Abstract
The catabolic process of autophagy is an essential cellular function that allows for the breakdown and recycling of cellular macromolecules. In recent years, the impact of epigenetic regulation of autophagy by non-coding microRNAs (miRNAs) has been recognized in human cancer. In colorectal cancer, Autophagy plays critical roles in cancer progression as well as resistance to chemotherapy, and recent evidence demonstrates that miRNAs are directly involved in mediating these functions. In this review, we will focus on the recent advancements in the field of miRNA regulation of autophagy in colorectal cancer.
Keywords: Autophagy, colorectal cancer, microRNA, chemotherapy, cancer stem cell
Introduction
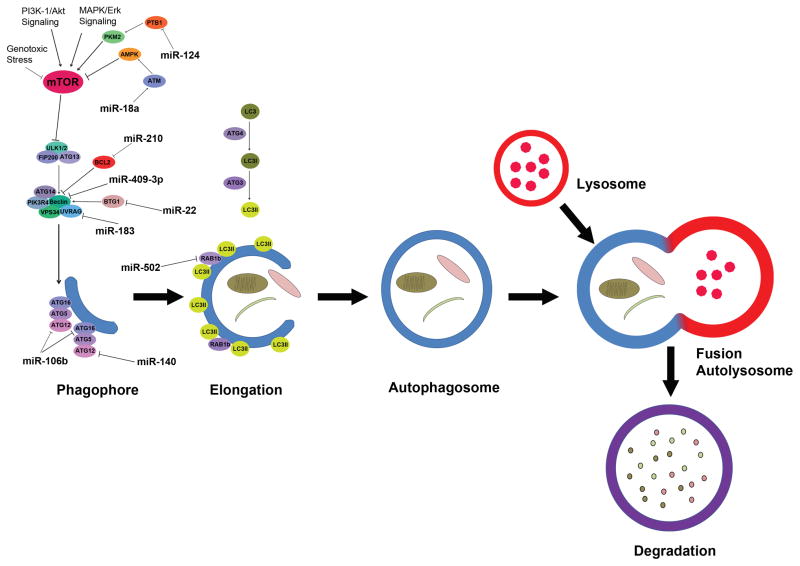
Twenty three years after its initial discovery in 1993, the significance of autophagy is being recognized with this year’s Nobel Prize in Physiology and Medicine. In the time since its discovery, extensive research efforts have been made to expand our fundamental understanding of autophagy in basic biology and human diseases including cancer (1). Autophagy is a conserved, essential and tightly regulated biological process in which cells undergo self-destruction (2). The process of autophagy involves the formation of double-membrane autophagic vacuoles (autophagosomes) around cytoplasmic components (3). Many of the important genes and pathways that are involved in the process and regulation of autophagy have been identified. These include the autophagy related genes (ATGs) family of over 30 genes, including ATG1, ATG4, LC3/ATG8 and beclin-1 (4–6). In mammalian cells, starvation conditions as well as other signals lead to inhibition of mTOR, which results in activation of autophagy initiation kinases ULK1 and ULK2, which phosphorylate ATG13 and FIP200. The ULK1 complex is recruited to the phagopore assemble site and is essential for the initiation of autophagy (7). The PtdIns3K complex which includes beclin-1, VPS34, ATG14, PIK3R4, URAG and AMBRA is also essential for vesicle nucleation (7, 8). Formation of the ATG16, ATG5, ATG12 conjugation complex promotes the elongation of the phagophore. The LC3 conjugation system which involves ATG4, a cysteine protease, also promotes phagophore elongation (9). Once formed, the mature autophagosome fuses with the lysosome to allow for the degradation of the contents of the autophagosome (10–13). The autophagy regulatory circuits and key proteins involved in such process as well as the miRNAs that are involved in the regulation of some of these genes related to colorectal cancer are illustrated in Figure 1.
Figure 1.
Schematic illustration of autophagy under stress. The autophagy process is divided into phagophore formation, elongation, autophagosome formation, fusion with lysosome, and degradation. Following the initiation stress signaling through the mTOR kinase, the hypophosphorylated ATG13 is maintained, and the autophagic vesicles are formed from the phagophore/isolation membrane to the autophagosome and autolysosome. ATG-11, -13, -17 and ULK complex mediate the early process followed by Beclin-1-hVPS-34-protein complex. hVPS-34/Beclin-1 complex converts PI to PI3P followed by ATG5-ATG12 conjugation and interaction with ATG16L. Following the LC3 processing events, the nascent complex further undergoes elongation and expansion to form a double membrane autophagosome. The Autolysosome is formed through the fusion of lysosomes docking and fusion with autophagosomes where cargo is degraded to generate amino and fatty acids. miRNAs that involved in colorectal cancer progression and resistance by mediating the initiation and enlongation of autophagy processes are illustrated in the pathway map.
