Abstract
As the first step to discover protein disease biomarkers from saliva, global analyses of the saliva proteome have been carried out since the early 2000s, and more than 3,000 proteins have been identified in human saliva. Recently, ethnic differences in the human plasma proteome have been reported, but such corresponding studies on human saliva in this aspect have not been previously reported. Thus, here, in order to determine ethnic differences in the human saliva proteome, a Korean whole saliva (WS) proteome catalogue indexing 480 proteins was built and characterized through nLC-Q-IMS-TOF analyses of WS samples collected from eleven healthy South Korean male adult volunteers for the first time. Identification of 226 distinct Korean WS proteins, not observed in the integrated human saliva protein dataset, and significant gene ontology distribution differences in the Korean WS proteome compared to the integrated human saliva proteome strongly support ethnic differences in the human saliva proteome. Additionally, the potential value of ethnicity-specific human saliva proteins as biomarkers for diseases highly prevalent in that ethnic group was confirmed by finding 35 distinct Korean WS proteins likely to be associated with the top 10 deadliest diseases in South Korea. Finally, the present Korean WS protein list can serve as the first level reference for future proteomic studies including disease biomarker studies on Korean saliva.
Introduction
Saliva is secreted from salivary glands, including three major glands (parotid, submandibular, sublingual glands) and minor glands. Saliva has various functions. It maintains oral cavity homeostasis, lubricates oral tissues, promotes chewing, swallowing, digestion, and speaking, and protects the oral cavity against microorganisms [1–4]. It is composed of water, proteins, peptides, lipids, other small molecules, and minerals. Healthy adults are known to produce 500–1500 mL of saliva daily at a rate of about 0.5 mL/min [1, 4]. Most organic compounds in saliva are produced in the salivary glands, but some are transferred from blood through various mechanisms, including diffusion, active transport, and ultrafiltration [4]. Moreover, its collection is non-invasive and it is easy to collect and store saliva samples [5]. Thus, saliva can be a good alternative to blood for diagnosis due to its characteristics mentioned above. For example, major systemic infections of viruses such as human immunodeficiency virus, hepatitis C virus, and human papillomavirus have been successfully tested by saliva-based diagnostic methods [6]. Thus, clinical diagnosis using saliva specimens is an emerging field. Among various constituents of saliva, proteins have gained the most interest as probable disease biomarkers because numerous proteins are known to be present in the saliva and many of them are believed to represent the progress of diseases [4].
As the first step to discover protein disease biomarkers from saliva, global analyses of the saliva proteome have been carried out since the early 2000s. As a result, more than 3000 proteins have been identified in human saliva [1, 7–11]. Some of them are accessible through public databases such as Human Salivary Proteome Central Repository (1,166 proteins) and Sys-BodyFluid Database (2,161 proteins) [12, 13]. Additionally, systematic comparisons of human saliva and plasma proteomes have been carried out and several interesting points have been reported in the saliva proteome [1, 2]. First, only about 27% of proteins identified in human whole saliva (WS) are found in plasma, indicating that it is possible to discover totally novel biomarkers from saliva [2]. In addition, human saliva and plasma proteomes are over-represented in the categories of response-to-stimulus and response-to-stress compared to total human proteome. This indicates that both fluids (saliva and plasma) might play important roles of in the defense system of the human body and their probable potential for disease diagnosis [1, 2]. These points have been supported by the discovery of many protein disease biomarker candidates from saliva for oral diseases and systemic diseases [2, 5, 14–17]. Moreover, about 58% of immunoglobulins (Igs) identified in human saliva are found in plasma, and the abundance of these overlapping Igs in saliva and plasma shows a high correlation (r of at least 0.87) [1, 2]. This indicates that it is possible to transform antibody-based diagnostic methods using blood to methods employing saliva; an excellent example is the commercial saliva HIV test kit [6].
Recently, ethnic differences in human plasma proteome have been reported. Jeong et al. confirmed 100 unique proteins out of 185 proteins in Korean plasma compared to 3,380 proteins in the HUPO Plasma Proteome Project dataset [18], and Kim et al. observed plasma level differences of some cardiovascular disease protein marker candidates between African-American and non-Hispanic White ethnicity [19]. These results indicate that there is a fundamental need to determine ethnic difference in human saliva proteome. Unique proteins might be found only in ethnicity-specific saliva samples and they might be useful as novel biomarkers for diseases prevalent in that ethnic group.
Therefore, the Korean WS proteome catalogue indexing 480 proteins was built and characterized in this study through proteomic analyses of WS samples collected from eleven healthy Korean male adult volunteers for the first time. It was then compared to the integrated human saliva proteome including 3,449 proteins to determine ethnic differences in human saliva proteome. Confirmed differences of protein identities and GO category distributions between the two proteomes strongly support that there are ethnic differences in the human saliva proteome. In addition, some distinct proteins in the Korean WS are likely to be associated with highly prevalent diseases in South Korea, demonstrating the high diagnostic potential of ethnicity-specific human saliva proteins for diseases highly prevalent within an ethnic group. Finally, the present list of Korean WS proteins can serve as the first level reference for future proteomic studies including disease biomarker studies on Korean saliva.
Materials and methods
Sample collection and Preparation
This study was approved by Dankook University Institutional Review Board. The participants of this study were recruited between August 1, 2014 and Augst 31, 2014 by posting its notices including the brief summary of this study around Dankook University, Cheonan, Chungnam, South Korea. A total of 15 volunteers wanted to participate this study, but 4 of them were excluded based on their history of diseases informed through their introductory screening surveys. Finally, eleven healthy South Korean male adults (25.9±2.3 years old; 22–30 years old) were decided to be the participants and each participant signed an informed consent form. Any specific baseline demographic characteristics of the study populations were not available in this study, because only basic personal information and history of diseases were obtained through the introductory screening survey. WS (15 mL/person) was collected from volunteers at 9:30 am prior to eating and after rinsing the mouth with water. A protease inhibitor cocktail solution (Sigma-Aldrich, St. Louis, MO) was spiked (the final volume ratio of 1:100) to WS samples immediately after sample collection. These protease-spiked samples were centrifuged at 12,000 rpm and 4°C for 10 min. Each supernatant was stored at -70°C until use. Prior to protein digestion, 4 mL of the thawed protease-spiked sample supernatant was applied to a 3 kDa cutoff filter unit (Amicon Ultra-4, Merck Millipore, Billerica, MA) for buffer exchange with water. The filter unit was centrifuged at 3,500 rpm and the retentate was dried by vacuum centrifugation. The dried residue was resuspended with 500 μL of water and its total protein concentration was determined by BCA assay (Pierce BCA Protein Assay Kit, Thermo Scientific, Waltham, MA). An appropriate portion of the resuspended solution (equivalent to 1 mg of total protein) was then dried by vacuum centrifugation again, and the resulting residue was applied to procedures described previously with slight modifications (S1 Appendix) [20, 21]. A portion of the final form of the sample solution was subjected to nanoliquid chromatography-quadrupole-ion mobility spectroscopy-time of flight (nLC-Q-IMS-TOF) analysis. In the case of the pooled Korean WS sample, 1 mL of each thawed protease-spiked sample supernatant was mixed and the mixture was applied to the same method mentioned above. A portion of the final form of the pooled sample solution was subjected to nLC-Q-IMS-TOF analysis and nLC-Q-orbitrap analysis.
Separation and analysis
All nLC-Q-IMS-TOF analyses were carried out on a Waters nanoACQUITY UPLC system (Waters, Milford, MA) and a Waters SYNAPT G2-S HDMS system. The prepared sample was injected into a Waters nanoACQUITY UPLC Symmetry C18 trap column (5 μm, 0.18×20 mm). It was desalted with 99% mobile phase A (0.1% formic acid in water) and 1% mobile phase B (0.1% formic acid in acetonitrile) for 5 min at a flow rate of 10 μL/min. Trap column-retained peptides were eluted into a Waters nanoACQUITY UPLC BEH300 C18 column (1.7 μm, 0.075×250 mm) and separated by a linear gradient of mobile phase B from 1 to 60% for 120 min at a flow rate of 250 nL/min. Peptides eluted from the analytical column were delivered into the mass spectrometer through a nanoelectrospray ionization (nESI) source operating in positive ion mode. Mass spectrometry of peptide ions was performed in resolution data-independent acquisition mode (MSE). Prior to fragmentation processes, IMS was carried out to separate similar precursor ions. Parameters related with mass spectrometry are listed in S1 Appendix.
All nLC-Q-orbitrap analyses were carried out on a Thermo Scientific Easy-nLC 1000 system (Waltham, MA) and a Thermo Scientific Q Exactive system. The prepared sample was desalted by Top Tip (Glygen, Columbia, MD) following the direction by the manufacturer and the desalted sample was separated on an in-house analytical column (0.075×250 mm), packed with C18 resin (Jupiter, 3 μm, Phenomenex, Torrance, CA), by a linear gradient of mobile phase B from 1 to 80% for 120 min at a flow rate of 300 nL/min. Peptides eluted from the column were delivered into the mass spectrometer through a nESI source operating in positive ion mode. Mass spectrometry of peptide ions was performed in data-dependent product ion scan (MS2). Parameters related with mass spectrometry are listed in S1 Appendix.
Protein identification and bioinformatics
Raw data from nLC-Q-IMS-TOF and nLC-Q-orbitrap were analyzed with Waters ProteinLynx Global Server (PLGS) v3.0.2 and Thermo Proteome Discoverer v2.1, respectively. For the identification of peptides and proteins, database search against the IPI human database v3.87 was performed and database search parameters are listed in S1 Appendix. All database search results were verified manually.
