SUMMARY
Multidrug transporters are ubiquitous efflux pumps that provide cells with defense against various toxic compounds. In bacteria, which typically harbor numerous multidrug transporter genes, the majority function as secondary multidrug/proton antiporters. Proton-coupled secondary transport is a fundamental process that is not fully understood, largely owing to the obscure nature of proton-transporter interactions. Here we analyzed the substrate/proton coupling mechanism in MdfA, a model multidrug/ proton antiporter. By measuring the effect of protons on substrate binding and by directly measuring proton binding and release, we show that substrates and protons compete for binding to MdfA. Our studies strongly suggest that competition is an integral feature of secondary multidrug transport. We identified the proton-binding acidic residue and show that, surprisingly, the substrate binds at a different site. Together, the results suggest an interesting mode of indirect competition as a mechanism of multi-drug/proton antiport.
INTRODUCTION
The introduction and widespread use of antibiotics has created selective pressure for the emergence of bacterial strains that would persist despite antibiotic toxicity. One important class of genes that mediate resistance against many drugs simultaneously (termed multidrug resistance) encode membrane proteins called multidrug transporters; each recognizes and expels a broad spectrum of chemically unrelated drugs from the cell (Fluman and Bibi, 2009). These transporters constitute a defense mechanism shared by all kingdoms of life. Interestingly, many species of bacteria seem to accommodate a large repertoire of multidrug transporter genes in their genomes (Fluman and Bibi, 2009).
Many of these transporters belong to the major facilitator super-family (MFS) and utilize the proton-motive force to energize transport, by employing an H+/multidrug antiport mechanism (Fluman and Bibi, 2009). In addition to their drug transport activity, the multispecific nature of MFS multidrug transporters enables them to mediate other important functions (Krulwich et al., 2005). Two such examples are MdfA from E. coli, which plays a role in alkaline tolerance, possibly via a K+/ H+ exchange mechanism (Lewinson et al., 2004), and the mammalian vesicular monoamine transporter, which pumps neurotransmitter into vesicles by exchanging it with protons (Liu et al., 1992). Even though proton translocation is essential for all known functions of these transporters, its underlying molecular mechanisms have remained poorly understood. Yet knowledge about other, well-characterized proton translocators, such as bacteriorhodopsin and LacY (Lanyi, 2006; Smirnova et al., 2009, 2008), indicates that membrane-embedded proton-titratable residues play essential roles, probably in proton binding and/or during proton conduction. Indeed, previous studies have indicated that membrane-embedded carboxylic residues in MFS multidrug transporters are crucial for transport and may play important roles in substrate binding and/or proton conduction (Adler et al., 2004; Bapna et al., 2007; Schaedler and van Veen, 2010; Sigal et al., 2009).
How is secondary antiport coupling achieved? In one characterized example, the small multidrug transporter EmrE, counter-transported substrates bind to the transporter in a mutually exclusive manner (Adam et al., 2007; Soskine et al., 2004). This principle, combined with the ability of the transporter to switch between inward- and outward-facing conformations only in the substrate-bound state, is thought to facilitate coupling (Law et al., 2008). However, this is not the only plausible mechanism. Indeed, in the CLC family-related Cl−/H+ exchangers, it was suggested that protons and chloride ions bind simultaneously and, consequently, a different mechanism has been proposed (Picollo et al., 2012).
In MdfA, a model MFS proton/drug antiporter from E. coli, two carboxylic residues, E26 and D34, are predicted to lie within the membrane (Sigal et al., 2009, 2005). Previous studies have indicated that for active transport of cationic substrates, both carboxyls are essential, but for electrically neutral substrates they may compensate for one another’s deficiencies (Adler et al., 2004; Edgar and Bibi, 1999; Sigal et al., 2009, 2006). Additionally, it was shown that mutating E26 into a nonprotonatable glutamine preserves high-affinity substrate binding but fails to support active transport of positively charged substrates (Adler et al., 2004), further supporting a more general role for E26 in the transport mechanism. D34 is also expected to play a major role, however it has not yet been characterized in detail. Acidic residues were shown to play one or more important roles in the multidrug transporter LmrP, a well-characterized MFS member (Schaedler and van Veen, 2010).
In this report, we directly analyzed proton and substrate binding by MdfA and several mutants. We show that protons and substrates compete for binding to MdfA and that a membrane-embedded aspartate at position 34 serves as the proton-binding site of MdfA. Surprisingly, it appears that protons and substrates do not share a single binding site, suggesting that competition is indirect.
RESULTS
Protons Inhibit Drug Binding by MdfA
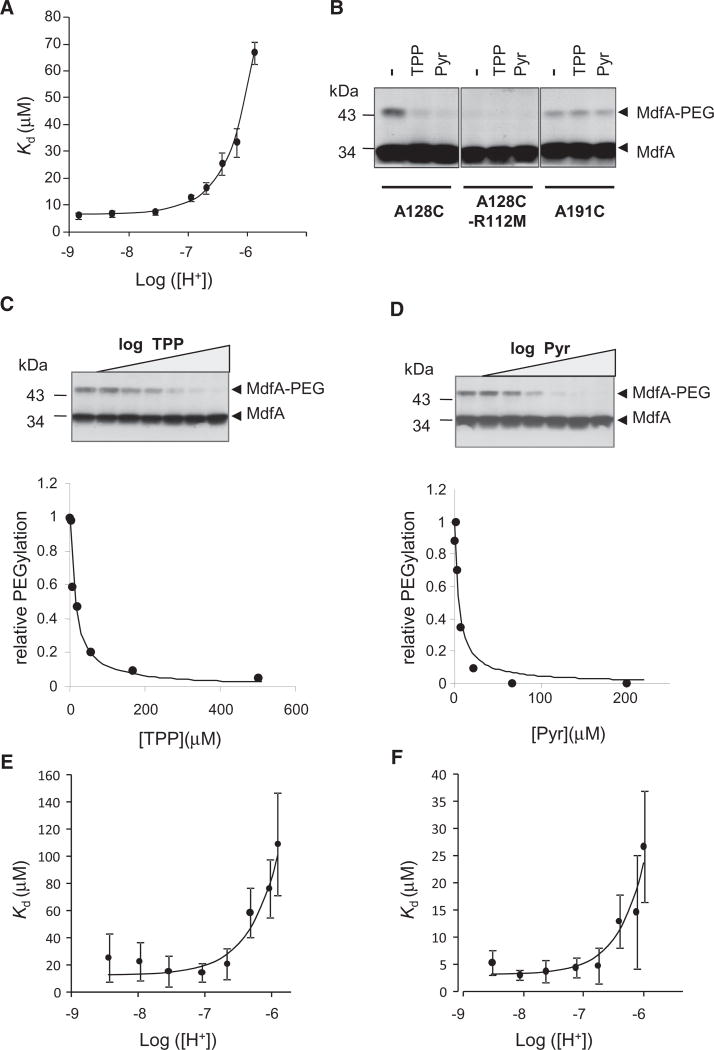
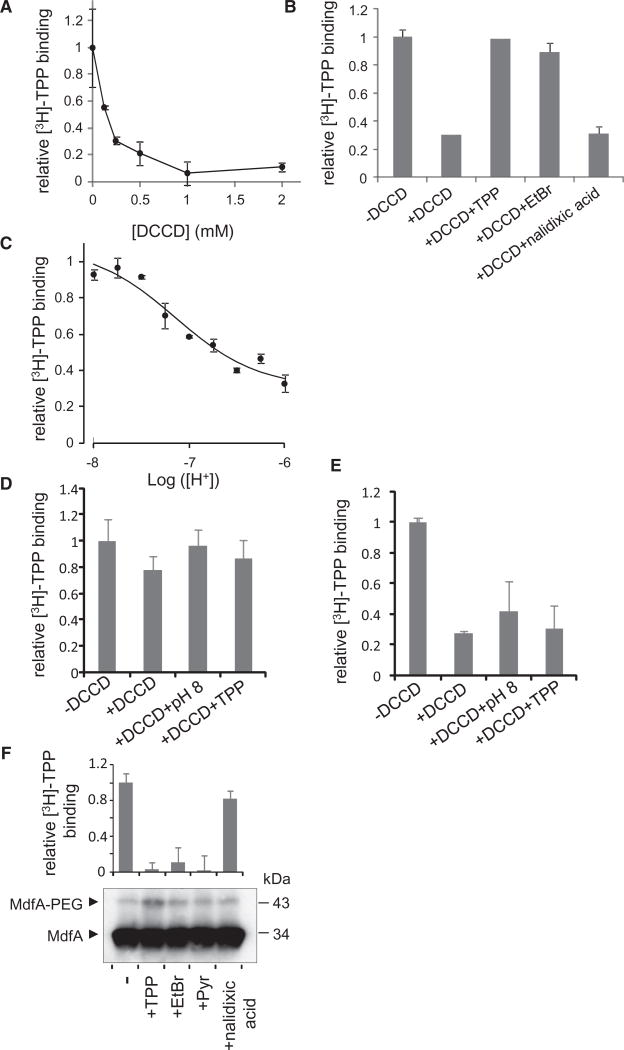
As a first step to study the interplay between protons and drugs in MdfA, we studied how protons affect drug binding. To this end, the affinity of purified, detergent-solubilized MdfA for its substrate tetraphenylphosphonium (TPP) was measured under various pH values, utilizing an intrinsic fluorescence assay that probes TPP-binding by its quenching effect on MdfA’s fluorescence (Fluman et al., 2009) (Figure S1A). Under increasing proton concentrations, the affinity for TPP was reduced (Figure 1A), indicating that high proton concentrations interfere with substrate binding. Importantly, acidifying the solution (from pH 8.5 to pH 6) did not affect the structural integrity of the solubilized transporter (Figure S1B, S1C, and S1D).
