Abstract
Introduction
Disc herniation is a complex spinal disorder associated with disability and high healthcare cost. Lumbar disc herniation is strongly associated with disc degeneration. Candidate genes of the aggrecan metabolic pathway may associate with the severity of lumbar disc herniation.
Objectives
This study evaluated the association of single nucleotide variants (SNVs) of the candidate genes of the aggrecan metabolic pathway with the severity of lumbar disc herniation in patients with chronic mechanical low back pain. In addition, we assessed the in-silico functional analysis of the significant SNVs and association of their haplotypes with the severity of lumbar disc herniation.
Methods
A descriptive cross sectional study was carried out on 106 patients. Severity of disc herniation and disc degeneration were assessed on T2-weighted mid sagittal lumbar MRI scan. Sixty two exonic SNVs of ten candidate genes of aggrecan metabolic pathway (ACAN, IL1A, IL1B, IL6, MMP3, ADAMTS4, ADAMTS5, TIMP1, TIMP2 and TIMP3) were genotyped on a Sequenom MassARRAY iPLEX platform. Multivariable linear regression analysis was carried out using PLINK 1.9 software adjusting for age, gender, body mass index and severity of disc degeneration. Four online bioinformatics tools (Provean, SIFT, PolyPhen and Mutation Taster) were used for in-silico functional analysis.
Results
Mean age was 52.42 ± 9.42 years and 69.8% were females. The mean severity of disc herniation was 2.81 ± 1.98. The rs2272023, rs35430524, rs2882676, rs2351491, rs938609, rs3825994, rs1042630, rs698621 and rs3817428 variants and their haplotypes of ACAN were associated with the severity of lumbar disc herniation. However, only the rs35430524, rs938609 and rs3817428 variants of ACAN were detected as pathogenic by in-silico functional analysis.
Conclusions
SNVs of ACAN and their haplotypes are associated with the severity of lumbar disc herniation. Functional genetic studies are necessary to identify the role of these significant SNVs in the pathogenesis of disc herniation.
Introduction
Lumbar disc herniation is a complex multifactorial spinal condition associated with disability, work time loss and high health related costs [1]. Lumbar disc degeneration is strongly associated with disc herniation [2]. In traditional view, lumbar disc degeneration and herniation are mainly related to age, gender, body mass index (BMI), smoking, physical activity and heavy loading of the spine. Significant associations were identified in our previous study between single nucleotide variants (SNVs) of candidate genes of the aggrecan metabolic pathway and lumbar disc degeneration [3]. Therefore, it is worthwhile to explore the association of SNVs of respective candidate genes (ACAN, IL1A, IL1B, IL6, MMP3, ADAMTS4, ADAMTS5, TIMP1, TIMP2 and TIMP3) with the severity of disc herniation.
Aggrecan is the main proteoglycan of the intervertebral disc and provides osmotic properties which assist the disc in resisting mechanical compressive loads transmitted along the spine [4]. With aging, the disc undergoes dehydration due to loss of proteoglycan with aging and the degenerative process is further augmented by repetitive damages. This transforms the disc into a degenerative state and probably disc herniation [5]. Interleukins (ILs), matrix metalloproteinases (MMPs), a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTSs) and tissue inhibitor of metalloproteinases (TIMPs) are important molecules in the aggrecan metabolic pathway [6].
Abnormal genetic variants of the candidate genes in the aggrecan metabolic pathway may alter the structural and functional properties of the aggrecan molecule. Although there are several studies which have identified associations of a Variable Number of Tandem Repeats (VNTR) polymorphism of ACAN with lumbar disc degeneration and symptomatic lumbar disc herniation [7], there is a lack of studies which have explored the association of SNVs of ACAN with the severity of disc herniation. The “T” allele of the rs1042631 variant of ACAN is associated with signal intensity of the disc and increased the frequency of disc herniation among the Finnish population [8]. Although the MMP1 and MMP3 are associated with disc herniation [9, 10], relationships of SNVs of the ADAMTS4, ADAMTS5, TIMP1, TIMP2 and TIMP3 with the severity of disc herniation need to be explored.
Early detection of genetically predisposed individuals for disc herniation might provide opportunity to take necessary precautions in lifestyle related risk factors. Identification of significant SNVs/haplotypes provides probable therapeutic targets for gene therapy in the future. Considering the findings of the previous genetic study on lumbar disc degeneration [3] and current evidence available in the literature, we extended our research to investigate the associations of candidate genes of the aggrecan metabolic pathway with the severity of lumbar disc herniation. The main objective of this study was to determine the associations of single nucleotide variants of the candidate genes of the aggrecan metabolic pathway with the severity of disc herniation in patients with chronic mechanical low back pain. In addition, we assessed in-silico functional analysis of significant SNVs of the candidate genes and associations of their haplotypes with the severity of lumbar disc herniation.
Methods
Study design, setting and participants
A descriptive cross-sectional study was carried out on patients with chronic mechanical low back pain who attended the rheumatology clinic, National Hospital of Sri Lanka, Colombo from May 2012 to October 2014. 368 consecutive patients with chronic mechanical low back pain who fulfilled the eligibility criteria were assessed with lateral x-rays of lumbar spine after obtaining written informed consent. Male and female patients (20 to 69 years) having pain/muscle tension or stiffness localised below the costal margin and inferior gluteal folds [11], during day time and worsening in the latter part of the day due to movements [12] for at least three months duration [13] were included in the study. Patients with back pain due to inflammatory causes, visceral origin, systemic infections affecting spine, metabolic bone diseases, fractures in the vertebral column, past surgeries in the spine, and spinal tumours were excluded. 120 patients were selected to undergo MRI scan of lumbar spine and genotyping based on the severity of disc related degenerative changes (disc space narrowing and anterior osteophyte) in the x-ray of lumbar spine [3, 14]. The study was carried out in accordance with the Declaration of Helsinki and with the approval of the Ethics Review Committee of the Faculty of Medicine, University of Colombo.
Clinical evaluation
An interviewer administered questionnaire was used to record age, gender and clinical examination was used to record body mass index (BMI). Height (cm) and weight (kg) of the patients were assessed with light clothing and without shoes to the nearest 0.1 cm and 0.1 kg, respectively, and BMI was calculated (kg/m2) [15]. International cut off values were used for categorisation of BMI (normal, overweight and obese) [16].
X-ray assessment
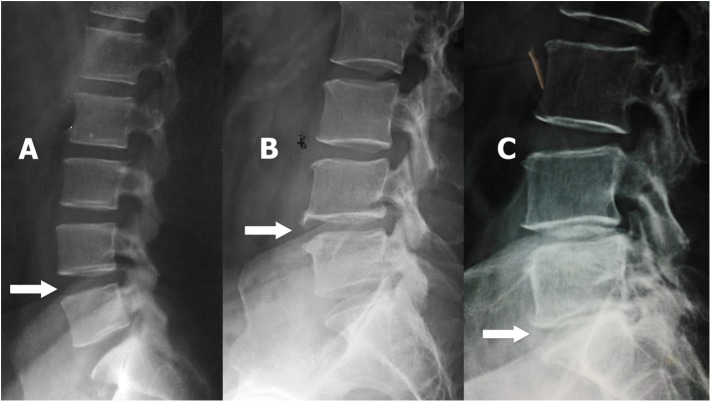
Lateral lumbar x-rays were carried out while patients were in lateral recumbent position on the table flexing the knees and hips just enough to achieve comfortable position [17]. The intervertebral disc spaces (L1/L2 to L5/S1) in lateral lumbar x-rays were assessed for disc space narrowing and anterior osteophytes by a consultant radiologist blinded to clinical details of the patients and overall lumbar disc degeneration was calculated (grade 0–2) according to a scoring system defined by Lane et al. 1993 (Fig 1) [14].
Fig 1. Assessment of the x-ray features of lumbar disc degeneration—lateral x-ray of lumbar spine.
Arrows—A–no disc space narrowing/anterior osteophyte (grade 0 lumbar disc degeneration), B–mild disc space narrowing and small anterior osteophyte (grade 1 lumbar disc degeneration), C–small anterior osteophyte and moderate disc space narrowing (grade 2 lumbar disc degeneration) [3].
