Abstract
Protein AMPylation – the covalent attachment of an adenosine 5’-monophosphate (AMP) residue to amino acid side chains using ATP as the donor – is a post-translational modification increasingly appreciated as relevant for both normal and pathological cell signaling. In metazoans, single copies of fic-domain-containing AMPylases, the enzymes responsible for AMPylation, preferentially modify a set of dedicated targets and contribute to the perception of cellular stress and its regulation. Pathogenic bacteria can exploit AMPylation of eukaryotic target proteins to rewire host cell signaling machinery in support of their propagation and survival. We review endogenous as well as parasitic protein AMPylation in metazoans and summarize current views of how Fic-domain containing AMPylases contribute to cellular proteostasis.
Keywords: Adenylylation, Heat shock protein, Grp78/BiP, Endoplasmic reticulum (ER), Protein aggregation
The regulation of protein function by AMPylation
Post-translational modifications (PTMs) of proteins have been likened to how the umlaut or the tilde modify the basic alphabet used to compose the written word: they can change use and meaning of the underlying structure. In like manner, every single polypeptide, no matter its genetic specification, can receive further modifications that are not template-encoded, yet with far reaching functional consequences. When key signaling proteins toggle between the phosphorylated and the unmodified state, they change the compendium of proteins they interact with, or adjust their enzymatic activity. Protein methylation, lipidation and glycosylation are yet other examples of modifications that reflect functional diversification of the underlying polypeptide backbone. While modifications such as phosphorylation are often transient and highly dynamic, N-linked glycosylation or protein methylation – as seen for histones – are far more stable PTMs. The cellular PTM universe is rich: while the importance of protein glycosylation, lipidation and phosphorylation has long been recognized, other PTMs remain to be more fully appreciated. Protein AMPylation (see glossary) – also referred to as adenylylation or adenylation – has emerged as a PTM that can regulate or sabotage eukaryotic cell signaling (Fig. 1A) (Itzen et al., 2011, Woolery et al., 2010). All three kingdoms of life (archaea, bacteria, eukaryota) have enzymes capable of protein AMPylation, as do certain viruses. The AMPylases belong to three distinct protein families, which include Glutamine Synthetase Adenylyl transferase (GS-ATase), DrrA and Fic (Filamenation induced by cAMP) (Khater and Mohanty, 2015) (Fig. 1B–C). Protein AMPylation as catalyzed by these enzymes represents a stable PTM that is based on the covalent linkage of AMP to amino acid side chains. In contrast, transient AMPylation is a frequent, energetically favorable modification in several biosynthetic pathways (Text Box I). In the early 1970s, protein AMPylation was described in Escherichia coli as a mechanism to control activity of glutamine synthetase (GS) through AMPylation by GS-ATase of up to 12 tyrosine residues (Anderson et al., 1970). Following years of rather limited interest in protein AMPylation, GS-ATase finally lost its orphan status with the discovery of a secreted bacterial AMPylase, VopS, from Vibrio parahaemolyticus. VopS modifies members of the Rho GTPase family upon its translocation by a type-III secretion system into human host cells (Yarbrough et al., 2009). VopS-mediated AMPylation relies on the catalytic activity of the so-called filamentation induced by cAMP (fic) domain, a domain present in more than 24,000 proteins (InterPro domain IPR00381), most of which are found in bacteria (approximately 21’500) and some 1700 representatives encoded by eukaryotes. The human genome has a single copy of a fic domain-containing gene (FICD/HYPE) (Finn et al., 2017). All fic domains contain a signature motif near their C-terminus (HxFx[D/E]GN[G/K]RxxR) that is directly involved in catalysis, not strictly restricted to AMP transfer (Text Box II) (Engel et al., 2012). DrrA proteins were the last family of AMPylases to be discovered. So far, only a few members in Legionella pneumophila spp. have been described (Müller et al., 2010). DrrA proteins do not contain a fic domain but share certain structural features with GS-ATases from bacteria (Müller et al., 2010). These enzymes – together with yet-to-be-identified de-AMPylases – specify the stable AMPylome in eukaryotic cells, believed to be involved in the regulation of cellular proteostasis.
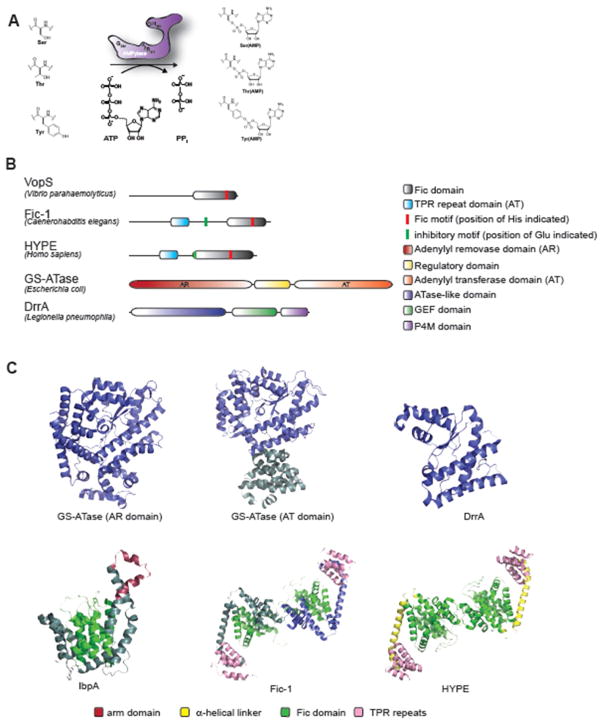
Figure 1. Organization and structure of prokaryotic and eukaryotic AMPylases.
(A) Schematic representation of AMP transfer to Serine (Ser), Threonine (Thr) and Tyrosine (Tyr) catalyzed by a Fic-domain containing AMPylase. (B) Proportional representation of domain organization of fic-domain containing AMPylases VopS (V. parahaemolyticus), Fic-1 (C. elegans) and HYPE (H. sapiens) as well as non-fic AMPylases DrrA (L. pneumophila) and GS-ATase (E. coli). Key amino acids of the active site motif (His) and inhibitory motif (Glu) are highlighted in red and green, respectively. (C) Structures of five representative AMPylases. GS-ATase (AR domain PDB ID: 1V4A; AT domain PDB ID: 3K7D) (Xu et al., 2010, Xu et al., 2004), DrrA (PDB ID: 3NKU) (Müller et al., 2010) (both non-fic) as well as IbpA (PDB: 3N3U) (Xiao et al., 2010) are presented as monomers. Fic-1 (PDB ID: 5JJ7) (Truttmann et al., 2016) and HYPE (PDB ID: 4U07) (Bunney et al., 2014) are shown as inverted homo-dimers (as found in solution). Dimerization is mediated exclusively through interactions between the two fic domains while TPR domains do not participate and extend from the individual monomers.
Text Box I. Transient AMPylation.
The covalent attachment of AMP to proteins by GS-ATase, DrrA or Fic proteins results in a stable phosphodiester-linked modification that requires dedicated enzymes for its removal. However, transient AMPylation is widespread as carboxylate-activating modification on reaction intermediates in biosynthetic pathways. It contributes to fatty acid oxidation and ribosomal as well as non-ribosomal peptide synthesis (Babbitt et al., 1992, Stachelhaus et al., 1999, Trivedi et al., 2004, Turgay et al., 1992). AMPylated carboxylates sites (carboxylate adenylate) represent a high-energy acid anhydride prone to hydrolysis. To avoid its decomposition, dedicated enzymes – often the AMPylases themselves – catalyse the covalent linkage of the reaction intermediate to a nucleophile (alcohol, thiol, amine), liberating AMP from the carboxylate. Transient carboxylate adenylate-forming enzymes include the closely related non-ribosomal peptide synthetases (NRPS) (reviewed in: (Challis, 2005)), acyl- or aryl ChA synthetases as well as luciferase oxidoreductases (reviewed in (Gulick, 2009)) and aminoacyl-tRNA synthetases (reviewed in (Francklyn, 2008)), polynucleotide ligases (reviewed in (Shuman, 2009)) and enzymes contributing to siderophore synthesis (reviewed in (Balhara et al., 2016)).
