Abstract
It is increasingly recognized that microbiota affect host health and physiology. However, it is unclear what factors shape microbiome community assembly in nature, and how microbiome assembly can be manipulated to improve host health. All plant leaves host foliar endophytic fungi, which make up a diverse, environmentally acquired fungal microbiota. Here, we experimentally manipulated assembly of the cacao tree (Theobroma cacao) fungal microbiome in nature and tested the effect of assembly outcome on host health. Using next-generation sequencing, as well as culture-based methods coupled with Sanger sequencing, we found that manipulating leaf litter exposure and location within the forest canopy significantly altered microbiome composition in cacao. Exposing cacao seedlings to leaf litter from healthy conspecific adults enriched the seedling microbiome with Colletotrichum tropicale, a fungal endophyte known to enhance pathogen resistance of cacao seedlings by upregulating host defensive pathways. As a result, seedlings exposed to healthy conspecific litter experienced reduced pathogen damage. Our results link processes that affect the assembly and composition of microbiome communities to their functional consequences for host success, and have broad implications for understanding plant–microbe interactions. Deliberate manipulation of the plant–fungal microbiome also has potentially important applications for cacao production and other agricultural systems in general.
Keywords: community assembly, foliar endophytic fungi, Janzen–Connell hypothesis, Theobroma cacao, vertical stratification
1. Background
Exploring the incredible diversity and effects of host-associated microorganisms is a cornerstone of modern biology. We now know that macroorganisms are colonized by diverse microbial communities that play an integral role in their host's biology. These discoveries have been catalysed by recent advances in sequencing technologies, which, coupled with traditional culture-based approaches, are now allowing us to move beyond simple description of the microbiome to understand its role in host health and physiology. Plant leaves, for example, are colonized by a remarkably diverse fungal microbiome. These cryptic symbionts, known as foliar endophytic fungi (FEF), share strong similarities with other microbiota, such as bacteria, in their ecological organization and functional importance [1]. FEF communities can exhibit complex dynamics, including host specificity [2], priority effects [3], and temporal and spatial variability [4], and component FEF species can fulfil various symbiotic functions for their host, including increasing plant vigour [5], drought resistance [6], and enemy defence [7–10]. A growing body of literature suggests that many FEF are modifiers of host plant disease severity [11], and multiple mechanisms of disease suppression by certain endophyte species have been identified, including by secreting antimicrobial substances [12], out-competing pathogens [13], or increasing expression of host defences [14]. Although artificial inoculation studies have revealed the functional roles of some component members of the microbiome, the connections between natural FEF community assembly and their effects on host health have not been investigated previously.
Like other diverse microbiota, FEF are primarily horizontally transmitted by spores that land on compatible leaf surfaces, germinate, and penetrate to form localized infections [1]. This transmission mode allows the fungal microbiome to be easily manipulated. For instance, FEF-free plants can be grown in growth chambers and greenhouses [14] and inoculated with particular fungal species [7,15]. FEF-free seedlings can also be transplanted into the field to examine how FEF communities form in nature. For instance, a previous study placed FEF-free cacao tree (Theobroma cacao) seedlings in a forest where the presence and abundance of leaf litter within approximately 20 m of the seedlings had been manipulated [16]. Exposure to leaf litter increased FEF colonization of tree seedlings, suggesting that FEF recolonize living tissues of the host or its neighbours from nearby senesced leaves [16,17]. If local leaf litter is an important source of FEF colonization, litter identity (i.e. the host species and its associated microbiota) is also likely to affect FEF community structure. Moreover, while local leaf litter may be an important source of FEF near the forest floor, vertical changes in abiotic conditions potentially filter which microbes successfully colonize host tissues at different microsites within the canopy [17,18], as fungal spore densities are influenced by abiotic conditions such as humidity, temperature, wind, light, and canopy drip [19]. However, even if local environmental factors alter FEF community composition, it is unclear whether such changes affect host health or if differential outcomes of community assembly are functionally redundant.
Here, we investigated whether variation in local leaf litter and canopy microsite altered the outcome of microbiome assembly in cacao (T. cacao). Specifically, we used endophyte-free T. cacao seedlings as sentinels for FEF colonization to assess differences in FEF community assembly. We then tested whether changes in microbiome composition affected host resistance to the pathogenic oomycete, Phytophthora palmivora, which can infect all cacao tissues and causes black pod, cacao's most economically important and widespread disease [20]. We found that manipulating leaf litter exposure and location within the forest canopy significantly altered microbiome composition in cacao, and that seedlings exposed to the leaf litter of healthy adults exhibited reduced pathogen damage. These results are similar to results from other microbiome systems [21] and suggest that there are opposing forces (e.g. pathogens and mutualists) acting synchronously within the predictions of the Janzen–Connell framework for distance-dependent seedling performance in tropical forests [22].
2. Material and methods
(a). Generation of endophyte-free seedlings
Endophyte-free T. cacao seedlings were generated at the Smithsonian Tropical Research Institute in Gamboa, Panama, as previously described [7,14]. Seeds were collected from T. cacao trees accession UF12, which were grown in a plantation in Charagre, Bocas del Toro province, Panama. Cacao seeds were surface sterilized by submerging them in 0.5% sodium hypochlorite for 3 min, rinsed with sterile water and then placed in plastic trays of sterilized soil (2 : 1 mixture of clay-rich soil from Barro Colorado Island, Panama, and rinsed river sand), and germinated in growth chambers. After one month, seedlings were transplanted into individual 600 ml pots containing the same soil mixture and returned to growth chambers. Both seed germination and seedling growth took place in Percival growth chambers (model I35LL, 115 volts, 1/4Hp, series: 8503122.16, Percival Scientific, Inc., Perry, IA) with a 12 L : 12 D photoperiod and temperatures of 30°C and 26°C, respectively. Germination in growth chambers has been shown to prevent endophytic colonization of plant tissue [14]. Prior to experimental manipulation, leaves were tested for FEF colonization. Only 2% of the 4 mm2 leaf fragments tested positively for FEF colonization, confirming that seedlings were essentially endophyte-free, consistent with previous studies. Theobroma cacao hosts no known seed-transmitted fungi.
(b). Experimental treatments and field placement
Potted seedlings were transported in sterile plastic containers to Barro Colorado Island, Panama, for experimental manipulation. Seedlings (n = 54) received one of three litter treatments: mixed species litter (n = 18), T. cacao litter (n = 18), or no litter as a control (n = 18). Mixed litter was collected from a previously established long-term ‘litter addition’ experiment on the nearby peninsula of Gigante. These plots receive bulk compilations of litter from other experimental plots on the peninsula, and are relatively well mixed and contain no T. cacao plant material. Theobroma cacao litter was collected from a healthy, disease-free, isolated T. cacao tree in Gamboa, Panama. As a result of high neotropical pathogen pressure, we only had access to one healthy cacao tree growing in nature, as opposed to a highly managed plantation. ‘No litter’ plants did not receive a litter treatment. All pots were covered with clean plastic mesh screening at the base of the plant stem in order to secure the litter treatment (if present) and to exclude foreign litter and other debris from the pot.
