Abstract
Rationale
Ataxia telangiectasia and Rad3-related (ATR) threonine serine kinase is one of the key elements in orchestrating the DNA damage response (DDR). As such, inhibition of ATR can amplify the effects of chemo- and radiation-therapy, and several ATR inhibitors (ATRi) have already undergone clinical testing in cancer. For more accurate patient selection, monitoring and staging, real-time in vivo imaging of ATR could be invaluable; the development of appropriate imaging agents has remained a major challenge.
Methods
3-amino-N-(4-[18F]phenyl)-6-(4-(methylsulfonyl)phenyl)pyrazine-2-carboxamide ([18F]-ATRi), a close analogue of Ve-821, (a clinical ATRi candidate), was readily accomplished similarly to already established synthetic procedures. Structurally, 18F was introduced at the 4-position of the aromatic ring of Ve-821 for generating a labeled ATR inhibitor. In vitro experiments were conducted in U251MG glioblastoma cell lines and ex vivo biodistribution were performed in subcutaneous U251 MG xenograft bearing athymic nude mice following microPET imaging.
Results
[18F]-ATRi has a similar pharmacokinetic profile to that of Ve-821. Using an U251 MG glioblastoma mouse model, we evaluated the in vivo binding efficiency of [18F]-ATRi. Blood and tumor showed a statistically significant difference between mice injected with only the probe or following blocking experiment with Ve-821 (1.48 ± 0.40%ID/g vs. 0.46 ± 0.12%ID/g in tumor and 1.85 ± 0.47%ID/g vs. 0.84 ± 0.3%ID/g in blood respectively).
Conclusions
[18F]-ATRi represents the first 18F positron emission tomography (PET) ATR imaging agent, and is designed on a low nanomolar and clinically relevant ATR inhibitor.
Keywords: ATR, Cell cycle, PET imaging, U251 mg, Ve-821, 18F
1. Introduction
One of the most fundamental functions of cells is to maintain the integrity of their genomes [1–5]. Sophisticated cell cycle checkpoint pathways are in place to respond and remediate DNA damage quickly, and to efficiently control cycle progression and DNA replication [5,6]. The overall function of these checkpoints is to detect damaged or abnormally structured DNA, and to coordinate cell-cycle progression with DNA repair. Fundamentally speaking, cell-cycle checkpoints control cell-cycle phase transitions and ensure the correct completion of prior events [7]. ATR protein kinase and ataxia telangiectasia mutated (ATM) are two central kinases of such checkpoint pathways [7–10]. Both are activated by DNA damage and DNA replication stress, but their DNA-damage specificities are distinct and their functions are not redundant [7,8,11]. The serine/threonine-protein kinase ATR is involved in the DNA damage repair (DDR) mechanism and is activated by single stranded DNA structures, which may occur at resected DNA double strand breaks (DSBs) or stalled replication forks [12–14]. When DNA polymerases stall during DNA replication, replicative helicases continue to unwind the DNA ahead of the replication fork, leading to the generation of long stretches of single stranded DNA (ssDNA), which are then bound by the single-strand binding protein complex Replication protein A (RPA) [7]. Once activated, ATR triggers Checkpoint-1 (Chk1) and other downstream targets to promote DNA repair, stabilization, restart of stalled replication forks and transient cell cycle arrest [9,15,16]. A growing body of data, however, now shows roles for ATR far beyond the activation of Chk1 [17–20] where ATR also plays a role in normal replication of undamaged DNA [12]. Over the last decade, a variety of studies have also suggested that checkpoint inhibition might indeed selectively sensitize cancer cells [17,19,21–26], and many current cancer treatments, including chemotherapeutic agents and ionizing radiation, induce DNA damage and replication fork stalling, thereby activating cell cycle checkpoint pathways [6,11,17]. Following this, efficient inhibition of ATR would enhance and amplify the chemotherapeutic effects of such treatments, and variety of studies have indeed shown that selective inhibition of ATR and Chk1 enhances the killing of tumor cells [17,19,25,27]. The ATR/Chk1 cell cycle pathway is often upregulated, and loss or mutations in human cancers is low [8,9,11,25]. Therefore, ATR could be ideal target for PET imaging agent development. Noninvasive imaging of ATR is of high clinical relevance, not only for patient selection, but potentially also for measuring target engagement during combination therapies. With this goal in mind, we set out to develop [18F]-ATRi, a PET radiolabeled version of the ATR inhibitor Ve-821, which non-invasively images kinase expression in animal model of cancer (Fig. 1). Specifically, our goal was to determine if: i) [18F]-ATRi has suitable pharmacokinetic properties for imaging ATR, and ii) whether the tracer is selective. Herein, we report on the chemical synthesis, radiolabeling and preclinical evaluation of this new ATR based Ve-821 imaging agent. For both in vitro as well as in vivo evaluation we used U251MG cells, a human glioblastoma cell line. We show that our imaging agent could be a valuable probe targeting ATR for tumor imaging and also demonstrating that ATR activation as a biological process could have a strong prognostic value in the future.
Fig. 1.
ATM and ATR are activated by DNA damage as well as DNA replication stress. ATR triggers Chk1 and other downstream targets to promote DNA repair, stabilization, restart of stalled replication forks and transient cell cycle arrest.