MRI assessment
Patients with grade 2 or more lumbar disc degeneration in the lateral lumbar x-rays were selected to undergo MRI scan of the lumbar spine. In addition age and gender matched samples from patients with grade 0 and grade 1 lumbar disc degeneration were selected to undergo MRI scans of the lumbar spine. T2 weighted sagittal MRI scans of lumbar spine were carried out in supine position using a GE 1.5T MRI Scanner (Signa, General Electric, Milwaukee, Wisconsin) and assessed for the severity of lumbar disc degeneration and disc herniation.
Severity of lumbar disc degeneration
The severity of lumbar disc degeneration was graded on T2-weighted mid sagittal MRI images using modified Pfirrmann grading system and each lumbar level was graded from grade 1 to 5 (Table 1) [18, 19]. Grade 2 and 3 was defined as mild lumbar disc degeneration, while grades 4 and 5 were defined as severe lumbar disc degeneration.The grades of lumbar disc degeneration of the five lumbar levels were summed to calculate the severity of lumbar disc degeneration.
Table 1. Scoring system to assess the severity of lumbar disc degeneration in T2 weighted midsagittal MRI of lumbar spine.
| Grade | Structure | Distinction of nucleus pulposus and annulus fibrosus | Signal Intensity | Height of the disc |
|---|---|---|---|---|
| 1 | Homogeneous shape, no horizontal bands | Clear | Hyperintense, isointense | Normal |
| 2 | Nonhomogeneous shape with horizontal bands | Some blurring | Hyperintense, isointense | Normal |
| 3 | Nonhomogeneous shape | Blurring, but annulus shape still recognizable | Intermediate | Normal to slightly decreased |
| 4 | Nonhomogeneous shape | Annulus shape not intact and distinction impossible | Hypointense | Usually decreased |
| 5 | Nonhomogeneous shape | Annulus shape not intact and distinction impossible | Hypointense | Collapse disc space |
Severity of lumbar disc herniation
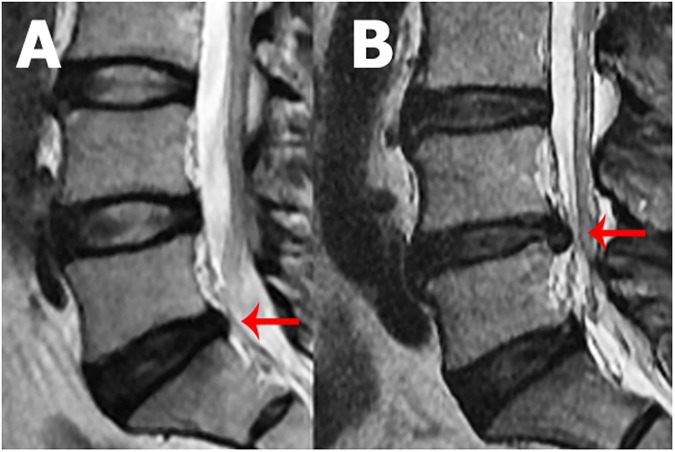
Disc herniation was considered present when the disc material is displaced beyond the disc space. It was further categorised as disc protrusion when the distance between the edges of the herniated disc material extending outside the disc space is less than the distance between the edges of the base of that disc material. Disc extrusion was considered present when the distance between the edges of the herniated disc material extending outside the disc space was more than the distance between the edges of the base of that disc material (Fig 2) [20]. Presence of disc protrusion in a disc was given a score of 1 and presence of disc extrusion in a disc was given a score of 2. The scores of each lumbar level were summed to calculate the severity of disc herniation.
Fig 2.
Sagittal MRI of the lumbar spine showing disc protrusion at L5/S1 (A) and disc extrusion at L4/L5 (B).
SNV selection, DNA extraction and genotyping
The selected molecules of the aggrecan metabolic pathway are aggrecan, interleukin 1α, interleukin 1β, interleukin 6, matrix metalloproteinases 3, a disintegrin and metalloproteinase with thrombospondin motifs 4 and 5, tissue inhibitor of metalloproteinase 1, 2 and 3. Their functions and respective gene symbols are summarised in the Table 1 in the previous article [3]. Common SNVs in the exonic regions of these ten candidate genes in the aggrecan metabolic pathway (ACAN, IL1A, IL1B, IL6, MMP3, ADAMTS4, ADAMTS5, TIMP1, TIMP2 and TIMP3) were identified from 25 Sri Lankan exomes and Gujarati Indians in Houston (GIH) in the Hapmap database [3]. Sixty two SNVs in exonic regions of the candidate genes in aggrecan metabolic pathway were selected for genotyping (Table 2). DNA was extracted from 200 ml of venous blood using QIAamp DNA Mini Kit in accordance to the blood and body fluid protocol (spin protocol) [21] http://dx.doi.org/10.17504/protocols.io.ihfcb3n. Extracted DNA was quantified using the Quantus fluorometer (Promega) with QuantiFluor® Double stranded DNA system [22] http://dx.doi.org/10.17504/protocols.io.ihgcb3w. Selected variants were genotyped using Sequenom iPLEX MassARRAY system (Sequenom, San Diego, CA) [23] at the Australian Genome Research Facility, Australia. Although there is a VNTR polymorphism of ACAN which is associated with the severity of disc herniation, this study focused only on exonic SNVs.
Table 2. Selected single nucleotide variants of the candidate genes, their allele frequencies and Hardy–Weinberg equilibrium.
| No. | CHR | Position GRCh37/hg19 | Gene | Region | SNV | Minor allele | Major allele | MAF % | HWE p value |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 161160872 | ADAMTS4 | 3’UTR | rs34884997 | C | T | 0.12 | 0.68 |
| 2 | 1 | 161161284 | ADAMTS4 | nonsynonymous | rs41270041 | C | G | 0.18 | 0.52 |
| 3 | 1 | 161163037 | ADAMTS4 | nonsynonymous | rs4233367 | T | C | 0.18 | 0.76 |
| 4 | 1 | 161168004 | ADAMTS4 | synonymous | rs33941127 | T | C | 0.40 | 0.85 |
| 5 | 1 | 161168189 | ADAMTS4 | nonsynonymous | rs34448954 | T | C | 0.13 | 0.42 |
| 6 | 2 | 113532083 | IL1A | 3’UTR | rs2856836 | G | A | 0.27 | 0.06 |
| 7 | 2 | 113532236 | IL1A | 3’UTR | rs1304037 | C | T | 0.28 | 0.06 |
| 8 | 2 | 113537223 | IL1A | nonsynonymous | rs17561 | A | C | 0.27 | 0.06 |
| 9 | 2 | 113542960 | IL1A | 5’UTR | rs1800587 | A | G | 0.27 | 0.06 |
| 10 | 2 | 113587121 | IL1B | downstream | rs2853550 | A | G | 0.29 | 0.65 |
| 11 | 2 | 113590390 | IL1B | synonymous | rs1143634 | A | G | 0.13 | 0.69 |
| 12 | 7 | 22766246 | IL6 | 5upstream | rs1800796 | C | G | 0.46 | 0.59 |
| 13 | 7 | 22766645 | IL6 | 5upstream | rs1800795 | C | G | 0.13 | 0.10 |
| 14 | 7 | 22771156 | IL6 | synonymous | rs2069849 | T | C | 0.05 | 1.00 |
| 15 | 11 | 102709425 | MMP3 | synonymous | rs520540 | A | G | 0.33 | 0.10 |
| 16 | 11 | 102713465 | MMP3 | synonymous | rs602128 | A | G | 0.33 | 0.10 |
| 17 | 11 | 102713620 | MMP3 | nonsynonymous | rs679620 | T | C | 0.33 | 0.21 |
| 18 | 15 | 89382027 | ACAN | synonymous | rs372041880 | C | A | 0.07 | 0.10 |
| 19 | 15 | 89382129 | ACAN | nonsynonymous | rs16942318 | A | C | 0.05 | 0.05 |
| 20 | 15 | 89386652 | ACAN | nonsynonymous | rs34949187 | A | G | 0.11 | 1.00 |
| 21 | 15 | 89388894 | ACAN | nonsynonymous | rs148070768 | G | A | 0.14 | 0.26 |
| 22 | 15 | 89388905 | ACAN | synonymous | rs16942341 | T | C | 0.07 | 1.00 |