The ligation of ubiquitin and ubiquitin-like proteins (UBLs) to target proteins also involves a transient AMPylation step (Hochstrasser, 2009). There, the AMPylase E1, also referred to as E1-like enzyme, attaches AMP to the carboxyl group of the C-terminal glycine of Ubl. The high-energy Ubl-AMP intermediate is subsequently attacked by the catalytic cysteine of E1, followed by transfer of Ubl onto the Ubl-conjugating enzyme E2 (Scheffner et al., 1995).
Text Box II. Alternative catalytic functions of FIC proteins.
Most characterized FIC proteins support the covalent attachment of an AMP moiety to a proteinaceous target. A number of FIC proteins have evolved alternative catalytic functions. The bacteriophage-encoded FIC protein Doc acts as a kinase and phosphorylates translation elongation factor EF-Tu in on Tyr378 (Castro-Roa et al., 2013, Cruz et al., 2014). This modification impedes translational activity and inhibits E.coli cell division and propagation. Another pair of FIC proteins that catalyze alternative reactions are the two bacterial proteins AnkX (from Legionella pneumophila) and CBU2078 (from Coxiella burnetti). Both proteins are translocated into human host cells by a dedicated type IV secretion apparatus, where they phosphorylcholinate host proteins using CDP-choline as donor (Campanacci et al., 2013, Mukherjee et al., 2011). AnkX targets the small GTPases Rab1 and Rab35 and adds a phosphorylcholine moiety to a Tyr residue in the switch II region, thereby trapping the GTPase in the GDP-bound state. While not lethal, this modification impairs intracellular vesicle trafficking and causes Golgi fragmentation (Mukherjee et al., 2011). CBU2078 phosphorylcholinates yet to be identified human target proteins but does not obviously affect vesicular trafficking or Golgi integrity.
The FIC protein AvrAC, a translocated effector protein of the plant pathogen Xanthomonas campestris covalently attaches uridine 5’-monophosphate (UMP) to Ser/Thr residues of kinases BIK1, PBL2 and RIPK, thereby occluding phosphorylation sites in the activation loop, thus limiting kinase activity (Feng et al., 2012). UMPylation-dependent inhibition of BIK1, PBL2 and RIPK attenuates the plant cell’s ability to launch an innate immune response against the bacterial intruder.
In addition to Fic enzymes dedicated to the transfer non-AMP moieties, several bona fide bacterial AMPylases accept different nucleotide triphosphates in vitro. IbpA from Histophilus somnii modifies Cdc42, its prime target, with CMP and also has a limited ability to transfer UMP and TMP (Mattoo et al., 2011). Similarly, VopS of Vibrio parahaemolyticus was shown to attach GMP, CMP as well as UMP to Cdc42 (Mattoo et al., 2011). Among metazoan AMPylases, the constitutively active version of the C. elegans AMPylase Fic-1 (E274G) can undergo auto-CMPylation and auto-UMPylation cycles in vitro (Truttmann et al., 2016). The positioning of adenosine in the catalytic cleft of HYPE suggests a rather loose fit, supporting the possible binding of other nucleotide substrates or cofactors in this site. Indeed, HYPE efficiently binds not only ATP but also GTP and to a lesser extent CTP and UTP (Bunney et al., 2014). Whether these enzymes in fact use these alternative nucleotide substrates in vivo remains to be demonstrated.
Here we discuss AMPylases that modify eukaryotic proteins and the consequences of such modification. We shall not cover eukaryotic adenylyl transferases that modify small, non-peptide molecules i.e. nicotinamide mononucleotide adenylyltransferase (reviewed in (Petrelli et al., 2011)), bacterial AMPylases that modify endogenous bacterial targets (reviewed in (Garcia-Pino et al., 2014, Harms et al., 2016, Itzen et al., 2011, Woolery et al., 2010)) or methods to study protein AMPylation (reviewed in (Hedberg and Itzen, 2015, Müller et al., 2014, Westcott and Hang, 2014).
Bacterial AMPylases that modify eukaryotic proteins
Eukaryotic proteins are AMPylated either by endogenous AMPylases or by bacterial toxins translocated into eukaryotic host cells in the course of infection (see also table 1). AMPylation of cytosolic host proteins appears to be an efficient strategy of hijacking cellular signaling machinery as a means of maximizing pathogen fitness and survival. Most of these AMPylases are evolutionarily related and rely on the presence of a fic domain endowed with AMP transferase activity. The basic catalytic machinery is shared among the distinct representatives, yet the modification-receiving host proteins vary for the different AMPylases.
Table 1.
Metazoan target proteins of AMPylases
| Species | Protein | Target(s) | Site(s) of modification | Consequence on cell signaling |
|---|---|---|---|---|
| Histophilus somnii | IbpA | Rho GTPases (Rac1, Cdc42, RhoA, RhoB, RhoC, RhoG, TC10) | Tyr32 of switch I region | cytotoxic; interferes with regulation of cytoskeleton |
| Pasteurella multocida | PfhB2 | Rho GTPases (Cdc42, Rac1, RhoA, RhoG, TC10) | Tyr32 of switch I region | cytotoxic; interferes with regulation of cytoskeleton |
| Vibrio parahaemolyticus | VopS | Rho GTPases (Rac1, Cdc42, RhoA) | Thr35 of switch I region | cytotoxic; interferes with regulation of cytoskeleton |
| Legionella pneumophila | DrrA | Rab-1b | Tyr77 of switch II region | inhibits proper Rab- 1b downstream signaling |
| Bartonella henselae | BepA | 40kDa protein / 50kDa protein | n.d. | increase in cellular cAMP levels |
| Bartonella rochalimae | Bep2 | vimentin | Tyr53 | n.d. |
| Homo sapiens | HYPE/FICD | BiP, Hsp70, Eef1A, Hsp40 | BiP: Ser365/Thr3 66 or Thr518; Hsp70: n.d.; Eef1A:Thr26 1; HSP40: n.d. | BiP AMPylation negatively regulates UPR activation in the ER; HSP40 and HSP70 AMPylation interferes with chaperoning activities |
| Drosophila melanogaster | dfic | BiP | Thr366 | Absence of dfic renders flies blind; BiP AMPylation negatively regulates UPR activation in ER |
| Caenerobhaditis elegans | Fic-1 | Hsp-1, Hsp-3, Eef-1A.2 | HSP-1: n.d.; Hsp-3: Thr176; eEF1A: Thr269, Thr432 | AMPylation levels directly correlate to pathogen tolerance; HSP-1 and HSP-3 AMPylation interferes with cellular chaperoning machineries (cytosol, ER) |
In bacteria, Fic proteins are often the toxic unit of toxin-antitoxin systems, attaching AMP to bacterial type II topoisomerases to reversibly stall their enzymatic activities and thus halt bacterial growth (Engel et al., 2012, Harms et al., 2015, Lu et al., 2016). The presence of a potent antitoxin, usually a small peptide or protein that directly binds to the toxin, inhibits the AMPylase and allows bacterial propagation (Harms et al., 2015). A subset of pathogenic bacteria has evolved Fic proteins that enter eukaryotic cells in an attempt to rewire cellular signaling cascades in the host.
Among the best-studied examples is VopS of V. parahaemolyticus, a secreted bacterial AMPylase that modifies Thr35 in the switch I loop of small Rho GTPases (Yarbrough et al., 2009). VopS-mediated AMPylation of Rho, Rac1 and Cdc42 impairs cellular signaling in several ways: the AMP moiety not only interferes with the binding of direct interaction partners, but also prevents E3 ubiquitin ligases from targeting these now non-functional AMPylated GTPases for proteolytic degradation (Luong et al., 2010, Woolery et al., 2014, Yarbrough et al., 2009). VopS also alters cellular immunity through inhibition of the pro-inflammatory NFκB signaling cascade, limits the generation of superoxide and attenuates Erk and JNK signaling (Woolery et al., 2014). AMPylation of Rho GTPases further activates the pyrin-dependent inflammasome while inhibiting NLRC4-dependent inflammasome activation (Higa et al., 2013, Woolery et al., 2014, Xu et al., 2014). As a direct consequence, the actin cytoskeleton collapses and cells rapidly die (Yarbrough et al., 2009).