2. Material and methods
2.1. Reagents and instrumentation
Reagents were purchased from Acros and Sigma-Aldrich Co. and used without further purification unless otherwise noted. Ve-821 was obtained from Aldrich (Sigma-Aldrich Co., St. Louis, MO). Proton and carbon nuclear magnetic resonance (1H and 13C NMR) spectra were recorded on a Bruker Daltonics (600MHz) spectrometer (Bruker, Billerica, MA). Chemical shift of protons and carbons were analyzed against the DMSO lock signal and reported as parts per million (ppm). Phosphate buffered saline (PBS) and Dulbecco’s Modified Eagle Medium (DMEM) was purchased from the media preparation facility at Memorial Sloan Kettering Cancer Center (New York, NY, USA). U251MG, a human glioblastoma cancer cell line was purchased from ATCC (Manassas, VA). Semi-preparative high performance liquid chromatography (HPLC) purification was achieved using a Luna C18 250 mm × 10 mm (Phenomenex, Torrance, CA) (Method A: eluents A = 0.1% TFA in water and B = 0.1% TFA in MeCN; gradient 0–17 min, 5–80% B; 17–21 min, 80–95% B; 21–24 min, 95% B; 24–25 min, 95–5% B; 25–30 min, 5% B; 3 mL min−1). Analytical HPLC analyses were performed using a Gemini C18 250 mm × 4.6 mm reversed-phase column (Phenomenex, Torrance, CA) (Method B: eluents A=0.1% TFA in water and B = 0.1% TFA in MeCN; gradient 0–17 min, 5–95% B; 17–21 min, 95% B; 21–22 min, 95–5% B; 22–25 min, 5% B; 1 mL min−1). Both semi-preparative and analytical HPLC purifications were performed on a Shimadzu UFLC HPLC system (Shimadzu Scientific Instruments, Inc., Columbia, MD) equipped with a DGU-20A degasser, an SPD-M20A UV detector, an LC-20AB pump system, and a Scan-RAM radio-TLC/HPLC-detector from LabLogic (Brandon, FL) with a dual-wavelength UV–V is detector followed by a flow-through γ detector connected in series. Semi-preparative and analytical HPLC analyses of 18F–labeled compounds were calibrated with the corresponding 19F analogues. Microwave syntheses were conducted with a Biotage® Initiator+ synthesizer (Charlotte, NC). Radioactivity in blood half-life, cell uptake, competitive displacement and biodistribution studies was quantified with a WIZARD2 automatic γ-counter from Perkin Elmer (Boston, MA). Small animal PET imaging data were recorded on a Focus 120 MicroPET (Concorde Microsystems, Knoxville, TN) and reconstructed using ASI Pro VM™ MicroPET Analysis software (SiemensMedical Solutions, Knoxville, TX). [18F]-ATRi, 19F–ATRi and Ve-821 were formulated in 20% PEG300/80% PBS and used without any further filtration unless otherwise described.
2.2. Preparation of 3-amino-6-(4-(methylsulfonyl)phenyl)pyrazine-2-carboxylic acid (3)
To a reaction tube containing 3-amino-6-bromo-N-phenyl-pyrazine-2-carboxamide 1a (88 mg, 0.3 mmol) in DMF (4 mL), 2-(4-methanesulfonylphenyl)-4,4,5,5-tetramethyl-1,3,2-dioxaborolane 1b (0.3 mmol) was added, followed by Pd(PPh3)2Cl2 (5 mol%) and 2 M aqueous Na2CO3 solution (2 mL). The tubes were then flushed with N2 and heated to 110 °C for 1 h in a microwave. After this time, the reaction was allowed to cool to ambient temperature, filtered, and purified by reverse phase HPLC (method A) to give the desired product as solid after lyophilization (80 ± 10% yield). The lyophilized product methyl 3-amino-6-(4-(methylsulfonyl)phenyl)pyrazine-2-carboxylate 2 (50 mg, 0.15mmol)was further dissolved in MeOH/H2O (3 mL) and reacted overnight in the presence of LiOH (11 mg, 0.45 mmol) under reflux conditions to yield 3-amino-6-(4-(methylsulfonyl)phenyl)pyrazine-2-carboxylic acid 3 (65 ± 7% yield). The reaction was allowed to cool to ambient temperature, filtered, and purified by reverse phase HPLC to give the desired products as solids after lyophilization (75 ± 10% yield). HRMS(+) m/z = 294.0178 [M + H]+, 292.0267 [M - H]-; 1H–NMR (600 MHz, DMSO-d6) δ = 9.02 (s, 1H), 8.35 (d, 3JHH = 8.5Hz, 2H), 7.99 d, 3JHH = 8.5 Hz, 2H), 7.70 (m, 2H), 3.25 (s, 3H).
2.3. Preparation of 3-amino-N-(4-fluorophenyl)-6-(4-(methylsulfonyl)-phenyl)pyrazine-2-carboxamide (5)
To a solution of 3-amino-6-(4-(methylsulfonyl)phenyl)pyrazine-2-carboxylic acid in DMSO (6 mg, 0.04 mmol), 4-fluoroaniline (0.2 mmol), HBTU (0.12 mmol) and triethylamine (0.12 mmol) were added and the solution reacted at 150 °C for 10 min. The resulting solution was filtered and purified by reverse phase HPLC (method A) to the desired amide derivative (yields ranged from 65% to 74%), dried and lyophilized to yield the final product 5. HRMS(+) m/z = 308.0887 [M + H]+, 306.0267 [M - H]-; 1H–NMR (600 MHz, DMSO-d6) δ = 10.52 (s, 1H), 9.03 (s, 1H), 8.50 (d, 3JHH = 8.6 Hz, 2H), 7.99 (d, 3JHH = 8.6 Hz, 2H), 7.86 (m, 2H), 7.82 (dd, 3JHH = 9.1 Hz, 3JHF = 5.0 Hz, 2H), 7.25 (t, 3JHH = 8.9 Hz, 2H), 3.26 (s, 3H).
