Abstract
BACKGROUND
Deficiency of the lysosomal enzyme acid α-glucosidase (GAA) causes Pompe disease. Newborn screening for Pompe disease is ongoing, and improved methods for distinguishing affected patients from those with pseudodeficiency, especially in the Asian population, would substantially reduce the number of patient referrals for clinical follow-up.
METHODS
We measured the enzymatic activity of GAA in dried blood spots on newborn screening cards (DBS) using a tandem mass spectrometry (MS/MS) assay. The assay displayed a relatively large analytical range compared to the fluorimetric assay with 4-methylumbelliferyl-α-glucoside. DBS from newborns confirmed to have infantile-onset Pompe disease (IOPD, n=11) or late-onset Pompe (LOPD, n=12) Pompe disease and those from patients bearing pseudodeficiency alleles with or without Pompe mutations, or Pompe disease carriers (n=230) were studied.
RESULTS
Using the MS/MS GAA assay in DBS, 96% of the pseudodeficiency newborns and all of the Pompe disease carriers were well separated from the IOPD and LOPD newborns. The fluorometric assay separated <10% of the pseudodeficiencies from the IOPD/LOPD group.
CONCLUSIONS
The relatively large analytical range MS/MS GAA assay but not the fluorimetric assay in DBS provides a robust approach to reduce the number of referrals and should dramatically facilitate newborn screening of Pompe disease.
Keywords: Pompe disease, lysosomal storage disease, newborn screening, tandem mass spectrometry, inborn errors of metabolism, pseudodeficiency, biochemical genetics
INTRODUCTION
Pompe disease is a lysosomal storage disease that results in the buildup of glycogen-derived short polymers of glucose especially in skeletal muscle (1). The ability to screen newborns for Pompe disease by measurement of the deficient enzyme, acid α-glucosidase (GAA), was first made possible with the use of the natural polysaccharide acarbose to block the interfering enzyme maltase-glucoamylase present in blood (2, 3). This development combined with pilot studies showing the feasibility of newborn screening for Pompe disease based on measurement of GAA activity in dried blood spots on newborns screening cards (DBS) (4, 5) and the importance of early diagnosis for optimal treatment with enzyme replacement therapy (4) contributed to the recent addition of Pompe disease to the Recommended Newborn Screening Panel in the USA. Taiwan is also screening most newborns for Pompe disease, and other countries are expected to add Pompe disease to their national newborn screening panels in the near future.
The first large scale Pompe disease newborn screening pilot study was carried out at the National Taiwan University Hospital (covering about one-third of the newborns in Taiwan) using a multi-tier assay to measure three enzymatic activites: i) GAA, measured at low pH in the presence of acarbose; ii) neutral α-glucosidase, measured at elevated pH in the absence of acarbose; and iii) total α-glucosidase, measured at low pH in the absence of acarbose (4, 6). All three enzymatic activities were measured with a plate-reader fluorimeter using the fluorimetric substrate 4-methylumbelliferyl-α-glucoside. An algorithm involving ratios of the 3 enzymatic activities was developed to help distinguish Pompe afffected newborns from those bearing pseudodeficiency alleles. The latter reduce the activity of GAA but do not lead to Pompe disease. Pseudodeficiency alleles are especially common in the Asian population; for example, the allele p.G576S occurs with a frequency of 14.5% in Taiwan (94 in 650 chromosomes) (4). This adds substantially to the challenge of newborn screening for Pompe disease.
The second large scale Pompe disease newborn screening pilot was carried out in the USA in the Washington state newborn screening laboratory using a 3-plex tandem mass spectrometry (MS/MS) assay to simultaneously measure GAA and the enzymes relevant to Fabry disease and mucopolysaccharidosis-I using a single punch from a DBS and a single assay cocktail (5). Among 111,544 newborns tested, the number of Pompe screen positives (GAA activity below the cutoff) was 17, 4 of which were predicted to develop Pompe disease based on genoytyping, with the remainder bearing mutations only on one allele (carriers, n=4) or bearing pseudodeficiency alleles (n=9). A more recent pilot study was carried out with a MS/MS assay to measure 6 lysosomal enzymes in a single assay mixture (7). Of the ~44,000 newborns tested, 8 were screen positive for Pompe disease using a low enzymatic activity cutoff of 15% of the daily mean GAA activity. In the New York newborn screening laboratory, MS/MS analysis of GAA in several hundred thousand DBS yielded a virtually identical screen positive rate as in Washington using the same cutoff value of 15% of the daily mean GAA activity (7). A newborn screening study using digital microfluidics fluorimetry with 4-methylumbelliferyl-α-glucoside to measure GAA activity carried out in the Missouri state newborn screening laboratory yielded 84 screen positives out of 175,000 newborns (8), a screen positive rate that was 2.5-fold higher than the value obtained by MS/MS in Washington and New York. All 3 studies were carried out with equivalent cutoff values, chosen to be a GAA activity just above that measured in an identical set of DBS from patients clinically confirmed to have Pompe disease (7). A recent pilot study showed that it is readily feasible to carry out newborn screening for 6 lysosomal storage diseases including Pompe disease in a multiplex fashion with the biomarker for X-linked adrenoleukodystrophy (9). The latter has been recently added to the Recommended Uniform Screening Panel in the USA.