Figure 1. [H+]-Dependence of Substrate Affinity.
(A) Affinity of solubilized, purified MdfA for TPP as represented by dissociation constants. The line represents a nonlinear regression fit to an equation describing competitive binding between substrates and protons.
(B) Western blot showing the effect of 0.5 mM substrates on cysteine PEGylation of selected single-Cys mutants in membrane vesicles. Pyr = Pyronin. The PEGylated protein appears as a heavier adduct.
(C and D) Upper panel, substrate concentration dependence of the protection against A128C PEGylation in membrane vesicles. Lower panel: quantification of upper panel by densitometry. The line represents a nonlinear regression fit to a saturation binding function.
(E and F) [H+]-dependent affinity of single Cys MdfA(A128C) for TPP and pyronin, respectively, as represented by dissociation constants determined from the PEGylation of the protein in membrane vesicles. The lines represent a nonlinear regression fit to an equation describing competitive binding between substrates and protons. All the experiments were repeated at least three times and the error bars represent SD. See also Figure S1.
One possible explanation for the inhibitory effect of protons on TPP binding is that they compete for binding to MdfA. To examine this possibility, we analyzed the affinity data by fitting it to a competition-based equation (see Supplemental Experimental Procedures). The data fitted well and yielded a KI of 0.14 ± 0.02 µM for protons, corresponding to a pKa of 6.84 ± 0.06 (Figure 1A, solid line). However, given the numerous potential titratable acidic residues in MdfA, alternative mechanisms could also account for the impact of protons on its affinity for a substrate.
To further examine whether proton/substrate competition plays an important role in MdfA function, we tested how protons affect the binding of TPP and an additional substrate, pyronin, utilizing a different approach. We utilized membrane vesicles containing the single-Cys mutant MdfA(A128C). Residue 128 is found in the putative substrate-binding pocket of MdfA, and substrates can protect this residue against chemical modification (Adler and Bibi, 2004). Our results show that substrates indeed inhibit modification by the sulfhydryl reagent maleimide-polyethyleneglycol (mal-PEG) (Figure 1B). Notably, substrates did not affect the modification of a Cys residue engineered in another location of the transporter (A191C). In addition, the unfolded protein MdfA(A128C/R112M) (Figure S1E) could not be modified by mal-PEG (Figure 1B).
As shown in Figures 1C and 1D, by examining the concentration dependence of substrate-protection against mal-PEG, one can measure the affinity of MdfA for various substrates. This assay enabled us to characterize the effect of protons on substrate binding affinity. Figure 1E shows that the apparent affinity for TPP is decreased under elevated proton concentrations. The data are fitted well by a competition-based equation, yielding a Kd of ~20 µM for TPP and a pKa of ~6.7 for the competing protons (Figure 1E, solid line). To test the generality of the competition effect, we used another substrate, pyronin, and as shown for TPP, its binding was also inhibited by protons, with a similar pKa (~6.7) (Figure 1F), demonstrating that the proton effect on substrate binding is not restricted to TPP.
Substrate Binding Releases a Stoichiometric Proton from MdfA
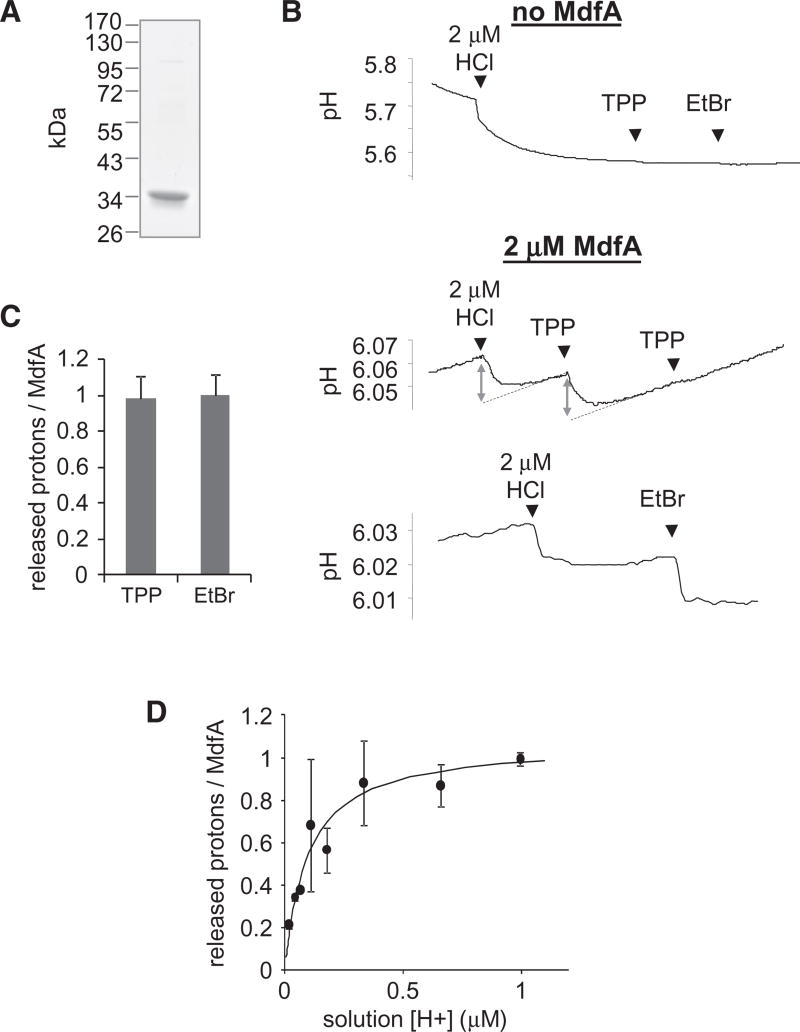
If protons and substrates indeed compete, then substrates should block proton binding to MdfA at high enough concentrations. To examine this possibility, we adapted the method of Soskine et al. (2004) to test whether substrate binding triggers the release of protons from MdfA. Briefly, purified MdfA (Figure 2A) was dialyzed against an unbuffered solution, and the preparation was challenged with substrates while measuring the solution’s pH. Notably, the absence of a buffering agent sometimes generates a spontaneous slow drift in the pH, which creates a background signal. The substrate-induced changes can be distinguished from the background by their much higher rate of change (Figure 2B). In the absence of MdfA, we observed that adding 2 µM protons (in the form of HCl) rapidly reduced the solution’s pH, whereas adding the substrates TPP or ethidium bromide (EtBr) did not appreciably alter the pH (Figure 2B, upper panel). In contrast, when 2 µM MdfA was present in the solution, adding TPP or EtBr caused rapid acidification (Figure 2B, lower panels), which is comparable to an MdfA-equivalent addition of protons, suggesting that each substrate triggered the release of a single stoichiometric proton from MdfA (Figure 2C). Moreover, adding TPP a second time failed to release additional protons, indicating that the effect is saturable.
