Abstract
Gastric cancer is a leading cause of cancer-related deaths worldwide. The Tff1 knockout (KO) mouse model develops gastric lesions that include low-grade dysplasia (LGD), high-grade dysplasia (HGD), and adenocarcinomas. In this study, we used Affymetrix microarrays gene expression platforms for analysis of molecular signatures in the mouse stomach (Tff1-KO (LGD) and Tff1 wild-type (normal)) and human gastric cancer tissues and their adjacent normal tissue samples. Combined integrated bioinformatics analysis of mouse and human datasets indicated that 172 genes were consistently deregulated in both human gastric cancer samples and Tff1-KO LGD lesions (P<0.05). Using Ingenuity pathway analysis, these genes mapped to important transcription networks that include MYC, STAT3, β-catenin, RELA, NFATC2, HIF1A, and ETS1 in both human and mouse. Further analysis demonstrated activation of FOXM1 and inhibition of TP53 transcription networks in human gastric cancers but not in Tff1-KO LGD lesions. Using real-time RT-PCR, we validated the deregulated expression of several genes (VCAM1, BGN, CLDN2, COL1A1, COL1A2, COL3A1, EpCAM, IFITM1, MMP9, MMP12, MMP14, PDGFRB, PLAU, and TIMP1) that map to altered transcription networks in both mouse and human gastric neoplasia. Our study demonstrates significant similarities in deregulated transcription networks in human gastric cancer and gastric tumorigenesis in the Tff1-KO mouse model. The data also suggest that activation of MYC, STAT3, RELA, and β-catenin transcription networks could be an early molecular step in gastric carcinogenesis.
Keywords: Gastric cancer, TFF1, MYC, STAT3, β-catenin
Introduction
Gastric cancer is the third leading cause of cancer mortality globally, responsible for 723,000 deaths, 8.8% of the total cancer-related deaths worldwide 1. Diagnosis of gastric cancer is usually made at advanced stages of the disease due to lack of symptoms in early stages of gastric tumorigenesis. Therefore, most gastric cancer patients have poor prognosis 2. Comprehensive understanding of molecular alterations in gastric cancer is necessary for early detection, treatment, and prevention. Trefoil factor 1 (TFF1) is a small-secreted peptide that is predominantly expressed in human normal gastric mucosa and its silencing and reduced expression level are observed in a majority of human gastric cancers 3–6. Of note, our previous studies demonstrated that knockout of trefoil factor 1 (Tff1) gene expression in mouse successfully induced a cascade of gastric lesions that include low-grade dysplasia (LGD), high-grade dysplasia (HGD), and gastric adenocarcinoma 7. Genetically engineered mouse models provide an excellent platform to study human diseases including cancer 8. Although several molecular studies of gastric cancer have been reported 9, there is a crucial need for molecular characterization of mouse models of this disease to assess their similarity to human gastric cancer and suitability for in vivo studies. Studies of animal models can provide a better understanding of early changes in gastric tumorigenesis that can possibly improve our current diagnostic, prognostic, and possibly therapeutic approaches in gastric cancer.
Activation of WNT/β-catenin, MYC, and STAT transcription factors plays an important role in initiation and progression of several cancers 10–13. The role of β-catenin activation and cancer is best exemplified in the cascade of colon carcinogenesis 14,15. The aberrant activation of the Wnt/β-catenin signaling pathway has been described in 30% to 60% of gastric cancer tissues and in gastric cancer cell lines 16,11,17–19. The conserved Wnt/β-Catenin pathway regulates stem cell pluripotency and cell fate decisions during development 11. A recent study has suggested that Wnt/β-catenin signaling may be involved in the self-renewal of gastric cancer stem cells (GCSC) 12. The oncogenic MYC family encodes N-myc, c-myc, and L-myc transcription factors’ proteins 20. MYC plays a fundamental role in several cellular functions, including regulation of cell growth, proliferation, metabolism, differentiation, apoptosis, and angiogenesis 21,22. MYC activation has been reported in different types of cancers that include gastric cancer 10,23,24. Similar to MYC, the signal transducer and activator of transcription protein 3 (STAT3) participates in a series of tumorigenic processes including cell proliferation, cell survival, anti-apoptosis, angiogenesis, drug resistance, immune evasion, and inflammation 25,26. STAT3 is constitutively activated in several human cancers including thyroid, lung, ovarian, breast, colon, and gastric cancer 27,28. Inhibition of STAT3 has anti-tumor effects in several human cancer models 26,29,30.
In this study, we investigated the aberrant gene expression signature and transcription networks in early dysplastic gastric lesions from mouse and human gastric cancer tissue samples. Using integrated bioinformatics analysis approaches, we identified similar molecular signatures and transcription networks in mouse and human neoplastic lesions. The observed similarities suggest that activation of these pathways could be an early step in initiation of gastric tumorigenesis. The results also denote that the Tff1 KO mouse is an excellent model for in vivo studies of molecular mechanisms in gastric tumorigenesis.
Materials and Methods
Mouse and human gastric tissue samples
In this study, we used gastric tissue samples from Tff1 knockout (KO) and wild-type (WT) C57BL/6J/129/Svj mice. Tissue samples from the glandular antrum region of the stomach were collected from 4 Tff1 KO and 6 Tff1 WT mice of similar background and matching ages (Supplementary Table S1). All vertebrate animal studies were approved by the Institutional Animal Care and Use Committee at Vanderbilt University. Following euthanasia, animals were dissected through midline incision of the abdomen. Stomachs were removed, cut along the greater curvature, washed with ice cold PBS, and opened to lie flat. The stomachs were examined visually for abnormalities and for number and size of individual gastric tumors and photographed. The stomach was cut into symmetrical halves. One half was submerged in 10% buffered formalin solution, embedded in paraffin, and processed for standard H&E staining for histopathology evaluation. The remaining half of the stomach was snap-frozen and stored at −80°C for further use. Based on histological evaluation, we selected tissue samples that showed low-grade dysplasia (LGD) from the Tff1 KO mice. Tissue samples from the Tff1 WT mice showed normal gastric mucosa histology. The histology and age are included in Supplementary Table S1. Eighteen de-identified human tissue samples from gastric cancers and their matching histologically normal non-tumor tissue samples were collected from the National Cancer Institute Cooperative Human Tissue Network (CHTN) and the pathology archives at Vanderbilt University Medical Center (Nashville, TN). All tissue samples were obtained coded and de-identified in accordance with the Vanderbilt University Institutional Review Board-approved protocols.
Gene expression microarray analysis
RNeasy mini kit (Qiagen, Germantown, MD) was used to isolate total RNAs from antrum region of gastric mucosa of 10 Tff1 KO (LGD) and Tff1 WT (normal) mice and 9 human gastric cancer and 9 adjacent histologically normal tissues (Table 1). RNA quality was evaluated by a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). RNA samples with an RNA integrity number greater than 7 were reverse transcribed and amplified using a WT-Ovation Pico RNA amplification kit and labeled with FL-Ovation cDNA Biotin module v2 (NuGen, San Carlos, CA). Amplified products from mice were hybridized to Affymetrix Mouse 430 2.0 microarrays (Affymetrix, Santa Clara, CA). RNAs from 18 human gastric tissues (Table 1) were analyzed using Affymetrix GeneTitan WT Human Gene 1.0 ST arrays, following the manufactures’ recommendations, by the Vanderbilt Functional Genomics Shared Resource. Gene expression was compared between Tff1 KO mice (n=4) and Tff1 WT mice (n=6) or between human gastric cancer samples (n=9) and human normal gastric tissues (n=9). The raw gene expression data (.cel files) were preprocessed and normalized by using the robust multiarray average (RMA) expression measure, with RMA function in Bioconductor affy package (http://www.bioconductor.org/packages/release/bioc/html/affy.html) 31. The expression values were in log2 format after RMA 31. Bioconductor limma package was used for array data analysis (http://www.bioconductor.org/packages/release/bioc/html/limma.html) 32. A linear model was fitted to the expression data for each probe. Moderated t statistics were computed by empirical Bayes shrinkage of the standard errors toward a common value. The P values corresponded to the moderated t statistics. We used both P values as well as fold change to determine candidate probe list by requiring at least 1.5-fold change and P≤0.05, using R software version 2.10.0 (https://cran.r-roject.org/bin/windows/base/old/2.10.0/). Last, the data sets of normalized expression values plus their associated gene identifiers were uploaded into IPA software (Ingenuity Systems) to generate biological networks. Gene expression data were analysed using Ingenuity® Pathway Analysis (IPA®, QIAGEN, Redwood City, CA www.qiagen.com/ingenuity) tools to predict signaling pathways and upstream transcription networks that explain the observed gene expression changes in our dataset.
Table 1.