For GO analysis of saliva proteomes, the Generic GO term mapper was used [22]. Significance of difference in individual GO categories between the Korean WS proteome and the integrated human saliva proteome was tested by the chi-square method [23].
Database of disease-related biomarkers was used to check probable association between distinct proteins observed in Korean WS proteome but not in the integrated human saliva proteome and diseases [24].
Results
The Korean WS proteome
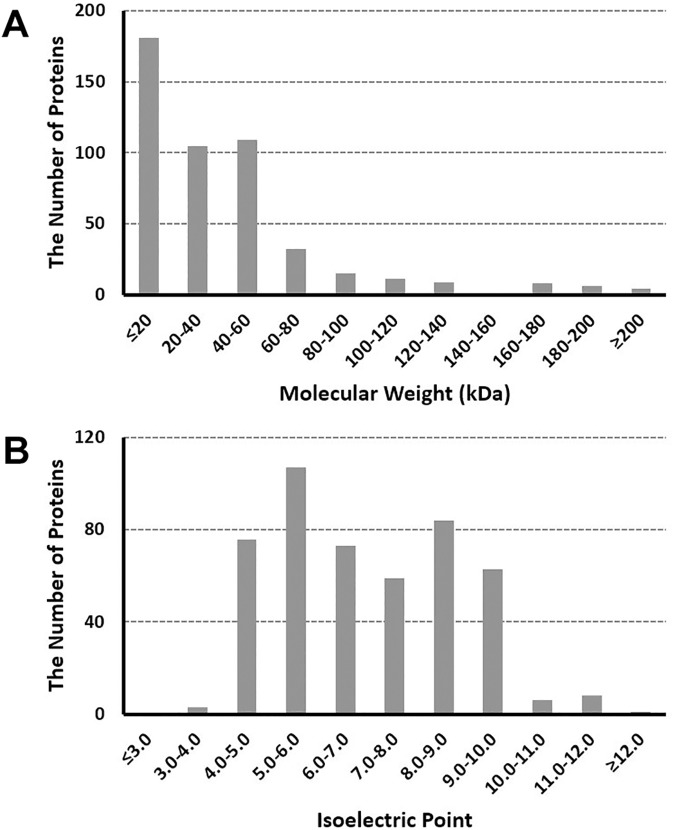
Based on nLC-Q-IMS-TOF analyses of Korean WS samples, the Korean WS proteome was built successfully for the first time. In order to enhance the credibility of protein identification results, the following criteria were set: 1) any identification derived from only one unique peptide was rejected, 2) FDR was kept at no more than 1%, 3) only protein identification with at least 95% probability from PLGS results were accepted, and 4) all results which passed the above criteria were verified manually. These criteria were applied to all downstream protein identifications. As a result, a total of 480 proteins were identified (S1 Table). Also, the distribution of theoretical molecular weight and isoelectric point (pI) of the Korean WS proteome were examined (Fig 1A for molecular weight and Fig 1B for pI). As shown in Fig 1A, a large portion (82.3%) of the Korean WS proteome is composed of proteins with molecular weight of less than 60 kDa. There is a roughly inverse correlation between distribution proportions and molecular weights of component proteins at range of 60–160 kDa. In the case of pI distribution, the Korean WS proteome is composed of 16.5, 37.5, 30.0, and 16.0% of proteins with pI values lower than 5.0, between 5.0 and 7.0, between 7.0 and 9.0, and higher than 9.0, respectively (Fig 1B). The average molecular weight and pI value of the Korean WS proteome were calculated to 42 kDa and 6.95, respectively.
Fig 1.
Theoretical molecular weight (A) and isoelectric point (B) distribution of the Korean whole saliva proteome.
Comparison of protein lists from the Korean WS proteome and the integrated human saliva proteome
To determine ethnic differences in the human saliva proteome, the present Korean WS proteome was compared to P. Sivadasan et al.'s updated human saliva protein list (a total of 3,449 proteins) built by the integration of their own and previously-reported five human saliva protein lists [1, 7–11]. For accurate comparison between proteomes, all available information of a protein, including IPI accession number, SwissProt number, gene symbol, amino acid sequence, molecular weight, and brief description, was used for its UniProt KB search. Then, search results from similar proteins in various proteomes were carefully compared to one another to determine if they are the same. As shown in Fig 2, the Korean WS protein list has 226 out of 480 (47.1%) proteins not included in the integrated human saliva protein list. These distinct Korean WS proteins are summarized in Table 1 and S2 Table.
Fig 2. Venn diagram illustrating the total number of proteins specific to either the Korean whole saliva proteome or the integrated human saliva proteome and those observed in both proteomes.
Table 1. Distinct proteins observed in Korean whole saliva but not in other human saliva.
1, response to stimulus; 2, response to stress; 3, cell communication; 4, protein metabolic; 5, other primary metabolic; 6, transport; 7, organization and biogenesis; 8, catabolic process; 9, cell homeostasis; 10, regulation of biological process; 11, nucleic acid binding; 12, protein binding; 13, other binding; 14, transporter activity; 15, signal transducer activity; 16, catalytic activity; 17, motor activity; 18, structural regulator; 19, transcription regulator; 20, antioxidant activity; 21, enzyme regulator activity.
| Accession number | Description | Biological process | Molecular function | Biomarker candidates |
|---|---|---|---|---|
| IPI00000149 | ISOFORM 1 OF CASPASE-8 | 1, 2, 3, 4, 6, 7, 8, 10 | 12, 13, 15, 16 | o |
| IPI00003842 | ISOFORM 1 OF MICROTUBULE-ASSOCIATED PROTEIN 2 | 7 | 12, 18 | o |
| IPI00006079 | ISOFORM 1 OF BCL-2-ASSOCIATED TRANSCRIPTION FACTOR 1 | 2, 3, 4, 5, 7, 10 | 11, 12 | x |
| IPI00011528 | CLEAVAGE STIMULATION FACTOR SUBUNIT 1 | 4, 5 | 11, 12 | x |
| IPI00011651 | ISOFORM 1 OF RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE GAMMA | 3, 4, 10 | 12, 15, 16, 18 | x |
| IPI00012865 | ISOFORM PLD1A OF PHOSPHOLIPASE D1 | 1, 3, 4, 5, 6, 7, 8, 10 | 12, 13, 16 | x |
| IPI00015894 | CDC42 EFFECTOR PROTEIN 4 | 3, 7, 10 | 12, 21 | x |
| IPI00016472 | ISOFORM 2 OF ZINC FINGER CCCH DOMAIN-CONTAINING PROTEIN 13 | unknown | 12, 13 | x |
| IPI00017290 | ISOFORM 5 OF SERINE/THREONINE-PROTEIN PHOSPHATASE 4 REGULATORY SUBUNIT 3A | unknown | unknown | x |
| IPI00018214 | ISOFORM 1 OF PROTEIN MAX | 1, 2, 3, 4, 5, 7, 10 | 11, 12, 19 | o |
| IPI00019226 | ISOFORM 1 OF BROMODOMAIN-CONTAINING PROTEIN 8 | 3, 4, 5, 7, 10 | 11, 12, 15, 18, 19 | o |
| IPI00019794 | SH3 AND MULTIPLE ANKYRIN REPEAT DOMAINS PROTEIN 3 | 1, 3, 4, 6, 7, 9, 10 | 12, 13, 18 | x |
| IPI00020019 | ADIPONECTIN | 1, 2, 3, 4, 5, 6, 7, 8, 10 | 12, 13 | o |
| IPI00022434 | UNCHARACTERIZED PROTEIN (SIMILAR TO SERUM ALBUMIN) | unknown | unknown | o |
| IPI00024134 | IG KAPPA CHAIN V-I REGION WALKER | 3, 4, 6, 10 | 13, 16 | x |
| IPI00026126 | MAMMAGLOBIN-B | 1, 3 | 12 | o |
| IPI00027838 | RNA-BINDING PROTEIN 4B | 1, 5, 10 | 11, 12, 13 | x |
| IPI00029403 | SORTING NEXIN-4 | 1, 2, 3, 6, 7, 10 | 12, 13 | x |
| IPI00030144 | PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A-LIKE 4A/B/C | 4 | 16 | x |
| IPI00032896 | ISOFORM 1 OF DISRUPTED IN SCHIZOPHRENIA 1 PROTEIN | 1, 3, 4, 7, 8, 9, 10 | 12, 18 | x |
| IPI00033892 | GDNF-INDUCIBLE ZINC FINGER PROTEIN 1 | 4, 5, 7, 10 | 11, 13, 19 | x |
| IPI00064474 | ZINC FINGER CCCH DOMAIN-CONTAINING PROTEIN 10 | unknown | 12, 13 | x |
| IPI00102979 | ORAL CANCER-OVEREXPRESSED PROTEIN 1 | unknown | unknown | x |
| IPI00103636 | ISOFORM 2 OF WAP FOUR-DISULFIDE CORE DOMAIN PROTEIN 2 | 4, 10 | 21 | o |
| IPI00107502 | ISOFORM 1 OF WD REPEAT AND SOCS BOX-CONTAINING PROTEIN 1 | 3, 10 | 12, 16 | x |
| IPI00150554 | UNCHARACTERIZED PROTEIN (SIMILAR TO CELL DIVISION CYCLE-ASSOCIATED 7-LIKE PROTEIN) | unknown | unknown | o |