| 23 | 15 | 89391160 | ACAN | synonymous | rs2272023 | C | A | 0.34 | 0.42 |
| 24 | 15 | 89392689 | ACAN | nonsynonymous | rs144501729 | A | C | 0.14 | 0.26 |
| 25 | 15 | 89398105 | ACAN | synonymous | rs2351491 | T | C | 0.37 | 0.84 |
| 26 | 15 | 89398553 | ACAN | nonsynonymous | rs35430524 | A | C | 0.05 | 0.05 |
| 27 | 15 | 89398553 | ACAN | nonsynonymous | rs3743399 | G | A | 0.28 | 0.65 |
| 28 | 15 | 89398631 | ACAN | nonsynonymous | rs938609 | A | T | 0.37 | 0.70 |
| 29 | 15 | 89400339 | ACAN | nonsynonymous | rs2882676 | A | C | 0.47 | 0.46 |
| 30 | 15 | 89400680 | ACAN | nonsynonymous | rs28407189 | G | A | 0.07 | 1.00 |
| 31 | 15 | 89400963 | ACAN | nonsynonymous | rs79925540 | T | G | 0.16 | 0.48 |
| 32 | 15 | 89401109 | ACAN | nonsynonymous | rs4932439 | A | G | 0.28 | 0.65 |
| 33 | 15 | 89401615 | ACAN | synonymous | rs3825994 | G | T | 0.44 | 0.85 |
| 34 | 15 | 89401616 | ACAN | nonsynonymous | rs76282091 | C | G | 0.16 | 0.48 |
| 35 | 15 | 89402051 | ACAN | nonsynonymous | rs1042630 | A | G | 0.41 | 0.45 |
| 36 | 15 | 89402239 | ACAN | synonymous | rs1042631 | T | C | 0.35 | 1.00 |
| 37 | 15 | 89402596 | ACAN | synonymous | rs698621 | G | T | 0.38 | 0.85 |
| 38 | 15 | 89415247 | ACAN | nonsynonymous | rs3817428 | G | C | 0.05 | 1.00 |
| 39 | 15 | 89417238 | ACAN | nonsynonymous | rs1126823 | G | A | 0.36 | 0.84 |
| 40 | 17 | 76867017 | TIMP2 | synonymous | rs2277698 | T | C | 0.24 | 0.80 |
| 41 | 21 | 28291455 | ADAMTS5 | 3’UTR | rs1444269 | G | A | 0.26 | 1.00 |
| 42 | 21 | 28291846 | ADAMTS5 | 3’UTR | rs2298657 | C | T | 0.05 | 1.00 |
| 43 | 21 | 28292581 | ADAMTS5 | 3’UTR | rs3746836 | A | G | 0.25 | 0.81 |
| 44 | 21 | 28293095 | ADAMTS5 | 3’UTR | rs229072 | T | A | 0.48 | 0.27 |
| 45 | 21 | 28293117 | ADAMTS5 | 3’UTR | rs229073 | G | A | 0.48 | 0.27 |
| 46 | 21 | 28293924 | ADAMTS5 | 3’UTR | rs11700721 | T | C | 0.12 | 1.00 |
| 47 | 21 | 28294090 | ADAMTS5 | 3’UTR | rs16979423 | G | T | 0.14 | 0.70 |
| 48 | 21 | 28294143 | ADAMTS5 | 3’UTR | rs9978597 | G | T | 0.04 | 1.00 |
| 49 | 21 | 28296135 | ADAMTS5 | 3’UTR | rs229078 | T | G | 0.22 | 1.00 |
| 50 | 21 | 28296324 | ADAMTS5 | 3’UTR | rs151065 | A | G | 0.19 | 0.56 |
| 51 | 21 | 28296389 | ADAMTS5 | synonymous | rs3746839 | G | A | 0.08 | 1.00 |
| 52 | 21 | 28302355 | ADAMTS5 | nonsynonymous | rs226794 | A | G | 0.10 | 0.60 |
| 53 | 21 | 28338298 | ADAMTS5 | nonsynonymous | rs457947 | G | C | 0.07 | 0.46 |
| 54 | 21 | 28338423 | ADAMTS5 | synonymous | rs55933916 | G | C | 0.08 | 0.50 |
| 55 | 22 | 33253280 | TIMP3 | synonymous | rs9862 | T | C | 0.50 | 0.46 |
| 56 | 22 | 33253292 | TIMP3 | synonymous | rs11547635 | T | C | 0.07 | 0.42 |
| 57 | 22 | 33257322 | TIMP3 | 3’UTR | rs1427384 | C | T | 0.19 | 0.53 |
| 58 | 22 | 33258050 | TIMP3 | 3’UTR | rs2267184 | T | C | 0.16 | 0.48 |
| 59 | 22 | 33258288 | TIMP3 | 3’UTR | rs1065314 | C | T | 0.17 | 0.32 |
| 60 | 23 | 47444879 | TIMP1 | nonsynonymous | rs5953060 | C | G | 0.43 | 0.66 |
| 61 | 23 | 47444985 | TIMP1 | synonymous | rs4898 | C | T | 0.43 | 0.66 |
| 62 | 23 | 47445286 | TIMP1 | nonsynonymous | rs6609533 | G | A | 0.43 | 0.66 |
CHR–chromosome number, SNV—single nucleotide variant, MAF–minor allele frequency, HWE—Hardy Weinberg equilibrium, UTR–untranslated region
Statistical analysis
Descriptive statistics were used to summarise the sample characteristics. Chi-square test was used to assess the association between two categorical variables while Pearson’s correlation test was used to assess association between two continuous variables. SNVs with Hardy–Weinberg equilibrium (HWE) ≥ 0.05 were included in the genetic association analysis. Quantitative trait association of SNVs was carried out with the multivariable linear regression analysis using PLINK 1.9 software [24]. Severity of disc herniation was used as the quantitative outcome. The genotype of the respective SNV was used as the main independent variable and was treated as a quantitative variable and coded 0, 1 or 2 to represent the number of variant allele, consistent with an additive genetic model. Haplotype frequencies of significant SNVs were performed using PLINK 1.07 software [24], which uses the expectation maximization algorithm to determine common haplotypes. Multivariable linear regression model was used to assess the quantitative trait association of haplotypes of significant SNVs of the candidate genes with the severity of disc herniation. Age, gender, BMI and highest grade of lumbar disc degeneration out of five lumbar levels were used as the additional covariates. Permutation function in PLINK was used with 10,000 permutations to generate the significance levels empirically. It helps to relax the assumptions of normality and correct the errors due to small sample size. P value < 0.05 was used as the level of significance. Four free online bioinformatics prediction tools (Provean [25], SIFT [26], PolyPhen [27] and Mutation taster [28]) were used to conduct the in-silico functional analysis.
Results
Although 120 patients were selected to undergo MRI scans of lumbar spine, only 106 patients attended for MRI scans of lumbar spine. Characteristics of the patients who underwent both the MRI scan of lumbar spine and genotyping (106 patients) are summarised in Table 3. The proportion of patients increased up to the 50–59 years age group and declined after age of 60 years. The majority of patients were females. Most of the patients were overweight and obese. Mean severity of disc herniation was 2.81 ± 1.98. The majority of patients had disc herniations with 45 (42.4%) having disc extrusions. Among a total of 530 lumbar levels evaluated, 72 (13.6%) and 69 (13.0%) of the discs had severe lumbar disc degeneration and disc extrusions, respectively. Severe lumbar disc degeneration and disc extrusions were more frequent in lower lumbar levels (65.3% and 66.7%, respectively). Severity of disc herniation was positively correlated with the severity of disc degeneration (correlation coefficient = 0.49, p = <0.001). Furthermore, percentage of disc extrusions were higher in discs with severe disc degeneration (33.3%) compared to the discs with mild disc degeneration (9.8%) (χ2 (1) = 30.36, p < 0.001). Overall genotyping efficiency of these patients was 98.68%. Minor allele frequency ranged from 0.05 to 0.50 and all the variants were in HWE (Table 2).