In May 2014, seedlings were placed in the secondary forest of Barro Colorado Island, Panama, at three heights within the canopy: 0 m (ground level), 2 m (low understorey), and 30 m (upper canopy). Six plants from each litter treatment were placed at each height, representing a full factorial experimental design. Vertical stratification was achieved by securing pots to Lutz Tower, a 48 m canopy tower that sits on a concrete base within the forest. All litter and debris present at each of the three heights was cleared away before seedling placement, and cleared daily during the duration of the experiment. Plants remained in the field for two weeks and were watered daily at soil level to avoid artificially wetting the foliage. It did rain regularly over the course of the experiment, typical of lowland moist tropical forests in the rainy season. After two weeks, seedlings were collected and placed in a covered greenhouse for a pathogen challenge experiment and endophyte community analysis.
(c). Pathogen challenge
To test whether litter exposure and canopy microsite affected host pathogen resistance, a subset of leaves from experimental plants and FEF-free control plants (n = 40 leaves distributed across 30 seedlings, including nine FEF-free control leaves distributed across seven control seedlings) were inoculated with a strain of P. palmivora previously isolated from symptomatic T. cacao in Panama. To control for possible effects of leaf age and development, we only experimentally infected leaves at certain stages (specifically stages B, C, or D [23]), which are intermediate stages of development, and have been shown to be most receptive to experimental infection by P. palmivora [23]. To infect leaves, agar plugs of P. palmivora with mycelia from the margin of the culture were placed on leaves, which were pricked with a sterile needle and received an application of 10 µl of sterile water to facilitate infection. After inoculation, plants were enclosed in a plastic chamber with wet paper towels to create a humid environment promoting colonization. After 21 days, pathogen damage was measured by photographing leaves (electronic supplementary material, figure S1) and then measuring the damaged area using ImageJ.
(d). Endophyte isolation
To determine whether FEF community composition explained differences in pathogen damage, one mature leaf per seedling was harvested for microbiome characterization following the two-week field exposure. A fully expanded leaf was removed from each seedling, rinsed under tap water, and processed within 12 h of collection. Thirty-two 4 mm2 tissue fragments were obtained haphazardly from each leaf and surface sterilized as follows: tissue fragments were submerged in 70% ethanol for 3 min, 0.525% sodium hypochlorite for 2 min, and then sterile water for 1 min. Sixteen tissue fragments were stored at −20°C until used for culture-independent analysis of fungal communities. The other 16 pieces of tissue were immediately placed in a grid in 10 cm diameter Petri plates containing 2% malt extract agar. Plates were sealed with Parafilm and incubated in a growth chamber with 12 L : 12 D cycle at a constant temperature of 22°C. Plates were monitored daily for fungal growth. Emergent hyphae were subcultured to new plates and allowed to grow until the colony covered the agar plate. Vouchers of living mycelia were suspended in sterile water and stored in the laboratory building at Barro Colorado Island.
(e). Molecular identification: culture-independent methods
Total genomic DNA was extracted directly from sterilized plant tissue using a QIAGEN DNeasy Plant Mini Kit. Primers NSA3 and NLC2 were used to amplify an approximately 1 000 bp region surrounding the entire internal transcribed spacer (ITS) region of fungal DNA [24]. Each 25 μl PCR reaction included: 13.38 µl Milli-Q water, 5 µl 5X Green GoTaq® Reaction Buffer, 3 µl MgCl2 (25 mM), 0.5 µl dNTPs (0.2 mM each dNTP), 1 µl each primer (5 µM), 0.125 µl GoTaq® DNA Polymerase, and 1 µl template. The amplification was run in an MJ Research Tetrad PTC-225 Thermal Cycler (2.5 min at 95°C, followed by 25 cycles (30 s at 95°C, 30 s at 60.2°C, 45 s at 72°C), 5 min at 72°C). PCR products were cleaned using an Omega Bio-Tek MicroElute® Cycle-Pure Kit, and sent to the Biosciences Division (BIO) Environmental Sample Preparation and Sequencing Facility (ESPSF) at Argonne National Laboratory for sequencing on the Illumina MiSeq platform. At Argonne National Laboratory, genomic DNA was amplified using modified versions of primers ITS1F and ITS2 [25]. The reverse amplification primer also contained a 12 base barcode sequence that supports pooling of up to 2 167 different samples in each lane [26,27]. Each 25 µl PCR reaction consisted of 9.5 µl of MO BIO PCR Water (Certified DNA-Free), 12.5 µl of QuantaBio AccuStart II PCR ToughMix (2× concentration, 1× final), 1 µl Golay barcode tagged Forward Primer (5 µM concentration, 200 pM final), 1 µl Reverse Primer (5 µM concentration, 200 pM final), and 1 µl of template DNA. Amplification was performed as follows: 3 min at 94°C, followed by 35 cycles (45 s at 94°C, 60 s at 50°C, 90 s at 72°C), 10 min at 72°C. Amplicons were quantified using PicoGreen (Invitrogen) and a plate reader. Once quantified, products were pooled into a single tube at equal concentration. The pool was cleaned up using AMPure XP Beads (Beckman Coulter), and then quantified using a fluorometer (Qubit, Invitrogen). After quantification, the molarity of the pool was determined and diluted to 2 nM, denatured, and then diluted to a final concentration of 6.75 pM with a 10% PhiX spike for paired 251-nucleotide read sequencing on the Illumina MiSeq platform. Reads were demultiplexed, chimaeras were removed, and reads were clustered at 97% sequence identity. Three samples had a very low number of reads and were removed prior to clustering. Identification of consensus sequences was performed using the Ribosomal Database Project (RDP) Bayesian Classifier with the Warcup ITS training set [28], and archived at GenBank under accession numbers MF148556-MF148849.
(f). Molecular identification: culture-dependent methods
Total genomic DNA was extracted directly from fungal mycelia of each fungal isolate using a QIAGEN DNeasy Plant Mini Kit. Primers ITS5 and ITS4 were used to amplify the ITS region of fungal DNA. PCR amplifications were achieved using a Thermo Scientific Phire Plant Direct PCR Kit. Each 20 μl PCR reaction included: 7.1 µl Milli-Q water, 10 µl 2× Phire Plant PCR Buffer (which included dNTPs and MgCl2), 1 µl each primer (5 µM), 0.4 µl Phire Hot Start II DNA Polymerase, and 0.5 µl template DNA. The amplification was run in an MJ Research Tetrad PTC-225 Thermal Cycler (30 s at 98°C, followed by 30 cycles (5 s at 98°C, 5 s at 62°C, 20 s at 72°C), 1 min at 72°C). Gel electrophoresis using SYBR Safe produced single bands for all products, and no bands in negative controls. PCR products were cleaned using an Omega Bio-Tek MicroElute® Cycle-Pure Kit and Sanger sequenced for both forward and reverse reads (primers ITS5 and ITS4, respectively) on an ABI3730 at the Indiana Molecular Biology Institute.
