Summary
Homeostatic scaling allows neurons to maintain stable activity patterns by globally altering their synaptic strength in response to changing activity levels. Suppression of activity by blocking action potentials increases synaptic strength through an upregulation of surface AMPA receptors. Although this synaptic up-scaling was shown to require transcription, the molecular nature of the intrinsic transcription program underlying this process and its functional significance have been unclear. Using RNA-seq, we identified 73 genes that were specifically upregulated in response to activity suppression. In particular, Neuronal pentraxin-1 (Nptx1) increased within 6 h of activity blockade, and knockdown of this gene blocked the increase in synaptic strength. Notably, Nptx1 induction is mediated by calcium influx through the T-type Voltage-Gated Calcium Channel, as well as two transcription factors, SRF and ELK1. Taken together, these results uncover a transcriptional program that specifically operates when neuronal activity is suppressed, to globally coordinate the increase in synaptic strength.
Introduction
Traditional forms of Hebbian synaptic plasticity have the potential to modify synaptic strength too far in either direction, potentially leading to runaway activity in a circuit. Homeostatic plasticity provides a way to globally scale up or down synapses to maintain the relative strengths of individual synapses without maxing out the system. Neurons are able to homeostatically regulate synaptic strength through controlling AMPA receptor (AMPAR) surface expression. This can be seen by measuring changes in the amplitudes of AMPAR mediated miniature excitatory postsynaptic currents (mEPSCs) as well as staining of surface AMPARs (Thiagarajan et al., 2005; Turrigiano et al., 1998). These changes are inducible in culture using pharmacological reagents that increase or decrease activity levels. TTX, a sodium channel blocker, blocks the action potentials in the culture, decreasing neuronal activity. Conversely, bicuculline (Bic), a GABAA receptor antagonist, blocks the major inhibitory input in the cultures, leading to a burst of synaptic activity. Deficits in the homeostatic regulation of synaptic strength (synaptic scaling) have been identified in many models of psychiatric disorders, such as autism spectrum disorders and schizophrenia (Wondolowski and Dickman, 2013), suggesting that the tight control of network activity is crucial for proper neuronal function.
Several gene products have been shown to function in scaling (Turrigiano, 2011). For example, BDNF, TNF-α, integrin beta-3 (β3), PICK1, and PSD95 have all been implicated in up-regulating the synaptic strength in response to chronic activity deprivation mainly via increasing AMPAR accumulation at synapses. Polo-like kinase 2 (Plk2), CDK5, ARC, Homer1a and EphA4 mediate the opposite effect (scaling down) in response to a chronic increase in neural activity. Therefore despite the fact that synaptic scaling occurs bi-directionally depending on the level of intracellular calcium, different sets of effector molecules mediate each direction of the scaling process.
Although increases in AMPAR levels can be seen as early as 4 – 6 h of activity silencing, this increase in synaptic strength can continue for up to 24 h or 48 h (Ibata et al., 2008). This time window of synaptic up-scaling suggests that nuclear signaling and transcription might be involved. Consistently, addition of the transcription inhibitor, Actinomycin D in cortical neuronal cultures blocked the TTX-induced increase in mEPSC amplitude (Ibata et al., 2008), suggesting the importance of gene transcription in synaptic up-scaling in response to activity suppression. However, the mechanism and function of activity-regulated transcription in neurons have mostly been studied in conditions during which activity is increased, as with Bic, rather than when activity is absent. Upon an increase in neural activity, calcium influx through the NMDAR and/or L-type voltage-gated calcium channel (VGCC) triggers activation of distinct calcium-dependent signaling pathways, which then modulate the activity of various transcription factors (e.g., CREB, MEF2, SRF) to induce expression of a specific gene set (Flavell and Greenberg, 2008). Gene products from the transcription induced by increased neural activity then mediate the cell-wide adaptive responses important for proper neuronal development and plasticity. However, little is known about whether there is a neuronal transcription program that is inherently designed to mediate the adaptive response to the opposite condition, when neuronal activity is suppressed.
In order to determine the transcription program responsible for gene induction during the quiescent state that invokes synaptic up-scaling, we performed RNA-seq as well as global run-on sequencing (GRO-seq) in mouse cortical neurons before and after TTX treatment and identified 73 genes whose expression is reproducibly upregulated by TTX-mediated activity blockade. One gene in particular, Neuronal pentraxin-1 (Nptx1) was found to be necessary for the TTX-induced scaling effect, fitting with its known role in promoting the clustering of AMPARs (Sia et al., 2007; Xu et al., 2003). Further characterization of the Nptx1 induction mechanism revealed that calcium-regulated transcription factors appear to mediate this pathway, and we discovered that the T-type VGCCs initiate this pathway, providing insight into a role for this channel in neuronal activity-dependent gene regulation.
Results
Genome-wide identification of genes that are upregulated in response to TTX
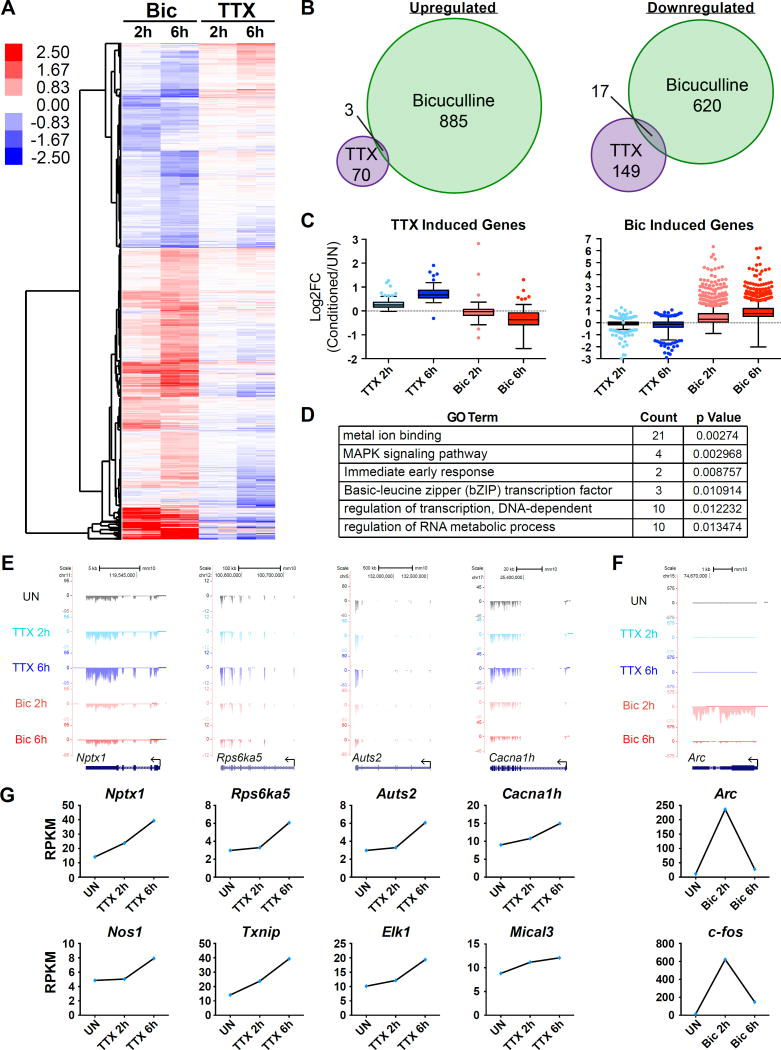
To examine the transcription program that specifically mediates TTX-induced homeostatic scaling, we performed RNA-seq analysis from neuron cultures treated with TTX for 2 h or 6 h at days in vitro (DIV) 12. We reasoned that since Actinomycin D blocked TTX-induced homeostatic up-scaling (Ibata et al., 2008), genes whose expression is increased upon activity suppression might be functionally implicated in the process. As a control, we also treated neurons with Bic for the same time periods to ensure the neurons are transcriptionally responsive to bi-directional activity manipulations (Figure 1A). We took the data from two biological replicates and identified differentially regulated genes at FDR < 0.05 and a fold change cutoff of 1.25 (see methods). A total of 73 genes were reproducibly induced by TTX whereas the Bic-mediated synaptic activity burst induced 888 genes at either time point. These two gene sets showed little overlap, suggesting that gene induction in response to TTX and Bic are mediated by distinct pathways. (Figure 1B). However, a large fraction of each gene set showed bi-directional changes, such that genes induced by TTX tend to be downregulated by Bic and vice versa (Figure 1C). Under our filtering criteria, 28 out of 73 TTX-induced genes are significantly down-regulated by Bic (Fig. S1C). If TTX-induced genes were to actively mediate synaptic up-scaling through promoting AMPAR expression and/or trafficking to the surface, their expression should be decreased when neuronal activity is chronically increased, otherwise their gene products would functionally interfere with the synaptic down-scaling process. In principle, gene expression changes in any direction could be functionally relevant to homeostatic plasticity. Our data identifies members of the cadherin family, such as Chd10 and Pcdh20, to be bidirectionally regulated, with their expression decreasing in response to TTX and increasing in response to Bic (Table S1), and cell adhesion molecules, including N-cadherin (Chd2), have been previously implicated in scaling (Fernandes and Carvalho, 2016). Stargazin (Cacng2), a gene that is downregulated in response to Bic, has previously been identified as having a role in scaling in the visual cortex (Louros et al., 2014).
Figure 1. RNA-Seq identifies a subset of genes upregulated in response to TTX.
(A) A heatmap demonstrating the expression of genes that are regulated by either TTX or Bic at 2 h or 6 h of indicated treatments in cortical neurons. The 2 h and 6 h columns each contain two lanes showing replicates of the indicated treatments. (B) Venn diagram showing the overlap between TTX and Bic upregulated (left) and downregulated (right) genes. (C) Box plots of the Log2 values for TTX upregulated genes (left) or Bic upregulated genes (right) in each different condition normalized to the unstimulated (UN) condition. A grey dotted line is present at 0 to represent no change. (D) Select GO terms identified for the TTX upregulated genes. (E) Genome browser views of example genes that are upregulated by TTX: Nptx1, Rps6ka5, Auts2, Cacna1h. (F) Genome browser views of an example gene that is upregulated in response to Bic: Arc. (G) Graphs displaying the reads per kilobase of transcript per million mapped reads (RPKM) values of the example genes for the indicated conditions. See also Figure S1 and S2.