Figure 2. Substrate-Induced Proton Release.
(A) Purity of MdfA as judged by SDS-PAGE stained with Coomassie.
(B) Time-dependent pH measurements; effect of adding 2 µM HCl or saturating amounts of substrates on the pH of an unbuffered solution with or without 2 µM MdfA. EtBr = ethidium bromide. To quantify the amount of proton released, the magnitude of pH changes elicited by substrate was compared with the effect of HCl (double arrows). Linear regression was utilized to compensate for pH drifts (dotted lines).
(C) Stoichiometry of protons released from MdfA upon binding TPP or EtBr. Error bars indicate SD from 15 (TPP) or 7 (EtBr) independent measurements.
(D) Effect of [H+] on the amount of protons released from MdfA upon addition of 0.3 mM TPP. The line represents a nonlinear regression fit to an equation describing saturable proton binding. The experiment was repeated three times and the error bars represent SD. See also Figure S2.
To evaluate these results further, the substrate-induced proton release was measured by another method. We utilized fluorescein, a fluorescent pH indicator that produces large signals, as a reporter for changes in the solution’s acidity. This method reproduced the results obtained with pH measurements (Figure S2).
To further characterize the site from which a proton is released, we determined its pKa. To this end, we utilized the amount of protons that were released from MdfA upon TPP binding as a measure for its protonation state under various pHs. As shown in Figure 2D, the amount of released protons increased with the solution’s H+ concentration, saturating at ~1 proton per MdfA molecule (1.08 ± 0.09). When the proton concentration was below 0.5 µM (corresponding to pH > 6.3), the released protons were a fraction of MdfA molecules, suggesting that the other fraction of the transporter was deprotonated even before substrate binding and therefore unable to release protons. Importantly, the concentration dependence suggests a pKa of 7.05 ± 0.12 (Figure 2D, solid line), in agreement with the pKa calculated from the substrate competition experiments (Figure 1A). This indicates that the protonatable site involved in inhibiting substrate binding is probably the same site from which protons are released upon substrate binding. All together, the results demonstrate that substrates and protons compete for binding to MdfA.
Neutralized E26 or D34 Mutants Bind Substrates but the Mutations Abolish Proton/Substrate Competition and Coupling
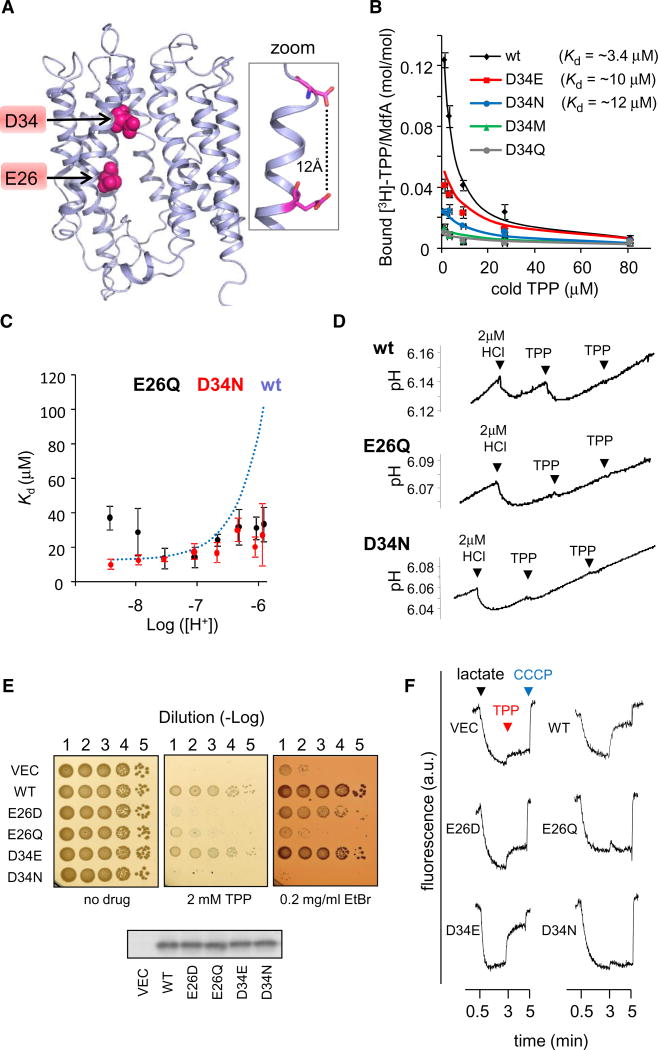
To elucidate the mechanism underlying proton binding, it is essential to locate the functionally relevant proton-binding site in MdfA. Previous studies have suggested that membrane-embedded carboxyl residues are important for the function of secondary multidrug transporters, including MdfA (e.g., Seeger et al., 2009; Sigal et al., 2009; Soskine et al., 2004). MdfA contains 2 carboxyls, E26 and D34, which are proposed to lie within the membrane (Figure 3A) (Sigal et al., 2005). Calculation based on the predicted MdfA model suggests pKas of 7 and 6 for E26 and D34, respectively (Li et al., 2005). This is a substantial deviation from the pKa of solvent-exposed acidic residues, consistent with their proposed hydrophobic, membrane-embedded location. The model-based calculations are likely inaccurate, yet they suggest E26 and D34 as plausible candidates to bind a proton under physiological pH.
Figure 3. Characterization of E26 and D34 Mutants.
(A) Location of E26 and D34 in the 3D-structural model of MdfA. The 11th transmembrane segment is not shown for clarity. The inset on the right shows the calculated distance between E26 and D34.
(B) Inhibition by cold TPP of [3H]-TPP binding to the various D34X mutants. Dissociation constants obtained from nonlinear regression fitting (solid line) are indicated. Error bars indicate SD.
(C) [H+]-dependence of the affinity of MdfA(E26Q) or MdfA(D34N) for TPP, as determined by PEGylation protection in membrane vesicles. The dependence of the wild-type, as determined in Figure 1E is shown for comparison (dotted line). Error bars indicate SD.
(D) Time-dependent pH measurements of proton release by MdfA(E26Q) and MdfA(D34N) (see Figure 2B).
(E) Drug-resistance of E. coli cells harboring the indicated MdfA mutant (or empty vector). Serial dilutions of cells were spotted on drug-supplemented media. Western blotting analysis shows that the expression of the various MdfA mutants is comparable (lower panel).
(F) Proton transport by everted membrane vesicles containing different MdfA mutants. The trans-membrane ΔpH was followed by measuring ACMA fluorescence. Lactate (added after 0.5 min) and CCCP (after 5 min) were used to generate or dissipate ΔpH, respectively. TPP was added after 3 min.
Since D34 has been much less characterized compared with E26, we first addressed the importance of the negative charge for interaction with the substrate TPP by measuring the binding affinity of several purified MdfA(D34X) mutants. This was carried out by testing the ability of unlabeled TPP to compete with the binding of radiolabeled TPP to MdfA (Figure 3B). The results suggested that D34 is not essential for substrate binding since both asparagine and glutamate could replace it while preserving substantial binding affinity to MdfA.
