Summary
Candidate phyla (CP) are broad phylogenetic clusters of organisms that lack cultured representatives. Included in this fraction is the candidate Parcubacteria superphylum. Specific characteristics that have been ascribed to the Parcubacteria include reduced genome size, limited metabolic potential, and exclusive reliance on fermentation for energy acquisition. The study of new environmental niches, such as the marine versus terrestrial subsurface, often expands the understanding of the genetic potential of taxonomic groups. For this reason we analyzed twelve Parcubacteria single amplified genomes (SAGs) from sediment samples collected within the Challenger Deep of the Mariana Trench, obtained during the Deepsea Challenge (DSC) Expedition. Many of these SAGs are closely related to environmental sequences obtained from deep-sea environments based on 16S rRNA gene similarity and BLAST matches to predicted proteins. DSC SAGs encode features not previously identified in Parcubacteria obtained from other habitats. These include adaptation to oxidative stress, polysaccharide modification, and genes associated with respiratory nitrate reduction. The DSC SAGs are also distinguished by relative greater abundance of genes for nucleotide and amino acid biosynthesis, repair of alkylated DNA and the synthesis of mechanosensitive ion channels. These results present an expanded view of the Parcubacteria, among members residing in an ultra-deep hadal environment.
Introduction
Ocean sediments harbor a large number of diverse microorganisms, including the presence of numerous candidate phyla (CP) within the large candidate phyla radiation of the tree of life (Whitman et al., 1998; Schauer et al., 2010; Nunoura et al, 2012; Hug et al. 2016). Underexplored areas of ocean sediment include hadal oceanic trenches, such as the Challenger Deep, the deepest ocean location on earth. It is located in the western Pacific Ocean, extending to a depth of approximately 10,920 m (Nakanishi and Hashimoto, 2011), corresponding to about 110 megapascals (MPa) of hydrostatic pressure. This environment has been shown to have high microbial activity in surface sediments based on oxygen utilization (Glud et al., 2013) but little is known about the diversity of its microbial community (Kato et al., 1998; Pathom-Aree et al., 2006; Yoshida et al., 2013; Tarn et al., 2016). During the Deepsea Challenge (DSC) Expedition a sediment core was obtained by the manned submersible Deepsea Challenger, and from it single cell-derived genomes were obtained, including a number derived from members of the candidate Parcubacteria (OD1) superphylum. These genomes provide the opportunity to examine the genome characteristics of the Parcubacteria CP in the context of an extreme habitat.
Currently, the Parcubacteria stands out among CP as one of the most studied due to its abundance in many different anoxic marine and terrestrial environments (Harris et al., 2004; Elshahed et al, 2005; Gihring et al., 2011; Peura et al., 2012). Metabolic information has been acquired from metagenomic composite genomes and by using single cell genomics (Rinke et al., 2013; Kantor et al., 2013; Wrighton et al., 2012; Wrighton et al., 2014; Brown et al., 2015). Candidatus Paceibacter normanii (single cell AAA255-P19) was designated as a Candidatus type species for the Parcubacteria superphylum (Rinke et al., 2013). It was recovered from brackish water present at 120 m depth in Sakinaw Lake, British Columbia, Canada. Its genome is 0.6 Mbp in size and estimated to be 70% complete (Rinke et al., 2013). Candidatus Paceibacter normanii appears to have very limited metabolic potential highlighted by the lack of genes involved in sugar and amino acid degradation, complete TCA cycle, the pentose phosphate pathway, pyruvate metabolism and the electron transport chain.
Additional Parcubacteria genomes have been recovered from metagenomes of an acetate-amended aquifer sediment, including the sublineage OD1-i, which also possess a relatively reduced genome in terms of metabolic potential, lacking the tricarboxylic acid cycle (TCA) cycle and oxidative phosphorylation components (Wrighton et al., 2012). With a mostly fermentative metabolism, this sub-lineage is predicted to utilize acetyl-CoA synthetase for ATP generation and to reoxidize NADH produced during glycolysis by converting pyruvate to D-lactate and acetyl-CoA to ethanol. Another nearly complete Parcubacteria genome sequence, designated RAAC4, was also recovered via genome reconstruction from a metagenome of acetate-amended sediments (Kantor et al., 2013). As with other Parcubacteria, RAAC4 lacks a TCA cycle and respiratory chain enzymes and appears to be a strictly fermentative anaerobic organism. Although it does contain genes associated with the pentose phosphate pathway and a modified Embden-Meyerhof-Parnas (EMP) pathway, and appears to be able to utilize complex organic carbon and perhaps to create biofilms, this genome sequence information has reinforced the conclusion that members of the Parcubacteria have limited numbers of metabolic genes (Kantor et al., 2013).
Most recently the Parcubacteria were described as a superphylum following the reconstruction and annotation of 429 member genomes from an acetate amended groundwater aquifer at Rifle, CO (Brown et al., 2015). One the most significant findings derived from this massive dataset was the discovery of a great phylogenetic radiation within the Parcubacteria (14 phyla described, more undescribed). In addition, many of the genomes encode self-splicing introns within their 16S rRNA genes and lack a number of ribosomal protein genes (Brown et al., 2015). The difference in ribosomal structure suggests that Parcubacteria may have evolved alternative mechanisms for ribosome regulation and function; and as a result of such differences have been likely under sampled in environmental surveys (Brown et al., 2015). In addition to having small genomes, microscopic analyses suggest that Parcubacteria cells have extremely small cell volume size of 0.009±0.002 µm3 (Luef et al, 2015). Another recent study proposed the Parcubacteria to be symbiotic organisms based on their small genomes and lack of central biosynthetic pathways, although some novel metabolic genes were also described associated with aerobic respiration, particularly genomes C7867-007 and C7867-008 (Nelson & Stegen, 2015). The potential symbiotic nature of the OD1 CP was also highlighted by the discovery of a fresh water protist, Paramecium bursaria, in association with bacteria from the Parcubacteria superphylum (Gong et al., 2014). Most recently the description of the phylum Parcunitrobacteria suggested that the genome Candidatus Parcunitrobacter nitroensis is a member of the Parcubacteria that may have the potential to oxidize hydroxylamine (NH2OH) to nitric oxide (NO) coupled to NO reduction (Castelle et al., 2017). Castelle and colleagues reported the first genome with respiratory potential based on genes encoding a modified oxidative phosphorylation pathway. This finding shows further variability in metabolic potential exists among members of the Parcubacteria superphylum.
Here we present a comparative genomic analysis of twelve Parcubacteria single cell genomes from cells collected within Challenger Deep sediment. The results indicate that deep-sea sediment Parcubacteria belong to diverse phylogenetic groups within the superphylum and while they share many metabolic pathways with previously analyzed genomes, they also expand the known metabolic potential of Parcubacteria.
Results and Discussion
Genomic properties
From the recovered sample, 3,520 total cells were sorted, 704 cells were subjected to MDA, and 494 MDA reactions were positively amplified as identified by subsequent 16S rRNA gene amplification. Among the known phyla, Cyanobacteria from the genus Prochlorococcus and Alphaproteobacteria, were the most abundant lineages detected (Figure S1). This trench axis is known to host greater amounts and younger and more labile organic matter than sediment located along the trench wall of the incoming plate (Glud et al., 2013). The cyanobacterial sequences are interpreted as evidence of the recent vertical transport of fresh organic matter from sunlit shallow waters (Agusti et al., 2015).
The CP with the most number of cells detected, Parcubacteria, had a total of 20 cells identified by 16S rRNA gene sequences with >85% identity to Parcubacteria representatives. This amounted to approximately 5.4% of the total single cells analyzed for the study (Figure S1). Of these, twelve genomes were further analyzed (Table 1). Sequences recovered from SAGs ranged from 0.4 to 1.0 Mbp and genome completeness ranged from 25% (0.40Mbp) to 66% (0.47 Mbp) based on recovery of conserved, single copy marker genes including various ribosomal proteins (Parks et al., 2015). The estimated genome size for these cells averaged 1.39 Mbp, which is at the higher end of the range known for Parcubacteria (Rinke et al., 2013). Percent GC ranged from 34.1% to 45.6%, with an average of 39.2% GC, similar to that of the proposed type species Candidatus Paceibacter normanii (39%).
Table 1.