Gene ontology analysis of TTX-induced genes did not show strong enrichment of any particular functional category, but instead distributed throughout diverse functional categories such as MAPK signaling, RNA metabolism, and transcription (Figure 1D). Notably, when compared to a well-curated list of Autism susceptibility genes, 7 out of the 73 TTX-induced genes have been implicated in ASDs: Auts2, Cacna1h, Bbs4, Fan1, Ogt, Smg6, and Upf2, and previously published studies identified associations of another 5 genes, including Nptx1, with disorders like schizophrenia and bipolar disorder (Table S2)(Fernandez-Enright et al., 2014; Girirajan et al., 2013; Howerton et al., 2013; Ionita-Laza et al., 2014; Kalscheuer et al., 2007; Nguyen et al., 2013; Rajkumar et al., 2015; Sanders et al., 2013; Shinkai et al., 2002; Splawski et al., 2006; Sultana et al., 2002). The observed association of TTX-induced genes with psychiatric disorders suggests that activity suppression-induced gene expression might be important for proper cognitive function.
Among the 73 TTX-induced genes, we have chosen 8 candidate genes (Auts2, Cacna1h, Rps6ka5, Nos1, Elk1, Mical3, Txnip, and Nptx1) for further analysis, in parallel with traditional immediate early genes like Arc and c-fos that show strong induction in response to Bic (Figure 1E–1G). Manual reverse transcription qPCR (RT-qPCR) was able to confirm the changes seen in the genome-wide data (Figure S1). A more extensive time course experiment revealed that there are roughly two groups of genes based on their induction kinetics, with one group peaking at about 6 h of TTX treatment, while another increases until 24 h (Figure S2). While the significance of the different kinetics of the two groups is unknown, this is reminiscent of the transcription induction profiles in response to an increase in neuronal activity (e.g., Bic or KCl treated condition), where rapid induction of immediate early genes is followed by a second wave of transcription of slowly induced genes (Flavell and Greenberg, 2008). The slower overall expression kinetics of TTX-induced genes also fit nicely with the previously shown time window for TTX-induced scaling of 4 – 48 h.
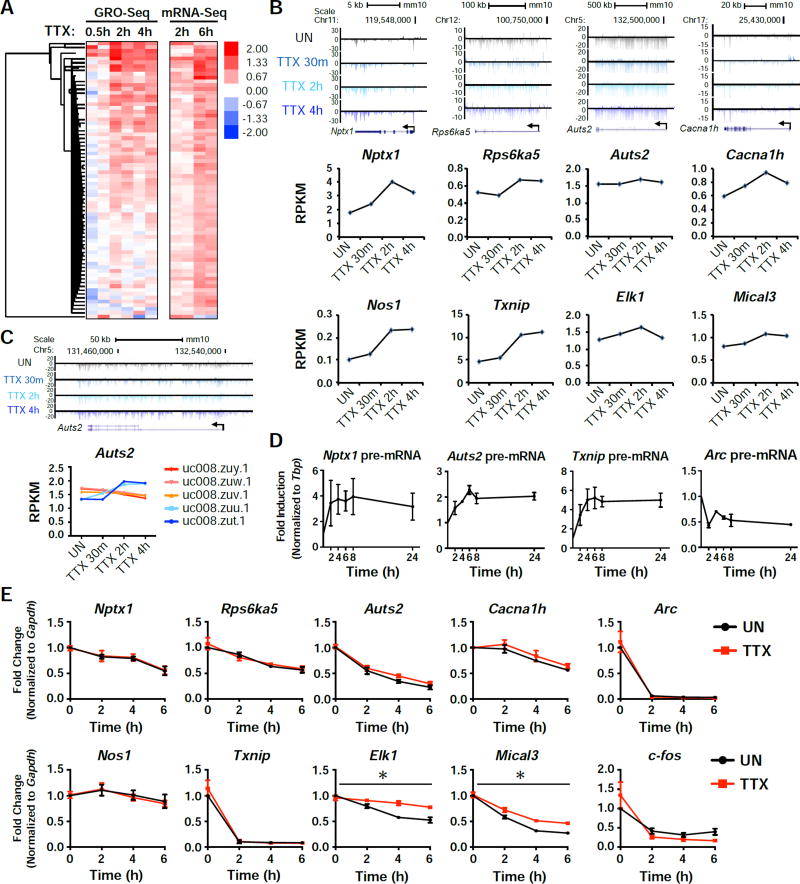
To examine the transcriptional response to activity suppression more directly, we used GRO-seq to globally measure actively transcribed genes in response to TTX treatment. For the 73 genes that we identified as TTX-inducible based on mRNA-seq, we find that most of them also show a similar increase in transcription levels determined by GRO-seq (Figure 2A and 2B). As GRO-seq only detects nascent transcription activity of RNA polymerase II (RNAPII) during the nuclear run-on (NRO) procedure, it generally shows faster expression kinetics than mRNA-seq. Of note, several genes identified in our list are extremely long genes with multiple isoforms. One such gene, Auts2, showed only a slight increase by GRO-seq when looking at the longest isoform, but two of the shorter isoforms (uc008zut.1 and uc008zuu.1) showed much higher induction (Figure 2C), suggesting isoform specific changes in expression in response to activity suppression. Additionally, by using primers targeting intronic regions to detect nascent transcripts, we measured the levels of Nptx1, Auts2, and Txnip pre-mRNAs and saw induction upon treatment with TTX (Figure 2D). In parallel, we examined whether changes in mRNA stability would also contribute to the TTX-induced increase in mRNA levels (Figure 2E). We blocked RNAPII transcription in TTX-pretreated or untreated control neurons by adding Actinomycin D, and monitored the decay rate of several candidate RNAs. Among tested genes, Elk1 and Mical3 showed a noticeable increase in mRNA stability when neurons were treated with TTX, but the rest of the tested genes showed little change in their mRNA stability. Taken together, our combined analysis of RNA-seq and GRO-seq strongly suggest that neurons have an intrinsic transcription network that can be induced in the absence of action potentials.
Figure 2. GRO-Seq confirms an active transcription program in response to activity-suppression.
(A) Heatmap of the 73 TTX-upregulated genes identified by mRNA-seq, showing their expression normalized to unstimulated. Each time point shows the data of two biological replicates. (B) Genome browser views showing the global nuclear run-on sequencing (GRO-Seq) tracks aligned with 4 of the candidate genes in response to 30 min, 2h, or 4h TTX treatment. Below the tracks are graphs displaying the RPKM values for each condition. (C) A genome browser view of the shorter isoforms of Auts2 (top), along with isoform specific RPKM values for the indicated time points (bottom). (D) qRT-PCR results from cells that were treated with TTX for the indicated time points. pre-mRNA levels were measured for the indicated genes. n = 3 biological replicates. (E) RNA levels of the indicated genes after treatment with Actinomycin D (Act D) for the indicated time points, with or without a 15 min pretreatment with TTX. n = 3 biological replicates. Error bars represent SEM. *, p<0.05. p values were determined by a two-way ANOVA.
Nptx1 is necessary for homeostatic upregulation of synaptic strength
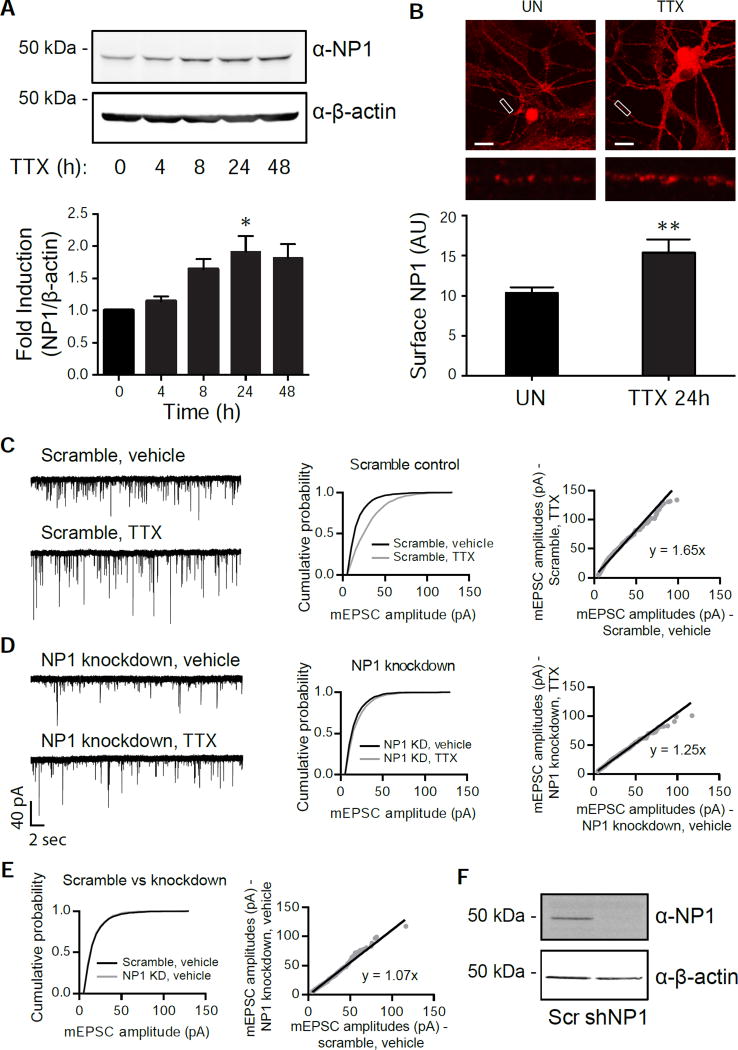
To evaluate the functional significance of the transcriptional response in neurons under activity blockade, we tested one of our candidate genes, Nptx1 for its role in synaptic up-scaling. NPTX1 protein (also known as NP1) plays a role in clustering AMPARs at the cell surface, which may imply its function in synaptic scaling. In addition to an increase in RNA levels, the total protein level of NP1 increased over time, but peaked at about 24 h of TTX treatment (Figure 3A), supporting the idea that this protein could be playing a role in increasing synaptic strength. We further observed an increase in surface expression of NP1 protein in neurons treated with TTX for 24 h (Figure 3B). To directly test the functional role of NP1 in homeostatic scaling, we measured AMPAR mEPSC amplitudes and frequencies in dissociated hippocampal cultures after knocking down Nptx1. While TTX treatment of neurons with the scrambled control for 24 h significantly increased AMPA-mediated mEPSC amplitude without a significant change in mEPSC frequency (Scr, veh: 6.04 ± 1.41; Scr, TTX: 8.83 ± 2.60; n.s. via Tukey’s multiple comparisons test), knockdown of Nptx1 blocked this increase in mEPSC amplitude (Figure 3C and 3D). However, there was no difference in the cumulative distributions of mEPSC amplitude between neurons infected with the scrambled control and Nptx1 knockdown in the vehicle condition, illustrating that the knockdown does not change baseline AMPAR surface expression (Figure 3E and 3F, p = 0.17 via K-S test. D = 0.4). Conversely, sustained over-expression of NP1 protein alone caused a significant increase in AMPAR amplitude, occluding the effect of TTX treatment (Figure S3A and S3B). There were no significant changes in mEPSC frequency when NP1 was knocked down or overexpressed (Scr, vehicle: 6.04 ± 1.41; KD, veh: 7.72 ± 2.37; OE, veh: 9.88 ± 1.36; n.s. via Tukey’s multiple comparisons test). These results suggest that the TTX-induced increase in NP1 is functionally important for the homeostatic synaptic up-scaling process and also that the precise level of NP1 within the cell is critical for maintaining proper AMPAR levels at the surface.