Next, we studied the possible role of the negative charges of E26 or D34 in proton binding by studying the proton dependent activities of MdfA(E26Q) or MdfA(D34N). As shown in Figure 3C, both mutants preserved TPP binding with affinities similar to wild-type MdfA at ~pH 7. Interestingly, however, neutralizing either E26 or D34 abolished the competition between protons and TPP, as revealed by the [H+]-independent affinity of the mutants for TPP (Figure 3C) and by their inability to release protons upon TPP binding (Figure 3D). These results suggest that both residues play an important role in the observed substrate/proton competition. Indeed, although the proposed protonation-incapable mutants MdfA(E26Q) and MdfA(D34N) bind TPP at least as well as wild-type MdfA (Figures 3B, 3C, and 3D) or the respective protonation-capable mutants MdfA(E26D) and MdfA(D34E) (Figure 3B; see also Adler et al. [2004]), they do not confer multidrug resistance (Figure 3E). This implies an important role for the acidic groups of E26 and D34 that extends beyond substrate binding.
To further study the role of the acidic residues, we followed MdfA-catalyzed proton transport in everted membrane vesicles, by utilizing the membrane ΔpH-sensitive fluorescent probe ACMA (9-Amino-6-Chloro-2-Methoxyacridine). Oxidation of lactate by everted vesicles generated an inward proton current, as observed by quenching of AMCA fluorescence (Figure 3F). Addition of TPP caused dequenching, indicating an outward proton current, consistent with H+/TPP antiport. The effect of TPP was much more notable in membranes prepared from wt-MdfA-expressing cells than in cells harboring an empty vector. However, some H+/TPP antiport was also detected in these control membranes, likely because of other E. coli H+/ TPP antiporters. Membranes expressing the neutralized mutants MdfA(E26Q) and MdfA(D34N) showed no response to TPP, whereas the mutants MdfA(E26D) and MdfA(D34E) showed moderate or high responses, respectively (Figure 3F), indicating active H+/TPP antiport. Thus, the carboxylic groups of E26 and D34 are important for antiport coupling and/or proton transport by MdfA.
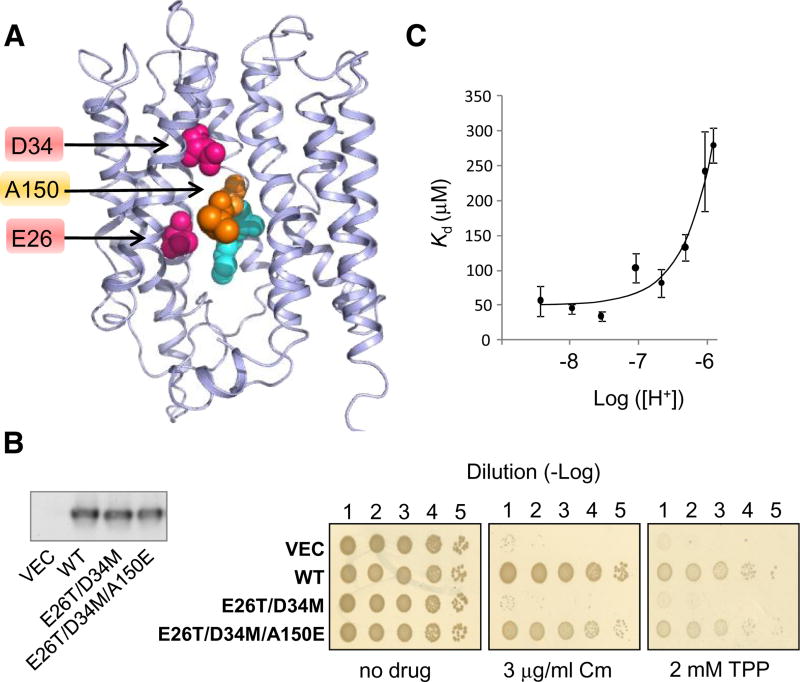
A Suppressive A150E Mutation Restores Competition to the Inactive MdfA(E26T/D34M) Double Mutant
We have previously shown that a double mutant of MdfA in which both E26 and D34 have been neutralized (MdfA[E26T/D34M]) retained no detectable activities. Intriguingly, a third mutation that reintroduces a negative charge at position 150 (A150E) rescued MdfA(E26T/D34M) (Figures 4A and 4B), suggesting that the essential acidic residue(s) can be functionally relocated in MdfA (Sigal et al., 2009). Notably, as shown above for the wild-type transporter (Figures 1B and S1E), mutating the essential residue R112 inactivates the triple mutant, suggesting that the role of R112 is probably not related to E26 and D34 (Figure S3).
Figure 4. Rescue of MdfA(E26T/D34M) by A150E.
(A). Selected residues viewed on the 3D–structural model of MdfA. The studied residues are indicated by an arrow. M146 and A147 are colored cyan and teal, respectively.
(B) Drug resistance of E. coli cells harboring the indicated MdfA mutant (or empty vector) (See Figure 3E). Chloramphenicol = Cm. Left panel: Western blotting analysis shows that the expression of the various MdfA mutants is comparable.
(C) Affinity of MdfA(A128C) harboring the triple mutation E26T/D34M/A150E for TPP. The lines represent a nonlinear regression fit to an equation describing competitive binding between substrates and protons. The experiment was repeated three times and the error bars represent SD. See also Figure S3.
The triple mutant MdfA(E26T/D34M/A150E) is interesting because it replaces both E26 and D34 from TM1 by a single acidic residue in a completely different location in the pocket (position 150 in TM4) (Figure 4A). If proton competition is an important feature of active drug transport, then the rescuing mutation might have re-established competition. Briefly, the A128C mutation was introduced into the triple mutant, and the effect of protons on the affinity for TPP was assessed by mal-PEG modification (see Figure 1C and 1E). Remarkably, the results indicate that the A150E mutation restored also the proton-TPP competition (Figure 4C). Protons inhibited TPP binding by the triple mutant; analysis of the inhibition suggests a pKa of ~6.5 for the proton binding site and a somewhat reduced TPP binding affinity (Kd ~ 60 µM, compared with wild-type, ~20 µM) (Figure 4C, solid line). Notably, we could not detect binding of TPP to the double mutant MdfA(E26T/D34M) by this method (data not shown) . These results suggest that proton-substrate competition constitutes an essential mechanistic aspect of MdfA function and that a single glutamate at position 150 can fulfill the function(s) of both E26 and D34, in mechanistic terms.
Chemical Modification by Carbodiimide Suggests a Carboxyl-Mediated Proton Binding
To further investigate whether acidic residues interact functionally with protons during transport, we modified MdfA by dicyclohexylcarbodiimide (DCCD), a reagent that reacts with protonated carboxyls in a hydrophobic environment under physiological pH conditions. Such carboxyls are rare in proteins, because most carboxyls are deprotonated in aqueous environments. Therefore, DCCD is a powerful reagent for exploring membrane-embedded acidic residues involved in proton transport (Seeger et al., 2009; Weinglass et al., 2005, 2003).
DCCD-modification at pH 6 inhibited [3H]-TPP binding by MdfA in a concentration-dependent manner (Figure 5A). Although the inhibition mechanism is not clear, it demonstrates that DCCD can react with the transporter. Notably, similar conditions (1 hr incubation at 30°C with ~0.25 mM DCCD) have specifically labeled membrane-embedded acidic residues in other transporters (Seeger et al., 2009; Weinglass et al., 2005, 2003). The observed inhibitory effect of DCCD was utilized to further characterize its reaction with MdfA. We observed that adding TPP or EtBr, but not the nonsubstrate nalidixic acid, blocked the reaction with DCCD (Figure 5B), strongly suggesting that the modification occurred at a functionally relevant site. To investigate the effect of pH on the carboxyls’ protonation state, the reactivity with DCCD, which labels only protonated carboxyls, was characterized. Decreasing the proton concentration during reaction restricted the reactivity and therefore the inhibitory effect of DCCD (Figure 5C). Analyzing the data according to acid titration yielded a pKa of 6.99 ± 0.1 (Figure 5C, line). Taken together, the data indicate that a critical, protonated carboxyl with a pKa near neutral reacts with DCCD and that substrates confer protection of this carboxyl, either by sterically hindering the reaction or by rendering the carboxyl deprotonated.
Figure 5. DCCD Modification of MdfA and Mutants as Probed by Inhibition of [3H]-TPP Binding.