Clinicopathological information of human gastric cancer tissue samples used in Affymetrix gene expression microarrays.
| Sample ID | Site | Histology | TNM | Stage | Age | Sex |
|---|---|---|---|---|---|---|
| 1 | Stomach | Adenocarcinoma | T4N2Mx | IV | 58 | F |
| 2 | Stomach | Adenocarcinoma | T3N2Mx | IIIB | 57 | M |
| 3 | Stomach | Adenocarcinoma | T4N0M0 | IIIA | 61 | F |
| 4 | Stomach | Adenocarcinoma | T2N1Mx | IIA | Missing | Missing |
| 5 | Stomach | Adenocarcinoma | T2N0Mx | IB | Missing | Missing |
| 6 | Stomach | Adenocarcinoma | T3N0Mx | IIA | 72 | F |
| 7 | Stomach | Adenocarcinoma | T4N2Mx | IV | 55 | F |
| 8 | Stomach | Adenocarcinoma | T3N2Mx | IIIB | 55 | M |
| 9 | Stomach | Adenocarcinoma | T4N1Mx | IV | 46 | F |
TNM, TNM cancer staging system
Quantitative Real-time RT-PCR validation of downstream target genes in mice and human gastric tissues
This analysis was performed using independent tissue samples from mice and human. Mouse glandular stomach tissue samples included 10 Tff1 WT, 10 Tff1 KO with LGD, and 9 Tff1 KO with high-grade dysplasia. The histology and age information are provided in Supplementary Table S1. De-identified human stomach tissue samples included 19 with normal histology and 22 showing gastric cancer (Supplementary Table S2). Total RNA was purified using the RNeasy mini kit (Qiagen). Total RNA (1 μg) was reverse transcribed by an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). The quantitative real-time PCR (qRT-PCR) was performed using a Bio-Rad CFX Connect Real-time System with the threshold cycle number determined by Bio-Rad CFX manager software version 3.0. Primers that detect mouse and human genes were ordered from Integrated DNA Technologies (IDT, Coralville, IA). The genes and sequences of qRT-PCR primers are given in Supplementary Table S3. Results of target genes were normalized to mouse Hprt1 or human HPRT1. Expression fold changes were calculated by using the formula; 2 (Rt–Et)/2 (Rn–En), as previously described 33,34.
Statistical analyses
Data were demonstrated as mean ± standard deviation of 3 independent experiments. Statistical significance of the studies was analyzed by a Student’s t test, One way ANOVA, analysis of variance, and Mann-Whitney U test. Differences with P values ≤0.05 were considered significant.
Results
Significant alterations in gene expression are detectable in low-grade dysplasia
Analysis of gene expression data from mouse gastric LGD lesions, as compared to normal tissues, demonstrated significant deregulation of 395 genes, using cutoffs of ratio change ≥1.5 or ≤0.75 and P≤.01 (Supplementary Table S4). Among 328 genes that were overexpressed, 13 genes exhibited a fold change greater than 10 in LGD lesions. These genes included matrix metallopeptidase 10 (Mmp10), Mmp13, Mmp3, chemokine (C-X-C motif) ligand 1 (Cxcl1), Cxcl2 inhibin beta-A (Inhba), and prostaglandin-endoperoxide synthase 2 (Ptgs2, also known as Cox2) (Table 2). On the other hand, 67 genes were down-regulated in gastric LGD tissues. Notably, gastrin (Gast) and solute carrier family 5 (Slc5a5) were among the top down-regulated genes in LGD tissues. Of note, Tff1 expression was almost undetectable, as expected, in the Tff1 KO LGD lesions (Table 2). Supplementary Table S4 lists all deregulated genes in gastric LGD lesions, as compared to normal gastric mucosa.
Table 2.
Top 30 significantly de-regulated genes in low-grade dysplasia gastric tissue samples from Tff1 knockout mouse model
| Symbol | Fold change | P value | PFDR | Cytoband | Gene Name |
|---|---|---|---|---|---|
| Saa3 | 59.84 | 1.0E−06 | 0.0E+00 | 7 B4 | serum amyloid A 3 |
| Chi3l4 | 41.41 | 6.1E−04 | 5.5E−02 | 3 F3 | chitinase-like 4 |
| Mmp10 | 40.22 | 1.0E−06 | 1.9E−03 | 9 A1 | matrix metallopeptidase 10 |
| Cxcll | 31.12 | 3.3E−06 | 3.1E−03 | 5 E–F | chemokine (C-X-C motif) ligand 1 |
| Cxcl2 | 20.88 | 6.0E−05 | 1.6E−02 | 5 E1 | chemokine (C-X-C motif) ligand 2 |
| S100a9 | 19.75 | 1.6E−03 | 9.4E−02 | 3 F1–F2 | S100 calcium binding protein A9 (calgranulin B) |
| Reg3b | 17.80 | 6.1E−04 | 5.4E−02 | 6 | regenerating islet-derived 3 beta |
| Reg3g | 16.98 | 3.2E−04 | 4.0E−02 | 6 C3 | regenerating islet-derived 3 gamma |
| Inhba | 15.43 | 1.0E−06 | 1.1E−03 | 13 A1 | inhibin beta-A |
| Mmp13 | 15.37 | 1.0E−05 | 5.2E−03 | 9 | matrix metallopeptidase 13 |
| Il1b | 12.78 | 7.0E−05 | 1.7E−02 | 2 F | interleukin 1 beta |
| Cxcl5 | 10.97 | 3.0E−05 | 1.1E−02 | 5 E1 | chemokine (C-X-C motif) ligand 5 |
| Mmp3 | 10.03 | 1.0E−06 | 3.0E−04 | 9 A1 | matrix metallopeptidase 3 |
| Serpine2 | 9.43 | 1.0E−06 | 0.0E+00 | 1 C4 | serine (or cysteine) peptidase inhibitor, clade E, member 2 |
| Slpi | 9.13 | 6.0E−05 | 1.5E−02 | 2 H | secretory leukocyte peptidase inhibitor |
| Ereg | 8.91 | 1.0E−06 | 1.2E−03 | 5 E1 | epiregulin |
| Mcpt2 | 8.47 | 1.0E−05 | 4.1E−03 | 14 C3 | mast cell protease 2 |
| Sftpd | 8.17 | 1.0E−06 | 3.0E−04 | 14 B | surfactant associated protein D |
| Ptgs2 | 8.07 | 1.5E−04 | 2.4E−02 | 1 H1 | prostaglandin-endoperoxide synthase 2 |
| Capn9 | 0.13 | 5.0E−05 | 5.2E−02 | 8 E2 | calpain 9 |
| Maob | 0.12 | 1.0E−06 | 1.6E−02 | X 11.88 | monoamine oxidase B |
| Pln | 0.12 | 5.0E−04 | 7.9E−02 | 10 B3 | phospholamban |
| Pcp4 | 0.11 | 4.0E−05 | 4.7E−02 | 16 C4 | Purkinje cell protein 4 |
| Ankrd35 | 0.099 | 1.0E−06 | 1.5E−02 | 3 F2.1 | ankyrin repeat domain 35 |
| Rpp25 | 0.08 | 6.0E−05 | 5.5E−02 | 9 B | ribonuclease P 25 subunit (human) |
| Bex1 | 0.08 | 7.0E−05 | 5.6E−02 | X F1 | brain expressed gene 1 |
| Muc5ac | 0.07 | 1.0E−06 | 1.6E−02 | 7 F5 | mucin 5, subtypes A and C, tracheobronchial/gastric |
| Slc5a5 | 0.04 | 1.0E−06 | 1.6E−02 | 8 C1 | solute carrier family 5 (sodium iodide symporter), member 5 |
| Tff1 | 0.008 | 1.0E−06 | 0.0E+00 | 17 A3.3 | trefoil factor 1 |
| Gast | 0.006 | 1.0E−05 | 1.6E−02 | 11 D | gastrin |
Symbol, approved gene symbol; Fold change, relative gene expression fold change; P value, P value of ≤.05 was considered significant; PFDR, positive false discovery rate; Cytoband, gene location on subregions of a chromosome; Gene Name, full name of the gene.
Human gastric cancers demonstrate recurrent significant changes in gene expression
Analysis of human gastric cancer gene expression data demonstrated deregulation of 783 genes; 469 genes were overexpressed while 314 genes were down-regulated, using cutoffs of ratio change ≥1.5 or ≤0.75 and P≤.01, (Supplementary Table S5). The top up-regulated genes included matrix metallopeptidase 7 (MMP7), MMP12, MMP9, and chemokine (C-X-C motif) ligand 9 (CXCL9). The down-regulated genes included ATPase, H+/K+ exchanging, beta polypeptide (ATP4B), ATPase, H+/K+ exchanging, alpha polypeptide (ATP4A), chitinase acidic (CHIA), and carboxypeptidase A2 (CPA2). The top 30 deregulated genes are listed in Table 3. Supplementary Table S5 lists significantly deregulated genes in human gastric cancer tissue samples.
Table 3.