The bacterial surface antigen IbpA of Histophilus somnii contains a pair of fic domains, together with a YopT domain at its C-terminus (Worby et al., 2009). Following attachment to the host cell, a C-terminal portion of IbpA is internalized (presumably by pinocytosis) and transfers AMP to up to seven distinct Rho family GTPases on a conserved Tyr residue in the switch I region (Mattoo et al., 2011, Worby et al., 2009, Xiao et al., 2010). The consequences of these modifications largely mirror the toxicity associated with VopS activity, as manifest from the collapse of cytoskeletal architecture and cell death. Co-crystallization of IbpAFic2 with AMPylated Cdc42 showed that IbpA simultaneously grips the switch1 and switch2 regions of Cdc42, an interaction that involves both the Fic as well as the arm domain of IbpA (Xiao et al., 2010) (Fig. 1C). The IbpAFic2-Cdc42 complex structurally mimics the guanosine nucleotide dissociation inhibitor (GDI)-bound state of the Rho GTPase. The less well characterized protein PfhB2 of Pasteurella multocida, a close homolog of IbpA, likewise possesses in vitro AMPylation activity and transfers AMP to Rho, Rac1 and Cdc42 (Mattoo et al., 2011).
Bartonellae spp. comprise a number of human pathogens that transfer Bartonella effector proteins (Beps) into host cells (Rhomberg et al., 2009, Truttmann et al., 2011). A sizable subset of these translocated effectors comes equipped with an N-terminal fic domain and engages in host target modifications (Engel et al., 2011, Schmid et al., 2004, Schulein et al., 2005). BepA of Bartonella haenselae not only binds to Gαs and raises cellular cAMP levels but also efficiently attaches AMP to two yet to be identified proteins of ~ 40–50 kDa (Palanivelu et al., 2011, Pulliainen et al., 2012, Schmid et al., 2006). Furthermore, Bep2 of Bartonella rochalimae AMPylates Tyr53 in mouse vimentin, an integral component of intermediate filaments (Pieles et al., 2014). Additional fic domain-containing Beps remain to be investigated.
The secreted Legionella pneumophila effector DrrA (also referred to as SidM) is a unique example of a bacterial non-Fic enzyme that AMPylates proteins upon translocation into host cells. DrrA was first described as a membrane-anchored guanosine nucleotide exchange factor (GEF), with an additional N-terminal domain that – when expressed individually in cells – exhibits cytotoxicity (Brombacher et al., 2009, Murata et al., 2006). This domain was recently assigned AMPylase activity, modifying Tyr77 in the switch II region of the small Rab GTPase Rab1b (Müller et al., 2010). AMPylation of Rab1b restricts binding of GTPase activating proteins (GAPs) and subsequent activation of Rab1b, thus locking Rab1b in the GTP-bound state. Simultaneously, AMPylation of Rab1b blocks downstream interactions with binding partners such as MICAL-3 and enhances retention of Rab1b at Legionella-containing vacuoles (LCVs) during infection (Hardiman and Roy, 2014). Unlike Fic domain-containing effectors, the catalytic cleft of DrrA’s AMPylase domain shares structural similarities with the bacterial AMPylase GS-ATase, including the catalytically important GxDxD motif, highlighting a different evolutionary origin of this effector as compared to FIC proteins (Khater and Mohanty, 2015).
Eukaryotic AMPylases
The assignment of an enzymatic function to fic domains in bacterial toxins and to eukaryotic Fic proteins fueled the notion of endogenous protein AMPylation in eukaryotes. Work on metazoan AMPylases has focused on the human (HYPE), Drosophila melanogaster (CG9523) and Caenorhabditis elegans (Fic-1) enzymes. As the number of sequenced eukaryotic genomes grows, we now know that numerous metazoans contain only a single functional fic gene in their genome (Finn et al., 2017, Finn et al., 2016). Phylogenetic analysis suggests that these fic genes were acquired repeatedly and independently in the course of evolution, probably in horizontal gene transfer events. This has led to a classification of metazoan fic genes into four distinct groups, with small differences in their conserved Fic motifs (Khater and Mohanty, 2015). Their overall architecture is remarkably similar: its hallmarks are the presence of an N-terminal transmembrane anchor, followed by – usually two – tetratricopeptide repeats (TPRs) linked to the C-terminal fic domain by a series of α helices (Bunney et al., 2014, Truttmann et al., 2016). Target AMPylation is catalyzed by the fic domain, whose core consists of four α helices, a shared feature among all metazoan AMPylases. The TPR domains presumably orchestrate target recognition and selectivity.
The activity of eukaryotic AMPylases is tightly regulated, with little or no activity under standard growth conditions (Engel et al., 2012). Accordingly, over-expression of endogenous wild-type AMPylases has a limited effect on cellular signaling. Recombinantly produced wild-type AMPylases are comparatively poor at modifying target proteins in vitro (Engel et al., 2012, Ham et al., 2014, Sanyal et al., 2015, Truttmann et al., 2016, Worby et al., 2009). Such intra- or intermolecular inhibition – purified recombinant AMPylases behave as non-covalent dimers upon size exclusion chromatography (Bunney et al., 2014, Truttmann et al., 2016) – results from the positioning of an inhibitory α helix (αinh) that limits access to the ATP binding site (Engel et al., 2012). In its “off” state, interactions between an Arg residue embedded within the catalytic fic motif (HxFx[D/E]GN[G/K]RxxR) and a Glu residue of the αinh helix prevent ATP from entering the catalytic cleft (Engel et al., 2012). Significant conformational changes are required to weaken the interaction between αinh and the catalytic core and to allow a transition of the enzyme to its “on” state (Bunney et al., 2014, Truttmann et al., 2016). However, the nature and cause for such changes remain elusive. The αinh helix is absent from secreted bacterial AMPylases, yet shares remarkable similarities to bacterial AMPylase-specific antitoxins, including a canonical fic inhibition motif [S/T]xxxE[G/N], suggesting a common evolutionary origin of the two elements (Engel et al., 2012, Khater and Mohanty, 2015). Substitution of the αinh-embedded glutamine residue with a glycine residue relieves structural constraints (vide supra) and renders the enzyme constitutively active (Engel et al., 2012, Goepfert et al., 2013). Mutating the conserved histidine to alanine within the fic motif (HxFx[D/E]GN[G/K]RxxR) prevents AMP transfer activity, even when the enzyme is in its “on” state (αinh glutamine to glycine / fic motif histidine to alanine double mutant) (Worby et al., 2009, Yarbrough et al., 2009). This particular histidine residue acts as a proton sink and accepts the hydrogen atom from the hydroxyl group of the targeted amino acid side chain (Engel et al., 2012, Xiao et al., 2010). The constitutively-active (HYPE E234G, Fic-1 E274G, CG9523 E247G) as well as the catalytically dead (HYPE H363A, Fic-1 H404A, CG9523 H375A) enzyme versions have been the workhorses with which to investigate the fundamentals of AMPylation in metazoans. Recent advances in the understanding of these enzymes allow us to study this PTM in its proper cellular context.
Auto-AMPylation is a feature shared among fic-domain containing AMPylases (Bunney et al., 2014, Engel et al., 2012, Goepfert et al., 2013, Kinch et al., 2009, Lu et al., 2016, Luong et al., 2010, Palanivelu et al., 2011, Pieles et al., 2014, Sanyal et al., 2015, Truttmann et al., 2016, Xiao et al., 2010). Auto-AMPylation in cis of the Neisseria meningitidis AMPylase NmFic relieves auto-inhibition. This is a prerequisite for target modification, while preventing the formation of an inhibitory tetrameric NmFic complex (Stanger et al., 2016). Self- modification of Tyr 183 and Tyr188, two residues within the αinh helix, leads to partial unfolding of αinh and enhances access to the active site. In metazoans, auto-AMPylation events have been mapped for HYPE (Thr183, Ser79 and Thr80) as well as for Fic-1 (Thr352, Thr476) (Sanyal et al., 2015, Truttmann et al., 2016). However, none of these auto-AMPylation sites lie within the αinh, nor does substitution of these sites materially change the enzymes’ auto-modification and target modification properties.
In what follows, we review current literature on eukaryotic AMPylases. We highlight similarities as well as differences that arose between species.