Single cell genome descriptions
| Group | Study Name | Genome Size | % Complete | Est. Genome Size | # contigs | Coding Base Count % | Gene Count | HTGs (#per genome) |
% of Parcubacteria protiens |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| OD1-L1 | OD1DSC1 | 825670 | 58 | 1423568 | 68 | 88 | 881 | 8 | 83% |
| OD1DSC3 | 473888 | 66 | 718912 | 18 | 88 | 524 | 4 | 88% | |
| OD1DSC4 | 600239 | 39 | 1539074 | 48 | 87 | 601 | 8 | 83% | |
| OD1DSC8 | 380175 | 41 | 927256 | 38 | 86 | 395 | 4 | 80% | |
| OD1DSC9 | 473882 | 31 | 1528651 | 40 | 86 | 495 | 6 | 81% | |
| OD1DSC10 | 546935 | 66 | 828689 | 37 | 88 | 581 | 7 | 88% | |
| OD1DSC11 | 599599 | 39 | 1537433 | 82 | 83 | 651 | 6 | 82% | |
| OD1DSC12 | 690602 | 50 | 1381204 | 79 | 86 | 658 | 8 | 80% | |
|
| |||||||||
| OD1-DSC | OD1DSC5 | 406664 | 25 | 1626656 | 47 | 88 | 397 | 10 | 67% |
| OD1DSC6 | 1028328 | 59 | 1742928 | 68 | 86 | 1048 | 12 | 76% | |
| OD1DSC7 | 758051 | 41 | 1848904 | 69 | 86 | 770 | 11 | 85% | |
|
| |||||||||
| Uhrbacteria | OD1DSC2 | 520375 | 32 | 1626171 | 45 | 86 | 595 | 10 | 77% |
Statistics are based on CheckM (Parks et al., 2015)
Open reading frame predictions ranged from 395 to 1048 genes; of these 32–56% were of uncharacterized function (Table 1). 16S rRNA genes were recovered from the Illumina HiSeq 2500 sequencing for all but one genome and tRNA gene counts ranged from 15 to 40 per genome. Pairwise average nucleotide identity (ANI) relationships among the DSC SAGs ranged from 64 –93%, with the scores of relatedness averaging in the low 70s and only DSC5 and DSC8 showing 93% relatedness (Table S1). This suggests that none of the genomes are related enough to be the same species, although DSC5 and DSC8 are near the proposed cutoff (Konstantinidis and Tiedje, 2005). The presence or absence of ribosomal proteins, including ribosomal protein L1, has been found to be a major distinguishing factor between Parcubacteria, creating OD1 and OD1-L1 lineages (Brown et al., 2015). The DSC genomes, while incomplete, are all missing ribosomal protein L30, and most lack ribosomal protein L1 and GTPase Der (except DSC5, 7 for L1 and DSC6 for Der). The loss of these genes has previously been noted in other Parcubacteria (Brown et al., 2015). Although the genomes are incomplete, DSC2 and DSC6 may be the first example of genomes that cluster within the OD1 group, but lack the ribosomal protein L1 gene. Interestingly, none of the DSC genomes recovered encoded catalytic RNAs within their 16S rRNA genes (Brown et al., 2015).
Phylogenetic relationships
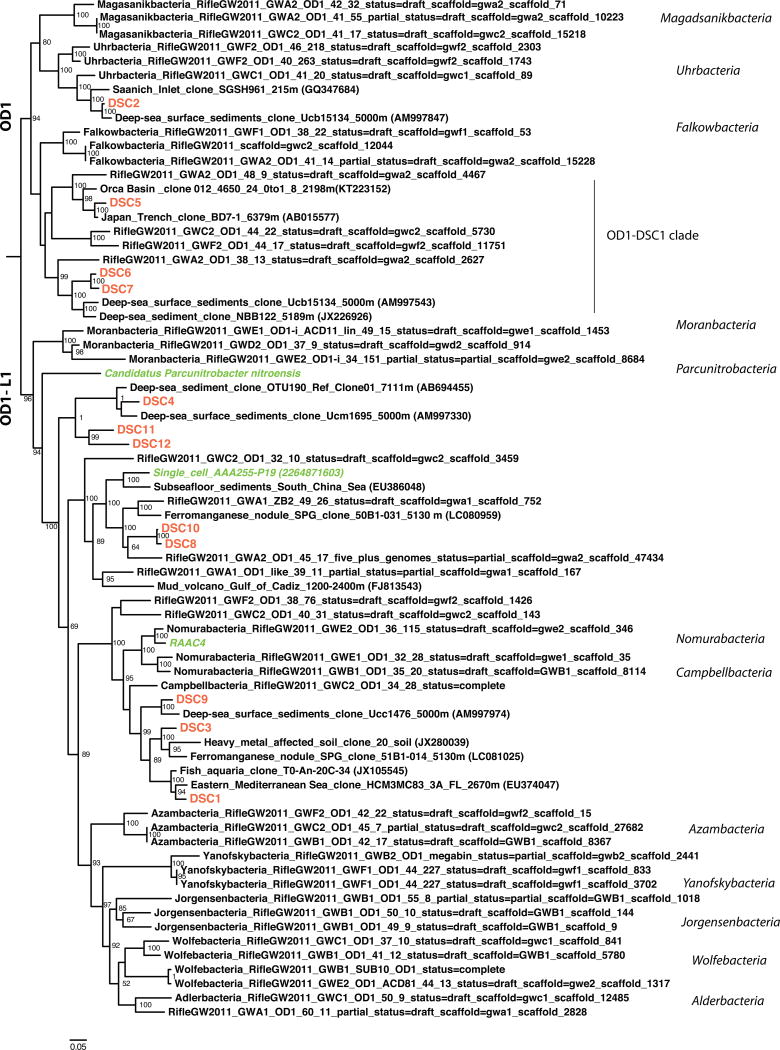
When comparing the 16S rRNA genes to the NCBI nt database using BLASTN, the top hit percent identity for each DSC SAG 16S rRNA gene ranged from 83 – 97% similarity to environmental clone sequences, suggesting that DSC genomes are distantly related to other previously described Parcubacteria. Phylogenetic analyses were performed for the eleven genomes for which 16S rRNA genes were recovered in the sequenced genome and one of sequences from the preliminary MDA screen (Figure 1).
Figure 1. 16S rRNA gene-based phylogenetic tree of the DSC OD1 SAGs and additional Parcubacteria superphylum members.
Maximum likelihood phylogenetic tree of 16S rRNA gene sequences obtained from eleven single amplified genomes (orange), one MDA screen (DSC9), draft genomes recently described from amended subterranean aquifers as part of the Parcubacteria superphylum (rRNA genes >1000bp, Brown et al., 2015) and previously described single cell and composite genomes (green; Rinke et al., 2013, Wrighton et al., 2012). The OD1-DSC clade falls into a separated unnamed/unclassfied lineage, a division supported by additional phylogenies (figures S3–S5). Scale bar represents 0.04 changes per position. Confidence values above 50% are shown at the tree nodes. The tree was rooted using Escherichia coli K12.
Nine DSC Parcubacteria 16S rRNA gene sequences are closely related to environmental sequences associated with deep-sea environments. Homology to known 16S rRNA genes was generally low, with the only 97% identity score matching a deep-sea sediment sample obtained at a water column depth greater than 5000 m (DSC2, Schauer et al., 2010). Four of the SAG 16S rRNA genes are most closely related to sequences obtained from deep-sea sediments in the South Atlantic, Guinea Basin (DSC2 (97% similarity), 6 (89%), 7 (88%) and 9 (92%); Schauer et al., 2010; >5000m), two others are closely related to samples from sub-seafloor sediment of the South China Sea (DSC 8 (87%) and 10 (84%); Tao et al., 2008; 3697 m below the sea surface and 0.1m below the seafloor), DSC1 is related to samples recovered from a canyon slope in the Eastern Mediterranean Sea at 3603m (93%; Polymenakou et al., 2009 ) and DSC4 (93%) and 5 (96%) are closely related to sediments within the Japan Trench at depths of 7111m and 6379m respectively (Hori et al., 2013; Li et al., 1999). No previous suggestion of deep sea relatedness across Parcubacteria has been noted, yet these new sequences from the Mariana Trench clearly show significant relationships to deep sea relatives. Although pairwise ANI analysis suggested that DSC5 and DSC8 may be within species level relatedness, their phylogeny suggests they are likely separate species.
Following the proposed nomenclature by Brown et al. (2015), based on 16S rRNA genes most of the DSC genomes belong to unknown or unclassified clades/phyla, with one exception, as genome DSC2 falls within the Uhrbacteria phylum (Figure 1). The Parcubacteria superphylum is separated into two major groups, designated as OD1 and OD1-L1 (lack ribosomal protein L1). The DSC genomes fall within both major groups, eight of them within the OD1-L1 group (DSC1, 3, 4, 8, 9, 10, 11, 12) and four of them within the OD1 group (DSC2, 5, 6, 7) (Figure 1). OD1-L1 is a large subgroup from the Parcubacteria subphylum encompassing most of the initially described Parcubacteria genomes including those from Candidatus Paceibacter normanii and RAAC4. Genomes DSC8 and 10 are the SAGs whose 16S rRNA gene sequences are most similar to Candidatus Paceibacter normanii (Rinke et al., 2013). Of the four DSC genomes that reside within the other major subgroup of the Parcubacteria superphylum (OD1), 16S rRNA genes from DSC5, 6 and 7 form a distinct cluster associated with other deep sea environmental 16S rRNA gene sequences of unknown lineage, herein called the OD1-DSC1 clade. Phylogenetic associations of the OD1-DSC1 clade were further investigated by analyzing other conserved phylogenetic gene markers, including DNA gyrase B, recombinase A and RNA polymerase B (see Supplementary Figures S3, S4 and S5), and the results support the 16S rRNA-based relationships (Figure 1). However, a clear separation between subclades of the OD1-DSC1 clade was not always reproduced when using the marker genes. This was due, in part, to missing marker genes in the OD1-DSC1 genomes.