The lower rate of Pompe disease false positives using MS/MS versus fluorimetry may be due to the relatively large analytical range of the MS/MS GAA assay (7). The latter is defined as the GAA assay response (MS/MS ion counts for the GAA product divided by that for the internal standard) with the quality control HIGH DBS (typical of a healthy newborn) divided by the assay response due to the blank (all GAA-independent processes) (10). Assays with a large analytical range spread the enzymatic activity values over a large range, and this is expected to increase the accuracy of the activity measurement. The analytical range of the MS/MS assay of GAA is >10-fold greater than that for the fluorimetric assay (8); the difference is due to the fact that the 4-methylumbellifery-α-glucosidase substrate is intrinsically fluorescent and thus substantially contributes to the non-enzymatic blank (10). The analytical range of the MS/MS assay is limited by a small amount of product that is generated by non-enzymatic breakdown of the substrate in the heated electrospray ionization source of the mass spectrometer (10).
The goal of the present study was to test a variant of the MS/MS GAA assay with an exceptionally large analytical range of 187 on a set of DBS in Taiwan from newborns confirmed to be affected with infantile-onset Pompe disease IOPD, those predicted to develop late-onset Pompe disease (LOPD), those with pseudodeficiency alleles with or without Pompe mutations, and Pompe disease carriers.
METHODS
Newborn DBS were collected and shipped in Taiwan within 3 d of birth at ambient temperature. DBS were submitted to GAA analysis on the day of arrival in the newborn screening laboratory. After that they were stored at 4°C for 2–3 weeks before being transferred to −20°C storage for 2–3 y. MS/MS assays of GAA in DBS was carried out using a modification of the previously reported method (11). Fluorimetric assay of GAA in DBS was carried out as described (12). The preparation of the substrates and internal standards standards for both assays is provided in the Supplemental file. The study was approved by the Institutional Review Board of the Taipei Veterans General Hospital.
Infants with a GAA activity <0.50 µmol/L/h (normal activity >1.6 µmol/L/h) were referred immediately to a hospital for diagnostic confirmation. IOPD cases were defined by the following criteria: i) general weakness; ii) extremely low initial GAA activity (<0.50 mmol/L/h); iii) increased creatine kinase activity (>250 U/L); and iv) increased left ventricular mass index (LVMI; >80 g/m2) (13). In addition, for all IOPD patients, thighs were examined by electron microscopy. The pathological findings all showed glycogen pools and lysosomal accumulation of glycogen granules. PAS and D-PAS staining were all positive for glycogen compatible with IOPD.
GAA genotyping also suggested IOPD (all genotypes provided in Supplemental Tables 1 and 2). After parental informed consent was obtained, quadriceps muscle biopsies and GAA gene sequencing were performed. GAA gene sequencing was performed and analyzed as previously described (14). Biopsy and creatine kinase data is provided in the Supplemental file. We defined LOPD cases as non-IOPD newborns with at least one mutation on one allele and at least one mutation on the other allele. Such mutations included known Pompe disease pathogenic mutations and novel variants excluding pseudodeficiencies. As in all previous Pompe disease pilot studies, most of these LOPD newborns did not yet have Pompe disease symptoms, and we could not know at that time if and when they would develop Pompe disease.