Mounting evidence has revealed that autophagy has important effects on tumor progression. Autophagy has a role in various stages of tumor progression including primary tumor initiation and expansion, invasion, tumor dormancy, metastasis, and resistance to therapy (14). Autophagy provides critical building blocks for tumor cells to survive during hypoxia and nutrient deprivation (15). It helps cancer cells gain metabolic fitness during intravasation and invasion (14, 16, 17). It also prolongs the survival of quiescent tumor cells during tumor dormancy (18). In addition, autophagy aids tumor cells in adapting to a new tissue microenvironment during metastasis to distant organs (19, 20). Autophagy also promotes tumor stem cell survival in response to acute stress and environmental changes due to chemotherapy and radiation treatment (14). As a result, autophagy maybe a double edge sword in cancer treatment (21). Autophagy triggered by chemotherapy and radiation can effectively eliminate bulk tumors and stromal tissue (22). At the same time, it also provides the survival advantage for the cancer stem cells during anticancer therapeutic treatment (23).
Recently it has been recognized that non-coding miRNAs play key roles in autophagy (24, 25). miRNAs are non-coding RNA molecules, 18–25 nucleotides in length, that regulate the expression of their target genes by mRNA degradation or translational inhibition. This regulation occurs mainly through interaction at the 3′-UTRs of the target mRNAs (26–28). Dysregulation of miRNAs has been associated with cancer development and progression, and miRNAs have emerged as a new research frontier for understanding cancer development at the post-transcriptional and translational level (29). miRNAs have been found to regulate many cellular processes including apoptosis (30–33), differentiation (27, 34, 35), cell proliferation (30, 35–37) and autophagy (24, 25). Some miRNAs important in colorectal cancer and their targets in the autophagy pathway are shown in Figure 1. In this review, we will highlight some of the recent research efforts on the mechanism and impact of autophagy mediated by miRNAs in colorectal cancer.
miRNAs regulation of autophagy and colorectal cancer progression
Several miRNA have been found to have important roles in the regulation of autophagy in colorectal cancer, and this regulation can influence cancer progression. Recent studies from our group show that miR-140 directly targets Smad2 and through repression of Smad2, overexpression of miR-140 in colon cancer cell lines inhibits invasion, proliferation and induces cell cycle arrest (38). Ectopic expression of miR-140 in colorectal cancer stem cells leads to the disruption of autophagy, inhibiting tumor stem cell growth and sphere formation (38). We have identified ATG12 as one of the main targets of miR-140 involved in autophagy. ATG12 is important in autophagosome elongation (Figure 1) (Table 1) (10). Furthermore, overexpression of miR-140 in colon cancer stem cells abolished tumor formation and metastasis in vivo. In addition, there is a progressive loss of miR-140 expression from normal colorectal mucosa to primary tumor tissues, with further reduction in liver metastatic tissues (39). Higher miR-140 expression is significantly correlated with better survival in stage III and IV colorectal cancer patients (38). This suggests that miR-140 is a key regulator in colorectal cancer progression and metastasis, and miR-140 disrupts colon cancer stem cell growth through interrupting autophagy (38).
Table 1.
miRNA and there important autophagy related targets in colorectal cancer.
| miRNA | Important Target(s) | Effect on Autophagy |
|---|---|---|
| miR-140 | ATG-12 | Inhibition |
| miR-210 | BCL2 | Promotes |
| miR-502 | RAB1B | Inhibition |
| miR-124 | PTB1 | Promotes |
| miR-18a | ATM, mTORC1, hnRNP A1 | Promotes |
| miR-106a/miR-106b | ATG16L and ATG12 | Inhibition |
| miR-183 | UVRAG | Inhibition |
| miR-409-3p | Beclin-1 | Inhibition |
| miR-22 | BTG1 | Inhibition |
We also discovered that miR-502 directly suppresses autophagy by targeting the small GTPase, RAB1B, in colon cancer cell lines (40). RAB1B modulates autophagic activity through the regulation of autophagosome formation (Figure 1) (41). Rab1B regulates vesicle trafficking and directly impacts autophagy (42, 43). Ectopic expression of miR-502 in HCT-116 cells interrupted autophagic flux under acute and prolonged nutrient starvation and significantly decrease colon cancer cell growth (40). Inhibition of both p53 and RAB1B, the mediators for autophagy, can reproduce this phenotype, suggesting autophagy plays an important role in its tumor suppressive function. Profiling of human colon cancer samples reveals that miR-502 is downregulated in tumor tissue as compared to normal tissue. Ectopic expression of miR-502 in human colon cancer xenografts can significantly reduce tumor size, indicating miR-502 has potential as an adjuvant treatment for colon cancer patients (40).