Top significantly de-regulated genes in human gastric cancer tissues as compared with normal human gastric tissue samples.
| Symbol | Fold change | P value | PFDR | Cytoband | Gene Name |
|---|---|---|---|---|---|
| UBD | 9.95 | 1.0E−06 | 1.9E−03 | 6p21.3 | ubiquitin D |
| PLEKHS1 | 8.60 | 1.0E−06 | 2.3E−03 | 10q25.3 | pleckstrin homology domain containing, family S member 1 |
| MMP1 | 8.12 | 8.8E−04 | 5.6E−02 | 11q22.3 | matrix metallopeptidase 1 (interstitial collagenase) |
| PLA2G7 | 7.72 | 1.0E−05 | 6.4E−03 | 6p21.2-p12 | phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| MMP7 | 7.39 | 9.0E−05 | 2.2E−02 | 11q21-q22 | matrix metallopeptidase 7 (matrilysin, uterine) |
| CCL20 | 7.37 | 1.7E−04 | 2.8E−02 | 2q33-q37 | chemokine (C-C motif) ligand 20 |
| CXCL9 | 7.25 | 1.91−3 | 7.4E−02 | 4q21 | chemokine (C-X-C motif) ligand 9 |
| C4BPA | 6.43 | 1.5E−04 | 2.6E−02 | 1q32 | complement component 4 binding protein, alpha |
| MMP12 | 6.25 | 5.0E−05 | 1.4E−02 | 11q22.3 | matrix metallopeptidase 12 (macrophage elastase) |
| IDO1 | 6.24 | 1.7E−03 | 7.1E−02 | 8p12-p11 | indoleamine 2,3-dioxygenase 1 |
| LOC93432 | 6.19 | 2.1E−04 | 3.0E−02 | 7q34 | maltase-glucoamylase (alpha-glucosidase) pseudogene |
| SAA2 | 5.90 | 2.5E−03 | 8.5E−02 | 11p15.1-p14 | serum amyloid A2 |
| CFTR | 5.03 | 2.0E−05 | 1.1E−02 | 7q31.2 | cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| MMP9 | 4.42 | 1.2E−03 | 6.2E−02 | 20q11.2-q13.1 | matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| IGSF6 | 4.08 | 3.7E−04 | 4.0E−02 | 16p12.2 | immunoglobulin superfamily, member 6 |
| VIL1 | 4.02 | 1.3E−03 | 6.5E−02 | 2q35 | villin 1 |
| TRIM31 | 3.97 | 1.3E−03 | 6.3E−02 | 6p21.3 | tripartite motif containing 31 |
| BGN | 3.94 | 2.7E−03 | 8.7E−02 | Xq28 | biglycan |
| C2 | 3.91 | 1.0E−05 | 7.6E−03 | 6p21.3 | complement component 2 |
| PSCA | 0.13 | 9.5E−05 | 2.0E−02 | 8q24.2 | prostate stem cell antigen |
| ESRRG | 0.13 | 5.9E−04 | 4.8E−02 | 1q41 | estrogen-related receptor gamma |
| AKR1B10 | 0.12 | 3.7E−03 | 9.8E−02 | 7q33 | aldo-keto reductase family 1, member B10 (aldose reductase) |
| AQP4 | 0.12 | 2.0E−04 | 3.0E−02 | 18q11.2-q12.1 | aquaporin 4 |
| CCKAR | 0.11 | 1.1E−03 | 5.8E−02 | 4p15.1-p15.2 | cholecystokinin A receptor |
| MAL | 0.10 | 7.0E−05 | 1.9E−02 | 2q11.1 | mal, T-cell differentiation protein |
| KCNE2 | 0.08 | 5.9E−04 | 4.8E−02 | 21q22.12 | potassium voltage-gated channel, Isk-related family, member 2 |
| CPA2 | 0.07 | 1.7E−03 | 7.1E−02 | 7q32 | carboxypeptidase A2 (pancreatic) |
| CHIA | 0.07 | 3.8E−04 | 4.0E−02 | 1p13.2 | chitinase, acidic |
| ATP4A | 0.07 | 1.6E−03 | 7.1E−02 | 19q13.1 | ATPase, H+/K+ exchanging, alpha polypeptide |
| ATP4B | 0.06 | 1.7E−03 | 7.1E−02 | 13q34 | ATPase, H+/K+ exchanging, beta polypeptide |
Symbol, approved gene symbol; Fold change, relative gene expression fold change; P value, P value of ≤.05 was considered significant; PFDR, positive false discovery rate; Cytoband, gene location on subregions of a chromosome; Gene Name, full name of the gene.
Integrated analysis of mouse gastric LGD and human gastric cancer datasets identifies activation of common transcription networks
Our integrated bioinformatics analysis indicated that 172 genes were consistently and significantly deregulated in the same direction in both datasets (P≤0.05, FC ±1.5). Among the 172 differentially expressed genes, 122 genes were overexpressed, whereas 50 genes were down-regulated (Supplementary Table S6). The commonly deregulated genes included overexpression of phospholipase A2, group VII (PLA2G7), complement component 4 binding protein, alpha (C4BPA), matrix metallopeptidase 12 (MMP12) and down-regulation of aquaporin 4 (AQP4), prostate stem cell antigen (PSCA), alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide (ADH7).
We also determined signaling pathways that are likely to be early drivers of gastric carcinogenesis by performing network analysis of mouse LGD and human gastric cancer data sets using QIAGEN’s Ingenuity® Pathway Analysis (IPA®, QIAGEN, www.qiagen.com/ingenuity). Pathway Analysis of downstream targets demonstrated activation of STAT3, RELA, CTNNB1, HIF1A, ETS1, and NFATC2 transcription networks in mouse LGD lesions (Table 4). Of note, analysis of human gastric cancer samples data predicated activation of the same pathways, suggesting that they are required for initiation and early steps of gastric tumorigenesis. In addition, human gastric cancers also demonstrated activation of FOXM1 and inhibition of TP53 transcription networks (Table 5 and Supplementary Figure S1). A representative diagram showing co-activated networks that include MYC, STAT3 and β-catenin signaling pathways in mouse LGD and human gastric cancer is shown in Figure 1.
Table 4.
Upstream transcription networks and their target genes in the Tff1 knockout gastric low-grade dysplasia tissue samples.
| Upstream Regulator |
Predicted Activation State |
p-value of overlap |
Target molecules in dataset |
|---|---|---|---|
| STAT3 | Activated | 2.76E−13 | ARG1,BCL3,CD9,CEBPB,CXCL3,CXCL6,DDIT3,FN1,FST,GLIPR 1,HIF1A,HK2,ICAM1,IGFBP5,IL13RA2,IL1B,IL1RN,MUC1,NF ATC2,PDIA4,PTGS2,PTPN2,SLFN12L,TIMP1,TNFSF11,VEGFA |
| RELA | Activated | 1.91E−12 | APOE,CCL11,Ccl2,CD44,CEBPB,COL1A1,CXCL1,CXCL2,CXCL3, CXCL6,CXCR4,DDIT3,ICAM1,IL1B,IL1RN,PTGS2,REG3A,Reg 3g,Saa3,SDC4,SERPINE2,VEGFA |
| CTNNB1 | Activated | 2.14E−11 | BGN,CAPNS1,CCND2,CD44,CELSR1,CLDN2,COL1A1,COL4A1, COL4A2,CTGF,FN1,FOXC1,GAST,GPX2,HSD17B2,HTRA1,IFI TM1,IGFBP5,IL1B,IL1R1,INHBB,MFGE8,MMP23B,MMP3,PS AP,PTGS2,RUNX2,Saa3,SERPINA3,SERPINE2,SOX4,TIMP1,V EGFA |
| MYC | 3.21E−10 | APEX1,ARG1,CCND2,CD44,Clu,CNP,COL1A1,COL1A2,COL4A 1,COL4A2,COL5A2,DDIT3,FN1,GJA1,HIF1A,HK2,ME2,PDGF RB,PROM1,SAT1,SCEL,SERPINE1,SERPINE2,SHMT1,SPARC,T HBS1,TNFSF11,VEGFA | |
| HIF1A | Activated | 5.02E−08 | APOE,CTGF,CXCR4,CYP2S1,FHL1,FLT1,GJA1,HIF1A,HK2,IGF BP3,IL1B,INHBB,ITGAV,KIAA1199,MUC1,PROM1,PTGS2,SE RPINE1,VEGFA |
| ETS1 | Activated | 6.69E−04 | Ccl2,CTGF,FCGR2A,FLT1,FN1,MMP13,RUNX2,SERPINE1,ZE B2 |
| NFATC2 | Activated | 3.42E−03 | ABCA1,CXCL3,DAB2,INHBA,PTGS2,REL,SLFN12L |
Upstream Regulator, the upstream regulator for the target genes in dataset. Predicted Activation State, signaling pathway status is predicted based on the downstream target gene expression levels. P value of overlap, p value computed using Fisher’s Exact Test. This p-value measures the significance of overlap between our gene set and genes that are regulated by a transcriptional regulator stored in the Ingenuity Knowledge Base. Target molecules in dataset, genes that are regulated by a transcription upstream regulator based on the Ingenuity Knowledge Base.