MS/MS assay for assay of GAA in DBS
A 3.2 mm diameter punch from a DBS (Whatman 903 filter paper) was placed into a 96-well PS plate (Greiner Bio-one, Germany) using a BSD 700 puncher (BSD technologies, Australia). For sample extraction, 70 µL of extraction buffer (20 mM sodium phosphate, pH 7.1) was added to each well. The plate was then sealed with plastic sealing film (Fasson S-695), centrifuged at 800×g for 2 min, and mixed in an orbital shaker at 875 rpm at 37°C for 1 h. After extraction, the plate was centrifuged as above for 2 min. Ten µL of DBS extract was added to 15 µL of GAA assay cocktail (Supplemental File) in a 96-well plate. To a separate plate was added 10 µL of DBS extract to 15 µL of GLA assay cocktail, and in the same way for the ABG and IDUA plates.
The 96-well plates were sealed with aluminum film (FocusBio Cat. 2396) and incubated on a shaker at 225 rpm and 37°C for 20 h. The enzyme reactions were quenched by adding 100 µl of 1:1 ethyl acetate:methanol to each well (see (7) for information about sources of ethyl acetate). After pipetting up and down 3 times, the four assay reactions were then combined into a single deep well plate, and 400 µL of ethyl acetate and 400 µL of water were added by a liquid handler (Tecan Freedom EVO, Austria). The plates were sealed with 96 well Piercable Plate Seals (Phenix, MPI-P06) and shaken for 5 min at 600 rpm at room temperature. The deep-well plates were centrifuged at 3,000×g for 5 min, and a 300 µL aliquot of the top organic layer was transferred into a new 96-well PP plate (Greiner Bio-one, Germany). The solvent was evaporated under a steam of nitrogen, and residues were reconstituted in 100 µL of a 19:1 of ethyl acetate/methanol. The plates were shaken for 5 min at 200 rpm at room temperature. After reconstitution, the samples were subjected to a solid phase extraction step performed with a 96-well 0.45 µm filter plate (Acroprep 96 Filter Plate, Pall, Life Sciences) containing 100 mg silica gel per well (SILICYCLE, R12030B, added as a dry powder using a plate that held 100 mg silica per well). The silica plate was rinsed with 0.25 mL of 19:1 ethyl acetate:methanol before use. After the silica rinse, the reconstituted solution was added by pipet to wells with silica. Then the silica was washed 4 times with 0.4 mL of 19:1 ethyl acetate/methanol using suction and a plate vacuum manifold. During sample application and 4 washings, the eluent was collected into a deep-well plate.
The solvent was removed with jets of nitrogen. Samples were reconstituted with 300 µL of mobile phase (80% acetonitrile:20% water containing 0.2% formic acid). After shaking for 5 min, 20 µL was injected into the mass spectrometer by flow injection (0.12 mL/min, Dionex Ultimate 3000 UPLC system) and analyzed with MS/MS system (TSQ Vantage triple-quadruple, Thermo Scientific, Waltham, USA). The sample plate was cooled to 10°C in the autosampler during data collection to minimize solvent loss. Electrospray source and multiple reaction monitoring parameters are provided in Supplemental Table 3. Multiple reaction monitoring ion peak areas were obtained using the X-calibur software from TSQ Vantage. Blanks were carried out exactly as above except that a 3.2 mm punch of filter without blood was used instead of a DBS punch. The blank was subtracted from all DBS runs. The GAA activities (µmole/hr/L) were calculated by multiplying the GAA product-to-GAA internal standard ion peak area ratio by the micromoles of internal standard in the assay (0.0001 µmoles) and dividing by the incubation time (20 h) and the volume of blood in the 3 mm DBS punch (3.2 (µl).
MS/MS assay of GAA in DBS using 6-plex substrate/internal standard mix
A 3.2 mm diameter punch from a DBS (Whatman 903 filter paper) was placed into a 96-well PS plate (Greiner Bio-one, Germany) using a BSD 700 puncher (BSD technologies, Australia). For sample extraction, 70 µL of extraction buffer (20 mM sodium phosphate, pH 7.1) was added to each well. The plate was then sealed with plastic sealing film (Fasson S-695), centrifuged at 3,000 × g for 2 min, and mixed in an orbital shaker at 875 rpm at 37°C for 1 h. After extraction, the plate was centrifuged at 3,000×g for 2 min. Ten µL of DBS extract was added to 15 µL of PerkinElmer assay cocktail (Supplemental File) in a 96-well plate.
