Abstract
Purpose
A goal of this study was to identify and investigate previously unrecognized components of the remodeling process in the progression to heart failure by comparing protein expression in ischemic failing (F) and nonfailing (NF) human hearts.
Experimental design
Protein expression differences were investigated using multidimensional protein identification and validated by Western analysis. This approach detected basal lamina (BL) remodeling, and further studies analyzed samples for evidence of structural BL remodeling. A rat model of pressure overload (PO) was studied to determine whether nonischemic stressors also produce BL remodeling and impact cellular adhesion.
Results
Differential protein expression of collagen IV, laminin α2, and nidogen-1 indicated BL remodeling develops in F versus NF hearts Periodic disruption of cardiac myocyte BL accompanied this process in F, but not NF heart. The rat PO myocardium also developed BL remodeling and compromised myocyte adhesion compared to sham controls.
Conclusions and clinical relevance
Differential protein expression and evidence of structural and functional BL alterations develop during heart failure. The compromised adhesion associated with this remodeling indicates a high potential for dysfunctional cellular integrity and tethering in failing myocytes. Therapeutically targeting BL remodeling could slow or prevent the progression of heart disease.
Keywords: Basal lamina, Heart failure, Myocardium, Proteins, Remodeling
1 Introduction
Myocardial infarction increases the risk of developing end-stage heart failure [1] and continues to be a major contributor to morbidity and mortality in the United States [2]. Effective treatment modalities for treating heart failure are limited [2], although significant mechanistic insight into the role(s) played by individual genes and proteins in the progression of heart failure continue to emerge from research on genetic mouse models [3–5]. Animal studies together with epidemiological data also indicate multiple approaches ranging from systems based to molecular biology further improve our understanding of this complex disease process.
Several systems-based approaches have led to insights into heart disease including, genome-wide association studies, microarray-based gene expression, single nucleotide variations, and epigenetic analysis of human cardiac tissues [3,6,7]. These approaches revealed links between specific genes or genetic polymorphisms and heart disease, as well as key differences between nonfailing (NF) and end-stage failing (F) hearts [8, 9]. Technical and bioinformatic improvements in high-throughput approaches continue to accelerate the pace of gene identification and discovery [10]. There is also evidence these methods can be adapted to evaluate the efficacy of current therapies, such as ventricular assist devices [11, 12]. Proteomic analysis is often a desired high throughput approach when the need arises to identify a group of proteins within a complex or pathway to understand their collective impact [13]. For example, protein stoichiometry within the cardiac sarcomere is tightly regulated regardless of the gene expression profile [14], but any changes in this stoichiometry can significantly disrupt myofilament function. In addition, proteomic analysis can provide insights into protein network modifications and alterations in PTMs during end-stage heart failure.
Both up- and downregulation of proteins are reported in diseased hearts analyzed by 2DE or gel-free separation of partially purified subfractions from F and NF human heart homogenates [15–18]. High throughput, gel-free approaches utilizing LC-MS and/or protein microarray are available for studying human tissue [19, 20], but few studies have utilized this MS-based approach to analyze ischemic human heart failure. The present study utilizes multidimensional protein identification technology (MudPIT) to compare protein expression in ischemic F versus NF human heart tissue. In this analysis, repetitive (ten times) LC/MS/MS analysis of protein lysates from F and NF human hearts were analyzed for differential expression of proteins known to change during heart failure and to identify lesser or unknown networks of proteins. A select group of biologically and statistically significant protein expression differences were then chosen for further validation by Western blot analysis of human tissue explants from multiple patients, and compared to gene expression microarray analysis of F and NF human tissue.
An important outcome from this analysis was the identification of significant remodeling within the basal lamina (BL) of ischemic heart failure tissue. Architectural remodeling coincides with the changes in protein expression, and was also detected in an animal model of heart failure along with evidence of functional deficits in myocyte adhesion. Taken together, the current proteomic analysis revealed alterations in protein expression, which are shown to coincide with previously unrecognized structural and functional deficits in F myocardium.
2 Materials and methods
2.1 Tissue collection and sample processing
Cardiac tissue collected from F and NF hearts was approved by the Internal Review Board at the University of Michigan, and the consent and protocol for collecting nonmatched, NF hearts is approved by Gift of Life—Michigan. Samples were analyzed from patients diagnosed with ischemic cardiomyopathy with a left ventricular (LV) ejection fraction (EF) ≤ 25% and no record of assist device support. NF hearts were structurally normal and an EF >50%, and were explanted at the time other organs were procured. Both F and NF hearts were flushed with ice-cold cardioplegia prior to explant, and small samples were rapidly flash frozen in liquid N2 and stored at −80°C immediately after explant [21]. Relevant quantitative echocardiography findings, age, diagnosis, duration of heart failure (in years), and medication were recorded at the time of tissue collection and prior to sample deidentification. Tissue collected from five NF donor hearts and ten F hearts was analyzed in the present studies (Supporting Information Table 1).
Within this tissue set, proteomic analysis was carried out on explants from the LV anterior wall remote from the is-chemic scar of a 59-year-old patient diagnosed with ischemic cardiomyopathy 5.9 years prior to transplantation (EF<20%; max. LV wall thickness = 7 mm). Medication prior to transplant included an angiotensin converting enzyme inhibitor, diuretic, aldosterone inhibitor, and beta blocker therapy to provide inotropic support as well as nitrates and amioderone for arrhythmia. A similar sample from a 67-year-old non-matched NF donor (EF 60%) was used for the MudPIT comparison. The NF patient had a history of pulmonary hypertension and aortic/mitral valve regurgitation prior to an ischemic cerebral hemorrhage.
2.2 Preparation of samples for peptide separation
Frozen tissue samples were lyophilized and ground to a powder, and then stored at −20°C prior LC/MS/MS analysis. Proteins were extracted from each sample in HEPES- EGTA buffer (2 mM HEPES, 2.5 mM EGTA; pH 7.4) supplemented with protease inhibitor cocktail (Roche) and PhosStop phosphatase inhibitor. Due to their high content, homogenates were depleted of myofilaments, by brief sonication on ice, vortexing, and then centrifugation at 40 000 x g for 1 h at 4°C, as described earlier [22]. Supernatants were stored on ice and the remaining pellet was reextracted twice in HEPES-EGTA buffer. Proteins were precipitated from pooled supernatants in ten volumes ice cold acetone overnight at −20°C. Protein precipitates were pelleted prior to their removal under a gentle stream of helium gas. Each protein pellet was resuspended in 10 mM DTT for 1 h at 60°C, cooled to room temperature, incubated with 5 mM iodoacetamide in the dark for 30 min, and then treated overnight at 37°C with L-(tosylamido-2-phenyl) ethyl chloromethyl ketone-modified sequencing grade porcine trypsin (1:50) (Promega). Digestion and reduction were terminated with 1 µL TFA, and peptides were then concentrated and desalted on a trap column (Michrom BioResources). Prior to peptide loading, trap columns were washed with 5–10 volumes of solvent B (90% ACN: 10% dH2O: 0.1% TFA), and equilibrated with 5–10 volumes of solvent A (2/98/0.1%, ACN/H2O/TFA). After adding a peptide sample, the trap column was washed with five volumes of solvent A, and peptides were eluted with 1–2 volumes of 65–90% solvent B. Samples were then stored at −20°C for LC/MS/MS.
2.3 Analysis of complex mixture of protein digests by MudPIT
Peptides from the protein digest were analyzed by the MudPIT approach adopted from Kline and colleagues [23, 24]. Briefly, peptides were identified with a nano-LC/MS/MS system consisting of an ultra-performance liquid chromatography (Waters; USA) and a hybrid linear quadrapole ion trap—Fourier transform ICR (LTQ-FT) MS (ThermoFinnigan; USA) equipped with a nano-ESI source. Peptide solution aliquots (10 µL) were loaded onto a modified vented column consisting of strong cation exchange (SCX; 5 µm, 3 cm) followed by a C18 trap (Aqua; particle size 5 µm, 1.5 cm; [23]). Peptides displaced from the SCX phase by a salt gradient were introduced into the C18 phase column through an auto-sampler loop and then desalted for 10 min at a flow rate of 5 µL/min. Trapped peptides were separated on a custom-made 75 µm silica capillary column (6 µm id) packed with C18 (3 µm, 12 cm) in an ACN gradient. A nine-step salt gradient was performed using 2 µL of 0–500 mM ammonium acetate with 0.1% formic acid in H2O and then a 3-step gradient of 2–5 µL of 500 mM ammonium acetate with 0.1% formic acid in 30% ACN. Mobile phases A and B contained 0.1% formic acid in 0 and 100% ACN, with an initial gradient of 5% B and then a ramp up to 15% B over 10 min, 55% B over the next 60 min, then 95% B over 20 min, and maintenance at 95% B for 5 min [25]. Electrospray voltage was set at 2.1 kV. One high mass resolution (100 000) MS spectra was acquired by the LTQ-FT analyzer followed by five data-dependent MS/MS scans with the linear ion trap analyzer in each mass analysis duty circle. A normalized collision energy of 35% was used throughout the CID phase. Ten replicate analyses/sample were obtained to optimize reproducibility and protein expression coverage [23].