| IPI00158280 | FORMYLTETRAHYDROFOLATE DEHYDROGENASE ISOFORM A VARIANT | unknown | unknown | x |
| IPI00158847 | DNAJ HOMOLOG SUBFAMILY C MEMBER 14 | 6 | 13 | x |
| IPI00160396 | ISOFORM 4 OF PUTATIVE HOMEODOMAIN TRANSCRIPTION FACTOR 2 | 4, 5, 10 | 11 | x |
| IPI00169307 | RHO GTPASE-ACTIVATING PROTEIN 21 | 3, 6, 7 | 12, 21 | x |
| IPI00170867 | ISOFORM 1 OF DEATH DOMAIN-ASSOCIATED PROTEIN 6 | 1, 2, 3, 4, 5, 7, 10 | 12, 13, 15, 21 | x |
| IPI00181753 | CDNA FLJ13286 FIS, CLONE OVARC1001154, HIGHLY SIMILAR TO HOMO SAPIENS CLONE 24720 EPITHELIN 1 AND 2 MRNA | unknown | unknown | o |
| IPI00185027 | ISOFORM 1 OF ARGININE-GLUTAMIC ACID DIPEPTIDE REPEATS PROTEIN | 4, 5, 6, 7, 10 | 11, 12, 13, 19 | x |
| IPI00216773 | UNCHARACTERIZED PROTEIN (SIMILAR TO SERUM ALBUMIN) | unknown | unknown | o |
| IPI00217345 | ISOFORM 2 OF UDP-GLCNAC:BETAGAL BETA-1,3-N- ACETYLGLUCOSAMINYLTRANSFERASE 2 | 1, 4, 5, 7 | 16 | x |
| IPI00217979 | UNCHARACTERIZED PROTEIN MLL5 | 3, 4, 5, 7, 10 | 12, 13, 16 | x |
| IPI00218573 | ISOFORM 3 OF INTERLEUKIN-1 RECEPTOR ANTAGONIST PROTEIN | 1, 2, 3, 5, 6, 7, 10 | 12, 13 | o |
| IPI00218824 | UBIQUITOUSLY TRANSCRIBED TETRATRICOPEPTIDE REPEAT PROTEIN Y-LINKED TRANSCRIPT VARIANT 29 | 4, 5, 7, 10 | 13, 16 | x |
| IPI00220644 | ISOFORM M1 OF PYRUVATE KINASE ISOZYMES M1/M2 | 1, 2, 4, 5, 7, 8, 10 | 11, 12, 13, 16 | o |
| IPI00238220 | UNCHARACTERIZED PROTEIN KIAA2022 | 7 | unknown | x |
| IPI00246842 | ISOFORM 1 OF TATA BOX-BINDING PROTEIN-ASSOCIATED FACTOR RNA POLYMERASE I SUBUNIT C | 4, 5, 10 | 11, 12 | x |
| IPI00291064 | ISOFORM 1 OF AN1-TYPE ZINC FINGER PROTEIN 1 | unknown | 13 | x |
| IPI00292753 | ISOFORM 6 OF GTPASE-ACTIVATING PROTEIN AND VPS9 DOMAIN-CONTAINING PROTEIN 1 | 3, 6, 7 | 12, 21 | x |
| IPI00299749 | ZINC FINGER PROTEIN 16 | 4, 5, 7, 10 | 11, 12, 13, 19 | x |
| IPI00300502 | MYOZENIN-1 | 7 | 12 | x |
| IPI00305457 | PRO2275 | unknown | unknown | o |
| IPI00328712 | UNCHARACTERIZED PROTEIN C2ORF53 | unknown | unknown | x |
| IPI00329002 | SEC14 DOMAIN AND SPECTRIN REPEAT-CONTAINING PROTEIN 1 | unknown | 12, 13 | x |
| IPI00329688 | PROTEIN YIPF3 | 7 | 12 | x |
| IPI00334190 | STOMATIN-LIKE PROTEIN 2 | 2, 3, 4, 5, 6, 7, 9, 10 | 12, 13 | x |
| IPI00376170 | PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A-LIKE | 4, 5, 6, 7, 10 | 12, 13, 16 | o |
| IPI00382577 | KAPPA 1 LIGHT CHAIN VARIABLE REGION | unknown | unknown | x |
| IPI00384361 | ISOFORM 3 OF TRIGGERING RECEPTOR EXPRESSED ON MYELOID CELLS 2 | 1, 2, 3, 4, 6, 7, 10 | 12, 13, 18 | x |
| IPI00384397 | MYOSIN-REACTIVE IMMUNOGLOBULIN LIGHT CHAIN VARIABLE REGION | unknown | unknown | x |
| IPI00384404 | RHEUMATOID FACTOR RF-ET9 | unknown | unknown | x |
| IPI00385533 | ISOFORM 4 OF IODOTYROSINE DEHALOGENASE 1 | 5 | 12, 16, 20 | x |
| IPI00386134 | IG LAMBDA CHAIN V-I REGION BL2 | 3, 4, 6, 10 | 13, 16 | x |
| IPI00386136 | IG LAMBDA CHAIN V-VI REGION WLT | 3, 4, 6, 10 | 13, 16 | x |
| IPI00411765 | ISOFORM 2 OF 14-3-3 PROTEIN SIGMA | 2, 3, 4, 6, 7, 10 | 12, 21 | o |
| IPI00412407 | SERPIN PEPTIDASE INHIBITOR, CLADE B (OVALBUMIN), MEMBER 4, ISOFORM CRA_A | unknown | unknown | o |
| IPI00412701 | ZINC FINGER PROTEIN 517 | 4, 5, 10 | 11, 13 | x |
| IPI00418164 | ISOFORM 1 OF COILED-COIL DOMAIN-CONTAINING PROTEIN 30 | unknown | unknown | x |
| IPI00418780 | ISOFORM 3 OF REGULATOR OF G-PROTEIN SIGNALING 3 | 3, 4 | 12, 21 | x |
| IPI00419253 | ISOFORM 1 OF NCK-ASSOCIATED PROTEIN 5 | unknown | unknown | x |
| IPI00423464 | PUTATIVE UNCHARACTERIZED PROTEIN DKFZP686K03196 | unknown | unknown | o |
| IPI00433833 | UNCHARACTERIZED PROTEIN (SIMILAR TO THO COMPLEX SUBUNIT 3) | unknown | unknown | x |
| IPI00445778 | CDNA FLJ60358, HIGHLY SIMILAR TO REGULATOR OF G-PROTEIN SIGNALING 3 | unknown | unknown | x |
| IPI00451401 | ISOFORM 2 OF TRIOSEPHOSPHATE ISOMERASE | 4, 5, 7, 8 | 12, 16 | o |
| IPI00477003 | COILED-COIL DOMAIN-CONTAINING PROTEIN KIAA1407 | unknown | unknown | x |
| IPI00478115 | ISOFORM 2 OF DELETED IN MALIGNANT BRAIN TUMORS 1 PROTEIN | 1, 2, 3, 4, 6, 7 | 12, 15, 18 | o |
| IPI00479295 | GOLGIN SUBFAMILY A MEMBER 8-LIKE PROTEIN 2 | unknown | unknown | x |
| IPI00514669 | UNCHARACTERIZED PROTEIN (SIMILAR TO SH3 DOMAIN-BINDING GLUTAMIC ACID-RICH-LIKE PROTEIN 3) | unknown | unknown | x |
| IPI00556287 | PUTATIVE UNCHARACTERIZED PROTEIN | unknown | unknown | x |
| IPI00556391 | ACTIN-LIKE PROTEIN | unknown | unknown | x |
| IPI00641229 | IG ALPHA-2 CHAIN C REGION | 1, 2, 3, 4, 5, 6, 7, 10 | 13 | x |
| IPI00643174 | GATA-TYPE ZINC FINGER PROTEIN 1 | 4, 5, 7, 10 | 11, 12, 13, 19 | x |
| IPI00644220 | CDNA FLJ60139, HIGHLY SIMILAR TO HOMO SAPIENS HIV TAT SPECIFIC FACTOR 1 (HTATSF1), MRNA | unknown | unknown | x |
| IPI00646265 | 58 KDA PROTEIN (SIMILAR TO AMYLASE) | 5 | 12, 13, 16 | x |
| IPI00646771 | ISOFORM 4 OF DISKS LARGE HOMOLOG 2 | 3, 5, 7, 10 | 12, 13, 16 | x |
| IPI00657660 | UNCHARACTERIZED PROTEIN (SIMILAR TO HEMOGLOBIN SUBUNIT DELTA) | 6 | 13, 14 | o |
| IPI00657911 | UNCHARACTERIZED PROTEIN (SIMILAR TO HEMOGLOBIN SUBUNIT GAMMA-2) | 6 | 13, 14 | o |
| IPI00719452 | IGL@ PROTEIN | unknown | unknown | x |
| IPI00738655 | POTE ANKYRIN DOMAIN FAMILY MEMBER J | unknown | unknown | x |
| IPI00739539 | POTE ANKYRIN DOMAIN FAMILY MEMBER F | unknown | unknown | o |
| IPI00740545 | POTE ANKYRIN DOMAIN FAMILY MEMBER I | unknown | unknown | x |
| IPI00742996 | RING FINGER PROTEIN 214 | unknown | 13 | x |
| IPI00743194 | KAPPA LIGHT CHAIN VARIABLE REGION | unknown | unknown | x |
| IPI00745468 | SIMILAR TO TRANSMEMBRANE PROTEIN 90A | 1 | unknown | x |
| IPI00746516 | PEJVAKIN | unknown | unknown | x |
| IPI00759479 | ISOFORM 1 OF IQ MOTIF AND SEC7 DOMAIN-CONTAINING PROTEIN 2 | 3, 7 | unknown | x |
| IPI00759832 | ISOFORM SHORT OF 14-3-3 PROTEIN BETA/ALPHA | 3, 4, 5, 6, 7, 10 | 12 | o |
| IPI00761051 | ISOFORM 1 OF ERYTHROID DIFFERENTIATION-RELATED FACTOR 1 | 4, 5, 10 | 12 | x |
| IPI00783184 | IMMUNGLOBULIN HEAVY CHAIN VARIABLE REGION | unknown | unknown | x |
| IPI00783287 | IMMUNGLOBULIN HEAVY CHAIN VARIABLE REGION | unknown | unknown | x |
| IPI00783393 | IMMUNGLOBULIN HEAVY CHAIN VARIABLE REGION | unknown | unknown | x |
| IPI00783909 | IMMUNGLOBULIN HEAVY CHAIN VARIABLE REGION | unknown | unknown | x |