The 96-well plate was sealed with aluminum film (FocusBio Cat. 2396) and incubated on a shaker at 225 rpm and 37°C for 20 hr. The enzyme reactions were quenched by adding 100 µl of 1:1 ethyl acetate:methanol to each well. After pipetting up and down 3 times, the GAA assay reactions were then transfer into a single deep well plate, and 400 µL of ethyl acetate and 400 µL of water were added by a liquid handler (Tecan Freedom EVO, Austria). Additional steps for sample processing and MS/MS was as described above.
Fluorimetric assay of GAA in DBS
A 3.2 mm diameter punch from a DBS (Whatman 903 filter paper) was placed into a 96-well PS plate (Greiner Bio-one, Germany) using a BSD 700 puncher (BSD technologies, Australia). Two hundred microliter of distilled water was added to each well, and the plate was sealed with plastic sealing film (Fasson S-695) and centrifuge at 3,000×g for 2 min. Samples were shaken at 37°C with orbital shaking at 875 rpm for 1 h.
A 70 mM stock solution of 4-methylumbelliferyl-α-D-glucoside (Calbiochem, San Diego, CA) in dimethyl sulfoxide was prepared in advance. Substrate solution (1.4 mM) was prepared by diluting the DMSO stock with 40 mM aqueous sodium acetate buffer (pH 3.8). Enzyme reactions at pH 3.8 were composed of 50 µL of substrate solution, 20 µL of aqueous acarbose (80 µM), and 40 µL of DBS extract. The plate was sealed and shaken at 225 rpm at 37°C for 20h. Reactions were quenched by addition of 140 µL of 150 mM EDTA (pH 11.4) to each well. A 4-methylumbelliferone calibration curve was prepared for every plate by mixing aqueous standards in the range of 0.00 to 1.00 µM with the reaction buffer include 40 µL of DBS extract, 50 µL of 40 mM aqueous sodium acetate buffer (pH 3.8), 20 µL of aqueous acarbose (80 µM), and 150 µL EDTA solution per well. Eight different calibrators per curve were measured in duplicate. Fluorescence was measured on a on a SpectraMax M3 plate reader (Molecular device) with excitation at 360 nm and emission at 450 nm. Blanks were made by incubating 40 µL of DBS extract in 1 well and 50 µL substrate solution + 20 µL aqueous acarbose (80 µM) in a second well at 37°C for 20 hrs. Then the contents of the wells were combined and processed as above.
RESULTS AND DISCUSSION
To maximize the analytical range of the MS/MS we used solid-phase extraction with silica gel for sample work-up after incubation of the DBS punch in buffer with GAA substrate and before analysis by MS/MS. Solid-phase extraction removed most of the GAA-substrate, which stuck tightly to the silica gel, while allowing high yield recovery of GAA product and internal standard. The internal standard was chemically identical to the product but substituted with deuterium so that both could be quantified by MS/MS in separate multiple reaction monitoring channels. Since the analytical range of the MS/MS assay was limited by the non-enzymatic formation of product from substrate in the heated electrospray ionization source of the mass spectrometer (10), we expected that removal of the substrate by solid-phase extraction would increase the analytical range of the assay. The analytical range was calculated as the MS/MS product ion peak area divided by that for the internal standard obtained with the quality control HIGH DBS divided by the corresponding ratio for the blank in which a filter paper punch (no blood) was incubated with assay cocktail and processed for MS/MS. The analytical range was found to be 187. The analytical range for the MS/MS assay, in which the solid-phase extraction is substituted with liquid-liquid extraction using ethyl acetate, was found to be lower at 59. This was because a substantial portion of the GAA substrate extracted into the ethyl acetate layer, and thus there was more in-source substrate-to-product conversion. The analytical range for the fluorimetric assay of GAA in DBS with 4-methylumbelliferyl-α-glucoside was found to be 12.3 (10), thus 15-fold lower than that for the MS/MS assay using solid-phase extraction. The calculation of the analytical range for both assays required careful consideration of factors that contributed to the assay response for all processes that were independent of GAA activity, and the full details have been reported (10).
