Abstract
DNA interstrand crosslinks (ICLs) covalently join the two strands of a DNA duplex and block essential processes such as DNA replication and transcription. Several important anti-tumor drugs such as cisplatin and nitrogen mustards exert their cytotoxicity by forming ICLs. However, multiple complex pathways repair ICLs and these are thought to contribute to the development of resistance towards ICL-inducing agents. While the understanding of many aspects of ICL repair is still rudimentary, studies in recent years have provided significant insights into the pathways of ICL repair. In this perspective we review the recent advances made in elucidating the mechanism of ICL repair with a focus on the role of TLS polymerases. We describe the emerging models for how these enzymes contribute to and are regulated in ICL repair, discuss the key open questions and examine the implications for this pathway in anti-cancer therapy.
Keywords: DNA polymerases, translesion synthesis, inter-strand crosslink repair, cisplatin, nitrogen mustard
1. Introduction
DNA interstrand crosslinks (ICLs) are formed by bifunctional agents and covalently link two strands of a DNA duplex. They can be formed by endogenous sources such as products of lipid metabolism and abasic sites, as well as by a number of antitumor agents such as nitrogen mustards, cisplatin and mitomycin C [1]. Despite the clinical success of crosslinking agents in antitumor therapy, multiple cellular pathways, including repair mechanisms that remove ICLs from DNA, cause resistance to such treatment [2–4]. There are multiple ICL repair pathways operating throughout the cell cycle, but the majority of ICLs are processed during replication, where they provide an absolute block for replicative helicases and polymerases. A common step for all ICL repair pathways is believed to be the unhooking of the ICL from one of the strands followed by translesion synthesis past the unhooked ICL. Here we review the role of DNA polymerases in ICL repair and discuss how the nature of the unhooking step as well as ICL structures influence activity of DNA polymerases during ICL repair.
2. Replication-dependent ICL repair pathways
The majority of ICLs are repaired in a replication-coupled manner in vertebrates (Fig. 1). There appear to be multiple replication-associated repair pathways, and several are still not well understood. We know most about a pathway initiated by two converging replication forks and have gotten some first glimpses at a replication traverse pathway. These pathways are discussed here with a focus on how DNA polymerases engage with unhooked ICLs.
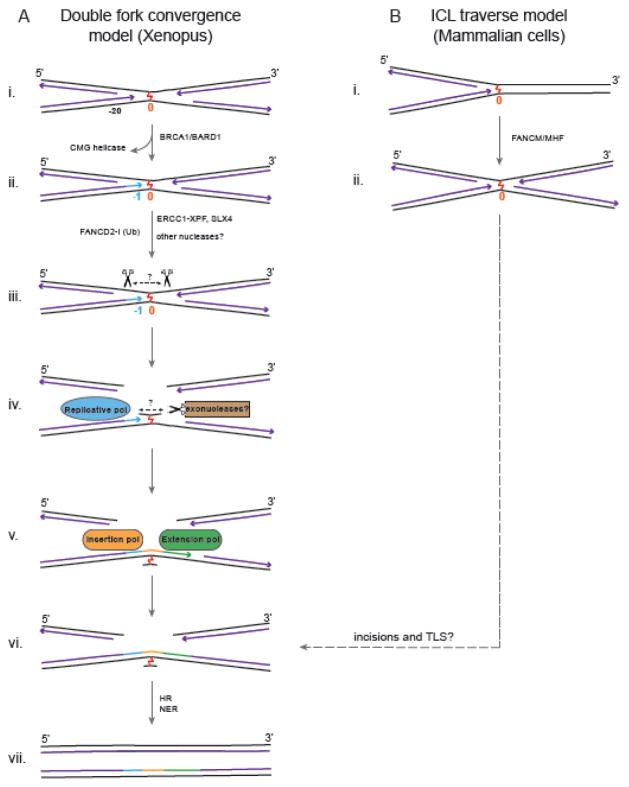
Fig. 1. Models for Replication-Dependent ICL Repair Pathways.
A: Double Fork Convergence Model. (i) Two replication forks converge on an ICL and stall 20–40 nt away from it. (ii) Removal of the CMG helicase by BRCA1 allows one of the leading strands to approach within 1 nt of the ICL (−1). (iii) Activation of the FA pathway leads to ubiquitylation of FANCD2-I, which is required for unhooking of the ICL by SLX4/ERCC1-XPF and possibly other nucleases. The position of these incisions have not been determined, and the amount of duplex surrounding the ICL is unknown. (iv) The unhooked ICL could then be further processed by exonucleases to trim the duplex around the ICL, making it more amenable to bypass by DNA polymerases. (v, vi) An insertion polymerase inserts nucleotide(s) opposite the ICL and an extension polymerase extends the insertion product further. (vi) Ligation to downstream Okazaki fragments restores one daughter duplex, and (vii) is used to restore the other duplex by HR. The ICL remnant on one strand is likely removed by NER to complete the repair of both daughter duplexes. B: ICL traverse model. (i) A single fork collides with the ICL, and (ii) in a FANCM/MHF dependent manner ‘traverses’ the ICL to continue replication on the other side of the crosslink without unhooking it. The later steps of this pathway are not known, but could involve incisions and TLS for post-replicative repair.
2.1 Dual-fork convergence pathway
The most well defined mechanism has been elucidated by the Walter group using plasmids containing site-specific ICLs with replication-competent Xenopus egg extracts [5]. In this system, two replication forks converge on an ICL and their leading strands pause 20 to 40 nucleotides from the crosslink due to presence of the CMG helicase [6] (Fig. 1A, i). The stalled CMG helicase is removed from the vicinity of the ICL by BRCA1/BARD1 [7], allowing one of the leading strands to proceed to one nucleotide before the ICL. At this point, the Fanconi anemia pathway is activated, resulting in the ubiquitylation of FANCD2-I [8], followed by unhooking of the ICL by incisions involving ERCC1-XPF-SLX4 on the strand opposite the approached replication fork (Fig. 1A, ii) [9, 10]. The unhooking incisions on one parental strand lead to formation of a double-stranded break (DSB) in that daughter duplex, while the unhooked ICL remains attached to the other parental strand (Fig. 1A iii–iv). Translesion synthesis across the unhooked ICL allows the leading strand to be extended past the ICL and eventually be ligated to the downstream lagging strand to generate a template for repair of the DSB by homologous recombination (HR, Fig. 1A, v–vii). The bypassed ICL remnant is no longer very toxic and is eventually likely removed by NER.
2.2. Replication fork ICL traverse pathway
A study by Seidman and colleagues revealed another replication coupled pathway (Fig. 1B). They used an ICL repair-specific fiber assay with fluorescently labeled psoralen ICLs and dual labeling to map the progression of a replication fork around the ICL in mammalian cells [11]. Surprisingly, although dual fork convergence (~20% of the events) was observed, the main pathway observed (~50% of the events) was different. The replication fork was found to ‘traverse’ the ICL without unhooking, leaving an intact ICL behind (Fig. 1B, i–ii). Although the mechanism is unknown, this traverse pathway is believed to involve continuation or re-initiation of replication just past the ICL. These observations suggest that ICLs may also be repaired in a post-replicative manner. Notably, both ICL traverse and the dual fork convergence pathways ultimately lead to an X-shaped structure around the ICL (Figs. 1A ii, 1B ii), which could undergo similar unhooking and ICL bypass steps to complete repair.