| IPI00794543 | CDNA FLJ75174, HIGHLY SIMILAR TO HOMO SAPIENS CALMODULIN 1 (PHOSPHORYLASE KINASE, DELTA), MRNA | unknown | unknown | x |
| IPI00816314 | PUTATIVE UNCHARACTERIZED PROTEIN DKFZP686I15196 | unknown | unknown | o |
| IPI00816799 | RHEUMATOID FACTOR D5 LIGHT CHAIN | unknown | unknown | x |
| IPI00827510 | HRV FAB 026-VL | unknown | unknown | x |
| IPI00827560 | HRV FAB N27-VL | unknown | unknown | x |
| IPI00827581 | VARIABLE IMMNOGLOBULIN ANTI-ESTRADIOL HEAVY CHAIN | unknown | unknown | x |
| IPI00827618 | VH6DJ PROTEIN | unknown | unknown | x |
| IPI00827690 | VH-3 FAMILY (VH26)D/J PROTEIN | unknown | unknown | x |
| IPI00827789 | HRV FAB N6-VL | unknown | unknown | x |
| IPI00827826 | COLD AGGLUTININ FS-2 L-CHAIN | unknown | unknown | x |
| IPI00828061 | ANTI-MUCIN1 HEAVY CHAIN VARIABLE REGION | unknown | unknown | x |
| IPI00828088 | VH6DJ PROTEIN | unknown | unknown | x |
| IPI00828099 | UGA8H | unknown | unknown | x |
| IPI00829590 | 13 KDA PROTEIN (SIMILAR TO IMMUNOGLOBULIN HEAVY VARIABLE 3–72) | unknown | unknown | x |
| IPI00829600 | 13 KDA PROTEIN (SIMILAR TO IMMUNOGLOBULIN KAPPA VAIRABLE 2D-26) | unknown | unknown | x |
| IPI00829701 | 13 KDA PROTEIN (SIMILAR TO IMMUNOGLOBULIN HEAVY VARIABLE 1–18) | unknown | unknown | x |
| IPI00829752 | MYOSIN-REACTIVE IMMUNOGLOBULIN HEAVY CHAIN VARIABLE REGION | unknown | unknown | x |
| IPI00829812 | 13 KDA PROTEIN (SIMILAR TO IMMUNOGLOBULIN HEAVY VARIABLE 3–13) | 3, 4, 6, 10 | 13, 16 | x |
| IPI00829841 | 13 KDA PROTEIN (SIMILAR TO IMMUNOGLOBULIN HEAVY VARIABLE 3–73) | unknown | unknown | x |
| IPI00829845 | IMMUNGLOBULIN HEAVY CHAIN VARIABLE REGION | unknown | unknown | o |
| IPI00829896 | HEMOGLOBIN LEPORE-BALTIMORE | unknown | unknown | o |
| IPI00829956 | RHEUMATOID FACTOR C6 LIGHT CHAIN | unknown | unknown | x |
| IPI00844168 | INTERFERON-INDUCIBLE PROTEIN AIM2 | 1, 2, 3, 4, 5, 6, 7, 10 | 11, 12 | x |
| IPI00844239 | IMMUNOBLOBULIN G1 FAB HEAVY CHAIN VARIABLE REGION | unknown | unknown | x |
| IPI00847618 | UNCHARACTERIZED PROTEIN (SIMILAR TO SH3 AND MULTIPLE ANKYRIN REPEAT DOMAINS PROTEIN 3) | 1, 3, 4, 6, 7, 9, 10 | 12, 13, 18 | x |
| IPI00853045 | ANTI-RHD MONOCLONAL T125 KAPPA LIGHT CHAIN | unknown | unknown | x |
| IPI00853455 | PROTEIN | unknown | unknown | x |
| IPI00853525 | UNCHARACTERIZED PROTEIN (Apolipoprotein A) | 4, 6 | 13 | o |
| IPI00853641 | UNCHARACTERIZED PROTEIN (SIMILAR TO HEMOGLOBIN SUBUNIT EPSILON) | 6 | 13, 14 | x |
| IPI00854644 | IG KAPPA CHAIN V-III REGION TI | 3, 4, 6, 10 | 13, 16 | x |
| IPI00854732 | SIMILAR TO MYOSIN-REACTIVE IMMUNOGLOBULIN HEAVY CHAIN VARIABLE REGION | unknown | unknown | x |
| IPI00854743 | IMMUNGLOBULIN HEAVY CHAIN VARIABLE REGION | unknown | unknown | x |
| IPI00854841 | MYOSIN-REACTIVE IMMUNOGLOBULIN HEAVY CHAIN VARIABLE REGION | unknown | unknown | x |
| IPI00871622 | PUTATIVE ZINC-ALPHA-2-GLYCOPROTEIN-LIKE 1 | unknown | unknown | x |
| IPI00871681 | PUTATIVE UNCHARACTERIZED PROTEIN ENSP00000344348 | unknown | unknown | x |
| IPI00878173 | CDNA FLJ39583 FIS, CLONE SKMUS2004897, HIGHLY SIMILAR TO ACTIN, ALPHA SKELETAL MUSCLE | unknown | unknown | x |
| IPI00878282 | 23 KDA PROTEIN (SIMILAR TO SERUM ALBUMIN) | 6 | unknown | o |
| IPI00884092 | ANTI-HER3 SCFV | unknown | unknown | x |
| IPI00888712 | PUTATIVE BETA-ACTIN-LIKE PROTEIN 3 | 6 | 11 | x |
| IPI00888954 | IMMUNGLOBULIN HEAVY CHAIN VARIABLE REGION | unknown | unknown | x |
| IPI00890703 | CRYOCRYSTALGLOBULIN CC1 KAPPA LIGHT CHAIN VARIABLE REGION | unknown | unknown | x |
| IPI00890754 | CRYOCRYSTALGLOBULIN CC1 HEAVY CHAIN VARIABLE REGION | unknown | unknown | x |
| IPI00892724 | CELL DIVISION CYCLE-ASSOCIATED 7-LIKE PROTEIN ISOFORM 2 | unknown | unknown | o |
| IPI00893862 | SFI1 HOMOLOG, SPINDLE ASSEMBLY ASSOCIATED | unknown | unknown | x |
| IPI00894523 | UNCHARACTERIZED PROTEIN (SIMILAR TO ACTIN, CYTOPLASMIC 1) | unknown | unknown | o |
| IPI00896380 | ISOFORM 2 OF IG MU CHAIN C REGION | 1, 2, 3, 4, 6, 7, 10 | 11, 12, 13 | x |
| IPI00903112 | CDNA FLJ36533 FIS, CLONE TRACH2004428, HIGHLY SIMILAR TO LACTOTRANSFERRIN | unknown | unknown | x |
| IPI00908653 | CDNA FLJ60121, HIGHLY SIMILAR TO UNC-13 HOMOLOG B | unknown | unknown | x |
| IPI00909257 | CDNA FLJ50149, HIGHLY SIMILAR TO HOMO SAPIENS ATTRACTIN-LIKE 1 (ATRNL1), MRNA | unknown | unknown | x |
| IPI00909649 | IG KAPPA CHAIN C REGION | 1, 2, 3, 4, 6, 7, 10 | 13, 16 | x |
| IPI00910380 | CDNA FLJ54278, HIGHLY SIMILAR TO SPARC-LIKE PROTEIN 1 | unknown | unknown | x |
| IPI00910779 | CDNA FLJ52141, HIGHLY SIMILAR TO 14-3-3 PROTEIN GAMMA | unknown | unknown | x |
| IPI00915869 | MALATE DEHYDROGENASE, CYTOPLASMIC ISOFORM 3 | 5 | 16 | x |
| IPI00916434 | ANTI-(ED-B) SCFV | unknown | unknown | x |
| IPI00921079 | CDNA FLJ61647, HIGHLY SIMILAR TO NUCLEOLAR COMPLEX PROTEIN 2 HOMOLOG | unknown | unknown | o |
| IPI00922693 | CDNA FLJ53662, HIGHLY SIMILAR TO ACTIN, ALPHA SKELETAL MUSCLE | unknown | unknown | o |
| IPI00924681 | UNCHARACTERIZED PROTEIN (SIMILAR TO ZINC FINGER PROTEIN 568) | unknown | unknown | x |
| IPI00924820 | SIMILAR TO KAPPA LIGHT CHAIN VARIABLE REGION | unknown | unknown | x |
| IPI00924948 | UNCHARACTERIZED PROTEIN (SIMILAR TO ZINC-ALPHA-2-GLYCOPROTEIN) | unknown | 13 | o |
| IPI00925547 | UNCHARACTERIZED PROTEIN (SIMILAR TO LACTOTRANSFERRIN) | 1, 2, 4, 10 | 13, 16 | x |
| IPI00930072 | PUTATIVE UNCHARACTERIZED PROTEIN DKFZP686E23209 | unknown | unknown | x |
| IPI00930124 | PUTATIVE UNCHARACTERIZED PROTEIN DKFZP686C11235 | unknown | unknown | o |
| IPI00930351 | HBBM FUSED GLOBIN PROTEIN | unknown | unknown | o |
| IPI00930404 | ISOFORM 2 OF KALLIKREIN-1 | 4 | 16 | o |
| IPI00930442 | PUTATIVE UNCHARACTERIZED PROTEIN DKFZP686M24218 | 1, 2, 3, 4, 6, 7, 10 | 13, 16 | x |
| IPI00937669 | 10 KDA PROTEIN | unknown | unknown | x |
| IPI00939278 | UNCHARACTERIZED PROTEIN (SIMILAR TO ZINC-ALPHA-2-GLYCOPROTEIN) | unknown | 13 | o |
| IPI00940245 | IMMUNOGLOBULIN HEAVY CHAIN VARIANT | unknown | unknown | x |
| IPI00940727 | 10 KDA PROTEIN | unknown | unknown | x |
| IPI00943106 | UNCHARACTERIZED PROTEIN (SIMILAR TO GOLGIN SUBFAMILY A MEMBER 8N) | unknown | unknown | x |
| IPI00953352 | ISOFORM 10 OF PROTEIN SFI1 HOMOLOG | 5, 10 | 12 | x |
| IPI00956140 | HCG1652138, ISOFORM CRA_A | unknown | unknown | x |
| IPI00956602 | ANTI-STREPTOCOCCAL/ANTI-MYOSIN IMMUNOGLOBULIN LAMBDA LIGHT CHAIN VARIABLE REGION | unknown | unknown | x |