(A) Effect of DCCD on [3H]-TPP binding by purified wild-type MdfA.
(B) Effect of substrates on the DCCD reactivity, as probed by inhibition of [3H]-TPP binding. The nonsubstrate nalidixic acid is shown as control.
(C) Effect of altering the proton concentration during reaction with DCCD on its reactivity, probed by inhibition of [3H]-TPP binding. The line represents a nonlinear regression fit to an equation of acid-base equilibrium-dependent modification.
(D and E) (D) MdfA(D34N) and (E) MdfA(E26Q): Effect of pH and TPP on DCCD reactivity.
(F) Single Cys mutant MdfA(D34C): Effect of substrates on mal-PEGylation (lower panel) and [3H]-TPP binding (upper panel). The nonsubstrate nalidixic acid was used as a control. All the experiments were repeated at least three times and the error bars represent SD. See also Figure S4. All modification reactions were carried at pH 6 unless indicated otherwise.
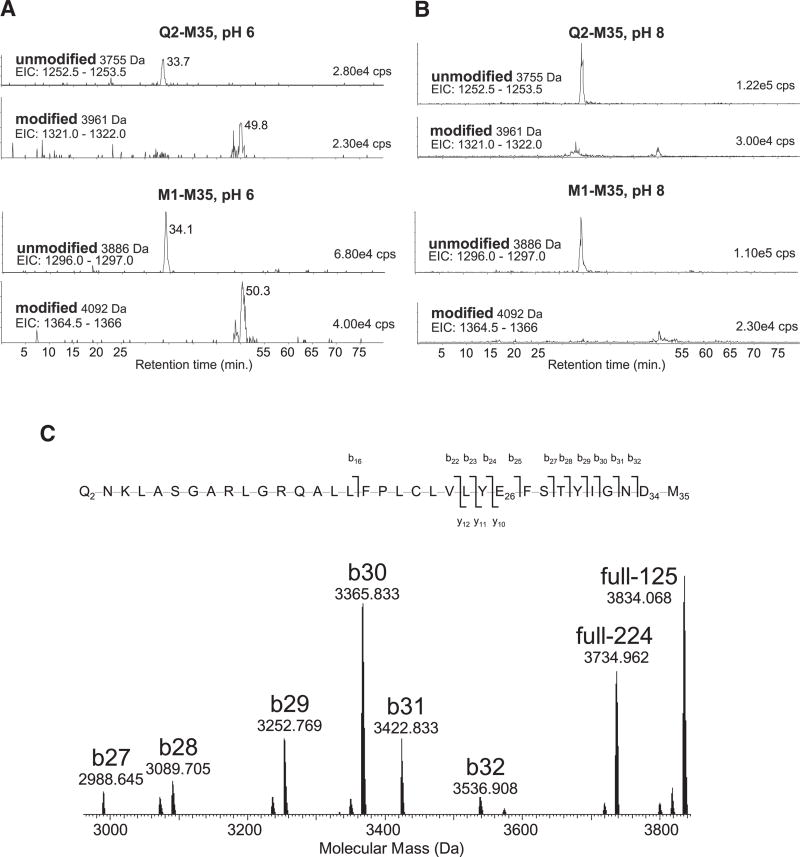
Mass Spectrometry of CNBr Cleaved MdfA Identifies D34 as the DCCD-Modified Residue
To identify the one or more crucial protonatable acidic residues in MdfA that react with DCCD, we utilized mass spectrometry, after cyanogen bromide (CNBr) cleavage of the DCCD-treated MdfA. Unmodified and modified peptides were separated by chromatography with mass spectrometry readout (LC-MS+) and the data interrogated for specific peptide species. The N-terminal peptides ending at M35 and thus containing both E26 and D34 were found in two forms, one of which retained the first methionine (M1-M35) due to incomplete cleavage by CNBr, and the second peptide started at Q2 (Q2-M35). The hydrophobic DCCD-modified forms of these peptides (~50 min) were more highly retained than the unmodified forms (~34 min) in reverse-phase chromatography (Figure 6A). Comparison of intensities of these peaks suggests a little under 50% of the peptide is modified when the DCCD labeling was performed at pH 6, assuming the recovery and ionization efficiency of modified and unmodified peptides is similar. When the DCCD labeling was performed at pH 8 there was a significant drop in intensity of the modified peptide species (Figure 6B) with most of the peptide in the unmodified state, consistent with the above-described loss of DCCD-reactivity at pH 8 (Figure 5C). High-resolution (tandem) mass spectrometry of the DCCD-modified peptide showed that fragmented ions containing both E26 and D34 were modified by DCCD (ions full −224 and full −125 and y10 – y12), whereas ions containing only E26 were not (ions b25, b27 – b32) (Figure 6C and Table S1). This indicates predominant modification of D34, rather than E26, and thus strongly supports a conclusion that D34 is the critical protonatable residue in MdfA.
Figure 6. Mass Spectrometry of Peptide Q2-M35.
(A–C) CNBr-generated peptides were separated by reverse-phase chromatography with online electrospray-ionization mass spectrometry. The MdfA N-terminal peptides M1-M35 and Q2-M35 containing E26 and D34 have a mass of 3755 or 3886 Da, respectively. Single modification with DCCD resulted in the respective peptides of mass 3961 or 4092 Da. (A) and (B) show extracted ion chromatograms (EIC: m/z range) for the triply charged ions that predominate after ESI of these four peptide species. DCCD treatment at pH 6.0 (A) and DCCD treatment at pH 8.0 (B) are shown. The intensity of each peak is given in the top right of each panel in cps (counts per second, arbitrary units). The traces shown are scaled to the most abundant species. In (C), the 3961 Da DCCD modified peptide (A) was analyzed by high-resolution FT-ICR tandem mass spectrometry with collisionally activated dissociation (CAD). The higher mass range of the deconvoluted molecular mass spectrum is shown for the ions indicated. The identified b- and y-ions are shown mapped onto the primary sequence (b-ions, of N-terminal origin are indicated by a slash rising to the left; y-ions, of C-terminal origin, with a slash dropping to the right). Explanation: DCCD modification adds 206.178 Da, and M35 modification to homoserine lactone reduces the mass by 48.003 Da. Mass accuracy of the ions is provided in Table S1. The ions b25 and b27–b32, which carry E26 but not D34, also do not carry the DCCD modification. Unmodified product ions could arise from a modified parent ion only if neutral loss of intact DCCD (−206 Da) occurred during CAD. The ions shown at 3734.965 (full −224) and 3834.068 (full −125) correspond to neutral losses of −224 and −125 Da, respectively, from the precursor ion, in agreement with previous tandem mass spectrometry analysis of DCCD-modified peptides (Seeger et al., 2009). Thus the only explanation for the intense b27–b32 unmodified product ions is that the DCCD modification is located predominantly on D34. See also Table S1.
Substrate Induces D34 Deprotonation Indirectly
Although both D34 and E26 were found critical for the competition between substrates and protons, only D34 seems to be protonated and thus DCCD-sensitive at low pH. To gain a deeper understanding on the roles of D34 and E26 in MdfA, we characterized MdfA mutated at these positions. In agreement with D34 being the DCCD-sensitive proton binding site, we found that DCCD hardly affected the binding of TPP to MdfA(D34N) (Figure 5D).
To evaluate further the possible importance of D34 and E26, we examined their evolutionary conservation. To this end, we aligned 57 sequences of homologs that share at least 20% identity and 40% similarity with MdfA. To remove redundancy, we excluded proteins that are 90% identical to any of the other sequences (for a detailed description, see Supplemental Experimental Procedures). This analysis shows that D34 is completely conserved (Figure S4), strongly suggesting that the conserved aspartate is the most important acidic residue in these transporters, as it is directly involved in proton binding and release. E26 was also conserved, but to a lesser extent (65% identical), with 12.3% of the homologs having a conservative D in this position and 23% harboring other residues.