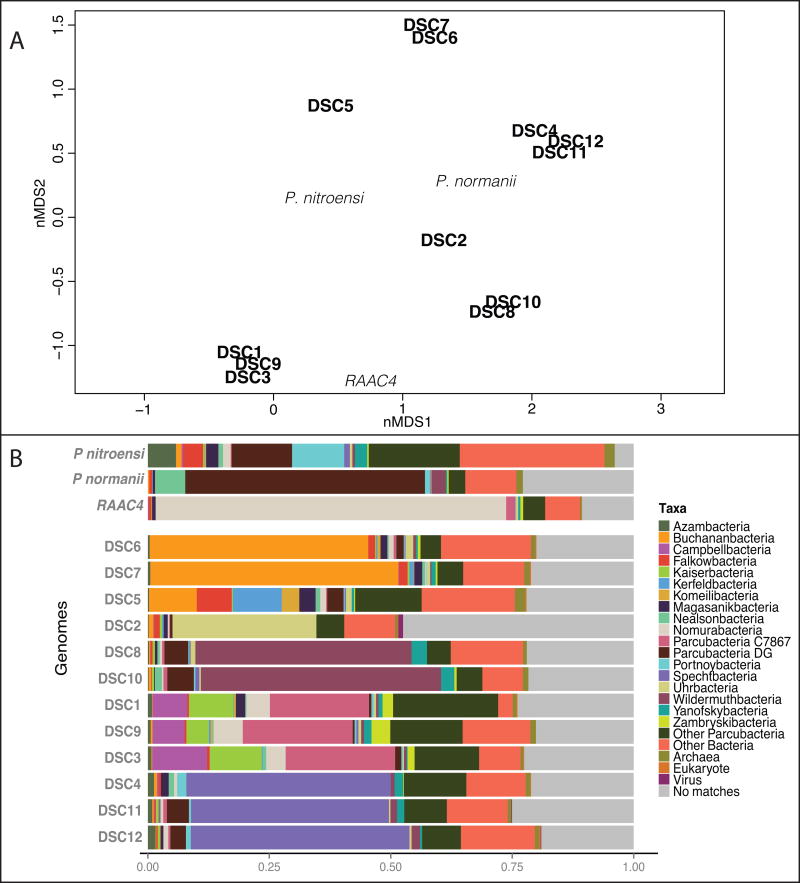
To assess the relationship of the SAGs from a protein relatedness perspective within the Parcubacteria superphylum and among each other, taxonomic classification of DSC predicted proteins were recovered via DarkHorse BLASTP analysis (Podell and Gaasterland, 2007). This analysis specifically targeted proteins that have taxonomic matches to previously characterized Parcubacteria phyla using the NCBI nr database of December 2016 (Adlerbacteria, Andersenbacteria, Azambacteria, Buchananbacteria, Brennerbacteria, Campbellbacteria, Colwellbacteria, Falkowbacteria, Giovannonibacteria, Harrisonbacteria, Jacksonbacteria, Jorgensenbacteria, Kaiserbacteria, Kerfeldbacteria, Komeilibacteria, Kuenenbacteria, Liptonbacteria, Llyodbacteria, Magasanikbacteria, Moranbacteria, Nealsonbacteria, Niyogibacteria, Nomurabacteria, Other Parcubacteria, Portnoybacteria, Ryanbacteria, Spechtbacteria, Staskawiczbacteria, Sungbacteria, Tagabacteria, Taylorbacteria, Terrybacteria, Uhrbacteria, Veblenbacteria, Vogelbacteria, Wildermuthbacteria, Wolfebacteria, Yanofskybacteria, Yonathbacteria and Zambryskibacteria). The number of predicted proteins in the DSC genomes that are associated with Parcubacteria ranges from 67% (DSC5) to 88% (DSC10) of the total predicted proteins of each genome (Table 1). Based on the percent of shared predicted proteins with Parcubacteria, DSC genomes cluster into groups that are similar to those predicted by the 16S rRNA gene phylogeny, yet are clearly distinct from each other, reinforcing the 16SrRNA gene-based data suggesting that large phylogenetic distances exist between these groups (Figure 2A). The taxonomic distribution of protein BLAST matches varies between the different DSC groups (Figure 2B). The analysis suggests that the DSC2 genome shares the most number of proteins with the Uhrbacteria phylum and DSC8 and 10 match the Wildermuthbacteria phylum (Figure 2B). DSC5, 6 and 7 are more similar to Buchanbacteria phylum, although DSC5 also shares numerous proteins with the Kerfeldbacteria phylum, clustering farther away from DSC6 and 7 (Figure 2A). DSC4, 11 and 12 appear to belong to the Spechtbacteria (Figure 2B). The DSC1, 3 and 9 genomes protein matches are not dominated by any particular group, but all share similar relative abundance to the Campbellbacteria, Kaiserbacteria and Nomurabacteria phyla, as well as Parcubacteria genomes C7867 analyzed by Nelson and Stegen (Nelson & Stegen, 2015). As such, the metabolic analysis of these new DSC genomes provides especially valuable new insights about these poorly understood microbial groups.
Figure 2. Taxonomic classification of annotated protein within the Parcubacteria superphylum.
A) Non-metric multidimensional scaling (nMDS) of the taxonomic classification of annotated proteins within the Parcubacteria superphylum were generated from the DarkHorse analysis. DSC protein annotated as Parcubacteria per SAG range from 67% (DSC5) to 88% (DSC10) of the total of protein in the genomes. DSC genomes (in bold black) and comparison genomes (in light black; Kantor et al., 2013, Brown et al 2015, Castalle et al, 2017) are ordinated in the 2D plot. Based on protein classifications the DSC Parcubacteria reinforces the 16S data, as they group similarly when looking the taxonomic classifications. Stress value = 0.01183975. B) Bar plot shows the distribution of Parcubacteria taxa in SAGs. Parcubacteria taxa shown make up 5% or more than the total protein matches and include proteins identified as archaeal, viral, non-Parcubacteria bacterial matches and un-assigned proteins
Metabolic comparisons
Considering that even when all the new DSC genomes are combined they still only yield 92.8% genome completion based on conserved marker genes, inferences about metabolism and cell structure are limited by the possibility of missing characteristics due to the incompleteness of this dataset. KEGG pathway metabolic reconstruction of the DSC genomes suggests that the cells are heterotrophic microbes with limited substrate utilization ability. As a community, the DSC genomes have the potential for glycolysis via the Embden–Meyerhof–Parnas (EMP) pathway, which is the main pathway for the conversion of glucose to pyruvate in order to generate energy. All EMP genes but one are present within the annotated proteins of the total dataset, the exception being phosphofructokinase (PFK). Bacterial PFK protein sequences retrieved from the NCBI conserved domains were searched against the DSC genomes by BLASTP and TBLASTN (data not shown), but no homologs were found in any of the SAGs. PFK is also absent from OD1-i and Candidatus Paceibacter normanii genomes, whereas RAAC4 has all of the EMP enzymes. Among the available Parcubacteria genomes in the Integrated Microbial Genomes web platform (IMG, Markowitz et al., 2014, 448 genomes at the time of the analysis), only 6 possess PFK. The lack of PFK in many of the Parcubacteria could reflect a primary EMP role for gluconeogenesis rather than glycolysis, using this anabolic pathway to synthesize sugar molecules from pyruvate. However, many previously described Parcubacteria genomes also lack other EMP genes whose products act early in the pathway, while possessing those responsible for the second phase of the pathway (the energetic payoff phase), although it is important to keep in mind that the great majority of the genomes analyzed are incomplete. Given that the DSC genomes possess other essentially irreversible enzymes involved in the first phase in the EMP pathway such as glucokinase (DSC1, 3, 7 and 10) and pyruvate kinase (DSC2, 6 and 11), it may be that the DSC Parcubateria accomplish fructose 6 phosphate conversion to fructose 1, 6 bisphosphate using a novel, yet to be characterized PFK. With this in mind, we probed the DSC genomes for proteins from the PFK superfamily using the most diverse sequences from the conserved domains, particularly ribokinase/pfkB superfamily (cd00287). Homology search by BLASTP revealed that DSC genomes (DSC1, 5, 6, 8, 10, 11) possess genes for a sugar kinase with a pfkB pfam domain (e-values 2e-07 to 1e-103; Figure S8). These genes could be candidates for a novel PFK enzyme found in the Parcubacteria, as homologs are also found annotated as a hypothetical or sugar kinase gene in other Parcubateria genomes in IMG.
It has previously been proposed that the Parcubacteria superphylum is strictly fermentative and obligately anaerobic (Wrighton et al., 2012). The capacity for fermentative metabolism is evident in the DSC genomes, as reflected in the presence of genes for lactate and alcohol dehydrogenases, glyoxylate reductase and acetate kinase (DSC1, 5, 6, 7, 8 and 10). Genes coding for enzymes in the tricarboxylic acid (TCA) cycle are less abundant, especially in the OD1-DSC1 clade. The TCA cycle is the process by which pyruvate is converted to carbon dioxide (CO2) to generate energy and reducing equivalents are created along with precursors for biosynthesis. The cycle enzymes encoded within the DSC genomes include 2-oxoglutarate: ferredoxin oxidoreductase (KorA and KorB; DSC5 and 11) and succinyl-CoA synthetase, citryl-CoA synthetase and citryl-CoA lyase (DSC6) (Table S2). These enzymes, although not present in most of the genomes, have been reported to be present in 3–8% of the recently described Parcubacteria genomes (Brown et al., 2015). Many other enzymes that are required for a complete TCA cycle were not found in any of the DSC genomes. While all of the DSC single cell genomes are incomplete, they are unlikely to encode an entire TCA cycle, in agreement with other Parcubacteria genomes.