Table 5.
Upstream transcription networks and their target genes in human gastric cancer tissue samples.
| Upstream Regulator |
Predicted Activation State |
p-value of overlap |
Target molecules in dataset |
|---|---|---|---|
| TP53 | Inhibited | 2.60E−16 | ACAT1,ACOT11,ALDH4A1,ANXA3,ARPC1B,AURKA,BCL2 L1,BID,BNIP3,BRCA1,BUB1,BUB1B,C2,CDC7,CDK4,CDK N3,CENPF,CHEK1,CKAP2,CKB,CTSB,CXCL1,CYB5A,DNM T1,DSN1,EPHX1,EXO1,F2R,F5,FAT1,FOS,FOXP3,GLUL,G NAI1,GPD1L,GPX1,GPX3,GSN,HBEGF,HMGB1,IDH2,IFI3 0,IFI35,IGF1R,IGFBP2,IGFBP3,IL2RA,IPO9,KAT2B,KIF23, KIFC1,KPNA2,LIF,LSS,MCM3,MCM6,MCM7,ME1,MMP 9,NCAPG,NCAPH,NEK2,NFKB2,NOTCH1,PARK2,PCCA,P ML,PRC1,PTPN6,PYCARD,RBL1,RFC3,RPRM,SERPINH1, SGK1,SH3BGRL2,SLC2A12,SLC6A6,SNX5,SPC25,STAT1,S TMN1,TAP1,TDO2,TMSB10/TMSB4X,TNFRSF10B,TNFR SF9,TOP2A,TPD52L1,TPP1,TPX2,TYMS,UBE2C,UBL3,XP O1 |
| RELA | Activated | 7.52E−08 | B2M,BCL2L1,CCL11,CCL20,CXCL1,CXCL11,CXCL9,CYP2B 6,FOS,ICAM1,IFNGR2,KIT,LYN,MMP1,MMP9,NFKB2,N MI,PSMB9,PTPN6,SAA2,SLC1A2,SLC2A4,TAP1,TAPBP |
| STAT3 | Activated | 4.52E−07 | BCL2L1,CCL20,CD80,CHI3L1,CLN6,CTLA4,CXCL9,ECT2,F OS,FOXP3,FST,HLA- B,ICAM 1,ICOS, IFI30,IFI35,IKBKE,I L2RA, KAT2B,MMP9,P SMB8,PSMB9,SGK1,STAT1,TAP1,TIMP1 |
| MYC | Activated | 2.28E−05 | ANGPT2,BAG1,BUB1,BUB1B,CAD,CCNB2,CCT3,CDC25B, CDK4,CHEK1,DLEU2,EFTUD2,FOXM1,HIST1H4A (includes others),MCM6,MCM7,MMP7,MMP9,PARP1,PDCD4,PL AU,PML,SERPINH1,SGK1,SNRPD1,TFRC,TMSB10/TMSB 4X,TNFRSF10B,TYMS,UBE2C,XPO1 |
| FOXM1 | Activated | 2.753 | ANLN,ASPM,ATP6V0D2,BID,CCNB2,CENPF,CKMT2, DEPDC1,DLGAP5,FOS,FOXM1,GK,KIF18A,ME1,MM P1, MMP9,NCAPG,NEK2,NUSAP1, PLSCR1, PRC1, PS MB8,SCARB1, SGK1, SLC2A4,SPC25,STAT2,TDO2 |
| NFATC2 | Activated | 3.92E−04 | ABCA1,CDK4,CTLA4,E2F5,FOXP3,ICOS,IFIT2,IL2RA,NMI, PML,STAT1,STAT2 |
| HIF1A | Activated | 7.64E−02 | AURKA,BCL2L1,BNIP3,CYB5A,ESRRG,FOS,HIST1H4A (includes others), IGFBP3,LIFR,MMP9,NOTCH1,SLC16A4,TFRC |
| CTNNB1 | 1.27E−04 | ALDH3A2,AQP4,BCL2L1,BGN,COL4A5,CTSZ,CXCL9,CYB 5A,CYP2E1,EPHB2,F2R,FCER1G,GLUL,GPR137B,ID4,IDO 1,IFITM1,KIF23,KIFC1,LAP3,LEF1,LIPA,ME1,MMP1,MM P14,MMP7,MMP9,NCAM1,PCCA,PLAU,SGK1,TIMP1,W IF1 | |
| ETS1 | Activated | 2.21E−02 | CCR8,FOXP3,IL2RA,MMP1,MMP9,PARP1,PLAU,PML, SP100,TBXAS1 |
Upstream Regulator, the upstream regulator for the target genes in dataset. Predicted Activation State, signaling pathways’ status predicted based on the downstream target gene expression levels. P value of overlap, this is the p value computed using Fisher’s Exact Test. This p-value measures the significance of overlap between our gene set and genes that are regulated by a transcriptional regulator stored in the Ingenuity Knowledge Base. Target molecules in dataset, genes that are regulated by a transcription upstream regulator based on the Ingenuity Knowledge Base.
Figure 1. Transcriptional network analysis demonstrates activation of MYC, STAT3 and β-catenin.
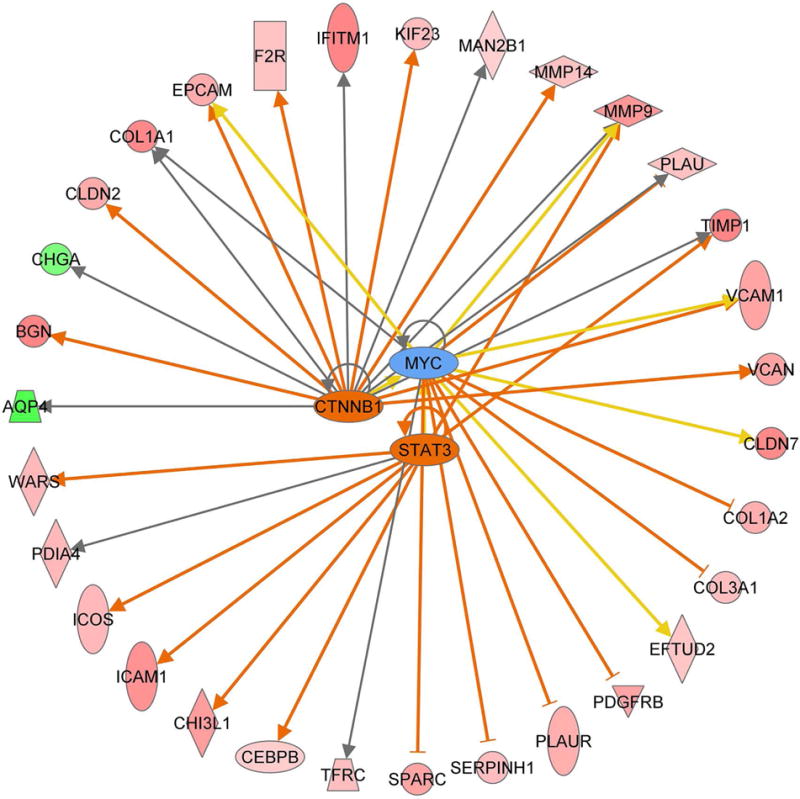
Pathway analysis of gene expression data using Ingenuity online tools indicated activation of MYC, STAT3 and β-catenin (CTNNB1) transcription networks in both Tff1 KO mice gastric LGD and human gastric cancer samples.
qPCR validation of representative transcription network target genes
To confirm the observed consistent changes in gene expression in mouse and human, we selected 14 differentially expressed genes based on their known functions in gastric tumorigenesis (Table 6). Using qRT-PCR, we validated several MYC, STAT3 and β-catenin downstream target genes in both mouse and human gastric tissue samples. Our results indicated that Timp1, Epcam, Cldn2, Vcam1, Mmp9, Mmp12, Pdgfrb, Bgn, and Plau were significantly overexpressed in both mouse LGD and HGD gastric tissue samples, as compared to normal gastric tissue samples (P<0.05, Figure 2). Notably, Mmp14 was significantly overexpressed only in mouse HGD tissues (P<0.01), while Iftm1 was up-regulated exclusively in mouse LGD tissues (P<0.01). In addition, our data showed a non-significant trend of overexpression of Col1a1, Col1a2 and Col3a1 in LGD or HGD gastric tissue samples, as compared to normal gastric tissue samples (Figure 2). On the other hand, our data indicated that all 14 genes that we tested were significantly overexpressed in human gastric cancer tissues, as compared to normal gastric samples (P<0.05, Figure 3). Of note, TFF1 mRNA expression level was significantly down-regulated in human gastric cancer tissue samples (Supplementary Figure S2). Collectively, our results validate the microarray data and suggest that these validated genes are possibly involved in early stages of gastric tumorigenesis and their deregulated expression persists in advanced stages of gastric cancer.
Table 6.