Of the 335 isolates, 313 high-quality ITS sequences were obtained. CodonCode Aligner v. 5.0.1 (CodonCode Aligner Company) was used to make base calls, perform quality assessments, and assemble consensus sequences according to 97% ITS sequence similarity, with a minimum of 40% overlap [2]. Identification of consensus sequences was performed using the RDP Bayesian Classifier with the Warcup ITS training set [28], and archived at GenBank under accession numbers MF148497-MF148555.
(g). Statistical analyses
The area of pathogen damage (cm2) experienced by each treatment group was compared with baseline pathogen damage in endophyte-free plants using ANOVA, with litter treatment, height treatment, and leaf stage as fixed effects. Height was not significant, nor was there a significant litter × height interaction, so height was removed from the model. Analysis of pathogen damage was performed using R v. 3.1.2 [29].
For both the culture-independent and culture-dependent datasets, operational taxonomic units (OTUs), designated by the ITS region, were used for ecological analyses. Species accumulation curves and estimates of total richness were inferred using EstimateS 9.0.1 [30]. Rarefaction curves were scaled by the number of accumulated samples (i.e. number of host plants sampled) to depict species density for culture-independent (electronic supplementary material, figure S2a) and culture-dependent (electronic supplementary material, figure S2b) methods [31].
All other community analyses were performed using R v. 3.1.2 [29]. ANOVA was used to compare culture-based isolation frequency across height and litter treatments. Assumptions of normality and homogeneity of variance were tested and met. To examine the effects of experimental treatments on FEF community composition, permutational multivariate analysis of variance (PERMANOVA), using distance matrices was used with the Bray–Curtis dissimilarity index (VEGAN Package, function adonis). For the culture-dependent dataset, PERMANOVA was performed on all non-singleton OTUs, and for the culture-independent dataset, data were Hellinger-transformed prior to ordination and diversity analyses. The ordination goodness-of-fit was measured by the stress value [32]. Non-metric multidimensional scaling (NMDS) ordinations were created in VEGAN to visualize community similarities across height and litter treatments [33]. The Shannon diversity index was calculated for both the culture-dependent and culture-independent datasets using the VEGAN Package. Patterns of co-occurrence of endophytes were analysed within individual plants in both datasets using C-score analysis, which indicates if species co-occur more or less often than predicted by a null model (VEGAN Package, function oecosimu). A structured community (i.e. significant C-score statistic) implies ecological mechanisms of community assembly as opposed to purely random processes. Indicator species analysis was performed on the culture-independent dataset (INDICSPECIES Package, function multipatt). For the culture-independent dataset, the number of reads of the dominant OTU (best hit: Colletotrichum tropicale) was compared across litter treatments using ANOVA. Height and total number of reads were included as fixed effects. The model was also run combining read counts for all OTUs that had a best match of C. tropicale, but this did not affect the significance of the model. Thus, we report results from the model that included read counts for only the dominant OTU as the response variable, in order to maintain a conservative estimate of how this one OTU affected disease outcomes. For the culture-dependent dataset, the percentage of isolates of the dominant OTU (best hit: C. tropicale) was compared across litter treatments using ANOVA. Height treatment was included as a fixed effect. Additionally, for the culture-independent dataset, a regression analysis was performed on log-transformed data to test if the number of reads of the dominant OTU correlated with pathogen damage across treatments with plant individual as a random effect. The R2 value for the mixed effects model was calculated using the MuMIn Package (function r.squaredGLMM).
3. Results and discussion
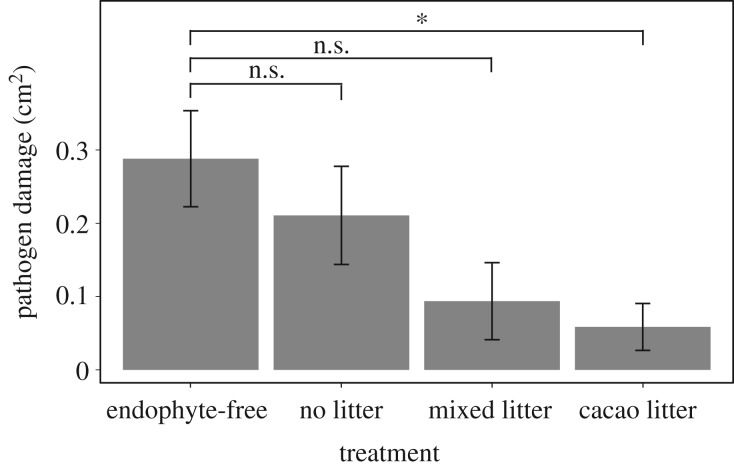
Three weeks after infection by P. palmivora, we measured leaf necrosis (electronic supplementary material, figure S1), which differed significantly among litter treatments (d.f. = 3, F = 3.133, p = 0.038; figure 1). Plants exposed to cacao litter experienced significantly less pathogen damage than FEF-free controls (p = 0.027). By contrast, damage was not significantly reduced in the no litter, (p = 0.947) and mixed litter (p = 0.071) treatments. Further, exposure to conspecific litter reduced pathogen damage to approximately 50% of the damage on seedlings exposed to mixed litter (figure 1). The ANOVA included leaf development stage as a fixed effect (see Material and methods), which was significant (d.f. = 2, F = 6.720, p = 0.003). Phytophthora palmivora has a wide range of dispersal mechanisms, and can be transmitted through rain, soil, or insects to infect pod, leaf, or seedling tissues [20]. While it is feasible that pathogen pressure from P. palmivora may vary throughout the canopy, canopy microsite did not significantly affect plant response to pathogen damage (d.f. = 2, F = 2.101, p = 0.139) and was removed from the full model.
Figure 1.
Exposure to cacao litter reduced pathogen damage. Compared with endophyte-free controls, plants exposed to cacao litter experienced significantly less pathogen damage, plants treated with mixed litter experienced marginally significantly less damage, and no litter plants did not experience reduced damage. Canopy microsite was not significant. Error bars represent standard error of the mean.