2.3. Unhooking in replication-coupled ICL repair
One of the central steps in ICL repair is unhooking, in which incisions around the ICL release it from one of the two crosslinked strands (Fig. 1A, iii–iv, for an excellent review on this topic in DNA Repair see [12]). From the point of view of translesion synthesis across the ICL, a critical concern is where the incisions take place during unhooking, and whether an unhooked ICL is processed further by exonucleases. These factors will determine the structures of intermediates DNA polymerases encounter and, consequently, which polymerase(s) may act on an ICL.
Determining the identity of the nucleases involved in ICL unhooking has been challenging. Genetic deletion of several nucleases, including ERCC1-XPF, MUS81, FAN1, SNM1A and SLX1 renders cells sensitive to ICL forming agents to various degrees [12]. Experiments in Xenopus extracts showed that ERCC1-XPF is essential for unhooking and that this activity requires SLX4 and ubiquitylated FANCD2-I [8, 9]. This suggests a model in which ERCC1-XPF makes the incision on the 3′ side of the by association with SLX4 and possibly other proteins [13]. The identity of the endonuclease making the cut on the other side has not been established. Possible candidates are SLX1 or again XPF, which could be making both cuts [10, 12, 14].
Currently, there is no experimental system available to determine where on the parental strand the unhooking incisions occur and what the length of the duplex surrounds an unhooked ICL in the product of this reaction. Additionally, the unhooked ICL may be further processed by an exonuclease such as SNM1A, which can digest DNA past an ICL [15–17]. SNM1A may also act on intermediates nicked only on one side of the ICL, digesting DNA past the ICL leaving an unhooked lesion behind. Although nothing is known about how ICLs are eventually unhooked in the traverse pathway, it is possible that the X-shaped intermediates are processed similarly by the same group of nucleases (Fig. 1 A&B, ii). MUS81-EME1 and FAN1 are likely to operate in different pathways, perhaps in situations where replication forks stall at some distance from the ICL or where fork regression occurs. As it is unclear how this relates to eventual polymerase activity, this scenario will not be further discussed here.
Multiple in vitro studies have shown that resection of the duplex around an ICL is crucial for translesion synthesis by TLS polymerases, with effective bypass only occurring with ICLs embedded in short (< 6 base pairs) duplexes (see below) [18–22]. Using a synthetic model nitrogen mustard ICL in its most resected form a single crosslinked base a recent study showed that such a fully processed ICL was efficiently bypassed by the bacterial replicative Klenow polymerase, apparently providing only a minimal obstacle [23]. It is therefore tempting to think that some ICLs may are also be bypassed by mammalian replicative polymerases, suggesting that TLS poly unhooked may also be bypassed by mammalian replicative polymerases and that TLS polymerases may not be absolutely required for the repair of all ICLs. Therefore, the degree of processing of unhooked ICLs is crucial for how DNA polymerases interact with them.
3. Replication-independent ICL repair
3.1 Overview
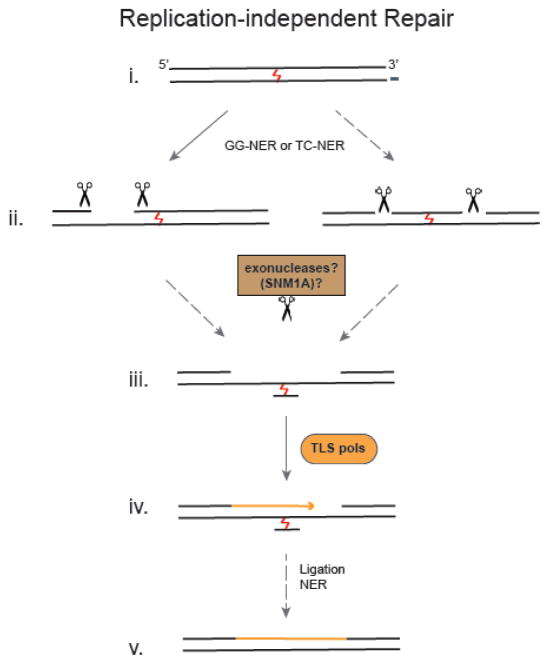
Although ICLs are especially deleterious during replication in S-phase, the replication-independent repair (RIR) of ICLs in G0/G1 is likely to play an important role in post-mitotic cells where endogenous ICLs could block transcription of essential genes. ICL repair in G1 has furthermore been shown to diminish the burden of ICLs before a cell enters S-phase [24]. Much of what we know about RIR ICL repair is based on studies with reporter plasmids harboring a site-specific ICL in mammalian cells [24–28] and Xenopus egg extracts [29, 30]. Nucleotide excision repair (NER) proteins have been shown to be involved in this ICL repair pathway (Fig. 2). Depending on the ICL and assay conditions, both branches of NER, global genome NER (GG-NER) and transcription-coupled NER (TC-NER), (which differ in the damage recognition step, but otherwise share the same set of common factors [31]) were shown to be involved. The recruitment of NER proteins to ICLs has been directly demonstrated in G1 cells, lending support to the studies conducted with reporter plasmids [32]. In addition to NER factors, mismatch repair (MMR) and HMG proteins have been shown to interact with NER proteins at ICLs and may therefore also contribute to replication-independent repair of ICLs [33, 34].
Fig. 2. Model for Replication-Independent ICL Repair.
(i) Global genome NER (GG-NER) as well as transcription coupled NER (TC-NER) proteins are involved in replication independent repair of ICLs. (ii) Although dual incisions 5′ to the ICL have been observed, unhooking incisions on either side of the ICL may also occur. (iii) These intermediates may be further processed by exonucleases like SNM1A to facilitate translesion synthesis. (iv) TLS polymerases carry out gap filling and (v) the ICL remnant is likely removed by NER. Solid arrows indicate details obtained from experimental observations and dashed arrows indicate indirect evidence and/or speculation.
3.2 Unhooking in replication-independent ICL repair
Although the involvement of NER in RIR has been clearly demonstrated, how incisions around the ICL occur is not immediately obvious. NER incisions require the opening of the DNA duplex around the lesion a step that would be blocked by ICLs [35]. Consistent with this idea, it has been shown that NER-proficient extracts incise psoralen and alkyl ICLs with both incisions occurring 5′ to the ICL [36–38] (Fig. 2 ii). As incisions on one side of the ICL do not enable a polymerase to bypass the ICL, another incision on the 3′ side of the ICL would be required to generate an unhooked substrate. Although it is not yet known how this happens, it has been shown that ERCC1-XPF together with RPA [37], and the exonucleases SNM1A and FAN1 can degrade one strand of an ICL-containing duplex [15, 39, 40]. In support of such a scenario, a recent study found CSB and SNM1A to interact at trioxsalen ICLs [41] raising the possibility that SNM1A may be directly recruited to TC-NER complexes for processing of these lesions. Interestingly, NER-independent incisions around an ICL have also been observed, but the factors responsible have not yet been identified [38].