| IPI00964049 | UNCHARACTERIZED PROTEIN (SIMILAR TO SPECKL-TYPE POZ PROTEIN) | unknown | unknown | x |
| IPI00965653 | 12 KDA PROTEIN (SIMILAR TO IMMUNOGLOBULIN KAPPA VARIABLE 2D-40) | 3, 4, 6, 10 | 13, 16 | x |
| IPI00966829 | 69 KDA PROTEIN (SIMILAR TO SERUM ALBUMIN) | 6 | unknown | o |
| IPI00966961 | PROTEIN (SIMILAR TO CENTROSOMAL PROTEIN 192 KDA) | 7, 10 | unknown | x |
| IPI00969456 | IGKC PUTATIVE UNCHARACTERIZED PROTEIN | unknown | unknown | x |
| IPI00969620 | LIGHT CHAIN FAB | unknown | unknown | x |
| IPI00972963 | LAMBDA LIGHT CHAIN OF HUMAN IMMUNOGLOBULIN SURFACE ANTIGEN-RELATED PROTEIN | unknown | unknown | x |
| IPI00973016 | UBIQUITOUSLY TRANSCRIBED TETRATRICOPEPTIDE REPEAT PROTEIN Y-LINKED TRANSCRIPT VARIANT 27 | unknown | unknown | x |
| IPI00973032 | V1-17 PROTEIN | unknown | unknown | x |
| IPI00973424 | IGLC1 PUTATIVE UNCHARACTERIZED PROTEIN | unknown | unknown | x |
| IPI00973474 | PUTATIVE UNCHARACTERIZED PROTEIN (SIMILAR TO IG GAMMA-3 CHAIN C REGION) | 1, 2, 3, 4, 6, 7, 10 | 13, 16 | o |
| IPI00973513 | IMUNOGLOBULIN HEAVY CHAIN | unknown | unknown | x |
| IPI00973531 | IGLC2 PUTATIVE UNCHARACTERIZED PROTEIN | unknown | unknown | x |
| IPI00974544 | ISOFORM SV OF 14-3-3 PROTEIN EPSILON | 1, 2, 3, 4, 6, 7, 10 | 12 | o |
| IPI00976299 | UNCHARACTERIZED PROTEIN (SIMILAR TO UTEROGLOBIN) | 3 | unknown | o |
| IPI00976559 | UNCHARACTERIZED PROTEIN (SIMILAR TO DIPHTHAMIDE BIOSYNTHESIS PROTEIN 2) | 4 | unknown | x |
| IPI00976928 | SIMILAR TO MYOSIN-REACTIVE IMMUNOGLOBULIN HEAVY CHAIN VARIABLE REGION | unknown | unknown | x |
| IPI00977221 | HYPOTHETICAL PROTEIN LOC100291917 | unknown | unknown | x |
| IPI00977405 | SIMILAR TO IG KAPPA CHAIN V-III REGION VG PRECURSOR | 3, 4, 6, 10 | 13, 16 | x |
| IPI00977733 | SIMILAR TO VH-7 FAMILY (N54P3)D/J PROTEIN | unknown | unknown | x |
| IPI00977788 | SIMILAR TO HEPATITIS B VIRUS RECEPTOR BINDING PROTEIN | unknown | unknown | x |
| IPI00978208 | PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A-LIKE | 4, 5, 6, 7, 10 | 12, 13, 16 | o |
| IPI00978930 | IG ALPHA-1 CHAIN C REGION | 1, 2, 3, 4, 5, 6, 7, 10 | 13 | x |
| IPI00979730 | SIMILAR TO IG KAPPA CHAIN V-II REGION GM607 PRECURSOR | 3, 4, 6, 10 | 13, 16 | x |
| IPI00980227 | CONSERVED HYPOTHETICAL PROTEIN MUC5B | 1, 2, 4, 10 | 12 | x |
| IPI00981659 | SIMILAR TO COLD AGGLUTININ FS-1 H-CHAIN | unknown | unknown | x |
| IPI00982101 | UNCHARACTERIZED PROTEIN | unknown | 12 | o |
| IPI00983257 | NEURONAL ACETYLCHOLINE RECEPTOR ALPHA-4 SUBUNIT | unknown | unknown | x |
| IPI00983475 | SIMILAR TO HEPATITIS B VIRUS RECEPTOR BINDING PROTEIN | unknown | unknown | x |
| IPI00985150 | 11 KDA PROTEIN | unknown | unknown | x |
| IPI00985211 | SIMILAR TO VH-3 FAMILY (VH26)D/J PROTEIN | unknown | unknown | x |
| IPI00985251 | PROTEIN (SIMILAR TO UPF0317 PROTEIN C14ORF159) | unknown | unknown | x |
| IPI01010056 | 58 KDA PROTEIN (SIMILAR TO AMYLASE) | 5 | 12, 13, 16 | x |
| IPI01011189 | MUC5B UNCHARACTERIZED PROTEIN | unknown | unknown | x |
| IPI01011784 | UNCHARACTERIZED PROTEIN (SIMILAR TO E3 UBIQUITIN-PROTEIN LIGASE MDM2) | unknown | unknown | o |
| IPI01012504 | 6-PHOSPHOGLUCONATE DEHYDROGENASE, DECARBOXYLATING | 4, 5 | 16 | o |
| IPI01013214 | RHEUMATOID FACTOR RF-IP14 | unknown | unknown | x |
| IPI01014621 | UNCHARACTERIZED PROTEIN (SIMILAR TO ZINC FINGER PROTEIN 705A) | 4, 5, 10 | 11 | x |
| IPI01014804 | UNCHARACTERIZED PROTEIN | unknown | unknown | x |
| IPI01015266 | PUTATIVE UNCHARACTERIZED PROTEIN DKFZP686O16217 | unknown | unknown | x |
| IPI01017938 | IG LAMBDA-6 CHAIN C REGION | 1, 2, 3, 4, 6, 7, 10 | 13, 16 | x |
| IPI01018060 | IG LAMBDA-3 CHAIN C REGIONS | 1, 2, 3, 4, 6, 7, 10 | 13, 16 | x |
| IPI01018161 | PYRUVATE KINASE ISOZYMES M1/M2 ISOFORM C | 1, 2, 4, 5, 7, 8, 10 | 11, 12, 13, 16 | o |
| IPI01018257 | HYPOTHETICAL PROTEIN (SIMILAR TO POLYMERIC IMMUNOGLOBULIN RECEPTOR) | unknown | unknown | o |
| IPI01018712 | ACTA2 PROTEIN | unknown | unknown | x |
| IPI01018716 | TOSPEAK-5 | unknown | unknown | x |
| IPI01018887 | UBIQUITOUSLY TRANSCRIBED TETRATRICOPEPTIDE REPEAT PROTEIN Y-LINKED TRANSCRIPT VARIANT 8 | unknown | unknown | x |
| IPI01018897 | MUCIN-5AC | unknown | unknown | o |
| IPI01018949 | CDNA FLJ51266, HIGHLY SIMILAR TO VITRONECTIN | unknown | unknown | x |
| IPI01022126 | UNCHARACTERIZED PROTEIN (SIMILAR TO KAT8 REGULATORY NSL COMPLEX SUBUNIT 2) | unknown | unknown | x |
| IPI01022319 | UNCHARACTERIZED PROTEIN (SIMILAR TO TWINFILIN-1) | unknown | unknown | x |
| IPI01022662 | UNCHARACTERIZED PROTEIN (SIMILAR TO CARBONIC ANHYDRASE 6) | 5 | 13, 16 | x |
| IPI01022818 | 15 KDA PROTEIN (SIMILAR TO LYSOZYME) | 1, 2, 4 | unknown | o |
| IPI01022820 | AMYLOID LAMBDA 6 LIGHT CHAIN VARIABLE REGION SAR | unknown | unknown | x |
| IPI01024840 | PROTEIN (SIMILAR TO X-LINKED RETINITIS PIGMENTOSA GTPASE REGULATOR-INTERACTING PROTEIN 1) | 7 | unknown | x |
| IPI01025882 | MYOSIN-REACTIVE IMMUNOGLOBULIN LIGHT CHAIN VARIABLE REGION | unknown | unknown | x |
| IPI01026053 | PROTEIN (SIMILAR TO EPIDIDYMAL SECRETORY PROTEIN E1) | unknown | unknown | x |
For the determination of the inter-platform variability in the nLC-Q-IMS-TOF system used in this study and the validation of the identities of proteins, especially, the distinct Korean WS proteins, results of the analyses of the pooled Korean WS sample by the nLC-Q-IMS-TOF system and a nLC-Q-orbitrap system were compared. As a result, 141 and 208 proteins were identified from the nLC-Q-TOF platform and the nLC-Q-orbitrap platform, respectively, and 98 out of 141 proteins (69.5%) from the nLC-Q-TOF platform were overlapped with those from the nLC-Q-orbitrap platform. Among proteins identified in the pooled sample, 130 proteins from the nLC-Q-TOF platform and 147 proteins from the nLC-Q-orbitrap platform were found to be within the Korean WS proteome index. Additionally, among those proteins overlapped with the Korean WS proteome, 22 out of 130 proteins (16.9%) and 29 out of 147 proteins (19.7%) were confirmed to belong to the distinct Korean WS proteins from the nLC-Q-TOF platform and the nLC-Q-orbitrap platform, respectively. Finally, the portion of the distinct proteins from the nLC-Q-IMS-TOF platform, which overlaps with those from the nLC-Q-orbitrap plarform was 68.2% (S3 and S4 Tables and S1 Fig).