Figure 3. Nptx1 is functionally important for homeostatic scaling.
(A) Western blot of NP1 and β-actin expression in cortical neurons that were treated with TTX for the indicated time points (top) and the quantification of the induction (Bottom). n = 3 biological replicates. (B) Staining of surface NP1 levels on neuronal dendrites in unstimulated (UN) or TTX treated (24h) conditions, with the quantification below. n = 13 and 15 dendrites for UN and TTX conditions, respectively. (C,D) Example traces, cumulative probability histograms, and rank order plots of AMPA-mediated mEPSC amplitudes from cells treated with either vehicle or TTX after infection with the scrambled control (C) or NP1 knockdown (D). Scramble vehicle (n = 9) vs Scramble TTX (n = 8) p = 1.5×10−74, D = 0.31. Knockdown vehicle (n = 7) vs Knockdown TTX (n = 8) p = 7.3×10−5, D = 0.08. (E) The cumulative probability histograms from vehicle-treated conditions of either the scrambled or knockdown cells from (C) and (D) overlaid. p = 0.03, D = 0.17. See also Figure S3. (F) Representative western blot of the efficiency of NP1 knockdown in the scaling experiments. *, p < 0.05; **, p < 0.01. p values determined by an unpaired Student’s t-test. Statistical significance between cumulative probability graphs was determined by the Kolmogorov-Smirnov test. Error bars represent SEM.
SRF and ELK1 mediate TTX-dependent Nptx1 induction
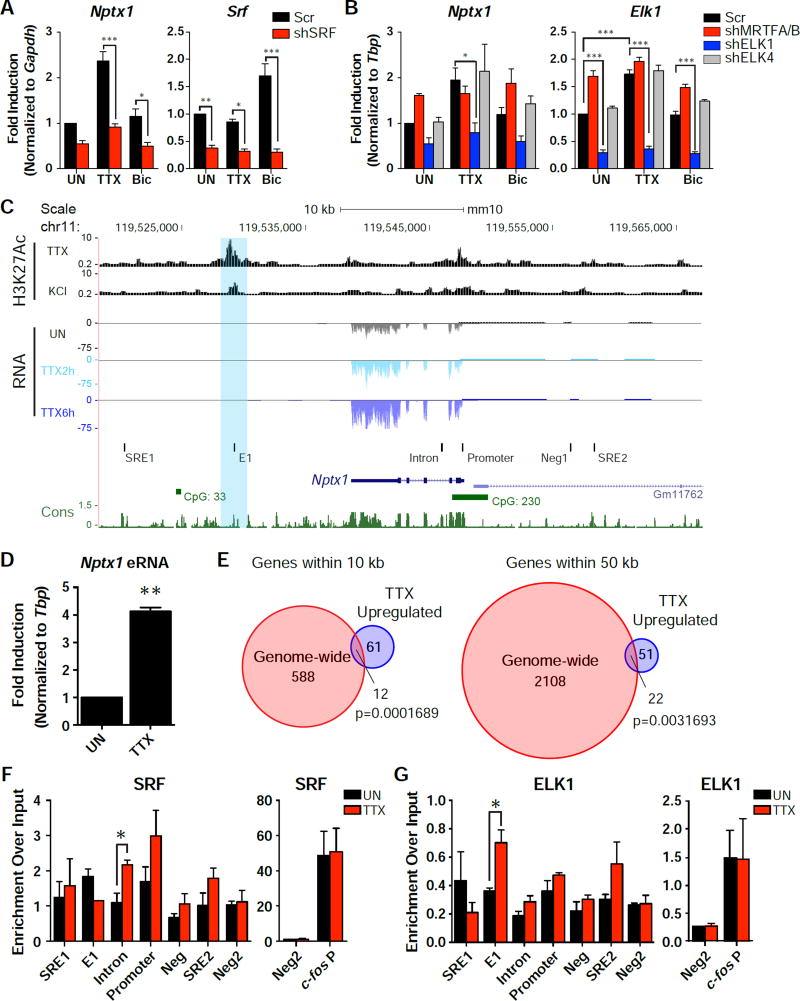
We next conducted a screen of several activity-regulated transcription factors (SRF, CREB, MEF2A, or MEF2D) in cortical neurons to see if any were necessary for Nptx1 induction. While knocking down CREB and MEF2A individually showed a slight decrease in the TTX-dependent induction of Nptx1, MEF2D knockdown had no effect (Figure S4A and S4B). Notably, knockdown of SRF significantly impaired the induction of several candidate genes, Nptx1, Rps6ka5 and Auts2 (Figure 4A and S5A). These activity-regulated transcription factors have been extensively characterized for their roles in transcription when neural activity is increased (Flavell and Greenberg, 2008). Their activities are tightly regulated by various calcium-dependent signaling pathways triggered by influx of extracellular calcium through the NMDAR and/or L-VGCC. However our finding suggests that at least some of these transcription factors might also promote transcription under the opposite condition when neural activity is chronically suppressed.
Figure 4. SRF and its cofactor ELK1 regulate the expression of Nptx1.
(A) qRT-PCR results from cells that were infected with the scrambled control or shSRF knockdown. Cells were left untreated (UN) or treated with TTX or Bic. n = 3 biological replicates. See also Figure S4. (B) qRT-PCR results from cells that were infected with an shRNA targeting the indicated co-factor family member (shMRTFA/B, shElk1, or shElk4) compared to a scrambled control. Cells were either left untreated or treated with TTX or Bic. n = 3 biological replicates. *, p < 0.05; **, p< 0.01; ***, p < 0.001. p values determined by a two-way ANOVA with Tukey’s multiple comparisons test. (C) A UCSC genome browser view of the Nptx1 locus aligned with H3K27Ac ChIP-seq from Malik et al., 2014 and the RNA-Seq data from TTX treated cells. The qPCR primer sets used in the ChIP experiments below are labeled with black bars. SRE1 and 4 are potential Serum Response Elements (SREs) based on SRF’s consensus sequence. E1 aligns with the H3K27Ac peak, with the blue bar highlighting the decreasing peak. Intron amplifies an intronic conserved region. Neg1 is a negative control region upstream of Nptx1, while Neg2 is a negative control on another chromosome. See also Figure S6. (D) qRT-PCR results of an eRNA expressed from the potential enhancer region of Nptx1 in response to 6 h TTX treatment. n = 3 biological replicates. (E) Venn diagrams demonstrating the overlap of genes throughout the genome that were within 10 kb (left) or 50 kb (right) of a H3K27Ac peak that decreased in response to KCl and the 73 genes found to be significantly upregulated in response to TTX. p values determined by the hypergeometric test. (F and G) ChIP results of SRF binding (F) or ELK1 binding (G) at the indicated loci around the Nptx1 gene. n = 4 biological replicates for the SRF ChIP, n = 3 biological replicates for the ELK ChIP. *, p < 0.05. p values determined by a two-way ANOVA with Tukey’s multiple comparisons test, comparing the primer set of interest with the negative region (Neg2). Error bars represent SEM.
SRF has been known to work with other co-factors to convert specific signal inputs into differential gene activation (Knoll and Nordheim, 2009). The two best characterized co-factor families are the ternary complex factors (TCFs) and members of the myocardin family (MRTF). Both MAPK and Ca2+ signaling have been shown to regulate the activity of TCFs through extensive phosphorylation. Actin signaling dynamically controls MRTF trafficking in which monomeric G-actins sequester them in the cytoplasm whereas filamentous F-actins trigger translocation of MRTFs into the nucleus to drive transcription with SRF. MRTFs are mainly composed of MRTF-A and MRTF-B, and Ternary complex factors (TCFs) are made up of ELK family members. Based on our RNA-Seq data, MRTF-A (Mkl-1), MRTF-B (Mkl-2), ELK1 (Elk1), and ELK4 (Elk4) are highly expressed in cortical neurons, while ELK-3 (Elk3) is expressed at an extremely low level (Figure S5B). In order to determine which co-factor(s) is required for SRF-mediated transcription of the Nptx1 gene, knockdown of both MRTF-A and MRTF-B with an shRNA targeting a common region between the two genes, and knockdown of ELK1 or ELK4 was performed in cortical cultures. Despite similar knockdown efficiency of each of the factors (Figure 4B and S5C), only knockdown of ELK1 impaired the TTX-mediated induction of Nptx1, suggesting that the activating signal to increase Nptx1 expression works through the SRF cofactor ELK1 (Figure 4B). Interestingly, Elk1 is the only co-factor gene that shows increased expression in response to TTX, which might be due to an increase in mRNA stability (Figure 1E, 2E, and 4B). ELK1 knockdown also blocked induction of Auts2 and Cacna1h (Figure S5D). Therefore, ELK1 also participates in the TTX-dependent transcriptional induction possibly in conjunction with SRF.
