, SMK, MF, YP, MC, JJW, YJZ, JW, HSS, DA, PSM, XWW, performed experiments and data analyses; JC, AB, HD, SMK, XWW interpreted omics data; VB, NL, AC, CP, CA, TS, KP, SS, CAL, PH, JBA, SST, TS, and NH, supervised and/or monitored patient recruitment, clinical data collection and pathological assessment; KY performed TMA construction, IHC assay development and performance; SMH performed TMA design, IHC evaluation, data interpretation; XWW, JC, AB, SMK wrote the manuscript; all authors read, edited and approved the manuscript.
References
- Andersen JB, Spee B, Blechacz BR, Avital I, Komuta M, Barbour A, Conner EA, Gillen MC, Roskams T, Roberts LR, et al. Genomic and genetic characterization of cholangiocarcinoma identifies therapeutic targets for tyrosine kinase inhibitors. Gastroenterology. 2012;142:1021–1031. doi: 10.1053/j.gastro.2011.12.005. e1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banales JM, Cardinale V, Carpino G, Marzioni M, Andersen JB, Invernizzi P, Lind GE, Folseraas T, Forbes SJ, Fouassier L, et al. Expert consensus document: Cholangiocarcinoma: current knowledge and future perspectives consensus statement from the European Network for the Study of Cholangiocarcinoma (ENS-CCA) Nature reviews Gastroenterology & hepatology. 2016;13:261–280. doi: 10.1038/nrgastro.2016.51. [DOI] [PubMed] [Google Scholar]
- Bridgewater J, Galle PR, Khan SA, Llovet JM, Park JW, Patel T, Pawlik TM, Gores GJ. Guidelines for the diagnosis and management of intrahepatic cholangiocarcinoma. Journal of hepatology. 2014;60:1268–1289. doi: 10.1016/j.jhep.2014.01.021. [DOI] [PubMed] [Google Scholar]
- Budhu A, Roessler S, Zhao X, Yu Z, Forgues M, Ji J, Karoly E, Qin LX, Ye QH, Jia HL, et al. Integrated metabolite and gene expression profiles identify lipid biomarkers associated with progression of hepatocellular carcinoma and patient outcomes. Gastroenterology. 2013;144:1066–1075. doi: 10.1053/j.gastro.2013.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calle EE, Rodriguez C, Walker-Thurmond K, Thun MJ. Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. NEnglJ Med. 2003;348:1625–1638. doi: 10.1056/NEJMoa021423. [DOI] [PubMed] [Google Scholar]
- Chen J, Xia H, Zhang X, Karthik S, Pratap SV, Ooi LL, Hong W, Hui KM. ECT2 regulates the Rho/ERK signalling axis to promote early recurrence in human hepatocellular carcinoma. Journal of hepatology. 2015;62:1287–1295. doi: 10.1016/j.jhep.2015.01.014. [DOI] [PubMed] [Google Scholar]
- Chung JY, Hong SM, Choi BY, Cho H, Yu E, Hewitt SM. The expression of phospho-AKT, phospho-mTOR, and PTEN in extrahepatic cholangiocarcinoma. Clin Cancer Res. 2009;15:660–667. doi: 10.1158/1078-0432.CCR-08-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JC, Horton JD, Hobbs HH. Human fatty liver disease: old questions and new insights. Science. 2011;332:1519–1523. doi: 10.1126/science.1204265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DR, Rossman KL, Der CJ. Rho guanine nucleotide exchange factors: regulators of Rho GTPase activity in development and disease. Oncogene. 2014;33:4021–4035. doi: 10.1038/onc.2013.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross SM, Sanchez CA, Morgan CA, Schimke MK, Ramel S, Idzerda RL, Raskind WH, Reid BJ. A p53-dependent mouse spindle checkpoint. Science. 1995;267:1353–1356. doi: 10.1126/science.7871434. [DOI] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Antonopoulos DA, Jabri B, Chang EB. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10 −/− mice. Nature. 2012;487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto GP, Rustgi AK. Squamous Cell Cancers: A Unified Perspective on Biology and Genetics. Cancer cell. 2016;29:622–637. doi: 10.1016/j.ccell.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Serag HB. Hepatocellular carcinoma. The New England journal of medicine. 2011;365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, Calderaro J, Bioulac-Sage P, Letexier M, Degos F, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. NatGenet. 2012;44:694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleday T, Eshtad S, Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nature Reviews Genetics. 