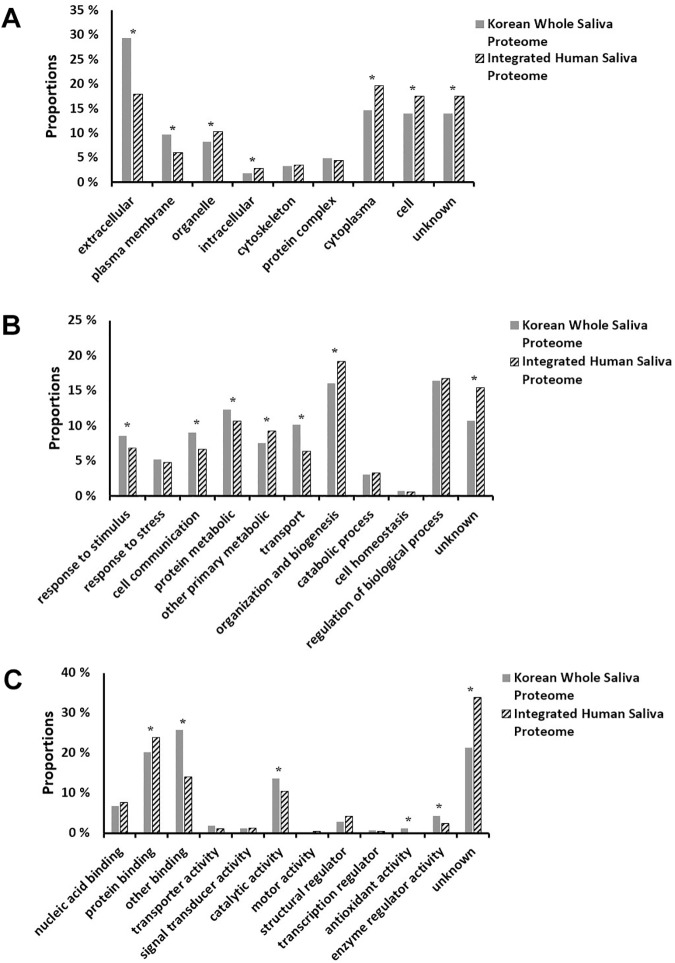
In addition to the comparison of protein identities, GO annotation in terms of cellular component, biological process, and molecular function between the Korean WS proteome and the integrated human saliva proteome was compared (Fig 3). First, in GO cellular component categories, the Korean WS proteome was significantly over-represented in extracellular space and the plasma membrane but under-represented in organelle, intracellular, cytoplasma, and the cell compared to the integrated human saliva proteome (p < 0.05). GO biologic process categories also showed higher portions of proteins for response to stimulus, cell communication, protein metabolism, and transport in the Korean WS proteome than those in the integrated human saliva proteome (p < 0.05). However, the opposite tendency was observed in proteins for other primary metabolic and organization and biogenesis (p < 0.05). Finally, in the case of GO molecular function categories, over-representation of the Korean WS proteome was observed in other binding, catalytic activity, antioxidant activity, and enzyme regulatory activity with under-representation in protein binding compared to the integrated human saliva proteome were found (p < 0.05). Allocation of proteins observed in the Korean WS proteome according to their GO annotation can be found in S5–S7 Tables.
Fig 3.
Relative allocation and comparison of proteins observed in the Korean whole saliva proteome and the integrated human saliva proteome according to their gene ontology annotations in terms of cellular component (A), biological process (B), and molecular function (C). *, p < 0.05.
Distinct Korean WS proteins and diseases
To evaluate the clinical applicability of ethnicity-specific human saliva proteome, 226 proteins observed in the Korean WS proteome, but not in the integrated human saliva proteome, were searched against the Database of disease-related biomarkers [24]. As shown in Table 1 and S2 Table, 50 of 226 distinct Korean WS proteins (22.1%) were found to be disease biomarker candidates. Also, Table 2 and S2 Table indicate that 7–21 of these distinct Korean WS proteins are probably associated with individual conditions representing the top 10 deadliest diseases in South Korea, 2015 (cerebrovascular disease, lung cancer, ischemic heart disease, liver cancer, diabetes mellitus, stomach cancer, colorectal cancer, pancreatic cancer, hypertension, and dementia) [25].
Table 2. Distinct Korean WS proteins probably associated with the top 10 deadliest diseases in South Korea, 2015.
| Rank | Disease (A) | The distinct Korean whole saliva proteins probably associated with A |
|---|---|---|
| 1 | Cerebrovascular disease | IPI00000149, IPI00003842, IPI00022434, IPI00216773, IPI00878282, IPI00966829, IPI00150554, IPI00892724, IPI00218573, IPI00220644, IPI01018161, IPI00305457, IPI00853525, IPI00930404 |
| 2 | Lung cancer | IPI00000149, IPI00003842, IPI00020019, IPI00022434, IPI00216773, IPI00878282, IPI00966829, IPI00103636, IPI00150554, IPI00892724, IPI00220644, IPI01018161, IPI00376170, IPI00978208, IPI00478115, IPI00976299, IPI01018257, IPI01011784, IPI01018897, IPI01022818, IPI00930404 |
| 3 | Ischemic heart disease | IPI00000149, IPI00020019, IPI00022434, IPI00216773, IPI00878282, IPI00966829, IPI00103636, IPI00150554, IPI00892724, IPI00218573, IPI00305457, IPI00376170, IPI00978208, IPI00411765, IPI00451401, IPI00739539, IPI00853525, IPI01011784, IPI00930404 |
| 4 | Liver cancer | IPI00000149, IPI00022434, IPI00216773, IPI00878282, IPI00966829, IPI00150554, IPI00892724, IPI00181753, IPI00220644, IPI01018161, IPI00305457, IPI00451401, IPI00739539, IPI00853525, IPI01018257, IPI01011784, IPI01018897, IPI01022818 |
| 5 | Diabetes mellitus | IPI00000149, IPI00018214, IPI00020019, IPI00022434, IPI00216773, IPI00878282, IPI00966829, IPI00150554, IPI00892724, IPI00305457, IPI00853525, IPI00976299, IPI00930404 |
| 6 | Stomach cancer | IPI00022434, IPI00216773, IPI00878282, IPI00966829, IPI00305457, IPI01018897, IPI01022818 |
| 7 | Colorectal cancer | IPI00000149, IPI00018214, IPI00019226, IPI00022434, IPI00216773, IPI00878282, IPI00966829, IPI00026126, IPI00150554, IPI00892724, IPI00220644, IPI01018161, IPI00739539, IPI01018257, IPI01011784, IPI01018897, IPI01022818 |
| 8 | Pancreatic cancer | IPI00022434, IPI00216773, IPI00878282, IPI00966829, IPI00103636, IPI00220644, IPI01018161, IPI00411765, IPI01018897, IPI01022818, IPI00930404 |
| 9 | Hypertension | IPI00018214, IPI00022434, IPI00216773, IPI00878282, IPI00966829, IPI00150554, IPI00892724, IPI00218573, IPI00657911, IPI00853525, IPI00976299, IPI01022818, IPI00930404 |
| 10 | Dementia | IPI00000149, IPI00003842, IPI00022434, IPI00216773, IPI00878282, IPI00966829, IPI00150554, IPI00892724, IPI00181753, IPI00218573, IPI00220644, IPI01018161, IPI00305457, IPI00376170, IPI00978208, IPI00739539, IPI00853525, IPI00924948, IPI00939278, IPI00930351, IPI00930404 |
Discussion
As the initial step to determine ethnic difference of human saliva proteome, the Korean WS proteome was constructed for the first time due to the fact that Korea is the most ethnically homogenous country in the world [26]. A total of 480 proteins are catalogued in the Korean WS proteome (S1 Table), including most of commonly observed saliva proteins (amylase, cystantins, acidic proline rich proteins, basic proline rich proteins, mucins, lactotransferrin, carbonic anhydrase, lysozymes, peroxidases, albumin, and statherines) [11, 27]. This observation indicated that the analytical method employed in the present study was performed properly. However, three groups of common saliva proteins (thymosins, defensins, and histatins) were not observed in the present study. Although an exact explanation on their absence cannot be provided with certainty, loss during sample preparation, the under-sampling issue of mass spectrometry brought by the complexity of a sample, their cleavage into small peptides, and/or binding of the resulting peptides to tissues may contribute to their absence [11, 27, 28].