In order to determine if the Nptx1 gene is a direct target of SRF, we examined the promoter and enhancer regions of the Nptx1 gene for SRF binding. We used the histone H3 lysine 27 acetylation (H3K27Ac) profiles recently determined for neuron cultures treated with 55 mM KCl or TTX to identify potential enhancer regions near the Nptx1 gene, as H3K27Ac is a marker for active enhancers (Malik et al., 2014). Based on these profiles, we identified one potential enhancer downstream of Nptx1 (E1; Figure 4C). The H3K27Ac level was high at this enhancer under TTX-mediated activity suppression (overnight treatment of TTX) but then decreased when neurons were depolarized by KCl, implying that the enhancer became activated by activity blockade. Further supporting this idea, qRT-PCR primers directed against this region detected enhancer RNAs (eRNAs) that were induced in response to TTX (Figure 4D). As eRNAs have been shown to be a reliable marker of active enhancers within a given cell type (Kim and Shiekhattar, 2015), this also suggests that this region is an enhancer that is activated in response to activity suppression. To address whether this was a feature common to enhancers of TTX-induced genes, we compared the peaks that decreased in response to KCl genome-wide with our list of TTX-induced genes. Out of ~2000 H3K27Ac peaks that decreased in response to KCl, 600 genes were found within 10 kb of a peak, including 12 of our TTX upregulated genes (Figure 4E, left). Expanding the distance to within 50 kb of an H3K27Ac peak, there were 2130 genes identified, 22 of which overlapped with our 73 TTX-upregulated genes (Figure 4E, right), including Txnip, Nos1, and Syt13 (Figure S6A). The overlaps at both distances are significant (p=0.0001689 and p=0.0031693, respectively). This analysis suggests that there might be a genome-wide epigenetic mechanism to regulate the activity of a large number of enhancers, which in turn promotes transcriptional induction of specific gene sets in response to low activity levels.
We also looked for Serum Response Elements (SREs) containing the motif of CC(A/T)6GG in the genomic regions surrounding the Nptx1 locus, using the Regulatory Sequence Analysis Tool (RSAT) (Miano, 2003; Thomas-Chollier et al., 2011). There are two canonical SRE sites, the first is ~20 kb downstream of Nptx1 (SRE1), and the second ~10 kb upstream of Nptx1 within an H3K27Ac peak present in the forebrain early in development (SRE2) (Figure 4C and S6B) (Nord et al., 2013). When allowing for 1 mismatch in the motif, there is another potential SRE found within an intron (Intron; Figure 4C) of Nptx1 that is highly conserved evolutionarily. Our ChIP analysis revealed that the SRF binding level increases ~ 2 fold in response to TTX at the site within the intron of Nptx1 (Figure 4F). These results suggest that SRF is directly regulating the transcriptional induction of Nptx1 in response to activity suppression. We also observed TTX-induced ELK1 binding at the potential downstream enhancer (E1), suggesting that this cofactor is also directly affecting Nptx1 transcription (Figure 4G).
Calcium-mediated signaling through T-type VGCC is critical for TTX-induced transcription
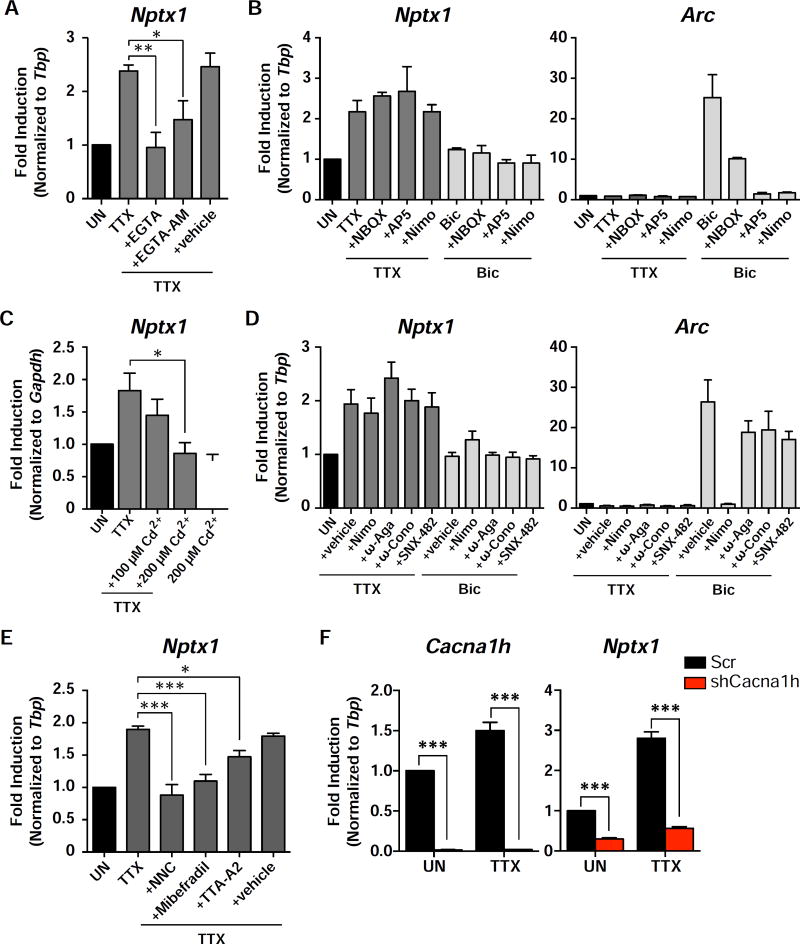
Previous studies suggested that TTX-induced synaptic up-scaling can be mimicked by a reduction in calcium (Ca2+) influx through the L-type channel or by reduced activity of CaMKK and CaMKIV (Ibata et al., 2008; Turrigiano et al., 1998). However, we found that influx of extracellular calcium is necessary to activate the TTX-specific nuclear signaling. Pre-treatment of neurons with the calcium chelator, EGTA, or a cell permeable form of the compound, EGTA-AM, that only becomes active once inside the cell, was able to block the induction of Nptx1 in response to TTX (Figure 5A).
Figure 5. The T-type VGCC mediates Nptx1 induction in response to TTX treatment.
(A) DIV14 cortical neurons were pretreated with EGTA or EGTA-AM before being treated with TTX for 4 h. Levels of Nptx1 were measured by qRT-PCR. The vehicle control for EGTA-AM is DMSO. (B) Expression levels of Nptx1 and Arc were measured with qRT-PCR from cortical neurons that were pretreated with NBQX, AP-V, or Nimodipine (Nimo) before treatment with TTX or Bic. (C) Cortical neurons were pretreated with 100 µM or 200 µM Cadmium (Cd2+) before treatment with TTX, or treated with 200 µM Cd2+ alone. n = 3 biological replicates. (D) Cortical neurons were pretreated with Nimodipine (Nimo), ω-Agatoxin IVA (ω-Aga), ω-Conotoxin GVIA (ω-Cono), or SNX-482 before treatment with TTX or Bic. Vehicle is DMSO. n = 3 biological replicates. (E) Blockers of the T-VGCC, NNC 55-0396 (NNC), Mibefradil, or TTA-A2, were used to pretreat cortical neurons for 15 min before treatment with TTX. n = 5 biological replicates. In all of the above bar graphs, black bars represent untreated cells. Dark grey bars represent cells treated with TTX and pretreated with the indicated compounds. Light grey bars represent cells treated with Bic and pretreated with the indicated compounds. See also Figure S7. (F) Cells were infected with a lentivirus containing either an shRNA targeting the CaV3.2 gene (shCacna1h) or a scrambled control. The levels of the indicated genes were measured by qRT-PCR. n = 3 biological replicates. *, p < 0.05; **, p< 0.01; ***, p < 0.001. p values determined by a one-way ANOVA using Dunnett’s multiple comparisons test, except for (F) where multiple t tests were performed and statistical significance was determined using the Holm-Sidak multiple comparison test.
