Abstract
Extracellular vesicles from eukaryotic cells and outer membrane vesicles (OMVs) released from gram-negative bacteria have been described as mediators of pathogen-host interaction and intercellular communication. Legionella pneumophila (L. pneumophila) is a causative agent of severe pneumonia. The differential effect of bacterial and host cell vesicles in L. pneumophila infection is unknown so far. We infected THP-1-derived or primary human macrophages with L. pneumophila and isolated supernatant vesicles by differential centrifugation. We observed an increase of exosomes in the 100 k pellet by nanoparticle tracking analysis, electron microscopy, and protein markers. This fraction additionally contained Legionella LPS, indicating also the presence of OMVs. In contrast, vesicles in the 16 k pellet, representing microparticles, decreased during infection. The 100 k vesicle fraction activated uninfected primary human alveolar epithelial cells, A549 cells, and THP-1 cells. Epithelial cell activation was reduced by exosome depletion (anti-CD63, or GW4869), or blocking of IL-1β in the supernatant. In contrast, the response of THP-1 cells to vesicles was reduced by a TLR2-neutralizing antibody, UV-inactivation of bacteria, or – partially – RNase-treatment of vesicles. Taken together, we found that during L. pneumophila infection, neighbouring epithelial cells were predominantly activated by exosomes and cytokines, whereas myeloid cells were activated by bacterial OMVs.
Introduction
Legionella pneumophila (L. pneumophila), a gram-negative bacterium, is described as a causative pathogen of pneumonia1. Alveolar macrophages, which protect the lung from inhaled microorganisms and other insults, are their main host cell in the human lung. Infection of macrophages with L. pneumophila leads to a broad activation of signalling pathways, triggered by both extracellular receptors, such as TLRs, and intracellular receptor molecules, such as RIG-I and NAIP52. The initial proinflammatory activation pattern that can be observed in macrophages upon infection is defined by the up-regulation of TNF-α, IL-6 and IL-1β3. L. pneumophila are phagocytosed and contained in vesicular cytosolic bodies, the phagosomes, that are bound for lysosomal degradation. Legionella actively blocks this mechanism of cellular defence by transmembrane secretion of effector proteins into the host cell via the dot/Icm type IV secretion system4. These factors are instrumental to formation of a replication niche, the Legionella-containing vacuole (LCV), which is recruited from ER-derived vesicles. In this way, the LCV gains structural resemblance to the ER, which is hypothesized to protect it from the endocytic pathway5. In the LCV, Legionella replicates for about 14 h until host cell lysis6. Of note, the secreted effectors are also recognized by cytosolic sensors such as NAIP5 and NLRC4, leading to further activation of the host cell by caspase-I cleavage7. While limiting intracellular Legionella growth8, the activation of the host cell by intracellular pathogens also leads to the release of stimulatory extracellular vesicles (EVs)9.
Eukaryotic cells can produce various kinds of EVs, which are classified by their sub-cellular origin. Apoptotic cells secrete larger apoptotic bodies, EVs shedding from the plasma membrane are named microparticles, and endosomal compartment-derived EVs are defined as exosomes. EVs are composed of a lipid bilayer, they are carrying transmembrane proteins, and they are transporting intraluminal cargo (RNA and proteins) to recipient cells10. Exosomes and microparticles can be distinguished by certain marker proteins, such as tetraspanins (e.g. CD9, CD63, CD81), Alix and Tsg101, which are present on exosomes due to their endosomal origin. EVs are found in various body fluids and are an important means of communication between eukaryotic cells11. Their uptake by recipient cells can be induced via receptor-ligand interaction, endocytosis, phagocytosis or membrane fusion to release the cargo12. The release of EVs depends on the physiological state of the donor cell such as the condition of the microenvironment13, 14. Under inflammatory conditions, the secretion of EVs can increase, as shown for sarcoidosis, asthma or Mycobacterium bovis infection9, 15, 16.
The secretion of EVs seems to be conserved throughout evolution, as not only higher eukaryotes are producing EVs, but also prokaryotes17. Gram-negative bacteria form spheroid, nano-sized EVs, the so called outer membrane vesicles (OMV)18. These allow the transport of diverse virulence factors (e.g. toxins and enzymes) as well as lipopolysaccharides (LPS), which are present on the OMV surface19. L. pneumophila OMVs are known to be pro-inflammatory activators of epithelial cells and macrophages20, 21. Additionally, they can modulate the course of infection in vitro. We recently published that Legionella pneumophila-derived OMVs promote bacterial replication in human and murine macrophages22.
We sought to better understand the impact of intercellular communication via EVs during the course of L. pneumophila infection and to distinguish the role of different EV subsets, namely OMVs and exosomes, on alveolar recipient cells.
Results
Infected cells release a heterogeneous EV population of exosomes and OMVs
THP-1 cells respond to L. pneumophila infection with increased secretion of pro-inflammatory cytokines (TNF-α, IL-1β, IL-6 and MCP-1, Supplementary Fig. 1) and notably EVs (Fig. 1a). These nano-sized vesicles were collected by a 100,000 xg ultracentrifugation step and then quantified by nanoparticle tracking analysis (NTA). The amount of particles/mL in this so called 100 k pellet increased with multiplicity of infection, which could also be observed upon administration of the sterile stimulant IL-1β. Similarly, primary human blood-derived macrophages responded to an infection with L. pneumophila with an increased secretion of EVs (Supplementary Fig. 2b). In contrast, the amount of the larger microparticles in the 16k pellet was found to be slightly reduced after infection (Fig. 1a). Within the 100 K EV fraction, we detected an increase of the exosomal markers Alix and Tsg101 after L. pneumophila infection Fig. 1b) which we could also show after IL-1β treatment (Supplementary Fig. 2a). We could further characterize this fraction by the presence of the tetraspanins CD63 and CD81, which are exosomal marker proteins, and the absence of cellular markers GRP78 and LaminA/C (Fig. 1b). EVs from the 100 k pellet were visualized by transmission electron microscopy, showing their spherical shapes of approximately 20–100 nm in diameter (Fig. 1c). The presence of Legionella-LPS in the 100 k pellet (Supplementary Fig. 2c) indicates that the 100 k pellet released from infected THP-1 cells consists of a mixed population of exosomes and microparticles (both secreted from eukaryotic cells), and of bacterial OMVs carrying LPS. The amount of bacterial OMVs in the 100 k pellet of L. pneumophila infected THP-1 cells was determined by relative LPS quantification and equalled 0.5 µg OMVs. As our approach lays a focus on the stimulatory potential of the EV fraction on uninfected recipient cells, we visualized the uptake of PKH67-stained EVs from A549 cells by THP-1 cells by immunofluorescence analysis (Fig. 1d).
Figure 1.