To determine whether FEF community composition explained differences in pathogen damage, we used next-generation sequencing (NGS) on the Illumina platform, as well as a culture-based approach coupled with Sanger sequencing. FEF provide an excellent opportunity to compare culture-independent and culture-dependent methods, allowing for a more detailed perspective of the plant microbiome. Most FEF are culturable, so culture-based approaches can inform abundance and proportion of different FEF species in tissue, while culture-independent approaches also improve sequencing depth [1]. Both of these methods are subject to inherent biases: in culture based-approaches, stronger competitors may emerge from tissue first, to the exclusion of weaker competitors. In culture-independent approaches, PCR bias may skew the representation of certain OTUs in the species pool. Given that we used nested PCR, the high number of amplification rounds is a potential source of bias. Using both culture-based and culture-independent methods helps counterbalance biases inherent to each individual method.
Following culturing, we obtained 335 isolates representing 59 OTUs (based on Sanger sequencing) from 864 tissue fragments, with at least one isolate recovered from 51 of 54 leaves (electronic supplementary material, table S1). Isolation frequency differed significantly among litter treatments (d.f. = 2, F = 3.841, p = 0.028), with more isolates obtained from no litter plants compared with plants exposed to cacao litter (p = 0.025) (figure 2a). Isolation frequency also varied among microsites (d.f. = 2, F = 6.164, p = 0.004), with fewer total isolates obtained from seedlings placed high in the canopy compared with low in the canopy (p = 0.036) or at ground level (p = 0.004; figure 2b). The most abundant OTU in the culture-based dataset comprised 35% of isolates (116/335), and its best taxonomic match was C. tropicale. Colletotrichum tropicale is the dominant species of FEF found in healthy T. cacao leaves in Panama, and has been previously reported to enhance pathogen resistance when inoculated as a pure culture into cacao hosts [15,23].
Figure 2.
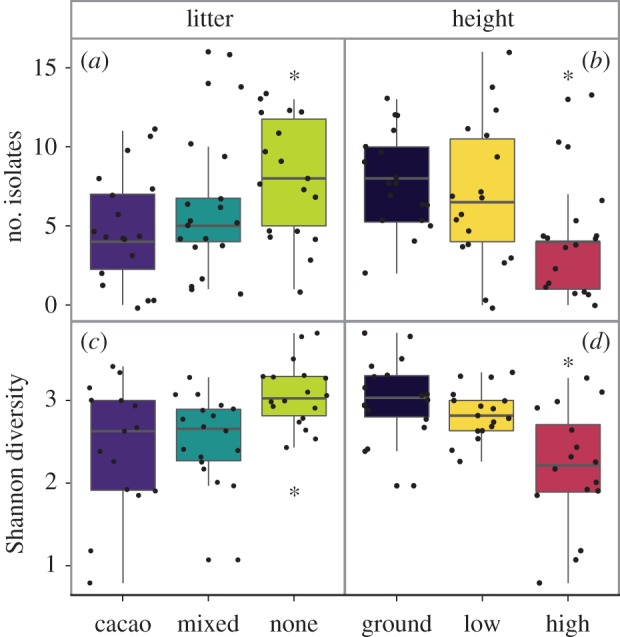
Litter and microsite affected density and diversity of endophytic fungi. (a) The number of isolated endophytes differed among litter treatments, with more isolates from no litter plants compared with plants exposed to cacao litter. (b) The number of isolated endophytes differed among microsites, with fewer isolates from seedlings placed high in the canopy compared with low in the canopy or at ground level. (c) Shannon diversity differed among litter treatments (pictured: NGS dataset). Endophyte diversity in no litter plants was significantly higher than plants exposed to cacao or mixed litter. (d) Shannon diversity differed significantly based on placement in the canopy (pictured: NGS dataset). Endophyte diversity was lower for seedlings placed high in the canopy compared with low in the canopy or at ground level. Dots represent individual hosts. Error bars represent standard error of the mean.
Independently, using NGS, we obtained 2 127 572 reads from the other 864 tissue fragments. This method identified five times more OTUs (294) than the culture-based method using the same amount of leaf tissue (electronic supplementary material, table S2). NGS was particularly useful for identifying rare members of the microbiome, and revealed a greater total number of OTUs. There was substantial taxonomic overlap between the datasets, with only 5% of isolates identified by Sanger sequencing unrepresented at the genus level in the NGS approach. Consistent with culture-based identification, the most abundant OTU produced by NGS again corresponded to C. tropicale (22% of reads).
Statistical analyses of FEF communities from both the NGS and culture-based approaches were qualitatively similar. Litter treatment, microsite, and their interaction significantly predicted FEF community composition in the NGS dataset (PERMANOVA; Litter: F2,50 = 1.888, R2 = 0.063, p = 0.003; Height: F2,50 = 4.359, R2 = 0.146, p = 0.001; Interaction: F4,50 = 1.313, R2 = 0.088, p = 0.033; figure 3a–c). Differences among microsites were qualitatively similar for the culture-based dataset for non-singleton OTUs (PERMANOVA; Height: F2,48 = 2.715, R2 = 0.106, p = 0.003), although the litter treatment (F2,48 = 0.982, R2 = 0.038, p = 0.447) and the height × litter interaction (F2,48 = 1.007, R2 = 0.078, p = 0.457) were not significant in the culture-based dataset. FEF co-occurred non-randomly compared with a null model (NGS: C-score = 17.67, z = −161.87, p < 0.001; Culture-based: C-score = 5.71, z = −34.06, p < 0.001), suggesting that treatments affected FEF community assembly. Shannon diversity also differed significantly among litter treatments (NGS: d.f. = 2, F = 6.668, p = 0.003; culture-based: d.f. = 2, F = 6.231, p = 0.004; figure 2c) and microsites (NGS: d.f. = 2, F = 21.342, p < 0.001); not significant for the culture-based dataset (d.f. = 2, F = 1.704, p = 0.192; figure 2d). Specifically, in the NGS dataset, plants in the no litter treatment harboured more diverse FEF communities than mixed (p = 0.019) and cacao (p = 0.007) litter treatments, and plants in the canopy had less diverse FEF communities than those in the understorey (p = 0.002) or at ground level (p < 0.001). There was a significant interaction between litter and microsite that affected FEF diversity in the culture-independent dataset (d.f. = 4, F = 5.013, p = 0.002). Specifically, FEF communities of no litter plants maintained high levels of diversity at 30 m, whereas plants exposed to cacao or mixed litter experienced a significant drop in FEF diversity at this height (electronic supplementary material, figure S3). This interaction was not significant for the culture-dependent dataset (d.f. = 4, F = 0.394, p = 0.812).
Figure 3.