4. TLS polymerases in ICL repair
In the preceding section, we have outlined how the unhooking step is key in determining the nature of substrate encountered by DNA polymerases. Translesion synthesis often involves multiple polymerases, with one carrying out the insertion of a nucleotide across the lesion, and another carrying out further extension [42]. Multiple polymerases have been implicated in the repair of ICLs based on genetic or biochemical studies. The existence of multiple ICL repair pathways, the vastly different structures of (unhooked) ICLs, potential redundancy among DNA polymerases and limited options in studying these pathways at a mechanistic level have made it difficult to definitely assign the roles of various DNA polymerases in ICL repair. The roles of various TLS polymerases in ICL repair has been reviewed previously [43, 44] and we provide an update on the status here with an emphasis on recent results from in vitro studies. We structure our observations based on the types of assays used and outline what the open questions and challenges are.
4.1. Sensitivity of cell lines with TLS polymerase deficiencies to crosslinking agents
Genetic studies involving treatment of cells with crosslinking agents have implicated multiple polymerases in ICL repair (Table 1). A limitation of such experiments is that they do not provide any information about which ICL pathway a polymerase is involved in. Results from such studies are further complicated by the fact that crosslinking agents can also form intrastrand crosslinks, which can have overlapping toxic effects with the ICLs, but are typically addressed by NER, not ICL repair.
TABLE 1.
Known roles of TLS polymerases in ICL repair
| Pol | Sensitivity | Replication dependent | Replication independent | In vitro assays |
|---|---|---|---|---|
| Pol ζ | Cisplatin MMC NM |
Cisplatin | Cisplatin Psoralen MMC |
n/d* |
| REV1 | Cisplatin | Cisplatin | Cisplatin Psoralen MMC |
n/d* |
| Pol η | Cisplatin Psoralen |
n/a | Psoralen MMC |
Cisplatin NM-like Acrolein-like** AP ICL*** |
| Pol κ | MMC | n/a | MMC Acrolein-like** |
Cisplatin NM-like Acrolein-like** |
| Pol ν | MMC Cisplatin |
n/a | n/a | N6-N6A (major groove) |
n/d : Current biochemical data is not consistent with known role in ICL repair
Acrolein-like: N2-N2 dG ICL
AP ICL: Oxidized abasic site ICL
Nonetheless, such assays have clearly revealed that deletion of the REV1 and REV3 genes leads to a clear hypersensitivity to cisplatin and mitomycin C exposure [44–49]. REV3 together with REV7 and the accessory subunits POLD2 and POLD3 constitutes Pol ζ, a B-family polymerase [50, 51], which is believed to frequently work together with REV1, a Y-family polymerase with dCMP transferase activity. The role of REV1 and Pol ζ seems to be important for both replication-dependent and -independent ICL repair, explaining the exquisite sensitivity of REV1 and REV3 deficient cell lines to crosslinking agents [5, 24, 28, 45, 46, 52].
The involvement of other TLS polymerases is less unequivocal. The main role of Pol η, deficient in XP-V (xeroderma pigmentosum-variant) cells, is to accurately bypass UV-lesions during replication [50]. Its open yet rigid active site also allows for the insertion of dNTPs opposite cisplatin intrastrand crosslinks [53, 54], and this active site architecture may be suitable for the bypass of certain ICLs. Pol η deficient cells are indeed sensitive to crosslinking agents such as cisplatin or psoralen [55–58], but this sensitivity is less pronounced than that for REV1 or REV3 [45], suggesting a less central role of Pol η in ICL repair.
Another Y-family TLS polymerase, Pol κ, is believed to be especially important for the bypass of minor groove DNA adducts [50]. Consistent with this property, Pol κ−/− cells are sensitive to exposure to MMC, which forms ICLs in the minor groove [18, 30, 59, 60]. Interestingly, Pol κ −/− cells were found to be more sensitive to both MMC and cisplatin in the G0/G1 phase of the cell cycle, suggesting a more important role for Pol κ in replication-independent repair processes [30].
The last polymerase that appears to be involved in the repair of ICLs based on cellular sensitivity is Pol ν. Knock down of Pol ν renders cells hypersensitive to MMC and cisplatin [19, 61], although the main activity of the enzyme in vitro seems to be on major groove ICLs [20] (see below). Apart from a role in translesion synthesis, Pol ν could also be functioning at later stages of ICL repair, such as homologous recombination, as depletion of Pol ν sensitized cells to DSB forming agents and also leads to reduced rates of homologous recombination [61].
4.2. Roles of TLS polymerases in ICL repair derived from functional assays
The information on the role of DNA polymerases in ICL repair based on functional ICL repair assays is rather limited, relying mainly on studies in Xenopus extracts for replication-dependent repair and on plasmid reporter assays for replication-independent repair (Table 1). The results from these studies have provided some initial valuable information about the involvement of TLS polymerases in ICL repair.
Experiments in replication-competent Xenopus egg extracts have shown that REV1 and Pol ζ are required for extension (but not insertion) of the leading strand past a cisplatin ICL, while they are not essential for repair of non-distorting nitrogen mustard-like ICLs [5, 62]. Pol ζ has a well-known role as an extension polymerase, with the ability to efficiently extend mismatched primer termini of insertion products of a variety of lesions [47, 51, 63], while REV1 is known to serve as a hub protein to coordinate the activities of multiple TLS polymerases [64, 65]. The unique roles of these two TLS enzymes are likely also critical for ICL repair. The role of other TLS polymerases in replication-dependent ICL repair, particularly in insertion opposite the ICL, remains to be elucidated. Due to the possible redundancy of polymerases in some of the steps, dissecting the role of each polymerase has not been straightforward. Interestingly, the repair of ICLs has been found to exhibit a mutagenicity rate of a few percent, with various mutations clustered around the site of the ICL [62]. Depletion of REV1 reduced this mutation rate, consistent with a role for TLS in lesion-induced mutagenesis.
The requirement of polymerases for replication-independent repair has been mainly determined by reporter plasmid systems and has revealed that the involvement of polymerases, at least in part, is dependent on the structure of the ICL. REV1 and Pol ζ were found to be required for replication-independent repair of cisplatin, psoralen and MMC ICLs in mammalian cells and nitrogen mustard ICLs in yeast [24, 28, 52], while Pol η was involved in the replication-independent repair of psoralen and MMC, but not cisplatin ICLs [24–26]. A definitive role for other TLS polymerases has not yet been established in this system. In a replication-independent in vitro system in Xenopus extracts, the repair of a minor groove acrolein-like N2-N2 trimethylene crosslink was specifically dependent on Pol κ [30]. Immunodepletion of Pol κ greatly diminished repair efficiency of an ICL-containing plasmid, whereas depletion of Pol ζ did not have any effect. While these studies have shed some light on how different TLS polymerases are required for ICLs with different structures, it remains to be seen what influence the different assay systems and organisms used have on these results.