2.4 Proteomic data analysis
MS/MS spectra were searched with the in-house software ProLucid [26] against the IPI Human protein database (released May, 2009) concatenated to a decoy database in which each entry sequence in the original database was reversed, and then the protein identifier cross-reference conversion tool was used to convert to UniProt accession numbers (2014_05 release; [27]). ProLuCID was used with monoisotopic mass selected, a precursor mass error of 50 ppm, and a fragment ion mass error of 1 Da. Full and half tryptic peptides were selected with one potential missed cleavage for each of the ten analyses performed per sample. The variable modifications chosen were oxidized methionine and carbamidomethylated cysteine. Maximum Sp rank was set as 1000, minimum Sp score set to −1, and DTASelect 2.0 was used to assemble and filter the resulting spectral matches with a peptide false positive rate of 1% and X corr: +1 ion >1.5, +2 ion >2.5, +3 ion >3.5. All peptides that mapped to a single UniProt accession number were counted as a single protein. Frequently, it was found that multiple, different, peptides mapped to the same UniProt accession number. These peptides originated from different regions of the identified protein and, as such, were all counted as a single protein. Protein information such as isoelectric point and molecular weight associated with protein identification were further utilized for initial validation. If this information was not based on the MudPIT method, then this identification was not selected for the protein. An average of 59 981 and 59 524 protein entries was searched during each replicate in the ten analyses of NF and F samples, respectively. Selected protein isoforms were further confirmed by Western blot analysis.
Spectral counts for listed proteins were uploaded to compare differences in expression levels between F and NF proteins. A unique protein identified in ≥ three runs for the same sample was considered significantly different, while the criteria for differential, highly expressed (HiE) proteins was a ≥twofold expression difference between F and NF samples. Ingenuity pathway analysis software (IPA) and Babelomics Fatigo (4.2; [28]) were used to determine pathway/molecular function differences. Subcellular protein distribution in F and NF samples was analyzed with a χ2 test (p < 0.05), and a right-tailed Fisher’s exact test (p < 0.05) was used to determine significance for IPA-generated networks/pathways.
2.5 Western blot analysis
Noninfarcted anterior-free wall tissue samples (<0.5 g) were ground to a fine powder in liquid N2, homogenized in ice-cold sample buffer, and stored at −80°C. Proteins were quantitated in a protein assay (Thermo Scientific) prior to separation by SDS-PAGE, and transfer to PVDF membranes. Individual proteins were then detected with primary and conjugated secondary antibodies (Abs; see [21, 29]). Primary Abs utilized included collagen IV (1:250; Sigma-Aldrich), pan-laminin (1:500; Sigma-Aldrich), laminins α2 (1:500; Millipore), β1 (1:500; Millipore), β2 (1:200; Thermo), and γ1 (1:500; Millipore), plus nidogen-1 (nid-1; 1:500; Millipore), and nidogen-2 (nid-2; 1:200; R&D Systems). These primary Abs were detected using goat anti-rabbit (GAR), goat anti-mouse (GAM), or donkey anti-goat (DAG) secondary Abs conjugated to HRP (1:1000–1:2000; Cell Signaling; DAG 1:1000; Santa Cruz) or Alexa-Fluor 680 GAM or GAR Abs (1:5000; Invitrogen). Proteins labeled with Alexa-Fluor secondary Abs were detected with an Odyssey infrared imaging system (LiCor), while HRP-conjugated secondary Abs were detected by ECL [29]. Protein expression was quantitated with BioRad Quantity One and normalized to a consistently expressed ~225 kDa protein band detected by silver stain or to actin on blots. Protein expression is expressed as mean ± SEM and was compared using an unpaired Student’s t-test or one-way ANOVA, and when appropriate, post-hoc Neuman–Keuls tests (p < 0.05).
2.6 Rat pressure overload (PO) model
Pressure overload was produced by abdominal coarctation in 6–8-weeks old Sprague–Dawley rats. The protocol was approved by the University of Michigan Institutional Animal Care and Use Committee and followed the PHS Policy on Humane Care and Use of Laboratory Animals. The abdominal aorta was exposed via a midline incision, and partially ligated just above the left renal artery with a 30-g needle, as described earlier [30]. After closing the abdominal wall, individually housed animals received analgesic (buprenorphine, ip), prior to and as needed up to 24 h postsurgery. Oxytetracycline (800 mg/mL) was added to drinking H2O for 7 days postsurgery. An equal number of sham-operated rats underwent the same procedure without coarctation. Cardiac function was monitored by echocardiography prior to surgery, 1–2 weeks and then every 4–8 weeks postsurgery.
2.7 Light and electron microscopy analysis of failing heart tissue
Hearts from sham and PO rats were removed, weighed, fixed in 3% paraformaldehyde, and then embedded in paraffin 18–28 weeks postsurgery. Sections (5 µm) were stained with H&E to examine tissue morphology or with Picrosirus Red to evaluate fibrosis, as described earlier [31].
Septal samples from F and NF human hearts were fixed overnight in Karnovsky’s fixative (Polysciences), postfixed in osmium tetroxide, dehydrated, and then embedded in Spurr’s resin by the University of Michigan Microscopy and Image Analysis core. Ultra-thin sections (70 nm) mounted on copper grids were prepared from screened semithin sections and poststained with uranyl acetate and lead citrate for each sample. Photomicrographs were prepared from images from a Philips CM-100 transmission electron microscope (TEM).
2.8 RNA isolation and gene array analysis
Gene expression was compared between four F and four non-matched NF human hearts. Tissue samples were homogenized in lysis buffer and RNA isolated using the Rneasy kit (Qiagen). Purified RNA was diluted to ≥100 ng/µL in RNase-free dH2O water and RNA quality was verified on an Agilent 2100 Bioanalyzer with RIN scores >9.0. Utilizing 200 ng of total RNA, first- and second-strand cDNAs were synthesized and amplified by in vitro transcription using the MessageAmp™ Premier RNA Amplification kit (Life Tech) following the standard manufacturer protocol. The biotinylated probes were then fragmented and subsequently hybridized to an U133Plus2.0 microarray. Arrays were scanned on a GeneChip Scanner 3000 7G with AGCC software following the standard array protocol provided by the manufacturer. Quality control was evaluated by analysis of RNA degradation and from standard error estimates after fitting a probe-level model. There were no differences in the slope of RNA degradation as a function of probe number, and there were no samples with elevated standard errors relative to other samples. Robust Multi-array Average [32], was used to convert intensity values from the GeneChip CEL data to log2 transformed expression values. Affy, affyPLM, and limma packages of Bioconductor implemented in the R statistical environment were used for data analysis. A final quality control analysis was performed using PCA, and then the data was fit to linear models [33] to determine the contrast of interest. Samples were weighted based on a gene-by-gene update algorithm [34], probe sets with a variance of <0.05 were filtered out, and then probe sets were selected with a ≥twofold change using p-values (≤0.05) adjusted for multiple comparisons using false discovery rates [35].
2.9 Adult rat myocyte isolation, adherence and protein localization
Myocytes were isolated from adult rats 37–45 weeks after sham or PO surgery. Briefly, adult rat hearts were heparinized, digested with collagenase and hyaluronidase [29]. Calcium-tolerant, isolated cells were resuspended to 4 × 103 rod-shaped myocytes/mL in DMEM supplemented with 5% FCS, 50 U/mL penicillin, and 50 µg/mL streptomycin (P/S) [29]. For adhesion studies, 100 µL myocyte aliquots were plated in triplicate onto 12 mm laminin-coated (0–40 µg/mL), glass coverslips. After 30 min, cells were gently washed twice with serum-free M199 media supplemented with P/S. Myocytes remaining attached to the coverslip after media replacement were counted under a light microscope to evaluate cell adhesion.