Examples of consistently deregulated genes in both human gastric cancer and Tff1 knockout mouse low-grade dysplasia tissue samples, as compared with normal gastric tissue samples.
| Human | Mouse | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Symbol | Cytoband | Fold change | P value | PFDR | Symbol | Cytoband | Fold change | P value | PFDR | Gene Name |
| MMP12 | 11q22.3 | 6.25 | 5.00E−05 | 1.43E−02 | Mmp12 | 9A1 | 3.42 | 1.20E−03 | 7.87E−02 | matrix metallopeptidase 12 (macrophage elastase) |
| MMP9 | 20q11.2-q13.1 | 4.42 | 1.20E−03 | 6.20E−02 | Mmp9 | 2 H1−H2 | 1.85 | 1.10E−02 | 2.40E−01 | matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| BGN | Xq28 | 3.9 | 2.73E−03 | 0.09 | Bgn | X B | 3 | 7.33E−05 | 0.02 | biglycan |
| TIMP1 | Xp11.3-p11.23 | 3.76 | 0.00E+00 | 6.20E−03 | Timp1 | X A1.3 | 3.14 | 1.30E−03 | 8.20E−02 | TIMP metallopeptidase inhibitor 1 |
| EPCAM | 2p21 | 3.33 | 1.00E−02 | 1.40E−01 | Epcam | 17 E4 | 1.64 | 6.40E−03 | 1.90E−01 | epithelial cell adhesion molecule |
| COL1A1 | 17q21.33 | 3 | 4.77E−03 | 0.11 | Col1a1 | 11 D | 3.8 | 5.00E−06 | 0 | collagen, type I, alpha 1 |
| IFITM1 | 11p15.5 | 2.91 | 8.30E−04 | 5.40E−02 | Ifitm1 | 7 | 3.96 | 0.00E+00 | 3.40E−03 | interferon induced transmembrane protein 1 |
| VCAM1 | 1p32-p31 | 2.6 | 9.86E−03 | 0.14 | Vcam1 | 3 G1 | 2.5 | 2.46E−02 | 0.34 | vascular cell adhesion molecule 1 |
| COL1A2 | 7q22.1 | 2.1 | 4.79E−02 | 0.27 | Col1a2 | 6 A1 | 2.5 | 1.05E−04 | 0.02 | collagen, type I, alpha 2 |
| COL3A1 | 2q31 | 2.1 | 4.60E−02 | 0.26 | Col3a1 | 1 C1.1 | 1.7 | 2.41E−03 | 0.11 | collagen, type III, alpha 1 |
| PDGFRB | 5q33.1 | 2.1 | 1.46E−02 | 0.16 | Pdgfrb | 18 E1 | 3 | 5.95E−04 | 0.05 | platelet-derived growth factor receptor, beta polypeptide |
| MMP14 | 14q11-q12 | 1.9 | 3.23E−03 | 0.09 | Mmp14 | 14 C2 | 1.9 | 1.34E−02 | 0.21 | matrix metallopeptidase 14 (membrane-inserted) |
| CLDN2 | Xq22.3-q23 | 1.78 | 1.70E−02 | 1.70E−01 | Cldn2 | X F1 | 3.11 | 4.80E−04 | 4.80E-02 | claudin 2 |
| PLAU | 10q24 | 1.7 | 1.33E−03 | 0.07 | Plau | 14 A3 | 2.1 | 1.47E−02 | 0.28 | plasminogen activator, urokinase |
Symbol, approved gene symbol; Fold change, relative gene expression fold change; P value, P value of ≤.05 was considered significant; PFDR, positive false discovery rate; Cytoband, gene location on subregions of a chromosome; Gene Name, full name of the gene.
Figure 2. Validation of representative MYC, STAT3 and β-catenin downstream target genes in gastric tissue samples from mice.
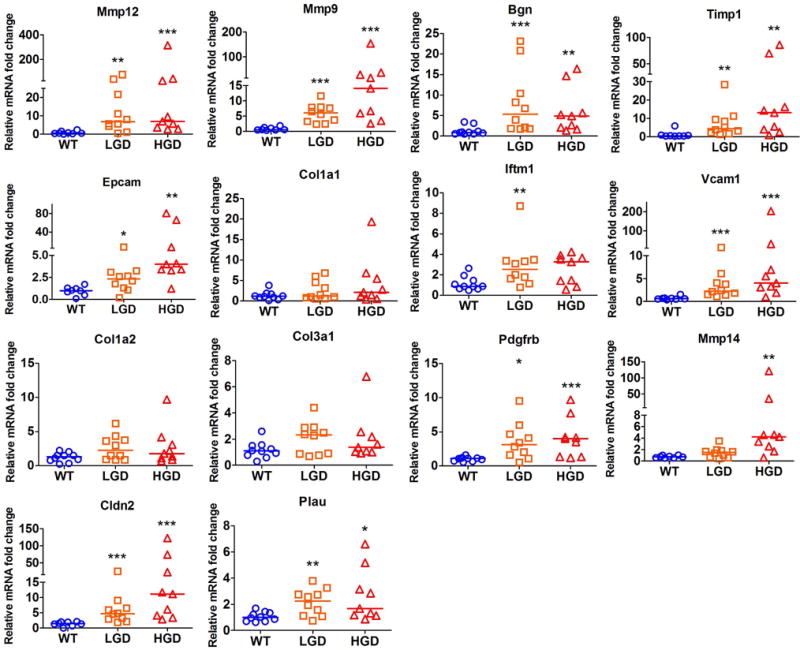
qRT-PCR analysis of expression of targets genes, as shown, was performed in normal glandular stomach of 10 wild-type (WT), 10 Tff1 KO with low-grade dysplasia (LGD), and 9 Tff1 KO with high-grade dysplasia (HGD). The horizontal line indicates the median. *P<0.05, **P<0.01, ***P<0.001, Mann-Whitney U test.
Figure 3. Validation of representative MYC, STAT3 and β-catenin downstream target genes in human gastric tissue samples.
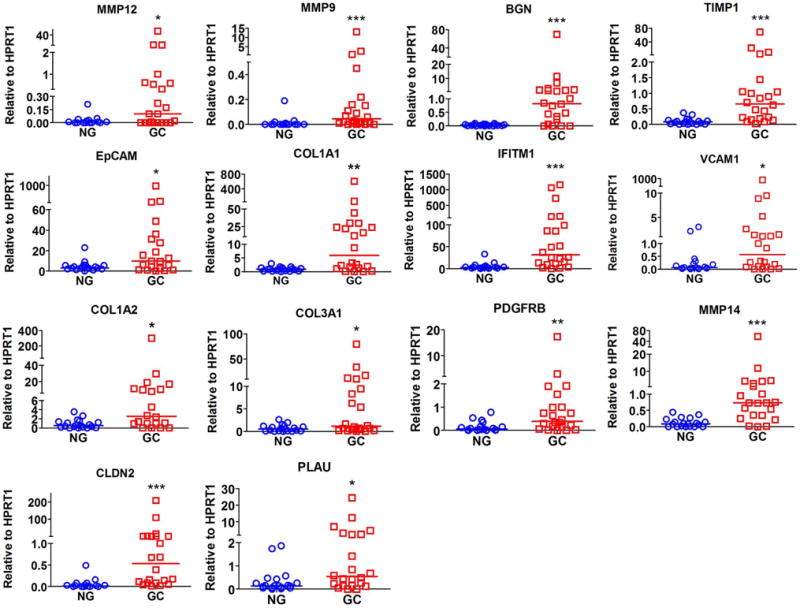
qRT-PCR analysis of expression of target genes, as shown, in 19 human normal stomach (NG) and 22 gastric cancer (GC) tissue samples. The horizontal line indicates the median *P<0.05, **P<0.01, ***P<0.001, Mann-Whitney U test
Discussion
Gastric cancer remains the third leading cause of cancer-associated death world-wide 35. Although the incidence of distal gastric cancer is declining 36, the incidence of proximal cancers that include gastric cardia and gastroesophageal junction continues to be on the rise. The late diagnosis of gastric cancer is a clinically challenging problem with low favorable response rates to current chemotherapeutics leading to poor prognosis and clinical outcome 1.
The use of genetic analysis of inbred mouse models of carcinogenesis has an important advantage of limited heterogeneity, therefore, allowing discovery of consistent genetic alterations that are related to the disease process. In addition, using mouse models offers an opportunity to overcome the difficulties associated with early diagnosis, procurement, and analysis of premalignant lesions in human.