Subunits of NADH-dehydrogenase, the enzyme responsible for the first step in the electron transport chain, are found in DSC genomes (Table S2; DSC4, 6, 11 and 12). Ubiquinol-cytochrome c reductase cytochrome c1 subunit, part of complex III of the electron transport chain, is found in one genome from the OD1-DSC1 clade (DSC6) (Table S2). A few DSC genomes also contain genes coding for heme/copper-type cytochrome/quinol oxidases (DSC1, 10 and 11), which are involved in oxidative phosphorylation and the utilization of oxygen as a terminal electron acceptor. The DSC6 and 7 genomes also encode cytochrome c oxidase subunit II (oxygen-reducing terminal oxidase) (Table S2). This has also been noted in other Parcubacteria genomes, however as with the previous genomes, genes encoding the other two subunits of cytochrome C oxidase, cytochrome c oxidase subunit I (containing the catalytic domain) and cytochrome c oxidase subunit III (whose activity is not fully understood), are not present (Brown et al., 2015).
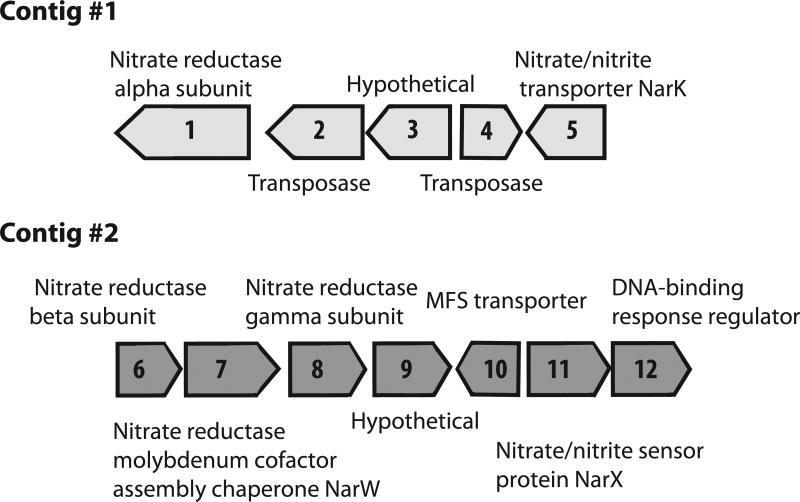
Intriguing aspects of nitrogen metabolism also exist within the OD1-DSC1 genomes. The DSC6 SAG contains a complete nitrate reductase operon in addition to a nitrous oxide reductase (Figure 3, Table S2). Phylogenetic analyses of the active subunit of the DSC6 nitrate reductase support a respiratory rather than assimilatory function (Figure S2). Most predicted proteins in the nitrate reductase operon are most closely related to genes present in Enhydrobacter aerosaccus, a member of the Gammaproteobacteria present in many environments, including marine (Staley et al., 1987; Irgens et al., 1989; Khandeparker and Anil, 2013, Leong et al., 2015; Figure 3, Table 2, Figure S2). The genes associated with nitrate reduction are present on two contigs from the DSC6 genome. Contig 1 contains the alpha subunit of the enzyme, a nitrate transporter and two transposable elements, suggesting a potential for horizontal transfer. Contig 2 contains the other nitrate reductase subunits and cofactors (Figure 3, Table 2). The DSC2 genome encodes subunits of a nitrite reductase nirD, used in the dissimilatory nitrite reduction pathway to ammonia. In addition, a number of nitrate/nitrite transporters are present, particularly in DSC5 and 6 (Table S2).
Figure 3. Scaffolds with nitrate reductase genes.
Two contigs are displayed showing each of the genes that are present and their respective annotated function. See Table 2 for associated gene information.
Table 2.
Nitrate reductase gene cluster homology information
| Protein | Start | End | Sense | Prokka ID | EC | Gene | Prokka annotation | NCBI BlastP annotation | Max score | Total score | Query cover | E value | Ident | Accession |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||
| 1 | 108 | 1367 | + | PROKKA_00706 | 1.7.99.4 | narH | Respiratory nitrate reductase 1 beta chain | nitrate reductase [Enhydrobacter aerosaccus] | 870 | 870 | 100% | 0 | 99% | WP_050324468.1 |
| 2 | 1369 | 2289 | + | PROKKA_00707 | narW | putative nitrate reductase molybdenum cofactor assembly chaperone NarW | nitrate reductase [Paenibacillus sophorae] | 574 | 574 | 91% | 0 | 98% | WP_051500173.1 | |
| 3 | 2286 | 2984 | + | PROKKA_00708 | 1.7.99.4 | narV | Respiratory nitrate reductase 2 gamma chain | nitrate reductase [Moraxella boevrei] | 361 | 361 | 99% | 6.00E-123 | 77% | WP_019519315.1 |
| 4 | 3455 | 4639 | + | PROKKA_00709 | 1.14.13.- | ubiI | 2-octaprenylphenol hydroxylase | hypothetical protein [Moraxella boevrei] | 543 | 543 | 98% | 0 | 64% | WP_019519414.1 |
| 5 | 4950 | 6188 | - | PROKKA_00710 | narT | putative nitrate transporter NarT | MFS transporter [Enhydrobacter aerosaccus] | 823 | 823 | 100% | 0 | 99% | WP_050324473.1 | |
| 6 | 6558 | 8633 | + | PROKKA_00711 | 2.7.13.3 | narX | Nitrate/nitrite sensor protein NarX | nitrate/nitrite two-component system sensor histidine kinase [Enhydrobacter aerosaccus] | 1396 | 1396 | 100% | 0 | 98% | WP_050324474.1 |
| 7 | 8795 | 9454 | + | PROKKA_00712 | narL | Nitrate/nitrite response regulator protein NarL | DNA-binding response regulator [Enhydrobacter aerosaccus] | 442 | 442 | 100% | 3.00E-155 | 99% | WP_050324475.1 | |
| 8 | 526 | 4293 | - | PROKKA_00732 | 1.7.99.4 | narG | Respiratory nitrate reductase 1 alpha chain | nitrate reductase [Enhydrobacter aerosaccus] | 2621 | 2621 | 100% | 0 | 99% | WP_050324469.1 |
| 9 | 4436 | 5542 | - | PROKKA_00733 | Putative transposase DNA-binding domain protein | transposase [Enhydrobacter aerosaccus] | 704 | 704 | 99% | 0 | 92% | WP_050323977.1 | ||
| 10 | 5682 | 6971 | - | PROKKA_00734 | NnrS protein | hypothetical protein [Enhydrobacter aerosaccus] | 840 | 840 | 100% | 0 | 99% | WP_050324471.1 | ||
| 11 | 7236 | 7475 | + | PROKKA_00735 | hypothetical protein | transposase [Enhydrobacter aerosaccus] | 103 | 103 | 64% | 2.00E-26 | 94% | WP_050324826.1 | ||
| 12 | 7390 | 8736 | - | PROKKA_00736 | narK | Nitrate/nitrite transporter NarK | nitrate/nitrite transporter NarK [Enhydrobacter aerosaccus] | 892 | 892 | 100% | 0 | 99% | WP_050324472.1 | |
Genes are also present in the DSC genomes for the F type ATPase, whose main role is to catalyze the synthesis of ATP using energy generated by cellular respiration (Yoshida et al., 2001; Table S2; DSC1, 4, 5, 7, 10 and 11). They are also found in many of the previously described Parcubacteria (Kantor et al., 2013; Brown et al., 2015). A V type ATPase is encoded within one genome belonging to OD1-DSC1 clade (Table S2; DSC6). In most cases these enzymes are thought to be used for ATP hydrolysis, but some microorganisms possess V type ATPases that are able to synthesize ATP (Toei et al, 2007). This type of respiratory potential has been previously reported for Candidatus Parcunitrobacter nitroensis (Castelle et al., 2017)
It is important to emphasize, taking all gene inventories described above into account, that only partial pathways of known respiration-linked systems exist within the DSC genomes. Nevertheless, members of the OD1-DSC1 clade (particularly DSC6) possess intriguing signs of respiration-related capacity, due to their high proportions of respiration genes in comparison to the majority of the publically available Parcubacteria genomes. In this regard they are similar to the Parcubacteria genomes C7867-007, C7867-008 and Candidatus Parcunitrobacter nitroensis (Nelson and Stegen, 2015; Castelle et al., 2017). If these gene systems in the DSC genomes are not used for energy-yielding respiration they may serve other functions involving electron/ion/solute transport or oxidative stress adaptation.
Several lines of evidence suggest that the genes connected to respiration in DSC6 are non-contaminants. For example, Enhydrobacter aerosaccus is not a common contaminant found in MDA amplified genomes, no other genes with homology to Enhydrobacter aerosaccus were found in DSC6 or in any of the other SAGs, and the lineage probability index (LPI) scores for the genes within the nitrate reductase contigs (0.487–0.558) are in the same range of scores from other non-Parcubacteria genes found through numerous contigs. Based on kmer frequency, scaffolds that encode for the nitrate reductases are closely clustered with the majority of scaffolds in the genome (Figure S6). The DSC6 V type ATPase has highest homology to that present in Clostridia-like organisms and its cytochrome C proteins have highest homology to those of Microgenomates-like organisms, perhaps reflecting additional sources of horizontal gene transfer or evolutionarily divergence from other Parcubacteria genomes.