Another subset of sham and pressure-overloaded myocytes were resuspended in DMEM plus 5% FCS and P/S (1 × 105 rod-shaped cells/mL), and plated on 25 mm2 laminin-coated coverslips [36]. Serum-free M199 media supplemented with P/S was added after 2 h. Myocytes were then fixed in 3% paraformaldehyde and immunostained with anti-nid-1 primary and Texas Red conjugated secondary Abs [21, 29]. Myocytes were imaged with a Nikon Ti-U fluorescent microscope equipped with a DS-U2 digital camera and NIS Elements software. Some myocytes also were collected in ice-cold sample buffer for nid-1 analysis using SDS-PAGE and Western blots, as described earlier.
3 Results
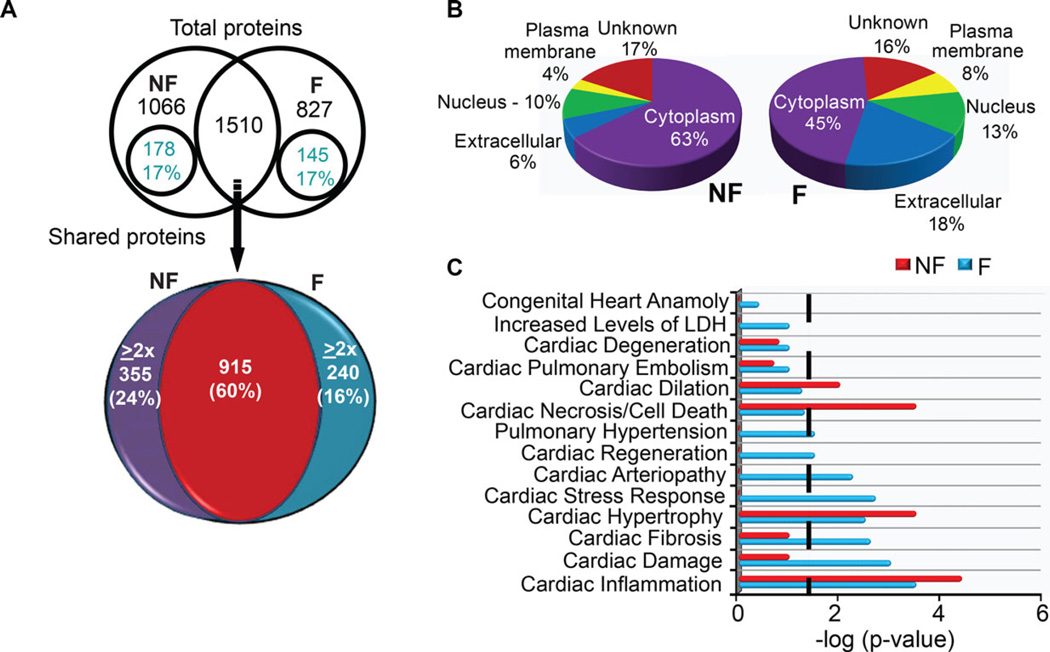
A total of 2339 and 2576 proteins were identified in the respective F and NF samples, after ten replicate analyses by MudPIT. This replicate number was chosen based on earlier work showing nine or more replicates saturate protein identification [23]. Among the identified proteins, similar numbers of unique proteins were identified in F (827) and NF hearts (1066), along with 1510 shared proteins (Fig. 1A). A total of 323 unique proteins and 595 HiE proteins met the differential expression criteria (unique = ≥3 runs; HiE = ≥ twofold), and the relative cellular compartment distribution of unique and HiE proteins was similar among F and NF samples (Fig. 1A, Supporting Information Table 2A–D). Differences detected in myofilament proteins were not further considered due to depletion prior to MS/MS analysis.
Figure 1.
Analysis summary of total identified proteins from nonfailing (NF) and ischemic failing (F) heart explants. (A) Venn diagram showing the distribution of 3405 proteins detected in ten replicate analyses of F and NF tissue (upper panel). Within this total protein pool, 56% were unique and 44% were shared proteins in the NF and F samples. A similar percentage of unique proteins (17%) met the significance criteria (detected ≥three times) within both NF and F protein pools. Among shared proteins, the pie chart in the lower panel illustrates that 40% were expressed at ≥ twofold higher (HiE) levels in one sample compared to the other (# of HiE proteins: 355 in NF, 240 in F). (B) Pie chart shows the subcellular distribution of significant differentially expressed proteins within NF and F heart tissue. These distributions were not different between NF and F heart tissue (χ2 test; p > 0.05). (C) Ingenuity pathway analysis (IPA) analysis of protein distribution within cardiac-specific functions for F and NF samples. A threshold p-value of <0.05 in this IPA analysis is indicated by the black line and the x-axis indicates the significance within each category. The length of each bar above the threshold indicates differentially expressed proteins related to this pathway within each sample.
The relative cellular distribution of proteins was not significantly different in F and NF samples, and the highest proportion of differentially expressed proteins (e.g. unique and HiE) was located in the cytosol for both F and NF samples (Fig. 1B). These findings also were examined with Protein Center software (Thermo, http://www.proxeon.com) analysis (Supporting Information Fig. 1A). IPA analysis also identified differential expression between F and NF samples among shared molecular and cellular functions and physiological processes (Supporting Information Fig. 1B). Among IPA-identified cardiac-specific disease pathways, there were significantly more proteins in the F versus NF sample related to heart disease-related arteriopathy, stress responses, fibrosis, and cardiac damage (Fig. 1C). More proteins contributing to cardiac dilation and cell death were identified in the NF tissue, and differentially expressed proteins associated with hypertrophy and inflammation were identified in both samples, which could be related to aging [37]. Elevated expression of individual proteins previously linked to cardiac remodeling and/or heart failure such as B2M, POSTN, ILK, FHL1, and RTN (Nogo) were detected at higher levels in F compared to NF tissue (Supporting Information Table 2; [38–41]). Several protein networks associated with heart disease were present in F, but not NF tissue when analyzed by FatiGO (Supporting Information Table 3; [4,42,43]).
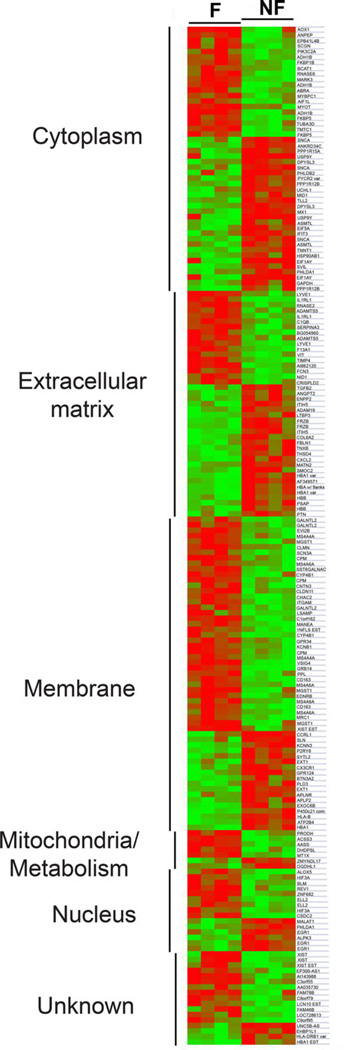
A similar direction of change for several differentially expressed proteins also agreed with altered gene expression previously reported for end-stage ischemic heart failure, including elevated HBA, HBB, MFAP4, SYNPO2L, OGN, and reduced CCT2, ANKRD2, SERPINB1, ANXA6, and SOD2 in F versus NF hearts (Supporting Information Table 3; [44–47]). However, gene array analysis of four F versus four NF hearts was performed for further comparisons to the proteomic results. Both up- (82) and downregulated (105) genes were identified for F compared to NF samples (Fig. 2), and there were several parallel changes in both gene and protein expression detected in F hearts (Supporting Information Tables 2 versus 4). However, a similar direction of gene and protein expression was only detected for nid-1 in the NF samples (Supporting Information Tables S2 versus S4).
Figure 2.
Heat map comparison of significant differential gene expression for F versus NF heart samples. Gene expression is clustered into GO-defined subcellular domains for four F (F3, F5, F6, F9) and four NF (NF, NF2, NF4, and NF5) samples. Relative decreases and increases in gene expression are indicated by green and red, respectively.