In this study, we performed integrated bioinformatics analysis of mouse and human molecular signatures to determine genes that are likely early drivers of gastric carcinogenesis. Our previous studies indicated that the Tff1 KO mice exhibit histological changes in the pyloric antrum of the stomach, progressing from gastritis to low-grade dysplasia, to high-grade dysplasia, and ultimately to malignant adenocarcinoma that are similar to human gastric tumorigenesis 7,19. Of note, loss of TFF1 expression is one of the most frequent molecular alterations in human gastric tumorigenesis due to epigenetic inactivation, loss of heterozygosity (LOH), or transcriptional regulation 3–6. Herein, we analyzed early premalignant LGD gastric lesions in the Tff1 KO mouse model and compared the results to human gastric cancer samples to eliminate the complex genetic and host heterogeneity factors that are present in human, and to overcome difficulties in acquiring human premalignant tissue samples. We postulated that if a gene is seen in mouse LGD lesions as well as in human gastric cancer samples, this gene is likely an early event and a driver of the disease process. Indeed, we have discovered several molecular similarities and validated several genes using mouse and human stomach tissues. For example, we have confirmed overexpression of MMP9, MMP12, MMP14, and EPCAM in both mouse and human. These genes play an important role in cellular invasion 37. We have also confirmed the overexpression of CLDN2 and PDGFRB, which are known to promote cellular transformation and survival 38,39. It is worth mentioning, our data have shown a progressive increase in expression level of several genes from LGD to HGD, and to adenocarcinoma. These observations provide confidence in our data and suggest their early roles in gastric tumorigenesis.
Our analysis of LGD lesions demonstrated deregulation of genes that mapped to key oncogenic transcription networks such as β-catenin, MYC, and STAT3. These transcription networks were similarly activated in human gastric cancer samples. Activation of WNT/β-catenin signaling cascade is a key step in several gastrointestinal malignancies. Studies in colon cancer have shown that activation of β-catenin occurs as early as in adenoma stage due to frequent mutations of its negative regulators, such as APC 40,41. While mutations of APC and β-catenin are rare in gastric cancer 18, non-mutational activation of β-catenin/TCF transcription network is common in gastric cancer 42. β-catenin can also be activated by AKT signaling pathway, which is predominantly active in gastric cancer 43. Furthermore, we and others have shown that molecular factors that inhibit GSK3β and PP2A activities play a role in activation of β-catenin in gastric cancer 43,44. In addition, regulation of β-catenin by miRNAs has been described in gastric cancer 45. β-catenin is an essential transcription network that regulates a wide spectrum of transcription target genes that control important cellular functions such as adhesion, proliferation, angiogenesis, and invasion 16. It has also been suggested that β-catenin could play a role in regulating gastric cancer stem cells and progenitor stem cells capacity 12,46. It is, therefore, possible that loss of TFF1 and activation of WNT/β-catenin signaling in early stages of gastric tumorigenesis 19 may regulate progenitor cells of gastric cancer. Based on our mouse to human data, we suggest that activation of β-catenin transcription network is conceivably related to initiation of the gastric carcinogenesis cascade.
MYC is one of the most recognized transcription factors in biological processes that regulates numerous oncogenic functions in gastric cancer 22. Although MYC overexpression alone is incapable of inducing neoplastic transformation of normal human cells 47,48, it is recognized as a potent oncogene that promotes tumor development and progression 49. We have found activation of MYC transcription network in LGD of mouse and human gastric cancer samples suggesting its importance in early stages of gastric tumorigenesis. This is particularly important given its known significant role in regulating cellular stemness, proliferation, and angiogenesis in cancer 22. Taken together, our findings suggest that activation of MYC transcription network is possibly another crucial early molecular event in gastric carcinogenesis.
Of note, we also detected activation of STAT3 transcription network in both mouse and human neoplastic gastric lesions. STAT3 is constitutively activated in several gastrointestinal malignancies that include colon cancer and gastric cancer 50,51. We have previously shown that STAT3 regulates angiogenesis, drug response, and cellular proliferation in gastric cancer 52,53. A number of studies have shown that STAT3 promotes stem-like properties and maintenance of cancer cells providing resistance to several chemotherapeutic drugs 13,54. It is worth mentioning that there is an overlap not only in the transcription targets of β-catenin, MYC, and STAT3, but also in their biological outcomes. Taken together, our finding of activation of β-catenin, MYC, and STAT3 networks suggests that together they can act in synergy and harmony to mediate oncogenic cellular functions in essential initiation and progression steps of gastric tumorigenesis.
Our data also suggested activation of NFATC2, HIF1A, and ETS1 in human gastric cancer and early stages of tumorigenesis in the Tff1 KO mice. NFATC2 plays an important role in regulating the development of cancer related inflammation, promoting colon cancer cell differentiation and proliferation 55. HIF1A is a master regulator of cell response to hypoxia by activating genes involved in angiogenesis, apoptosis, and energy metabolism 56–58. HIF1A expression has been recently reported in various human cancers including pancreatic cancer, esophageal cancer and breast cancer, promoting cancer angiogenesis, proliferation, and survival 58–60. Similarly, activation of ETS1, a member of the ETS family of transcription factors, is known to be involved in cancer progression in breast cancer, pancreatic cancer, prostate cancer, and gastric cancer 61–64. Recent studies suggested that ETS1 expression is linked to the cancers with higher invasive, angiogenic activity 61. The fact that we identified activation of these networks in LGD lesions of the Tff1 KO mouse imply that these transcription factors are not only involved in late stages of cancer but also possibly in early stages of initiation of gastric tumorigenesis.
We also detected transcription networks that were deregulated in human advanced gastric cancer but not in mouse LGD lesions. These included inactivation of TP53 and activation of FOXM1. Inactivation of p53 is caused by mutations in more than half of human cancers 65. Mutant p53 provides cancer cells gain-of-function properties, such as increased cell proliferation, metastasis and apoptosis resistance 65. Recent studies suggested that loss of p53 or the accumulation of mutant p53 were observed more in poorly-differentiated than in well-differentiated gastric carcinomas 66. The accumulation of p53 in immunohistochemical staining, an indicator of mutation of p53, was also significantly higher in large, advanced, and metastatic gastric cancers 67. On the other hand, FOXM1 plays a key role in tumor progression as noted in recent studies showing that cancer cell proliferation and tumor growth are significantly reduced when FOXM1 is deleted 68,69. In pancreatic cancer, matrix metalloproteinases (MMPs) were regulated by FOXM1, increasing cancer cell migration and invasion 69. Of note, FOXM1 expression in cancer cells can promote activation of DNA damage repair networks and confer resistance to chemotherapeutics 70. Collectively, these findings from earlier reports can explain the lack of inactivation of p53 and activation of FOXM1 in early neoplastic gastric lesions in the Tff1 KO mouse model.
Although our analysis and interpretation have focused on consistent changes between mouse LGD and human cancer, there were also examples of genes that did not overlap. A possible explanation of this apparent discrepancy may be attributed to the conserved nature of gene expression among different species (mouse and human). Alternatively, it is also plausible that some of these changes could be related to advanced stages of gastric cancer rather than early stages of tumorigenesis.
In conclusion, our study provided a comprehensive integrated molecular analysis of transcription networks in human and mouse models of gastric cancer. Our pilot data demonstrate that this is a powerful approach to study the molecular events and identify striking similarities such as activation of β-catenin, MYC, and STAT3 transcription networks. We acknowledge the limitations in our study due to lack of human dysplastic lesions. Nevertheless, our findings highlight the important role of mouse models of gastric cancer that provide an opportunity to overcome some of the inherent limitations in human studies.
Supplementary Material
Supplementary Figure S1. Overlapping genes in the P53 transcription network in LGD from Tff1 KO mice and human gastric cancer tissue samples. Ingenuity pathway analysis was used to generate the transcription network. TP53 in the center indicates deregulation of P53 signaling in both Tff1 KO mice LGD and human gastric cancer samples. Deregulated downstream targets of P53 are shown (P<0.05).
Supplementary Figure S2. TFF1 mRNA expression level in human gastric tissue samples. qRT-PCR analysis of expression of TFF1 in 19 human normal stomach (NG) and 22 gastric cancer (GC) tissue samples. The horizontal line indicates the median, ***P<0.001, Mann-Whitney U test.
Acknowledgments
This study was supported by a Research Career Scientist award (1IK6BX003787) and merit award (I01BX001179) from the U.S. Department of Veterans affairs (W. El-Rifai), and grants from the U.S. National Institutes of Health (R01CA93999), Vanderbilt SPORE in Gastrointestinal Cancer (P50 CA95103), Vanderbilt Ingram Cancer Center (P30 CA68485), and the Vanderbilt Digestive Disease Research Center (DK058404). The contents of this work are solely the responsibility of the authors and do not necessarily represent the official views of the Department of Veterans Affairs, National Institutes of Health, or Vanderbilt University.