Another important feature present in many of the DSC genomes concerns their biosynthetic capacity. All enzymes involved in the pentose phosphate pathways (PP) are represented in the OD1-L1 subgroup genomes, as is the case with up to 67% of the Parcubacteria genomes available in IMG (Table S2). The PP pathway generates NADPH and pentose, which are primarily utilized for anabolic purposes such as nucleotide synthesis. All of the DSC genomes share, along with 24% of other Parcubacteria, the presence of genes involved in the biosynthesis of purines and pyrimidines (Table S2). The coupled presence of PP and purine/pyrimidine biosynthetic pathways suggests that the DSC cells are capable of nucleotide biosynthesis, in contrast to the majority of Parcubacteria. It is interesting that DSC genomes have, among all 12 SAGs, 169 genes associated with amino acid metabolism, among those biosynthetic genes for glycine, serine, cysteine, lysine and glutamine (Table S2). Most other Parcubacteria genomes are missing the capability of synthesizing amino acids (Brown et al., 2015).
For metabolic comparisons at a whole genome level, the metabolic potential (all genes) of each DSC genome was compared against 232 Parcubacteria genomes that belong to one of the described phyla available in IMG (Figure S7, Table S3). The homology between the DSC SAGs and the genomes ascribed to the different Parcubacteria phyla varied. Metabolic potential from genomes in phyla Uhrbacteria and Falkowbacteria are more highly represented in the OD1-DSC1 clade. Clustering patterns based on metabolic relatedness do not exactly mirror the 16S rRNA gene phylogeny. For example, DSC3 and DSC4 are the most similar to each other, and to the Parcubacteria dataset from IMG, based on metabolic potential. DSC2 shares the least amount of metabolic potential with the Parcubacteria dataset, followed by the OD1-DSC1 clade. The metabolic processes shared most frequently (found to be present in 90% or more of the Parcubacteria genomes), were genes for ribosomal proteins, ATP-dependent Clp protease ATP-binding subunit ClpB, type II secretory pathway, type IV pilus assembly, cell division genes and peptidoglycan metabolism. These genes represent highly conserved metabolic processes that make up the core metabolic potential of the Parcubacteria. Some of the genes shared between less than 10% of all Parcubacteria genomes are abundant in these deep-sea SAGs. For example, the gene for DNA-3-methyladenine glycosylase I is shared by 8 of the DSC SAGs. This gene is involved in recognition and repair of alkylated DNA (Metz et al., 2007). The small-conductance mechanosensitive channel gene is another example, in this case shared across 6 of the SAGs. Small-conductance mechanosensitive channels are associated with protecting cells from increased turgor pressure (Lai et al., 2013), which we speculate could also facilitate cellular adaptation to elevated hydrostatic pressure (Bartlett, 2002). Other genes that are shared between five of the SAGs are predicted thiol-disulfide oxidoreductase YuxK, DCC family and NAD(P)H-dependent FMN reductase, involved in a variety of redox reactions involving extracellular enzyme modification (Ginalski et al., 2004, Ingelman et al., 1999).
These analyses also revealed a variety of genes uniquely present in the SAGs (Table 3). Over 88% of the unique genes present in each genome encode hypothetical proteins. Among the identifiable unique genes are a number of transporters and membrane proteins, suggesting that interactions with environmental factors may determine particular differences between the SAGs.
Table 3.
Description of unique genes found in SAGs
| SAG | % of unique hypoteticals | Product Name unique genes | BlastP |
|---|---|---|---|
|
| |||
| DSC1 | 90% | antitoxin ParD1/3/4 | hypothetical protein ETSY2_33695 [Candidatus Entotheonella sp. TSY2] |
| F-type H+-transporting ATPase subunit epsilon | hypothetical protein UV73_C0017G0003 [Microgenomates (Gottesmanbacteria) bacterium] | ||
| prevent-host-death family protein | hypothetical protein UW17_C0001G0027 [Candidatus Nomurabacteria bacterium GW2011_GWD1_44_10] | ||
| toxin ParE1/3/4 | Toxin ParE1 [Candidatus Accumulibacter sp. BA-93] | ||
| Putative DNA-binding domain-containing protein | DUF2063 domain-containing protein [Streptomyces roseoverticillatus] | ||
|
| |||
| DSC2 | 97% | Alkylated DNA repair dioxygenase AlkB | 2OG-Fe(II) oxygenase [Leptolyngbya sp. NIES-3755] |
| Helix-destabilising protein | helix destabilising protein [Enterobacteria phage M13] | ||
| phage/plasmid replication protein, gene II/X family | DNA replication protein [Psychromonas sp. PRT-SC03] | ||
|
| |||
| DSC3 | 100% | ||
|
| |||
| DSC4 | 93% | Glyoxalase/Bleomycin resistance protein/Dioxygenase superfamily protein | MULTISPECIES: hypothetical protein [Planktothrix] |
| indoleamine 2,3-dioxygenase | hypothetical protein [Streptomyces sp. MspMP-M5] | ||
| Lipoate-protein ligase B | octanoyltransferase [Alistipes sp. CAG:268] | ||
|
| |||
| DSC5 | 88% | Protein of unknown function (DUF3105) | hypothetical protein US72_C0008G0024 [Microgenomates group bacterium GW2011_GWC1_38_12] |
|
| |||
| DSC6 | 89% | anti-sigma B factor antagonist | anti-sigma factor antagonist [Rhodothermus marinus] |
| Putative vitamin uptake transporter | transporter [bacterium mt3] | ||
| Uncharacterized conserved protein | molybdenum metabolism regulator [Myxococcus hansupus] | ||
| YugN-like family protein | Uncharacterized conserved protein [Anoxybacillus flavithermus WK1] | ||
|
| |||
| DSC7 | 97% | archease family protein | hypothetical protein AMJ43_03100 [Coxiella sp. DG_40] |
| Protein of unknown function (DUF805) | membrane protein [Selenomonas sp. FOBRC6] | ||
|
| |||
| DSC8 | 100% | ||
|
| |||
| DSC9 | 95% | Helix-turn-helix | DNA-binding helix-turn-helix protein [Bacteroides sp. 2_2_4] |
| Xanthosine triphosphate pyrophosphatase | HAM1/NUDIX domain-containing protein [Microgenomates group bacterium GW2011_GWC1_38_12] | ||
|
| |||
| DSC10 | 92% | Acetyl/propionyl-CoA carboxylase, alpha subunit | hypothetical protein ACD_50C00024G0001 [uncultured bacterium] |
| Beta-glucosidase-related glycosidases | hypothetical protein [Bacillus bogoriensis] | ||
| Cupin domain protein | Cupin 2 conserved barrel domain protein [Candidatus Roizmanbacteria bacterium GW2011_GWC2_34_23] | ||
|
| |||
| DSC11 | 100% | ||
|
| |||
| DSC12 | 88% | Predicted DNA-binding protein, MmcQ/YjbR family | hypothetical protein AH06_02135 [candidate division TM6 bacterium Zodletone_IIa] |
| Protein of unknown function (DUF721) | hypothetical protein [Desulfuromonas acetoxidans] | ||
| Transcription factor WhiB | hypothetical protein [Rhodococcus sp. 114MFTsu3.1] | ||
Cell surface characteristics and Stress adaptation
Various SAGs encode the potential for polysaccharide modification, particularly that related to the synthesis of lipid A core found in lipopolysaccharides (Table S2). Among the genes present in the DSC cells are those for lipid A core-O-antigen ligase and related enzymes, 3-deoxy-manno-octulosonate cytidylyltransferase (CMP-KDO synthetase; EC:2.7.7.38; kdsB), 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase (KDO 8-P phosphatase; EC:3.1.3.45; kdsC), glycosyltransferases (pimB), as well as transport proteins for o-antigen and o-antigen ligase (Table S2). Out of 25 steps involved in the biosynthesis of the lipid A core, only the few mentioned above are present in the SAGs. Although, none of these genes are unique to the DSC SAGs, a few (e.g. 3-deoxy-manno-octulosonate cytidylyltransferase and 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase) are shared with 15% or less of all the available genomes in IMG and have not been previously documented. These results suggest that DSC OD1 cells have the potential to modify polysaccharides, perhaps facilitating membrane association.
Another feature not previously described within members of this CP, and that is shared between less than 10% of IMG available genomes, is the presence of genes associated with adaptation to oxidative stress. These features span all DSC OD1 genomes and include peroxiredoxin (DSC1, 3, 4, 6, 7, 10 and 11) and superoxide dismutase (DSC1, 2, 3, 4, 6, 7, 10 and 11) (Table S2). The presence of these enzymes implies some degree of tolerance to oxidizing conditions, perhaps making it possible for these cells to survive in the oxygen-exposed surficial sediment environment they were recovered from.