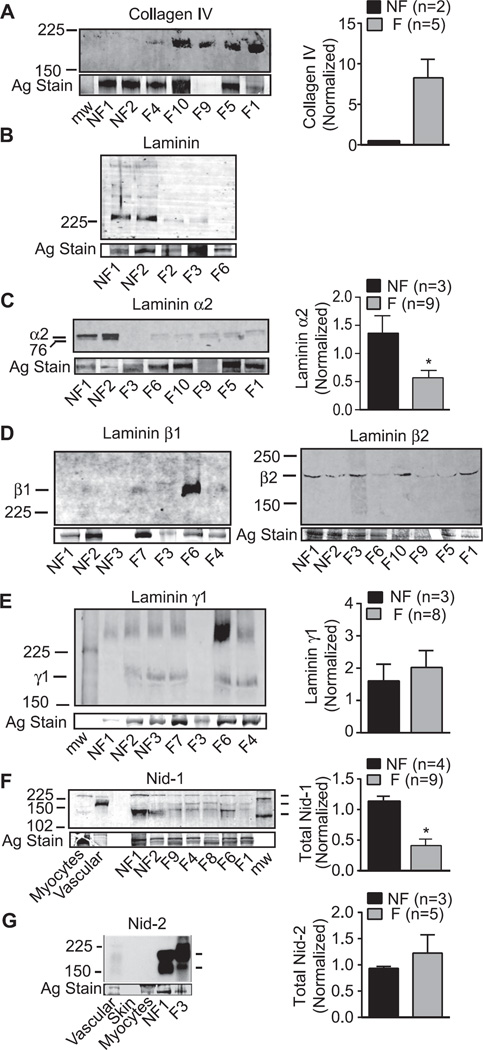
Proteomic data were further analyzed for differentially expressed proteins that have yet to be linked to heart failure. Western blot analysis of multiple F and NF samples verified the protein differences detected by MS/MS. One set of differentially expressed proteins and genes were clustered into the BL in the extracellular matrix, including collagen IV, multiple laminin isoforms, and nidogen isoforms (Fig. 2; Supporting Information Tables 2 and 5; [48]). Western analysis of multiple ischemic F and NF tissue samples confirmed the ~2.5-fold higher collagen IV expression detected in F hearts (Fig. 3A). In contrast, MS/MS analysis indicated laminin isoforms α2, β1, β2, and γ1 were expressed at lower levels in F compared to NF heart tissue (Supporting Information Fig. 2). Western detection with a polyclonal antibody recognizing laminin S (β1/γ1) confirmed the overall decrease in laminin expression in F versus NF hearts (Fig. 3B; [49]). Antibody detection of the carboxy-terminal 80 kDa fragment of laminin α2 [50] also indicated reduced expression of this domain in multiple ischemic F compared to NF samples (Fig. 3C). In contrast, the laminin β1 expression pattern was more variable across multiple F samples, and migrated slightly higher than the anticipated 200 kDa molecular weight (Fig. 3D). Variable glycosylation of one or more of 11 possible sites in this isoform could explain this migration pattern. Variable expression of the β2 and γ1 isoforms also were detected in multiple F and NF tissue samples (Fig. 3D, E).
Figure 3.
Representative Western and quantitative analyses of collagen IV (A), laminin isoform (B–E), and nidogen isoform (F, G) protein expression in multiple F versus NF heart tissues. (A) Collagen type IV α4 (P02462) was detected as a uniquely expressed protein in F tissue by MudPIT analysis. Enhanced expression of collagen IV was detected in F versus NF tissue by Western analysis (left panel), and quantitative comparison of Western analysis of multiple hearts (right panel). (B) Western detection of individual subunits (220–230 kDa; left panel) as well as residual intact laminin (400 kDa) using an antibody recognizing laminin β1γ1. (C) Representative Western detection (left) and quantitative (right) analyses of the 80 kDa laminin α2 fragment detected in multiple NF and F hearts. Proteomic analysis identified >threefold higher expression of laminin α2 in NF compared to F heart tissue (P24043; see Supporting Information Table 2B), and raw data is shown in Supporting Information Fig. 2A. Laminin α2 is a component of laminins 211 and 221. (D) Representative Western detection of laminin β1 (left panel) and β2 (right panel) expression in F and NF tissue. Expression of both isoforms was reduced in the MudPIT analysis for F versus NF tissue (see Supporting Information Fig. 2B and C; G3XAI2 and P55268, respectively). Western analysis indicated variable LV expression of these isoforms in multiple F tissue samples, which was not statistically different from NF samples. (E) Western detection of laminin γ1 isoform, which is variably expressed in F versus NF tissue. MudPIT analysis identified laminin γ1 expression (4.7-fold; P11047) as differentially reduced in F compared to NF tissue. (F) Representative Western blot demonstrates the reduced nidogen-1 (nid-1) expression in multiple F compared to NF hearts, which corroborates the MudPIT analysis (Supporting Information Fig. 2D). Expression is compared to isolated rat myocyte (1 and 5 × 104 cells) and vascular samples to demonstrate the expression of splice variants (136 and 122 kDa) for this isoform. Quantitative analysis of LV anterior wall nid-1 expression in F versus NF tissue (*p < 0.05) is shown in the right panel. (G) Western analysis of nidogen-2 (nid-2) expression shows the relatively high expression of this isoform in F and NF heart tissue compared to vasculature and skin. This isoform was not detected at significant levels in isolated adult rat myocytes (4 × 104 cells). Both F and NF heart tissue express two splice variants for this isoform. Quantitative analysis of LV anterior wall Nid-2 expression in F versus NF tissue (*p < 0.05) is shown in the right panel.
In addition to laminin, BL-associated, 130 kDa nid-1 was detected as a significant, unique protein in the NF sample (Supporting Information Fig. 2D). Western analysis confirmed nid-1 expression was consistently higher in NF compared to F tissue (Fig. 3F). A higher molecular weight nid-1 also was detected just above 225 kDa in rat myocytes and human heart tissue, which would be consistent with oligomers. However, the typical isoform detected at ~150 kDa in rat blood vessels was only sparsely present in the myocardium (Fig. 3F; [51]). The alternative isoform, nid-2 can compensate for the loss of nid-1 in knockout animal models [52], but nid-2 expression in F tissue was not different from NF in the proteomic analysis (Supporting Information Table 2). There are three possible nid-2 protein products [53], and two products (200, 160 kDa) were present in both F and NF tissue (Fig. 3G). Expression of these variants and total nid-2 expression in F samples was variable, but not significantly different than NF sample expression (Fig. 3G). Collectively, the analysis of BL-associated proteins suggests significant BL remodeling develops in end-stage heart failure.