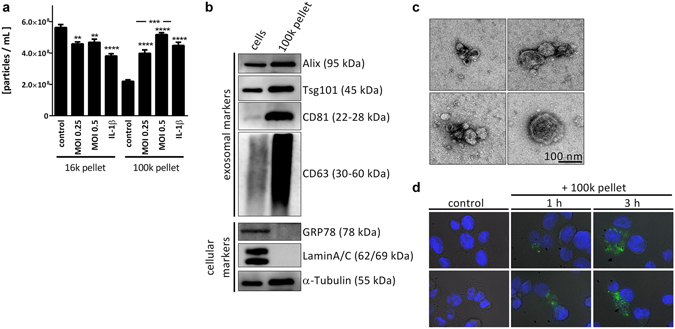
L. pneumophila infection increases the secretion of EVs in THP-1 cells. (a) Amount of EVs in response to L. pneumophila infection. THP-1 cells were treated with IL-1β (1 ng/mL) or infected with L. pneumophila (MOI 0.25 or 0.5, respectively) for 24 h. NTA was performed with the distinct particle fractions separated by differential centrifugation. (b) Western blot for exosomal marker proteins. Whole cell lysate or 100 k pellet derived from uninfected THP-1 cells were used. Equal protein amounts were loaded. (c) Transmission electron microscopy with 100 k pellet. Purified EVs from THP-1 cells were fixed and visualized after negative staining with uranyl acetate. Scale bar represents 100 nm. (d) Uptake of A549-derived EVs by THP-1 cells. A549 cells were stained with the membrane dye PKH67. EVs were collected (100 k pellet) and incubated with THP-1 cells for 1 or 3 h, respectively. Nuclei were stained by DAPI and pictures were taken with an original magnification of 630x. A: Data are shown as mean + SEM of three independent experiments. **p < 0.01, ***p < 0.001, ****p < 0.0001. B–D: representative results.
Recipient A549 cells and THP-1 cells respond differently to stimulation with the 100 k pellet or vesicle-free cytokine-containing supernatant of infected and control THP-1 donor cells
The stimulatory potential of the 100 K pellet on recipient cells needs to be carefully evaluated with respect to the variety of immune-stimulatory compounds that the pellet contains. To this end, the stimulatory capacity of EVs and cytokines that are both released from L. pneumophila-infected THP-1 cells was tested on different recipient cells. A549 cells responded with increased expression of CXCL8 and the anti-inflammatory microRNA-146a (miR-146a) to the cytokine-containing vesicle-free supernatant from infected THP-1 cells (Fig. 2a). Additionally, the secretion of IL-6, GM-CSF and MCP-1 was elevated in supernatant-treated A549 cells (Fig. 2b). In contrast, their response to stimulation with a 100 k pellet derived from infected THP-1 cells was significantly weaker, as assessed by the same parameters (Fig. 2a,b). This was correspondingly observed for primary human alveolar epithelial cells (Supplementary Fig. 3a,b). To establish a more profound transcriptional profile, we also measured the expression of SOD2, CXCL1, IFN-β, IL-1β and TNFAIP2 in A549 cells treated with supernatant or with the 100 K pellet (Supplementary Fig. 6), yielding the same pattern as observed before. As miRNAs can functionally be transferred via EVs23 and specifically miR-146a has been demonstrated to be transported between immune cells24, we measured the expression of the primary transcript of miR-146a (pri-mir-146a) in recipient cells. We observed a strong induction of pri-mir-146a in A549 cells after supernatant and EV treatment (Fig. 2a). As A549 cells responded to a stronger extend to the cytokine-containing supernatant of L. pneumophila-infected THP-1 cells, we aimed to discover the responsible cytokine. The response of A549 cells to the THP-1-derived supernatant could be blocked by pre-incubation of the recipient cells with an IL-1 receptor antagonist (IL-1RA), as illustrated by the significantly reduced expression of CXCL8 and miR-146a, while a TNFα-neutralizing antibody (α-TNF-α) had no effect on the expression of CXCL8 or miR-146a (Fig. 3a,b). Interestingly, when THP-1 cells were used as recipient cells, they responded significantly more to the EVs than to the supernatant when assessing the same parameters (Fig. 4a,b and Supplementary Fig. 7). We performed Ingenuity Pathway Analysis (Qiagen) on the identified markers and observed significant enrichment of relevant immune pathways and events in infection, such as attraction of leukocytes, apoptosis of leukocytes and organization of cytosceleton (Supplemental Fig. 8), indicating broad activation of the recipient cells. Thus, we tried to establish the response pattern of A549 and THP-1 cells to the EV fraction.
Figure 2.
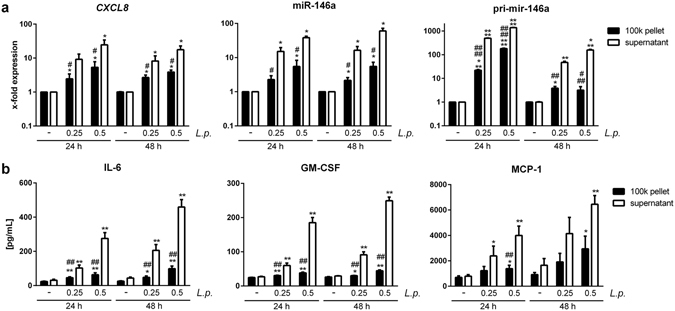
L. pneumophila induced cytokines elicit a pro-inflammatory response in non-infected A549 cells. (a,b) Response of A549 cells to vesicle-free supernatant or 100 k pellet from L. pneumophila-infected THP-1 cells. A549 cells were stimulated with vesicle-free supernatant or 100 k pellet from THP-1 cells infected with L. pneumophila (L.p.) at an MOI of 0.25 and 0.5, respectively, for 24 h. A549 cells were incubated as indicated for 24 or 48 h, respectively. RNA and supernatant were collected. (a) qPCR was performed for expression of CXCL8, miR-146a and pri-mir-146a. (b) Magnetic multiplex ELISA was performed. Results for IL-6, GM-CSF and MCP-1 are shown. Data are shown as mean + SEM of at least three independent experiments. *p < 0.05, **p < 0.01. *Compared to corresponding control, #compared to corresponding supernatant treated sample.
Figure 3.