For the actual determination of ethnic differences in human saliva proteome, the present Korean WS protein list was compared to the integrated human saliva protein list in a couple of ways. First, comparison of protein identities in each list revealed that 47.1% (226 out of 480) of proteins were unique in the Korean WS proteome. Discovering a large portion of Korean WS unique proteins from the Korean WS proteome was expected, because similar portion to that (54.1%, 100 out of 185 proteins) was already reported from distinct Korean plasma proteins compared to human plasma proteome [18]. However, there is a possibility of identifying common proteins for the first time by employing different analytical techniques, which would weaken the possibility of the connection between the distinct Korean WS proteins and ethnic differences in human saliva proteome. Thus, for the determination of the inter-platform variability in the nLC-Q-IMS-TOF system used in this study and the validation of the identities of proteins (especially, the distinct Korean WS proteins) simultaneously, results of the analyses of the pooled Korean WS sample by the nLC-Q-IMS-TOF system and a nLC-Q-orbitrap system, a platform widely used for proteomics were compared. As a result, 141 and 208 proteins were identified from the nLC-Q-TOF platform and the nLC-Q-orbitrap platform, respectively, and 98 out of 141 proteins (69.5%) from the nLC-Q-TOF platform were overlapped with those from the nLC-Q-orbitrap platform (S3 and S4 Tables and S1 Fig). If about 70–80% of the repeatability (the inner-system comparison) and about 60–80% of the reproducibility (the inter-system including inter-platform comparison) of a standardized analysis platform in protein identification are considered [29], no significant influence of the inter-platform variability in the nLC-Q-IMS-TOF system as well as the high credibility of protein identities in the Korean WS proteome can be urged. Among proteins identified in the pooled sample, 130 proteins from the nLC-Q-TOF platform and 147 proteins from the nLC-Q-orbitrap platform were found to be within the Korean WS proteome index. Additionally, among those proteins overlapped with the Korean WS proteome, 22 out of 130 proteins (16.9%) and 29 out of 147 proteins (19.7%) were confirmed to belong to the distinct Korean WS proteins from the nLC-Q-TOF platform and the nLC-Q-orbitrap platform, respectively (S3 and S4 Tables and S1 Fig). The numbers of every type of proteins identified from the analyses of the pooled sample by using each platform are smaller than the counter parts of the Korean WS proteome, likely the consequence of the dilution of individual proteins by saliva pooling and the analyses of a single sample. However, the portion of the distinct proteins from the nLC-Q-IMS-TOF platform, which overlaps with those from the nLC-Q-orbitrap platform, is still close to 70% (68.2%). Thus, no significant influence of the inter-platform variability in our system is observed, lending high credibility of the protein identities in the Korean WS proteome. Interestingly, the number of proteins from the nLC-Q-orbitrap platform, which belong to distinct protein in Korean WS is larger than that from the nLC-Q-IMS-TOF platform (22 proteins from the nLC-Q-IMS-TOF platform vs. 29 proteins from the nLC-Q-orbitrap platform; S3 and S4 Tables and S1 Fig). This observation provides additional evidence to support the high credibility of the identities of the distinct Korean WS proteins. Therefore, since the existence of the distinct Korean WS proteins are more confident and the possibility of their identification by the inter-platform variability can be significantly excluded, ethnic differences in the human saliva proteome, especially in the Korean WS proteome become more evident.
While it was observed that the nLC-Q-IMS-TOF system of this study did not bring higher performance than other proteomics platforms due to the relatively small number of proteins identified, the identification of the distinct proteins confirmed that the nLC-Q-IMS-TOF system still has good performance for the identification of distinct Korean WS proteins. Actually, to build a global protein list, most proteomic studies have employed multi-dimensional proteomics technique to include as many as possible proteins in their lists [1, 7–11, 30, 31]. However, such multi-dimensional proteomics technique demands enormous analysis time and computing power for protein identification. Therefore, we chose the combination of nLC-Q-TOF (a single dimensional technique) and IMS (an additional technique to separate ions based on their different mobility in a carrier gas) “on-line” instead of using the conventional multi-dimensional technique [28, 32]. To the best of our knowledge, this is the first study that applies IMS to saliva proteomics.
From comparison of GO annotations between the Korean WS proteome and the integrated human saliva proteome, some categories in the Korean WS proteome showed over-representation or under-representation (Fig 3). Regarding their applications to biomarker-related studies, over-represented categories might be more important than under-represented ones due to the probability of finding more meaningful information from more proteins belonging to over-represented categories. In the present study, over-represented GO categories in the Korean WS proteome are as follows: extracellular and plasma membrane of cellular components (Fig 3A), response to stimulus, cell communication, protein metabolic, and transport of biological processes (Fig 3B), and other binding, catalytic activity, antioxidant activity, and enzyme regulatory activity of molecular function (Fig 3C). Interestingly, most of them can provide substantial information on diseases due to their connectivity to disease-related features such as extracellular secretion for biological function (the extracellular category of cellular component), the defense system of the body (the response to stimulus category of biological processes), cellular signal transduction (the cell communication category of biological processes), chemical reactions and pathways involving a specific proteins (the protein metabolic category of biological processes), positioning of a substance or cellular entity (the transport category of biological processes), non-covalent interaction of a non-protein molecule with specific site(s) on another molecule (the other binding category of the biological processes), catalysis of a biochemical reaction (the catalytic activity category of the biological processes), inhibition of oxidation (the antioxidant activity category of the biological processes), and/or modulation (by direct binding) of the activity of an enzyme (enzyme regulator activity category for molecular function) [1, 2, 10, 33]. Additionally, over-representation of protein metabolic and catalytic activity categories in the Korean WS proteome compared with the integrated human saliva proteome may be consistent with its larger portion of proteins with molecular weight of less than 60 kDa (82.3%, Fig 1A), partially resulting from the cleavage of higher-molecular-weight proteins, than that of Yan et al.'s report (68%) [1]. In line with these findings, our results suggest another clue to discover ethnic differences in the human saliva proteome and the possibility of using such difference for early diagnosis and/or prognosis of diseases.
For further evaluation of the clinical applicability of ethnicity-specific human saliva proteome, 226 distinct proteins observed in Korean WS, but not in other human saliva, were searched through the Database of disease-related biomarkers. As a result, 22.1% (50 out of 226) of these distinct proteins were found to be disease biomarker candidates (Table 1 and S2 Table), firmly supporting the probable value of using ethnicity-specific human saliva proteome for disease biomarker applications. Also, all top 10 deadliest diseases in South Korea, 2015 (cerebrovascular disease, lung cancer, ischemic heart disease, liver cancer, diabetes mellitus, stomach cancer, colorectal cancer, pancreatic cancer, hypertension, and dementia) are found to have at least 7 disease biomarker candidates which belong to the distinct Korean WS proteins (Table 2 and S2 Table) [25]. The total number of distinct Korean WS proteins probably associated with the top 10 deadliest diseases in South Korea is 35, representing 70.0% (35 out of 50) of disease biomarker candidate proteins among distinct Korean WS proteins (Tables 1 and 2 and S2 Table). Thus, this result clearly shows that ethnicity-specific human saliva proteins have diagnostic potential for diseases highly prevalent in that ethnic group.
However, this study has a couple of limitations. First, as mentioned above, it did not employ any multi-dimensional separation technique, and, as a result, a relatively small number of proteins was catalogued in the Korean WS proteome index. Interestingly, however, its limited performance must have played an important role in supporting ethnicity-related differences in human saliva, because it did not seem to produce any significant platform-specific performance, the source of the inter-platform variability. Also, since WS samples were collected from only eleven young male adult volunteers, there would be concerns of gender bias as well as a lack of representativeness in the results because of the narrow age range of the participants. Thus, the expansion of the Korean WS proteome by analyzing more samples, including female WS and a broader range of participant ages, by using the nLC-Q-IMS-TOF system or a multi-dimensional proteomics technique is expected in the near future.
Conclusions
The Korean WS proteome catalogue indexing 480 proteins was built and characterized from nLC-Q-IMS-TOF analyses of WS samples collected from eleven healthy Korean male adult volunteers in this study for the first time. From comparison of the Korean WS proteome with the integrated human saliva proteome in terms of protein identities and GO annotations, evidences strongly support ethnic difference in human saliva proteome. Additionally, the potential value of ethnicity-specific human saliva proteins as biomarkers for diseases highly prevalent in that ethnic group was confirmed by finding 35 distinct Korean WS proteins probably associated with the top 10 deadliest diseases in South Korea. Finally, the present Korean WS protein list can serve as the first level reference for future proteomic studies including disease biomarker studies on Korean saliva.
Supporting information
Among multiple results on a certain protein from different sample and replicate analyses, only one with the highest PLGS score was selected for this table.
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Total proteins identified (A). Proteins which belong to the Korean whole saliva proteome (B). Proteins which belong to the distinct Korean whole saliva proteins (C).