CG9523 (dfic) (Drosophila melanogaster)
The D. melanogaster HYPE ortholog CG9523 is an endoplasmic reticulum (ER)-resident type II transmembrane protein that is N-glycosylated on Asn288, proteolytically processed and eventually released via the secretory pathway. Upon secretion, the enzyme localizes to the cell surface of capitate projections, the putative sites of neurotransmitter recycling (Rahman et al., 2012). CG9523 was the first metazoan AMPylase to be investigated in the context of an in vivo model. Knock-out flies were insensitive to light stimuli due to a failure to activate postsynaptic neurons. Expression of wild-type CG9523, but not catalytically dead CG9523 (H275A) in capitate projections of glia cells rescued this defect, suggesting a role for AMPylation in neurotransmitter recycling, at least in the fly (Rahman et al., 2012).
In vitro, CG9523 AMPylates the ER-resident HSP70 chaperone Grp78/BiP in a Ca2+-dependent manner (Ham et al., 2014). The modified site is Thr366, a residue embedded within Grp78/BiP’s ATPase domain. Grp78/BiP is a major regulator of the unfolded protein response (UPR) in the ER (UPRER) and its modification provided the first evidence for a link between intracellular AMPylation levels, stress signaling and proteostasis. Indeed, Grp78/BiP AMPylation levels are high at rest, but are reduced under stress conditions that lead to protein unfolding (Ham et al., 2014). AMPylation of Grp78/BiP is restricted to its inactive, ATPase “off” state. Thus, AMPylation is thought to lock BiP in its inactive state, either by affecting its intrinsic ATPase activity, by preventing structural rearrangements required for chaperoning function, or by interfering with the recruitment of co-chaperones of the DnaJ family (Ham et al., 2014). However, Grp78/BiP AMPylation is reversible and upon demand, Grp78/BiP is de-AMPylated to support protein refolding in the ER.
How CG9523 reaches the extracellular space, modifies neurotransmitter transporter activity in glia cells and whether Grp78/BiP AMPylation plays a role in this process remain to be investigated.
Huntingtin yeast interacting protein E (HYPE, FICD) (Homo sapiens)
HYPE was described in 1998 as one of 13 proteins found in a yeast two-hybrid screen to interact with the N-terminal portion of Huntingtin (Faber et al., 1998). However, this work did not attribute a function or enzymatic activity to HYPE. It was not until 2009 that HYPE was identified as a bona fide AMPylase (Worby et al., 2009). HYPE is composed almost entirely of α helices that build the TPR and FIC domains, with a linker consisting of a single α helix between them. Intramolecular interactions between the TPR motifs, linker and FIC domain impose a compact fold on HYPE-type proteins, with only limited flexibility (Bunney et al., 2014). HYPE forms a stable, asymmetric dimer in solution and crystallizes as a dimer. Antiparallel dimerization is achieved exclusively through interactions between the two FIC domains in the dimer. The TPR domains do not contribute to dimerization and protrude on opposite ends from HYPE dimers to engage independently in protein-protein interactions. HYPE dimerization is thought to affect its catalytic properties as well as its localization.
Localization
HYPE has the basic topology of a type II membrane protein and is N-glycosylated at Asn275 (Sanyal et al., 2015). When fused to GFP, HYPE’s N-terminal hydrophobic stretch (aa1-45) delivers GFP partially into the ER while excluding it from the nucleus, suggesting an embedded ER localization signal. Indeed, HYPE is at least in part an ER-resident AMPylase and is found predominantly in the ER-nuclear envelope continuum (Bunney et al., 2014, Sanyal et al., 2015, Truttmann et al., 2015). Recent studies identified cytosolic HYPE targets (see below). Moreover, HYPE activity is maximal in the presence of Mn2+ or Mg2+, whereas elevated Ca2+ concentrations – as found in the ER – did not enhance HYPE’s ability to AMPylate Grp78/BiP in vitro (Sanyal et al., 2015). The active site-embedded residue Asp367 (HPFIDGNGR) coordinates a Mg2+ ion that bridges the α- and β-phosphates of ATP during catalysis and is essential for catalysis (Bunney et al., 2014). ATP levels in the ER increase upon release of lumenal ER-resident Ca2+ pools, presumably generating favorable conditions for HYPE to AMPylate targets (Vishnu et al., 2014).
UPRER
The activity of metazoan AMPylases is tightly controlled and this is for a reason: knock-down of HYPE reduces cell survival under UPRER-inducing stress, while over-expression of constitutively active HYPE (E234G) is cytotoxic and triggers capase-dependent apoptosis (Preissler et al., 2015, Sanyal et al., 2015, Truttmann et al., 2015). Moreover, there is evidence for HYPE being a bi-functional enzyme, with both AMPylase and de-AMPylase activity, underscoring the need for its rigorous regulation (Text Box III) (Preissler et al., 2017). A growing body of evidence suggests that HYPE regulates the UPRER through modification of the ER-resident HSP70 family protein Grp78/BiP, a negative regulator of UPRER induction. Earlier studies suggested that Grp78/BiP is regulated by ADP-ribosylation, phosphorylation or both (Gaut, 1997, Ledford and Leno, 1994, Nakai et al., 1995). These conclusions were inferred from experiments that tracked the transfer of radiolabelled phosphate (32P) or adenosine (3H) to Grp78/BiP. Attempts to directly detect these PTMs failed (Chambers et al., 2012). It is now clear that BiP is AMPylated, rather than ADP-ribosylated or phosphorylated. However, the actual sites of modification and the immediate functional consequences of Bip AMPylation remain somewhat controversial. Two different models have been proposed as to how Grp78/BiP AMPylation may affect activation of the UPRER or deal with its consequences: one model proposes that HYPE-mediated AMPylation enhances its ATPase activity without affecting binding of denatured proteins (Sanyal et al., 2015). ER stress elevates intracellular HYPE levels and HYPE knock-down prevents the induction of the ATF-6 and PERK-dependent UPRER branches. This supports a mechanism in which HYPE-mediated BiP AMPylation is an activating modification, induced by ER stress. The alternative model postulates that AMPylation of ATP-bound, substrate-free Grp78/BiP is an inactivation modification that helps maintain a readily accessible yet inactive Grp78/BiP pool under low stress conditions (Ham et al., 2014, Preissler et al., 2015). AMPylation of Grp78/BiP is proposed to lock Grp78/BiP in an ATP-bound state characterized by reduced ATPase activity, non-responsiveness to J co-chaperone-dependent enhanced ATP hydrolysis and elevated substrate koff rates. Indeed, upon induction of ER stress, Grp78/BiP AMPylation decreases and reemerges once proteostasis is reestablished. Consequently, HYPE deficiency results in elevated levels of active Grp78/BiP in the ER. This not only boosts its intrinstic buffer capacity to deal with an increased load of unfolded proteins, but also attenuates induction of the UPR (Preissler et al., 2015) (Fig. 2).
Text Box III. HYPE as a bi-functional enzyme.
A recent study proposes that HYPE is primarily a de-AMPylase, tasked with removing AMP from modified BiP (Preissler et al., 2017). The presence of Glu234 in HYPE is critical for this process: interactions between the side chains of Glu234 and Arg371 sterically prevent proper alignment of ATP in the active site, thus preventing wild-type HYPE from target modifications. The flexibility of the Glu234 side chain could allow HYPE to accommodate alternative substrates such as ADP in its catalytic cleft, supporting target de-AMPylation. The model suggests that an increase in unfolded proteins in the ER stimulates de-AMPylation of BiP to increase the available pool of active BiP. Low levels of unfolded proteins would trigger a conformational change in HYPE to disengage Glu234 from its interactions with Arg371 and enable HYPE-mediated target AMPylation. Apart from HYPE’s suggested de-AMPylase activity, this model is in accordance with previous work on BiP AMPylation.
The concept of a single enzyme performing both target AMPylation/deAMPylation is not unique to Fic proteins: The very first described bacterial AMPylase, GS-ATase, not only attaches AMP to GS but can also remove it (Chock et al., 1980, Rhee et al., 1989). Although the AMPylation/de-AMPylation functionalities of GS-AT are linked to two different portions of the enzyme, the two domains are structurally similar and differ only in the positioning of flexible loop elements (Jaggi et al., 1997, Khater and Mohanty, 2015). The proposed de-AMPylase activity of wild-type HYPE needs to be reconciled with HYPE’s AMPylation activity in vitro and in vivo (Engel et al., 2012, Mattoo et al., 2011, Sanyal et al., 2015, Truttmann et al., 2015, Worby et al., 2009) and suggests an additional layer of regulatory complexity.