Footnotes
Authors’ contribution:
Zheng Chen: summarized data, prepared figures, and wrote a draft of the manuscript
Mohammed Soutto: assisted in in vivo experiments and interpretation of data
Bushra Rahman: assisted in validation of gene expression
Muhammad W Fazili: assisted in validation of gene expression
Dunfa Peng: assisted in preparation of samples and interpretation of data
Maria Blanca Piazuelo: histopathology analysis of mouse and human tissues
Heidi Chen: analyzed and interpreted microarray data
M. Kay Washington: histopathology analysis of mouse and human tissues
Yu Shyr: assisted in data analysis and interpretation
Wael El-Rifai: Study concept and design; study supervision; experimental troubleshooting; analysis and interpretation of data; drafting of the manuscript; critical revision of the manuscript
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Jin Z, Jiang W, Wang L. Biomarkers for gastric cancer: Progression in early diagnosis and prognosis (Review) Oncol Lett. 2015;9(4):1502–1508. doi: 10.3892/ol.2015.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carvalho R, Kayademir T, Soares P, et al. Loss of heterozygosity and promoter methylation, but not mutation, may underlie loss of TFF1 in gastric carcinoma. Lab Invest. 2002;82(10):1319–1326. doi: 10.1097/01.lab.0000029205.76632.a8. [DOI] [PubMed] [Google Scholar]
- 4.Khan ZE, Wang TC, Cui G, Chi AL, Dimaline R. Transcriptional regulation of the human trefoil factor, TFF1, by gastrin. Gastroenterology. 2003;125(2):510–521. doi: 10.1016/s0016-5085(03)00908-9. [DOI] [PubMed] [Google Scholar]
- 5.Sankpal NV, Moskaluk CA, Hampton GM, Powell SM. Overexpression of CEBPbeta correlates with decreased TFF1 in gastric cancer. Oncogene. 2006;25(4):643–649. doi: 10.1038/sj.onc.1209081. [DOI] [PubMed] [Google Scholar]
- 6.Tomita H, Takaishi S, Menheniott TR, et al. Inhibition of gastric carcinogenesis by the hormone gastrin is mediated by suppression of TFF1 epigenetic silencing. Gastroenterology. 2011;140(3):879–891. doi: 10.1053/j.gastro.2010.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soutto M, Belkhiri A, Piazuelo MB, et al. Loss of TFF1 is associated with activation of NF-kappaB-mediated inflammation and gastric neoplasia in mice and humans. J Clin Invest. 2011;121(5):1753–1767. doi: 10.1172/JCI43922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wroblewski LE, Piazuelo MB, Chaturvedi R, et al. Helicobacter pylori targets cancer-associated apical-junctional constituents in gastroids and gastric epithelial cells. Gut. 2015;64(5):720–730. doi: 10.1136/gutjnl-2014-307650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hessmann E, Schneider G, Ellenrieder V, Siveke JT. MYC in pancreatic cancer: novel mechanistic insights and their translation into therapeutic strategies. Oncogene. 2015 doi: 10.1038/onc.2015.216. [DOI] [PubMed] [Google Scholar]
- 11.Chiurillo MA. Role of the Wnt/beta-catenin pathway in gastric cancer: An in-depth literature review. World J Exp Med. 2015;5(2):84–102. doi: 10.5493/wjem.v5.i2.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mao J, Fan S, Ma W, et al. Roles of Wnt/beta-catenin signaling in the gastric cancer stem cells proliferation and salinomycin treatment. Cell Death Dis. 2014;5:e1039. doi: 10.1038/cddis.2013.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hajimoradi M, Mohammad Hassan Z, Ebrahimi M, et al. STAT3 is Overactivated in Gastric Cancer Stem-Like Cells. Cell J. 2016;17(4):617–628. doi: 10.22074/cellj.2016.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fonseca BF, Predes D, Cerqueira DM, et al. Derricin and derricidin inhibit Wnt/beta-catenin signaling and suppress colon cancer cell growth in vitro. PLoS One. 2015;10(3):e0120919. doi: 10.1371/journal.pone.0120919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shin HW, Choi H, So D, et al. ITF2 prevents activation of the beta-catenin-TCF4 complex in colon cancer cells and levels decrease with tumor progression. Gastroenterology. 2014;147(2):430–442 e438. doi: 10.1053/j.gastro.2014.04.047. [DOI] [PubMed] [Google Scholar]
- 16.Chiurillo MA. Role of the Wnt/beta-catenin pathway in gastric cancer: An in-depth literature review. World J Exp Med. 2015;5(2):84–102. doi: 10.5493/wjem.v5.i2.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ikenoue T, Ijichi H, Kato N, et al. Analysis of the beta-catenin/T cell factor signaling pathway in 36 gastrointestinal and liver cancer cells. Jpn J Cancer Res. 2002;93(11):1213–1220. doi: 10.1111/j.1349-7006.2002.tb01226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clements WM, Wang J, Sarnaik A, et al. beta-Catenin mutation is a frequent cause of Wnt pathway activation in gastric cancer. Cancer Res. 2002;62(12):3503–3506. [PubMed] [Google Scholar]
- 19.Soutto M, Romero-Gallo J, Krishna U, et al. Loss of TFF1 promotes Helicobacter pylori-induced beta-catenin activation and gastric tumorigenesis. Oncotarget. 2015;6(20):17911–17922. doi: 10.18632/oncotarget.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zimmerman KA, Yancopoulos GD, Collum RG, et al. Differential expression of myc family genes during murine development. Nature. 1986;319(6056):780–783. doi: 10.1038/319780a0. [DOI] [PubMed] [Google Scholar]
- 21.Mishra R, Watanabe T, Kimura MT, et al. Identification of a novel E-box binding pyrrole-imidazole polyamide inhibiting MYC-driven cell proliferation. Cancer Sci. 2015;106(4):421–429. doi: 10.1111/cas.12610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gabay M, Li Y, Felsher DW. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb Perspect Med. 2014;4(6) doi: 10.1101/cshperspect.a014241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calcagno DQ, Freitas VM, Leal MF, et al. MYC, FBXW7 and TP53 copy number variation and expression in gastric cancer. BMC Gastroenterol. 2013;13:141. doi: 10.1186/1471-230X-13-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu J, Chen Y, Olopade OI. MYC and Breast Cancer. Genes Cancer. 2010;1(6):629–640. doi: 10.1177/1947601910378691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan FH, Putoczki TL, Stylli SS, Luwor RB. The role of STAT3 signaling in mediating tumor resistance to cancer therapy. Curr Drug Targets. 2014;15(14):1341–1353. doi: 10.2174/1389450115666141120104146. [DOI] [PubMed] [Google Scholar]
- 26.Judd LM, Menheniott TR, Ling H, et al. Inhibition of the JAK2/STAT3 pathway reduces gastric cancer growth in vitro and in vivo. PLoS One. 2014;9(5):e95993. doi: 10.1371/journal.pone.0095993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uddin N, Kim RK, Yoo KC, et al. Persistent activation of STAT3 by PIM2-driven positive feedback loop for epithelial-mesenchymal transition in breast cancer. Cancer Sci. 2015;106(6):718–725. doi: 10.1111/cas.12668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schutz A, Roser K, Klitzsch J, et al. Lung Adenocarcinomas and Lung Cancer Cell Lines Show Association of MMP-1 Expression With STAT3 Activation. Transl Oncol. 2015;8(2):97–105. doi: 10.1016/j.tranon.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang H, Yamazaki T, Pietrocola F, et al. STAT3 Inhibition Enhances the Therapeutic Efficacy of Immunogenic Chemotherapy by Stimulating Type 1 Interferon Production by Cancer Cells. Cancer Res. 2015;75(18):3812–3822. doi: 10.1158/0008-5472.CAN-15-1122. [DOI] [PubMed] [Google Scholar]
- 30.Han Z, Wang X, Ma L, et al. Inhibition of STAT3 signaling targets both tumor-initiating and differentiated cell populations in prostate cancer. Oncotarget. 2014;5(18):8416–8428. doi: 10.18632/oncotarget.2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 32.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004:3. doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 33.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic acids research. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El-Rifai W, Moskaluk CA, Abdrabbo MK, et al. Gastric cancers overexpress S100A calcium-binding proteins. Cancer Res. 2002;62(23):6823–6826. [PubMed] [Google Scholar]
- 35.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 36.Bertuccio P, Chatenoud L, Levi F, et al. Recent patterns in gastric cancer: a global overview. International journal of cancer Journal international du cancer. 2009;125(3):666–673. doi: 10.1002/ijc.24290. [DOI] [PubMed] [Google Scholar]