miR-106a and miR-106b have been shown to inhibit starvation induced autophagy in colon cancer cells. This autophagy inhibition is the result of targeting of multiple important genes including ATG16L and ATG12 (44). Starvation induced autophagy is also inhibited by miR-183 targeting UVRAG, which normally promotes autophagosome formation in colon cancer (45).
As opposed to these miRNA that inhibit autophagy, miR-124 acts as tumor suppressor gene by inducing apoptosis and autophagy in colon cancer both in vitro and in vivo tumor xengrafts (46). miR-124 targeted polypyrimidine tract-binding protein 1 (PTB1), which is a splicer of pyruvate kinase muscles 1 and 2 (PKM1 and PKM2) and induced the switching of PKM isoform expression from PKM2 to PKM1. PKM2 is an activator of mTOR, so by switching expression to PKM1, mTOR activity is repressed (Figure 1). These findings suggest that miR-124 acts as a tumor-suppressor and a modulator of energy metabolism through a PTB1/PKM1/PKM2 feedback cascade in human colorectal tumor cells. miR-18a has been reported as a potential tumor suppressor by inducing apoptosis in colon cancer cells via the autophagolysosomal degradation of oncogenic heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) (47). The function of miR-18a provides another mode of miRNA action on the degradation of key targets such as hnRNP A1 via autophagy in cancer. miR-18a also induces autophagy through inhibition of ATM which activates AMPK, an inhibitor of mTOR (48). These miRNAs all have shown an ability to regulate autophagy in colon cancer. This regulation influences colon cancer growth and progression.
miRNA, autophagy and resistance mechanisms
In addition to these miRNA that regulate autophagy and influence cancer growth and progression, other miRNA have been found to influence resistance in colon cancer through their regulation of autophagy. It has been found that under hypoxia, HIF-1α induces miRNA-210 which in turn enhances autophagy and reduces radiosensitivity by downregulating Bcl-2 expression in colon cancer cells (49). This work provides experimental evidence that the expression of miR-210 in human colon cancer cell lines, SW480 and SW620, is significantly increased after hypoxia and forms a positive feedback loop with HIF-1α. As Bcl-2 is mediating both apoptosis and autophagy, downregulation of Bcl-2 by miR-210 reduces radiosensitivity in colon cancer cell lines. This study provides experimental evidence that autophagy may contribute to the reduction of radiosensitivity in hypoxic environments, and the process is mediated through the HIF-1α/miR-210/Bcl-2 pathway in human colon cancer cells (49).
miR-409-3p is a miRNA that inhibits autophagy. miR-409-3p inhibits autophagy by targeting Beclin-1. This autophagy regulation results in enhanced sensitivity to oxaliplatin (50). miR-22 is has also been shown to influence colon cancer cell resistance to chemotherapy. In colon cancer cells, miR-22 can inhibit autophagy and promote apoptosis. This is the result of targeting BTG1. Overexpression of miR-22 in colon cancer cells, increased sensitivity to 5-FU, one of the main chemotherapeutic agents used in the treatment of colorectal cancer (51). As chemoresistance is a major challenges faced in treatment of colorectal cancer, these miRNA and their regulation of autophagy have intriguing potential to improve patient outcomes. A summary of miRNA and their autophagy related targets in colorectal cancer is shown in Table 1.