4.3 Biochemical activity of TLS polymerases on ICL substrates
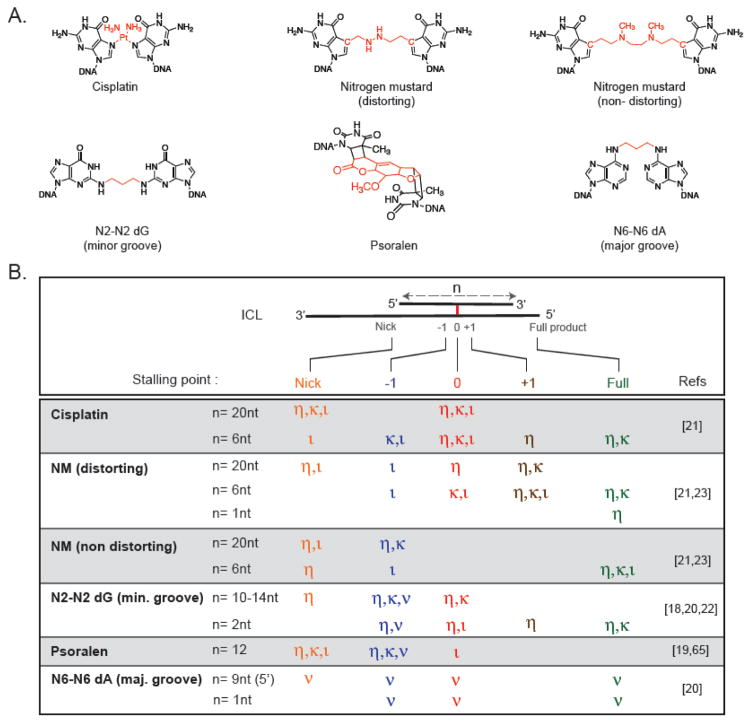
Biochemical assays using defined ICL substrates and purified DNA polymerases by contrast have provided more defined answers (Table 1). Although translesion synthesis can be studied at a single nucleotide resolution in this way, one limitation has been to design ICL substrates that are physiologically relevant. Given that the exact structure of unhooked ICLs that TLS polymerases encounter in cells are not known, a variety of different types of ICLs have been used. While some degree of selectivity for certain ICL structures was to be expected for various polymerases based on their substrate specificities for lesions on one strand of DNA, a key observation first made by Lloyd and coworkers was that the amount of duplex surrounding an ICL also dramatically affects the efficiency of bypass reaction [18]. While the bypass reactions are almost always completely inhibited by an ICL embedded in a long stable duplex, a certain amount of bypass is often possible if duplex is shortened to a few nucleotides [18, 20–22]. These findings highlight the importance of the unhooking step in ICL repair for polymerase bypass, as the position of the endonucleolytic incisions and possible further processing by exonucleases will determine what the unhooked ICL looks like when the polymerase encounters it. Biochemical assays have used a variety of unhooked ICL structures (Fig. 3), and while the diversity of ICL structures, polymerases and assay conditions used makes it challenging to compare the different studies, a number of general conclusions can be reached:
Fig. 3. Biochemical Activity of TLS Polymerases on ICL Substrates.
A: Chemical Structures of ICLs. The crosslinks between bases are highlighted in red. B: Primer Extension Activity of TLS Polymerases η,κ,ν and ι across diverse ICL substrates with varying amount of duplex (n) surrounding the crosslink. For polymerases that were tested with the substrates shown the main stalling points at the start of the duplex (nick, orange), 1 nt before the ICL (−1, blue), at the ICL (0, red), 1 nt after the ICL (+1, brown) as well as complete extension to full product (Full, green) are indicated for each ICL substrate. Although conditions used to generate the data listed in this figure varied greatly, ICLs embedded in a longer duplex were not bypassed efficiently by TLS polymerases.
Multiple TLS polymerases have the ability to insert dNTPs opposite ICLs and extend them to full length products, even if their involvement in ICL repair is not clear from genetic or functional studies. This could explain why deficiency in individual TLS polymerases does not always lead to hypersensitivity of cells to crosslinking agents.
The amount of duplex surrounding an ICL is a key parameter for the efficiency of insertion and extension. While some enzymes can insert a dNTP opposite an ICL in a long (12–20bp) duplex, none can extend these substrates to a full length product. By contrast, ICLs in a short duplex (2–6 bps) can be extended to full length product by a number of TLS polymerases. Intriguingly, a model for a fully unhooked and processed ICL (to a single nucleotide) was bypassed almost efficiently as non-damaged DNA by at least Pol η [23].
The structure of an ICL greatly influences the activity of TLS polymerases. In cases where insertion and extension is possible, polymerases approach helix-distorting ICLs more easily than non-distorting ones (likely because of ease of strand displacement), but hinder extension as multiple polymerases stall at or within a few bases past the ICLs. By contrast, for non-distorting ICLs, the approach is more challenging, while insertion and bypass occur more readily.
An overview of results obtained from polymerase studies with ICL-containing substrates is shown in Fig. 3 and the most important findings for each polymerase are summarized below.
REV1/Pol ζ
Several studies have investigated REV1/Pol ζ activity on ICLs, mostly using proteins purified from S. cerevisiae. Surprisingly, while experiments with purified REV1 or Pol ζ did reveal some insertion of dNTPs opposite ICLs, no extension or cooperation of the two enzymes was observed [21, 66]. Given the clear importance of REV1 and Pol ζ in ICL repair (see above), it is possible that proper in vitro activity might require the two additional subunits PolD2 and PolD3. The human four subunit Pol ζ complex was found to be more active in translesion synthesis across cisplatin intrastrand lesions [51] and testing the activity of this complex will likely be required to reveal the ability of Pol ζ to bypass ICLs.
Pol η
Pol η is the enzyme that has been most extensively studied with ICLs and is able to carry out insertion and in many cases extension across a diverse set of major and minor groove ICLs - including cisplatin, nitrogen mustard, acrolein mimics and ICLs formed at abasic sites (Fig. 3) [21, 22, 67]. Interestingly, the pattern of bypass by Pol η is similar for various ICLs, stalling predominantly at 0, +1 and +2 positions and the structure-function relationships mentioned above apply in particular to studies with Pol η. Interestingly, no stalling was observed at all when the duplex around an ICL is reduced to a single crosslinked nucleotide, emphasizing the dramatic influence of the unhooking step on polymerase activity on ICLs [23]. These observations are consistent with the structure of Pol η, which has the largest active site among Y-family polymerases and can therefore accommodate a variety of lesions [53]. The rigid molecular splint guiding the primer-template strands in the active of Pol η may also help the polymerase reaction in the presence of a bulky unhooked ICL.