2.4. Preparation of 3-amino-6-(4-(methylsulfonyl)phenyl)pyrazine-2-carbonyl chloride (8)
Thionyl chloride (1 μL, 50 μmol) was added to a solution of 3-amino-6-(4-(methylsulfonyl)phenyl)pyrazine-2-carboxylic acid in toluene (5 mg in 500 μL), and the solution reacted at 100 °C for 3 h. The final 3-amino-6-(4-(methylsulfonyl)phenyl)pyrazine-2-carbonyl chloride product (80 ± 15% yield) was stored at −80 °C until further use.
2.5. Radiochemistry
4-[18F]fluoroaniline was obtained analogously to what was reported before from Hendricks and coworkers [28]. Briefly, a QMA cartridge containing cyclotron-produced [18F] fluoride ion was eluted with a solution containing 9 mg Kryptofix [2.2.2] (4,7,13,16,21,24-hexaoxa-1,10-diazabicyclo[8.8.8]hexacosane), 0.08 mL 0.15 M K2CO3 and 1.92 mL MeCN into a 5 mL reaction vial. Water was removed azeotropically at 120 °C. 3 mg of 1,4-dinitrobenzene were dissolved in 300 μL of DMSO, transferred to a sealed reaction vial and finally heated to 120 °C in a microwave for 5 min and then cooled to room temperature. The reaction mixture was diluted with water (8 mL) and loaded onto a Sep.-Pak® Light C18 (Waters, Milford, MA) preconditioned with EtOH (10 mL) and water (10 mL). The cartridge was washed with 5 mL MilliQ water and 4-[18F]fluoronitrobenzene (6) was eluted from the cartridge with MeOH (1 mL). To (6), 3 mg of Pd/C and 28 mg NaBH4 were added and the reaction mixture stirred for 5 min, at which time unreacted NaBH4 was quenched by adding 300 μL 6 N HCl. The mixture was diluted with 1 N NaOH(aq) and passed through a Lichrolut EN cartridge (Merck, Billerica, MA) and a Na2SO4: Celite (3:2 w/w). The loaded 4-[18F]fluoroaniline (7) was finally eluted with THF (1 mL) in a vial containing 500 μg of 3-amino-6-(4-(methylsulfonyl)phenyl)pyrazine-2-carbonyl chloride (8) and the mixture was stirred at room temperature for 5 min. Thereafter, the reaction mixture was purified by semi-preparative HPLC (method A) yielding 28.7 ± 7% dcRCY in 120 ± 15 min and a 98 ± 2% radiochemical purity over 3 steps preparation. The specific activity was determined to be 0.7 Ci/μmol.
2.6. Cell culture
U251 MG cells were grown in Eagle’s Minimal Essential Medium (MEM) containing 10% (vol/vol) heat inactivated fetal bovine serum, 100 IU penicillin, and 100 μg/mL streptomycin as purchased from the culture media preparation facility at MSK (New York, NY).
2.7. Mouse model
16 Female athymic nude CrTac:NCr-Foxn1nu mice were purchased from Taconic Laboratories (Hudson, NY). 8 mice received subcutaneous injections with 106 U251 MG cancer cells in Matrigel® (BD Biosciences, San Jose, CA) into each left shoulder and were allowed to grow for approximately two weeks until the tumors reached 5–10mmin diameter. Mice were anesthetized (isoflurane 1.5%, 2 L/min medical air) during tumor implantation and microPET imaging. All animal experiments were conducted in accordance with protocols approved by the Institutional Animal Care and Use Committee of MSK and followed NIH guidelines for animal welfare.
2.8. Blood stability
[18F]-ATRi (approximately 300 μCi, 0.7 Ci/μmol) was injected in healthy athymic nude mice (n = 5) via tail injection. Mice were sacrificed at different time points (0, 60, 120, 180, 240 min p.i.) and blood was collected. 750 μL of MeCN were added to the collected blood and centrifuged (5 min at 5000 rpm) to pellet blood cells and proteins. The supernatant was collected and prepared for HPLC injection by adding 750 μL mQ H2O and filtering. The blood stability was measured by analytical HPLC analysis (method B).
2.9. Blood half-life
The blood half-life of [18F]-ATRi was calculated by measuring the activity of blood samples collected at different time points (5, 15, 30, 45, 60, 90 and 120 min p.i.). Female nude mice (n = 3) were injected via tail vein with [18F]-ATRi (20% PEG300/80% PBS) and blood samples obtained by retro-orbital bleed using tared capillary tubes. Samples were weighed, and activity was measured by γ counter. The blood half-life was calculated with Graph Prism 7 (GraphPad Software, La Jolla, CA) using a two-phase decay least squares fitting method and expressed as %IA/g.
2.10. Chemical hydrophobicity index
The Chemical Hydrophobicity Indices (CHI) were measured using a previously developed procedure [29,30]. Briefly, reverse phase HPLC was used to measure the retention times of a set of standards with known CHI. A standard curve was then created to calculate the CHI of 19F–ATRi based on the HPLC retention time.