2014;15:585–598. doi: 10.1038/nrg3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshida Y. Nearest template prediction: a single-sample-based flexible class prediction with confidence assessment. PLoS One. 2010;5:e15543. doi: 10.1371/journal.pone.0015543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshida Y, Brunet JP, Tamayo P, Golub TR, Mesirov JP. Subclass mapping: identifying common subtypes in independent disease data sets. PLoS One. 2007;2:e1195. doi: 10.1371/journal.pone.0001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshida Y, Nijman SM, Kobayashi M, Chan JA, Brunet JP, Chiang DY, Villanueva A, Newell P, Ikeda K, Hashimoto M, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69:7385–7392. doi: 10.1158/0008-5472.CAN-09-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Pawlik TM, Anders RA, Selaru FM, Streppel MM, Lucas DJ, Niknafs N, Guthrie VB, Maitra A, Argani P, et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. NatGenet. 2013 doi: 10.1038/ng.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H, Chung JY, Ylaya K, Conway C, Takikita M, Fukuoka J, Doki Y, Hanaoka J, Hewitt SM. Profiling of phospho-AKT, phospho-mTOR, phospho-MAPK and EGFR in non-small cell lung cancer. J Histochem Cytochem. 2014;62:335–346. doi: 10.1369/0022155414523022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nature medicine. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T, Durnez A, Demetris AJ, Thorgeirsson SS. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40:667–676. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- Lee JS, Heo J, Libbrecht L, Chu IS, Kaposi-Novak P, Calvisi DF, Mikaelyan A, Roberts LR, Demetris AJ, Sun Z, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. NatMed. 2006;12:410–416. doi: 10.1038/nm1377. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R Genome Project Data Processing S. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti S, Savage KJ, Kutok JL, Feuerhake F, Kurtin P, Mihm M, Wu B, Pasqualucci L, Neuberg D, Aguiar RC, et al. Molecular profiling of diffuse large B-cell lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood. 2005;105:1851–1861. doi: 10.1182/blood-2004-07-2947. [DOI] [PubMed] [Google Scholar]
- Nakamura H, Arai Y, Totoki Y, Shirota T, Elzawahry A, Kato M, Hama N, Hosoda F, Urushidate T, Ohashi S, et al. Genomic spectra of biliary tract cancer. Nat Genet. 2015;47:1003–1010. doi: 10.1038/ng.3375. [DOI] [PubMed] [Google Scholar]
- Nault JC, De Reynies A, Villanueva A, Calderaro J, Rebouissou S, Couchy G, Decaens T, Franco D, Imbeaud S, Rousseau F, et al. A hepatocellular carcinoma 5-gene score associated with survival of patients after liver resection. Gastroenterology. 2013;145:176–187. doi: 10.1053/j.gastro.2013.03.051. [DOI] [PubMed] [Google Scholar]
- Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nature methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niiya F, Tatsumoto T, Lee KS, Miki T. Phosphorylation of the cytokinesis regulator ECT2 at G2/M phase stimulates association of the mitotic kinase Plk1 and accumulation of GTP-bound RhoA. Oncogene. 2006;25:827–837. doi: 10.1038/sj.onc.1209124. [DOI] [PubMed] [Google Scholar]
- Oishi N, Kumar MR, Roessler S, Ji J, Forgues M, Budhu A, Zhao X, Andersen JB, Ye QH, Jia HL, et al. Transcriptomic profiling reveals hepatic stem-like gene signatures and interplay of miR-200c and epithelial-mesenchymal transition in intrahepatic cholangiocarcinoma. Hepatology. 2012;56:1792–1803. doi: 10.1002/hep.25890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong CK, Subimerb C, Pairojkul C, Wongkham S, Cutcutache I, Yu W, McPherson JR, Allen GE, Ng CC, Wong BH, et al. Exome sequencing of liver fluke-associated cholangiocarcinoma. NatGenet. 2012;44:690–693. doi: 10.1038/ng.2273. [DOI] [PubMed] [Google Scholar]
- Park EJ, Lee JH, Yu GY, He G, Ali SR, Holzer RG, Osterreicher CH, Takahashi H, Karin M. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell. 2010;140:197–208. doi: 10.1016/j.cell.2009.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler S, Jia HL, Budhu A, Forgues M, Ye QH, Lee JS, Thorgeirsson SS, Sun Z, Tang ZY, Qin LX, et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer research. 2010;70:10202–10212. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler S, Lin G, Forgues M, Budhu A, Hoover S, Simpson RM, Wu X, He P, Qin LX, Tang ZY, et al. Integrative genomic and transcriptomic characterization of matched primary and metastatic liver and colorectal carcinoma. International journal of biological sciences. 2015;11:88–98. doi: 10.7150/ijbs.10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler S, Long EL, Budhu A, Chen Y, Zhao X, Ji J, Walker R, Jia HL, Ye QH, Qin LX, et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology. 2012;142:957–966. doi: 10.1053/j.gastro.2011.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–1817. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- Sia D, Hoshida Y, Villanueva A, Roayaie S, Ferrer J, Tabak B, Peix J, Sole M, Tovar V, Alsinet C, et al. Integrative molecular analysis of intrahepatic cholangiocarcinoma reveals 2 classes that have different outcomes. Gastroenterology. 