To study the role of E26 further, we challenged MdfA(E26Q) with DCCD. The results indicate that TPP binding to this mutant was inhibited by DCCD, confirming that E26 is not the DCCD-sensitive site. Interestingly, unlike with the wild-type transporter, the DCCD reactivity of MdfA(E26Q) was pH and TPP independent (Figure 5E). Thus, the results suggest that functional protonation/deprotonation of D34 requires the acidic residue at position 26. Since MdfA(E26Q) binds TPP efficiently, its modification in the presence of substrate indicates that substrates cannot protect against DCCD by steric hindrance. Instead, the substrate appears to protect against modification indirectly by inducing D34 deprotonation. Importantly, this suggests that in spite of the observed competition, substrates bind to a site that is distinct from D34.
To further investigate this possibility, we studied how substrates affect accessibility of a single Cys introduced at position 34, by utilizing the mal-PEG modification assay. Notably, the single Cys mutant MdfA(D34C) retains substrate-binding activity; upon purification, we observed binding of radiolabeled substrate ([3H]-TPP) by this mutant, and the substrates TPP, EtBr, and pyronin could compete with [3H]-TPP for binding (Figure 5F, upper panel). Nevertheless, these substrates do not confer any protection against mal-PEG modification of residue D34C (Figure 5F, lower panel), indicating that they do not sterically hinder residue 34 from chemical modification. Interestingly, TPP binding appears to stimulate modification of D34C, suggesting that this substrate may cause exposure of this residue. Taken together, the results indicate that substrates bind to a site that is distinct from D34.
DISCUSSION
In recent years there has been significant advancement in our understanding of how transporters function, especially as their structures in different conformational states are revealed (for example, Abramson et al., 2003; Dang et al., 2010; and Seeger et al., 2006). Yet, despite this progress, proton-binding sites remained relatively elusive, and their location is not readily understood from moderate-resolution X-ray crystal structures.
Secondary antiporters, which transport substrates and ions in opposing directions, have been proposed to couple transport by utilizing a “sequential binding and translocation” mechanism, through which the substrate must be released prior to binding and translocation of the counter-transported ion (and vice versa) (Law et al., 2008). By preventing binding of the counter substrates together, this putative “Ping-Pong”-like mechanism prevents cotransport of both substrates in the same direction, which is an essential prohibition for antiport coupling. In our studies, we tested this proposal and we provide empirical evidence that, indeed, MdfA interacts with drugs and protons in a mutually exclusive manner. Furthermore, we were able to demonstrate the importance of this principle for active transport; namely, disrupting the competition abolishes active transport and rescuing the transporter revives competition.
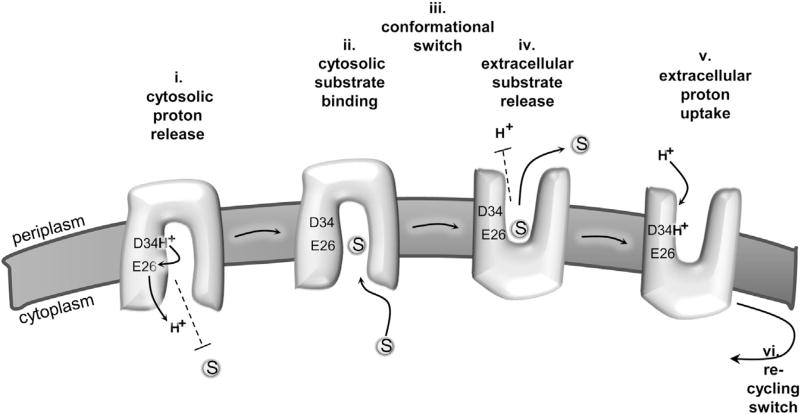
As described, MdfA uses a competition-based general anti-port mechanism as proposed for many antiporters. Our study indicates that MdfA utilizes a different molecular flavor of competition. In many antiporters, the counter-transported substrates are similar regarding their chemistry and especially regarding their charge. A few well-characterized examples are the chloride/bicarbonate anion exchanger (AE1), the glycerol-3-phosphate/Pi exchanger GlpT, the Na+/H+ antiporter NhaA, and the (cationic) drug/H+ exchanger EmrE (Huang et al., 2003; Padan et al., 2009; Soskine et al., 2004; Tanner, 1997). This prevalent design suggests that competition might be achieved by a common binding site for the counter-transported substrates, as indeed was proposed (e.g., Padan et al., 2009) or demonstrated (Soskine et al., 2004) for several systems. In contrast, our results suggest that MdfA deviates from this group and that it binds the counter-transported substrates at different sites without compromising the competitive nature of their binding, which converge at the same antiport mechanism (Figure 7). Mimicking the D34 protonated state by mutagenesis (D34N) does not abolish cationic substrate binding. This indicates that the proton-binding residue D34 does not interact electrostatically with the substrate. Importantly, when E26 is neutralized (E26Q), substrate binding no longer protects D34 against DCCD. This suggests that (i) E26 is required for substrate-induced D34 deprotonation, and (ii) the substrate does not interact directly with D34. The latter notion is strongly supported by the apparent inability of substrates to protect a cysteine at position 34 against mal-PEG modification. If so, then where is the substrate binding site in MdfA? In a previous study, 19 residues putatively involved in substrate recognition were identified using a single genetic screen (Adler and Bibi, 2004). When viewed in the 3D–structural model of the protein (see Figure 2 in Fluman and Bibi, 2009), these residues are located in a putative substrate-binding pocket formed in the middle of the protein and penetrating the membrane. As judged from the biochemical data, residues that line the pocket interact differently with different substrates (Adler and Bibi, 2004). In conclusion, unlike the defined proton-binding site of wild-type MdfA (D34), which is located on the periplasmic leaflet of the membrane, the proposed substrate-binding pocket is large, involves many residues, and is located on the cytoplasmic side of the membrane.
Figure 7. Model for MdfA-Catalyzed Substrate/H+ Antiport.
(i) Proton is released from D34 to the cytosol in an E26-mediated manner. Substrates are prevented from binding to MdfA in the proton bound state (dashed line).
(ii) Upon spontaneous deprotonation, substrates bind from the cytosol, to a site that is distinct from D34.
(iii) Substrate binding triggers a conformational switch that exposes the multidrug-binding pocket to the extracellular side (periplasm). (iv) Protons cannot bind to MdfA (dashed line) until substrate is released to the extracellular side. (v) A proton binds to D34 from the extracellular side.
(vi) The proton-binding event triggers a conformational switch that recycles MdfA to the inward-facing conformation.
As shown in previous studies (Adler et al., 2004; Edgar and Bibi, 1999), E26 plays an important role in MdfA, and it also participates (but is not essential) in interactions with substrates. Notably, it was shown that a carboxyl in this position is crucial for active transport of cationic drugs but not for their binding. Our results indicate that aside from its nonessential role in substrate binding, E26 facilitates deprotonation of D34. Although speculative, the titratable nature of E26 makes it tempting to suggest that this residue facilitates deprotonation by serving as a transient “proton-transfer” site, capable of accepting the proton transiently before its release to the cytoplasm. Upon examining MdfA’s homologs, we found that although E26 is not completely conserved (65% of homologs contain E in this position), most other homologs (26%) harbor proton titratable residues (D/S/T) in this location (Figure S4). Notably, E26 and D34 are too far apart for direct proton transfer between them (Figure 3A, inset). Thus, any possible transfer event must be mediated by additional hydrogen-bonding residues and/or water molecules that may serve, together with E26, as a proton relay (Lanyi, 2006). Interesting pairs of acidic residues were also observed in other proton-coupled transporters, (e.g., FucP; Dang et al., 2010). Thus, a “pair arrangement” might be a common architecture facilitating proton conduction.
The observation that the ion and substrate do not share a single binding site and that a negative charge in D34 is not necessary for binding cationic substrates indicates that competition is unlikely to be driven by electrical repulsion of the counter-transported substrates (or by electrical interaction with the transporter), as is often assumed for antiporters that exchange substrates of the same charge. Indeed, unlike most characterized antiporters, MdfA is permissive regarding the charge of its substrates. Whereas many of its known substrates are cationic, MdfA can also transport neutral or zwitterionic substrates such as chloramphenicol and ciprofloxacin. Thus, a mechanism that does not rely on direct competition might impose fewer restrictions on the counter-transported substrate and thus enable a broader substrate spectrum for MdfA.