Table 3. Summary of the sample characteristics of 106 patients who underwent both MRI scan of lumbar spine and genetic association analysis.
| Variable | N (%) | |
|---|---|---|
| Total patients | 106 | |
| Socio demographics | ||
| Age | Mean | 52.42 ± 9.42 |
| 20–29 years | 3 (2.8) | |
| 30–39 years | 7 (6.6) | |
| 40–49 years | 22 (20.8) | |
| 50–59 years | 51 (48.1) | |
| 60–69 years | 23 (21.7) | |
| Gender | Female | 74 (69.8) |
| Male | 32 (30.2) | |
| Body mass index | Normal (18–24.9 kg/m2) | 37 (34.9) |
| Overweight (25–29.9 kg/m2) | 42 (39.6) | |
| Obese (≥ 30 kg/m2) | 27 (25.5) | |
| MRI assessment | ||
| Lumbar disc degeneration | Grade 1 | 0 (0.0) |
| (maximum grade) | Grade 2 | 7 (6.6) |
| Grade 3 | 56 (52.8) | |
| Grade 4 | 36 (34.0) | |
| Grade 5 | 7 (6.6) | |
| Lumbar disc herniation | Yes | 97 (91.5) |
| No | 9 (8.5) | |
| Lumbar disc extrusion | Yes | 45 (42.4) |
| No | 61 (57.6) | |
Associations of SNVs of ACAN with severity of lumbar disc herniation
Nine SNVs of ACAN were significantly associated with the severity of disc herniation (Table 4). Presence of each additional “C” allele of the rs2272023 variant, “A” allele of the rs35430524 variant, “A” allele of the rs2882676 variant and “A” allele of the rs1042630 variant of ACAN were associated with a progressive increase in the severity of disc herniation. Furthermore, the presence of each additional “T” allele of the rs2351491 variant, “A” allele of the rs938609 variant, “G” allele of the rs3825994 variant and “G” allele of the rs698621 variant of ACAN were associated with progressive reduction in the severity of disc herniation. SNVs, rs2351491, rs938609 and rs698621 variants were in linkage disequilibrium (R2 ≥ 0.93, D’ ≥ 0.98). The rs3817428 variant of ACAN did not have homozygous genotype for the minor allele and the presence of “G” allele was associated with reduction in the severity of disc herniation. However, there was no significant association between the SNVs of the other candidate genes and the severity of disc herniation (Table 4).
Table 4. Severity of disc herniation tabulated according to genotype and the results of multiple linear regression–additive genetic model.
| No. | Gene | SNV | Variable | A1 | MAF | A2/A2 | A1/A2 | A1/A1 | Standardised coefficient (β) |
Adjusted p value |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ADAMTS4 | rs34884997 | C | 0.12 | ||||||
| Genotypes | T/T | C/T | C/C | |||||||
| N | 80 | 23 | 2 | |||||||
| DH mean (SD) | 2.69 (2.01) | 3.09 (1.88) | 3.00 (1.41) | 0.08 | 0.397 | |||||
| 2 | ADAMTS4 | rs41270041 | C | 0.18 | ||||||
| Genotypes | G/G | C/G | C/C | |||||||
| N | 69 | 35 | 1 | |||||||
| DH mean (SD) | 2.78 (1.92) | 2.83 (2.09) | 1.00 (0.00) | -0.06 | 0.502 | |||||
| 3 | ADAMTS4 | rs4233367 | T | 0.18 | ||||||
| Genotypes | C/C | T/C | T/T | |||||||
| N | 72 | 29 | 4 | |||||||
| DH mean (SD) | 2.81 (1.95) | 2.79 (1.99) | 2.25 (2.63) | 0.03 | 0.764 | |||||
| 4 | ADAMTS4 | rs33941127 | T | 0.40 | ||||||
| Genotypes | C/C | T/C | T/T | |||||||
| N | 39 | 48 | 18 | |||||||
| DH mean (SD) | 3.03 (2.18) | 2.6 (1.85) | 2.72 (1.81) | -0.08 | 0.377 | |||||
| 5 | ADAMTS4 | rs34448954 | T | 0.13 | ||||||
| Genotypes | C/C | T/C | T/T | |||||||
| N | 81 | 20 | 3 | |||||||
| DH mean (SD) | 2.72 (1.87) | 3.2 (2.46) | 2.00 (1.00) | 0.03 | 0.753 | |||||
| 6 | IL1A | rs2856836 | G | 0.27 | ||||||
| Genotypes | A/A | G/A | G/G | |||||||
| N | 58 | 38 | 9 | |||||||
| DH mean (SD) | 2.95 (2.06) | 2.66 (1.95) | 2.22 (1.30) | -0.08 | 0.379 | |||||
| 7 | IL1A | rs1304037 | C | 0.28 | ||||||
| Genotypes | T/T | C/T | C/C | |||||||
| N | 58 | 37 | 10 | |||||||
| DH mean (SD) | 2.95 (2.06) | 2.73 (1.92) | 2.00 (1.41) | -0.10 | 0.290 | |||||
| 8 | IL1A | rs17561 | A | 0.27 | ||||||
| Genotypes | C/C | A/C | A/A | |||||||
| N | 58 | 38 | 9 | |||||||
| DH mean (SD) | 2.95 (2.06) | 2.66 (1.95) | 2.22 (1.30) | -0.08 | 0.379 | |||||
| 9 | IL1A | rs1800587 | A | 0.27 | ||||||
| Genotypes | G/G | A/G | A/A | |||||||
| N | 59 | 38 | 9 | |||||||
| DH mean (SD) | 3.00 (2.08) | 2.71 (1.90) | 2.00 (1.50) | -0.09 | 0.301 | |||||
| 10 | IL1B | rs2853550 | A | 0.29 | ||||||
| Genotypes | G/G | A/G | A/A | |||||||
| N | 57 | 38 | 10 | |||||||
| DH mean (SD) | 2.58 (1.66) | 3.08 (2.29) | 2.80 (2.30) | 0.07 | 0.475 | |||||
| 11 | IL1B | rs1143634 | A | 0.13 | ||||||
| Genotypes | G/G | A/G | A/A | |||||||
| N | 78 | 27 | 1 | |||||||
| DH mean (SD) | 2.95 (2.08) | 2.48 (1.67) | 1.00 (0.00) | -0.07 | 0.457 | |||||
| 12 | IL6 | rs1800796 | C | 0.46 | ||||||
| Genotypes | G/G | C/G | C/C | |||||||
| N | 30 | 56 | 20 | |||||||
| DH mean (SD) | 2.60 (2.11) | 2.80 (1.85) | 3.15 (2.18) | 0.03 | 0.718 | |||||
| 13 | IL6 | rs1800795 | C | 0.13 | ||||||
| Genotypes | G/G | C/G | C/C | |||||||
| N | 76 | 20 | 4 | |||||||
| DH mean (SD) | 2.68 (1.91) | 3.55 (2.21) | 1.00 (0.82) | -0.04 | 0.708 | |||||
| 14 | IL6 | rs2069849 | T | 0.05 | ||||||
| Genotypes | C/C | T/C | T/T | |||||||
| N | 94 | 11 | 0 | |||||||
| DH mean (SD) | 2.74 (2.02) | 3.09 (1.51) | NA | 0.06 | 0.500 | |||||
| 15 | MMP3 | rs520540 | A | 0.33 | ||||||
| Genotypes | G/G | A/G | A/A | |||||||
| N | 51 | 38 | 16 | |||||||
| DH mean (SD) | 2.88 (2.02) | 2.74 (1.96) | 2.56 (1.93) | -0.05 | 0.618 | |||||
| 16 | MMP3 | rs602128 | A | 0.33 | ||||||
| Genotypes | G/G | A/G | A/A | |||||||
| N | 51 | 38 | 16 | |||||||
| DH mean (SD) | 2.88 (2.02) | 2.74 (1.96) | 2.56 (1.93) | -0.05 | 0.618 | |||||
| 17 | MMP3 | rs679620 | T | 0.33 | ||||||
| Genotypes | C/C | T/C | T/T | |||||||
| N | 51 | 39 | 15 | |||||||
| DH mean (SD) | 2.88 (2.02) | 2.72 (1.93) | 2.60 (1.99) | -0.05 | 0.565 | |||||
| 18 | ACAN | rs372041880 | C | 0.07 | ||||||
| Genotypes | A/A | C/A | C/C | |||||||
| N | 92 | 11 | 2 | |||||||