Pol κ
Pol κ can bypass diverse ICL structures in vitro including cisplatin, nitrogen mustard and acrolein ICLs and, consistent with genetic findings, is particularly effective in bypassing the minor groove acrolein lesions. [18, 21].
Pol ν
Pol ν is proficient in bypassing a variety of major groove ICLs, but not minor groove lesions or psoralen ICLs [19, 20]. While the biological importance of Pol ν in ICL repair is still elusive it might be a complement to the preference of Pol κ for minor groove adducts.
Pol ι
Pol ι was able to insert a base across psoralen, cisplatin and N2-dG ICLs derived from acrolein, but could only carry out an extension reaction on the non-distorting nitrogen mustard like ICL. [21, 22, 68]. Therefore, Pol ι could act as an insertion polymerase for some ICLs, although its in vivo role in ICL repair remains to be demonstrated.
5. Recruitment of TLS polymerases to ICL repair pathways
The activity of TLS polymerases needs to be a highly regulated process, as these enzymes are much more error prone than replicative polymerases. The mechanisms by which the activity of TLS polymerases is regulated at lesions on one strand of DNA has been studied in some detail. Uncoupling of the helicase and replicative polymerase at ssDNA lesions leads to long stretches of ssDNA, which are covered by RPA and lead to the activation of the E2–E3 ubiquitin ligase RAD6-RAD18 and PCNA ubiquitination [69]. This mono-ubiquitination of PCNA at Lys164 leads to recruitment of TLS polymerases which interact with ubiquitinated PCNA via their PCNA and ubiquitin binding motifs [50]. An important difference in replication-coupled ICL repair is that both the helicase and polymerases stall at ICLs, preventing their uncoupling and precluding the formation of long ssDNA stretches. Consistent with this difference, it has been shown that damaging agents like UV (which mostly form intrastrand lesions) elicited strong PCNA monoubiquitination, whereas MMC treatment (which mostly forms ICLs) did not [70]. Furthermore, REV1 was recruited to cellular foci induced by MMC independently of PCNA ubiquitination.
So how are TLS polymerases recruited to sites of ICL repair? An obvious candidate would be ubiquitinated FANCD2/FANCI, which is required for the incision and TLS steps in Xenopus egg extracts. However, it has been shown that while mutation rates are reduced in cells with deficiencies in the FA core complex, they are increased in FANCD2-deficient cells, suggesting that the core complex, but not FANCD2 promotes TLS [71]. More recent evidence suggests that the FA core complex directly interacts with REV1 [72, 73]. These findings are also supported by functional studies in Xenopus egg extracts, where the depletion of FANCA led to a reduction in REV1 binding to ICLs, while FANCD2-I depletion did not [62]. Given that REV1 interacts with the other Y-family polymerases [65, 74] this recruitment could then facilitate the formation of a TLS complex involving multiple polymerases at the site of the lesion.
The situation is likely to be more complex however. For example, unlike REV1, Pol η does not appear to be regulated by FA in response to ICLs [70, 72]. It has recently been shown that Pol η interacts with the Pol δ subunit POLD2 [75]. Furthermore, Pol ζ has also been shown to share subunits with Pol δ and this interaction enhances the TLS activity of Pol ζ [51, 76]. It is therefore possible that these connections with a replicative polymerase may also facilitate TLS activity in ICL repair. The regulation of the activity of TLS polymerases during ICL repair therefore appears to be complex and the elucidation of the underlying mechanisms will require many additional studies.
6. TLS polymerases in chemotherapy
ICL-inducing agents are widely used in anti-cancer therapy, however clinical efficacy is limited by the development of resistance and well as secondary tumors [3]. Error-prone translesion synthesis during ICL repair is a key step contributing to resistance as well as increased therapy-induced mutation load and has been implicated in the emergence of secondary tumors. Studies have shown that knock-down of REV3 and REV1 not only lead to increased cisplatin and cyclophosphamide sensitivity in experimental tumor models, but also led to a significant reduction in drug-induced mutagenesis and hence a reduction in acquired resistance and possibly induction of secondary malignancies [77, 78].
Consistent with this, increased expression of TLS polymerases in multiple cancers has been correlated with a poor prognosis and response to chemotherapy. Elevated levels of Pol η and Pol ζ were correlated with resistance to cisplatin treatment in ovarian cancer stem cells and cervical cancer cells, respectively [79, 80]. Pol η levels were also found to be elevated in head and neck squamous cell carcinomas and lower Pol η level was significantly correlated with better response to cisplatin and gemcitabine therapy in patients [81]. Altogether, these studies suggest TLS polymerase levels could be a useful predictor for therapeutic outcomes and that inhibition of TLS polymerase in cancer therapy could lead to a dual benefit reduced occurrence of resistance and reduced secondary tumor formation. A more detailed understanding of the roles of individual TLS polymerases in the contribution to ICL repair to specific agents will be an important guide to determine which polymerases may be specifically targeted for treatment modalities involving a variety of crosslinking agents.
Acknowledgments
This work was supported by NCI grant R01 CA165911 to O.D.S.