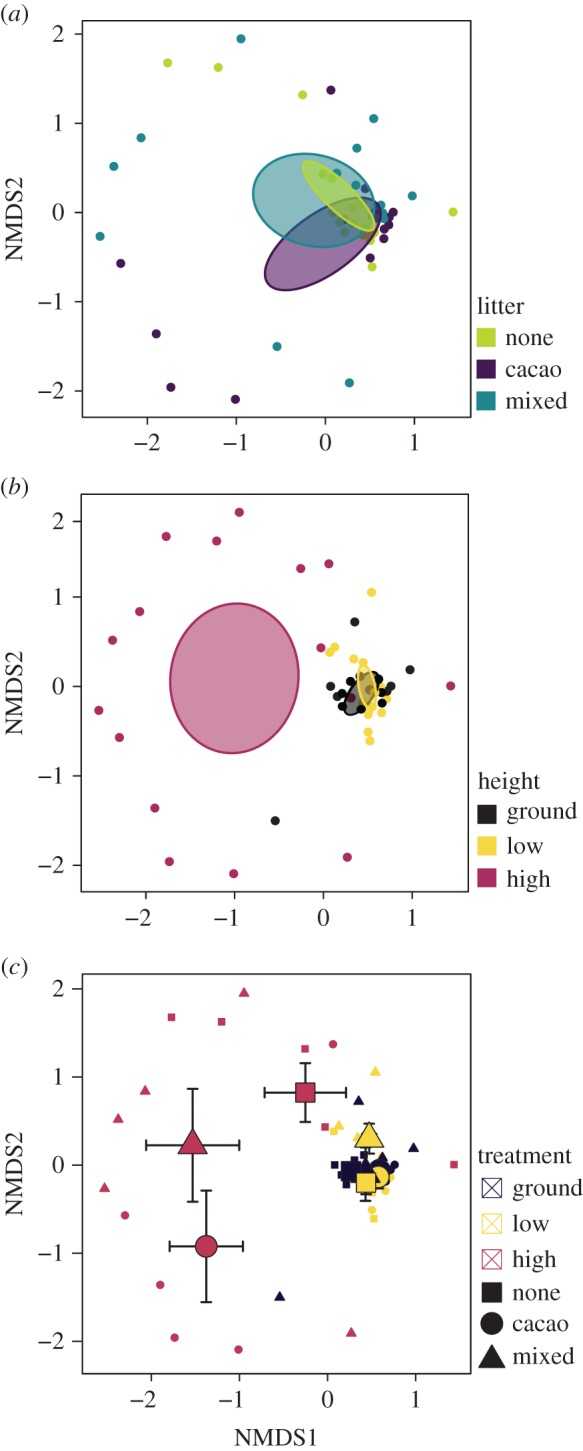
Biotic and abiotic factors affected endophyte community composition. (a) Litter exposure, (b) canopy microsite, and (c) their interaction predicted endophyte community composition. NMDS plots are based on the NGS dataset, but differences were qualitatively similar for the culture-based dataset for non-singleton OTUs. Error bars represent standard error of centroids.
Few studies have analysed vertical distribution of FEF in forests [34], but our results are consistent with a recent study reporting lower FEF abundance and richness in the canopy of European Ash [17]. The negative relationship between height and FEF density and diversity observed here is likely caused by hotter, drier, and brighter canopy conditions that constrain which FEF colonize and grow, despite the presence of litter in some treatments [19]. While previous work investigating vertical stratification of FEF communities has focused on surveying established FEF communities in adult canopy trees [35], it is not feasible to experimentally manipulate the endophyte status of individual leaves on adult trees. Instead, we placed FEF-free seedlings at varying heights in the canopy as sentinels for FEF colonization. This approach allowed us to standardize leaf material and control for potential physiological differences that could influence de novo endophyte colonization and pathogen resistance, while also testing the effect of abiotic conditions on the assembly of FEF communities. Moreover, P. palmivora infects cacao pods and leaves throughout a tree [36], so controlling for leaf age and development allowed us to test the hypothesis that microbiomes that assemble at different heights may be more or less effective at defending plants from pathogens. Our results suggest that position within the canopy had no effect on seedling pathogen damage, despite large changes in microbiome composition. However, the effect of height may have been obscured in litter-treated plants given that litter was added to pots.
It was unexpected that plants not exposed to litter were colonized by more abundant and diverse FEF than litter-treated seedlings, as this contrasts with previous studies [16], but may reflect differences in the spatial scale of litter manipulation. While previous work compared FEF colonization rates following litter manipulation across large forest plots, we manipulated litter at the extremely local scale of a single pot. Our results suggest that when local leaf litter is present, FEF from the litter quickly colonize nearby host plants and exert an inhibitory priority effect on later colonizers. Conversely, without local litter inocula as a source of FEF common to healthy adults, seedlings were colonized by a greater density and diversity of weedy, highly dispersible species. This was supported by indicator species analysis, which revealed more unique taxa associated with no litter seedlings and seedlings at 30 m (electronic supplementary material, table S3). Despite strong effects of vertical stratification on FEF, differences in pathogen damage were only attributable to litter treatment. Exposure to litter could have affected host pathogen response in several ways, including changing soil nutrient content or leaf chemistry. However, we found that exposure to conspecific litter significantly changed the relative abundance of component FEF species, which significantly correlated with host pathogen resistance.