| DH mean (SD) | 2.73 (1.99) | 3.36 (1.86) | 2 (1.41) | 0.06 | 0.498 | |||||
| 19 | ACAN | rs16942318 | A | 0.05 | ||||||
| Genotypes | C/C | A/C | A/A | |||||||
| N | 95 | 7 | 2 | |||||||
| DH mean (SD) | 2.85 (2.03) | 2.57 (0.79) | 1.50 (0.71) | -0.03 | 0.771 | |||||
| 20 | ACAN | rs34949187 | A | 0.11 | ||||||
| Genotypes | G/G | A/G | A/A | |||||||
| N | 84 | 20 | 1 | |||||||
| DH mean (SD) | 2.87 (2.04) | 2.45 (1.70) | 2.00 (0.00) | -0.13 | 0.173 | |||||
| 21 | ACAN | rs148070768 | G | 0.14 | ||||||
| Genotypes | A/A | G/A | G/G | |||||||
| N | 75 | 25 | 4 | |||||||
| DH mean (SD) | 2.84 (2.01) | 2.56 (1.45) | 3.75 (3.59) | 0.06 | 0.553 | |||||
| 22 | ACAN | rs16942341 | T | 0.07 | ||||||
| Genotypes | C/C | T/C | T/T | |||||||
| N | 90 | 15 | 0 | |||||||
| DH mean (SD) | 2.91 (2.02) | 2.00 (1.46) | NA | -0.16 | 0.084 | |||||
| 23 | ACAN | rs2272023 | C | 0.34 | ||||||
| Genotypes | A/A | C/A | C/C | |||||||
| N | 49 | 41 | 15 | |||||||
| DH mean (SD) | 2.67 (1.93) | 2.42 (1.86) | 4.13 (1.92) | 0.22 | 0.019* | |||||
| 24 | ACAN | rs144501729 | A | 0.14 | ||||||
| Genotypes | C/C | A/C | A/A | |||||||
| N | 75 | 25 | 4 | |||||||
| DH mean (SD) | 2.81 (2.04) | 2.56 (1.45) | 3.75 (3.59) | 0.06 | 0.546 | |||||
| 25 | ACAN | rs2351491 | T | 0.37 | ||||||
| Genotypes | C/C | T/C | T/T | |||||||
| N | 44 | 46 | 15 | |||||||
| DH mean (SD) | 3.39 (2.26) | 2.48 (1.67) | 1.93 (1.34) | -0.35 | <0.001* | |||||
| 26 | ACAN | rs35430524 | A | 0.05 | ||||||
| Genotypes | C/C | A/C | A/A | |||||||
| N | 96 | 7 | 2 | |||||||
| DH mean (SD) | 2.73 (1.92) | 2.71 (1.98) | 5.50 (3.54) | 0.19 | 0.040* | |||||
| 27 | ACAN | rs3743399 | G | 0.28 | ||||||
| Genotypes | A/A | G/A | G/G | |||||||
| N | 57 | 38 | 10 | |||||||
| DH mean (SD) | 2.84 (2.01) | 2.37 (1.84) | 4.00 (1.83) | 0.13 | 0.169 | |||||
| 28 | ACAN | rs938609 | A | 0.37 | ||||||
| Genotypes | T/T | A/T | A/A | |||||||
| N | 44 | 45 | 15 | |||||||
| DH mean (SD) | 3.37 (2.26) | 2.53 (1.65) | 1.93 (1.34) | -0.34 | <0.001* | |||||
| 29 | ACAN | rs2882676 | A | 0.47 | ||||||
| Genotypes | C/C | A/C | A/A | |||||||
| N | 32 | 47 | 26 | |||||||
| DH mean (SD) | 2.41 (1.78) | 2.51 (1.69) | 3.73 (2.38) | 0.30 | 0.002* | |||||
| 30 | ACAN | rs28407189 | G | 0.07 | ||||||
| Genotypes | A/A | G/A | G/G | |||||||
| N | 90 | 15 | 0 | |||||||
| DH mean (SD) | 2.91 (2.02) | 2.00 (1.46) | NA | -0.16 | 0.084 | |||||
| 31 | ACAN | rs79925540 | T | 0.16 | ||||||
| Genotypes | G/G | T/G | T/T | |||||||
| N | 74 | 27 | 4 | |||||||
| DH mean (SD) | 2.80 (2.05) | 2.59 (1.42) | 3.75 (3.59) | 0.05 | 0.633 | |||||
| 32 | ACAN | rs4932439 | A | 0.28 | ||||||
| Genotypes | G/G | A/G | A/A | |||||||
| N | 57 | 38 | 10 | |||||||
| DH mean (SD) | 2.84 (2.01) | 2.37 (1.84) | 4.00 (1.83) | 0.13 | 0.169 | |||||
| 33 | ACAN | rs3825994 | G | 0.44 | ||||||
| Genotypes | T/T | G/T | G/G | |||||||
| N | 34 | 51 | 20 | |||||||
| DH mean (SD) | 3.32 (2.24) | 2.53 (1.72) | 2.50 (1.96) | -0.23 | 0.012* | |||||
| 34 | ACAN | rs76282091 | C | 0.16 | ||||||
| Genotypes | G/G | C/G | C/C | |||||||
| N | 74 | 27 | 4 | |||||||
| DH mean (SD) | 2.80 (2.05) | 2.59 (1.42) | 3.75 (3.59) | 0.05 | 0.633 | |||||
| 35 | ACAN | rs1042630 | A | 0.41 | ||||||
| Genotypes | G/G | A/G | A/A | |||||||
| N | 40 | 45 | 20 | |||||||
| DH mean (SD) | 2.72 (2.02) | 2.40 (1.60) | 3.75 (2.34) | 0.20 | 0.031* | |||||
| 36 | ACAN | rs1042631 | T | 0.35 | ||||||
| Genotypes | C/C | T/C | T/T | |||||||
| N | 45 | 47 | 14 | |||||||
| DH mean (SD) | 2.93 (2.16) | 2.62 (1.75) | 3.07 (2.20) | 0.05 | 0.555 | |||||
| 37 | ACAN | rs698621 | G | 0.38 | ||||||
| Genotypes | T/T | G/T | G/G | |||||||
| N | 41 | 50 | 14 | |||||||
| DH mean (SD) | 3.24 (2.21) | 2.64 (1.82) | 1.93 (1.38) | -0.30 | 0.001* | |||||
| 38 | ACAN | rs3817428 | G | 0.05 | ||||||
| Genotypes | C/C | G/C | G/G | |||||||
| N | 96 | 9 | 0 | |||||||
| DH mean (SD) | 2.94 (1.95) | 1.11 (1.27) | NA | -0.23 | 0.013* | |||||
| 39 | ACAN | rs1126823 | G | 0.36 | ||||||
| Genotypes | A/A | G/A | G/G | |||||||
| N | 41 | 51 | 12 | |||||||
| DH mean (SD) | 2.76 (2.02) | 2.88 (1.97) | 2.42 (1.98) | 0.02 | 0.860 | |||||
| 40 | TIMP2 | rs2277698 | T | 0.24 | ||||||
| Genotypes | C/C | T/C | T/T | |||||||
| N | 60 | 40 | 5 | |||||||
| DH mean (SD) | 2.82 (2.08) | 2.82 (1.74) | 2.00 (2.55) | -0.11 | 0.266 | |||||
| 41 | ADAMTS5 | rs1444269 | G | 0.26 | ||||||
| Genotypes | A/A | G/A | G/G | |||||||
| N | 55 | 43 | 6 | |||||||
| DH mean (SD) | 2.82 (2.14) | 2.79 (1.74) | 2.83 (1.94) | -0.03 | 0.732 | |||||
| 42 | ADAMTS5 | rs2298657 | C | 0.05 | ||||||
| Genotypes | T/T | C/T | C/C | |||||||
| N | 94 | 9 | 0 | |||||||
| DH mean (SD) | 2.79 (1.93) | 2.44 (2.30) | NA | -0.04 | 0.671 | |||||
| 43 | ADAMTS5 | rs3746836 | A | 0.25 | ||||||
| Genotypes | G/G | A/G | A/A | |||||||
| N | 58 | 41 | 6 | |||||||
| DH mean (SD) | 2.74 (2.12) | 2.83 (1.77) | 2.83 (1.94) | 0.01 | 0.950 | |||||
| 44 | ADAMTS5 | rs229072 | T | 0.48 | ||||||
| Genotypes | A/A | T/A | T/T | |||||||
| N | 31 | 45 | 27 | |||||||
| DH mean (SD) | 2.58 (2.08) | 3.11 (2.01) | 2.26 (1.58) | -0.03 | 0.775 | |||||
| 45 | ADAMTS5 | rs229073 | G | 0.48 | ||||||
| Genotypes | A/A | G/A | G/G | |||||||
| N | 31 | 45 | 27 | |||||||
| DH mean (SD) | 2.71 (2.16) | 3.11 (2.01) | 2.26 (1.58) | -0.05 | 0.613 | |||||
| 46 | ADAMTS5 | rs11700721 | T | 0.12 | ||||||
| Genotypes | C/C | T/C | T/T | |||||||
| N | 81 | 23 | 1 | |||||||
| DH mean (SD) | 2.69 (2.03) | 3.09 (1.78) | 3.00 (0.00) | 0.04 | 0.673 | |||||
| 47 | ADAMTS5 | rs16979423 | G | 0.14 | ||||||
| Genotypes | T/T | G/T | G/G | |||||||
| N | 78 | 24 | 3 | |||||||
| DH mean (SD) | 2.85 (2.02) | 2.63 (1.88) | 2.33 (1.53) | -0.06 | 0.544 | |||||
| 48 | ADAMTS5 | rs9978597 | G | 0.05 | ||||||
| Genotypes | T/T | G/T | G/G | |||||||