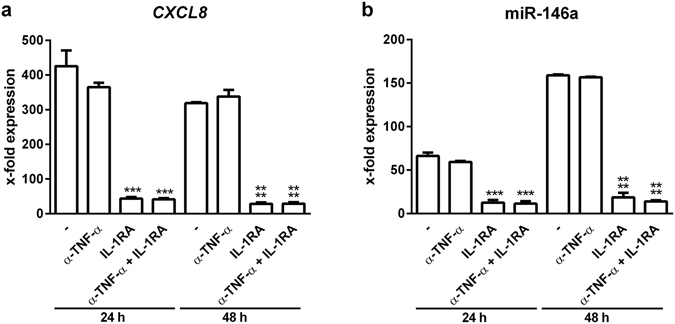
THP-1-derived IL-1β induces pro-inflammatory response in A549 cells. (a,b) Response of A549 cells to vesicle-free supernatant from L. pneumophila-infected THP-1 cells. THP-1 cells were infected with L. pneumophila (MOI 0.5) for 24 h. A549 cells were pre-incubated for 1 h with a neutralizing TNF-α antibody (α-TNF-α) or an IL-1 receptor antagonist (IL-1RA) alone or in combination, respectively. Cells were then incubated with the vesicle-free supernatant for 24 or 48 h, respectively. RNA was collected and used for expression analysis of CXCL8 (a) and miR-146a (b). Data are shown as mean + SEM of three independent experiments. ***p < 0.001, ****p < 0.0001 compared to corresponding supernatant treated cells.
Figure 4.
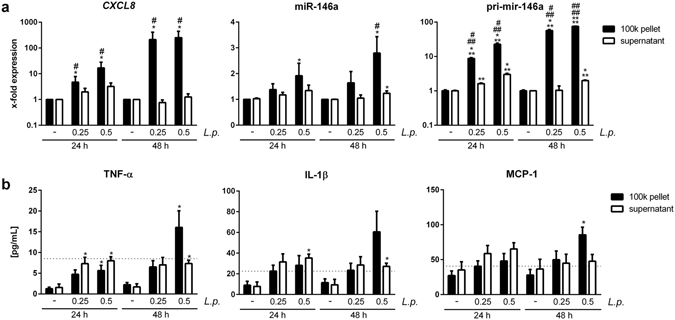
L. pneumophila induced EVs elicit a pro-inflammatory response in non-infected THP-1 cells. (a,b) Response of THP-1 cells to vesicle-free supernatant or 100 k pellet from L. pneumophila-infected THP-1 cells. THP-1 cells were stimulated with vesicle-free supernatant or 100 k pellet from THP-1 cells infected with L. pneumophila (MOI 0.25 and 0.5, respectively) for 24 h. Recipient THP-1 cells were incubated for 24 or 48 h, respectively. (a) qPCR was performed for expression of CXCL8, miR-146a and pri-mir-146a. (b) Magnetic multiplex ELISA was performed. Results for TNF-α, IL-1β and MCP-1 are shown. Dashed lines indicate the input of the cytokine contained in the vesicle-free supernatant of infected THP-1 cells (MOI 0.5) before adding it to the recipient cells. Data are shown as mean + SEM of at least three independent experiments. *p < 0.05. *Compared to corresponding control, # compared to corresponding supernatant treated sample.
While THP-1 cells respond to the bacterial fraction of the 100 k pellet, A549 cells respond to the exosome fraction
As the 100 K pellet is heterogeneous, its stimulatory effect on recipient THP-1 or A549 cells was addressed. Therefore, specific subfractions were targeted for deletion. A reduction of exosome secretion can be achieved by GW4869, a neutral sphingomyelinase (nSMase) inhibitor25, 26. GW4869 was applied to the donor THP-1 cells and reduced exosome secretion was validated by western blot for the exosomal protein Alix (Fig. 5a). It is noteworthy that L. pneumophila replication or OMV secretion was not affected by GW4869 treatment of the infected THP-1 cells (Supplementary Fig. 4a,b). Recipient THP-1 cells responded with CXCL8 mRNA expression to the exosome-depleted 100 k pellet (Fig. 5b). Conversely, recipient A549 cells showed a significant reduction of CXCL8 mRNA expression upon incubation with the 100 k pellet from GW4869 treated THP-1 cells (Fig. 5c).
Figure 5.

Block of exosome release does not reduce the THP-1 response to the 100 k pellet. (a) THP-1 cells were incubated with GW4869 (10 or 15 µM) or DMSO as a solvent control for 1 h before infection with L. pneumophila (MOI 0.5) for 24 h. A 100 k pellet was generated and Alix expression was analysed by western blot. Densitometric quantification of western blot bands is shown. Exosome secretion is reduced after GW4869 treatment. (b,c) Donor THP-1 cells were pre-incubated with GW4869 (15 µM) or the solvent control for 1 h before infection with L. pneumophila for 24 h. EVs were purified by differential centrifugation and added to THP-1 (b) or A549 (c) cells. CXCL8 expression was analysed by qPCR after 24 or 48 h of incubation. Data are shown as mean + SEM of three independent experiments. *p < 0.05, **p < 0.01, ns: not significant.
Stimulation of A549 and THP-1 cells with enriched 100 k pellet subfractions reveals differential response patterns
In order to pinpoint the described effects of the EVs more specifically to the bacterial or human component of the 100 k pellet, the CD63-positive exosome fraction was depleted by anti-CD63 immunoprecipitation. L. pneumophila OMVs were still present in the unbound fraction after CD63 immunoprecipitation, and naturally in the complete 100 k pellet, but not in the CD63-postitive exosome fraction, as shown by LPS dot blot (Supplementary Fig. 4c). Recipient THP-1 cells responded to the exosome-depleted fraction (CD63−) in the same manner as they did to the entire 100 k pellet, as illustrated on the level of CXCL8 mRNA expression and CXCL8 release (Fig. 6a,b), pointing towards the stimulatory effect of OMVs on macrophages.
Figure 6.
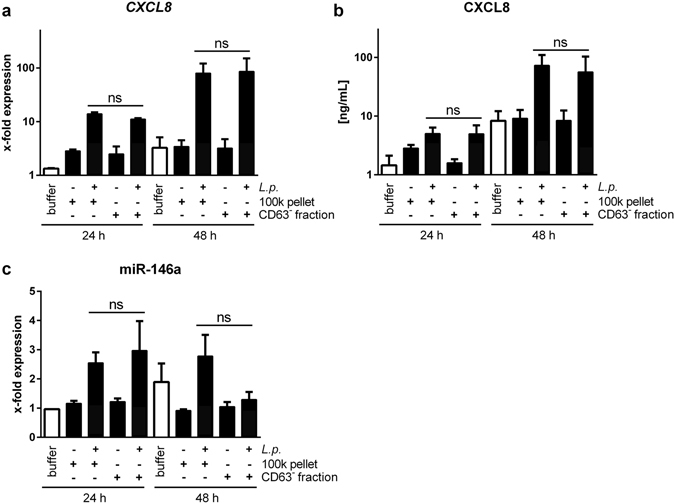
THP-1 cells respond to the CD63-negative EV fraction. (a,c) Pro-inflammatory THP-1 response depends on OMVs. 100 k pellets from L. pneumophila-infected or control THP-1 cells were generated. CD63-positive exosomes were removed by immunoprecipitation. Entire 100 k pellet or unbound EVs (CD63− fraction) were used for stimulation of THP-1 cells for 24 and 48 h. Expression of CXCL8 (a), CXCL8 secretion (b) and expression of miR-146a (c) were analysed. Data are shown as mean + SEM of three independent experiments. ns: not significant.