To determine the source of calcium entry, we then tested inhibitors of calcium channels important for nuclear gene expression upon an increase in neural activity: NBQX, a selective inhibitor of AMPARs, AP-5, an inhibitor of NMDARs, and nimodipine, a blocker of the L-type VGCC. Interestingly, none of these compounds were able to block TTX-mediated Nptx1 induction, even though they were all, to varying degrees, able to block Arc induction by Bic (Figure 5B). Next, we pretreated neurons with cadmium (Cd2+), a general VGCC blocker, before addition of TTX, and observed a dose-dependent decrease in Nptx1 induction (Figure 5C). To elucidate which VGCC is regulating the induction of genes during TTX treatment, we tested inhibitors of all known VGCCs (L-, N-, P/Q-, R-, and T-type VGCCs), and found that only T-Type VGCC blockers, NNC 55–0396 dihydrochloride, Mibefradil, and TTA-A2 significantly inhibited the Nptx1 induction in response to TTX (Figure 5D and 5E). All three subtypes of the T-VGCC are expressed in our cortical neuronal culture: CaV3.1 (Cacna1g), CaV3.2 (Cacna1h), and CaV3.3 (Cacna1i). One in particular, Cacna1h is also induced in response to TTX, and has been implicated in ASD (Figures 1, 2, and S7A, and Table S2). Knockdown of Cacna1h or treatment with the T-type blocker TTA-A2 was able to block induction of several TTX-induced genes in addition to Nptx1 (Figure 5F, Figure S7B, and S7C)
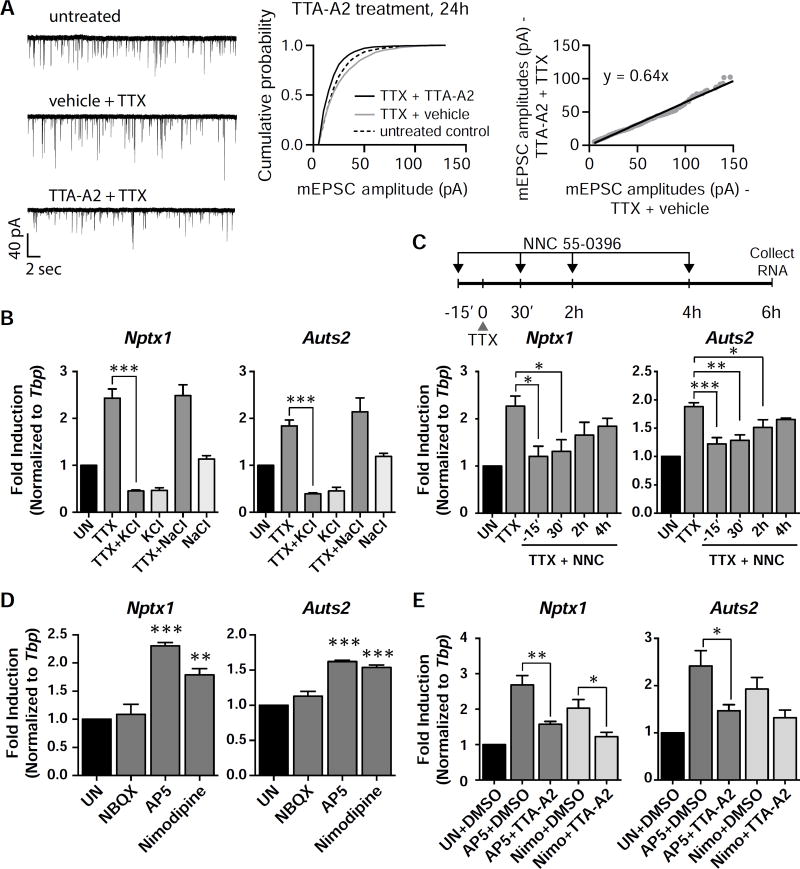
Furthermore, TTA-A2 blocked the TTX-induced increase in AMPA-mediated current, suggesting that the T-VGCC is also functionally required for homeostatic synaptic up-scaling (Figure 6A). Unlike other VGCCs (L-, R-, N-, P/Q-types), the T-VGCC is a low-voltage activated (LVA) channel that can operate even when Na+ channels are blocked by TTX (Iftinca and Zamponi, 2009). The T-VGCC only opens within a small voltage window near resting membrane potential (Iftinca and Zamponi, 2009; Tsien, 1998). If the T-VGCC activity is required for mediating gene induction in the TTX treated condition, mild depolarization of neurons to potentials past the channel’s window current should block the induction seen with TTX. To test this idea, the external K+ concentration was raised from 5 mM to 11 mM KCl during the TTX exposure, and we found that indeed, this was able to completely block the induction of Nptx1 and Auts2, while there was no effect with an increase of NaCl (Figure 6B). This result is consistent with the premise that T-VGCC channels are responsible for this induction. We next performed a time course experiment with the T-VGCC blocker NNC 55–0396 which revealed that the inhibitory effects of T-VGCC block decrease with greater delay in drug treatment (Figure 6C). This suggests that prolonged channel activity throughout the duration of activity suppression mediates the upregulation of TTX-induced genes. This is in contrast to the gene induction in response to an increase in activity, in which an immediate early phase of calcium signaling through the L-VGCC and/or NMDAR following the activity increase seems critical for ensuring late long-term potentiation (LTP) (Deisseroth et al., 1996; Saha et al., 2011; West et al., 2002).
Figure 6. The T-VGCC is necessary for scaling.
(A) Example traces, cumulative probability histograms, and rank order plots of cells that were pretreated with either TTA-A2 or vehicle, and then treated with TTX for 24 h. n = 9 cells for the untreated condition, n = 10 cells for both TTX treated conditions. TTA-A2/TTX vs vehicle/TTX (n = 10 cells each) p = 2.11×10−27, D = 0.185. Vehicle/TTX vs untreated (n = 9 cells) p = 4.79×10−6, D = 0.08. Statistical significance between cumulative probability graphs was determined by the Kolmogorov-Smirnov test. (B) Cells were pretreated with 6 mM KCl or 6 mM NaCl before the addition of TTX for 6 h. (C) The T-VGCC blocker was added either 15 min before the addition of TTX, or 30 min, 2 h, or 4 h after the addition of TTX. The total time of TTX treatment was 6 h. Nptx1 and Auts2 RNA levels were measured with qRT-PCR. (D) Neurons were treated with the indicated blockers for 6 h, and the RNA levels of the indicated genes were measured with qRT-PCR. (E) Cells were pretreated with the VGCC blocker TTA-A2, before the addition of the NMDAR or L-VGCC blocker. RNA levels of the indicated genes were measured with qRT-PCR. For bar graphs, *, p < 0.05; **, p < 0.01. p values determined by a one-way ANOVA using Dunnett’s multiple comparisons test. Error bars represent SEM.
To further characterize the calcium signaling pathway, we tested the necessity of components of the MAPK signaling pathways, as MAPK signaling is known to regulate ELK1/SRF transcription (Hill et al., 1993), and components of this pathway were identified in our GO analysis (Figure 1D). However, different inhibitors of this pathway did not show significant effects on Nptx1 induction (Figure S8A). A previous study showed that inhibition of CaMKIV, through blocking its upstream kinase CaMKK with STO-609, or a drop in somatic calcium influx through the L-VGCC, was sufficient to induce synaptic up-scaling in a manner similar to TTX (Ibata et al., 2008). However addition of STO-609 alone or together with TTX had no significant effect on the expression of Nptx1 and other TTX-induced genes (Figure S8B). On the other hand, the CaMKII inhibitor KN-62 alone was sufficient to cause Nptx1 induction to a level similar to TTX and also occluded the effect of TTX (Figure S8C). Three additional TTX-induced genes showed a similar increase in their mRNA levels by KN-62 alone, suggesting KN-62 alone is sufficient to mimic these TTX-induced transcriptional changes. Blocking the L-VGCC and NMDAR individually induces Nptx1 and Auts2 expression as well, similar to when activity is suppressed by TTX (Figure 6D). The gene expression driven by these treatments is also commonly inhibited by blocking the T-VGCC, which suggests a common mechanism (Figure 6E). Therefore, the TTX-induced transcription program we have defined is related to the signaling pathway previously characterized for synaptic up-scaling (Ibata et al., 2008; Turrigiano et al., 1998), and T-VGCC activity is required for this TTX-induced transcription program.
Gene induction occurs during activity suppression in vivo
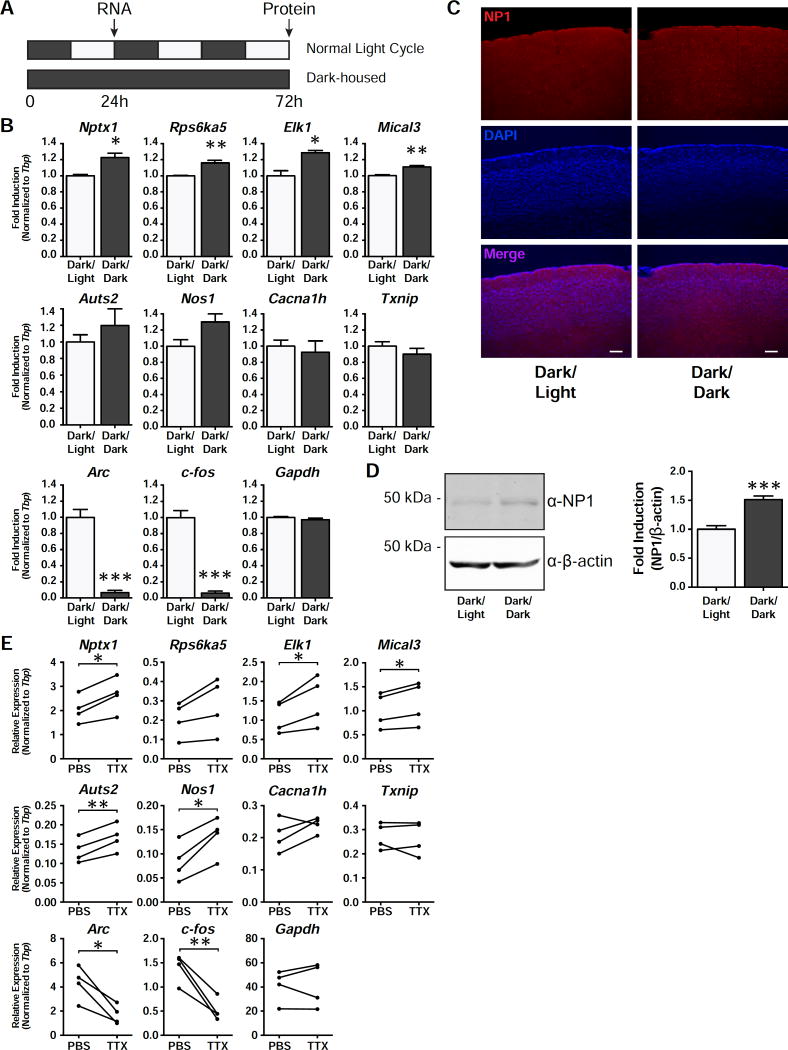
Homeostatic scaling has been shown to occur in layer 2/3 (L2/3) pyramidal neurons of the visual cortex upon visual deprivation (Goel and Lee, 2007; Lambo and Turrigiano, 2013; Maffei and Turrigiano, 2008). In order to see whether the synaptic up-scaling observed in the visual cortex is accompanied by a transcriptional response during dark rearing (DR), we housed mice in the dark for 24 h and then measured RNA levels in the visual cortex compared to mice that were housed in a normal 12 h light/dark cycle (Figure 7A). While the DR condition led to a significant decrease in the expression of activity-induced genes such as c-fos and Arc, a subset of TTX-induced genes, including Nptx1, were upregulated (Figure 7B). In addition to an increase in RNA levels, we also observed an increase in NP1 protein levels by both western blot and immunohistochemistry in the visual cortex following 3 d of DR (Figure 7C and 7D). Though dark rearing represents a physiological method to induce synaptic scaling, it does not suppress all spontaneous action potentials in the retina (Hengen et al., 2016). In order to better reproduce the in vitro findings, we performed intraocular injections of TTX, which silence all activity coming from the retina, and has previously been shown to trigger scaling up of AMPAR activity in the visual cortex (Desai et al., 2002; Frenkel and Bear, 2004) This alternative visual deprivation protocol was also able to show an increase in most of the tested TTX-induced genes in a more reliable manner than the dark-rearing paradigm (Figure 7E). Taken together, these results indicate the existence of an intrinsic transcription program that can induce expression of specific genes in response to activity suppression in vivo.