| N | 94 | 9 | 0 | |||||||
| DH mean (SD) | 2.80 (1.98) | 2.89 (2.09) | NA | 0.04 | 0.657 | |||||
| 49 | ADAMTS5 | rs229078 | T | 0.22 | ||||||
| Genotypes | G/G | T/G | T/T | |||||||
| N | 64 | 35 | 5 | |||||||
| DH mean (SD) | 2.84 (1.99) | 2.94 (19.70) | 1.00 (0.71) | -0.03 | 0.721 | |||||
| 50 | ADAMTS5 | rs151065 | A | 0.19 | ||||||
| Genotypes | G/G | A/G | A/A | |||||||
| N | 66 | 36 | 3 | |||||||
| DH mean (SD) | 2.81 (1.99) | 2.72 (1.89) | 2.66 (3.06) | -0.09 | 0.322 | |||||
| 51 | ADAMTS5 | rs3746839 | G | 0.08 | ||||||
| Genotypes | A/A | G/A | G/G | |||||||
| N | 88 | 16 | 0 | |||||||
| DH mean (SD) | 2.75 (1.96) | 3.12 (1.96) | NA | 0.04 | 0.679 | |||||
| 52 | ADAMTS5 | rs226794 | A | 0.10 | ||||||
| Genotypes | G/G | A/G | A/A | |||||||
| N | 84 | 21 | 0 | |||||||
| DH mean (SD) | 2.75 (2.04) | 2.91 (1.70) | NA | -0.03 | 0.772 | |||||
| 53 | ADAMTS5 | rs457947 | G | 0.07 | ||||||
| Genotypes | C/C | G/C | G/G | |||||||
| N | 92 | 12 | 1 | |||||||
| DH mean (SD) | 2.78 (1.99) | 2.92 (1.88) | 1.00 (0.00) | -0.01 | 0.890 | |||||
| 54 | ADAMTS5 | rs55933916 | G | 0.08 | ||||||
| Genotypes | C/C | G/C | G/G | |||||||
| N | 90 | 13 | 1 | |||||||
| DH mean (SD) | 2.77 (2.00) | 3.08 (1.85) | 1.00 (0.00) | -0.03 | 0.726 | |||||
| 55 | TIMP3 | rs9862 | T | 0.50 | ||||||
| Genotypes | C/C | T/C | T/T | |||||||
| N | 23 | 58 | 23 | |||||||
| DH mean (SD) | 2.70 (1.66) | 2.76 (1.99) | 2.96 (2.27) | -0.03 | 0.731 | |||||
| 56 | TIMP3 | rs11547635 | T | 0.07 | ||||||
| Genotypes | C/C | T/C | T/T | |||||||
| N | 93 | 11 | 1 | |||||||
| DH mean (SD) | 2.73 (1.89) | 3.09 (2.66) | 4.00 (0.00) | 0.07 | 0.464 | |||||
| 57 | TIMP3 | rs1427384 | C | 0.19 | ||||||
| Genotypes | T/T | C/T | C/C | |||||||
| N | 67 | 29 | 4 | |||||||
| DH mean (SD) | 3.05 (2.11) | 2.14 (1.53) | 3.25 (1.89) | -0.09 | 0.358 | |||||
| 58 | TIMP3 | rs2267184 | T | 0.16 | ||||||
| Genotypes | C/C | T/C | T/T | |||||||
| N | 76 | 26 | 3 | |||||||
| DH mean (SD) | 2.94 (2.07) | 2.38 (1.72) | 2.33 (0.58) | -0.05 | 0.601 | |||||
| 59 | TIMP3 | rs1065314 | C | 0.17 | ||||||
| Genotypes | T/T | C/T | C/C | |||||||
| N | 74 | 27 | 4 | |||||||
| DH mean (SD) | 2.99 (2.06) | 2.15 (1.59) | 3.25 (1.89) | -0.06 | 0.548 | |||||
| 60 | TIMP1 | rs5953060 | C | 0.43 | ||||||
| Genotypes | G/G | C/G | C/C | |||||||
| N | 43 | 34 | 28 | |||||||
| DH mean (SD) | 2.54 (1.75) | 2.94 (2.06) | 2.96 (2.19) | 0.14 | 0.129 | |||||
| 61 | TIMP1 | rs4898 | C | 0.43 | ||||||
| Genotypes | T/T | C/T | C/C | |||||||
| N | 43 | 34 | 28 | |||||||
| DH mean (SD) | 2.54 (1.75) | 2.94 (2.06) | 2.96 (2.19) | 0.14 | 0.129 | |||||
| 62 | TIMP1 | rs6609533 | G | 0.43 | ||||||
| Genotypes | A/A | G/A | G/G | |||||||
| N | 43 | 34 | 28 | |||||||
| DH mean (SD) | 2.54 (1.75) | 2.94 (2.06) | 2.96 (2.19) | 0.14 | 0.129 |
SNV–single nucleotide variant, SD–standard deviation, A1 –minor allele, A2 –major allele, MAF–minor allele frequency, NA–not applicable, β –standardised regression coefficient, data were analysed by multiple linear regression on variant genotypes adjusting for age, gender, body mass index and severity of lumbar disc degeneration
*—p value < 0.05
In-silico functional analysis of the significant variants associated with the severity of lumbar disc herniation
Among the nine significant SNVs only three (rs35430524, rs938609 and rs3817428) were predicted as pathogenic by the in-silico functional analysis (Table 5). The rs3817428 variant of ACAN was identified as pathogenic by two prediction tools (Provean and PolyPhen) while the other two were detected by one of either Provean or PolyPhen prediction tools. Both the rs35430524 and rs3817428 variants were located in conserved regions of ACAN. In addition, the rs35430524 variant had a non-conservative amino acid substitution.
Table 5. Significant SNVs and their functional predictions.
| No | Gene | SNV | Region | AA change | Conservative substitution |
Conserved region | Provean | SIFT | Polyphen | Mutant Taster |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ACAN | rs2272023 | synonymous | 1 | Neutral | Harmless | ||||
| 2 | rs2351491 | synonymous | 1 | Neutral | Harmless | |||||
| 3 | rs35430524 | nonsynonymous | P913T | No | 0.99 | Deleterious | Tolerated | Benign | Harmless | |
| 4 | rs938609 | nonsynonymous | S939T | Yes | 0.32 | Neutral | Tolerated | Probably damaging | Harmless | |
| 5 | rs2882676 | nonsynonymous | E1508A | No | 0.001 | Neutral | Tolerated | Benign | Harmless | |
| 6 | rs3825994 | synonymous | 0.19 | Neutral | Harmless | |||||
| 7 | rs698621 | synonymous | 0.24 | Neutral | Harmless | |||||
| 8 | rs1042630 | nonsynonymous | I2079V | Yes | 0.08 | Neutral | Tolerated | Benign | Harmless | |
| 9 | rs3817428 | nonsynonymous | D2373E D2335E |
Yes | 1 | Deleterious | Tolerated | Probably damaging | Harmless |
SNV–single nucleotide variaton; AA–amino acid. Single nucleotide variants with non-conservative substitutions and ones located at conserved regions of a gene have a higher probability to become pathogenic.
Associations of haplotypes of ACAN with the severity of lumbar disc herniation
Among the nine significant SNVs, rs938609 and rs2882676 formed three haplotypes while rs3825994 and rs1042630 also formed three haplotypes. The haplotypes frequencies and results of multivariable linear regression analysis are summarised in Table 6. In haplotype analysis, the T-A haplotype of rs938609 and rs2882676 loci and T-A haplotype of rs3825994 and rs1042630 loci increased the severity of disc herniation. In contrast A-C haplotype of rs938609 and rs2882676 loci and G-G haplotype of rs3825994 and rs1042630 loci reduced the severity of disc herniation after adjusting for confounders (age, gender, BMI and the severity of lumbar disc degeneration).