Summary and Future Perspectives
In summary, it is clear that a number of miRNAs regulate autophagy under various acute genotoxic stress situations in colorectal cancer. Autophagy, regulated by miRNAs, provides tumor cells with an acute response mechanism to genotoxic stress conditions such as chemotherapy. Tumor cells also grow under stress conditions due to hypoxia and/or nutrient deprivation, as a result, miRNAs that can modulate autophagy will impact cancer cell survival. This may be of particular importance in chemoresistant cancer stem cells. As tumor cells are highly heterogeneous, it might be an ideal two hit strategy to eliminate bulk tumor cells by first using tumor suppressive miRNAs that can effectively eliminate tumor stem cells through autophagy. The second step will be to use the miRNAs that can disrupt autophagy to prevent cancer stem cells survival by eliminating the building blocks provided by autophagy. Therefore, it is quite conceivable that modulating miRNA will provide a new direction to change cancer cells in response to stress by altering the autophagy process, and in turn will provide new therapeutic strategies to overcome chemoresistance.
Acknowledgments
This study was supported by the funding from the National Institute of Health R01CA155019 (J. Ju), R01CA197098 (J. Ju)
References
- 1.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS letters. 1993;333(1–2):169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 2.Ohsumi Y. Historical landmarks of autophagy research. Cell research. 2014;24(1):9–23. doi: 10.1038/cr.2013.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12(1):1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou S, et al. Autophagy in tumorigenesis and cancer therapy: Dr. Jekyll or Mr. Hyde? Cancer Lett. 2012 doi: 10.1016/j.canlet.2012.02.017. [DOI] [PubMed] [Google Scholar]
- 5.Fu LL, Wen X, Bao JK, Liu B. MicroRNA-modulated autophagic signaling networks in cancer. Int J Biochem Cell Biol. 2012;44(5):733–736. doi: 10.1016/j.biocel.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 6.Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Current opinion in cell biology. 2010;22(2):132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 7.Fullgrabe J, Klionsky DJ, Joseph B. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nature reviews. Molecular cell biology. 2014;15(1):65–74. doi: 10.1038/nrm3716. [DOI] [PubMed] [Google Scholar]
- 8.Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18(4):571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu ZQ, et al. Dual roles of Atg8-PE deconjugation by Atg4 in autophagy. Autophagy. 2012;8(6):883–892. doi: 10.4161/auto.19652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parzych KR, Klionsky DJ. An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Signal. 2014;20(3):460–473. doi: 10.1089/ars.2013.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lippai M, Szatmari Z. Autophagy-from molecular mechanisms to clinical relevance. Cell Biol Toxicol. 2016 doi: 10.1007/s10565-016-9374-5. [DOI] [PubMed] [Google Scholar]
- 12.Russell RC, et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15(7):741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nature reviews. Cancer. 2007;7(12):961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Molecular biology of the cell. 2004;15(3):1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karantza-Wadsworth V, et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes & development. 2007;21(13):1621–1635. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin S, White E. Role of autophagy in cancer: management of metabolic stress. Autophagy. 2007;3(1):28–31. doi: 10.4161/auto.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brech A, Ahlquist T, Lothe RA, Stenmark H. Autophagy in tumour suppression and promotion. Molecular oncology. 2009;3(4):366–375. doi: 10.1016/j.molonc.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mowers EE, Sharifi MN, Macleod KF. Autophagy in cancer metastasis. Oncogene. 2016 doi: 10.1038/onc.2016.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharifi MN, et al. Autophagy Promotes Focal Adhesion Disassembly and Cell Motility of Metastatic Tumor Cells through the Direct Interaction of Paxillin with LC3. Cell reports. 2016;15(8):1660–1672. doi: 10.1016/j.celrep.2016.04.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Degenhardt K, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer cell. 2006;10(1):51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ. Potential therapeutic applications of autophagy. Nature reviews. Drug discovery. 2007;6(4):304–312. doi: 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
- 23.Martinez-Outschoorn UE, et al. The autophagic tumor stroma model of cancer or “battery-operated tumor growth”: A simple solution to the autophagy paradox. Cell cycle. 2010;9(21):4297–4306. doi: 10.4161/cc.9.21.13817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhai H, Fesler A, Ju J. MicroRNA: a third dimension in autophagy. Cell cycle. 2013;12(2):246–250. doi: 10.4161/cc.23273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frankel LB, Lund AH. MicroRNA regulation of autophagy. Carcinogenesis. 2012;33(11):2018–2025. doi: 10.1093/carcin/bgs266. [DOI] [PubMed] [Google Scholar]