2.11. Octanol/Water partition coefficient
The lipophilicity of the [18F]-ATRi was acquired by adding 2.5 μCi to a mixture of 0.5 mL of 1-octanol and 0.5 mL of 25 mM phosphate buffered saline (pH 7.4) and mixed for 5 min. Then, the mixture was centrifuged at 15.000 rpm for 5 min. 100 μL samples were obtained from organic and aqueous layers, and the radioactivity of the samples were measured in a γ-counter WIZARD2 automatic γ-counter (PerkinElmer, Boston, MA). The experiment was performed in triplicate, and the resulting logPo/w was calculated as the mean ± SD.
2.12. Plasma protein binding
The plasma protein fraction was determined using the Rapid Equilibrium Dialysis Device System (Life Technologies, Grand Island, NY) according to the manufacturer’s protocol. Membrane dialysis was performed with 500 μM 19F–ATRi in serum (500 μL) on one side of the membrane and PBS (700 μL) on the other side. The system was sealed with parafilm and incubated for 4 h at 37 °C on an orbital shaker set to 100 rpm. Thereafter, 50 μL of solution was taken from both sides, and samples were treated with 300 μL of precipitation buffer (90/10 acetonitrile/water with 0.1% formic acid) and vortexed to remove protein before HPLC analysis. After injection, the [18F]-ATRi peaks from each sample were integrated, and the protein bound fraction determined as % bound = [1 – (Concentration buffer chamber/Concentration plasma chamber)]×100. The final albumin binding was calculated as the mean ± SD.
2.13. Competitive displacement assay
For inhibition studies, U251Mgcells were seeded in a 96wells plate (105 cells 24 h prior the experiment). The next day, [18F]-ATRi at a final 0.667 nM concentration was added and co-incubated together with different Ve-821 concentrations (ranging from 0 to 1333 nM) at 37 °C for 1 h. After 1 h, cells were first washed twice with PBS, then lysed with 1 N NaOH, and finally collected and counted in a γ-counter for bound 18F ligand. The percentage of bound radioligand at each concentration was measured in triplicate and plotted against Ve-821 concentration. Competitive displacement curves were fitted using Graph Prism 7 (GraphPad Software Inc., La Jolla, CA).
2.14. In vitro cell uptake
One day prior to the assay, U251 MG cells were placed in 6-well plates (0.5 million cells/well). On the day of the experiment, cells were washed twice with PBS and further incubated with [18F]-ATRi (6 μCi/well) at 37 °C for 0, 15, 30, 45, 60, 120 and 240 min in MEM medium to allow for internalization. 100-fold excess of Ve-821 was coincubated with [18F]-ATRi as blocking agent for ATR in the blocking study. At each of the reported time points, cells were first washed twice with PBS and then lysed by incubation with 1 M NaOH (10 min at 37 °C). Then, the resulting lysate in each well was collected and the radioactivity measured in a γ-counter.
2.15. MicroPET imaging and ex vivo biodistribution
MicroPET imaging studies were performed in subcutaneous U251 MG xenograft bearing athymic nude mice (n = 8). Mice were divided in two groups (blocked and unblocked) and administered with [18F]-ATRi (approximately 200 μCi, 0.7 Ci/μmol) via tail vein injection. The blocked group was pre-injected 30 min before with a 100-fold excess of Ve-821. Approximately 5 min prior to PET acquisition, mice were anesthetized by inhalation of a mixture of isoflurane (Baxter Healthcare, Deerfield, IL, USA; 2% isoflurane, 2 L/min medical air) and positioned on the scanner bed. Anesthesia was maintained using a 1% isoflurane/O2 mixture. PET data for each mouse were recorded starting at 30 min p.i. using dynamic scans of 5 min and acquired for the following 90 min. After microPET imaging acquisition was concluded, mice were sacrificed by CO2 asphyxiation (120 min p.i.) and major organs were collected, weighed, and counted in a γ-counter. The radiopharmaceutical uptake was expressed as a %ID/g using the following formula: [(activity in the target organ/g of tissue)/injected dose] × 100% and plotted as the mean ± SD.