Abbreviations
- DSB
DNA double strand break
- ICL
DNA interstrand crosslink
- NER
nucleotide excision repair
- RIR
replication-independent repair
- TLS
translesion synthesis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Clauson C, Schärer OD, Niedernhofer L. Advances in understanding the complex mechanisms of DNA interstrand cross-link repair. Cold Spring Harb Perspect Biol. 2013;5:a012732. doi: 10.1101/cshperspect.a012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer. 2007;7:573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 3.Deans AJ, West SC. DNA interstrand crosslink repair and cancer. Nat Rev Cancer. 2011;11:467–480. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kottemann MC, Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493:356–363. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Räschle M, Knipscheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Schärer OD, Walter JC. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134:969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J, Dewar JM, Budzowska M, Motnenko A, Cohn MA, Walter JC. DNA interstrand cross-link repair requires replication-fork convergence. Nat Struct Mol Biol. 2015;22:242–247. doi: 10.1038/nsmb.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Long DT, Joukov V, Budzowska M, Walter JC. BRCA1 promotes unloading of the CMG helicase from a stalled DNA replication fork. Mol Cell. 2014;56:174–185. doi: 10.1016/j.molcel.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knipscheer P, Räschle M, Smogorzewska A, Enoiu M, Ho TV, Schärer OD, Elledge SJ, Walter JC. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326:1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein Douwel D, Boonen RA, Long DT, Szypowska AA, Raschle M, Walter JC, Knipscheer P. XPF-ERCC1 acts in Unhooking DNA interstrand crosslinks in cooperation with FANCD2 and FANCP/SLX4. Mol Cell. 2014;54:460–471. doi: 10.1016/j.molcel.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hodskinson MR, Silhan J, Crossan GP, Garaycoechea JI, Mukherjee S, Johnson CM, Schärer OD, Patel KJ. Mouse SLX4 is a tumor suppressor that stimulates the activity of the nuclease XPF-ERCC1 in DNA crosslink repair. Mol Cell. 2014;54:472–484. doi: 10.1016/j.molcel.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang J, Liu S, Bellani MA, Thazhathveetil AK, Ling C, de Winter JP, Wang Y, Wang W, Seidman MM. The DNA translocase FANCM/MHF promotes replication traverse of DNA interstrand crosslinks. Mol Cell. 2013;52:434–446. doi: 10.1016/j.molcel.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, Walter JC. Mechanism and regulation of incisions during DNA interstrand cross-link repair. DNA Repair (Amst) 2014;19:135–142. doi: 10.1016/j.dnarep.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manandhar M, Boulware KS, Wood RD. The ERCC1 and ERCC4 (XPF) genes and gene products. Gene. 2015;569:153–161. doi: 10.1016/j.gene.2015.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuraoka I, Kobertz WR, Ariza RR, Biggerstaff M, Essigmann JM, Wood RD. Repair of an interstrand DNA crosslink initiated by ERCC1-XPF repair/recombination nuclease. J Biol Chem. 2000;275:26632–26636. doi: 10.1074/jbc.C000337200. [DOI] [PubMed] [Google Scholar]
- 15.Wang AT, Sengerová B, Cattell E, Inagawa T, Hartley JM, Kiakos K, Burgess-Brown NA, Swift LP, Enzlin JH, Schofield CJ, Gileadi O, Hartley JA, McHugh PJ. Human SNM1A and XPF ERCC1 collaborate to initiate DNA interstrand cross-link repair. Genes Dev. 2011;25:1859–1870. doi: 10.1101/gad.15699211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allerston CK, Lee SY, Newman JA, Schofield CJ, McHugh PJ, Gileadi O. The structures of the SNM1A and SNM1B/Apollo nuclease domains reveal a potential basis for their distinct DNA processing activities. Nucleic Acids Res. 2015;43:11047–11060. doi: 10.1093/nar/gkv1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sengerová B, Allerston CK, Abu M, Lee SY, Hartley J, Kiakos K, Schofield CJ, Hartley JA, Gileadi O, McHugh PJ. Characterization of the human SNM1A and SNM1B/Apollo DNA repair exonucleases. J Biol Chem. 2012;287:26254–26267. doi: 10.1074/jbc.M112.367243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minko IG, Harbut MB, Kozekov ID, Kozekova A, Jakobs PM, Olson SB, Moses RE, Harris TM, Rizzo CJ, Lloyd RS. Role for DNA polymerase κ in the processing of N2-N2-guanine interstrand cross-links. J Biol Chem. 2008;283:17075–17082. doi: 10.1074/jbc.M801238200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zietlow L, Smith LA, Bessho M, Bessho T. Evidence for the involvement of human DNA polymerase N in the repair of DNA interstrand cross-links. Biochemistry. 2009;48:11817–11824. doi: 10.1021/bi9015346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamanaka K, Minko IG, Takata K-i, Kolbanovskiy A, Kozekov ID, Wood RD, Rizzo CJ, Lloyd RS. Novel enzymatic function of DNA polymerase ν in translesion DNA synthesis past major groove DNA–peptide and DNA–DNA cross-links. Chem Res Tox. 2010;23:689–695. doi: 10.1021/tx900449u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho TV, Guainazzi A, Derkunt SB, Enoiu M, Schärer OD. Structure-dependent bypass of DNA interstrand crosslinks by translesion synthesis polymerases. Nucleic Acids Res. 2011;39:7455–7464. doi: 10.1093/nar/gkr448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klug AR, Harbut MB, Lloyd RS, Minko IG. Replication bypass of N2-deoxyguanosine interstrand cross-links by human DNA polymerases η and ι. Chem Res Tox. 2012;25:755–762. doi: 10.1021/tx300011w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roy U, Mukherjee S, Sharma A, Frank EG, Schärer OD. The Structure and Duplex Context of DNA Interstrand Crosslinks Affects the Activity of DNA Polymerase η. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Enoiu M, Jiricny J, Schärer OD. Repair of cisplatin-induced DNA interstrand crosslinks by a replication-independent pathway involving transcription-coupled repair and translesion synthesis. Nucleic Acids Res. 2012;40:8953–8964. doi: 10.1093/nar/gks670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Peterson CA, Zheng H, Nairn RS, Legerski RJ, Li L. Involvement of nucleotide excision repair in a recombination-independent and error-prone pathway of DNA interstrand cross-link repair. Mol Cell Biol. 2001;21:713–720. doi: 10.1128/MCB.21.3.713-720.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng H, Wang X, Warren AJ, Legerski RJ, Nairn RS, Hamilton JW, Li L. Nucleotide excision repair- and polymerase η-mediated error-prone removal of mitomycin C interstrand cross-links. Mol Cell Biol. 2003;23:754–761. doi: 10.1128/MCB.23.2.754-761.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hlavin EM, Smeaton MB, Noronha AM, Wilds CJ, Miller PS. Cross-link structure affects replication-independent DNA interstrand cross-link repair in mammalian cells. Biochemistry. 2010;49:3977–3988. doi: 10.1021/bi902169q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen X, Jun S, O’Neal LE, Sonoda E, Bemark M, Sale JE, Li L. REV3 and REV1 play major roles in recombination-independent repair of DNA interstrand cross-links mediated by monoubiquitinated proliferating cell nuclear antigen (PCNA) J Biol Chem. 2006;281:13869–13872. doi: 10.1074/jbc.C600071200. [DOI] [PubMed] [Google Scholar]