- 37.Gao J, Liu X, Yang F, Liu T, Yan Q, Yang X. By inhibiting Ras/Raf/ERK and MMP-9, knockdown of EpCAM inhibits breast cancer cell growth and metastasis. Oncotarget. 2015;6(29):27187–27198. doi: 10.18632/oncotarget.4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stalker L, Pemberton J, Moorehead RA. Inhibition of proliferation and migration of luminal and claudin-low breast cancer cells by PDGFR inhibitors. Cancer Cell Int. 2014;14(1):89. doi: 10.1186/s12935-014-0089-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dhawan P, Ahmad R, Chaturvedi R, et al. Claudin-2 expression increases tumorigenicity of colon cancer cells: role of epidermal growth factor receptor activation. Oncogene. 2011;30(29):3234–3247. doi: 10.1038/onc.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morin PJ, Sparks AB, Korinek V, et al. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275(5307):1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 41.Wen F, Liu Y, Wang W, et al. Adenomatous polyposis coli genotype-dependent toll-like receptor 4 activity in colon cancer. Oncotarget. 2016;7(7):7761–7772. doi: 10.18632/oncotarget.6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oue N, Sentani K, Sakamoto N, Yasui W. Clinicopathologic and molecular characteristics of gastric cancer showing gastric and intestinal mucin phenotype. Cancer Sci. 2015;106(8):951–958. doi: 10.1111/cas.12706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu MH, Lee WJ, Hua KT, Kuo ML, Lin MT. Macrophage Infiltration Induces Gastric Cancer Invasiveness by Activating the beta-Catenin Pathway. PLoS One. 2015;10(7):e0134122. doi: 10.1371/journal.pone.0134122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soutto M, Peng D, Katsha A, et al. Activation of beta-catenin signalling by TFF1 loss promotes cell proliferation and gastric tumorigenesis. Gut. 2015;64(7):1028–1039. doi: 10.1136/gutjnl-2014-307191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yanaka Y, Muramatsu T, Uetake H, Kozaki K, Inazawa J. miR-544a induces epithelial-mesenchymal transition through the activation of WNT signaling pathway in gastric cancer. Carcinogenesis. 2015;36(11):1363–1371. doi: 10.1093/carcin/bgv106. [DOI] [PubMed] [Google Scholar]
- 46.Ishimoto T, Oshima H, Oshima M, et al. CD44+ slow-cycling tumor cell expansion is triggered by cooperative actions of Wnt and prostaglandin E2 in gastric tumorigenesis. Cancer Sci. 2010;101(3):673–678. doi: 10.1111/j.1349-7006.2009.01430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoffman B, Liebermann DA. Apoptotic signaling by c-MYC. Oncogene. 2008;27(50):6462–6472. doi: 10.1038/onc.2008.312. [DOI] [PubMed] [Google Scholar]
- 48.Nilsson JA, Cleveland JL. Myc pathways provoking cell suicide and cancer. Oncogene. 2003;22(56):9007–9021. doi: 10.1038/sj.onc.1207261. [DOI] [PubMed] [Google Scholar]
- 49.Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19(1):1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang SW, Sun YM. The IL-6/JAK/STAT3 pathway: potential therapeutic strategies in treating colorectal cancer (Review) Int J Oncol. 2014;44(4):1032–1040. doi: 10.3892/ijo.2014.2259. [DOI] [PubMed] [Google Scholar]
- 51.Banerjee K, Resat H. Constitutive activation of STAT3 in breast cancer cells: A review. Int J Cancer. 2015 doi: 10.1002/ijc.29923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Z, Zhu S, Hong J, et al. Gastric tumour-derived ANGPT2 regulation by DARPP-32 promotes angiogenesis. Gut. 2015 doi: 10.1136/gutjnl-2014-308416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu S, Belkhiri A, El-Rifai W. DARPP-32 increases interactions between epidermal growth factor receptor and ERBB3 to promote tumor resistance to gefitinib. Gastroenterology. 2011;141(5):1738–1748. e1731–1732. doi: 10.1053/j.gastro.2011.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Z, Guo L, Liu D, et al. Acquisition of resistance to trastuzumab in gastric cancer cells is associated with activation of IL-6/STAT3/Jagged-1/Notch positive feedback loop. Oncotarget. 2015;6(7):5072–5087. doi: 10.18632/oncotarget.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gerlach K, Daniel C, Lehr HA, et al. Transcription factor NFATc2 controls the emergence of colon cancer associated with IL-6-dependent colitis. Cancer Res. 2012;72(17):4340–4350. doi: 10.1158/0008-5472.CAN-11-4155. [DOI] [PubMed] [Google Scholar]
- 56.Bakker WJ, Harris IS, Mak TW. FOXO3a is activated in response to hypoxic stress and inhibits HIF1-induced apoptosis via regulation of CITED2. Mol Cell. 2007;28(6):941–953. doi: 10.1016/j.molcel.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 57.Tennant DA. PK-M2 Makes Cells Sweeter on HIF1. Cell. 2011;145(5):647–649. doi: 10.1016/j.cell.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 58.Shi CY, Fan Y, Liu B, Lou WH. HIF1 contributes to hypoxia-induced pancreatic cancer cells invasion via promoting QSOX1 expression. Cell Physiol Biochem. 2013;32(3):561–568. doi: 10.1159/000354460. [DOI] [PubMed] [Google Scholar]
- 59.Goscinski MA, Nesland JM, Giercksky KE, Dhakal HP. Primary tumor vascularity in esophagus cancer. CD34 and HIF1-alpha expression correlate with tumor progression. Histol Histopathol. 2013;28(10):1361–1368. doi: 10.14670/HH-28.1361. [DOI] [PubMed] [Google Scholar]
- 60.Pawlus MR, Wang L, Hu CJ. STAT3 and HIF1alpha cooperatively activate HIF1 target genes in MDA-MB-231 and RCC4 cells. Oncogene. 2014;33(13):1670–1679. doi: 10.1038/onc.2013.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jung HH, Lee SH, Kim JY, Ahn JS, Park YH, Im YH. Statins affect ETS1-overexpressing triple-negative breast cancer cells by restoring DUSP4 deficiency. Sci Rep. 2016;6:33035. doi: 10.1038/srep33035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Z, Lin P, Gao C, et al. Integrin beta6 acts as an unfavorable prognostic indicator and promotes cellular malignant behaviors via ERK-ETS1 pathway in pancreatic ductal adenocarcinoma (PDAC) Tumour Biol. 2016;37(4):5117–5131. doi: 10.1007/s13277-015-4353-7. [DOI] [PubMed] [Google Scholar]
- 63.Zheng L, Qi T, Yang D, et al. microRNA-9 suppresses the proliferation, invasion and metastasis of gastric cancer cells through targeting cyclin D1 and Ets1. PLoS One. 2013;8(1):e55719. doi: 10.1371/journal.pone.0055719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith AM, Findlay VJ, Bandurraga SG, et al. ETS1 transcriptional activity is increased in advanced prostate cancer and promotes the castrate-resistant phenotype. Carcinogenesis. 2012;33(3):572–580. doi: 10.1093/carcin/bgs007. [DOI] [PubMed] [Google Scholar]
- 65.Mizuarai S, Yamanaka K, Kotani H. Mutant p53 induces the GEF-H1 oncogene, a guanine nucleotide exchange factor-H1 for RhoA, resulting in accelerated cell proliferation in tumor cells. Cancer Res. 2006;66(12):6319–6326. doi: 10.1158/0008-5472.CAN-05-4629. [DOI] [PubMed] [Google Scholar]
- 66.Feng CW, Wang LD, Jiao LH, Liu B, Zheng S, Xie XJ. Expression of p53, inducible nitric oxide synthase and vascular endothelial growth factor in gastric precancerous and cancerous lesions: correlation with clinical features. BMC Cancer. 2002;2:8. doi: 10.1186/1471-2407-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rajnakova A, Moochhala S, Goh PM, Ngoi S. Expression of nitric oxide synthase, cyclooxygenase, and p53 in different stages of human gastric cancer. Cancer Lett. 2001;172(2):177–185. doi: 10.1016/s0304-3835(01)00645-0. [DOI] [PubMed] [Google Scholar]
- 68.Kim IM, Ackerson T, Ramakrishna S, et al. The Forkhead Box m1 transcription factor stimulates the proliferation of tumor cells during development of lung cancer. Cancer Res. 2006;66(4):2153–2161. doi: 10.1158/0008-5472.CAN-05-3003. [DOI] [PubMed] [Google Scholar]
- 69.Wang Z, Banerjee S, Kong D, Li Y, Sarkar FH. Down-regulation of Forkhead Box M1 transcription factor leads to the inhibition of invasion and angiogenesis of pancreatic cancer cells. Cancer Res. 2007;67(17):8293–8300. doi: 10.1158/0008-5472.CAN-07-1265. [DOI] [PubMed] [Google Scholar]
- 70.Li X, Liang J, Liu YX, et al. Knockdown of the FoxM1 enhances the sensitivity of gastric cancer cells to cisplatin by targeting Mcl-1. Pharmazie. 2016;71(6):345–348. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. Overlapping genes in the P53 transcription network in LGD from Tff1 KO mice and human gastric cancer tissue samples. Ingenuity pathway analysis was used to generate the transcription network. TP53 in the center indicates deregulation of P53 signaling in both Tff1 KO mice LGD and human gastric cancer samples. Deregulated downstream targets of P53 are shown (P<0.05).
Supplementary Figure S2. TFF1 mRNA expression level in human gastric tissue samples. qRT-PCR analysis of expression of TFF1 in 19 human normal stomach (NG) and 22 gastric cancer (GC) tissue samples. The horizontal line indicates the median, ***P<0.001, Mann-Whitney U test.