3. Results and discussion
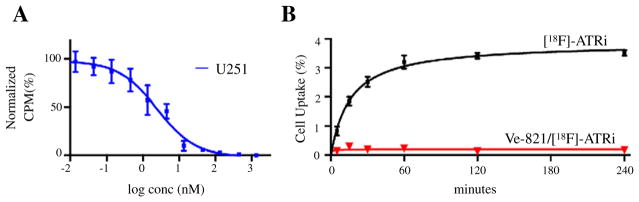
In this study, we report on the first in vivo [18F]-ATRi ([18F]-9) as a PET imaging agent with strong similarities in pharmacology, potency, and isoform selectivity to Ve-821, a clinically relevant ATR inhibitor [27]. Structurally, introduction of an 18F label at the 4-position of the aromatic ring of Ve-821 (4) appeared to be a viable approach for generating a labeled ATR inhibitor. Probing whether this functionalization could negatively impact the compound’s biological activity, we synthesized and profiled the cold fluorinated analogue of our desired probe, 19F–ATRi 5. The synthesis was readily accomplished and similar to already established synthetic procedures [31], including some minor modifications (Suppl. Figs. S1 and S2). Synthesis started from two commercially available precursors, 3-amino-6-bromo-N-phenyl-pyrazine-2-carboxamide (1a) and 2-(4-methanesulfonylphenyl)-4,4,5,5-tetramethyl-1,3,2-dioxaborolane(1b), which were coupled using standard Suzuki reaction conditions [32,33]. The reaction yielded an intermediate 2 in 80% yield under microwave-heating at 110 °C for 1 h. Subsequent reaction with aniline or fluoroaniline in basic conditions yielded the conversion of the carboxylic acid 3 to the target compound 4 and 5 respectively under microwave-heating at 150 °C for 10 min. Our goal was to develop a reliable synthetic strategy for the routine production of [18F]-ATRi with sufficiently high isolated radiochemical yields to permit in vivo studies. As reported by Hendricks and coworkers and Liang et al., direct fluorination of 1,4-dinitrobenzene with 18F (no carrier added, [n.c.a.]) under phase transfer conditions and microwave heating afforded 1-[18F]fluoro-4-nitrobenzene 6 after 5 min at 120 °C [28,34]. This intermediate was consecutively reduced by catalytic hydrogenation with sodium borohydride and Pd/C to yield 4-[18F]fluoroaniline 7 (Fig. 2A and B). The 18F–labeled aniline was subsequently converted to the desired product [18F]-ATRi ([18F]-9) via coupling with precursor 8 in toluene and after stirring the reaction mixture at RT for 5 min (Fig. 2A) (Suppl. Fig. 3). Overall, this three-step synthesis is remarkably clean (Fig. 2B) (Suppl. Fig. S4), can be carried out in less than 2 h, and affords [18F]-9 with an overall isolated decay corrected radiochemical yield of 30 ± 10% (after HPLC purification), a radiochemical purity greater than 97% (Fig. 2C) and a specific activity of 0.7 Ci/μmol (Suppl. Fig. 5). Following scale-up synthesis and characterization of the radiotracer, we next tested the radiolabeled tracer [18F]-9 both in vitro and in vivo. Initially, we determined the ex vivo blood stability of [18F]-9 (Fig. 3A and B), the in vivo blood half-life (Fig. 3C), and defined the main pharmacokinetic values for 5 (Fig. 3D). The ex vivo blood stability of [18F]-9 showed that more than 97% of the tracer was stable 4 h post injection with no major metabolites observed (Fig. 3B). The blood half-life, following a single bolus intravenous injection via tail vein, showed a biphasic pharmacokinetic profile with rapid elimination of [18F]-9 during the first 10 min, similarly to other small molecule inhibitors [28,35]. The weighted blood half-life was determined to be 6.2 min. Contrary to the rapid initial elimination, redistribution of the agent is slow, which is likely due to ATR being expressed in immune cells, which would consequently lead to specific retention of the tracer, explaining the observed plateau [9,11,36]. The basic pharmacokinetic profile of 5 demonstrated that, similarly to Ve-821, 19F–ATRi has a lipophilic behavior, as proven by its CHI value (76.8) and high plasma protein binding (Plasma Free Fraction ~10%). The LogPO/W of the radiofluorinated [18F]-9 tracer was measured to be 1.6, corroborating the CHI values obtained with 19F–ATRi (Fig. 3D). Together with multiple studies reporting expression of ATR in glioma cell lines [37–40] we chose U251 MG, a cell line with high ATR mRNA expression [41,42], as a mouse model for our in vitro/in vivo studies. The mRNA level of ATR in GBM, although significantly higher in comparison to healthy brain tissues [41,42], are lower than other major solid tumors such as squamous cell lung, breast, colorectal, pancreatic, prostate, and skin cancer, as well as most of the remaining cancer types (Suppl. Fig. 6). This is corroborated by Nadkarni et al. [39] and Ramirez et al. [40] who measured ATR downstream signaling and ATR expression in U251 MG cells. Both groups used glioblastoma cell lines as a model to study the effect of ATM/ATR inhibitors. Further, we showed the possibility to use Ve-821 to study the in vitro cell uptake of [18F]-9. The ability to suppress [18F]-9 uptake by competitive displacement in U251 MG cells was an indication of the similar inhibition effect between Ve-821 and [18F]-9 (Fig. 4A). In fact, the tracer appears to have an affinity for ATR comparable to Ve-821 (competitive displacement value of [18F]-ATRi by Ve-821=38 nM). Cell uptake rates were also significantly diminished in U251 MG cells blocked by co-incubation with Ve-821 (3.5 ± 0.1% at 240min incubation with [18F]-ATRi only vs. 0.16±0.1% inU251MG cells blocked with Ve-821 and co-incubated with [18F]-ATRi for 240 min) (Fig. 4B). These results taken together suggest that replacement of the para proton on the aniline ring with an 18Fatom did not fundamentally perturb the binding properties of [18F]-9 in comparison to Ve-821 (Ve-821: IC50 = 26 nM, [24]) and that Ve-821 and [18F]-9 presumably bind the same target. However, a more accurate and thorough IC50 study should be performed to determine the real inhibition of the ATR target and thus the measure of the activity of [18F]-9 in the presence of a known concentration of ATP. The uptake (%ID/g) and the biodistribution of [18F]-9 are shown in Fig. 5A–C and Suppl. Fig. S7. For each experiment, two groups of mice were sacrificed at 2 h following intravenous injection of the probe. Tissues were then excised, weighed and 18F activity was measured. [18F]-9 had the highest concentration in the kidney, liver, intestines and blood, with relatively low distribution in other tissues (Fig. 5B and C). The significant blocking effect in blood (from 1.85 ± 0.47 to 0.84 ± 0.30%ID/g) supports our hypothesis that [18F]-9 is retained in the blood pool selectively, rather than absorbed non-specifically by blood plasma components. [18F]-9 was also observed to specifically accumulate in tumors as proven by TAC (Fig. 5B) and blocking experiment (from 1.48 ± 0.39 to 0.46 ± 0.12%ID/g) (Fig. 5C and Suppl. Fig. S7). The tracer washes out via the hepatobiliary route as evident by microPET image reconstruction and biodistribution data. Kidney and liver showed uptake values of 3.51 ± 0.23 and 3.68± 1.39%ID/g, respectively, in biodistribution experiments. As expected for selective competition, a high tumor-to-background ratio was observed inmost of the non-targeting tissues (Fig. 5D). This tracer shows remarkable tumor-to-brain ratios (>15), leaning evidence toward the possibility that [18F]-9 could be used for brain tumor imaging. In conclusion, our data support that [18F]-ATRi could be a great resource to image ATR, together with all its implications for better understanding ATR therapy, target engagement and treatment monitoring.