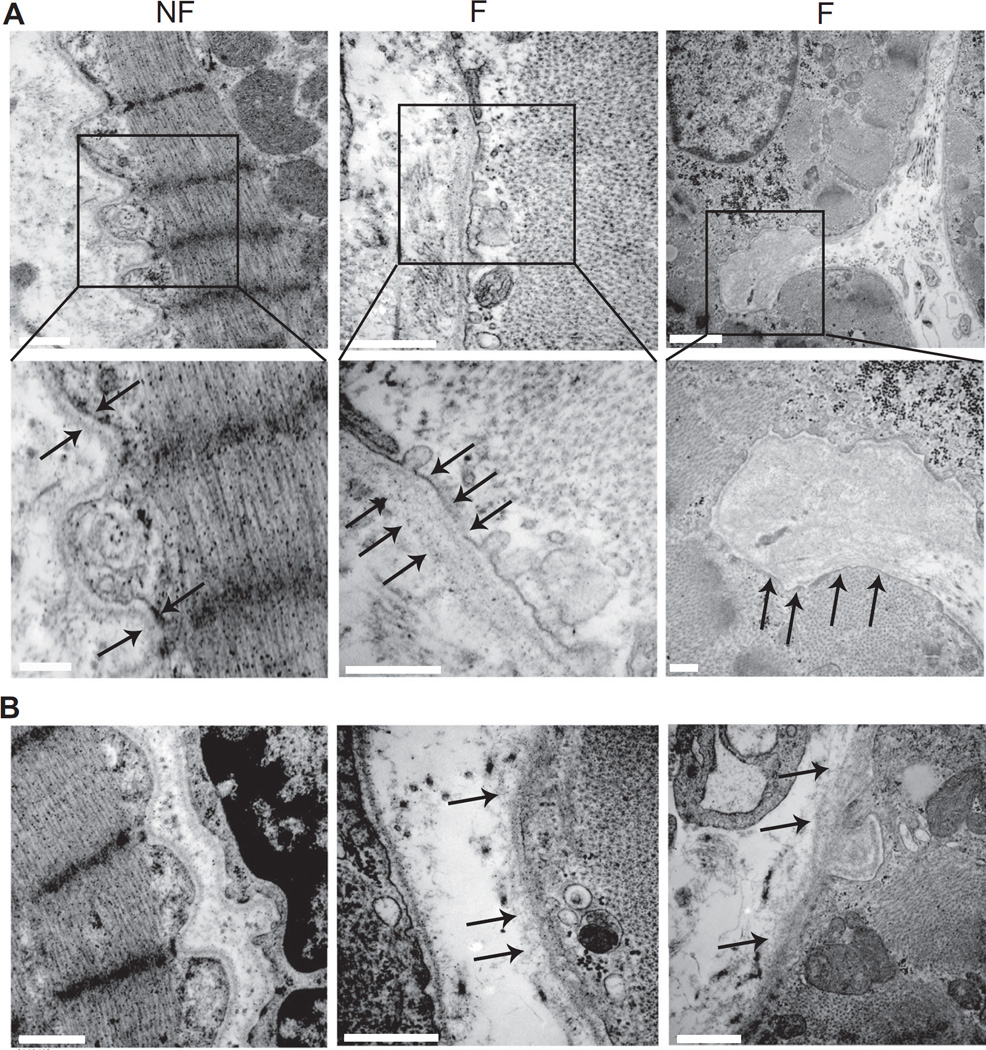
Variable changes in expression observed for some laminin and nidogen isoforms could indicate this remodeling is a dynamic and progressive process. To address this possibility, noninfarcted regions of human septum from human NF and F tissue were imaged with TEM to determine whether there was visual evidence of structural BL remodeling. The well organized and distinct BL was consistently present around myocytes in NF septa, while a more dispersed, amorphous protein layer with periodic disruptions was frequently visible around myocytes in F samples (Fig. 4). These images show the BL undergoes significant remodeling during heart failure, which could contribute to changes in cellular structure and/or function.
Figure 4.
Representative electron micrographs (TEM) from noninfarcted septum demonstrate BL remodeling in F versus NF hearts. (A) TEM images of NF and F human heart tissue. Boxes in the upper panels indicate the area shown in the expanded view shown in the lower panel. Arrows point to BL in each sample. Upper and lower panel scale bars are 500 and 250 nm, respectively. (B) Additional TEM micrographs from human heart tissue. Left panel. Micrograph shows a fibroblast adjacent to a myocyte. The maintenance of a defined BL in fibroblasts is observed in both F and NF tissue. Scale bar = 1 µm. Middle and right panels. These panels further illustrate the periodic loss of the defined BL in F, but not NF tissue. Scale bars = 500 nm (middle panel) or 1 µm (left panel).
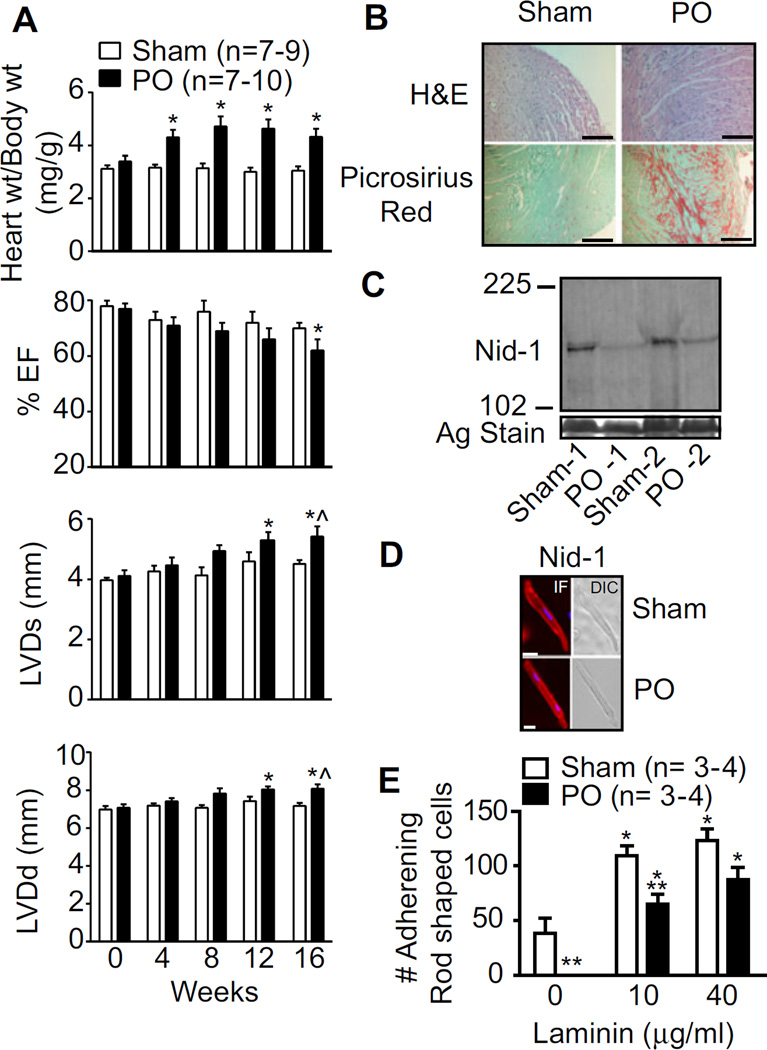
To begin addressing whether there are functional repercussions, myocardial BL structure, and function was evaluated in a rat PO model. This model was selected to establish whether BL remodeling is a process exclusively found in response to myocardial ischemia and/or also accompanies remodeling found with heart failure caused by other factors. The PO model produced significant organ-level hypertrophy by 4 weeks compared to sham controls (Fig. 5A), and caused cellular hypertrophy, fibrosis, and impaired cardiac function at 16 weeks postsurgery and beyond (Fig. 5A and B). Reduced nid-1 expression was detected with PO compared to the sham controls, although its cellular localization remained similar for the two groups (Fig. 5C and D). To determine whether this evidence for BL remodeling produced functional consequences, cellular adhesion to an exogenous laminin matrix was evaluated and found to be significantly compromised in cells isolated from PO compared to sham rats (Fig. 5E). Reduced myocyte adhesion also develops in pacing-induced cardiomyopathy animal models [54]. Collectively, the present data support the idea that BL remodeling is associated with significant functional impairments in adhesion to the myocyte matrix.
Figure 5.
Cardiac structure and function in sham and pressure overload (PO) rats. (A) Echocardiographic analysis of sham- and PO- rat hearts over time (n = number of rats). M-mode ejection fraction (EF%), LV diameter at end systole, and diastole show significant cardiac dysfunction along with hypertrophy by 16–20 weeks postsurgery in PO hearts compared to sham controls. (B) Sham and PO heart tissue stained with H&E are further com pa red using Picrosirius red staining to detect fibrosis. (C) Analysis of nid-1 expression by Western blot in sham and PO rats relative to a silver (Ag)-stained gel band. (D) Left panel: Detection of nid-1 cellular localization by immunofluorescence in isolated myocytes from sham and PO rats 16–20 weeks postsurgery. Right panel: A differential interference contrast image of the same myocyte. (E) Functional analysis of adhesion by cardiac myocytes isolated from sham- and PO- rats (n = number of hearts).
4 Discussion
Proteomic comparison of ischemic F versus NF tissue using MudPIT analysis revealed previously unrecognized alterations in BL proteins in addition to known changes in other proteins linked to heart failure (Supporting Information Table 2). The BL together with proteoglycans and other filamentous proteins form the basement membrane (BM), to act as a barrier between the sarcolemma and extracellular matrix. The BM maintains cellular integrity, adhesion, and proximity to adjacent cells, while also acting as a scaffold to modulate access and activity of protease networks and extracellular signaling cascades [48]. Our studies demonstrated structural remodeling in tissue explants from end-stage ischemic heart failure accompanied alterations in the expression of key BL proteins (Figs. 3 and 4, Supporting Information Fig. 2). Evidence of a similar remodeling process was detected in a rat PO model, and functional deficits in myocyte adhesion accompanied this remodeling process (Fig. 5). The reduced in vitro adhesion of myocytes from PO rats suggests cellular integrity, cell–cell coupling and/or extracellular signaling are likely compromised by this cardiac BM remodeling.