Figure 7. Nptx1 is induced in vivo.
(A) A schematic timeline of the dark rearing behavior experiment. Mice were either housed in normal 12 h dark-light cycle or placed in a completely dark chamber for 24 h. (B) qRT-PCR results of the indicated genes that are induced in the visual cortex in response to dark housing for 24 h. n = 3 biological replicates. (C) Staining of the visual cortex with an antibody against NP1, costained with the nuclear marker, DAPI. Images taken at 10X magnification. (D) Western blot results from the visual cortices of mice that were dark reared for 3 d compared to those reared in a normal light/dark cycle. Quantification of the western blot results of NP1 normalized to β-actin. n = 5 biological replicates. (E) qRT-PCR results of the visual cortices of mice that were injected intraocularly with either PBS or TTX. n = 4 biological replicates. *, p < 0.05; **, p < 0.01; ***, p < 0.001. p values were determined with an unpaired Student’s t-test for dark rearing experiments. p values for the intraocular injections were determined using a paired t-test. Error bars represent SEM.
Discussion
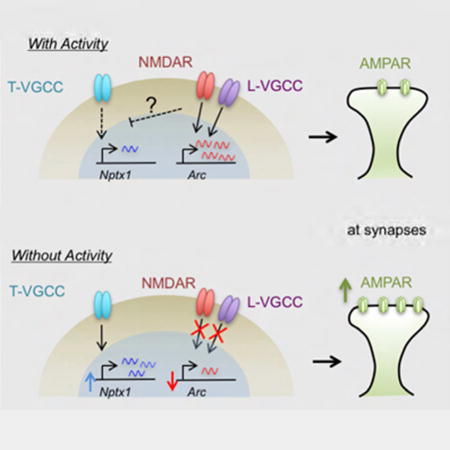
In this study, we have identified a transcriptional program that is activated in response to neuronal activity-suppression and drives a homeostatic increase in synaptic strength. This program requires T-VGCC-dependent Ca2+ signaling and the function of activity-regulated transcription factors like SRF and ELK1. Transcriptional induction during neuronal inactivity appears to be an important process for homeostatic plasticity as one of the genes induced by activity blockade, Nptx1 is necessary and sufficient to increase AMPAR amplitude.
Homeostatic scaling is crucial for proper neuronal function and maintaining balance between excitation and inhibition. This is highlighted by the fact that several models of psychiatric disorders show deficits in scaling (Wondolowski and Dickman, 2013). Importantly, multiple genes in our list of TTX-induced genes have also been linked to neurological disorders such as ASD and schizophrenia (Table S2), which further implies a functional relevance of this program. Auts2 has gained attention recently as an autism susceptibility gene (Gao et al., 2014), as well as Cacna1h, which encodes a subtype of T-VGCC that we have shown to be the critical VGCC mediating this response. Other genes in our list have also been implicated previously in homeostatic scaling. Rps6ka5 encodes a kinase (MSK1) that is necessary for TTX-induced scaling, and Nos1, a gene linked to schizophrenia, was recently identified in a study looking at transcriptional changes after long-term (48 h) treatment of TTX, although no functional studies for this gene were performed (Correa et al., 2012; Lee and Chung, 2014).
Recent studies suggest that activity-dependent changes in DNA methylation levels mediated by enzymes such as DNA methyltransferase (DNMT) or the Ten-eleven translocation (Tet) family protein, Tet3 are a critical mechanism of synaptic scaling (Meadows et al., 2015; Nelson et al., 2008; Yu et al., 2015). However, how these enzymes selectively alter the expression of genes related to synaptic scaling have not been demonstrated. When neural activity is suppressed, many of the activity-induced genes show a slightly dampened expression compared to the basal culture condition, which could also be related to the neuron’s compensatory response to maintain a stable firing rate. For example, chronic activity blockade of hippocampal neurons resulted in an overall reduction of potassium current with a concomitant decrease in expression levels of voltage-gated KV1 and KV7 potassium channels, which was explained as a mechanism for the homeostatic increase in intrinsic excitability (Lee et al., 2015).
The transcription program characterized in our study appears to be related to the previously defined signaling pathway as we observed that pharmacological inhibition of the L-VGCC or NMDAR alone could mimic the gene induction mediated by TTX (Figure 6D). A time course experiment of the T-VGCC inhibitor, NNC 55–0396 further elucidates that the continuous activity of T-VGCC for a prolonged period of time might underlie gene induction (Figure 6C). These results could further imply that the tonic activity or signaling of the T-VGCC at rest might be suppressed or overridden by other calcium channel activities such as the L-VGCC and NMDAR. The T-VGCC signaling to the nucleus might then become effective when the other calcium channel activities are directly inhibited by specific blockers or when neuronal firing is silenced by TTX.
Knockdown of Nptx1 was able to block the TTX-induced scaling up of synaptic strength, which suggests a functional relevance of gene induction. A previous study proposed that Nptx1 might be a proapoptotic gene based on the findings that it is overexpressed in cerebellar granule cells (CGNs) cultured with a low potassium concentration that causes cell death and that reducing Nptx1 mRNA significantly attenuated cell death (DeGregorio-Rocasolano et al., 2001). A subsequent study in rat cortical neurons described that knockdown of Nptx1 increases the number of excitatory synapses and neuronal excitability, suggesting a function in the negative regulation of excitatory synaptic plasticity (Figueiro-Silva et al., 2015). These functions of Nptx1 are ostensibly incompatible with the previously defined role of Nptx1 in AMPAR clustering and potentiating postsynaptic response, as well as our finding in homeostatic synapse up-scaling. We speculate that the previous studies and ours examine different stages of physiological consequences that might result from alterations in Nptx1 levels. Our study investigates the early stage (12 – 24 h) of homeostatic synapse plasticity initiated by activity blockade at which point we only observed increased AMPAR amplitude and ~ 2 – 2.5 fold increases in Nptx1 mRNA and protein levels peaking at 6 h and 24 h, respectively. During this period, Nptx1 knockdown blocked the TTX-induced scaling up of synaptic strength. On the other hand, a prolonged period of activity suppression (e.g., TTX treatment for 2 d or even longer) or Nptx1 knockdown might force neurons to trigger additional, more drastic homeostatic responses such as changes in intrinsic excitability, presynaptic release probability or even synapse numbers (Bacci et al., 2001; Burrone et al., 2002; Murthy et al., 2001), in which Nptx1 might also be functionally involved. Chronic activity blockade by TTX has been widely used and does not cause any significant cell death for at least a few days. The previously shown proapoptotic function of Nptx1 was studied in CGNs under low potassium concentration which caused 40% cell death within 24 hours and also induced a much more dramatic increase of Nptx1 mRNA and protein. We think that a mild increase in Nptx1 levels during the early period of activity blockade is a physiologically important process for a neuron’s homeostatic synaptic plasticity.
Nptx1 induction was mediated through the activity of the transcription factors SRF and ELK1, which have a well-known role in transcription in the opposite scenario when activity is increased. Although to a lesser degree, other activity-regulated transcription factors such as CREB and MEF2A appear to participate in the transcription program in response to activity blockade as well (Figure S4). These findings provide important insight into the understanding of the activity-regulated transcription program in neurons, showing that at least some transcription factors can engage in transcriptional induction in two opposing conditions – when neural activity is high or low. Despite the bi-directional function of transcription factors, our RNA-seq data shows that each activity condition induces a distinct set of genes, suggesting that those bi-directional transcription factors might coordinate with condition-specific factors to confer the target gene specificity. Therefore, it would be interesting to determine a more comprehensive list of transcription factors for their unique or specialized role in the transcription program at rest.
Our findings suggest a key function of the T- VGCC in homeostatic synaptic plasticity through the regulation of nuclear transcription. T-VGCCs are abundant in hippocampal neuronal dendrites along with other key VGCCs (Kavalali et al., 1997; Magee and Johnston, 1995). However, they are also unique among the VGCCs as these channels are low-voltage activated (LVA) channels, while the other types (L-, R-, N-, P/Q-types) are high-voltage activated (HVA) (Randall and Tsien, 1997). The T-VGCC can operate even when Na+ channels are blocked by TTX (Iftinca and Zamponi, 2009). Compared to HVAs, T-VGCCs open in response to smaller depolarizations, have smaller conductances, and exhibit rapid gating kinetics. After opening, these channels rapidly inactivate such that they only operate over a small voltage window near resting membrane potential. Recovery from inactivation is facilitated by a brief hyperpolarization, at which point they may exhibit bursting activity. These properties make T-type calcium channels ideal candidates to produce calcium signals near resting potential (Iftinca and Zamponi, 2009; Tsien, 1998). Our findings further suggest that these properties would also make the T-VGCC ideal for transmitting the signal to the nucleus when neural activity is suppressed. The identification of this channel as a regulator of gene expression opens up a new avenue of research in understanding the role of the T-VGCC in neuronal function.
Experimental Procedures
Cell culture
All experiments carried out with the use of animals were reviewed and approved by the IACUC committee at University of Texas Southwestern Medical Center. Primary cortical cultures were made from embryonic day 16–18 (E16/18) mice, and they were grown in Neurobasal media supplemented with B-27 and Glutamax. AraC was added at DIV6 to prevent glial proliferation. Cells were used in experiments on days in vitro (DIV) 12–15, as indicated. Hippocampal cultures were made from postnatal day 1–3 (P1–3) rat pups, and cultured as described previously (Kavalali et al., 1999).