Fig. 2.
Synthesis and analysis of [18F]-ATRi. (A) Shows the radiochemical synthesis of [18F]-ATRi. 4-[18F]fluoroaniline (7)was obtained by the reduction of 4-[18F]fluoronitrobenzene (6) in the presence of Pd/C and NaBH4. Following, 4-[18F]fluoroaniline was coupled with the acid chloride precursor (8) to obtain [18F]-ATRi (9) (room temperature for 5 min); (B) analytical radioactivity traces of the three reaction step with relative products: 4-[18F]fluoronitrobenzene (6), 4-[18F]fluoroaniline (7) and [18F]-ATRi (9). (C) Quality control shows that the radiochemical purity for [18F]-ATRi (9) is >97% and perfectly coelutes with the cold reference synthesized compound 19F–ATRi (5).
Fig. 3.
Stability and basic pharmacokinetics of [18F]-ATRi and 19F–ATRi. (A) Radiochemical stability of [18F]-ATRi. Blood was collected from healthy mice after injection of [18F]-ATRi (200 μCi/mouse). 5 time points after tracer i.v. injection were evaluated (0, 60, 120, 180, 240 min). (B) Tabulation of tracer stability after 4 h. (C) In vivo blood half-life of [18F]-ATRi. Healthy mice (n = 4) were injected with [18F]-ATRi and blood collected at different time points. Results are plotted as % injected activity/g over 120 min as a 2-phase decay curve indicating a fast clearance from bloodstream (<10 min). (D) Lipophilicity profiling and plasma free fraction of 19F–ATRi and [18F]-ATRi.
Fig. 4.
Competitive displacement of [18F]-ATRi by Ve-821 and [18F]-ATRi cell uptake. A) Shows the displacement for [18F]-ATRi by increasingly high concentrations of Ve-821 (IC50 = 26 nM) in U251 MG cells (n = 3). (B) Shows the uptake rate of [18F]-ATRi in a blocked vs. non-blocked cell uptake study. Cells were incubated for a maximum of 240 min with [18F]-ATRi only or with [18F]-ATRi + Ve-821, washed twice and lysed. The collected activity was quantified in a γ-counter. Results indicate a specific blocking effect of cells co-incubated with the ATR inhibitor Ve-821 (n = 4).
Fig. 5.
MicroPET imaging and ex vivo biodistribution. (A) Representative coronal microPET image of U251 MG bearing-mouse after injection of [18F]-ATRi (unblocked, left panel) and after pre-injection of blocking agent Ve-821/[18F]-ATRi (100-fold excess, 30 min before, right panel). (B) Shows %ID/g calculated from ROI activity curves (5 min time frames over a 90 min dynamic scan) for tumor, liver, kidney, muscle, heart and brain. (C) Ex vivo biodistribution of [18F]-ATRi. [18F]-ATRi was tested in U251 MG mice xenografts sacrificed at 120 min p.i. Effects of blocking were observed after injection of 100-fold excess of Ve-821 (n = 4 per group). Values are plotted as %ID/g (n = 3). (D) Tumor to non-tumor ratios
4. Conclusions
Here we report a novel fluorinated [18F]-ATR inhibitor ([18F]-9) that was synthesized starting from Ve-821, a clinically relevant ATR inhibitor. [18F]-9 was obtained in good yield (radiochemical corrected yield ~ 30% overall), and was efficiently labeled with 18F–fluoride (<2 h;> 97% radiochemical purity). Our data suggest that [18F]-ATRi is the first in vivo ATR PET imaging agent potentially leading to better understanding of ATR therapy, target engagement and treatment monitoring. This pilot study thus shows proof of principle that an imaging agent such as [18F]-ATRi could be a first step toward a clinically translatable diagnostic tool. It encourages further investigations with the ultimate aim of non-invasively sensing and quantifying ATR kinase.