Our studies indicate that indirect competition is based on substrate-dependent deprotonation of D34. Indeed, D34 is absolutely conserved in homologs of MdfA, suggesting that MdfA may not be able to tolerate mutations in this residue. Surprisingly, recent studies have shown the contrary. Although neutralizing mutations in D34 abolished the activity against most substrates, chloramphenicol (and its analog thiamphenicol) can still be actively transported by such mutant (Sigal et al., 2006). We have recently proposed that for active transport of neutral substrates MdfA can utilize alternatively either E26 or D34, demonstrating an unexpected degree of permissibility (Sigal et al., 2009). Remarkably, this notion is supported by our observation that active transport can rely on a single membrane carboxyl at position 150, which functionally replaces both E26 and D34 (Sigal et al., 2009). As shown here, this mutation (A150E, in the background of MdfA[E26T/D34M]) restores the proton/substrate competition of the otherwise inactive MdfA(E26T/D34M), suggesting that competition is essential for active transport in MdfA. We suggest that in the triple mutant MdfA(E26T/D34M/A150E) a simple mode of competition emerged, based on sharing a proton/drug-binding site. This proposal is in accordance with previous findings that implicate residues 146, 147, and 150 as involved in substrate recognition (Adler and Bibi, 2004). The structural model suggests that these residues are on the same helical face, pointing toward the substrate-binding site and E26 (Figure 4A). The ability of the proton-binding site to undergo displacements and/or possible changes in the competition mode (direct or indirect) suggests that proton/multidrug antiporters have a simple design, and this can explain their ability to evolve multiple times in evolution.
Interestingly, the proposal that MFS multidrug transporters may utilize both direct and indirect mechanisms of competition has gained support from studies on the multidrug transporter LmrP. In this transporter, which works with a stoichiometry of two or three protons per drug, two acidic residues were shown to affect both substrate binding and proton stoichiometry, potentially representing direct competition sites. A third carboxylate, however, which affects only the stoichiometry and is dispensable for substrate binding, might represent a dedicated proton-binding site (Schaedler and van Veen, 2010).
Importantly, both in wild-type MdfA and in the rescued mutant MdfA(E26T/D34M/A150E), the pKa of the proton-binding site is nearly neutral. This site must be able to pick up a proton from the periplasm and release it in the cytoplasm. The intracellular pH in E. coli is kept near 7.6, whereas the usually acidic periplasmic pH drifts with the external pH of the medium (Wilks and Slonczewski, 2007). Thus, the near neutral pKa of the proton-binding site is probably crucial, enabling protons to bind and be released spontaneously under physiological pH. The low proton affinity, also observed in the proton/drug antiporter EmrE, probably represents an important factor in the mechanism underlying proton/multidrug antiport, since it is entirely different in, for example, the proton/lactose symporter LacY (Adam et al., 2007; Smirnova et al., 2008).
To conclude, our studies revealed an interesting mechanism to explain the competition between drugs and protons during the MdfA-catalyzed antiport cycle (Figure 7). In light of previous studies on MdfA and its mutants, our results provide insight on how proton-coupled secondary multidrug transporters might have evolved and, particularly, how acidic residues shape the function and evolution of secondary multidrug transporters.
EXPERIMENTAL PROCEDURES
Plasmids
See Supplemental Experimental Procedures.
MdfA Expression and Purification
MdfA-His6 was expressed under the arabinose promoter in E. coli. Cells were washed and disrupted by passage through a liquidizer. Membranes collected by centrifugation were kept frozen. For purification, membranes were solubilized by 1.1% β-dodecyl maltopyranoside. The solubilized extract was bound to immobilized metal affinity beads and MdfA was eluted with imidazole and dialyzed against desired buffer. Protein concentration was determined by measuring OD280 (1 mg/ml ~ 2.1 OD280) (Sigal et al., 2007). Further information is in Supplemental Experimental Procedures.
TPP Binding Measured by Fluorescence
TPP binding was measured as described (Fluman et al., 2009), with minor modifications described in Supplemental Experimental Procedures.
Substrate Binding Measurements in Membrane Vesicles Utilizing Maleimide-Polyethyleneglycol
Membrane vesicles expressing MdfA(A128C) were supplemented with 50 mM BTP buffer in the indicated pH. The membranes were incubated with increasing substrate concentrations and reacted with maleimide-polyethyleneglycol (mal-PEG) for several hours on ice. Reaction adducts were resolved and identified by SDS-PAGE and western blotting. Bands were quantified by densitometry and the effect of substrate on PEGylation was analyzed by nonlinear regression. More information is in Supplemental Experimental Procedures.
Measurement of Substrate-Induced Proton-Release
Purified MdfA was dialyzed four times against 100 volumes of unbuffered solution. The protein (2 µM), titrated to the desired pH, was continuously measured by a pH electrode, while adding substrates (200–300 µM TPP or 500 µM EtBr) or HCl (2 µM). A detailed description is in Supplemental Experimental Procedures.
Drug Resistance Assay
The growth of E. coli harboring empty vector or expressing different MdfA mutants on drug-containing LB-Agar plates was assayed as described (Sigal et al., 2009).
Measurement of [3H]-TPP Binding
Radiolabeled TPP-binding was carried out by immobilizing purified MdfA-His6 on Ni2+ beads, allowing it to bind [3H]-TPP, and counting the MdfA-associated radioactivity as described (Sigal et al., 2007).
DCCD Labeling
Purified protein samples with the indicated drugs/buffers were reacted with DCCD and subsequently bound to Ni2+ beads and washed four times. [3H]-TPP binding was measured as above. For further information see Supplemental Experimental Procedures.
For mass spectrometric analysis, CNBr-cleaved peptides were subjected to reverse-phase chromatography with mass spectrometry and fraction collection (LC-MS+) as described (Weinglass et al., 2003; Whitelegge et al., 2002). Selected peptides were analyzed by high-resolution mass spectrometry and fragmented using collisionally activated dissociation (CAD) (Ryan et al., 2010; Whitelegge et al., 2006). For more details see Supplemental Experimental Procedures.
Proton Transport in Everted Membrane Vesicles
Everted membrane vesicles prepared from cells expressing MdfA or carrying an empty vector were prewarmed (30°C) and supplemented with 1 µM ACMA. The fluorescence of samples was measured while continuously stirring them. At the indicated times, lactate (2 mM), TPP (0.25 mM), or CCCP (2 µM) were added. For more details see Supplemental Experimental Procedures.
Acknowledgments
We thank Anna M. Mallett and Alexander J. Griffel for their help in characterizing the D34C mutant. This work was supported by a grant from the Israel Science Foundation (1128/06).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes four figures, one table, Supplemental Experimental Procedures, and Supplemental References and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2012.06.018.