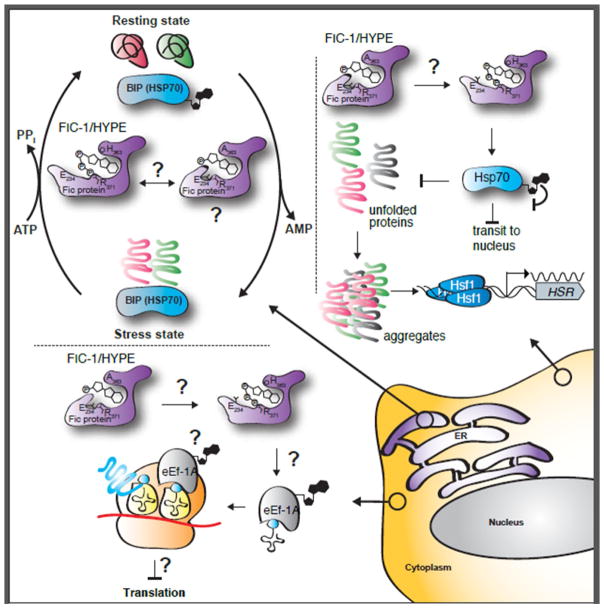
Figure 2. Consequences of HYPE-mediated protein AMPylation in eukaryotic cells.
The three zoom-in circles show cellular events affected by AMPylation in the indicated cellular compartments (cytoplasm, ER). Reaction arrows / AMPylated proteins marked with a question mark represent probable, yet non-described links.
Initial work has mapped the AMPylation site on Grp78/BiP to Thr366, a conserved residue in the ATPase domain of Grp78/BiP (Sanyal et al., 2015). Substitution of Thr366 eliminated Grp78/BiP as a substrate for HYPE (E234G) in vitro. More recent work suggests that Grp78/BiP is uniquely AMPylated on Thr518 in vitro and in vivo, a conserved residue on a connecting loop between β strands 7 and 8 of Grp78/BiP (Preissler et al., 2015). This loop is stabilized by six polar interactions in the ADP-bound conformation of Grp78/BiP. An exchange of ADP for ATP renders Thr518 on Grp78/BiP accessible to the catalytic cleft of HYPE. Indeed, ATP-locked BiP mutants such as BiP (E201G) and BiP (T229A) are preferentially AMPylated, whereas mutants unable to adopt the ATP-bound conformation such as BiP (G226D) do not serve as substrates in vitro.
AMPylation of non-ER targets
HYPE modifies a series of targets in vitro, including Rho family GTPases, core histones, heat shock proteins, nuclear envelope proteins, tubulins, components of the ATP synthase machinery as well as factors that regulate transcription and translation (Broncel et al., 2015, Engel et al., 2012, Ham et al., 2014, Mattoo et al., 2011, Preissler et al., 2015, Sanyal et al., 2015, Truttmann et al., 2016, Truttmann et al., 2015, Truttmann et al., 2017, Worby et al., 2009). The ability of recombinant HYPE and HYPE (E234G) to AMPylate purified proteins or putative targets offered in the context of cell lysates must be viewed critically, not unlike other enzyme-substrate combinations, such as protein kinases, when examined under artificial in vitro conditions. Early work on HYPE’s target preference highlighted its ability to modify Rho family GTPases Cdc42, Rac1 and RhoA in vitro, in a manner similar to VopS (Engel et al., 2012, Mattoo et al., 2011, Worby et al., 2009). These results were later shown to depend largely on the version of recombinant HYPE or HYPE (E234G) used in the assays. Full-length GST-HYPE fusions as well as 6xHIS-tagged HYPE181-458 (E234G) reliably modify Cdc42, Rac1 and RhoA in vitro (Engel et al., 2012, Mattoo et al., 2011, Worby et al., 2009). In contrast, 6xHIS-tagged HYPE45-458 (E234G) or HYPE103-445 (E234G) do not AMPylate Rho GTPase family members at detectable levels (Bunney et al., 2014, Sanyal et al., 2015). Removal of the TPR domains reduced AMPylase activity of recombinant HYPE proteins, suggesting that HYPE truncations may reveal promiscuous activities absent from the full-length enzyme (Bunney et al., 2014). Despite the concern that in vitro AMPylation assays might be prone to produce false positives, there is supporting evidence for HYPE-mediated modification of non-ER proteins: In addition to Grp78/BiP, ATP synthase subunits α and β, tubulin and translation elongation factor 1 directly interact with wild-type HYPE in vivo (Broncel et al., 2015). Expression of HYPE (E234G) in S. cerevisiae – an organism that lacks an endogenous Fic-type AMPylase – triggers a cytosolic heat shock response and evokes cytotoxic effects that limit growth (Truttmann et al., 2017). The primary target for HYPE (E234G) in S.cerevisiae is Ssa2, a major cytoplasmic HSP70 family-type chaperone critical for maintenance of proteostasis. Given that active AMPylation in yeast compromises integrity of its proteome, AMPylation of Ssa2/HSP70 appears to inhibit Ssa2/HSP70’s ability to deal with unfolded proteins in the cytoplasm. This leads to the formation of protein aggregates. If expressed in human cells, HYPE (E234G)-mediated target AMPylation likewise triggers a heat-shock response, attenuates translational activities and interferes with HSP70’s ability to cycle between the cytoplasm and the nucleus, recapitulating the findings made in S. cerevisiae (Truttmann et al., 2017). HYPE-mediated target AMPylation outside of the ER has obvious implications for regulation of cellular signaling cascades (Fig. 2). Nonetheless, how HYPE avoids or escapes the ER to modify cytoplasmic targets remains to be clarified.
Fic-1 (Caenorhabditis elegans)
Fic-1 is the sole FIC protein encoded by the nematode C. elegans. Despite being only 38% identical in amino acid sequence to HYPE, the overall structure of the two proteins is well-conserved. Like HYPE, Fic-1 forms asymmetric dimers through the interactions of two discrete interfaces that are embedded within the Fic domain (Truttmann et al., 2016). Mutations that render Fic-1 strictly monomeric interfere with both auto- and target AMPylation.
Fic-1 is expressed at low levels throughout the worm body, but is most pronounced in the adult germline and in embryonic cells(Truttmann et al., 2016). Within cells, Fic-1 predominantly localizes to the ER-nuclear envelope continuum, yet a fraction of the Fic-1 pool is also found in the cytoplasm.
The role of Fic-1 in animal fitness and cellular signaling has been studied using fic-1 knock-out mutants as well as in animals that express constitutively active Fic-1 (E274G) (Truttmann et al., 2016). Alterations in AMPylation levels did not obviously affect C. elegans physiology, survival, reproduction, or behavior. If exposed to acute or chronic ER stress, neither enhancement nor abrogation of target AMPylation affected the animals’ capacity to cope with it. This suggests a limited role for Fic-1 mediated AMPylation in the regulation of ER homeostasis. However, Fic-1 knock-out worms are more susceptible to infection by Pseudomonas aeruginosa, while animals that express the constitutively active Fic (E274G) mutant show enhanced tolerance to the pathogen. These findings may indicate a link between AMPylation of cellular targets and innate immunity in the nematode.
Fic-1 (E274G) AMPylates core histones, eEF1A-type translation elongation factors as well as heat shock (HSP) 40 and 70 family proteins in vitro (Truttmann et al., 2016, Truttmann et al., 2017). Among the endogenous HSP70 proteins modified are the two C. elegans BiP orthologs (HSP-3, HSP-4) as well as the major cytosolic HSP70 representative, HSP-1. Fic-1’s enzymatic activity is not restricted to nematode targets, and can also AMPylate human (HSPA1A) and yeast (SSA2) cytosolic HSP70 proteins. When expressed in Saccharomyces cerevisiae, Fic-1 (E274G) reliably modifies Ssa2 and interferes with its function (Truttmann et al., 2017).