Table 6. Haplotype frequencies and their associations with the severity of lumbar disc herniation.
| Haplotypes | Frequency | BETA coefficient | Empirical p value |
|---|---|---|---|
| rs938609-rs2882676 | |||
| TA | 0.48 | 0.77 | 0.001* |
| AC | 0.37 | -0.96 | <0.001* |
| TC | 0.16 | 0.15 | 0.650 |
| rs3825994-rs1042630 | |||
| TA | 0.40 | 0.52 | 0.040* |
| GG | 0.43 | -0.66 | 0.009* |
| TG | 0.16 | 0.17 | 0.617 |
*—p value < 0.05
Discussion
The study assessed associations of SNVs of candidate genes of the aggrecan metabolic pathway with the severity of lumbar disc herniation in patients with chronic mechanical low back pain. In addition, we assessed the in-silico functional analysis of significant SNVs and associations of their haplotypes with the severity of lumbar disc herniation. In our results, minor allele of the rs2272023 (C), rs35430524 (A), rs2882676 (A), rs2351491 (T), rs938609 (A), rs3825994 (G), rs1042630 (A), rs698621 (G) and rs3817428 (G) variants of ACAN were associated with the severity of lumbar disc herniation. Furthermore, the rs35430524, rs938609 and rs3817428 variants were detected as pathogenic by the in-silico functional analysis. In addition, haplotypes of rs938609 and rs2882676 loci, and haplotypes of rs3825994 and rs1042630 loci of ACAN were associated with the severity of lumbar disc herniation.
In disc degeneration, the concentration of aggrecan is reduced due to increased breakdown and reduced synthesis of aggrecan molecules leading to disc dehydration [6]. This process is further augmented by concurrent changes of the proteins involved in the aggrecan metabolic pathway (interleukins, matrix metalloproteinases, a disintegrin and metalloproteinase with thrombospondin motifs and tissue inhibitors of metalloproteinases). Annular fissures appear in the disc with the loss of integrity of annulus fibrosus and disc dehydration. Nuclear material can leak through these fissures and displace beyond the disc space margins leading to disc herniation. Disc herniation is categorised as disc protrusion and extrusion based on the extension of herniated disc material beyond the disc space [20]. In our sample, almost all the patients had disc herniations with 42.4% having disc extrusions.
Out of sixty two selected SNVs of ten candidate genes, only nine SNVs of ACAN were associated with the severity of disc herniation which included five nonsynonymous variants (rs35430524, rs938609, rs2882676, rs1042630 and rs3817428) and four synonymous variants (rs2272023, rs2351491, rs3825994 and rs698621). A nonsynonymous variant alters the amino acid sequence of the respective protein. This altered amino acid sequence may affect the function of the respective protein. In contrast, synonymous variants do not alter the amino acid sequence of the respective protein, but it can cause changes in long non-coding RNA, microRNA and promoters causing the disease by affecting the protein expression, conformation and function. Furthermore any nonsynonymous or synonymous variant may act as a marker of a functional SNV in the same gene or nearby gene [29]. VNTR polymorphisms reported as significantly associated with the disease are likely to be found as such, because they are in linkage disequilibrium with pathogenic SNVs [30]. Therefore, the focus of this study was to assess the SNVs associated with the severity of disc herniation.
There are a limited number of studies which have explored the association of SNVs of the ACAN with the severity of disc herniation. The presence of each “T” allele of the rs1042631 variant of ACAN increased the frequency of disc herniation among the Finnish population [8]; it also increased the risk of annular tears in Indian patients who attended a spine unit of a tertiary care hospital [31]. However, this variant was not associated with the severity of disc herniation in our sample. The rs2351491, rs938609 and rs698621 variants of ACAN were in strong linkage disequilibrium and each additional minor allele progressively reduced the severity of disc herniation (Table 4). The rs938609 variant is located in exon 12 of ACAN and causes a Serine to Threonine substitution at position 939 of the amino acid sequence. This substitution is a very highly conservative substitution, but only the Polyphen bioinformatics tool predicted it as pathogenic (Table 5). The rs2351491 and rs698621 variants are synonymous variants and functionally unremarkable.
The presence of each additional minor allele of the rs35430524 (A), rs1042630 (A) and rs2882676 (A) variants of ACAN was associated with a progressive increase in the severity of disc herniation while the presence of an additional “G” allele of the rs3817428 variant of ACAN reduced the severity of disc herniation. All these four variants were nonsynonymous, but only the rs35430524 and rs3817428 variants were predicted as pathogenic. The rs35430524 variant is located in exon 12 of ACAN and causes a Proline to Threonine substitution at position 913 of the amino acid sequence. It is located at a highly-conserved region of ACAN and is not a conservative substitution. Therefore there is a high probability that the rs35430524 variant is pathogenic and which was identified as such by the Provean prediction tool (Table 5). The rs3817428 variant located in exon 15 of ACAN causes an Aspartic acid to Glutamic acid substitution at position 2373 of the amino acid sequence. This is located in a highly-conserved area in ACAN, but its amino acid substitution is a highly conservative substitution. Two of the four bioinformatics tools (Provean and Polyphen) predicted that the rs3817428 variant is pathogenic (Table 5). The rs3825994 and rs2272023 variants of ACAN are synonymous variants and the results of the functional prediction tools were unremarkable.
Our findings on the associations of SNVs of ACAN with the severity of lumbar disc herniation were further strengthened by the results of the haplotype analysis. In haplotype analysis, we found that T-A (rs938609—rs2882676) and T-A (rs3825994—rs1042630) haplotypes increased the severity of disc herniation while A-C (rs938609-rs2882676) and G-G (rs3825994- rs1042630) haplotypes reduced the severity of disc herniation. This suggests that the severity of disc herniation is influenced by single nucleotide variation and also by haplotype differences.
Several studies reported significant associations of SNVs of IL6, IL10 [32], MMP1 and MMP3 [9] with disc herniation. Some of these variants were intronic variants [32] and some variants were not polymorphic in our population. We have assessed only the exonic variants and the SNVs of the candidate genes of inflammatory, catabolic, anti-catabolic molecules were not associated with the severity of disc herniation (Table 4). In our previous study, SNVs of IL1A (inflammatory gene), ADAMTS4 and ADAMTS5 (catabolic genes) were found to be associated with the severity of disc degeneration [3], but these SNVs were not associated with the severity of disc herniation. Thus, findings of the two studies suggest that SNVs of candidate genes in inflammatory and catabolic pathways are responsible for the variation in the severity of disc degeneration while SNVs of ACAN (structural gene) are responsible for the variation in the severity of disc herniation.
Quantitative traits like disc herniation may have significant effects from variants of several candidate genes and environmental factors. However, we have not corrected the genetic association analysis for the multiple variants we already assessed and other probable variants in other respective genes. Although the rs3817428, rs35430524 and rs938609 variants of ACAN were predicted as pathogenic by the in-silico functional analysis, overall results of the four prediction tools were inconsistent. Considering these facts, it is important to carry out genetic association analysis with a larger sample number using whole genome sequencing followed by functional genetic studies to identify the exact effect of the significant SNVs.
Limitations of the study
The results of this study are not generalizable as it was conducted on a specific group of patients with chronic mechanical low back pain. However, there were patients representing the three districts of the Western Province of Sri Lanka. Low sample number is another limitation of the study, but we used permutation function of the PLINK software tool to relax the assumptions of normality and correct the errors due to small sample size. There may be other confounding factors that we have not corrected for.
Conclusions
Single nucleotide variants of ACAN and their haplotypes are associated with the severity of lumbar disc herniation. The rs35430524, rs938609 and rs3817428 variants of ACAN are predicted as pathogenic by in-silico functional analysis. However functional involvement of these three SNVs should be confirmed with functional genetic studies. This study provides basic insight of genetic markers which could be used as probable therapeutic targets for novel treatment strategies for lumbar disc herniation.
Supporting information
(XLSX)
Acknowledgments
A special thanks to staff of the Rheumatology Clinic, National Hospital of Sri Lanka and all the patients who participated in the study.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by the University Grants Commission, Sri Lanka (UGC/ICD/2/RG2011/02/08) and the University of Colombo, Sri Lanka (AP/3/2012/PG/03). The funders provided funds for study design, data collection and analysis.