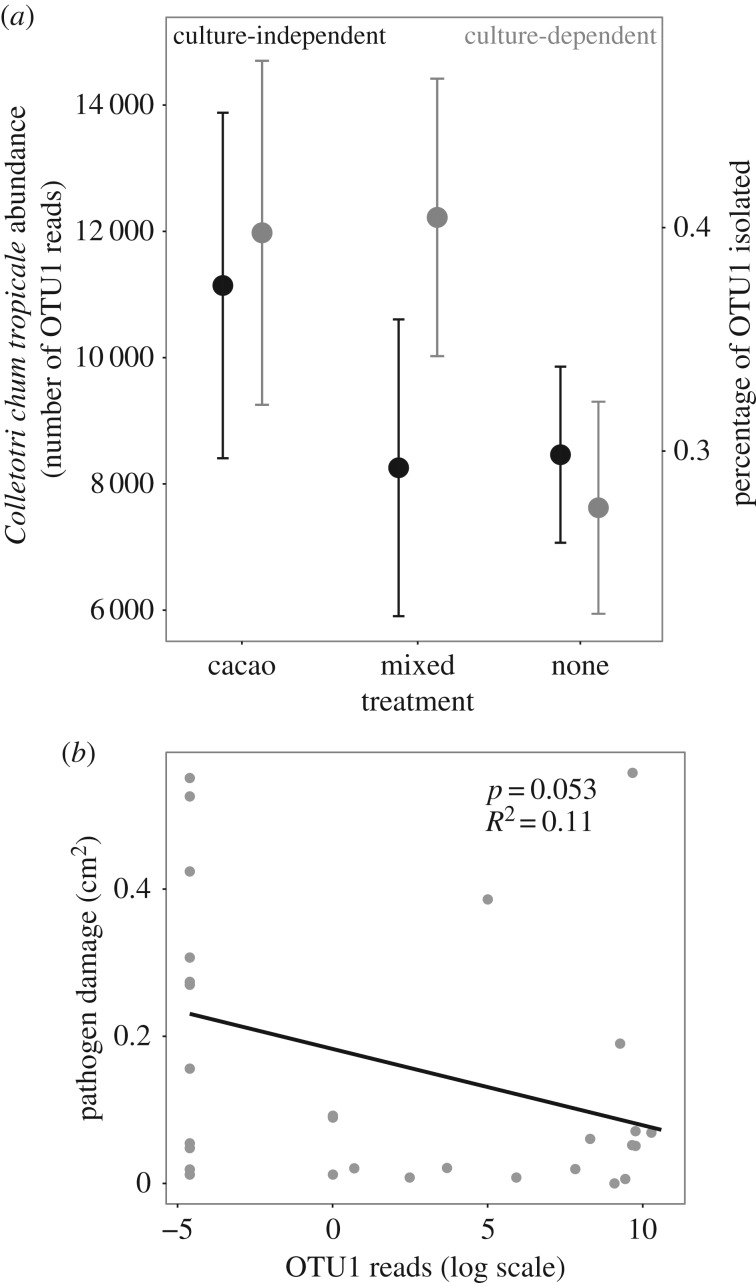
Colletotrichum tropicale was the most common OTU in both of our datasets, and has previously been identified as the most common fungal component of the healthy T. cacao microbiome in Panama [37]. FEF communities in seedlings exposed to cacao litter were characterized by increased dominance of C. tropicale (measured in the NGS dataset by the number of reads of OTU1; d.f. = 2, F = 3.674, p = 0.034; figure 4a), despite lower overall FEF density and diversity (figure 2a,c). Microsite and total number of reads were included in the ANOVA as fixed effects, and were both significant (Height: d.f. = 2, F = 11.405, p < 0.001; Total Number of Reads: d.f. = 1, F = 4.235, p = 0.046). For the culture-based dataset, this trend was qualitatively the same. Moreover, the number of reads of C. tropicale was negatively correlated with pathogen damage across all treatments (d.f. = 1, χ2 = 3.740, R2 = 0.11, p = 0.053; figure 4b). Taken together, these results suggest that C. tropicale was acting as a pathogen inhibitor, and that this effect was the largest in seedlings exposed to cacao litter, where C. tropicale abundance was the highest.
Figure 4.
The abundance of C. tropicale correlates with both litter treatment and pathogen damage. (a) Seedlings exposed to cacao litter were more strongly dominated by C. tropicale. There were significant differences in the number of reads of OTU1 (best match C. tropicale) across litter treatments (plotted in black: number of reads in the NGS dataset). For the culture-based dataset, this trend was qualitatively the same (plotted in grey: percentage of isolates identified as OTU1, for leaves with more than one isolate). Error bars represent standard error of the mean. (b) Across all treatments in the NGS dataset, the number of reads of OTU1 (best match C. tropicale) negatively correlated with pathogen damage.
We did not experimentally test the specific role of C. tropicale or other taxa as part of these experiments. However, previous work has repeatedly shown that artificial inoculations of C. tropicale reduced severity of P. palmivora damage in leaf [7] and fruit [15] tissues in cacao, due to upregulation of defensive pathways in cacao [14]. While infection by P. palmivora triggers an innate plant immune response by upregulating pathogenesis-related proteins [38], inoculation by C. tropicale enhances that immune response by inducing the upregulation of hundreds of cacao genes, including those related to defence [14]. Ultimately, this FEF-induced genetic response results in less severe Phytophthora damage than that of FEF-free plants [14]. Our results build upon the extensive previous research on the genetic pathways of T. cacao, and how they are affected by C. tropicale and P. palmivora, to link multiple levels of biological organization. Specifically, by identifying how the component members of the microbiome differentially affect host gene expression, we may be able to predict the functional outcomes of microbial community assembly in nature.
While there are other important members of the foliar microbiome besides fungi, the existing literature has not yet pointed to a comparable role of these microorganisms in plant leaves relative to the many well-documented studies of FEF that demonstrate their important functional roles for hosts [1]. Based on the results presented here, we suggest that future studies conduct similar experiments in other plant systems and with other phyllosphere components, such as bacteria, viruses, and microbiota with an epiphytic habit. Additionally, the ecological patterns found in this study are consistent with other more disparate systems. For example, our results suggest that litter from healthy conspecifics triggers priority effects during FEF community assembly, promoting colonization by species such as C. tropicale that are effective at inhibiting further microbial colonization, including infection by pathogens. Priority effects have been documented for seed-infecting fungal endophytes in some plant species (though not in T. cacao), which may host one endophyte per seed as a result of strong exclusionary interactions among endophytes [39]. Our results also directly parallel human microbiome research, which has demonstrated that vaginally delivered infants are colonized by a maternal, Lactobacillus-dominated microbiome, while infants delivered via C-section are more broadly colonized by the surrounding environment. These differences in colonization result in greater susceptibility to methicillin-resistant Staphylococcus aureus infections in C-section newborns [21].
Our results demonstrating increased pathogen resistance in seedlings following exposure to healthy conspecific litter appear to run counter to the predictions of the Janzen–Connell hypothesis. The Janzen–Connell hypothesis proposes that a tree's offspring have higher fitness when they are dispersed farther from the parent, due to localized accumulation of host-specific enemies [22,40]. Although cacao seedlings near conspecific adults may experience greater enemy pressure [41], we found that local FEF communities derived from healthy conspecific adults can also promote pathogen resistance in conspecific seedlings, potentially by mobilizing plant defences [14]. Similar patterns are also emerging in belowground studies of other plant systems. For instance, the rhizosphere microbiome can suppress soil-borne pathogens [42], and recent work showed that arbuscular mycorrhizal fungi (AMF) can neutralize the Janzen–Connell effect by mediating pathogen damage on seedlings [43]. Conversely, another recent study demonstrated that tree species associated with AMF have reduced conspecific seedling establishment because AMF associated with conspecific adults are not as effective at preventing pathogens compared with ectomycorrhizal hosts that inhibit pathogen damage to seedlings [44]. Thus, it is becoming increasingly clear that there are opposing forces (e.g. pathogens and mutualists) acting synchronously within the general Janzen–Connell framework. Moreover, while co-infections by mutualists and pathogens may occur commonly in nature, such interactions are an underappreciated nuance of the existing theoretical framework. These considerations may help to explain why the strength of Janzen–Connell effects varies across host species or geographical regions [22,41]. Our results demonstrate that the aboveground plant microbiome must also be considered when predicting how plant–microbe interactions affect host health and community interactions [7,14,16]. Thus, our results have far-reaching implications for host–microbe interactions, and provide strong impetus for future ecological research. Our results also suggest that the application of litter from healthy hosts could be an effective agricultural strategy to reduce crop losses with smaller economic and environmental costs than current practices [45].