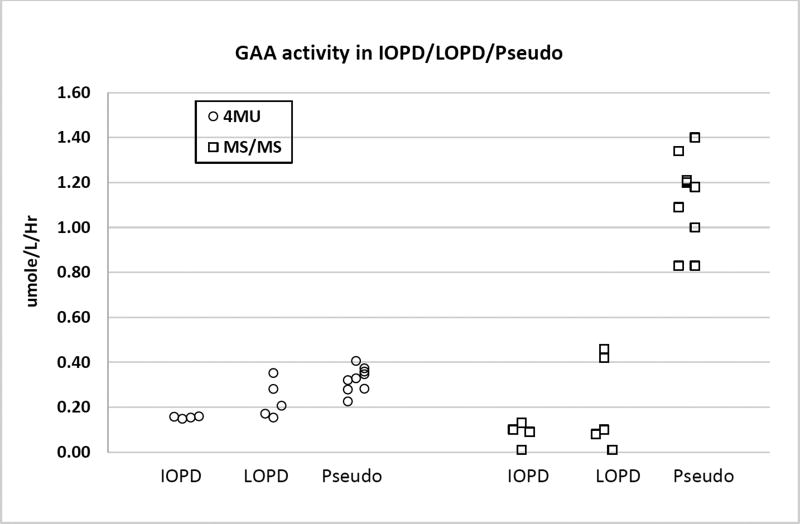
Figure 1 shows the data obtained with 4 IOPD, 5 LOPD and 9 pseudodeficiency/carrier DBS. Individual GAA activites (MS/MS and fluorescence), genotypes, and creatine kinase activities are provided in the Supplemental File. For this study we measured GAA activity using a plate reader fluorimeter with 4-methylumbelliferyl-α-glucoside as substrate as well as with the MS/MS assay. All DBS punches were analyzed at the same time using both methods so differential loss of GAA activity due to DBS storage was not a factor for the comparison of the fluorimetric versus the MS/MS method. The data clearly showed that the set of pseudodeficiency/carrier DBS were well separated from the IOPD/LOPD samples when GAA is measured with MS/MS, but there was essentially no separation with the fluorimetric assay.
Figure 1.
Identical set of DBS were analyzed by the to the fluorimetric and MS/MS GAA assays.
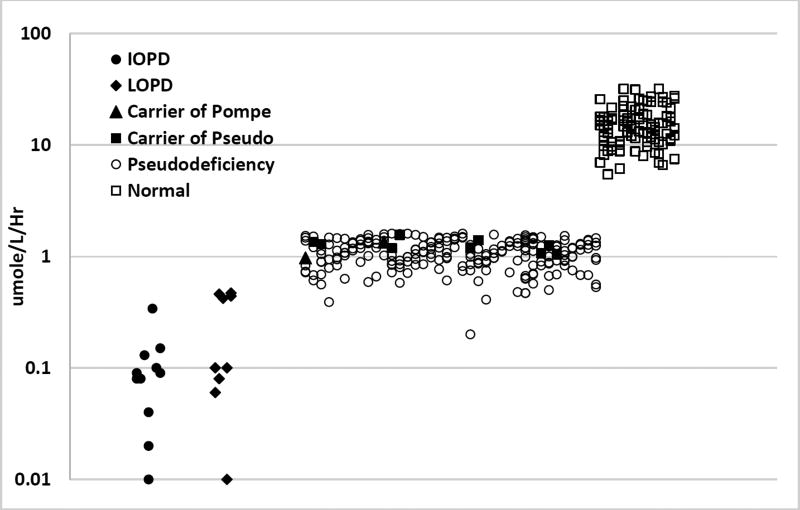
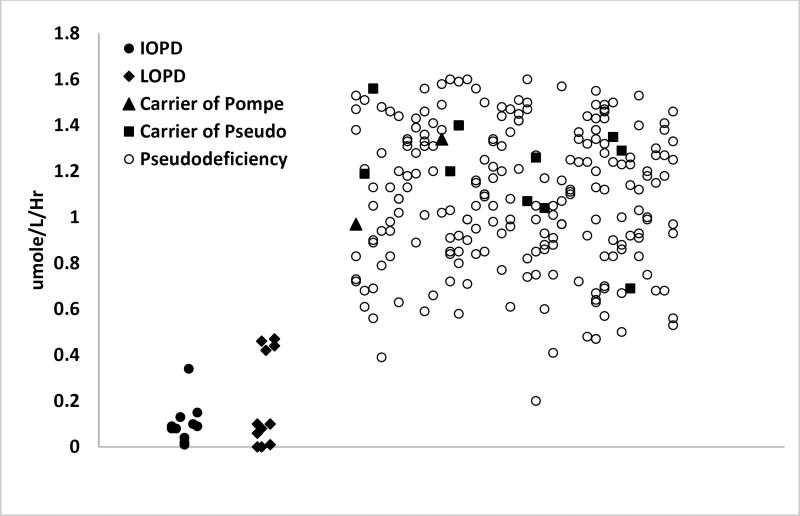
We then extended the MS/MS study by measuring GAA enzymatic activity in DBS from 11 IOPD patients, 12 LOPD patients, and 230 cases of non-Pompe disease affected newborns carrying the following DNA changes: one Pompe mutation (n=2); one Pompe mutation plus one pseudodeficiency allele (n=77); one Pompe mutation plus two pseudodeficiency alleles (n=79); one pseudodeficiency allele (n=10); two pseudodeficiency alleles (n=62) (Figure 2). Individual GAA activities (MS/MS), genotypes, and creatine kinase activities are provided in the Supplemental File. Figure 2 (top) also shows the GAA activity of 101 normal newborn DBS. We found that the GAA enzymatic activity of 96% of the pseudodeficiency samples and 100% of the carrier samples were separated from that of the IOPD/LOPD activities.