The DSC genomes encode both heat shock and cold shock proteins, which are also found in the majority of Parcubacteria genomes in IMG, among them the heat shock proteins involved in the sensing of misfolded proteins; DnaK, DnaJ and GrpE (Feder and Hofmann, 1999). Genes for cold shock proteins (CSP) appear to be less prevalent in previously sequenced Parcubacteria genomes (found in less than 50%), while CSP such as CspA, are found in the majority of DSC SAGs (DSC1, 2, 3, 4, 6, 7, 8, 10, 11 and 12; Table S2). CspA and its relatives are small proteins that bind to single-stranded polypyrimidine nucleic acids (Johnston et al., 2006) for the purpose of inhibiting potential deleterious mRNA secondary structures at low temperature (Jiang et al., 1997). This could be advantageous to the DSC cells growing at an in-situ temperature of ∼2–3°C.
Horizontally transferred genes
Each of the DSC OD1 genomes encodes some predicted proteins that have no apparent orthologs within the Parcubacteria, based on the LPI measurements for each gene using the DarkHorse program (Podell and Gaasterland, 2007). The LPI index evaluates the taxonomic similarity of BLAST matches over an entire genome, with lower values highlighting those genes matching only atypical lineages. In these cases the genes may have been horizontally transferred, or they may represent genes that are part of the non-core pan-genome of taxonomic groups that have only one or a few database representatives.
The relative abundance of these possible HTGs varies from 0.92% (DSC3) to 3.12% (DSC2) of predicted proteins in the DSC genomes (Table 4). The most frequently observed group of potential HTGs are archaeal in origin, mostly originating in the Methanomicrobia and Methanobacteria classes within the Euryarchaeota (Table S4a). Bacterial proteins in each DSC genome with highest homology to non-Parcubacteria genomes possess LPI scores higher than 0.4 but lower than 0.7, suggestive of HGT, but less definitive. Predicted proteins with eukaryotic taxonomic relationships were less abundant and mostly consisted of hypotheticals. Three DSC had closest matches to viral-like proteins (DSC2, 11 and 12; Table S4b). It is unlikely that these low LPI-scoring genes are artifacts associated with contaminating DNA that arose during single-cell isolation and DNA amplification, because most are located on contigs with high LPI scores in adjacent genes. HGT may provide varied metabolic potential to these deep-sea bacteria. It is also notable that over 51% of proposed archaeal HTGs are found in multiples per scaffold (two or more genes). GC% content of such scaffolds and individual genes also varied from the average genome GC content (Table S4a and b). The GC sigma variation from the mean GC supports the possibility of horizontal transfer for many of the suggested HTGs (Table S4a and b).
Table 4.
Numbers of horizontally transferred gene per SAG and LPI score range
| Genomes | Virus (#per genome) |
Eukarya (#per genome) |
Archaea (#per genome) |
Total | % of total HTG |
LPI scores |
|---|---|---|---|---|---|---|
|
| ||||||
| DSC1 | 0 | 2 | 6 | 8 | 1.12 | 0.010–0.189 |
| DSC2 | 6 | 1 | 3 | 10 | 3.12 | 0.020–0.076 |
| DSC3 | 0 | 1 | 3 | 4 | 0.92 | 0.010–0.091 |
| DSC4 | 0 | 1 | 7 | 8 | 1.41 | 0.013–0.071 |
| DSC5 | 0 | 1 | 9 | 10 | 3.05 | 0.029–0.076 |
| DSC6 | 0 | 2 | 10 | 12 | 1.41 | 0.005–0.059 |
| DSC7 | 0 | 0 | 11 | 11 | 1.75 | 0.014–0.025 |
| DSC8 | 0 | 0 | 3 | 3 | 0.94 | 0.012–0.018 |
| DSC9 | 0 | 0 | 6 | 6 | 1.48 | 0.016–0.031 |
| DSC10 | 0 | 0 | 7 | 7 | 1.43 | 0.015–0.042 |
| DSC11 | 1 | 0 | 5 | 6 | 1.21 | 0.008–0.019 |
| DSC12 | 1 | 2 | 5 | 8 | 1.61 | 0.005–0.284 |
Summary
Parcubacteria reside in diverse habitats, including deep-sea sediments. Here we report on Parcubacteria within the microbial population present in the surficial sediments of the Challenger Deep, Mariana Trench. The genome analyses presented here reinforce the view of the Parcubacteria as organisms with small genomes that are able to metabolize organics by fermentation. While considered to have a small genomic size, they also appear capable of expanding their genome. The significance of the newly identified features to the lifestyle and environmental contributions of this enigmatic but abundant super phylum awaits further investigation.
Experimental Procedures
Collection and sorting
Sediments were collected from a depth of 10,908 m and an in situ temperature of 2.5°C using a push-core apparatus controlled by a hydraulic arm attached to the manned submersible Deepsea Challenger. Sampling occurred on March 26, 2012 in the “East Deep” (Fujioka et al., 2002) of the Challenger Deep at 142.59° E, 11.37° N during the Deepsea Challenge Expedition. Recovered sediment was placed in glycerol/TE buffer (Rinke et al., 2014) and first stored in liquid nitrogen and later at −80°C prior to single cell sorting at the J. Craig Venter Institute (JCVI). The sediment sample was gently vortexed and allowed to settle briefly before filtering through a 35µm mesh (BD Biosciences, San Jose, CA, USA) to avoid larger sediment particles. Cells were stained with SYBR Green I nucleic acid stain (Invitrogen, Carlsbad, CA, USA) and sorted using a cooled FACS-Aria II flow cytometer (BD Biosciences, San Jose, CA) (McLean et al. 2013). Microtiter plates of sorted single cells were stored at −80°C.
Genome amplification and sequencing
DNA was amplified using a custom BioCel robotic system (Agilent Technologies, Santa Clara, CA) as described by McLean et al. (2013). Genomic material in the sorted microbial cells was amplified by multiple displacement amplification (MDA) in a 384-well format using GenomiPhi (GE Healthcare, Waukesha, WI, USA). 16S rRNA genes were PCR amplified from diluted MDA products using universal bacterial primers 27F and 1492R (Weisburg et al., 1991) as follows: 94 °C for 3 min, 35 cycles of 94 °C for 30 s, 55 °C for 30 s, 72 °C for 90 s, and 72 °C for 10 min. PCR products were cleaned with exonuclease I and shrimp alkaline phosphatase (Thermo Fisher Scientific Inc., Waltham, MA, USA) and sent for Sanger sequencing at the Joint Technology Center (JTC, J. Craig Venter Institute, Rockville, MD, USA). 16S rRNA gene trace files were analyzed with the CLC Workbench software program (CLC Bio, Cambridge, MA, USA). Gene sequences were evaluated for evidence of microbial DNA contamination associated with MDA reagents, based on a in-house JCVI database of common contaminants, including sequences related to the genera Escherichia, Propionibacterium, Shewanella and Pseudomonas (Table S5). Any single cells judged to be contaminated were removed from consideration for whole genome sequencing. These curated sequences were then compared to the NCBI nr/nt database using BLASTN (Altschul et al., 1990) for initial taxonomic screening of the sorted cells. DNA from thirteen separate Parcubacteria-classified cells was sent for Illumina HiSeq 2500 (101bp reads) sequencing at the JTC. Raw data is available from through the project number PRJEB10905.
Assembly, annotation and genome completion
Genomes were quality trimmed using Nesoni with mostly default parameter and q20 for quality score (www.vicbioinformatics.com/software.nesoni.shtml). Sequences were assembled using SPAdes 3 with flags for single cell genome assembly (--sc) and to reduce mismatches, short indels and error corrections (--careful) (Bankevich et al, 2012) and annotated by IMG-ER (https://img.jgi.doe.gov/cgi-bin/er/main.cgi, Markowitz et al, 2014). Annotated genomes are accessible through IMG (Genome IDs: 2547132480, 2547132497-98, 2547132500-1, 2547132503-4, 2548877123, 2548877145-6, 25488771458-9). Only contigs over 1000 basepairs are reported here. All metabolic comparisons were generated using the genomes that were assembled with SPAdes 3 and annotated via IMG. Later, genome DSC6 was re-assembled using SPAdes 3.6 and annotated using PROKKA (Seemann T, 2014). This assembly was performed as an attempt to utilize the latest assembly algorithm to potentially generate a better assembly; this data is only presented in Table 2 and Figure 3. Estimated genome size and completeness were calculated using CheckM (Parks et al., 2015). Annotations for genes of interest that are discussed in detail throughout the article were manually verified via BLASTP. Functional comparisons were performed using the IMG-ER platform (Markowitz et al., 2014). Protein sequences for DSC6 nitrate reductase gene, narG, were further compared to other nitrate reductase protein sequences in order to assess their potential role in assimilation or dissimilation of nitrate (Figure S2). Representative sequences from the NCBI database were aligned using muscle (Edgar 2004) and an unrooted maximum likelihood tree was generated with FastTree (Price et al., 2009).
Taxonomy assignments
16S rRNA gene sequences recovered from each SAG were analyzed by BLASTN against the NCBI nr/nt database (Altschul et al., 1990). Sequences with 85% or greater similarity to Parcubacteria superphylum 16S rRNA genes were extracted and used for phylogenetic reconstruction, along with reference sequences belonging to the Parcubacteria superphylum from previous publications (Wrighton et al., 2012; Kantor et al., 2013; Rinke et al., 2013, Brown et al., 2015). Sequences were aligned with the SINA aligner (http://www.arb-silva.de/aligner/; Pruesse et al, 2012) and a maximum-likelihood tree was created using FastTree (Price et al., 2009).