2013;144:829–840. doi: 10.1053/j.gastro.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sripa B, Kaewkes S, Sithithaworn P, Mairiang E, Laha T, Smout M, Pairojkul C, Bhudhisawasdi V, Tesana S, Thinkamrop B, et al. Liver fluke induces cholangiocarcinoma. PLoSMed. 2007;4:e201. doi: 10.1371/journal.pmed.0040201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strebhardt K. Multifaceted polo-like kinases: drug targets and antitargets for cancer therapy. Nat Rev Drug Discov. 2010;9:643–660. doi: 10.1038/nrd3184. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research, N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theise ND. Liver cancer (Lyon, France: International Agency for Research on Cancer) 2014 [Google Scholar]
- Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nat Rev Drug Discov. 2008;7:678–693. doi: 10.1038/nrd2619. [DOI] [PubMed] [Google Scholar]
- Totoki Y, Tatsuno K, Covington KR, Ueda H, Creighton CJ, Kato M, Tsuji S, Donehower LA, Slagle BL, Nakamura H, et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet. 2014;46:1267–1273. doi: 10.1038/ng.3126. [DOI] [PubMed] [Google Scholar]
- Vigil D, Cherfils J, Rossman KL, Der CJ. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat Rev Cancer. 2010;10:842–857. doi: 10.1038/nrc2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XW, Thorrgeirsson SS. The Biological and clinical challenge of liver cancer heterogeneity. Hepatic Oncology. 2014;1:5. doi: 10.2217/hep.14.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welzel TM, Graubard BI, Zeuzem S, El-Serag HB, Davila JA, McGlynn KA. Metabolic syndrome increases the risk of primary liver cancer in the United States: a study in the SEER-Medicare database. Hepatology. 2011;54:463–471. doi: 10.1002/hep.24397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HG, Lee JH, Yoon JH, Kim CY, Lee HS, Jang JJ, Yi NJ, Suh KS, Lee KU, Park ES, et al. Identification of a cholangiocarcinoma-like gene expression trait in hepatocellular carcinoma. Cancer research. 2010;70:3034–3041. doi: 10.1158/0008-5472.CAN-09-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HG, Park ES, Lee JS, Lee YH, Ishikawa T, Kim YJ, Thorgeirsson SS. Identification of potential driver genes in human liver carcinoma by genomewide screening. Cancer research. 2009;69:4059–4066. doi: 10.1158/0008-5472.CAN-09-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worns MA, Galle PR. HCC therapies--lessons learned. Nature reviews Gastroenterology & hepatology. 2014;11:447–452. doi: 10.1038/nrgastro.2014.10. [DOI] [PubMed] [Google Scholar]
- Yamashita T, Forgues M, Wang W, Kim JW, Ye Q, Jia H, Budhu A, Zanetti KA, Chen Y, Qin LX, et al. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res. 2008;68:1451–1461. doi: 10.1158/0008-5472.CAN-07-6013. [DOI] [PubMed] [Google Scholar]
- Ye QH, Qin LX, Forgues M, He P, Kim JW, Peng AC, Simon R, Li Y, Robles AI, Chen Y, et al. Predicting hepatitis B virus-positive metastatic hepatocellular carcinomas using gene expression profiling and supervised machine learning. NatMed. 2003;9:416–423. doi: 10.1038/nm843. [DOI] [PubMed] [Google Scholar]
- Yim H. Current clinical trials with polo-like kinase 1 inhibitors in solid tumors. Anticancer Drugs. 2013;24:999–1006. doi: 10.1097/CAD.0000000000000007. [DOI] [PubMed] [Google Scholar]
- Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, Iwakura Y, Oshima K, Morita H, Hattori M, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499:97–101. doi: 10.1038/nature12347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S7, related to Figure 5. Comparison of somatic mutation frequency in ICC and HCC cohorts.
Table S8, related to Figure 6. Comparison of somatic copy number alteration amplification or deletion in TIGER-LC and TCGA HCC patients.
Table S9, related to Figure 6. Driver genes.
Table S10, related to Figure 7. Metabolite analysis of TIGER-LC tumors.
Table S1, related to Figure 1. Demographic, clinical and laboratory characteristics of ICC and HCC patients in the TIGER-LC cohorts at the time of surgery.
Table S2, related to Figure 2. Significantly different pathways among subtypes from GSEA analysis (cut off NOM p <0.05).
Table S3, related to Figure 2. Gene signatures tested in the study.
Table S4, related to Figure 3. Demographic, clinical and laboratory characteristics of patients in the validation cohorts.
Table S5, related to Figure 4. List of genes sequenced in this study.
Table S6, related to Figure 4. OncoVar mutation summary of the TIGER-LC cohort.