References
- Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- Adam Y, Tayer N, Rotem D, Schreiber G, Schuldiner S. The fast release of sticky protons: kinetics of substrate binding and proton release in a multidrug transporter. Proc. Natl. Acad. Sci. USA. 2007;104:17989–17994. doi: 10.1073/pnas.0704425104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler J, Bibi E. Determinants of substrate recognition by the Escherichia coli multidrug transporter MdfA identified on both sides of the membrane. J. Biol. Chem. 2004;279:8957–8965. doi: 10.1074/jbc.M313422200. [DOI] [PubMed] [Google Scholar]
- Adler J, Lewinson O, Bibi E. Role of a conserved membrane-embedded acidic residue in the multidrug transporter MdfA. Biochemistry. 2004;43:518–525. doi: 10.1021/bi035485t. [DOI] [PubMed] [Google Scholar]
- Bapna A, Federici L, Venter H, Velamakanni S, Luisi B, Fan TP, van Veen HW. Two proton translocation pathways in a secondary active multidrug transporter. J. Mol. Microbiol. Biotechnol. 2007;12:197–209. doi: 10.1159/000099641. [DOI] [PubMed] [Google Scholar]
- Dang S, Sun L, Huang Y, Lu F, Liu Y, Gong H, Wang J, Yan N. Structure of a fucose transporter in an outward-open conformation. Nature. 2010;467:734–738. doi: 10.1038/nature09406. [DOI] [PubMed] [Google Scholar]
- Edgar R, Bibi E. A single membrane-embedded negative charge is critical for recognizing positively charged drugs by the Escherichia coli multi-drug resistance protein MdfA. EMBO J. 1999;18:822–832. doi: 10.1093/emboj/18.4.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluman N, Bibi E. Bacterial multidrug transport through the lens of the major facilitator superfamily. Biochim. Biophys. Acta. 2009;1794:738–747. doi: 10.1016/j.bbapap.2008.11.020. [DOI] [PubMed] [Google Scholar]
- Fluman N, Cohen-Karni D, Weiss T, Bibi E. A promiscuous conformational switch in the secondary multidrug transporter MdfA. J. Biol. Chem. 2009;284:32296–32304. doi: 10.1074/jbc.M109.050658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Lemieux MJ, Song J, Auer M, Wang DN. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science. 2003;301:616–620. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- Krulwich TA, Lewinson O, Padan E, Bibi E. Do physiological roles foster persistence of drug/multidrug-efflux transporters? A case study. Nat. Rev. Microbiol. 2005;3:566–572. doi: 10.1038/nrmicro1181. [DOI] [PubMed] [Google Scholar]
- Lanyi JK. Proton transfers in the bacteriorhodopsin photocycle. Biochim. Biophys. Acta. 2006;1757:1012–1018. doi: 10.1016/j.bbabio.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Law CJ, Maloney PC, Wang DN. Ins and outs of major facilitator superfamily antiporters. Annu. Rev. Microbiol. 2008;62:289–305. doi: 10.1146/annurev.micro.61.080706.093329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinson O, Padan E, Bibi E. Alkalitolerance: a biological function for a multidrug transporter in pH homeostasis. Proc. Natl. Acad. Sci. USA. 2004;101:14073–14078. doi: 10.1073/pnas.0405375101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Robertson AD, Jensen JH. Very fast empirical prediction and rationalization of protein pKa values. Proteins. 2005;61:704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- Liu Y, Peter D, Roghani A, Schuldiner S, Privé GG, Eisenberg D, Brecha N, Edwards RH. A cDNA that suppresses MPP+ toxicity encodes a vesicular amine transporter. Cell. 1992;70:539–551. doi: 10.1016/0092-8674(92)90425-c. [DOI] [PubMed] [Google Scholar]
- Padan E, Kozachkov L, Herz K, Rimon A. NhaA crystal structure: functional-structural insights. J. Exp. Biol. 2009;212:1593–1603. doi: 10.1242/jeb.026708. [DOI] [PubMed] [Google Scholar]
- Picollo A, Xu Y, Johner N, Bernèche S, Accardi A. Synergistic substrate binding determines the stoichiometry of transport of a prokaryotic H(+)/Cl(−) exchanger. Nat. Struct. Mol. Biol. 2012;19:525–531. S1. doi: 10.1038/nsmb.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan CM, Souda P, Bassilian S, Ujwal R, Zhang J, Abramson J, Ping P, Durazo A, Bowie JU, Hasan SS, et al. Post-translational modifications of integral membrane proteins resolved by top-down Fourier transform mass spectrometry with collisionally activated dissociation. Mol. Cell. Proteomics. 2010;9:791–803. doi: 10.1074/mcp.M900516-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaedler TA, van Veen HW. A flexible cation binding site in the multidrug major facilitator superfamily transporter LmrP is associated with variable proton coupling. FASEB J. 2010;24:3653–3661. doi: 10.1096/fj.10-156927. [DOI] [PubMed] [Google Scholar]
- Seeger MA, Schiefner A, Eicher T, Verrey F, Diederichs K, Pos KM. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science. 2006;313:1295–1298. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]
- Seeger MA, von Ballmoos C, Verrey F, Pos KM. Crucial role of Asp408 in the proton translocation pathway of multidrug transporter AcrB: evidence from site-directed mutagenesis and carbodiimide labeling. Biochemistry. 2009;48:5801–5812. doi: 10.1021/bi900446j. [DOI] [PubMed] [Google Scholar]
- Sigal N, Vardy E, Molshanski-Mor S, Eitan A, Pilpel Y, Schuldiner S, Bibi E. 3D model of the Escherichia coli multidrug transporter MdfA reveals an essential membrane-embedded positive charge. Biochemistry. 2005;44:14870–14880. doi: 10.1021/bi051574p. [DOI] [PubMed] [Google Scholar]
- Sigal N, Molshanski-Mor S, Bibi E. No single irreplaceable acidic residues in the Escherichia coli secondary multidrug transporter MdfA. J. Bacteriol. 2006;188:5635–5639. doi: 10.1128/JB.00422-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal N, Lewinson O, Wolf SG, Bibi E. E. coli multidrug transporter MdfA is a monomer. Biochemistry. 2007;46:5200–5208. doi: 10.1021/bi602405w. [DOI] [PubMed] [Google Scholar]
- Sigal N, Fluman N, Siemion S, Bibi E. The secondary multi-drug/proton antiporter MdfA tolerates displacements of an essential negatively charged side chain. J. Biol. Chem. 2009;284:6966–6971. doi: 10.1074/jbc.M808877200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova IN, Kasho V, Kaback HR. Protonation and sugar binding to LacY. Proc. Natl. Acad. Sci. USA. 2008;105:8896–8901. doi: 10.1073/pnas.0803577105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova I, Kasho V, Sugihara J, Choe JY, Kaback HR. Residues in the H+ translocation site define the pKa for sugar binding to LacY. Biochemistry. 2009;48:8852–8860. doi: 10.1021/bi9011918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soskine M, Adam Y, Schuldiner S. Direct evidence for substrate-induced proton release in detergent-solubilized EmrE, a multidrug transporter. J. Biol. Chem. 2004;279:9951–9955. doi: 10.1074/jbc.M312853200. [DOI] [PubMed] [Google Scholar]
- Tanner MJ. The structure and function of band 3 (AE1): recent developments (review) Mol. Membr. Biol. 1997;14:155–165. doi: 10.3109/09687689709048178. [DOI] [PubMed] [Google Scholar]
- Weinglass AB, Whitelegge JP, Hu Y, Verner GE, Faull KF, Kaback HR. Elucidation of substrate binding interactions in a membrane transport protein by mass spectrometry. EMBO J. 2003;22:1467–1477. doi: 10.1093/emboj/cdg145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinglass AB, Soskine M, Vazquez-Ibar JL, Whitelegge JP, Faull KF, Kaback HR, Schuldiner S. Exploring the role of a unique carboxyl residue in EmrE by mass spectrometry. J. Biol. Chem. 2005;280:7487–7492. doi: 10.1074/jbc.M413555200. [DOI] [PubMed] [Google Scholar]
- Whitelegge JP, Zhang H, Aguilera R, Taylor RM, Cramer WA. Full subunit coverage liquid chromatography electrospray ionization mass spectrometry (LCMS+) of an oligomeric membrane protein: cytochrome b(6)f complex from spinach and the cyanobacterium Mastigocladus laminosus. Mol. Cell. Proteomics. 2002;1:816–827. doi: 10.1074/mcp.m200045-mcp200. [DOI] [PubMed] [Google Scholar]
- Whitelegge J, Halgand F, Souda P, Zabrouskov V. Top-down mass spectrometry of integral membrane proteins. Expert Rev. Proteomics. 2006;3:585–596. doi: 10.1586/14789450.3.6.585. [DOI] [PubMed] [Google Scholar]
- Wilks JC, Slonczewski JL. pH of the cytoplasm and periplasm of Escherichia coli: rapid measurement by green fluorescent protein fluorimetry. J. Bacteriol. 2007;189:5601–5607. doi: 10.1128/JB.00615-07. [DOI] [PMC free article] [PubMed] [Google Scholar]