Supplementary Material
Supplementary Material
Acknowledgements
We are grateful to Posy Busby, Luke Henry, and Briana Whitaker for their feedback on earlier drafts of this manuscript. Raul Ruiz, Marta Vargas-Timchenko, Enith Rojas, Amanda Winters, Kristin Saltonstall, Adalberto Gomez, Justin Shaffer, James DeVore, Courtney Sullivan, and Noelle Visser assisted with the experiment. Betsy Arnold provided valuable advice. Illumina sequencing was performed at Argonne National Laboratory, and the Indiana University Center for Genomics and Bioinformatics assisted with bioinformatics. Permission to do this research was granted by the Autoridad Nacional del Ambiente de Panamá (ANAM).
Data accessibility
Supporting data are available in the electronic supplementary material. Raw sequence data are archived at GenBank under accession numbers MF148497-MF148849.
Authors' contributions
N.C. designed and conducted the experiments, analysed the data, and wrote the first draft. K.C., E.A.H., and L.C.M. advised on experimental design and provided important revisions. All authors gave final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
N.C. was supported by an NSF Graduate Research Fellowship. This research was funded by a Smithsonian Tropical Research Institute Short Term Fellowship (N.C.), and grants from the Mycological Society of America (N.C.), Garden Club of America (N.C.), and Indiana University (N.C.). This work was also supported by a grant from the Simons Foundation (429440, WTW).
References
- 1.Christian N, Whitaker BK, Clay K. 2015. Microbiomes: unifying animal and plant systems through the lens of community ecology theory. Front. Microbiol. 6, 1–15. ( 10.3389/fmicb.2015.00869) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Del Olmo-Ruiz M, Arnold AE. 2014. Interannual variation and host affiliations of endophytic fungi associated with ferns at La Selva, Costa Rica. Mycologia 106, 8–21. ( 10.3852/13-098) [DOI] [PubMed] [Google Scholar]
- 3.Adame-Álvarez R-M, Mendiola-Soto J, Heil M. 2014. Order of arrival shifts endophyte–pathogen interactions in bean from resistance induction to disease facilitation. FEMS Microbiol. Lett. 355, 100–107. ( 10.1111/1574-6968.12454) [DOI] [PubMed] [Google Scholar]
- 4.Christian N, Sullivan C, Visser ND, Clay K. 2016. Plant host and geographic location drive endophyte community composition in the face of perturbation. Microb. Ecol. 72, 621–632. ( 10.1007/s00248-016-0804-y) [DOI] [PubMed] [Google Scholar]
- 5.Ernst M, Mendgen KW, Wirsel SGR. 2003. Endophytic fungal mutualists: seed-borne Stagonospora spp. enhance reed biomass production in axenic microcosms. Mol. Plant. Microbe. Interact. 16, 580–587. ( 10.1094/MPMI.2003.16.7.580) [DOI] [PubMed] [Google Scholar]
- 6.Bae H, Sicher RC, Kim MS, Kim S-H, Strem MD, Melnick RL, Bailey BA. 2009. The beneficial endophyte Trichoderma hamatum isolate DIS 219b promotes growth and delays the onset of the drought response in Theobroma cacao. J. Exp. Bot. 60, 3279–3295. ( 10.1093/jxb/erp165) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arnold A, Mejía L, Kyllo D, Rojas EI, Maynard Z, Robbins N, Herre EA. 2003. Fungal endophytes limit pathogen damage in a tropical tree. Proc. Natl Acad. Sci. USA 100, 15 649–15 654. ( 10.1073/pnas.2533483100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Busby PE, Peay KG, Newcombe G. 2016. Common foliar fungi of Populus trichocarpa modify Melampsora rust disease severity. New Phytol. 209, 1681–1692. ( 10.1111/nph.13742) [DOI] [PubMed] [Google Scholar]
- 9.Clay K, Hardy T, Hammond A Jr. 1985. Fungal endophytes of grasses and their effects on an insect herbivore. Oecologia 66, 1–5. ( 10.1007/BF00378545) [DOI] [PubMed] [Google Scholar]
- 10.Estrada C, Degner EC, Rojas EI, Wcislo WT, Van Bael SA. 2015. The role of endophyte diversity in protecting plants from defoliation by leaf-cutting ants. Curr. Sci. 109, 19–25. [Google Scholar]
- 11.Busby PE, Ridout M, Newcombe G. 2015. Fungal endophytes: modifiers of plant disease. Plant Mol. Biol. 90, 645–655. ( 10.1007/s11103-015-0412-0) [DOI] [PubMed] [Google Scholar]
- 12.Rodriguez Estrada AE, Hegeman A, Corby Kistler H, May G. 2011. In vitro interactions between Fusarium verticillioides and Ustilago maydis through real-time PCR and metabolic profiling. Fungal Genet. Biol. 48, 874–885. ( 10.1016/j.fgb.2011.06.006) [DOI] [PubMed] [Google Scholar]
- 13.Alabouvette C, Olivain C, Migheli Q, Steinberg C. 2009. Microbiological control of soil-borne phytopathogenic fungi with special emphasis on wilt-inducing Fusarium oxysporum. New Phytol. 184, 529–544. ( 10.1111/j.1469-8137.2009.03014.x) [DOI] [PubMed] [Google Scholar]
- 14.Mejía LC, et al. 2014. Pervasive effects of an endophytic fungus on host genetic and phenotypic expression in a tropical tree. Front. Microbiol. 5, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mejía L., Rojas EI, Maynard Z, Van Bael SA, Arnold AE, Hebbar P, Samuels GJ, Robbins N, Herre EA. 2008. Endophytic fungi as biocontrol agents of Theobroma cacao pathogens. Biol. Control 46, 4–14. ( 10.1016/j.biocontrol.2008.01.012) [DOI] [Google Scholar]
- 16.Herre EA, Mejía LC, Kyllo DA, Rojas E, Maynard Z, Butler A, Van Bael SA. 2007. Ecological implications of anti-pathogen effects of tropical fungal endophytes and mycorrhizae. Ecology 88, 550–558. ( 10.1890/05-1606) [DOI] [PubMed] [Google Scholar]
- 17.Scholtysik A, Unterseher M, Otto P, Wirth C. 2013. Spatio-temporal dynamics of endophyte diversity in the canopy of European ash (Fraxinus excelsior). Mycol. Prog. 12, 291–304. ( 10.1007/s11557-012-0835-9) [DOI] [Google Scholar]
- 18.Griffin EA, Carson WP. 2015. The ecology and natural history of foliar bacteria with a focus on tropical forests and agroecosystems. Bot. Rev. 81, 105–149. ( 10.1007/s12229-015-9151-9) [DOI] [Google Scholar]
- 19.Gilbert G, Reynolds D, Bethancourt A. 2007. The patchiness of epifoliar fungi in tropical forests: host range, host abundance, and environment. Ecology 88, 575–581. ( 10.1890/05-1170) [DOI] [PubMed] [Google Scholar]