As L. pneumophila OMVs are known to activate macrophages via TLR222, we analysed the involvement of TLR-ligands in the 100 k pellet of Legionella-infected THP-1 cells on macrophage activation by applying the EVs to mouse bone marrow derived macrophages (mBMDM) from different genetic backgrounds. TLR2/4−/− macrophages did not respond with CXCL1 secretion to the 100 k pellet as wild type mBMDM did. While TLR3/7/9−/− cells showed a slightly stronger secretion of CXCL1 when compared to TLR2/4−/− cells, there was a substantial reduction when compared to wild type macrophage (Supplementary Fig. 5a). Irrespective of the genetic background, all three responded comparably to the stimulation with murine TNF-α (Supplementary Fig. 5b).
THP-1 cells respond to TLR2 ligands from viable bacteria
In order to analyse the active bacterial compounds in the 100 k pellet to which THP-1 cells respond, the donor THP-1 cells were either treated with UV-inactivated bacteria, to date the only way to block OMV release, or the 100 k pellet was treated with Triton X-100 in combination with RNase A before administration to recipient THP-1 cells. While viable bacteria were crucial for the activating properties of the 100 k pellet on THP-1 cells, the degradation of the pellet’s RNA content yielded inconclusive results, as shown by CXCL8 mRNA expression (Fig. 7a). Pre-treatment of the recipient THP-1 cells with a TLR2-blocking antibody also reduced the activation potential of the pellet as shown by a sharp drop in CXCL8 mRNA expression to 13.4% (Fig. 7b). Furthermore, blockage of TLR2 diminished miR-146a expression in the recipient cells (Fig. 7c). We could thus confirm the presence of bacterial components in the pellet that require infection of donor THP-1 cells with viable bacteria and activate recipient macrophages via TLR2.
Figure 7.
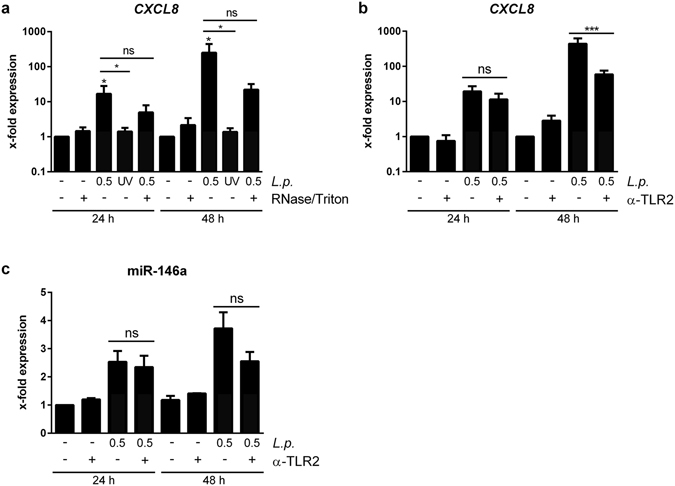
Viable L. pneumophila are critical for response of THP-1 cells to EVs. (a) Response of THP-1 cells to EVs from L. pneumophila-infected THP-1 cells. THP-1 cells were infected with L. pneumophila or stimulated with UV-inactivated L. pneumophila (both MOI 0.5 for 24 h). 100 k pellets were generated and left untreated or digested with RNase A (0.2 µg/µL) in the presence of 0.3% Triton X-100 for 1 h at 37 °C. The prepared EVs were added to THP-1 cells for 24 or 48 h, respectively, exposing the cells to a subcritical final Triton X-100 concentration of 0.003%. Expression of CXCL8 was determined. (b,c) TLR2 dependent response of THP-1 cells to EVs from L. pneumophila-infected THP-1. THP-1 cells were infected with L. pneumophila (MOI 0.5 for 24 h) and the 100 k pellet was generated. Recipient THP-1 cells were pre-incubated for 90 min with a TLR2 blocking antibody (α-TLR2,+) or a control antibody (−; both 20 µg/mL). The EVs were added for 24 or 48 h, respectively. Expression of CXCL8 (b) and miR-146a (c) was determined. Data are shown as mean + SEM of three independent experiments. *p < 0.05, ns: not significant.
Discussion
In this study, we have found that L. pneumophila infection of human macrophages induced the release of EVs and cytokines, which can activate uninfected bystander cells. Upon infection, we observed an increased release of smaller EVs (100 k pellet), while the larger microparticles (16 k pellet) were reduced upon infection with L. pneumophila. The 100 k pellet mainly contained exosomes, as shown by the enrichment of exosomal marker proteins on western blot and the observed size in electron microscopy (<150 nm). The increase in EV release is in line with the current literature on inflammatory conditions, as mycobacterial infection9, asthma16, sarcoidosis15 or acute lung injury27 also lead to an enhanced EV release. We further analysed which of the different uninfected recipient cells in the alveolus, namely alveolar epithelial cells or macrophages, respond to the released cytokines and the vesicle fraction upon L. pneumophila infection of neighbouring cells. We could demonstrate that alveolar epithelial cells mainly responded to the cytokine fraction by secretion of IL-6, CXCL8, GM-CSF and MCP-1 and to a weaker extend to the 100 k pellet. We hypothesized that IL-1β was the main stimulating cytokine, as it is known that macrophage derived IL-1β can prime alveolar epithelial cells, which is crucial for the recruitment of neutrophils in a murine pneumonia model28. To address this, blocking experiments with an IL-1 receptor antagonist were carried out, which confirmed that IL-1β was the main stimulating factor of the vesicle-free supernatant for alveolar epithelial cells. In contrast, resting macrophages responded with release of pro-inflammatory cytokines to EVs from infected donor cells, which was also observed for the infection with other intracellular pathogens elsewhere9. The release of CXCL8 can lead to recruitment of neutrophils, which are instrumental in clearing the infection29, 30. We furthermore measured the expression of additional inflammatory markers, such as SOD2, CXCL1, IFN-β, IL-1β and TNFAIP2. The marker pattern we observe indicates a broad pro-inflammatory activation pattern of the recipient cells.