Genome-encoded protein predictions of DNA fragments of >1000bp were obtained from IMG-ER and taxonomically classified using DarkHorse software, version 2.0 (http://darkhorse.ucsd.edu/, Podell and Gaasterland, 2007), with default settings. Annotated proteins from each of the genomes were compared against the NCBI nr protein database (December 2016) with Diamond (Buchfink et al., 2015). This data was used as input to the DarkHorse program, which identifies closest database relatives for individual proteins and constructs an overall statistical profile for the query genome. This profile is used to calculate a lineage probability index (LPI) score for each individual protein, indicating whether its taxonomic match is typical or atypical for the genome being investigated.
Darkhorse results were used initially to identify and exclude potential contaminating sequences among SAG contigs (Jones et al., 2011). Contamination was assessed based on whether or not taxonomic lineages associated with predicted proteins on each assembled contig were similar to or different from the JCVI in house database of common MDA and laboratory potential contaminants, as described above. Contamination levels were also verified by CheckM (Parks et al., 2015).
DarkHorse LPI scores were also used to discover patterns of taxonomic relatedness between the SAGS, and as well as to identify potential horizontally transferred genes. The DarkHorse software uses a unique, taxonomically weighted algorithm that explicitly compensates for potential database bias, and adjusts the stringency of match criteria for each individual input sequence based on relative variability of potential database orthologs as a proxy for evolutionary conservation rates (Podell et al 2007). The relative abundance of proteins predicted to be most similar to Parcubacteria phyla in the NCBI nr database were tallied for each SAG, and subjected to Non-Metric Multidimensional scaling (nMDS) analysis using the vegan R package (Oksanen et al., 2013) and plotted in R. Potential horizontally transferred genes were further evaluated based on nucleotide composition (percent GC) of individual coding sequences versus genomic averages using z-scores, as well as scaffold locations relative to other HGT candidates.
Metabolic Similarity Assessments
The whole genome metabolic potential of all SAGs was compared against 232 Parcubacteria genomes present within IMG (database accessed March 2016). Bray-Curtis similarity analysis was calculated with the vegan R package. A heatmap was generated using the R package ggplot2 to display the homology of each DSC genome against the other IMG Parcubacteria (Wickham H, 2009).
Supplementary Material
Originality and significance statement.
The work highlighted here presents an unprecedented opportunity to explore the microbial community of the deepest ocean region. Single cell genome technology permitted the use of a limited and precious sediment sample to describe the phylogeny and metabolic potential of cells from the candidate superphylum Parcubacteria. Genomic comparisons were generated to better understand how these organisms compare to previously described Parcubacteria in order to assess their connection to the deep-sea and develop hypotheses about potential adaptations.
Acknowledgments
We are grateful for the financial support provided by the National Science Foundation (0801973, 0827051, 1536776), a National Science Foundation Graduate Research Fellowship (068775), the National Aeronautics and Space Administration (NNX11AG10G), a National Institutes of Health Marine Biotechnology Training grant (T32GM067550) and a gift from Earthship Productions.
Footnotes
The authors of this publication declare no conflict of interest in relation to this article.
References
- Agusti S, González-Gordillo JI, Vaqué D, Estrada M, Cerezo MI, Salazar G, Gasol JM, Duarte CM. Ubiquitous healthy diatoms in the deep sea confirm deep carbon injection by the biological pump. Nature communications. 2015;6:7608. doi: 10.1038/ncomms8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich A, Dvorkin M, Kulikov A, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. Journal of Computational Biology. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett DH. Pressure effects on in vivo microbial processes. Biochimica et Biophysica Acta (BBA)-Protein Structure and Molecular Enzymology. 2002;1595(1):367–381. doi: 10.1016/s0167-4838(01)00357-0. [DOI] [PubMed] [Google Scholar]
- Brown CT, Hug LA, Thomas BC, Sharon I, Castelle CJ, Singh A, Wilkins MJ, Wrighton KC, Williams KH, Banfield JF. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature. 2015;523(7559):208–211. doi: 10.1038/nature14486. [DOI] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nature methods. 2015;12(1):59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- Castelle CJ, Brown CT, Thomas BC, Williams KH, Banfield JF. Unusual respiratory capacity and nitrogen metabolism in a Parcubacterium (OD1) of the Candidate Phyla Radiation. Scientific Reports. 2017;7:40101. doi: 10.1038/srep40101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshahed MS, Najar FZ, Aycock M, Qu C, Roe BA, Krumholz LR. Metagenomic Analysis of the Microbial Community at Zodletone Spring (Oklahoma): Insights into the Genome of a Member of the Novel Candidate Division OD1. Applied and Environmental Microbiology. 2005;71:7598–7602. doi: 10.1128/AEM.71.11.7598-7602.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: Evolutionary and Ecological Physiology. Annual Review of Physiology. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- Fujioka K, Okino K, Kanamatsu T, Ohara Y. Morphology and origin of the Challenger Deep in the Southern Mariana Trench. Geophysical Research Letters. 2002;29 10-11-10-14. [Google Scholar]
- Gihring TM, Zhang G, Brandt CC, Brooks SC, Campbell JH, Carroll S, Criddle Craig S, Green SJ, Jardine P, Kostka JE, Lowe K, Mehlhorn TL, Overholt W, Watson DB, Yang Z, Wu W, Schadt CW. A Limited Microbial Consortium Is Responsible for Extended Bioreduction of Uranium in a Contaminated Aquifer. Applied and Environmental Microbiology. 2011;77:5955–5965. doi: 10.1128/AEM.00220-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginalski K, Kinch L, Rychlewski L, Grishin NV. DCC proteins: a novel family of thiol-disulfide oxidoreductases. Trends in biochemical sciences. 2004;29(7):339–42. doi: 10.1016/j.tibs.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Glud RN, Wenzhofer F, Middelboe M, Oguri K, Turnewitsch R, Canfield DE, Kitazato H. High rates of microbial carbon turnover in sediments in the deepest oceanic trench on Earth. Nature Geosci. 2013;6:284–288. [Google Scholar]
- Gong J, Qing Y, Guo X, Warren A. “Candidatus Sonnebornia yantaiensis”, a member of candidate division OD1, as intracellular bacteria of the ciliated protist Paramecium bursaria (Ciliophora, Oligohymenophorea) Systematic and applied microbiology. 2014;37(1):35–41. doi: 10.1016/j.syapm.2013.08.007. [DOI] [PubMed] [Google Scholar]
- Harris JK, Kelley ST, Pace NR. New Perspective on Uncultured Bacterial Phylogenetic Division OP11. Applied and Environmental Microbiology. 2004;70:845–849. doi: 10.1128/AEM.70.2.845-849.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori S, Tsuchiya M, Nishi S, Arai W, Yoshida T, Takami H. Active Bacterial Flora Surrounding Foraminifera (Xenophyophorea) Living on the Deep-Sea Floor. Bioscience, biotechnology, and biochemistry. 2013;77(2):381–384. doi: 10.1271/bbb.120663. [DOI] [PubMed] [Google Scholar]
- Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, Butterfield CN, Hernsdorf AW, Amano Y, Ise K, Suzuki Y. A new view of the tree of life. Nature Microbiology. 2016;1:16048. doi: 10.1038/nmicrobiol.2016.48. [DOI] [PubMed] [Google Scholar]
- Ingelman M, Ramaswamy S, Nivière V, Fontecave M, Eklund H. Crystal structure of NAD (P) H: flavin oxidoreductase from Escherichia coli Biochemistry. 1999;38(22):7040–9. doi: 10.1021/bi982849m. [DOI] [PubMed] [Google Scholar]
- Irgens RL, Suzuki I, Staley JT. Gas vacuolate bacteria obtained from marine waters of Antarctica. Current Microbiology. 1989;18(4):261–5. [Google Scholar]
- Jiang W, Hou Y, Inouye M. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 1997;272:196–202. doi: 10.1074/jbc.272.1.196. [DOI] [PubMed] [Google Scholar]
- Johnston D, Tavano C, Wickner S, Trun N. Specificity of DNA binding and dimerization by CspE from Escherichia coli. J. Biol Chem. 2006;281:40208–40215. doi: 10.1074/jbc.M606414200. [DOI] [PubMed] [Google Scholar]