Metazoan AMPylases preferentially target highly conserved proteins. BiP and HSP-3 as well as eEF1A and eEF1A.2 share >80% amino acid sequence identity, respectively. Core histones are even more conserved and may differ in no more than a single amino acid (Baxevanis and Landsman, 1996). Therefore it is remarkable that HYPE (E234G) and Fic-1 (E274G) modify different residues on these evolutionarily conserved targets: Hsp-3 is AMPylated on Thr176 by Fic-1 (E274G). An exchange of the orthologous residues to BiP S365/T366 or T518 in HSP-3 did not alter HSP-3 AMPylation (Truttmann et al., 2016). Furthermore, eEF1A.2 modification by Fic-1 (E274G) was mapped to T269 as well as T432, while mutating T261, a conserved residue modified in human eEF1A by HYPE (E234G), was of no consequence (Broncel et al., 2015, Truttmann et al., 2016). Thus, it appears that target site preference depends on the enzyme examined and might reflect a distinct substrate fingerprint for each metazoan AMPylase.
Similar to HYPE (E234G), expression of Fic-1 (E274G) in S. cerevisiae induces protein aggregation, triggers a heat shock response and is highly toxic (Truttmann et al., 2017). Intracellular expression of a camelid antibody fragment that binds to Fic-1 (E274G) suppresses its toxicity. AMPylation of human HSP40 and HSP70 as well as HSP-1 (in vitro) and Ssa2 (both in vitro and in vivo) was conclusively attributed to Fic-1 (E274G) activity. Fic-1 co-localized with markers of the ER (Ire-1) as well as with cytoplasmic proteins (Ssa2) when expressed in S. cerevisiae, mirroring its localization pattern observed in its endogenous environment.
Concluding remarks
Our current understanding of protein AMPylation in eukaryotes underscores the importance of this modification as a regulator of its target proteins and co-dependent signaling cascades. Pathogen-secreted AMPylases preferentially modify small GTPases and rewire signaling networks to maximize pathogen survival and proliferation. It appears that metazoan AMPylases primarily contribute to the control of cellular stress responses, both in the cytoplasm and in the ER. However, our understanding of protein AMPylation is incomplete and many questions remain (see outstanding questions). A better understanding of the impact of protein AMPylation on cellular physiology, particularly under stress conditions, may well uncover targets that can be exploited to modulate cellular stress tolerance. From that perspective, the human AMPylase HYPE deserves validation as a therapeutic target.
Outstanding Questions.
What is/are the physiological trigger(s) that transform(s) HYPE from an auto-inhibited into a fully active AMPylase?
How is HYPE activity regulated and is/are there (a) proteinaceous modulator(s)?
How do plants and the majority of fungi compensate for the absence of Fic protein AMPylases in stress signaling?
Is there a dedicated eukaryotic de-AMPylase?
Can metazoan Fic proteins accept other nucleotide substrates in vivo and transfer UMP, GMP or CMP to target proteins instead?
What is the purpose served by auto-AMPylation and AMPylase dimerization?
Trends Box.
Protein AMPylation is a novel post-translational protein modification prevalent in all three kingdoms of life (archaea, bacteria, eukaryota)
The active sites of fic-domain containing AMPylases are structurally highly conserved
Following their translocation into eukaryotic host cells, bacterial AMPylases preferentially modify members of the small Rho or Rab GTPase families to abrogate or alter their function and facilitate bacterial invasion and intracellular trafficking.
Emerging data suggest that metazoan AMPylases contribute to cellular stress signaling and perception, as well as the maintenance of proteostasis.
Metzoan AMPylases preferentially modify heat shock protein 70 (HSP70) family proteins in the ER as well as the cytoplasm and influence their chaperoning activity.
Acknowledgments
The authors like to thank Leo Hanke, Tao Feng and Ross Cheloha for help with preparing figure 1A and 1C. This work was supported by awards from the National Institute of Health to H.L.P. M.C.T. is supported by a Young Investigator Award from Emerald Foundation, Inc.
Glossary
- AMPylation, Adenylation, Adenylylation
refers to the covalent attachment of an AMP residue to a free hydroxyl of an amino acid side chain.
- AMPylase, Adenylyl transferase
(also referred to as AMPylators) enzymes that covalently attach AMP residues to target proteins
- AMPylome
the complete universe of AMPylated proteins
- Target
refers to a protein receiving an AMP residue catalyzed by an AMPylase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Anderson WB, Hennig SB, Ginsburg A, Stadtman ER. Association of ATP: glutamine synthetase adenylyltransferase activity with the P1 component of the glutamine synthetase deadenylylation system. Proc Natl Acad Sci U S A. 1970;67:1417–1424. doi: 10.1073/pnas.67.3.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babbitt PC, Kenyon GL, Martin BM, Charest H, Slyvestre M, Scholten JD, … Dunaway-Mariano D. Ancestry of the 4-chlorobenzoate dehalogenase: analysis of amino acid sequence identities among families of acyl:adenyl ligases, enoyl-CoA hydratases/isomerases, and acyl-CoA thioesterases. Biochemistry. 1992;31:5594–5604. doi: 10.1021/bi00139a024. [DOI] [PubMed] [Google Scholar]
- 3.Balhara M, Chaudhary R, Ruhil S, Singh B, Dahiya N, Parmar VS, … Chhillar AK. Siderophores; iron scavengers: the novel & promising targets for pathogen specific antifungal therapy. Expert Opin Ther Targets. 2016;20:1477–1489. doi: 10.1080/14728222.2016.1254196. [DOI] [PubMed] [Google Scholar]
- 4.Baxevanis AD, Landsman D. Histone Sequence Database: a compilation of highly-conserved nucleoprotein sequences. Nucleic Acids Res. 1996;24:245–247. doi: 10.1093/nar/24.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brombacher E, Urwyler S, Ragaz C, Weber SS, Kami K, Overduin M, Hilbi H. Rab1 guanine nucleotide exchange factor SidM is a major phosphatidylinositol 4-phosphate-binding effector protein of Legionella pneumophila. J Biol Chem. 2009;284:4846–4856. doi: 10.1074/jbc.M807505200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broncel M, Serwa RA, Bunney TD, Katan M, Tate EW. Global profiling of HYPE mediated AMPylation through a chemical proteomic approach. Mol Cell Proteomics. 2015 doi: 10.1074/mcp.O115.054429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bunney TD, Cole AR, Broncel M, Esposito D, Tate EW, Katan M. Crystal structure of the human, FIC-domain containing protein HYPE and implications for its functions. Structure. 2014a;22:1831–1843. doi: 10.1016/j.str.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campanacci V, Mukherjee S, Roy CR, Cherfils J. Structure of the Legionella effector AnkX reveals the mechanism of phosphocholine transfer by the FIC domain. EMBO J. 2013;32:1469–1477. doi: 10.1038/emboj.2013.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castro-Roa D, Garcia-Pino A, De Gieter S, van Nuland NA, Loris R, Zenkin N. The Fic protein Doc uses an inverted substrate to phosphorylate and inactivate EF-Tu. Nat Chem Biol. 2013;9:811–817. doi: 10.1038/nchembio.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Challis GL. A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases. Chembiochem. 2005;6:601–611. doi: 10.1002/cbic.200400283. [DOI] [PubMed] [Google Scholar]
- 11.Chambers JE, Petrova K, Tomba G, Vendruscolo M, Ron D. ADP ribosylation adapts an ER chaperone response to short-term fluctuations in unfolded protein load. J Cell Biol. 2012;198:371–385. doi: 10.1083/jcb.201202005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chock PB, Rhee SG, Stadtman ER. Interconvertible enzyme cascades in cellular regulation. Annu Rev Biochem. 1980;49:813–843. doi: 10.1146/annurev.bi.49.070180.004121. [DOI] [PubMed] [Google Scholar]