The results focusing on BL-associated proteins also indicate further work is needed to evaluate this compartment during heart failure. The collagen IV upregulation observed here (Fig. 3A) agrees with elevated message levels observed in patients with aortic disorders [55], although our work found no significant differences in collagen IV subunit gene expression between F and NF tissue samples (Fig. 2). The current results also are in contrast to a report of reduced collagen IV α4 gene expression in end-stage human heart failure [56]. However, a more acute porcine ischemia/reperfusion (I/R) model of cardiac disease increased collagen IV protein expression, although this change was not statistically significant [57].
Myocardial BL laminin expression has been studied slightly more than collagen IV in animal models of cardiac dysfunction, although the results are similarly inconsistent from one study to another. The absence of change in myocardial laminin gene expression reported in explants from patients with idiopathic dilated cardiomyopathy and pressure overloaded rats [58] agrees with our results (Supporting Information Table 4). While this observation challenges the idea of BL remodeling during heart failure, the present proteomic results suggest BL protein levels may not necessarily correlate with gene expression. The reduced laminin α2 protein expression observed here (Fig. 3C) is in agreement with results from a porcine I/R model [57]. Changes in laminin β and γ isoform protein expression in the same I/R model depended on tissue handling protocols, but the current Western results (Fig. 4) suggest the outcome may depend on variability in temporal remodeling. In addition, the laminin β/γ protein results are consistent with the changes reported in posttranslational glycosylation of laminin in a rodent (I/R) model [59], and cardiomyopathy resulting from congenital laminin α2 deficiencies [60].
Nidogen, collagen IV, and laminin are essential components of the BL, but there are no known published studies examining nidogen protein expression (Fig. 3F and G) and alternative splicing during heart failure. Nidogen serves a critical coordinating role in BL assembly, and nidogen-laminin interactions are essential for BL stabilization [48, 51]. Nid-2 partially compensates for a loss of nid-1 in many organs, but there is evidence of subtle differences in laminin-nidogen interactions with these isoforms [52], and this compensation has not been studied in myocardium. Overall, technical replicate analysis by MudPIT provided a fundamental differential comparison list of cardiac peptides in a F versus NF heart. The anticipated ability of such a list to serve as a guide for a more targeted analysis [23] was demonstrated by Western analysis of nidogen, laminin, collagen IV expression in multiple human hearts as well as a rat model of PO. Due to the small number of biological replicates, future studies using more failing human hearts and animal models of heart failure are needed to target and or verify the other protein networks and pathways identified by MudPIT. The identification of cardiac BL remodeling during heart failure also raises further questions about spatial and temporal pattern of cardiac BL remodeling, the role BL remodeling plays in pump dysfunction, and the potential for targeting BL proteins to delay or prevent cardiac dysfunction.
Analysis of tissue architecture in earlier animal models indicated BL remodeling around the vasculature in the ischemic region in one model [61], but not in the ventricular laminin distribution in a rat PO model [62]. In contrast, our TEM imaging of human septum indicates the BL remodeling process develops in areas distant from the infarct site (Fig. 4). Work in other animal models offers some limited support, as comparable BL remodeling around cardiac myocytes was associated with cardiac hypertrophy and electrophysiological abnormalities in copper-deficient rats [63]. Moreover, in vitro BL formation and reorganization are critical for cultured myocyte attachment and survival [64, 65]. The impaired adhesion observed in cardiac myocytes from PO rats (Fig. 7) is consistent with this conclusion, and may indicate additional myocyte functions are compromised as a consequence of the reorganized BL. For example, remodeling may disrupt the contribution of BL to tethering and transmission of mechanical forces from the cellular cytoskeleton to fibrillar collagen in the extracellular matrix [64–66]. Differential expression patterns among proteins thought to play a role in mechanical force transmission [64–66] also are found in the proteomic comparison of F versus NF cardiac tissue, including elevated expression of the costameric proteins integrin α7, ILK, talin, paxillin, and myospryn (CMYA5) in F tissue compared to enhanced melusin, vinculin, and filamin Cγ expression in NF tissue (Supporting Information Table 2). In contrast, stretch-sensitive cytoskeleton sensor proteins linked to integrins and associated with sarcomeres [67], such as α-actinins, PDLIM5, telethonin, and myozenin (calsarcin) remained comparable in NF and F tissue. Cytoskeletal sensor elements with a less restricted expression and/or associated with the sarcomeric thick filament such as titin, PDLIM3 (ALP), and the enigma homolog are more highly expressed in NF tissue. While further studies to evaluate whether costameric and cytoskeletal sensor expression and function are differentially influenced in myocytes during heart failure, the present results indicate costameric proteins, which interact and anchor to the BL also could serve as therapeutic targets in heart failure patients.
4.1 Concluding remarks
Taken together, our studies in human and a rat PO model indicate BL plays in important role in the myocardial remodeling process during the development of heart failure. BL remodeling is likely a dynamic process, which progresses and/or changes from early compensatory to late-stage de-compensatory heart disease [58]. Further work is now needed to define the stressors contributing to BL remodeling, and determine the beneficial and/or detrimental impact of this remodeling on cellular anchoring, tethering, signaling, as well as cardiac performance.
Supplementary Material
Clinical Relevance.
Heart failure is a complex pathophysiological process that continues to be a leading cause of death in the United States. The present study utilized multidimensional protein identification to identify protein networks not previously linked to heart failure by comparing ischemic, failing versus nonfailing human hearts. The differential expression of multiple basal lamina proteins in failing heart tissue was validated by Western analysis of multiple samples. In additional studies, structural basal lamina disruption was observed in failing tissue, and reduced myocyte adhesion accompanied basal lamina remodeling in a rat model of pressure overload. Collectively, these results indicate basal lamina remodeling develops during heart failure and contributes to the structural and functional progression of this disease. Most importantly, agents targeting this remodeling process could therapeutically delay the progression of heart failure.
Acknowledgments
This work was supported by National Institutes of Health Grant R01-HL-067254, R21-HL89193 (M.V.W.), R01-CA140211 (D.E.M.), and the Functional Assessment Core in the Nathan Shock Center (NIH AG013283), and internal funding from the Department of Surgery at the University of Michigan. Studies in this manuscript also utilized the Core Facilities (Morphology and Image Analysis, and Cell and Molecular Biology Cores) of the Michigan Diabetes Research and Training Center (supported by National Institutes of Health Grant P60-DK20572). Affymetrix data is deposited on GEO (Series GSE76701).
Abbreviations
- Abs
antibodies
- BL
basal lamina
- BM
basement membrane
- DAG
donkey anti-goat
- EF
ejection fraction
- F
failing
- GAM
goat anti-mouse
- GAR
goat anti-rabbit
- HiE
highly expressed
- IPA
ingenuity pathway analysis
- I/R
ischemia/reperfusion
- LTQ-FT
hybrid linear quadrapole ion trap-Fourier transform ICR
- LV
left ventricle
- MudPIT
multidimensional protein identification technology
- NF
nonfailing
- nid-1
nidogen-1
- nid-2
nidogen-2
- PO
pressure overload
- P/S
penicillin/streptomycin
- SCX
strong cation exchange
- TEM
transmission electron microscopy
Footnotes
Additional supporting information may be found in the online version of this article at the publisher’s web-site
The authors have declared no conflict of interest.