- Jones AC, Monroe EA, Podell S, Hess WR, Klages S, Esquenazi E, Niessen S, Hoover H, Rothmann M, Lasken RS, Yates JR, Reinhardt R, Kube M, Burkart MD, Allen EE, Dorrestein PC, Gerwick WH, Gerwick L. Genomic insights into the physiology and ecology of the marine filamentous cyanobacterium Lyngbya majuscula. Proceedings of the National Academy of Sciences. 2011;108:8815–8820. doi: 10.1073/pnas.1101137108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantor RS, Wrighton KC, Handley KM, Sharon I, Hug LA, Castelle CJ, Thomas, Brian C, Banfield JF. Small Genomes and Sparse Metabolisms of Sediment-Associated Bacteria from Four Candidate Phyla. mBio. 2013;4(5):e00708–13. doi: 10.1128/mBio.00708-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato C, Li L, Nogi Y, Nakamura Y, Tamaoka J, Horikoshi K. Extremely barophilic bacteria isolated from the Mariana Trench, Challenger Deep, at a depth of 11,000 meters. Applied and environmental microbiology. 1998;64(4):1510–1513. doi: 10.1128/aem.64.4.1510-1513.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandeparker L, Anil AC. Association of bacteria with marine invertebrates: Implications for ballast water management. EcoHealth. 2013;10(3):268–276. doi: 10.1007/s10393-013-0857-z. [DOI] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(7):2567–72. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis KT, Braff J, Karl DM, DeLong EF. Comparative metagenomic analysis of a microbial community residing at a depth of 4,000 meters at station ALOHA in the North Pacific subtropical gyre. Appl Environ Microbiol. 2009;75:5345–5355. doi: 10.1128/AEM.00473-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai JY, Poon YS, Kaiser JT, Rees DC. Open and shut: crystal structures of the dodecylmaltoside solubilized mechanosensitive channel of small conductance from Escherichia coli and Helicobacter pylori at 4.4 Å and 4.1 Å resolutions. Protein Science. 2013;22(4):502–9. doi: 10.1002/pro.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauro FM, Bartlett DH. Prokaryotic lifestyles in deep sea habitats. Extremophiles. 2008;12:15–25. doi: 10.1007/s00792-006-0059-5. [DOI] [PubMed] [Google Scholar]
- Li L, Kato C, Horikoshi K. Bacterial diversity in deep-sea sediments from different depths. Biodiversity & Conservation. 1999;8(5):659–677. [Google Scholar]
- Luef B, Frischkorn KR, Wrighton KC, Holman HY, Birarda G, Thomas BC, Singh A, Williams KH, Siegerist CE, Tringe SG, Downing KH. Diverse uncultivated ultra-small bacterial cells in groundwater. Nature communications. 2015;6:6372. doi: 10.1038/ncomms7372. [DOI] [PubMed] [Google Scholar]
- Markowitz V, Chen I, Palaniappan K, Chu K, Szeto E, Pillay M, Ratner A, Huang JH, Woyke T, Huntemann M, Anderson I, Billis K, Varghese N, Mavromatis K, Pati A, Ivanova NN, Kyrpides NC. IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Research. 2014;42:D560–D567. doi: 10.1093/nar/gkt963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean JS, Lombardo M-J, Badger JH, Edlund A, Novotny M, Yee-Greenbaum J, Vyahhi N, Hall AP, Yang Y, Dupont CL, Ziegler MG, Chitsaz H, Allen AE, Yooseph S, Tesler G, Pevzner PA, Friedman RM, Nealson KH, Venter JC, Lasken RS. Candidate phylum TM6 genome recovered from a hospital sink biofilm provides genomic insights into this uncultivated phylum. Proceedings of the National Academy of Sciences. 2013;110:E2390–E2399. doi: 10.1073/pnas.1219809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz AH, Hollis T, Eichman BF. DNA damage recognition and repair by 3 methyladenine DNA glycosylase I (TAG) The EMBO journal. 2007;26(9):2411–20. doi: 10.1038/sj.emboj.7601649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi M, Hashimoto J. A precise bathymetric map of the world’s deepest seafloor, Challenger Deep in the Mariana Trench. Marine Geophysical Research. 2011;32:455–463. [Google Scholar]
- Nelson WC, Stegen JC. The reduced genomes of Parcubacteria (OD1) contain signatures of a symbiotic lifestyle. Frontiers in microbiology. 2015;6:713. doi: 10.3389/fmicb.2015.00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunoura T, Takaki Y, Kazama H, Hirai M, Ashi J, Imachi H, Takai K. Microbial Diversity in Deep-sea Methane Seep Sediments Presented by SSU rRNA Gene Tag Sequencing. Microbes, Environments. 2012;27:382–390. doi: 10.1264/jsme2.ME12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MH, Wagner H, Oksanen MJ, Package ‘vegan’ Community ecology package, version. 2013;2(9) [Google Scholar]
- Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathom-Aree W, Stach JE, Ward AC, Horikoshi K, Bull AT, Goodfellow M. Diversity of actinomycetes isolated from Challenger Deep sediment (10,898 m) from the Mariana Trench. Extremophiles. 2006;10(3):181–189. doi: 10.1007/s00792-005-0482-z. [DOI] [PubMed] [Google Scholar]
- Peura S, Eiler A, Bertilsson S, Nykanen H, Tiirola M, Jones RI. Distinct and diverse anaerobic bacterial communities in boreal lakes dominated by candidate division OD1. ISME Journal. 2012;6:1640–1652. doi: 10.1038/ismej.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podell S, Gaasterland T. DarkHorse: a method for genome-wide prediction of horizontal gene transfer. Genome Biology. 2007;8:R16. doi: 10.1186/gb-2007-8-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenakou PN, Lampadariou N, Mandalakis M, Tselepides A. Phylogenetic diversity of sediment bacteria from the southern Cretan margin, Eastern Mediterranean Sea. Systematic and Applied Microbiology. 2009;32(1):17–26. doi: 10.1016/j.syapm.2008.09.006. [DOI] [PubMed] [Google Scholar]
- Price M, Dehal P, Arkin A. FastTree: Computing Large Minimum Evolution Trees with Profiles instead of a Distance Matrix. Molecular Biology and Evolution. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Peplies J, Glockner F. SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinke C, Schwientek P, Sczyrba A, Ivanova N, Anderson I, Cheng J, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu WT, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- Rinke C, Lee J, Nath N, Goudeau D, Thompson B, Poulton N, Dmitrieff E, Malmstrom R, Stepanauskas R, Woyke T. Obtaining genomes from uncultivated environmental microorganisms using FACS-based single-cell genomics. Nature protocols. 2014;9:1038–1048. doi: 10.1038/nprot.2014.067. [DOI] [PubMed] [Google Scholar]
- Schauer R, Bienhold C, Ramette A, Harder J. Bacterial diversity and biogeography in deep-sea surface sediments of the South Atlantic Ocean. ISME J. 2010;4:159–170. doi: 10.1038/ismej.2009.106. [DOI] [PubMed] [Google Scholar]
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu153. btu153. [DOI] [PubMed] [Google Scholar]
- Staley JT, Irgens RL, Brenner DJ. Enhydrobacter aerosaccus gen. nov., sp. nov., a gas-vacuolated, facultatively anaerobic, heterotrophic rod. International Journal of Systematic and Evolutionary Microbiology. 1987;37(3):289–91. [Google Scholar]
- Tao L, Peng W, Pinxian W. Microbial diversity in surface sediments of the Xisha Trough, the South China Sea. Acta Ecologica Sinica. 2008;28(3):1166–1173. [Google Scholar]
- Tarn J, Peoples LM, Hardy K, Cameron J. Bartlett D.H. Identification of Free-Living and Particle-Associated Microbial Communities Present in Hadal Regions of the Mariana Trench. Frontiers in Microbiology. 2016;7:665. doi: 10.3389/fmicb.2016.00665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrash JC, Temperton B, Swan BK, Landry ZC, Woyke T, DeLong EF, Stepanauskas R, Giovannoni ST. Single-cell enabled comparative genomics of a deep ocean SAR11 bathytype. ISME J. 2014;8:1440–1451. doi: 10.1038/ismej.2013.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toei M, Gerle C, Nakano M, Tani K, Gyobu N, Tamakoshi M, Sone N, Yoshida M, Fujiyoshi Y, Mitsuoka K, Yokoyama K. Dodecamer rotor ring defines H+/ATP ratio for ATP synthesis of prokaryotic V-ATPase from Thermus thermophilus. Proceedings of the National Academy of Sciences. 2007;104:20256–20261. doi: 10.1073/pnas.0706914105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisburg W, Barns S, Pelletier D, Lane D. 16S ribosomal DNA amplification for phylogenetic study. Journal of Bacteriology. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman W, Coleman D, Wiebe W. Prokaryotes: The unseen majority. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. ggplot2: elegant graphics for data analysis. Springer; New York: 2009. [Google Scholar]
- Wrighton K, Castelle C, Wilkins M, Hug L, Sharon I, Thomas B, Handley KM, Mullin SW, Nicora CD, Singh A, Lipton MS, Long PE, Williams KH, Banfield JF. Metabolic interdependencies between phylogenetically novel fermenters and respiratory organisms in an unconfined aquifer. ISME Journal. 2014;8:1452–1463. doi: 10.1038/ismej.2013.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrighton KC, Thomas BC, Sharon I, Miller CS, Castelle CJ, VerBerkmoes NC, Wilkins MJ, Hettich RL, Lipton MS, Williams KH, Long PE, Banfield JF. Fermentation, Hydrogen, and Sulfur Metabolism in Multiple Uncultivated Bacterial Phyla. Science. 2012;337:1661–1665. doi: 10.1126/science.1224041. [DOI] [PubMed] [Google Scholar]
- Yoshida M, Muneyuki E, Hisabori T. ATP synthase—a marvellous rotary engine of the cell. Nature Reviews Molecular Cell Biology. 2001;2:669–677. doi: 10.1038/35089509. [DOI] [PubMed] [Google Scholar]
- Yoshida M, Takaki Y, Eitoku M, Nunoura T, Takai K. Metagenomic analysis of viral communities in (hado) pelagic sediments. PLoS One. 2013;8(2):e57271. doi: 10.1371/journal.pone.0057271. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.