(PPTX)
(DOCX)
Acknowledgments
The authors would like to thank Ms. Erica Oh (Waters Korea), Mr. Paul Park (Waters Korea), Mr. Hyo Chun Lee (Dankook University), and Mr. Dong Yoon Kim (Dankook University) for their technical support.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by a grant (2014R1A1A1004674 to YSC) of the National Research Foundation of Korea funded by the Korean Government. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Yan W, Apweiler R, Balgley BM, Boontheung P, Bundy JL, Cargile BJ, et al. Systematic comparison of the human saliva and plasma proteomes. Proteomics Clin Appl. 2009; 3: 116–134. doi: 10.1002/prca.200800140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loo JA, Yan W, Ramachandran P, Wong DT. Comparative human salivary and plasma proteomes. J Dent Res. 2010; 89: 1016–1023. doi: 10.1177/0022034510380414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mireya GB, Lu B, Liao L, Xu T, Bedi G, Melvin JE, et al. Characterization of the human submandibular/sublingual saliva glycoproteome using lectin affinity chromatography coupled to multidimensional protein identification technology. J Proteome Res. 2011; 10: 5031–5046. doi: 10.1021/pr200505t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfaffe T, Justin CW, Beyerlein P, Konstner K, Punyadeera C. Diagnostic potential of saliva: current state and future applications. Clin Chem. 2011; 57: 675–687. doi: 10.1373/clinchem.2010.153767 [DOI] [PubMed] [Google Scholar]
- 5.Csősz É, Kalló G, Márkus B, Deák E, Csutak A, Tőzsér J. Quantitative body fluid proteomics in medicine—A focus on minimal invasiveness. J Proteomics. 2017; 153: 30–43. doi: 10.1016/j.jprot.2016.08.009 [DOI] [PubMed] [Google Scholar]
- 6.Corstjens PLAM, Abrams WR, Malamud D. Detecting viruses by using salivary diagnostics. J Am Dent Assoc. 2012; 143: 12S–18S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie H, Rhodus NL, Griffin RJ, Carlis JV, Griffin TJ. A catalogue of human saliva proteins identified by free flow electrophoresis-based peptide separation and tandem mass spectrometry. Mol Cell Proteomics. 2005; 4: 1826–1830. doi: 10.1074/mcp.D500008-MCP200 [DOI] [PubMed] [Google Scholar]
- 8.Fang X, Yang L, Wang W, Song T, Lee CS, Devoe D, et al. Comparison of electrokinetics-based multidimensional separations coupled with electrospray ionization-tandem mass spectrometry for characterization of human salivary proteins. Anal Chem. 2007; 79: 5785–5792. doi: 10.1021/ac070611a [DOI] [PubMed] [Google Scholar]
- 9.Denny P, Hagen FK, Hardt M, Liao L, Yan W, Arellanno M, et al. The proteomes of human parotid and submandibular/sublingual gland salivas collected as the ductal secretions. J Proteome Res. 2008; 7: 1994–2006. doi: 10.1021/pr700764j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bandhakavi S, Stone MD, Onsongo G, Van Riper SK, Griffin TJ. A dynamic range compression and three-dimensional peptide fractionation analysis platform expands proteome coverage and the diagnostic potential of whole saliva. J Proteome Res. 2009; 8: 5590–5600. doi: 10.1021/pr900675w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sivadasan P, Gupta MK, Sathe GJ, Balakrishnan L, Palit P, Gowda H, et al. Human salivary proteome—a resource of potential biomarkers for oral cancer. J Proteomics. 2015; 127: 89–95. doi: 10.1016/j.jprot.2015.05.039 [DOI] [PubMed] [Google Scholar]
- 12.UCLA Dental Research Institute. Human Salivary Proteome Central Repository; 2005 [cited 2017 Feb 24]. Available from: http://www.skb.ucla.edu/cgi-bin/hspmscgi-bin/welcome_c.cgi/.
- 13.Shanghai Institutes for Biological Sciences. Sys-Body Fluid Database; 2008 [cited 2017 Feb 24]. Available from: http://lifecenter.sgst.cn/bodyfluid/fluid.jsp?bf=Saliva/.
- 14.Wang Q, Yu Q, Lin Q, Duan Y. Emerging salivary biomarkers by mass spectrometry. Clin Chim Acta. 2015; 438: 214–221. doi: 10.1016/j.cca.2014.08.037 [DOI] [PubMed] [Google Scholar]
- 15.Kuehl MN, Rodriguez H, Burkhardt BR, Alman AC. Tumor necrosis factor-α, matrix-metalloproteinases 8 and 9 levels in the saliva are associated with increased hemoglobin A1c in type 1 diabetes subjects. PLOS ONE. 2015; 10: e0125320 doi: 10.1371/journal.pone.0125320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacobs R, Maasdorp E, Malherbe S, Loxton AG, Kim S, der Spuy GV, et al. Diagnostic potential of novel salivary host biomarkers as candidates for the immunological diagnosis of tuberculosis disease and monitoring of tuberculosis treatment response. PLOS ONE. 2016; 11: e0160546 doi: 10.1371/journal.pone.0160546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Camisasca DR, Gonçalves LDR, Soares MR, Sandimb V, Nogueira FCS, Garcia CHS, et al. A proteomic approach to compare saliva from individuals with and without oral leukoplakia. J Proteomics. 2017; 151: 43–52. doi: 10.1016/j.jprot.2016.07.029 [DOI] [PubMed] [Google Scholar]
- 18.Jeong SK, Lee EY, Cho JY, Lee HJ, Jeong AS, Cho SY, et al. Data management and functional annotation of the Korean reference plasma proteome. Proteomics. 2010; 10: 1250–1255. doi: 10.1002/pmic.200900371 [DOI] [PubMed] [Google Scholar]
- 19.Kim CX, Bailey KR, Klee GG, Ellington AA, Liu G, Mosley TH Jr, et al. Sex and ethnic differences in 47 candidate proteomic markers of cardiovascular disease: the mayo clinic proteomic markers of arteriosclerosis study. PLOS ONE. 2010; 5: e9065 doi: 10.1371/journal.pone.0009065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi YS, Hou S, Choe LH, Lee KH. Targeted human cerebrospinal fluid proteomics for the validation of multiple Alzheimer’s disease biomarker candidates. J Chromatogr B. 2013; 930: 129–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi YS, Lee KH. Multiple reaction monitoring assay based on conventional liquid chromatography and electrospray ionization for simultaneous monitoring of multiple cerebrospinal fluid biomarker candidates for Alzheimer’s disease. Arch Pharm Res. 2016; 39: 390–397. doi: 10.1007/s12272-015-0663-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Princeton University. Generic gene ontology (GO) term mapper. 2017. Available from: http://go.princeton.edu/cgi-bin/GOTermMapper/.
- 23.GraphPad Software. QuickCalcs. 2017. Available from: https://graphpad.com/quickcalcs/contingency1.cfm/.
- 24.IMIM-UPF. Database of disease-related biomarkers; 2016 [cited 2017 Feb 24]. Available from: http://ibi.imim.es/biomarkers/.
- 25.Korean Statistical Information Service. Statistical database: cause of death in 2015; 2016 [cited 2017 Feb 24]. Available from: http://kosis.kr/eng/statisticsList/statisticsList_01List.jsp?vwcd=MT_ETITLE&parentId=D/.
- 26.Fearon JD. Ethnic and cultural diversity by country. J Econ Growth. 2003; 8: 195–222. [Google Scholar]
- 27.Castagnola M, Cabras T, Vitali A, Sanna MT, Messana I. Biotechnological implications of the salivary proteome. Trends Biotechnol. 2011; 29: 409–418. doi: 10.1016/j.tibtech.2011.04.002 [DOI] [PubMed] [Google Scholar]
- 28.Angel TE, Aryal UK, Hengel SM, Baker ES, Kelly RT, Robinson EW, et al. Mass spectrometry based proteomics: existing capabilities and future directions. Chem Soc Rev. 2012; 41: 3912–3928. doi: 10.1039/c2cs15331a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tabb DL, Vega-Montoto L, Rudnick PA, Variyath AM, Ham AJ, Bunk DM, et al. Repeatability and reproducibility in proteomic identifications by liquid chromatography-tandem mass spectrometry. J Proteome Res. 2010; 9: 761–776. doi: 10.1021/pr9006365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Righetti PG, Castagna A, Antonioli P, Boschetti E. Prefractionation techniques in proteome analysis: the mining tools of the third millennium. Electrophoresis. 2005; 26: 297–319. doi: 10.1002/elps.200406189 [DOI] [PubMed] [Google Scholar]
- 31.Choi Y.S. Reaching for the deep proteome: recent nano liquid chromatography coupled with tandem mass spectrometry-based studies on the deep proteome. Arch Pharm Res. 2012; 35: 1861–1870. doi: 10.1007/s12272-012-1102-y [DOI] [PubMed] [Google Scholar]
- 32.Shliaha PV, Bond NJ, Gatto L, Lilley KS. Effects of traveling wave ion mobility separation on data independent acquisition in proteomics studies. J Proteome Res. 2013; 12: 2323–2339. doi: 10.1021/pr300775k [DOI] [PubMed] [Google Scholar]
- 33.University of Southern California. PANTHER classification system. 2017. Available from: www.pantherdb.org/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Among multiple results on a certain protein from different sample and replicate analyses, only one with the highest PLGS score was selected for this table.
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Total proteins identified (A). Proteins which belong to the Korean whole saliva proteome (B). Proteins which belong to the distinct Korean whole saliva proteins (C).
(PPTX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.