- 13.Cruz JW, Rothenbacher FP, Maehigashi T, Lane WS, Dunham CM, Woychik NA. Doc toxin is a kinase that inactivates elongation factor Tu. J Biol Chem. 2014;289:19276. doi: 10.1074/jbc.M113.544429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engel P, Goepfert A, Stanger FV, Harms A, Schmidt A, Schirmer T, Dehio C. Adenylylation control by intra- or intermolecular active-site obstruction in Fic proteins. Nature. 2012;482:107–110. doi: 10.1038/nature10729. [DOI] [PubMed] [Google Scholar]
- 15.Engel P, Salzburger W, Liesch M, Chang CC, Maruyama S, Lanz C, … Dehio C. Parallel evolution of a type IV secretion system in radiating lineages of the host-restricted bacterial pathogen Bartonella. PLoS Genet. 2011;7:e1001296. doi: 10.1371/journal.pgen.1001296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faber PW, Barnes GT, Srinidhi J, Chen J, Gusella JF, MacDonald ME. Huntingtin interacts with a family of WW domain proteins. Hum Mol Genet. 1998;7:1463–1474. doi: 10.1093/hmg/7.9.1463. [DOI] [PubMed] [Google Scholar]
- 17.Feng F, Yang F, Rong W, Wu X, Zhang J, Chen S, … Zhou JM. A Xanthomonas uridine 5'-monophosphate transferase inhibits plant immune kinases. Nature. 2012;485:114–118. doi: 10.1038/nature10962. [DOI] [PubMed] [Google Scholar]
- 18.Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ, … Mitchell AL. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 2017;45:D190–D199. doi: 10.1093/nar/gkw1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, … Bateman A. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Francklyn CS. DNA polymerases and aminoacyl-tRNA synthetases: shared mechanisms for ensuring the fidelity of gene expression. Biochemistry. 2008;47:11695–11703. doi: 10.1021/bi801500z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Pino A, Zenkin N, Loris R. The many faces of Fic: structural and functional aspects of Fic enzymes. Trends Biochem Sci. 2014;39:121–129. doi: 10.1016/j.tibs.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 22.Gaut JR. In vivo threonine phosphorylation of immunoglobulin binding protein (BiP) maps to its protein binding domain. Cell Stress Chaperones. 1997;2:252–262. doi: 10.1379/1466-1268(1997)002<0252:ivtpoi>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goepfert A, Stanger FV, Dehio C, Schirmer T. Conserved inhibitory mechanism and competent ATP binding mode for adenylyltransferases with Fic fold. PLoS One. 2013;8:e64901. doi: 10.1371/journal.pone.0064901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gulick AM. Conformational dynamics in the Acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS Chem Biol. 2009;4:811–827. doi: 10.1021/cb900156h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ham H, Woolery AR, Tracy C, Stenesen D, Krämer H, Orth K. Unfolded protein response-regulated dFic reversibly AMPylates BiP during endoplasmic reticulum homeostasis. J Biol Chem. 2014 doi: 10.1074/jbc.M114.612515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hardiman CA, Roy CR. AMPylation is critical for Rab1 localization to vacuoles containing Legionella pneumophila. MBio. 2014;5:e01035–01013. doi: 10.1128/mBio.01035-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harms A, Stanger FV, Dehio C. Biological Diversity and Molecular Plasticity of FIC Domain Proteins. Annu Rev Microbiol. 2016;70:341–360. doi: 10.1146/annurev-micro-102215-095245. [DOI] [PubMed] [Google Scholar]
- 28.Harms A, Stanger FV, Scheu PD, de Jong IG, Goepfert A, Glatter T, … Dehio C. Adenylylation of Gyrase and Topo IV by FicT Toxins Disrupts Bacterial DNA Topology. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.07.056. [DOI] [PubMed] [Google Scholar]
- 29.Hedberg C, Itzen A. Molecular perspectives on protein adenylylation. ACS Chem Biol. 2015;10:12–21. doi: 10.1021/cb500854e. [DOI] [PubMed] [Google Scholar]
- 30.Higa N, Toma C, Koizumi Y, Nakasone N, Nohara T, Masumoto J, … Suzuki T. Vibrio parahaemolyticus effector proteins suppress inflammasome activation by interfering with host autophagy signaling. PLoS Pathog. 2013;9:e1003142. doi: 10.1371/journal.ppat.1003142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Itzen A, Blankenfeldt W, Goody RS. Adenylylation: renaissance of a forgotten post-translational modification. Trends Biochem Sci. 2011;36:221–228. doi: 10.1016/j.tibs.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 33.Jaggi R, van Heeswijk WC, Westerhoff HV, Ollis DL, Vasudevan SG. The two opposing activities of adenylyl transferase reside in distinct homologous domains, with intramolecular signal transduction. EMBO J. 1997;16:5562–5571. doi: 10.1093/emboj/16.18.5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khater S, Mohanty D. In silico identification of AMPylating enzymes and study of their divergent evolution. Sci Rep. 2015;5:10804. doi: 10.1038/srep10804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kinch LN, Yarbrough ML, Orth K, Grishin NV. Fido, a novel AMPylation domain common to fic, doc, and AvrB. PLoS One. 2009;4:e5818. doi: 10.1371/journal.pone.0005818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ledford BE, Leno GH. ADP-ribosylation of the molecular chaperone GRP78/BiP. Mol Cell Biochem. 1994;138:141–148. doi: 10.1007/BF00928456. [DOI] [PubMed] [Google Scholar]
- 37.Lu C, Nakayasu ES, Zhang LQ, Luo ZQ. Identification of Fic-1 as an enzyme that inhibits bacterial DNA replication by AMPylating GyrB, promoting filament formation. Sci Signal. 2016;9:ra11. doi: 10.1126/scisignal.aad0446. [DOI] [PubMed] [Google Scholar]
- 38.Luong P, Kinch LN, Brautigam CA, Grishin NV, Tomchick DR, Orth K. Kinetic and structural insights into the mechanism of AMPylation by VopS Fic domain. J Biol Chem. 2010;285:20155–20163. doi: 10.1074/jbc.M110.114884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mattoo S, Durrant E, Chen MJ, Xiao J, Lazar CS, Manning G, … Worby CA. Comparative analysis of Histophilus somni immunoglobulin-binding protein A (IbpA) with other fic domain-containing enzymes reveals differences in substrate and nucleotide specificities. J Biol Chem. 2011;286:32834–32842. doi: 10.1074/jbc.M111.227603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mukherjee S, Liu X, Arasaki K, McDonough J, Galán JE, Roy CR. Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature. 2011;477:103–106. doi: 10.1038/nature10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Müller MP, Albers MF, Itzen A, Hedberg C. Exploring adenylylation and phosphocholination as post-translational modifications. Chembiochem. 2014;15:19–26. doi: 10.1002/cbic.201300508. [DOI] [PubMed] [Google Scholar]
- 42.Müller MP, Peters H, Blümer J, Blankenfeldt W, Goody RS, Itzen A. The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science. 2010;329:946–949. doi: 10.1126/science.1192276. [DOI] [PubMed] [Google Scholar]
- 43.Murata T, Delprato A, Ingmundson A, Toomre DK, Lambright DG, Roy CR. The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat Cell Biol. 2006;8:971–977. doi: 10.1038/ncb1463. [DOI] [PubMed] [Google Scholar]
- 44.Nakai A, Kawatani T, Ohi S, Kawasaki H, Yoshimori T, Tashiro Y, … Nagata K. Expression and phosphorylation of BiP/GRP78, a molecular chaperone in the endoplasmic reticulum, during the differentiation of a mouse myeloblastic cell line. Cell Struct Funct. 1995;20:33–39. doi: 10.1247/csf.20.33. [DOI] [PubMed] [Google Scholar]
- 45.Palanivelu DV, Goepfert A, Meury M, Guye P, Dehio C, Schirmer T. Fic domain-catalyzed adenylylation: insight provided by the structural analysis of the type IV secretion system effector BepA. Protein Sci. 2011;20:492–499. doi: 10.1002/pro.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petrelli R, Felczak K, Cappellacci L. NMN/NaMN adenylyltransferase (NMNAT) and NAD kinase (NADK) inhibitors: chemistry and potential therapeutic applications. Curr Med Chem. 2011;18:1973–1992. doi: 10.2174/092986711795590048. [DOI] [PubMed] [Google Scholar]