Whole cell voltage clamp recordings
At DIV14-18, dissociated hippocampal cultures were voltage-clamped at −70 mV, as previously described (Reese and Kavalali, 2015), using an Axon Instruments Axopatch 200B amplifier. For detailed procedures, see the supplemental experimental methods.
RNA-seq
Total RNA was extracted using Trizol reagent (Life Technologies) and the library was prepared using the TruSeq RNA Library Preparation Kit (Illumina) according to the manufacturer’s instructions. The analysis procedure is described in the supplemental experimental methods.
GRO-seq
GRO-seq was carried out as previously described (Core et al., 2008; Hah et al., 2011) with modifications (Danko et al., 2013; Lam et al., 2013), using 10 million nuclei per sample. Detailed procedures are described in the supplemental experimental methods.
qRT-PCR
Total RNA was extracted from neurons using Trizol reagent (Life Technologies) according to the manufacturer’s protocol. cDNA made with the high-capacity reverse transcription kit (Life Technologies), and amplification was performed with SybrGreen (Life Technologies) using the primers indicated in the supplemental experimental methods.
Visual Cortex experiments
For all visual cortex experiments, male C57Bl6/J mice were used at P21–25. For detailed procedures, see the supplemental experimental methods.
Supplementary Material
Acknowledgments
We thank L. Farbiak for assistance with cell culture, and C. Green and J. Stubblefield for providing the dark controlled-environment chambers. This work was supported by the US National Science Foundation (NSF) BRAIN EAGER Award (IOS1451034), the US National Institute of Neurological Disorders and Stroke (NINDS) under award number R01NS085418 (T.-K.K.), and a National Institute of Mental Health award MH066198 (E.T.K). K.S. was supported by a National Institute of Mental Health Institutional Training Grant, T32-MH76690, and A.L.R. was supported by the Cellular Biophysics of the Neuron Training Program T32 NS069562.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
T.-K.K., K.S., and E.T.K. designed the project. K.S. performed RNA-seq, transcription factor knockdown, ChIP, and RNA analysis of pharmacological experiments and T-type channel knockdown. A.L.R. performed the whole cell patch clamping. S.-K.K. performed GRO-seq and nascent RNA analysis. G.K. performed the analysis for the sequencing data. J.-Y.J. provided the transcription factor knockdown constructs and helped with the visual cortex experiments. K.S. and T.-K.K. wrote the manuscript with input from E.T.K. and A.L.R. All authors discussed the results and commented on the manuscript.
Accession codes
The accession number for the data sets reported in this paper is GEO: GSE90988 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE90988).
References
- Abad MA, Enguita M, DeGregorio-Rocasolano N, Ferrer I, Trullas R. Neuronal pentraxin 1 contributes to the neuronal damage evoked by amyloid-beta and is overexpressed in dystrophic neurites in Alzheimer’s brain. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2006;26:12735–12747. doi: 10.1523/JNEUROSCI.0575-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacci A, Coco S, Pravettoni E, Schenk U, Armano S, Frassoni C, Verderio C, De Camilli P, Matteoli M. Chronic blockade of glutamate receptors enhances presynaptic release and downregulates the interaction between synaptophysin-synaptobrevin-vesicle-associated membrane protein 2. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2001;21:6588–6596. doi: 10.1523/JNEUROSCI.21-17-06588.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrone J, O’Byrne M, Murthy VN. Multiple forms of synaptic plasticity triggered by selective suppression of activity in individual neurons. Nature. 2002;420:414–418. doi: 10.1038/nature01242. [DOI] [PubMed] [Google Scholar]
- Chae M, Danko CG, Kraus WL. groHMM: a computational tool for identifying unannotated and cell type-specific transcription units from global run-on sequencing data. BMC Bioinformatics. 2015;16:222. doi: 10.1186/s12859-015-0656-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa SA, Hunter CJ, Palygin O, Wauters SC, Martin KJ, McKenzie C, McKelvey K, Morris RG, Pankratov Y, Arthur JS, et al. MSK1 regulates homeostatic and experience-dependent synaptic plasticity. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32:13039–13051. doi: 10.1523/JNEUROSCI.0930-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danko CG, Hah N, Luo X, Martins AL, Core L, Lis JT, Siepel A, Kraus WL. Signaling pathways differentially affect RNA polymerase II initiation, pausing, and elongation rate in cells. Mol Cell. 2013;50:212–222. doi: 10.1016/j.molcel.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGregorio-Rocasolano N, Gasull T, Trullas R. Overexpression of neuronal pentraxin 1 is involved in neuronal death evoked by low K(+) in cerebellar granule cells. The Journal of biological chemistry. 2001;276:796–803. doi: 10.1074/jbc.M007967200. [DOI] [PubMed] [Google Scholar]
- Deisseroth K, Bito H, Tsien RW. Signaling from synapse to nucleus: postsynaptic CREB phosphorylation during multiple forms of hippocampal synaptic plasticity. Neuron. 1996;16:89–101. doi: 10.1016/s0896-6273(00)80026-4. [DOI] [PubMed] [Google Scholar]
- Desai NS, Cudmore RH, Nelson SB, Turrigiano GG. Critical periods for experience-dependent synaptic scaling in visual cortex. Nature neuroscience. 2002;5:783–789. doi: 10.1038/nn878. [DOI] [PubMed] [Google Scholar]
- Fernandes D, Carvalho AL. Mechanisms of homeostatic plasticity in the excitatory synapse. Journal of neurochemistry. 2016 doi: 10.1111/jnc.13687. [DOI] [PubMed] [Google Scholar]
- Fernandez-Enright F, Andrews JL, Newell KA, Pantelis C, Huang XF. Novel implications of Lingo-1 and its signaling partners in schizophrenia. Transl Psychiatry. 2014;4:e348. doi: 10.1038/tp.2013.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiro-Silva J, Gruart A, Clayton KB, Podlesniy P, Abad MA, Gasull X, Delgado-Garcia JM, Trullas R. Neuronal pentraxin 1 negatively regulates excitatory synapse density and synaptic plasticity. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2015;35:5504–5521. doi: 10.1523/JNEUROSCI.2548-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Greenberg ME. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci. 2008;31:563–590. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel MY, Bear MF. How monocular deprivation shifts ocular dominance in visual cortex of young mice. Neuron. 2004;44:917–923. doi: 10.1016/j.neuron.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Gao Z, Lee P, Stafford JM, von Schimmelmann M, Schaefer A, Reinberg D. An AUTS2-Polycomb complex activates gene expression in the CNS. Nature. 2014;516:349–354. doi: 10.1038/nature13921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Dennis MY, Baker C, Malig M, Coe BP, Campbell CD, Mark K, Vu TH, Alkan C, Cheng Z, et al. Refinement and discovery of new hotspots of copy-number variation associated with autism spectrum disorder. Am J Hum Genet. 2013;92:221–237. doi: 10.1016/j.ajhg.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel A, Lee HK. Persistence of experience-induced homeostatic synaptic plasticity through adulthood in superficial layers of mouse visual cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27:6692–6700. doi: 10.1523/JNEUROSCI.5038-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hah N, Danko CG, Core L, Waterfall JJ, Siepel A, Lis JT, Kraus WL. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145:622–634. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengen KB, Torrado Pacheco A, McGregor JN, Van Hooser SD, Turrigiano GG. Neuronal Firing Rate Homeostasis Is Inhibited by Sleep and Promoted by Wake. Cell. 2016;165:180–191. doi: 10.1016/j.cell.2016.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CS, Marais R, John S, Wynne J, Dalton S, Treisman R. Functional analysis of a growth factor-responsive transcription factor complex. Cell. 1993;73:395–406. doi: 10.1016/0092-8674(93)90238-l. [DOI] [PubMed] [Google Scholar]
- Howerton CL, Morgan CP, Fischer DB, Bale TL. O-GlcNAc transferase (OGT) as a placental biomarker of maternal stress and reprogramming of CNS gene transcription in development. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:5169–5174. doi: 10.1073/pnas.1300065110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsen T, de Vlieg J, Alkema W. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibata K, Sun Q, Turrigiano GG. Rapid synaptic scaling induced by changes in postsynaptic firing. Neuron. 2008;57:819–826. doi: 10.1016/j.neuron.2008.02.031. [DOI] [PubMed] [Google Scholar]
- Iftinca MC, Zamponi GW. Regulation of neuronal T-type calcium channels. Trends Pharmacol Sci. 2009;30:32–40. doi: 10.1016/j.tips.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Ionita-Laza I, Xu B, Makarov V, Buxbaum JD, Roos JL, Gogos JA, Karayiorgou M. Scan statistic-based analysis of exome sequencing data identifies FAN1 at 15q13.3 as a susceptibility gene for schizophrenia and autism. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:343–348. doi: 10.1073/pnas.1309475110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo JY, Schaukowitch K, Farbiak L, Kilaru G, Kim TK. Stimulus-specific combinatorial functionality of neuronal c-fos enhancers. Nature neuroscience. 2015 doi: 10.1038/nn.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalscheuer VM, FitzPatrick D, Tommerup N, Bugge M, Niebuhr E, Neumann LM, Tzschach A, Shoichet SA, Menzel C, Erdogan F, et al. Mutations in autism susceptibility candidate 2 (AUTS2) in patients with mental retardation. Hum Genet. 2007;121:501–509. doi: 10.1007/s00439-006-0284-0. [DOI] [PubMed] [Google Scholar]