Figure 2.
(Top) GAA enzymatic activity measured in DBS by MS/MS (note the log scale). (Bottom) Same as top but covering the range 0-2 µmole/hr/L (linear scale). Carrier of Pompe were patients with one Pompe mutation allele and a wild type allele, carrier of pseudo were patients with at least one pseudodeficiency DNA change as one allele and wild type as the second allele, and pseudodeficiency were patients with at least one pseudodeficiency DNA change on both alleles.
We also measured GAA enzymatic activity using MS/MS on an identical set of DBS (3 IOPD and 12 normal) using both the liquid-liquid extraction method and the solid-phase extraction method (Supplemental Material Table 4). The ratios of the mean GAA activities for healthy individuals divided by the mean of the IOPD activities were 200 and 25 for the solid-phase extraction and liquid-liquid extraction methods, repsectively, showing that the solid-phase extraction method leads to a better separation of GAA activity values.
Most newborn screening laboratories carrying out lysosomal storage disease newborn screening by MS/MS will use the 6-plex method that makes use of liquid-liquid extraction with ethyl acetate (7) since this is the major method going forward. One option would be to use the method described here with solid-phase extraction through silica gel as a second-tier analysis on GAA screen-positive samples from the first-tier 6-plex assay. This could be done on the same DBS, and thus results would be available without the need to contact the family.
The GAA substrate and internal standard provided by CDC (Supplemental File) is being replaced by reagents provided by PerkinElmer. The results shown in Figures 1 and 2 were carried out by incubating the DBS with assay buffer containing only a single substrate and internal standard for GAA (CDC reagents, Supplemental File). PerkinElmer will provide a mixture of 6 different substrates and internal standards (for Pompe, Gaucher, Niemann-Pick-A/B, Krabbe, MPS-I and Fabry), but will also support customer requests for individual reagents. The GAA substrate and internal standard in the 6-plex mixture is chemically identical to the CDC reagents for GAA. The GAA activities measured in a 1-plex assay with the CDC reagents and those measured in a 6-plex assay with the PerkinElmer reagents correlated (Supplemental FIgure 1), and the separation in GAA activities between the IOPD/LOPD and pseudodeficiency cohorts seen with the PerkinElmer 6-plex reagents was as robust as that seen with the CDC reagents (Figures 1 and 2).
In summary, we have shown with a large collection of Pompe disease affected and pseudodeficiency DBS that the MS/MS GAA enzymatic activity is more powerful than the fluorimetric assay in distinguishing Pompe-affected patients from those carrying a single GAA mutation or one or more pseudodeficiency alleles. This result is important because newborn screening for Pompe disease is plagued by the relatively large number of pseudodeficiency samples especially in the Asian population. We believe that the MS/MS method is the method of choice for newborn screening for Pompe disease with the anticipation that the number of patients referred for followup and accompanying family anxiety can be greatly reduced.
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Institutes of Health (R01 DK067859).