- 26.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 27.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75(5):855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 28.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 29.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nature reviews. Cancer. 2010;10(6):389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113(1):25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 31.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Research. 2005;65(14):6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 32.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94(6):776–780. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghodgaonkar MM, et al. Abrogation of DNA vector-based RNAi during apoptosis in mammalian cells due to caspase-mediated cleavage and inactivation of Dicer-1. Cell Death Differ. 2009;16(6):858–868. doi: 10.1038/cdd.2009.15. [DOI] [PubMed] [Google Scholar]
- 34.Tang F. Small RNAs in mammalian germline: Tiny for immortal. Differentiation. 2010;79(3):141–146. doi: 10.1016/j.diff.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 35.Navarro F, Lieberman J. Small RNAs guide hematopoietic cell differentiation and function. J Immunol. 2010;184(11):5939–5947. doi: 10.4049/jimmunol.0902567. [DOI] [PubMed] [Google Scholar]
- 36.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson CD, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67(16):7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 38.Zhai H, Fesler A, Ba Y, Wu S, Ju J. Inhibition of colorectal cancer stem cell survival and invasive potential by hsa-miR-140-5p mediated suppression of Smad2 and autophagy. Oncotarget. 2015 doi: 10.18632/oncotarget.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhai H, Fesler A, Ba Y, Wu S, Ju J. Inhibition of colorectal cancer stem cell survival and invasive potential by hsa-miR-140-5p mediated suppression of Smad2 and autophagy. Oncotarget. 2015;6(23):19735–19746. doi: 10.18632/oncotarget.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhai H, Song B, Xu X, Zhu W, Ju J. Inhibition of autophagy and tumor growth in colon cancer by miR-502. Oncogene. 2012 doi: 10.1038/onc.2012.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zoppino FC, Militello RD, Slavin I, Alvarez C, Colombo MI. Autophagosome formation depends on the small GTPase Rab1 and functional ER exit sites. Traffic. 2010;11(9):1246–1261. doi: 10.1111/j.1600-0854.2010.01086.x. [DOI] [PubMed] [Google Scholar]
- 42.Plutner H, et al. Rab1b regulates vesicular transport between the endoplasmic reticulum and successive Golgi compartments. The Journal of cell biology. 1991;115(1):31–43. doi: 10.1083/jcb.115.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nature reviews. Molecular cell biology. 2009;10(8):513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 44.Zhai Z, Wu F, Chuang AY, Kwon JH. miR-106b fine tunes ATG16L1 expression and autophagic activity in intestinal epithelial HCT116 cells. Inflamm Bowel Dis. 2013;19(11):2295–2301. doi: 10.1097/MIB.0b013e31829e71cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huangfu L, et al. miR-183 regulates autophagy and apoptosis in colorectal cancer through targeting of UVRAG. Oncotarget. 2016;7(4):4735–4745. doi: 10.18632/oncotarget.6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taniguchi K, et al. MicroRNA-124 inhibits cancer cell growth through PTB1/PKM1/PKM2 feedback cascade in colorectal cancer. Cancer Lett. 2015;363(1):17–27. doi: 10.1016/j.canlet.2015.03.026. [DOI] [PubMed] [Google Scholar]
- 47.Fujiya M, et al. microRNA-18a induces apoptosis in colon cancer cells via the autophagolysosomal degradation of oncogenic heterogeneous nuclear ribonucleoprotein A1. Oncogene. 2014;33(40):4847–4856. doi: 10.1038/onc.2013.429. [DOI] [PubMed] [Google Scholar]
- 48.Qased AB, et al. MicroRNA-18a upregulates autophagy and ataxia telangiectasia mutated gene expression in HCT116 colon cancer cells. Molecular medicine reports. 2013;7(2):559–564. doi: 10.3892/mmr.2012.1214. [DOI] [PubMed] [Google Scholar]
- 49.Sun Y, et al. Hypoxia-induced autophagy reduces radiosensitivity by the HIF-1alpha/miR-210/Bcl-2 pathway in colon cancer cells. International journal of oncology. 2015;46(2):750–756. doi: 10.3892/ijo.2014.2745. [DOI] [PubMed] [Google Scholar]
- 50.Tan S, et al. miR-409-3p sensitizes colon cancer cells to oxaliplatin by inhibiting Beclin-1-mediated autophagy. Int J Mol Med. 2016;37(4):1030–1038. doi: 10.3892/ijmm.2016.2492. [DOI] [PubMed] [Google Scholar]
- 51.Zhang H, et al. MiR-22 regulates 5-FU sensitivity by inhibiting autophagy and promoting apoptosis in colorectal cancer cells. Cancer Lett. 2015;356(2 Pt B):781–790. doi: 10.1016/j.canlet.2014.10.029. [DOI] [PubMed] [Google Scholar]