To determine the EV component inducing the CXCL8 expression in recipient cells, the release of exosomes was blocked by administration of GW4869, a nSMase inhibitor25, 26. Application of GW4869 to donor THP-1 cells before infection reduced the exosome secretion, while it did not affect L. pneumophila replication. The obtained 100 k pellet, devoid of exosomes, was equally stimulatory to recipient THP-1 cells, while it was less stimulatory to A549 cells. As alveolar epithelial cells strongly respond to IL-1β and as exosomes can transport IL-1β31–33, the reduction of exosome release might in turn also reduce IL-1β in the 100 k pellet, which could explain the diminished A549 response after GW4869 application to the EV-producing macrophages. The observation that macrophages still responded to the 100 k pellet might be explained by the presence of OMVs, which are released by L. pneumophila during the intracellular growth in the macrophages20. This is in line with our detection of L. pneumophila LPS in the EV pellet, which was not affected by GW4869 treatment. The presence of bacterial LPS in the EV fraction was also observed for Salmonella typhimurium infection9, which also secrete OMVs34. In another study, the EV fraction was found to contain mycobacterial lipids which could be transferred to uninfected bystander cells35 and could induce a pro-inflammatory response36. Besides lipids and LPS, bacterial proteins37 or even viral RNA as demonstrated for Epstein-Barr virus38 or Hepatitis C virus39 infection have been detected in EVs. To further investigate the involvement of the different EV subsets present upon Legionella infection, CD63-positive exosomes were depleted from the 100 K pellet derived from infected THP-1 cells. The remaining unbound EVs were still stimulatory to recipient macrophages, which is in line with the results obtained by sphingomyelinase inhibition, as both treatments lead to an exosome-depleted 100 K pellet. As there is to date no Legionella mutant incapable of releasing OMVs, we performed infection experiments of donor THP-1 cells with UV-inactivated bacteria and used the obtained 100 k pellet for stimulation experiments. This reduced the CXCL8 expression in THP-1 cells in response to the EVs to baseline, suggesting that L. pneumophila OMVs are the main stimulating factor in the 100 k pellet for THP-1 macrophages. As previously published, L. pneumophila OMVs are strong pro-inflammatory stimulators for human und murine macrophages22, 40, which can explain the herein described results of the 100 k pellet on THP-1 cells. It has previously been published for mycobacterial infections that different EV populations have distinct stimulatory properties41. In this study, exosomes were not stimulatory on recipient THP-1 cells, while OMVs were. The involvement of bacterial LPS in the EV-mediated activation of THP-1 cells was shown by pre-incubation of recipient cells with a TLR2 blocking antibody, as Legionella LPS is recognized mainly via this membrane receptor42 and as the released OMVs also activate TLR222, 40. Blocking of TLR2 resulted in a reduced expression of CXCL8 in THP-1 macrophages. In accordance with this, mBMDM from TLR2/4 double knockout mice did not respond to the 100 k pellet. The involvement of TLR signalling in the pro-inflammatory response to EVs was equally observed for mycobacteria9, 43.
It is well described that EVs can transport different RNA species from one cell to another23. To address this, we tested the impact of the transported RNA cargo on CXCL8 induction in recipient macrophages with RNase A digestion experiments. As we only observed a partial reduction in CXCL8 response, we concluded that RNA is only an accessory stimulating factor in our system. Additionally, the induction of the anti-inflammatory miR-146a after EV stimulation of macrophages was due to a transcriptional induction and further processing of the primary transcript, as demonstrated by the increased expression of pri-mir-146a in the recipient cell. Furthermore, it has recently been demonstrated that OMVs can transport bacterial RNA44–46. Since EVs released during infection can contain pathogenic RNA38, 39, the involvement of endosomal TLRs in macrophage activation was assessed by stimulating mBMDM from TLR3/7/9 knockout animals. While mBMDM from wild type animals showed an increased secretion of CXCL1, the murine functional homologue to the human CXCL8, macrophages from mice lacking the endosomal TLRs 3, 7 and 9 were only slightly more responsive to the 100 k pellet than TLR2/4−/− cells. While mechanisms of EV susceptibility in mice and men might differ, this result suggests a similar contribution of TLR2/4 and TLR3/7/9 agonists in the activation of murine macrophages by the 100 k pellet.
We conclude that the infection of macrophages with L. pneumophila increases the release of exosomes along with the secretion of bacterial OMVs (Fig. 8). While OMVs are activating resting macrophages, leading to the secretion of pro-inflammatory cytokines, exosomes are stimulating alveolar epithelial cells. Macrophage-derived cytokines are additionally activating epithelial cells, which further enhances the amount of pro-inflammatory cytokines, potentially leading to the recruitment of alveolar macrophages and neutrophils in vivo. The observation that the recruitment of neutrophils and the subsequent bacterial clearance in Legionella pneumonia depend on IL-1R47, strengthens our hypothesis that the intercellular communication via cytokines and EVs is critical for the host immune response to bacterial infections.
Figure 8.
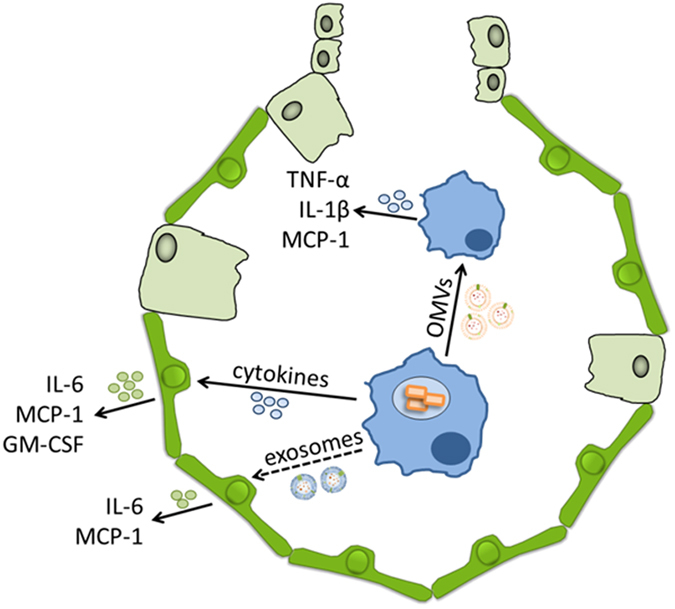
Differential microvesicles signalling in L. pneumophila infection of macrophages. The infection of macrophage-like THP-1 cells with L. pneumophila induces the release of pro-inflammatory cytokines and EVs, eukaryotic exosomes and prokaryotic OMVs. While alveolar epithelial cells mostly respond to the cytokines with a pro-inflammatory response and to a weaker extent to the exosomes, macrophages respond to the released EVs. Here, the bacterial OMVs carrying the LPS are the main stimulating agent.
Our study provides for the first time a detailed analysis of the differential effects of the released cytokines and EVs on different cell types in the context of L. pneumophila pneumonia. However, the influence of cytokines and EVs needs to be tested in vivo. This could help to further understand the pathogenesis of Legionella.
In summary, we report that intercellular communication in the context of L. pneumophila infection occurs via activation of surrounding alveolar epithelial cells by cytokines and exosomes. Bacterial OMVs activate resting bystander macrophages via TLR2. Culminating in an enhanced CXCL8 release, this might be beneficial to create a pro-inflammatory environment in order to mount an immune response, eventually clearing the infection by recruitment of neutrophils.