Abbreviations
- DBS
dried blood spot on a newborn screening card
- GAA
acid α-glucosidase
- IOPD
infantile-onset Pompe disease
- LOPD
predicted late-onset Pompe disease
- MS/MS
tandem mass spectrometry
References
- 1.Chen Y-T, Burchell A. Glycogen Storage Disease. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The Metabolic and Molecular Bases of Inherited Disease. New York: McGraw-Hill; 1995. [Google Scholar]
- 2.Li Y, Brockman K, Turecek F, Scott CR, Gelb MH. Tandem mass spectrometry for the direct assay of enzymes in dried blood spots: application to newborn screening for Krabbe disease. Clin Chem. 2004;50:638–40. doi: 10.1373/clinchem.2003.028381. [DOI] [PubMed] [Google Scholar]
- 3.Winchester B, Bali D, Bodamer O, Caillaud C, Christensen E, Cooper A, et al. Methods for a prompt and reliable laboratory diagnoisis of Pompe disease: Report from an international consensus meeting. Molec Genet Metabol. 2007;93:275–81. doi: 10.1016/j.ymgme.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Chiang SC, Hwu WL, Lee NC, Hsu LW, Chien YH. Algorithm for Pompe disease newborn screening: results from the Taiwan screening program. Mol Genet Metab. 2012;106:281–6. doi: 10.1016/j.ymgme.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 5.Scott CR, Elliott S, Buroker N, Thomas LI, Keutzer J, Glass M, et al. Identification of Infants at Risk for Developing Fabry, Pompe, or Mucopolysaccharidosis-I from Newborn Blood Spots by Tandem Mass Spectrometry. J Pediatr. 2013;163:498–503. doi: 10.1016/j.jpeds.2013.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dajnoki A, Fekete G, Keutzer J, Orsini JJ, De Jesus VR, Chien YH, et al. Newborn screening for Fabry disease by measuring GLA activity using tandem mass spectrometry. Clin Chim Acta. 2010;411:1428–31. doi: 10.1016/j.cca.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 7.Elliot S, Buroker N, Cournoyer J, Potier A, Trometer J, Elbin C, et al. Pilot study of newborn screening for six lysosomal storage diseases using Tandem Mass Spectrometry. Molec Genet Metab. 2016;118:304–9. doi: 10.1016/j.ymgme.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hopkins P. CDC/APHL Workshop on newborn screening for lysosomal storage diseases. 2015 https://www.aphl.org/programs/newborn_screening/Documents/LSDs-2/LSD-Screening-with-DMF_Patrick-Hopkins_4-17-15.pdf.
- 9.Tortorelli S, Turgeon CT, Gavrilov DK, Oglesbee D, Raymond KM, Rinaldo P, et al. Simultaneous Testing for 6 Lysosomal Storage Disorders and X-Adrenoleukodystrophy in Dried Blood Spots by Tandem Mass Spectrometry. Clin Chem. 2016;62:1248–54. doi: 10.1373/clinchem.2016.256255. [DOI] [PubMed] [Google Scholar]
- 10.Kumar AB, Spacil Z, Masi S, Ghomashchi F, Ito M, Scott CR, et al. Higher Analytic Range of Tandem Mass Spectrometric versus Fluorimetric Assays of Lysosomal Enzymes: Application to Newborn Screening and Diagnosis of Mucopolysaccharidosis Types IVA and VI. Clin Chem. 2015;61:1363–71. doi: 10.1373/clinchem.2015.242560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang XK, Elbin CS, Chuang WL, Cooper SK, Marashio CA, Beauregard C, et al. Multiplex Enzyme Assay Screening of Dried Blood Spots for Lysosomal Storage Disorders by Using Tandem Mass Spectrometry. Clin Chem. 2008;54:1725–8. doi: 10.1373/clinchem.2008.104711. [DOI] [PubMed] [Google Scholar]
- 12.Chien YH, Chiang SC, Zhang XK, Keutzer J, Lee NC, Huang AC, et al. Early detection of pompe disease by newborn screening is feasible: results from the Taiwan screening program. Pediatr. 2008;122:e39–45. doi: 10.1542/peds.2007-2222. [DOI] [PubMed] [Google Scholar]
- 13.Yang CF, Yang CC, Liao HC, Huang LY, Chiang CC, Ho HC, Lai CJ, Yang TF, Hsu TR, Soong WJ, Niu DM. Very early treatment for infantile-onset Pompe disease contributes to better outcomes. J Pediatr. 2016;169:174–80. doi: 10.1016/j.jpeds.2015.10.078. [DOI] [PubMed] [Google Scholar]
- 14.Ko TM, Hwu WL, Lin YW, Tseng LH, Hwa HL, Wang TR, et al. Hum Mutat. Vol. 13. John Wiley & Sons, Inc; 1999. Molecular genetic study of Pompe disease in Chinese patients in Taiwan; pp. 380–4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.