Supplementary Material
Acknowledgments
The authors thank Dr. Jason S. Lewis, Dr. NagaVara Kishore Pillarsetty and Dr. Edmund J. Keliher for helpful discussions, Dr. Christian Brand for help with experiments and Dave Gregory for editing the manuscript. Technical services provided by the MSK Small-Animal Imaging Core Facility, supported in part by NIH Cancer Center Support Grant No 2 P30 CA008748-48, are gratefully acknowledged. The authors thank the NIH (K25 EB016673, R01 CA204441 and R21 CA191679 for T.R.), the Brain Tumor Center of Memorial Sloan-Kettering Cancer (for T.R.), the Center for Molecular Imaging and Nanotechnology (for T.R.) and the American-Italian Cancer Foundation (AICF) Post-Doctoral Research Fellowship (for G.C.) for their generous support. The Tow Foundation Fellowship Program in Molecular Imaging and Nanotechnology (for B.C.), the National Science Foundation’s Integrative Graduate Education and Research Traineeship (IGERT 0965983 at Hunter College for B.C.).
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.nucmedbio.2017.01.002.
Footnotes
The authors declare no competing financial interests.
Author contributions
G. C and T.R. designed the experiments and analyzed the data. G.C., B.C., J.T., A.S., and A. V. carried out the experiments. G. C. and T. R. interpreted data and wrote the manuscript. All authors read, provided feedback on, and approved the manuscript.
References
- 1.Jackson SP, Helleday T. Drugging DNA repair. Science. 2016;352:1178–9. doi: 10.1126/science.aab0958. [DOI] [PubMed] [Google Scholar]
- 2.Da-Rè C, Halazonetis TD. DNA replication stress as an Achilles’ heel of cancer. Oncotarget. 2015;6(1) doi: 10.18632/oncotarget.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartek J, Mistrik M, Bartkova J. Thresholds of replication stress signaling in cancer development and treatment. Nat Struct Mol Biol. 2012;19:5–7. doi: 10.1038/nsmb.2220. [DOI] [PubMed] [Google Scholar]
- 4.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–8. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–23. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 6.Benada J, Macurek L. Targeting the checkpoint to kill cancer cells. Biomolecules. 2015:5. doi: 10.3390/biom5031912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–96. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 8.Manic G, Obrist F, Sistigu A, Vitale I. Trial watch: targeting ATM–CHK2 and ATR– CHK1 pathways for anticancer therapy. Mol Cell Oncol. 2015;2:e1012976. doi: 10.1080/23723556.2015.1012976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weber AM, Ryan AJ. ATM and ATR as therapeutic targets incancer. Pharmacol Ther. 2014 doi: 10.1016/j.pharmthera.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 10.Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–6. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karnitz LM, Zou L. Molecular pathways: targeting ATR in cancer therapy. Clin Cancer Res. 2015 doi: 10.1158/1078-0432.CCR-15-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodríguez-Bravo V, Guaita-Esteruelas S, Salvador N, Bachs O, Agell N. Different S/M checkpoint responses of tumor and non–tumor cell lines to DNA replication inhibition. Cancer Res. 2007;67:11648–56. doi: 10.1158/0008-5472.CAN-07-3100. [DOI] [PubMed] [Google Scholar]
- 13.Ruzankina Y, Schoppy DW, Asare A, Clark CE, Vonderheide RH, Brown EJ. Tissue regenerative delays and synthetic lethality in adult mice after combined deletion of Atr and Trp53. Nat Genet. 2009;41:1144–9. doi: 10.1038/ng.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruzankina Y, Pinzon-Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, et al. Deletion of the developmentally essential Gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell. 2007;1:113–26. doi: 10.1016/j.stem.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–27. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casper AM, Nghiem P, Arlt MF, Glover TW. ATR regulates fragile site stability. Cell. 2002;111:779–89. doi: 10.1016/s0092-8674(02)01113-3. [DOI] [PubMed] [Google Scholar]
- 17.Kwok M, Davies N, Agathanggelou A, Smith E, Oldreive C, Petermann E, et al. ATR inhibition induces synthetic lethality and overcomes chemoresistance in TP53- or ATM-defective chronic lymphocytic leukemia cells. Blood. 2016;127:582–95. doi: 10.1182/blood-2015-05-644872. [DOI] [PubMed] [Google Scholar]
- 18.Abdel-Fatah TMA, Middleton FK, Arora A, Agarwal D, Chen T, Moseley PM, et al. Untangling the ATR-CHEK1 network for prognostication, prediction and therapeutic target validation in breast cancer. Mol Oncol. 2015;9:569–85. doi: 10.1016/j.molonc.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huntoon CJ, Flatten KS, Wahner Hendrickson AE, Huehls AM, Sutor SL, Kaufmann SH, et al. ATR inhibition broadly sensitizes ovarian cancer cells to chemotherapy independent of BRCA status. Cancer Res. 2013;73:3683–91. doi: 10.1158/0008-5472.CAN-13-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown EJ, Baltimore D. Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes Dev. 2003;17:615–28. doi: 10.1101/gad.1067403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmed SU, Carruthers R, Gilmour L, Yildirim S, Watts C, Chalmers AJ. Selective inhibition of parallel DNA damage response pathways optimizes Radiosensitization of glioblastoma stem-like cells. Cancer Res. 2015;75:4416–28. doi: 10.1158/0008-5472.CAN-14-3790. [DOI] [PubMed] [Google Scholar]