Materials and Methods
Chemicals and reagents
RPMI-1640 and FCS were obtained from Thermo Fisher Scientific Inc. (Waltham, USA). Ham’s F12 was obtained from GE Healthcare Europe (Freiburg, Germany). PBS was from Biochrom GmbH (Berlin, Germany). Phorbol 12-myristate 13-acetate (PMA) and GW4869 were purchased from Sigma-Aldrich Chemie GmbH (Taufkirchen, Germany). 4′,6-diamidino-2-phenylindole (DAPI) was from AAT Bioquest (Sunnyvale, USA). TLR2 blocking antibody was acquired from eBioscience (T2.5, San Diego, USA). Anti-LPS antibody (L. pneumophila specific; ABIN235748) was obtained from antikörper-online.de (Aachen, Germany). All other antibodies were purchased from Santa Cruz: anti-Alix (sc-49268), anti-Tsg101 (sc-7964), anti-CD63 (sc-5275), anti-CD81 (sc-7637), anti-GRP78 (sc-1050), anti-LaminA/C (sc-20681) and anti-tubulin (sc-5286). All chemicals used were of analytical grade and obtained from commercial sources.
Bacterial strains and infection
L. pneumophila Corby wild type was kindly provided by A. Flieger, Wernigerode, Germany, and routinely grown as described48. Briefly, bacteria were plated on BCYE charcoal agar and incubated for 3 days at 37 °C and 5% CO2. For infection, bacteria were resuspended in PBS. Infection doses were calculated by OD600 measurement. THP-1 cells were infected for 24 h at multiplicity of infection (MOI) of 0.25 or 0.5.
Cell culture
THP-1 and A549 cells were obtained from the American Type Culture Collection (ATCC) and cultured at 37 °C and 5% CO2 in RPMI-1640 or Ham’s F12 with 10% FCS without antibiotics, respectively. For all experiments, THP-1 cells were differentiated by addition of 20 nM PMA for 24 h. Murine bone marrow-derived macrophages (mBMDM) were prepared from femurs and tibiae of C57BL/6 N wild type and TLR2/4−/− mice (kindly provided by M. Schnare, Marburg, Germany) and cultivated as described49. Briefly, bone marrow-derived cells were cultivated for 7 days in the consecutive presence of M-CSF and GM-CSF. Maturation was validated by microscopy.
Quantitative RT-PCR
RNA isolation, DNase I digestion and quantitative RT-PCR were performed as described22. Briefly, RNA was isolated from THP-1 and A549 cells by phenol-chloroform precipitation. For pri-mir-146a measurement, purified RNA was DNase digested to eliminate contaminating traces of genomic DNA. For quantitative PCR (qPCR), the SYBR Green and Taqman probe detection methods were used. The following custom made primers were used for SYBR Green based qPCR: RPS18: fwd: 5′-GCGGCGGAAAATAGCCTTTG-3′, rev: 5′-GATCACACGTTCCACCTCATC-3′; CXCL8: fwd: 5′-ACTGAGAGTGATTGAGAGTGGAC-3′, rev: 5′-AACCCTCTGCACCCAGTTTTC-3′. SOD2: fwd: 5′-ATGTTGAGCCGGGCAGTGTG-3′, rev: 5′-GCGCGTTGATGTGAGGTTCC-3′; TNFAIP2: fwd: 5′-GACTTGGGCTCACAGATAAAGC-3′, rev: 5′-CAGGCAGTTGTTGATGTTGG-3′; CXCL1: fwd: 5′-AAGTCATAGCCACACTCAAGAATGG-3′, rev: 5′-TTCAGGAACAGCCACCAGTGAG-3′; IL-1β: fwd: 5′-AGCTCGCCAGTGAAATGATGG-3′, rev: 5′-CAGGTCCTGGAAGGAGCACTTC-3′; IFN-β: fwd: 5′-CCGCATTGACCATCTATGAG-3′, rev: 5′-AGACATTAGCCAGGAGGTTC-3′. Commercial microRNA and pri-microRNA Taqman primers were purchased from Thermo Fisher Scientific Inc. (RNU48: 001006, miR-146a-5p: 000468, pri-mir-16–2: hs03303046_pri, pri-mir-146a: hs03303259_pri). All samples were processed on a ViiA7 qPCR device (Life Technologies).
Extracellular vesicle preparation and analysis
THP-1 cells were incubated in exosome-depleted medium and infected with L. pneumophila at indicated MOIs or left untreated for control. After 24 h, the supernatant was centrifuged at 1,000 × g for 15 min to remove dead cells. The supernatant was then centrifuged again at 16,000 × g for 30 min to pellet apoptotic bodies and microparticles, yielding the 16 K pellet. After sterile-filtration, the remaining supernatant was ultracentrifuged at 100,000 × g for 3 h to obtain the 100 K pellet, containing extracellular vesicles (EVs) such as exosomes and OMVs. Purified EVs were analysed by nanoparticle tracking analysis (NTA) in a NanoSight instrument (kindly provided by S. Dimmeler, Frankfurt, Germany). Transmission electron microscopic (TEM) analysis was performed to visualize vesicles and assess purity. After fixation of the 100 K sample with 0.5% glutaraldehyde and 2% formaldehyde for 15 min on ice and subsequent negative staining (2% uranyl acetate), samples were scanned using a JEOL 2100 TEM operated at 120 kV. Immunoprecipitation of CD63+ exosomes from the purified EV fraction was performed overnight on a rotator at 4 °C using the exosome-human CD63 isolation/detection reagent (Thermo Fisher Scientific Inc.). For an exosome-free EV fraction, THP-1 cells were incubated with the nSMase inhibitor GW4869 (10–15 µM) before infection and collection of supernatant after 24 h of infection.
Stimulation experiments
Purified EVs from 2 × 106 THP-1 cells were resuspended in RPMI and added to recipient cells (A549 or THP-1 cells). When indicated, recipient THP-1 cells were pre-incubated with TLR2 blocking antibody (α-TLR2, 20 µg/mL) for 90 min before administration of EVs. Cytokine-containing supernatant that was clear of EVs after 100,000 × g ultracentrifugation from control or infected THP-1 cells was added to the recipient cells at one sixth of the total volume. Recipient cells were incubated for 24 or 48 h, respectively. When indicated, TNF-α neutralizing antibody (α-TNF-α, 15 ng/mL) or IL-1 receptor antagonist (IL-1RA, 100 ng/mL; both R&D systems, Minneapolis, USA) were added 1 h before stimulation of recipient A549 cells with EV-free supernatant.
Immunofluorescence
For immunofluorescence analysis of EV uptake, the membrane of EV-donor A549 cells was stained with the PKH67 membrane dye (Sigma-Aldrich Chemie GmbH) according to the manufacturer’s recommendations. The resulting dyed EV pellet was prepared as described above and incubated with recipient THP-1 cells for 1 or 3 h, respectively. Cells were fixed for 15 min with 4% paraformaldehyde and nuclei were stained by addition of DAPI. Pictures were taken on an Axio Vert.A1 Fluorescence Microscope with an AxioCam MRm (Zeiss, Oberkochen, Germany).