- 20.Guest D. 2007. Black pod: diverse pathogens with a global impact on cocoa yield. Phytopathology 97, 1650–1653. ( 10.1094/PHP-2001-0709-01-RV) [DOI] [PubMed] [Google Scholar]
- 21.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R, Gordon JI. 2010. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl Acad. Sci. USA 107, 11 971–11 975. ( 10.1073/pnas.1002601107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Comita LS, Queenborough SA, Murphy SJ, Eck JL, Xu K, Krishnadas M, Beckman N, Zhu Y. 2014. Testing predictions of the Janzen–Connell hypothesis: a meta-analysis of experimental evidence for distance- and density-dependent seed and seedling survival. J. Ecol. 102, 845–856. ( 10.1111/1365-2745.12232) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mejía LC, Guiltinan MJ, Shi Z, Landherr L, Maximova SN. 2012. Expression of designed antimicrobial peptides in Theobroma cacao L. trees reduces leaf necrosis caused by Phytophthora spp. In Small wonders: peptides for disease control (eds Rajasekaran K, Cary JW, Jaynes JM, Montesinos E), pp. 379–395. Washington, DC: American Chemical Society. [Google Scholar]
- 24.Martin KJ, Rygiewicz PT. 2005. Fungal-specific PCR primers developed for analysis of the ITS region of environmental DNA extracts. BMC Microbiol. 5, 1–11. ( 10.1186/1471-2180-5-28) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith DP, Peay KG. 2014. Sequence depth, not PCR replication, improves ecological inference from next generation DNA sequencing. PLoS ONE 9, e90234 ( 10.1371/journal.pone.0090234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caporaso JG, et al. 2010. Correspondence QIIME allows analysis of high-throughput community sequencing data intensity normalization improves color calling in SOLiD sequencing. Nat. Publ. Gr. 7, 335–336. ( 10.1038/nmeth0510-335) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caporaso JG, et al. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. ( 10.1038/ismej.2012.8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deshpande V, Wang Q, Greenfield P, Charleston M, Porras-Alfaro A, Kuske CR, Cole JR, Midgley DJ, Tran-Dinh N. 2016. Fungal identification using a Bayesian classifier and the Warcup training set of internal transcribed spacer sequences. Mycologia 108, 1–5. ( 10.3852/14-293) [DOI] [PubMed] [Google Scholar]
- 29.R Core Development Team. 2008. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; see http://www.R-project.org. [Google Scholar]
- 30.Colwell RK. 2013. EstimateS: statistical estimation of species richness and shared species from samples. Version 9. User's Guide and application at http://purl.oclc.org/estimates .
- 31.Gotelli NJ, Colwell RK. 2001. Quantifying biodiversity: procedures and pitfalls in the measurement and comparison of species richness. Ecol. Lett. 4, 379–391. ( 10.1046/j.1461-0248.2001.00230.x) [DOI] [Google Scholar]
- 32.Legendre P, Legendre L. 1998. Numerical ecology, 2nd edn Amsterdam, The Netherlands: Elsevier Science BV. [Google Scholar]
- 33.Zimmerman NB, Vitousek PM. 2012. Fungal endophyte communities reflect environmental structuring across a Hawaiian landscape. Proc. Natl Acad. Sci. USA 109, 13 022–13 027. ( 10.1073/pnas.1209872109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gamboa MA, Bayman P. 2001. Communities of endophytic fungi in leaves of a tropical timber tree (Guarea guidonia: Meliaceae). Biotropica 33, 352–360. ( 10.1111/j.1744-7429.2001.tb00187.x) [DOI] [Google Scholar]
- 35.Harrison JG, Forister ML, Parchman TL, Koch GW. 2016. Vertical stratification of the foliar fungal community in the worlds tallest trees. Am. J. Bot. 103, 1–9. ( 10.3732/ajb.1600277) [DOI] [PubMed] [Google Scholar]
- 36.Vanegtern B, Rogers M, Nelson S. 2015. Black pod rot of cacao caused by Phytophthora palmivora. Plant Dis. 108, 1–5. [Google Scholar]
- 37.Rojas EI, et al. 2010. Colletotrichum gloeosporioides s.l. associated with Theobroma cacao and other plants in Panama: multilocus phylogenies distinguish host-associated pathogens from asymptomatic endophytes. Mycologia 102, 1318–1338. ( 10.3852/09-244) [DOI] [PubMed] [Google Scholar]
- 38.Fister AS, Mejia LC, Zhang Y, Herre EA, Maximova SN, Guiltinan MJ. 2016. Theobroma cacao L. pathogenesis-related gene tandem array members show diverse expression dynamics in response to pathogen colonization. BMC Genomics 17, 363 ( 10.1186/s12864-016-2693-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raghavendra AKH, Newcombe G, Shipunov A, Baynes M, Tank D. 2013. Exclusionary interactions among diverse fungi infecting developing seeds of Centaurea stoebe. FEMS Microbiol. Ecol. 84, 143–153. ( 10.1111/1574-6941.12045) [DOI] [PubMed] [Google Scholar]
- 40.Johnson DJ, Beaulieu WT, Bever JD, Clay K. 2012. Conspecific negative density dependence and forest diversity. Science 336, 904–907. ( 10.1126/science.1220269) [DOI] [PubMed] [Google Scholar]
- 41.Mangan SA, Schnitzer SA, Herre EA, Mack KML, Valencia MC, Sanchez EI, Bever JD. 2010. Negative plant-soil feedback predicts tree-species relative abundance in a tropical forest. Nature 466, 752–755. ( 10.1038/nature09273) [DOI] [PubMed] [Google Scholar]
- 42.Mendes R, et al. 2011. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332, 1097–1100. ( 10.1126/science.1203980) [DOI] [PubMed] [Google Scholar]
- 43.Liang M, Liu X, Etienne RS, Huang F, Wang Y, Yu S. 2015. Arbuscular mycorrhizal fungi counteract the Janzen–Connell effect of soil pathogens. Ecology 96, 562–574. ( 10.1890/14-0871.1) [DOI] [PubMed] [Google Scholar]
- 44.Bennett J, Maherali H, Reinhart K, Lekberg Y, Hart M, Klironomos J. 2017. Plant-soil feedbacks andmycorrhizal type influence temperate forest population dynamics. Science 355, 181–184. ( 10.1126/science.aai8212) [DOI] [PubMed] [Google Scholar]
- 45.Clay K. 2004. Fungi and the food of the gods. Nature 427, 401–402. ( 10.1038/427401a) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supporting data are available in the electronic supplementary material. Raw sequence data are archived at GenBank under accession numbers MF148497-MF148849.