- Kavalali ET, Klingauf J, Tsien RW. Activity-dependent regulation of synaptic clustering in a hippocampal culture system. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:12893–12900. doi: 10.1073/pnas.96.22.12893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavalali ET, Zhuo M, Bito H, Tsien RW. Dendritic Ca2+ channels characterized by recordings from isolated hippocampal dendritic segments. Neuron. 1997;18:651–663. doi: 10.1016/s0896-6273(00)80305-0. [DOI] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Shiekhattar R. Architectural and Functional Commonalities between Enhancers and Promoters. Cell. 2015;162:948–959. doi: 10.1016/j.cell.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll B, Nordheim A. Functional versatility of transcription factors in the nervous system: the SRF paradigm. Trends Neurosci. 2009;32:432–442. doi: 10.1016/j.tins.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Lam MT, Cho H, Lesch HP, Gosselin D, Heinz S, Tanaka-Oishi Y, Benner C, Kaikkonen MU, Kim AS, Kosaka M, et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498:511–515. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambo ME, Turrigiano GG. Synaptic and intrinsic homeostatic mechanisms cooperate to increase L2/3 pyramidal neuron excitability during a late phase of critical period plasticity. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:8810–8819. doi: 10.1523/JNEUROSCI.4502-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KY, Chung HJ. NMDA receptors and L-type voltage-gated Ca(2)(+) channels mediate the expression of bidirectional homeostatic intrinsic plasticity in cultured hippocampal neurons. Neuroscience. 2014;277:610–623. doi: 10.1016/j.neuroscience.2014.07.038. [DOI] [PubMed] [Google Scholar]
- Lee KY, Royston SE, Vest MO, Ley DJ, Lee S, Bolton EC, Chung HJ. N-methyl-D-aspartate receptors mediate activity-dependent down-regulation of potassium channel genes during the expression of homeostatic intrinsic plasticity. Mol Brain. 2015;8:4. doi: 10.1186/s13041-015-0094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Vasishtha M, Prywes R. Activation and repression of cellular immediate early genes by serum response factor cofactors. The Journal of biological chemistry. 2010;285:22036–22049. doi: 10.1074/jbc.M110.108878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louros SR, Hooks BM, Litvina L, Carvalho AL, Chen C. A role for stargazin in experience-dependent plasticity. Cell Rep. 2014;7:1614–1625. doi: 10.1016/j.celrep.2014.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons MR, West AE. Mechanisms of specificity in neuronal activity-regulated gene transcription. Prog Neurobiol. 2011;94:259–295. doi: 10.1016/j.pneurobio.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffei A, Turrigiano GG. Multiple modes of network homeostasis in visual cortical layer 2/3. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28:4377–4384. doi: 10.1523/JNEUROSCI.5298-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee JC, Johnston D. Characterization of single voltage-gated Na+ and Ca2+ channels in apical dendrites of rat CA1 pyramidal neurons. J Physiol. 1995;487(Pt 1):67–90. doi: 10.1113/jphysiol.1995.sp020862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik AN, Vierbuchen T, Hemberg M, Rubin AA, Ling E, Couch CH, Stroud H, Spiegel I, Farh KK, Harmin DA, et al. Genome-wide identification and characterization of functional neuronal activity-dependent enhancers. Nature neuroscience. 2014;17:1330–1339. doi: 10.1038/nn.3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows JP, Guzman-Karlsson MC, Phillips S, Holleman C, Posey JL, Day JJ, Hablitz JJ, Sweatt JD. DNA methylation regulates neuronal glutamatergic synaptic scaling. Sci Signal. 2015;8:ra61. doi: 10.1126/scisignal.aab0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miano JM. Serum response factor: toggling between disparate programs of gene expression. J Mol Cell Cardiol. 2003;35:577–593. doi: 10.1016/s0022-2828(03)00110-x. [DOI] [PubMed] [Google Scholar]
- Murthy VN, Schikorski T, Stevens CF, Zhu Y. Inactivity produces increases in neurotransmitter release and synapse size. Neuron. 2001;32:673–682. doi: 10.1016/s0896-6273(01)00500-1. [DOI] [PubMed] [Google Scholar]
- Nelson ED, Kavalali ET, Monteggia LM. Activity-dependent suppression of miniature neurotransmission through the regulation of DNA methylation. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28:395–406. doi: 10.1523/JNEUROSCI.3796-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LS, Kim HG, Rosenfeld JA, Shen Y, Gusella JF, Lacassie Y, Layman LC, Shaffer LG, Gecz J. Contribution of copy number variants involving nonsense-mediated mRNA decay pathway genes to neuro-developmental disorders. Hum Mol Genet. 2013;22:1816–1825. doi: 10.1093/hmg/ddt035. [DOI] [PubMed] [Google Scholar]
- Nord AS, Blow MJ, Attanasio C, Akiyama JA, Holt A, Hosseini R, Phouanenavong S, Plajzer-Frick I, Shoukry M, Afzal V, et al. Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell. 2013;155:1521–1531. doi: 10.1016/j.cell.2013.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkumar AP, Christensen JH, Mattheisen M, Jacobsen I, Bache I, Pallesen J, Grove J, Qvist P, McQuillin A, Gurling HM, et al. Analysis of t(9;17)(q33.2;q25.3) chromosomal breakpoint regions and genetic association reveals novel candidate genes for bipolar disorder. Bipolar Disord. 2015;17:205–211. doi: 10.1111/bdi.12239. [DOI] [PubMed] [Google Scholar]
- Randall AD, Tsien RW. Contrasting biophysical and pharmacological properties of T-type and R-type calcium channels. Neuropharmacology. 1997;36:879–893. doi: 10.1016/s0028-3908(97)00086-5. [DOI] [PubMed] [Google Scholar]
- Reese AL, Kavalali ET. Spontaneous neurotransmission signals through store-driven Ca(2+) transients to maintain synaptic homeostasis. Elife. 2015:4. doi: 10.7554/eLife.09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Gomez JA, Levitsky KL, Lopez-Barneo J. T-type Ca2+ channels in mouse embryonic stem cells: modulation during cell cycle and contribution to self-renewal. Am J Physiol Cell Physiol. 2012;302:C494–504. doi: 10.1152/ajpcell.00267.2011. [DOI] [PubMed] [Google Scholar]
- Saha RN, Wissink EM, Bailey ER, Zhao M, Fargo DC, Hwang JY, Daigle KR, Fenn JD, Adelman K, Dudek SM. Rapid activity-induced transcription of Arc and other IEGs relies on poised RNA polymerase II. Nature neuroscience. 2011;14:848–856. doi: 10.1038/nn.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders AR, Goring HH, Duan J, Drigalenko EI, Moy W, Freda J, He D, Shi J, Mgs Gejman PV. Transcriptome study of differential expression in schizophrenia. Hum Mol Genet. 2013;22:5001–5014. doi: 10.1093/hmg/ddt350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaukowitch K, Joo JY, Liu X, Watts JK, Martinez C, Kim TK. Enhancer RNA facilitates NELF release from immediate early genes. Mol Cell. 2014;56:29–42. doi: 10.1016/j.molcel.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkai T, Ohmori O, Hori H, Nakamura J. Allelic association of the neuronal nitric oxide synthase (NOS1) gene with schizophrenia. Mol Psychiatry. 2002;7:560–563. doi: 10.1038/sj.mp.4001041. [DOI] [PubMed] [Google Scholar]
- Sia GM, Beique JC, Rumbaugh G, Cho R, Worley PF, Huganir RL. Interaction of the N-terminal domain of the AMPA receptor GluR4 subunit with the neuronal pentraxin NP1 mediates GluR4 synaptic recruitment. Neuron. 2007;55:87–102. doi: 10.1016/j.neuron.2007.06.020. [DOI] [PubMed] [Google Scholar]
- Splawski I, Yoo DS, Stotz SC, Cherry A, Clapham DE, Keating MT. CACNA1H mutations in autism spectrum disorders. The Journal of biological chemistry. 2006;281:22085–22091. doi: 10.1074/jbc.M603316200. [DOI] [PubMed] [Google Scholar]
- Sultana R, Yu CE, Yu J, Munson J, Chen D, Hua W, Estes A, Cortes F, de la Barra F, Yu D, et al. Identification of a novel gene on chromosome 7q11.2 interrupted by a translocation breakpoint in a pair of autistic twins. Genomics. 2002;80:129–134. doi: 10.1006/geno.2002.6810. [DOI] [PubMed] [Google Scholar]
- Thiagarajan TC, Lindskog M, Tsien RW. Adaptation to synaptic inactivity in hippocampal neurons. Neuron. 2005;47:725–737. doi: 10.1016/j.neuron.2005.06.037. [DOI] [PubMed] [Google Scholar]
- Thomas-Chollier M, Defrance M, Medina-Rivera A, Sand O, Herrmann C, Thieffry D, van Helden J. RSAT 2011: regulatory sequence analysis tools. Nucleic Acids Res. 2011;39:W86–91. doi: 10.1093/nar/gkr377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien RW. Molecular physiology. Key clockwork component cloned. Nature. 1998;391:839, 841. doi: 10.1038/35981. [DOI] [PubMed] [Google Scholar]
- Turrigiano G. Too many cooks? Intrinsic and synaptic homeostatic mechanisms in cortical circuit refinement. Annu Rev Neurosci. 2011;34:89–103. doi: 10.1146/annurev-neuro-060909-153238. [DOI] [PubMed] [Google Scholar]
- Turrigiano GG, Leslie KR, Desai NS, Rutherford LC, Nelson SB. Activity-dependent scaling of quantal amplitude in neocortical neurons. Nature. 1998;391:892–896. doi: 10.1038/36103. [DOI] [PubMed] [Google Scholar]
- West AE, Griffith EC, Greenberg ME. Regulation of transcription factors by neuronal activity. Nat Rev Neurosci. 2002;3:921–931. doi: 10.1038/nrn987. [DOI] [PubMed] [Google Scholar]
- Wondolowski J, Dickman D. Emerging links between homeostatic synaptic plasticity and neurological disease. Frontiers in cellular neuroscience. 2013;7:223. doi: 10.3389/fncel.2013.00223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Hopf C, Reddy R, Cho RW, Guo L, Lanahan A, Petralia RS, Wenthold RJ, O’Brien RJ, Worley P. Narp and NP1 form heterocomplexes that function in developmental and activity-dependent synaptic plasticity. Neuron. 2003;39:513–528. doi: 10.1016/s0896-6273(03)00463-x. [DOI] [PubMed] [Google Scholar]
- Yu H, Su Y, Shin J, Zhong C, Guo JU, Weng YL, Gao F, Geschwind DH, Coppola G, Ming GL, et al. Tet3 regulates synaptic transmission and homeostatic plasticity via DNA oxidation and repair. Nature neuroscience. 2015;18:836–843. doi: 10.1038/nn.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.