- 22.Josse R, Martin SE, Guha R, Ormanoglu P, Pfister TD, Reaper PM, et al. The ATR inhibitors VE-821 and VX-970 sensitize cancer cells to topoisomerase I inhibitors by disabling DNA replication initiation and fork elongation responses. Cancer Res. 2014 doi: 10.1158/0008-5472.CAN-13-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prevo R, Fokas E, Reaper PM, Charlton PA, Pollard JR, McKenna WG, et al. The novel ATR inhibitor VE-821 increases sensitivity of pancreatic cancer cells to radiation and chemotherapy. Cancer Biol Ther. 2012;13:1072–81. doi: 10.4161/cbt.21093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reaper PM, Griffiths MR, Long JM, Charrier J-D, MacCormick S, Charlton PA, et al. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat Chem Biol. 2011;7:428–30. doi: 10.1038/nchembio.573. [DOI] [PubMed] [Google Scholar]
- 25.Li C-C, Yang J-C, Lu M-C, Lee C-L, Peng C-Y, Hsu W-Y, et al. ATR-Chk1 signaling inhibition as a therapeutic strategy to enhance cisplatin chemosensitivity in urothelial bladder cancer. Oncotarget. 2015;7(2) doi: 10.18632/oncotarget.6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lerner RG, Grossauer S, Kadkhodaei B, Meyers I, Sidorov M, Koeck K, et al. Targeting a Plk1-controlled polarity checkpoint in therapy-resistant glioblastoma-propagating cells. Cancer Res. 2015;75:5355–66. doi: 10.1158/0008-5472.CAN-14-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charrier J-D, Durrant SJ, Golec JMC, Kay DP, Knegtel RMA, MacCormick S, et al. Discovery of potent and selective inhibitors of ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase as potential anticancer agents. J Med Chem. 2011;54:2320–30. doi: 10.1021/jm101488z. [DOI] [PubMed] [Google Scholar]
- 28.Hendricks JA, Keliher EJ, Marinelli B, Reiner T, Weissleder R, Mazitschek R. In vivo PET imaging of histone Deacetylases by 18F-Suberoylanilide Hydroxamic acid (18F-SAHA) J Med Chem. 2011;54:5576–82. doi: 10.1021/jm200620f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valkó K. Application of high-performance liquid chromatography based measurements of lipophilicity to model biological distribution. J Chromatogr A. 2004;1037:299–310. doi: 10.1016/j.chroma.2003.10.084. [DOI] [PubMed] [Google Scholar]
- 30.Valkó K, Bevan C, Reynolds D. Chromatographic hydrophobicity index by fastgradient RP-HPLC: a high-throughput alternative to log P/log D. Anal Chem. 1997;69:2022–9. doi: 10.1021/ac961242d. [DOI] [PubMed] [Google Scholar]
- 31.Valeur E, Bradley M. Amide bond formation: beyond the myth of coupling reagents. Chem Soc Rev. 2009;38:606–31. doi: 10.1039/b701677h. [DOI] [PubMed] [Google Scholar]
- 32.Sindhuja E, Ramesh R, Liu Y. Palladium(ii) thiocarboxamide complexes: synthesis, characterisation and application to catalytic Suzuki coupling reactions. Dalton Trans. 2012;41:5351–61. doi: 10.1039/c2dt12243j. [DOI] [PubMed] [Google Scholar]
- 33.Thakur A, Zhang K, Louie J. Suzuki-Miyaura coupling of heteroaryl boronic acids and vinyl chlorides. Chem Commun. 2012;48:203–5. doi: 10.1039/c1cc15990a. [DOI] [PubMed] [Google Scholar]
- 34.Liang SH, Collier TL, Rotstein BH, Lewis R, Steck M, Vasdev N. Rapid microfluidic flow hydrogenation for reduction or deprotection of 18F-labeled compounds. Chem Commun. 2013;49:8755–7. doi: 10.1039/c3cc45166f. [DOI] [PubMed] [Google Scholar]
- 35.Carney B, Carlucci G, Salinas B, Di Gialleonardo V, Kossatz S, Vansteene A, et al. Noninvasive PET imaging of PARP1 expression in glioblastoma models. Mol Imaging Biol. 2016;18:386–92. doi: 10.1007/s11307-015-0904-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bauer M, Goldstein M, Heylmann D, Kaina B. Human monocytes undergo excessive apoptosis following Temozolomide activating the ATM/ATR pathway while dendritic cells and macrophages are resistant. PLoS One. 2012;7:e39956. doi: 10.1371/journal.pone.0039956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kao GD, Jiang Z, Fernandes AM, Gupta AK, Maity A. Inhibition of phosphatidylinositol-3-OH kinase/Akt signaling impairs DNA repair in glioblastoma cells following ionizing radiation. J Biol Chem. 2007;282:21206–12. doi: 10.1074/jbc.M703042200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rivera M, Sukhdeo K, Yu JS. Ionizing radiation in glioblastoma initiating cells. Front Oncol. 2013:3. doi: 10.3389/fonc.2013.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nadkarni A, Shrivastav M, Mladek AC, Schwingler PM, Grogan PT, Chen J, et al. ATM inhibitor KU-55933 increases the TMZ responsiveness of only inherently TMZ sensitive GBM cells. J Neurooncol. 2012;110:349–57. doi: 10.1007/s11060-012-0979-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramirez YP, Mladek AC, Phillips RM, Gynther M, Rautio J, Ross AH, et al. Evaluation of novel Imidazotetrazine analogues designed to overcome Temozolomide resistance and glioblastoma regrowth. Mol Cancer Ther. 2015;14:111. doi: 10.1158/1535-7163.MCT-14-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Tissue-based map of the human proteome. Science. 2015:347. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 42.Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, et al. Towards a knowledge-based human protein atlas. Nat Biotechnol. 2010;28:1248–50. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.