References
- 1.Friedly J, Standaert C, Chan L. Epidemiology of spine care: the back pain dilemma. Phys Med Rehabil Clin N Am. 2010;21(4):659–677. doi: 10.1016/j.pmr.2010.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kadow T, Sowa G, Vo N, Kang JD. Molecular basis of intervertebral disc degeneration and herniations: what are the important translational questions? Clin Orthop Relat Res. 2015;473(6):1903–1912. doi: 10.1007/s11999-014-3774-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perera RS, Dissanayake PH, Senarath U, Wijayaratne LS, Karunanayake AL, Dissanayake VH. Single Nucleotide Variants of Candidate Genes in Aggrecan Metabolic Pathway Are Associated with Lumbar Disc Degeneration and Modic Changes. PLoS One. 2017;12(1):e0169835 doi: 10.1371/journal.pone.0169835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sivan SS, Hayes AJ, Wachtel E, Caterson B, Merkher Y, Maroudas A, et al. Biochemical composition and turnover of the extracellular matrix of the normal and degenerate intervertebral disc. Eur Spine J. 2014;23 Suppl 3:S344–353. [DOI] [PubMed] [Google Scholar]
- 5.Adams MA, Dolan P. Intervertebral disc degeneration: evidence for two distinct phenotypes. J Anat. 2012;221(6):497–506. doi: 10.1111/j.1469-7580.2012.01551.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roughley PJ. Biology of intervertebral disc aging and degeneration: involvement of the extracellular matrix. Spine. 2004;29(23):2691–2699. [DOI] [PubMed] [Google Scholar]
- 7.Cong L, Zhu Y, Pang H, Guanjun TU. The interaction between aggrecan gene VNTR polymorphism and obesity in predicting incident symptomatic lumbar disc herniation. Connect Tissue Res. 2014;55(5–6):384–390. doi: 10.3109/03008207.2014.959117 [DOI] [PubMed] [Google Scholar]
- 8.Videman T, Saarela J, Kaprio J, Nakki A, Levalahti E, Gill K, et al. Associations of 25 structural, degradative, and inflammatory candidate genes with lumbar disc desiccation, bulging, and height narrowing. Arthritis Rheum. 2009;60(2):470–481. doi: 10.1002/art.24268 [DOI] [PubMed] [Google Scholar]
- 9.Eser B, Eser O, Yuksel Y, Aksit H, Karavelioglu E, Tosun M, et al. Effects of MMP-1 and MMP-3 gene polymorphisms on gene expression and protein level in lumbar disc herniation. Genetics and molecular research: GMR. 2016;15(3). [DOI] [PubMed] [Google Scholar]
- 10.Jacobsen LM, Schistad EI, Storesund A, Pedersen LM, Espeland A, Rygh LJ, et al. The MMP1 rs1799750 2G allele is associated with increased low back pain, sciatica, and disability after lumbar disk herniation. Clin J Pain. 2013;29(11):967–971. doi: 10.1097/AJP.0b013e31827df7fd [DOI] [PubMed] [Google Scholar]
- 11.Manek NJ, MacGregor AJ. Epidemiology of back disorders: prevalence, risk factors, and prognosis. Curr Opin Rheumatol. 2005;17(2):134–140. [DOI] [PubMed] [Google Scholar]
- 12.Walker BF, Williamson OD. Mechanical or inflammatory low back pain. What are the potential signs and symptoms? Man Ther. 2009;14(3):314–320. doi: 10.1016/j.math.2008.04.003 [DOI] [PubMed] [Google Scholar]
- 13.Omair A, Holden M, Lie BA, Reikeras O, Brox JI. Treatment outcome of chronic low back pain and radiographic lumbar disc degeneration are associated with inflammatory and matrix degrading gene variants: a prospective genetic association study. BMC Musculoskelet Disord. 2013;14:105 doi: 10.1186/1471-2474-14-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lane NE, Nevitt MC, Genant HK, Hochberg MC. Reliability of new indices of radiographic osteoarthritis of the hand and hip and lumbar disc degeneration. J Rheumatol. 1993;20(11):1911–1918. [PubMed] [Google Scholar]
- 15.Arambepola C, Ekanayake R, Fernando D. Gender differentials of abdominal obesity among the adults in the district of Colombo, Sri Lanka. Prev Med. 2007;44(2):129–134. doi: 10.1016/j.ypmed.2006.11.004 [DOI] [PubMed] [Google Scholar]
- 16.Katulanda P, Jayawardena MA, Sheriff MH, Constantine GR, Matthews DR. Prevalence of overweight and obesity in Sri Lankan adults. Obes Rev. 2010;11(11):751–756. doi: 10.1111/j.1467-789X.2010.00746.x [DOI] [PubMed] [Google Scholar]
- 17.Whitley AS, Sloane C, Hoadley G, Moore AD. Clark's Positioning in Radiography 12Ed: CRC Press; 2005. [Google Scholar]
- 18.Pfirrmann CW, Metzdorf A, Zanetti M, Hodler J, Boos N. Magnetic resonance classification of lumbar intervertebral disc degeneration. Spine. 2001;26(17):1873–1878. [DOI] [PubMed] [Google Scholar]
- 19.Takatalo J, Karppinen J, Niinimaki J, Taimela S, Nayha S, Jarvelin MR, et al. Prevalence of degenerative imaging findings in lumbar magnetic resonance imaging among young adults. Spine. 2009;34(16):1716–1721. doi: 10.1097/BRS.0b013e3181ac5fec [DOI] [PubMed] [Google Scholar]
- 20.Fardon DF, Williams AL, Dohring EJ, Murtagh FR, Gabriel Rothman SL, Sze GK. Lumbar disc nomenclature: version 2.0: Recommendations of the combined task forces of the North American Spine Society, the American Society of Spine Radiology and the American Society of Neuroradiology. The spine journal: official journal of the North American Spine Society. 2014;14(11):2525–2545. [DOI] [PubMed] [Google Scholar]
- 21.QIAGEN. QIAamp DNA Mini Blood Mini Handbook 2014 [cited 2014 10/02/]. Available from: https://www.qiagen.com/gb/shop/sample-technologies/dna/qiaamp-dna-mini-kit/#resources.
- 22.Promega Corporation. QuantiFluor® dsDNA System Technical Manual 2014 [cited 2014 15/03/]. Available from: https://worldwide.promega.com/resources/protocols/technical-manuals/101/quantifluor-dsdna-system-protocol/.
- 23.Gabriel S, Ziaugra L, Tabbaa D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Current protocols in human genetics / editorial board, Jonathan L Haines [et al]. 2009;Chapter 2:Unit 2.12. [DOI] [PubMed] [Google Scholar]
- 24.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7(10):e46688 doi: 10.1371/journal.pone.0046688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–1081. doi: 10.1038/nprot.2009.86 [DOI] [PubMed] [Google Scholar]
- 27.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nature methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Meth. 2014;11(4):361–362. [DOI] [PubMed] [Google Scholar]
- 29.Katsonis P, Koire A, Wilson SJ, Hsu TK, Lua RC, Wilkins AD, et al. Single nucleotide variations: biological impact and theoretical interpretation. Protein Sci. 2014;23(12):1650–1666. doi: 10.1002/pro.2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mill J, Asherson P, Craig I, D'Souza UM. Transient expression analysis of allelic variants of a VNTR in the dopamine transporter gene (DAT1). BMC Genet. 2005;6:3 doi: 10.1186/1471-2156-6-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rajasekaran S, Kanna RM, Senthil N, Raveendran M, Cheung KM, Chan D, et al. Phenotype variations affect genetic association studies of degenerative disc disease: conclusions of analysis of genetic association of 58 single nucleotide polymorphisms with highly specific phenotypes for disc degeneration in 332 subjects. The spine journal: official journal of the North American Spine Society. 2013;13(10):1309–1320. [DOI] [PubMed] [Google Scholar]
- 32.Huang X, Chen F, Zhao J, Wang D, Jing S, Li H, et al. Interleukin 6 (IL-6) and IL-10 Promoter Region Polymorphisms Are Associated with Risk of Lumbar Disc Herniation in a Northern Chinese Han Population. Genetic testing and molecular biomarkers. 2017;21(1):17–23. doi: 10.1089/gtmb.2016.0189 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.