- 47.Pieles K, Glatter T, Harms A, Schmidt A, Dehio C. An experimental strategy for the identification of AMPylation targets from complex protein samples. Proteomics. 2014;14:1048–1052. doi: 10.1002/pmic.201300470. [DOI] [PubMed] [Google Scholar]
- 48.Preissler S, Rato C, Chen R, Antrobus R, Ding S, Fearnley IM, Ron D. AMPylation matches BiP activity to client protein load in the endoplasmic reticulum. Elife. 2015:4. doi: 10.7554/eLife.12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Preissler S, Rato C, Perera LA, Saudek V, Ron D. FICD acts bifunctionally to AMPylate and de-AMPylate the endoplasmic reticulum chaperone BiP. Nat Struct Mol Biol. 2017;24:23–29. doi: 10.1038/nsmb.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pulliainen AT, Pieles K, Brand CS, Hauert B, Böhm A, Quebatte M, … Dehio C. Bacterial effector binds host cell adenylyl cyclase to potentiate Gαs-dependent cAMP production. Proc Natl Acad Sci U S A. 2012;109:9581–9586. doi: 10.1073/pnas.1117651109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rahman M, Ham H, Liu X, Sugiura Y, Orth K, Krämer H. Visual neurotransmission in Drosophila requires expression of Fic in glial capitate projections. Nat Neurosci. 2012;15:871–875. doi: 10.1038/nn.3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rhee SG, Chock PB, Stadtman ER. Regulation of Escherichia coli glutamine synthetase. Adv Enzymol Relat Areas Mol Biol. 1989;62:37–92. doi: 10.1002/9780470123089.ch2. [DOI] [PubMed] [Google Scholar]
- 53.Rhomberg TA, Truttmann MC, Guye P, Ellner Y, Dehio C. A translocated protein of Bartonella henselae interferes with endocytic uptake of individual bacteria and triggers uptake of large bacterial aggregates via the invasome. Cell Microbiol. 2009;11:927–945. doi: 10.1111/j.1462-5822.2009.01302.x. [DOI] [PubMed] [Google Scholar]
- 54.Sanyal A, Chen AJ, Nakayasu ES, Lazar CS, Zbornik EA, Worby CA, … Mattoo S. A Novel Link Between Fic (Filamentation induced by cAMP)-mediated Adenylylation/AMPylation and the Unfolded Protein Response. J Biol Chem. 2015 doi: 10.1074/jbc.M114.618348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scheffner M, Nuber U, Huibregtse JM. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature. 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- 56.Schmid MC, Scheidegger F, Dehio M, Balmelle-Devaux N, Schulein R, Guye P, … Dehio C. A translocated bacterial protein protects vascular endothelial cells from apoptosis. PLoS Pathog. 2006;2:e115. doi: 10.1371/journal.ppat.0020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmid MC, Schulein R, Dehio M, Denecker G, Carena I, Dehio C. The VirB type IV secretion system of Bartonella henselae mediates invasion, proinflammatory activation and antiapoptotic protection of endothelial cells. Molecular microbiology. 2004;52:81–92. doi: 10.1111/j.1365-2958.2003.03964.x. [DOI] [PubMed] [Google Scholar]
- 58.Schulein R, Guye P, Rhomberg TA, Schmid MC, Schröder G, Vergunst AC, … Dehio C. A bipartite signal mediates the transfer of type IV secretion substrates of Bartonella henselae into human cells. Proc Natl Acad Sci U S A. 2005;102:856–861. doi: 10.1073/pnas.0406796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shuman S. DNA ligases: progress and prospects. J Biol Chem. 2009;284:17365–17369. doi: 10.1074/jbc.R900017200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stachelhaus T, Mootz HD, Marahiel MA. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chemistry & biology. 1999;6:493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- 61.Stanger FV, Burmann BM, Harms A, Aragão H, Mazur A, Sharpe T, … Schirmer T. Intrinsic regulation of FIC-domain AMP-transferases by oligomerization and automodification. Proc Natl Acad Sci U S A. 2016;113:E529–537. doi: 10.1073/pnas.1516930113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trivedi OA, Arora P, Sridharan V, Tickoo R, Mohanty D, Gokhale RS. Enzymic activation and transfer of fatty acids as acyl-adenylates in mycobacteria. Nature. 2004;428:441–445. doi: 10.1038/nature02384. [DOI] [PubMed] [Google Scholar]
- 63.Truttmann MC, Cruz VE, Guo X, Engert C, Schwartz TU, Ploegh HL. The Caenorhabditis elegans Protein FIC-1 Is an AMPylase That Covalently Modifies Heat-Shock 70 Family Proteins, Translation Elongation Factors and Histones. PLoS Genet. 2016;12:e1006023. doi: 10.1371/journal.pgen.1006023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Truttmann MC, Rhomberg TA, Dehio C. Combined action of the type IV secretion effector proteins BepC and BepF promotes invasome formation of Bartonella henselae on endothelial and epithelial cells. Cell Microbiol. 2011;13:284–299. doi: 10.1111/j.1462-5822.2010.01535.x. [DOI] [PubMed] [Google Scholar]
- 65.Truttmann MC, Wu Q, Stiegeler S, Duarte JN, Ingram J, Ploegh HL. HypE-specific Nanobodies as Tools to modulate HypE-mediated Target AMPylation. J Biol Chem. 2015 doi: 10.1074/jbc.M114.634287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Truttmann MC, Zheng X, Hanke L, Damon JR, Grootveld M, Krakowiak J, … Ploegh HL. Unrestrained AMPylation targets cytosolic chaperones and activates the heat shock response. Proc Natl Acad Sci U S A. 2017;114:E152–E160. doi: 10.1073/pnas.1619234114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Turgay K, Krause M, Marahiel MA. Four homologous domains in the primary structure of GrsB are related to domains in a superfamily of adenylate-forming enzymes. Molecular microbiology. 1992;6:2743–2744. doi: 10.1111/j.1365-2958.1992.tb01451.x. [DOI] [PubMed] [Google Scholar]
- 68.Vishnu N, Jadoon Khan M, Karsten F, Groschner LN, Waldeck–Weiermair M, Rost R, … Malli R. ATP increases within the lumen of the endoplasmic reticulum upon intracellular Ca2+ release. Mol Biol Cell. 2014;25:368–379. doi: 10.1091/mbc.E13-07-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Westcott NP, Hang HC. Chemical reporters for exploring ADP- ribosylation and AMPylation at the host-pathogen interface. Current opinion in chemical biology. 2014;23:56–62. doi: 10.1016/j.cbpa.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Woolery AR, Luong P, Broberg CA, Orth K. AMPylation: Something Old is New Again. Front Microbiol. 2010;1:113. doi: 10.3389/fmicb.2010.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Woolery AR, Yu X, LaBaer J, Orth K. AMPylation of Rho GTPases Subverts Multiple Host Signaling Processes. J Biol Chem. 2014;289:32977–32988. doi: 10.1074/jbc.M114.601310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Worby CA, Mattoo S, Kruger RP, Corbeil LB, Koller A, Mendez JC, … Dixon JE. The fic domain: regulation of cell signaling by adenylylation. Mol Cell. 2009;34:93–103. doi: 10.1016/j.molcel.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xiao J, Worby CA, Mattoo S, Sankaran B, Dixon JE. Structural basis of Fic-mediated adenylylation. Nat Struct Mol Biol. 2010;17:1004–1010. doi: 10.1038/nsmb.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu H, Yang J, Gao W, Li L, Li P, Zhang L, … Shao F. Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome. Nature. 2014;513:237–241. doi: 10.1038/nature13449. [DOI] [PubMed] [Google Scholar]
- 75.Xu Y, Carr PD, Vasudevan SG, Ollis DL. Structure of the adenylylation domain of E. coli glutamine synthetase adenylyl transferase: evidence for gene duplication and evolution of a new active site. J Mol Biol. 2010;396:773–784. doi: 10.1016/j.jmb.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 76.Xu Y, Zhang R, Joachimiak A, Carr PD, Huber T, Vasudevan SG, Ollis DL. Structure of the N-terminal domain of Escherichia coli glutamine synthetase adenylyltransferase. Structure. 2004;12:861–869. doi: 10.1016/j.str.2004.02.029. [DOI] [PubMed] [Google Scholar]
- 77.Yarbrough ML, Li Y, Kinch LN, Grishin NV, Ball HL, Orth K. AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science. 2009;323:269–272. doi: 10.1126/science.1166382. [DOI] [PubMed] [Google Scholar]