- 29.Lu X, Zhang N, Vasquez K, Barton M, Legerski R. Repair of psoralen interstrand cross-links in Xenopus laevis egg extracts is highly mutagenic. Biochem Biophys Res Commun. 2005;336:69–75. doi: 10.1016/j.bbrc.2005.08.042. [DOI] [PubMed] [Google Scholar]
- 30.Williams Hannah L, Gottesman Max E, Gautier J. Replication-independent repair of DNA interstrand crosslinks. Mol Cell. 2012;47:140–147. doi: 10.1016/j.molcel.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JH. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol. 2014;15:465–481. doi: 10.1038/nrm3822. [DOI] [PubMed] [Google Scholar]
- 32.Muniandy PA, Thapa D, Thazhathveetil AK, Liu S-t, Seidman MM. Repair of laser-localized DNA interstrand cross-links in G1 phase mammalian cells. J Biol Chem. 2009;284:27908–27917. doi: 10.1074/jbc.M109.029025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao J, Jain A, Iyer RR, Modrich PL, Vasquez KM. Mismatch repair and nucleotide excision repair proteins cooperate in the recognition of DNA interstrand crosslinks. Nucleic Acids Res. 2009;37:4420–4429. doi: 10.1093/nar/gkp399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mukherjee A, Vasquez KM. HMGB1 interacts with XPA to facilitate the processing of DNA interstrand crosslinks in human cells. Nucleic Acids Res. 2016;44:1151–1160. doi: 10.1093/nar/gkv1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schärer OD. Nucleotide excision repair in eukaryotes. Cold Spring Harb Perspect Biol. 2013;5:a012609. doi: 10.1101/cshperspect.a012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bessho T, Mu D, Sancar A. Initiation of DNA interstrand cross-link repair in humans: the nucleotide excision repair system makes dual incisions 5′ to the cross-linked base and removes a 22- to 28-nucleotide-long damage-free strand. Mol and Cell Biol. 1997;17:6822–6830. doi: 10.1128/mcb.17.12.6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mu D, Bessho T, Nechev LV, Chen DJ, Harris TM, Hearst JE, Sancar A. DNA interstrand cross-links induce futile repair synthesis in mammalian cell extracts. Mol Cell Biol. 2000;20:2446–2454. doi: 10.1128/mcb.20.7.2446-2454.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smeaton MB, Hlavin EM, McGregor Mason T, Noronha AM, Wilds CJ, Miller PS. Distortion-dependent unhooking of interstrand cross-links in mammalian cell extracts. Biochemistry. 2008;47:9920–9930. doi: 10.1021/bi800925e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang R, Persky NS, Yoo B, Ouerfelli O, Smogorzewska A, Elledge SJ, Pavletich NP. DNA repair. Mechanism of DNA interstrand cross-link processing by repair nuclease FAN1. Science. 2014;346:1127–1130. doi: 10.1126/science.1258973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pizzolato J, Mukherjee S, Schärer OD, Jiricny J. FANCD2-associated nuclease 1, but not exonuclease 1 or flap endonuclease 1, is able to unhook DNA interstrand cross-links in vitro. J Biol Chem. 2015;290:22602–22611. doi: 10.1074/jbc.M115.663666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iyama T, Lee SY, Berquist BR, Gileadi O, Bohr VA, Seidman MM, McHugh PJ, Wilson DM., 3rd CSB interacts with SNM1A and promotes DNA interstrand crosslink processing. Nucleic Acids Res. 2015;43:247–258. doi: 10.1093/nar/gku1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shachar S, Ziv O, Avkin S, Adar S, Wittschieben J, Reissner T, Chaney S, Friedberg EC, Wang Z, Carell T, Geacintov N, Livneh Z. Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J. 2009;28:383–393. doi: 10.1038/emboj.2008.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ho TV, Schärer OD. Translesion DNA synthesis polymerases in DNA interstrand crosslink repair. Environ Mol Mutagen. 2010;51:552–566. doi: 10.1002/em.20573. [DOI] [PubMed] [Google Scholar]
- 44.Sharma S, Canman CE. REV1 and DNA polymerase zeta in DNA interstrand crosslink repair. Environ Mol Mutagen. 2012;53:725–740. doi: 10.1002/em.21736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nojima K, Hochegger H, Saberi A, Fukushima T, Kikuchi K, Yoshimura M, Orelli BJ, Bishop DK, Hirano S, Ohzeki M, Ishiai M, Yamamoto K, Takata M, Arakawa H, Buerstedde JM, Yamazoe M, Kawamoto T, Araki K, Takahashi JA, Hashimoto N, Takeda S, Sonoda E. Multiple repair pathways mediate tolerance to chemotherapeutic cross-linking agents in vertebrate cells. Cancer Res. 2005;65:11704–11711. doi: 10.1158/0008-5472.CAN-05-1214. [DOI] [PubMed] [Google Scholar]
- 46.Sonoda E, Okada T, Zhao GY, Tateishi S, Araki K, Yamaizumi M, Yagi T, Verkaik NS, van Gent DC, Takata M, Takeda S. Multiple roles of Rev3, the catalytic subunit of pol[zeta] in maintaining genome stability in vertebrates. EMBO J. 2003;22:3188–3197. doi: 10.1093/emboj/cdg308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gan GN, Wittschieben JP, Wittschieben BO, Wood RD. DNA polymerase zeta (pol [zeta]) in higher eukaryotes. Cell Res. 2008;18:174–183. doi: 10.1038/cr.2007.117. [DOI] [PubMed] [Google Scholar]
- 48.Okada T, Sonoda E, Yoshimura M, Kawano Y, Saya H, Kohzaki M, Takeda S. Multiple roles of vertebrate REV genes in DNA repair and recombination. Mol Cell Biol. 2005;25:6103–6111. doi: 10.1128/MCB.25.14.6103-6111.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simpson LJ, Sale JE. Rev1 is essential for DNA damage tolerance and non-templated immunoglobulin gene mutation in a vertebrate cell line. EMBO J. 2003;22:1654–1664. doi: 10.1093/emboj/cdg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol. 2012;13:141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee YS, Gregory MT, Yang W. Human Pol zeta purified with accessory subunits is active in translesion DNA synthesis and complements Pol eta in cisplatin bypass. Proc Natl Acad Sci USA. 2014;111:2954–2959. doi: 10.1073/pnas.1324001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sarkar S, Davies AA, Ulrich HD, McHugh PJ. DNA interstrand crosslink repair during G1 involves nucleotide excision repair and DNA polymerase zeta. EMBO J. 2006;25:1285–1294. doi: 10.1038/sj.emboj.7600993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Biertumpfel C, Zhao Y, Kondo Y, Ramon-Maiques S, Gregory M, Lee JY, Masutani C, Lehmann AR, Hanaoka F, Yang W. Structure and mechanism of human DNA polymerase eta. Nature. 2010;465:1044–1048. doi: 10.1038/nature09196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao Y, Biertümpfel C, Gregory MT, Hua YJ, Hanaoka F, Yang W. Structural basis of human DNA polymerase eta-mediated chemoresistance to cisplatin. Proc Natl Acad Sci USA. 2012;109:7269–7274. doi: 10.1073/pnas.1202681109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Misra RR, Vos JM. Defective replication of psoralen adducts detected at the gene-specific level in xeroderma pigmentosum variant cells. Molecular and Cellular Biology. 1993;13:1002–1012. doi: 10.1128/mcb.13.2.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Albertella MR, Green CM, Lehmann AR, O’Connor MJ. A role for polymerase η in the cellular tolerance to cisplatin-induced damage. Cancer Res. 2005;65:9799–9806. doi: 10.1158/0008-5472.CAN-05-1095. [DOI] [PubMed] [Google Scholar]