References
- 1.Jessup M, Brozena S. Heart failure. N. Engl. J. Med. 2003;348:2007–2018. doi: 10.1056/NEJMra021498. [DOI] [PubMed] [Google Scholar]
- 2.Roger VL, Go AS, Lloyd-Jones DM, Benjamin EJ, et al. Heart disease and stroke statistics — 2012 update: a report from the American Heart Association. Circulation. 2012;725:e2–e220. doi: 10.1161/CIR.0b013e31823ac046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kathiresan S, Srivastava D. Genetics of human cardiovascular disease. Cell. 2012;148:1242–1257. doi: 10.1016/j.cell.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosca MG, Tandler B, Hoppel CL. Mitochondria in cardiac hypertrophy and heart failure. J. Mol. Cell Cardiol. 2013;55:31–41. doi: 10.1016/j.yjmcc.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doetschman T, Azhar M. Cardiac-specific inducible and conditional gene targeting in mice. Circ. Res. 2012;770:1498–1512. doi: 10.1161/CIRCRESAHA.112.265066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sarwar R, Cook SA. Genomic analysis of left ventricular remodeling. Circulation. 2011;720:437–444. doi: 10.1161/CIRCULATIONAHA.108.797225. [DOI] [PubMed] [Google Scholar]
- 7.Zeller T, Blankenberg S, Diemert R. Genomewide association studies in cardiovascular disease—an update 2011. Clin. Chem. 2012;58:92–103. doi: 10.1373/clinchem.2011.170431. [DOI] [PubMed] [Google Scholar]
- 8.Tan FL, Moravec CS, Li J, Apperson-Hansen C, et al. The gene expression fingerprint of human heart failure. Proc. Natl. Acad. Sci. USA. 2002;99:11387–11392. doi: 10.1073/pnas.162370099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steenman M, Chen YW, Le Cunff M, Lamirault G, et al. Transcriptomal analysis of failing and nonfailing human hearts. Physiol. Genomics. 2003;72:97–112. doi: 10.1152/physiolgenomics.00148.2002. [DOI] [PubMed] [Google Scholar]
- 10.Adams KF. Systems biology and heart failure: Concepts, methods, and potential research applications. Heart Fail Rev. 2010;75:371–398. doi: 10.1007/s10741-009-9138-x. [DOI] [PubMed] [Google Scholar]
- 11.Felkin LE, Lara-Pezzi EA, Hall JL, Birks EJ, Barton PJ. Reverse remodelling and recovery from heart failure are associated with complex patterns of gene expression. J. Cardiovasc. Transl. Res. 2011;4:321–331. doi: 10.1007/s12265-011-9267-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Margulies KB, Matiwala S, Cornejo C, Olsen H, et al. Mixed messages: transcription patterns in failing and recovering human myocardium. Circ. Res. 2005;96:592–599. doi: 10.1161/01.RES.0000159390.03503.c3. [DOI] [PubMed] [Google Scholar]
- 13.Mayr M, Zhang J, Greene AS, Gutterman D, et al. Proteomics-based development of biomarkers in cardiovascular disease: mechanistic, clinical, and therapeutic insights. Mol. Cell Proteomics. 2006;5:1853–1864. doi: 10.1074/mcp.R600007-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.James J, Robbins J. Molecular remodeling of cardiac contractile function. Am. J. Physiol. 1997;273:H2105–H2118. doi: 10.1152/ajpheart.1997.273.5.H2105. [DOI] [PubMed] [Google Scholar]
- 15.Van Eyk JE. Overview: the maturing of proteomics in cardiovascular research. Circ. Res. 2011;70S:490–498. doi: 10.1161/CIRCRESAHA.110.226894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macri J, Rapundalo ST. Application of proteomics to the study of cardiovascular biology. Trends Cardiovasc. Med. 2001;77:66–75. doi: 10.1016/s1050-1738(01)00088-3. [DOI] [PubMed] [Google Scholar]
- 17.Ruse CI, Willard B, Jin JR, Haas T, et al. Quantitative dynamics of site-specific protein phosphorylation determined using liquid chromatography electrospray ionization mass spectrometry. Anal. Chem. 2002;74:1658–1664. doi: 10.1021/ac0157122. [DOI] [PubMed] [Google Scholar]
- 18.Kim N, Lee Y, Kim H, Joo H, et al. Potential biomarkers for ischemic heart damage identified in mitochondrial proteins by comparative proteomics. Proteomics. 2006;6:1237–1249. doi: 10.1002/pmic.200500291. [DOI] [PubMed] [Google Scholar]
- 19.Kislinger T, Gramolini AO, MacLennan DH, Emili A. Multidimensional protein identification technology (Mud-PIT): technical overview of a profiling method optimized for the comprehensive proteomic investigation of normal and diseased heart tissue. J. Am. Soc. Mass Spectrom. 2005;16:1207–1220. doi: 10.1016/j.jasms.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 20.Horn S, Lueking A, Murphy D, Staudt A, et al. Profiling humoral autoimmune repertoire of dilated cardiomyopathy (DCM) patients and development of a disease-associated protein chip. Proteomics. 2006;6:605–613. doi: 10.1002/pmic.200401293. [DOI] [PubMed] [Google Scholar]
- 21.Hwang H, Robinson DA, Stevenson TK, Wu HC, et al. PKCβ modulation of myocyte contractile performance. J. Mol. Cell Cardiol. 2012;53:176–186. doi: 10.1016/j.yjmcc.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neverova I, Van Eyk JE. Application of reversed phase high performance liquid chromatography for subproteomic analysis of cardiac muscle. Proteomics. 2002;2:22–31. [PubMed] [Google Scholar]
- 23.Kline KG, Frewen B, Bristow MR, Maccoss MJ, Wu CC. High quality catalog of proteotypic peptides from human heart. J. Proteome Res. 2008;7:5055–5061. doi: 10.1021/pr800239e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kline KG, Finney GL, Wu CC. Quantitative strategies to fuel the merger of discovery and hypothesis-driven shotgun proteomics. Brief Funct. Genomic Proteomic. 2009;8:114–125. doi: 10.1093/bfgp/elp008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim JY, Lee JR, Choi S, Kim EM, et al. Quantitative pattern analysis of the N-terminally processed isoforms of platelet factor-4 in serum. Rapid Commun. Mass Spectrom. 2013;27:521–530. doi: 10.1002/rcm.6480. [DOI] [PubMed] [Google Scholar]
- 26.Xu T, Venable JD, Park SK, Cociorva D, et al. ProLu-CID, a fast and sensitive tandem mass spectra-based protein identification program. Mol. Cell Proteomics. 2006;5:S174. [Google Scholar]
- 27.Wein SP, Cote RG, Dumousseau M, Reisinger F, et al. Improvements in the protein identifier cross-reference service. Nucleic Acids Res. 2012;40:W276–W280. doi: 10.1093/nar/gks338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Medina I, Carbonell J, Pulido L, Madeira S, et al. Babelomics: an integrative platform for the analysis of transcriptomics, proteomics and genomic data with advanced functional profiling. Nucleic Acids Res. 2010;38:W210–W213. doi: 10.1093/nar/gkq388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lang SE, Robinson DA, Wu HC, Herron TJ, et al. Myofilament incorporation and contractile function after gene transfer of cardiac troponin I Ser43/45Ala. Arch. Biochem. Biophys. 2013;535:49–55. doi: 10.1016/j.abb.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bayer AL, Heidkamp MC, Patel N, Porter M, et al. Alterations in protein kinase C isoenzyme expression and autophosphorylation during the progression of pressure overload-induced left ventricular hypertrophy. Mol. Cell Biochem. 2003;242:145–152. [PubMed] [Google Scholar]
- 31.Junqueira LCU, Bignolas G, Brentani RR. Picrosirius staining plus polarization microscopy: a specific method for collagen detection in tissue sections. Histochem. J. 1979;77:447–455. doi: 10.1007/BF01002772. [DOI] [PubMed] [Google Scholar]
- 32.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 33.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3:3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 34.Ritchie ME, Diyagama D, Nielson J, van Laar R, et al. Empirical array quality weights in the analysis of microarray data. BMC Bioinformatics. 2006;7:261–277. doi: 10.1186/1471-2105-7-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Royal Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 36.Lang SE, Westfall MV. Gene transfer into cardiac myocytes. Methods Mol. Biol. 2015;7299:177–190. doi: 10.1007/978-1-4939-2572-8_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simkhovich BZ, Marjoram P, Poizat C, Kedes L, et al. Age-related changes of cardiac gene expression following myocardial ischemia/reperfusion. Arch. Biochem. Biophys. 2003;420:268–278. doi: 10.1016/j.abb.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 38.Oka T, Xu J, Kaiser RA, Melendez J, et al. Genetic manipulation of periostin expression reveals a role in cardiac hypertrophy and ventricular remodeling. Circ. Res. 2007;707:313–321. doi: 10.1161/CIRCRESAHA.107.149047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu H, Fedak PWM, Dai X, Du C, et al. Integrin-linked kinase expression is elevated in human cardiac hypertrophy and induces hypertrophy in transgenic mice. Circulation. 2006;114:2271–2279. doi: 10.1161/CIRCULATIONAHA.106.642330. [DOI] [PubMed] [Google Scholar]