ELISA
CXCL8 from the supernatant of THP-1 cells was analysed with the OptEIA ELISA kit (BD Biosciences, Heidelberg, Germany). CXCL1 from supernatant of mBMDM was analysed with the DuoSet ELISA kit (R&D systems). All other cytokines (IL-1β, IL-6, IL-10, MCP-1, GM-CSF, TNF-α) were measured in a Bio-Plex Magpix (Luminex Corporation, Austin, USA) with a custom made Luminex Assay (R&D systems) according to the manufacturer’s recommendations.
Western Blot
Western blot was performed as described50. Briefly, samples were transferred by wet blot onto the membrane. After blocking and washing, immunodetection was carried out with the above-listed antibodies and visualized by HRP-mediated turnover of ECL substrate (GE Healthcare, Little Chalfont, UK) on a chemiluminescence imager (INTAS Science Imaging Instruments, Göttingen, Germany).
Ethical Statement
Animals were handled according to the EU council directive 86/609/EEC for the protection of animals. The performed protocols were approved by the responsible animal ethics committee (Philipps-University Marburg; permit number: EX-22-2013).
Statistical analyses
Data are presented as mean + SEM (standard error of the mean) of at least three independent experiments. Effects were statistically evaluated employing the non-parametric Mann-Whitney-U test or 2-way ANOVA. p-values < 0.05 were considered significant.
Electronic supplementary material
Acknowledgements
We thank Kerstin Hoffmann and Nadine Siebert (Philipps University Marburg, Germany) for excellent technical assistance. We thank Harshavardhan Janga (Philipps University Marburg, Germany) for providing us with primary alveolar epithelial cells. We are grateful to Professor Uwe Maier (Philipps University Marburg, Germany) for providing the electron microscopy facility. The work was in part supported by grants from the DFG (TR84), BMBF (e:bio 0316175A, e:Med 01X1304E) and HMWK (LOEWE UGMLC and LOEWE Medical RNomics) to B.S.
Author Contributions
A.L.J., A.S.S., W.B., and B.S. conceived experiments; A.L.J., C.E.H., C.S., K.B., K.S., and N.S. conducted experiments; A.L.J. and W.B. analysed the results, A.L.J., W.B., and B.S. wrote the manuscript; A.L.J. prepared the figures. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-06443-1
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fraser DW, et al. Legionnaires’ disease: description of an epidemic of pneumonia. The New England journal of medicine. 1977;297:1189–1197. doi: 10.1056/NEJM197712012972201. [DOI] [PubMed] [Google Scholar]
- 2.Massis LM, Zamboni DS. Innate immunity to legionella pneumophila. Frontiers in microbiology. 2011;2:109. doi: 10.3389/fmicb.2011.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benoit M, Desnues B, Mege JL. Macrophage polarization in bacterial infections. J Immunol. 2008;181:3733–3739. doi: 10.4049/jimmunol.181.6.3733. [DOI] [PubMed] [Google Scholar]
- 4.Franco IS, Shuman HA, Charpentier X. The perplexing functions and surprising origins of Legionella pneumophila type IV secretion effectors. Cell Microbiol. 2009;11:1435–1443. doi: 10.1111/j.1462-5822.2009.01351.x. [DOI] [PubMed] [Google Scholar]
- 5.Hubber A, Roy CR. Modulation of host cell function by Legionella pneumophila type IV effectors. Annual review of cell and developmental biology. 2010;26:261–283. doi: 10.1146/annurev-cellbio-100109-104034. [DOI] [PubMed] [Google Scholar]
- 6.Isaac DT, Isberg R. Master manipulators: an update on Legionella pneumophila Icm/Dot translocated substrates and their host targets. Future microbiology. 2014;9:343–359. doi: 10.2217/fmb.13.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zamboni DS, et al. The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat Immunol. 2006;7:318–325. doi: 10.1038/ni1305. [DOI] [PubMed] [Google Scholar]
- 8.Pereira MS, et al. Activation of NLRC4 by flagellated bacteria triggers caspase-1-dependent and -independent responses to restrict Legionella pneumophila replication in macrophages and in vivo. J Immunol. 2011;187:6447–6455. doi: 10.4049/jimmunol.1003784. [DOI] [PubMed] [Google Scholar]
- 9.Bhatnagar S, Shinagawa K, Castellino FJ, Schorey JS. Exosomes released from macrophages infected with intracellular pathogens stimulate a proinflammatory response in vitro and in vivo. Blood. 2007;110:3234–3244. doi: 10.1182/blood-2007-03-079152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annual review of cell and developmental biology. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 11.Tkach M, Thery C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell. 2016;164:1226–1232. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 12.Zaborowski MP, Balaj L, Breakefield XO, Lai CP. Extracellular Vesicles: Composition, Biological Relevance, and Methods of Study. Bioscience. 2015;65:783–797. doi: 10.1093/biosci/biv084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parolini I, et al. Microenvironmental pH is a key factor for exosome traffic in tumor cells. The Journal of biological chemistry. 2009;284:34211–34222. doi: 10.1074/jbc.M109.041152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andaloussi EL, Mager I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nature reviews. Drug discovery. 2013;12:347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 15.Qazi KR, et al. Proinflammatory exosomes in bronchoalveolar lavage fluid of patients with sarcoidosis. Thorax. 2010;65:1016–1024. doi: 10.1136/thx.2009.132027. [DOI] [PubMed] [Google Scholar]
- 16.Kulshreshtha, A., Ahmad, T., Agrawal, A. & Ghosh, B. Proinflammatory role of epithelial cell-derived exosomes in allergic airway inflammation. The Journal of allergy and clinical immunology131, 1194–1203, 1203 e1191–1114, doi:10.1016/j.jaci.2012.12.1565 (2013). [DOI] [PubMed]
- 17.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. The Journal of cell biology. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellis TN, Kuehn MJ. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiology and molecular biology reviews: MMBR. 2010;74:81–94. doi: 10.1128/MMBR.00031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuehn MJ, Kesty NC. Bacterial outer membrane vesicles and the host-pathogen interaction. Genes & development. 2005;19:2645–2655. doi: 10.1101/gad.1299905. [DOI] [PubMed] [Google Scholar]