- 57.Chen Y-w, Cleaver JE, Hanaoka F, Chang C-f, Chou K-m. A novel role of DNA polymerase η in modulating cellular sensitivity to chemotherapeutic agents. Molecular Cancer Research. 2006;4:257–265. doi: 10.1158/1541-7786.MCR-05-0118. [DOI] [PubMed] [Google Scholar]
- 58.Mogi S, Butcher CE, Oh DH. DNA polymerase η reduces the γ-H2AX response to psoralen interstrand crosslinks in human cells. Experimental Cell Res. 2008;314:887–895. doi: 10.1016/j.yexcr.2007.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ogi T, Shinkai Y, Tanaka K, Ohmori H. Polkappa protects mammalian cells against the lethal and mutagenic effects of benzo[a]pyrene. Proc Natl Acad Sci USA. 2002;99:15548–15553. doi: 10.1073/pnas.222377899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takeiri A, Wada NA, Motoyama S, Matsuzaki K, Tateishi H, Matsumoto K, Niimi N, Sassa A, Gruz P, Masumura K, Yamada M, Mishima M, Jishage K, Nohmi T. In vivo evidence that DNA polymerase kappa is responsible for error-free bypass across DNA cross-links induced by mitomycin C. DNA Repair (Amst) 2014;24:113–121. doi: 10.1016/j.dnarep.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 61.Moldovan GL, Madhavan MV, Mirchandani KD, McCaffrey RM, Vinciguerra P, D’Andrea AD. DNA polymerase POLN participates in cross-link repair and homologous recombination. Mol Cell Biol. 2010;30:1088–1096. doi: 10.1128/MCB.01124-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Budzowska M, Graham TG, Sobeck A, Waga S, Walter JC. Regulation of the Rev1-pol zeta complex during bypass of a DNA interstrand cross-link. EMBO J. 2015;34:1971–1985. doi: 10.15252/embj.201490878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haracska L, Prakash S, Prakash L. Yeast DNA polymerase zeta is an efficient extender of primer ends opposite from 7,8-dihydro-8-Oxoguanine and O6-methylguanine. Mol Cell Biol. 2003;23:1453–1459. doi: 10.1128/MCB.23.4.1453-1459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sharma S, Helchowski CM, Canman CE. The roles of DNA polymerase ζ and the Y family DNA polymerases in promoting or preventing genome instability. Mutation Res. 2013;743–744:97–110. doi: 10.1016/j.mrfmmm.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wojtaszek J, Lee CJ, D’Souza S, Minesinger B, Kim H, D’Andrea AD, Walker GC, Zhou P. Structural basis of Rev1-mediated assembly of a quaternary vertebrate translesion polymerase complex consisting of Rev1, heterodimeric polymerase (Pol) zeta, and Pol kappa. J Biol Chem. 2012;287:33836–33846. doi: 10.1074/jbc.M112.394841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Minko IG, Yamanaka K, Kozekov ID, Kozekova A, Indiani C, O’Donnell ME, Jiang Q, Goodman MF, Rizzo CJ, Lloyd RS. Replication bypass of the acrolein-mediated deoxyguanine DNA-peptide cross-links by DNA polymerases of the DinB family. Chem Res Tox. 2008;21:1983–1990. doi: 10.1021/tx800174a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu W, Ouellette A, Ghosh S, O’Neill TC, Greenberg MM, Zhao L. Mutagenic Bypass of an Oxidized Abasic Lesion-Induced DNA Interstrand Cross-Link Analogue by Human Translesion Synthesis DNA Polymerases. Biochemistry. 2015;54:7409–7422. doi: 10.1021/acs.biochem.5b01027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith LA, Makarova AV, Samson L, Thiesen KE, Dhar A, Bessho T. Bypass of a psoralen DNA interstrand cross-link by DNA polymerases beta, iota, and kappa in vitro. Biochemistry. 2012;51:8931–8938. doi: 10.1021/bi3008565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Davies AA, Huttner D, Daigaku Y, Chen S, Ulrich HD. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein A. Mol Cell. 2008;29:625–636. doi: 10.1016/j.molcel.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hicks JK, Chute CL, Paulsen MT, Ragland RL, Howlett NG, Gueranger Q, Glover TW, Canman CE. Differential roles for DNA polymerases eta, zeta, and REV1 in lesion bypass of intrastrand versus interstrand DNA cross-links. Mol Cell Biol. 2010;30:1217–1230. doi: 10.1128/MCB.00993-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Niedzwiedz W, Mosedale G, Johnson M, Ong CY, Pace P, Patel KJ. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol Cell. 2004;15:607–620. doi: 10.1016/j.molcel.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 72.Kim H, Yang K, Dejsuphong D, D’Andrea AD. Regulation of Rev1 by the Fanconi anemia core complex. Nat Struct Mol Biol. 2012;19:164–170. doi: 10.1038/nsmb.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mirchandani KD, McCaffrey RM, D’Andrea AD. The Fanconi anemia core complex is required for efficient point mutagenesis and Rev1 foci assembly. DNA Repair (Amst) 2008;7:902–911. doi: 10.1016/j.dnarep.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guo C, Fischhaber PL, Luk-Paszyc MJ, Masuda Y, Zhou J, Kamiya K, Kisker C, Friedberg EC. Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. EMBO J. 2003;22:6621–6630. doi: 10.1093/emboj/cdg626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baldeck N, Janel-Bintz R, Wagner J, Tissier A, Fuchs RP, Burkovics P, Haracska L, Despras E, Bichara M, Chatton B, Cordonnier AM. FF483–484 motif of human Poleta mediates its interaction with the POLD2 subunit of Poldelta and contributes to DNA damage tolerance. Nucleic Acids Res. 2015;43:2116–2125. doi: 10.1093/nar/gkv076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baranovskiy AG, Lada AG, Siebler HM, Zhang Y, Pavlov YI, Tahirov TH. DNA polymerase delta and zeta switch by sharing accessory subunits of DNA polymerase delta. J Biol Chem. 2012;287:17281–17287. doi: 10.1074/jbc.M112.351122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Doles J, Oliver TG, Cameron ER, Hsu G, Jacks T, Walker GC, Hemann MT. Suppression of Rev3, the catalytic subunit of Pol{zeta}, sensitizes drug-resistant lung tumors to chemotherapy. Proc Natl Acad Sci USA. 2010;107:20786–20791. doi: 10.1073/pnas.1011409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xie K, Doles J, Hemann MT, Walker GC. Error-prone translesion synthesis mediates acquired chemoresistance. Proc Natl Acad Sci USA. 2010;107:20792–20797. doi: 10.1073/pnas.1011412107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Srivastava AK, Han C, Zhao R, Cui T, Dai Y, Mao C, Zhao W, Zhang X, Yu J, Wang QE. Enhanced expression of DNA polymerase eta contributes to cisplatin resistance of ovarian cancer stem cells. Proc Natl Acad Sci USA. 2015;112:4411–4416. doi: 10.1073/pnas.1421365112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang L, Shi T, Liu F, Ren C, Wang Z, Li Y, Tu X, Yang G, Cheng X. REV3L, a promising target in regulating the chemosensitivity of cervical cancer cells. PLoS One. 2015;10:e0120334. doi: 10.1371/journal.pone.0120334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhou W, Chen YW, Liu X, Chu P, Loria S, Wang Y, Yen Y, Chou KM. Expression of DNA translesion synthesis polymerase eta in head and neck squamous cell cancer predicts resistance to gemcitabine and cisplatin-based chemotherapy. PLoS One. 2013;8:e83978. doi: 10.1371/journal.pone.0083978. [DOI] [PMC free article] [PubMed] [Google Scholar]