- 40.Bullard TA, Protack TL, Aguilar R, Bagwe S, et al. Identification of Nogo as a novel indicator of heart failure. Physiol. Genomics. 2008;32:182–189. doi: 10.1152/physiolgenomics.00200.2007. [DOI] [PubMed] [Google Scholar]
- 41.Amighi J, Hoke M, Mlekusch W, Schlager O, et al. β2 microglobulin and the risk for cardiovascular events in patients with asymptomatic carotid atherosclerosis. Stroke. 2011;42:1826–1833. doi: 10.1161/STROKEAHA.110.600312. [DOI] [PubMed] [Google Scholar]
- 42.Bugger H, Schwarzer M, Chen D, Schrepper A, et al. Proteomic remodelling of mitochondrial oxidative pathways in pressure overload-induced heart failure. Cardiovasc. Res. 2010;85:376–384. doi: 10.1093/cvr/cvp344. [DOI] [PubMed] [Google Scholar]
- 43.Liew CC, Dzau VJ. Molecular genetics and genomics of heart failure. Nat. Rev. Genet. 2004;5:811–825. doi: 10.1038/nrg1470. [DOI] [PubMed] [Google Scholar]
- 44.Kong SW, Hu YW, Ho JW, Ikeda S, et al. Heart failure-associated changes in RNA splicing of sarcomere genes. Circ. Cardiovasc. Genet. 2010;3:138–146. doi: 10.1161/CIRCGENETICS.109.904698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kittleson MM, Minhas KM, Irizarry RA, Ye SQ, et al. Gene expression analysis of ischemic and nonischemic cardiomyopathy: shared and distinct genes in the development of heart failure. Physiol Genomics. 2005;21:299–307. doi: 10.1152/physiolgenomics.00255.2004. [DOI] [PubMed] [Google Scholar]
- 46.Song G, Campos B, Wagoner LE, Dedman JR, Walsh RA. Altered cardiac annexin mRNA and protein levels in the left ventricle of patients with end-stage heart failure. J. Mol. Cell Cardiol. 1998;30:443–451. doi: 10.1006/jmcc.1997.0608. [DOI] [PubMed] [Google Scholar]
- 47.Lu Z, Xu X, Hu X, Zhu G, et al. Extracellular superoxide dismutase deficiency exacerbates pressure overload-induced left ventricular hypertrophy and dysfunction. Hypertension. 2008;51:19–25. doi: 10.1161/HYPERTENSIONAHA.107.098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yurchenco PD, Patton BL. Developmental and pathogenic mechanisms of basement membrane assembly. Curr. Pharm. Des. 2009;15:1277–1294. doi: 10.2174/138161209787846766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang X, Chaudhry A, Chintala SK. Inhibition of plasminogen activation protects against ganglion cell loss in a mouse model of retinal damage. Mol. Vis. 2003;9:238–248. [PubMed] [Google Scholar]
- 50.Cohn RD, Herrmann R, Sorokin L, Wewer UM, Voit T. Laminin α2 chain-deficient congenital muscular dystrophy: variable epitope expression in severe and mild cases. Neurology. 1998;51:94–100. doi: 10.1212/wnl.51.1.94. [DOI] [PubMed] [Google Scholar]
- 51.Katz A, Fish AJ, Pe’er J, Frucht-Pery J, et al. Entactin/nidogen: synthesis by bovine corneal endothelial cells and distribution in the human cornea. Invest Ophthalmol. Vis. Sci. 1994;35:495–502. [PubMed] [Google Scholar]
- 52.Miosge N, Sasaki T, Timpl R. Evidence of nidogen-2 compensation for nidogen-1 deficiency in transgenic mice. Matrix Biol. 2002;21:611–621. doi: 10.1016/s0945-053x(02)00070-7. [DOI] [PubMed] [Google Scholar]
- 53.Kohfeldt E, Sasaki T, Gohring W, Timpl R. Nidogen-2: a new basement membrane protein with diverse binding properties. J. Mol. Biol. 1998;282:99–109. doi: 10.1006/jmbi.1998.2004. [DOI] [PubMed] [Google Scholar]
- 54.Spinale FG, Zellner JL, Johnson WS, Eble DM, Munyer PD. Cellular and extracellular remodeling with the development and recovery from tachycardia-induced cardiomyopathy: changes in fibrillar collagen, myocyte adhesion capacity and proteoglycans. J. Mol. Cell Cardiol. 1996;28:1591–1608. doi: 10.1006/jmcc.1996.0150. [DOI] [PubMed] [Google Scholar]
- 55.Cotrufo M, DeSanto L, Della Corte A, Di Meglio F, et al. Basal lamina structural alterations in human assymmetic aneurismatic aorta. Eur. J. Histochem. 2005;49:363–370. doi: 10.4081/964. [DOI] [PubMed] [Google Scholar]
- 56.Bruggink AH, van Oosterhout MF, de Jonge N, Cleutjens JP, et al. Type IV collagen degradation in the myocardial basement membrane after unloading of the failing heart by a left ventricular assist device. Lab Invest. 2007;87:1125–1137. doi: 10.1038/labinvest.3700670. [DOI] [PubMed] [Google Scholar]
- 57.Barallobre-Barreiro J, Didangelos A, Schoendube FA, Drozdov I, et al. Proteomics analysis of cardiac extracellular matrix remodeling in a porcine model of ischemia/reperfusion injury. Circulation. 2012;725:789–802. doi: 10.1161/CIRCULATIONAHA.111.056952. [DOI] [PubMed] [Google Scholar]
- 58.Oliviero P, Chassagne C, Salichon N, Corbier A, et al. Expression of laminin α2 chain during normal and pathological growth of myocardium in rat and human. Cardiovasc. Res. 2000;46:346–355. doi: 10.1016/s0008-6363(00)00034-1. [DOI] [PubMed] [Google Scholar]
- 59.Parker BL, Palmisano G, Edwards AV, White MY, et al. Quantitative N-linked glycoproteomics of myocardial ischemia and reperfusion injury reveals early remodeling in the extracellular environment. Mol. Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.006833. M110.006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spyrou N, Philpot J, Foale R, Camici PG, Muntoni F. Evidence of left ventricular dysfunction in children with merosin-deficient congenital muscular dystrophy. Am. Heart J. 1998;736:474–476. doi: 10.1016/s0002-8703(98)70222-4. [DOI] [PubMed] [Google Scholar]
- 61.Begieneman MP, van de Goot FR, Krijnen PA, Fritz J, et al. The basement membrane of intramyocardial capillaries is thickened in patients with acute myocardial infarction. J. Vase. Res. 2010;47:54–60. doi: 10.1159/000231721. [DOI] [PubMed] [Google Scholar]
- 62.Contard F, Koteliansky V, Marotte F, Dubus I, et al. Specific alterations in the distribution of extracellular matrix components within rat myocardium during the development of pressure overload. Lab Invest. 1991;64:65–75. [PubMed] [Google Scholar]
- 63.Davidson J, Medeiros DM, Hamlin RL. Cardiac ultrastructural and electrophysiological abnormalities in postweanling copper-restricted and copper-repleted rats in the absence of hypertrophy. J. Nutr. 1992;722:1566–1575. doi: 10.1093/jn/122.7.1566. [DOI] [PubMed] [Google Scholar]
- 64.Lundgren E, Terracio L, Borg TK. Adhesion of cardiac myocytes to extracellular matrix components. Basic Res. Cardiol. 1985;80(S1):69–74. doi: 10.1007/978-3-662-11041-6_13. [DOI] [PubMed] [Google Scholar]
- 65.Lundgren E, Gullberg D, Rubin K, Borg TK, et al. In vitro studies on adult cardiac myocytes: attachment and biosynthesis of collagen type IV and laminin. J. Cell Physiol. 1988;736:43–53. doi: 10.1002/jcp.1041360106. [DOI] [PubMed] [Google Scholar]
- 66.Imanaka-Yoshida K, Enomoto-lwamoto M, Yoshida T, Sakakura T. Vinculin, talin, integrin alpha6beta1 and laminin can serve as components of attachment complex mediating contraction force transmission from cardiomyocytes to extracellular matrix. Cell Motil. Cytoskeleton. 1999;42:1–11. doi: 10.1002/(SICI)1097-0169(1999)42:1<1::AID-CM1>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 67.Cox L, Umans L, Cornelis F, Huylebroeck D, Zwijsen A. A broken heart: a stretch too far: an overview of mouse models with mutations in stretch-sensor components. Int. J. Cardiol. 2008;737:33–44. doi: 10.1016/j.ijcard.2008.06.049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.