- 20.Galka F, et al. Proteomic characterization of the whole secretome of Legionella pneumophila and functional analysis of outer membrane vesicles. Infection and immunity. 2008;76:1825–1836. doi: 10.1128/IAI.01396-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jager J, et al. Human lung tissue explants reveal novel interactions during Legionella pneumophila infections. Infection and immunity. 2014;82:275–285. doi: 10.1128/IAI.00703-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung AL, et al. Legionella pneumophila-Derived Outer Membrane Vesicles Promote Bacterial Replication in Macrophages. PLoS pathogens. 2016;12:e1005592. doi: 10.1371/journal.ppat.1005592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature cell biology. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 24.Alexander M, et al. Exosome-delivered microRNAs modulate the inflammatory response to endotoxin. Nature communications. 2015;6:7321. doi: 10.1038/ncomms8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo BB, Bellingham SA, Hill AF. The neutral sphingomyelinase pathway regulates packaging of the prion protein into exosomes. The Journal of biological chemistry. 2015;290:3455–3467. doi: 10.1074/jbc.M114.605253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trajkovic K, et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science (New York, N.Y.) 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 27.Soni S, et al. Alveolar macrophage-derived microvesicles mediate acute lung injury. Thorax. 2016 doi: 10.1136/thoraxjnl-2015-208032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marriott HM, et al. Interleukin-1 beta Regulates CXCL8 Release and Influences Disease Outcome in Response to Streptococcus pneumoniae, Defining Intercellular Cooperation between Pulmonary Epithelial Cells and Macrophages. Infect Immun. 2012;80:1140–1149. doi: 10.1128/IAI.05697-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ribeiro RA, Flores CA, Cunha FQ, Ferreira SH. IL-8 causes in vivo neutrophil migration by a cell-dependent mechanism. Immunology. 1991;73:472–477. [PMC free article] [PubMed] [Google Scholar]
- 30.Smith WB, Gamble JR, Clark-Lewis I, Vadas MA. Interleukin-8 induces neutrophil transendothelial migration. Immunology. 1991;72:65–72. [PMC free article] [PubMed] [Google Scholar]
- 31.MacKenzie A, et al. Rapid secretion of interleukin-1beta by microvesicle shedding. Immunity. 2001;15:825–835. doi: 10.1016/S1074-7613(01)00229-1. [DOI] [PubMed] [Google Scholar]
- 32.Pizzirani C, et al. Stimulation of P2 receptors causes release of IL-1beta-loaded microvesicles from human dendritic cells. Blood. 2007;109:3856–3864. doi: 10.1182/blood-2005-06-031377. [DOI] [PubMed] [Google Scholar]
- 33.Qu Y, Franchi L, Nunez G, Dubyak GR. Nonclassical IL-1 beta secretion stimulated by P2X7 receptors is dependent on inflammasome activation and correlated with exosome release in murine macrophages. Journal of immunology (Baltimore, Md.: 1950) 2007;179:1913–1925. doi: 10.4049/jimmunol.179.3.1913. [DOI] [PubMed] [Google Scholar]
- 34.Bai J, Kim SI, Ryu S, Yoon H. Identification and characterization of outer membrane vesicle-associated proteins in Salmonella enterica serovar Typhimurium. Infection and immunity. 2014;82:4001–4010. doi: 10.1128/IAI.01416-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beatty WL, et al. Trafficking and release of mycobacterial lipids from infected macrophages. Traffic (Copenhagen, Denmark) 2000;1:235–247. doi: 10.1034/j.1600-0854.2000.010306.x. [DOI] [PubMed] [Google Scholar]
- 36.Rhoades E, et al. Identification and macrophage-activating activity of glycolipids released from intracellular Mycobacterium bovis BCG. Molecular microbiology. 2003;48:875–888. doi: 10.1046/j.1365-2958.2003.03473.x. [DOI] [PubMed] [Google Scholar]
- 37.Giri PK, Kruh NA, Dobos KM, Schorey JS. Proteomic analysis identifies highly antigenic proteins in exosomes from M. tuberculosis-infected and culture filtrate protein-treated macrophages. Proteomics. 2010;10:3190–3202. doi: 10.1002/pmic.200900840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfeffer S, et al. Identification of virus-encoded microRNAs. Science (New York, N.Y.) 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 39.Dreux M, et al. Short-range exosomal transfer of viral RNA from infected cells to plasmacytoid dendritic cells triggers innate immunity. Cell host & microbe. 2012;12:558–570. doi: 10.1016/j.chom.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jager J, Keese S, Roessle M, Steinert M, Schromm AB. Fusion of Legionella pneumophila outer membrane vesicles with eukaryotic membrane systems is a mechanism to deliver pathogen factors to host cell membranes. Cellular microbiology. 2014 doi: 10.1111/cmi.12392. [DOI] [PubMed] [Google Scholar]
- 41.Athman JJ, et al. Bacterial Membrane Vesicles Mediate the Release of Mycobacterium tuberculosis Lipoglycans and Lipoproteins from Infected Macrophages. Journal of immunology (Baltimore, Md.: 1950) 2015;195:1044–1053. doi: 10.4049/jimmunol.1402894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Girard R, et al. Lipopolysaccharides from Legionella and Rhizobium stimulate mouse bone marrow granulocytes via Toll-like receptor 2. Journal of cell science. 2003;116:293–302. doi: 10.1242/jcs.00212. [DOI] [PubMed] [Google Scholar]
- 43.Bhatnagar S, Schorey JS. Exosomes released from infected macrophages contain Mycobacterium avium glycopeptidolipids and are proinflammatory. The Journal of biological chemistry. 2007;282:25779–25789. doi: 10.1074/jbc.M702277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sjostrom AE, Sandblad L, Uhlin BE, Wai SN. Membrane vesicle-mediated release of bacterial RNA. Scientific reports. 2015;5:15329. doi: 10.1038/srep15329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghosal A, et al. The extracellular RNA complement of Escherichia coli. MicrobiologyOpen. 2015 doi: 10.1002/mbo3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koeppen K, et al. A Novel Mechanism of Host-Pathogen Interaction through sRNA in Bacterial Outer Membrane Vesicles. PLoS pathogens. 2016;12:e1005672. doi: 10.1371/journal.ppat.1005672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mascarenhas DP, Pereira MS, Manin GZ, Hori JI, Zamboni DS. Interleukin 1 receptor-driven neutrophil recruitment accounts to MyD88-dependent pulmonary clearance of legionella pneumophila infection in vivo. The Journal of infectious diseases. 2015;211:322–330. doi: 10.1093/infdis/jiu430. [DOI] [PubMed] [Google Scholar]
- 48.Schmeck B, et al. Legionella pneumophila-induced NF-kappaB- and MAPK-dependent cytokine release by lung epithelial cells. The European respiratory journal. 2007;29:25–33. doi: 10.1183/09031936.00141005. [DOI] [PubMed] [Google Scholar]
- 49.Sester DP, et al. LPS regulates a set of genes in primary murine macrophages by antagonising CSF-1 action. Immunobiology. 2005;210:97–107. doi: 10.1016/j.imbio.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 50.Krull M, et al. Differences in cell activation by Chlamydophila pneumoniae and Chlamydia trachomatis infection in human endothelial cells. Infect Immun. 2004;72:6615–6621. doi: 10.1128/IAI.72.11.6615-6621.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


