Abstract
IκBζ, which is encoded by the Nfkbiz gene, is a member of the nuclear IκB family of proteins that act as transcriptional regulators via association with NF-κB. Nfkbiz-deficient (Nfkbiz −/−) mice develop spontaneous dermatitis; however, the underlying mechanism has yet to be elucidated. In our study, we found higher skin pathology scores and more serum IgE antibodies and trans-epidermal water loss in Nfkbiz −/− than in Nfkbiz-sufficient (Nfkbiz +/−) mice. There was also greater expansion of IFN-γ-, IL-17A-, and IL-22-secreting CD4+ T cells and of IL-17A-secreting γδ+ T cells in the skin of Nfkbiz −/− mice than in with Nfkbiz +/− mice. Pyrosequencing analysis showed decreased diversity of resident bacteria and markedly expanded Staphylococcus (S.) xylosus in the skin of Nfkbiz −/− mice. Oral administration of antibiotics including cephalexin and enrofloxacin ameliorated skin inflammation. Topical application of S. xylosus also resulted in the expansion of IL-17A-secreting CD4+ T cells along with high levels of pro-inflammatory cytokines and chemokines in the skin of Nfkbiz −/− mice. The expansion of commensal S. xylosus may be one cause of skin dysbiosis in Nfkbiz −/− mice and suggests that the Nfkbiz gene may play a regulatory role in the microbiota-skin immunity axis.
Introduction
The skin acts as an important physical barrier that is directly exposed to numerous pathogens in the environment. It maintains a symbiotic relationship with various commensal microorganisms that inhabit it1. Changes in microbial composition have been found in many skin diseases including atopic dermatitis (AD) and psoriasis2, 3. For instance, microbiome analysis in patients with AD revealed that the proportion of Staphylococcus (S.) aureus that increased during disease flares correlated with disease severity2. S. aureus was also isolated from skin lesions of STAT3-deficient mice and of ADAM17-deficient mice with spontaneous dermatitis, and antibiotic treatments were essential to control the disease progression4, 5. S. xylosus is a coagulase-negative gram-positive staphylococcal species and a common skin commensal bacterium of humans and other mammals6. Some cases of S. xylosus infection, including erythema nodosum7, pyelonephritis8, and corneal infections9 occur in humans. In laboratory mice, S. xylosus has been associated with spontaneous dermatitis in both NOS2−/− mice10 and athymic nude mice11 and with conjunctivitis in NADPH oxidase−/− mice12, implying that S. xylosus is an opportunistic pathogen. Although microorganisms are not the only inducers of skin disease, it seems likely that microbial alteration and the predominance of specific resident bacteria directly affect dermatitis severity.
As gut microbes are associated with gut immunity, commensal microbiota in the skin also play a pivotal role in local immunity to maintain homeostasis both in a steady state and during infection13. When the skin of germ-free mice was colonized with S. epidermidis and compared with that of conventional mice, the germ-free mice had fewer resident-effector T cells than the conventional mice and developed protective T-cell responses against the Leishmania major parasite14. In addition, S. epidermidis-induced IL-17A+CD8+ T cells enhanced innate barrier immunity and ameliorated pathogen infection15. However, when the expansion and aberration of commensal microbiota with pathogenic potential occurred under specific circumstances, such as infection and injury, it could disturb immune system homeostasis and worsen inflammation. Previous studies revealed that staphylococcal enterotoxin B or alpha-toxin derived by S. aureus induced IL-17A secretion, leading to the abnormal proliferation of keratinocytes and recruitment of inflammatory cells16, 17. Notably, patients with severe skin signs showed more S. aureus specific-IgE antibodies (Abs) than those with mild symptoms18. Because of the complex interplay among commensal microbes, immunity, and environmental factors in the skin mucosa, the underlying mechanisms have yet to be elucidated.
IκBζ, encoded by the Nfkbiz gene, is a member of the nuclear IκB family of proteins that act as transcriptional regulators via association with NF-κB. Several previous studies revealed a critical role of IκBζ signaling in the regulation of immune responses19–22. IκBζ−/− mice showed severe skin irritation in the face, neck, and periocular regions and several signs of Sjögren’s syndrome23, 24. IκBζ interacts with NF-κB and is associated with both positive and negative regulation of NF-kB transcriptional activity. Like other IκB proteins, it has inhibitory effects on the transcription of inflammatory genes regulated by NF-kB, such as TNF-α, but it can also induce pro-inflammatory cytokines, such as IL-6 and IL-12p4025. IκBζ also plays an important role in adaptive immunity by regulating Th17 development19. Although IL-6 and IL-17A production was significantly reduced in IκBζ-deficient cells in vitro 19, 23, highly elevated levels of these cytokines were detected in vivo 26. It is known that IκBζ deficiency in hematopoietic cells is not the major cause of spontaneous inflammation24. The specific depletion of IκBζ in lymphocytes or myeloid cells does not induce inflammation in the conjunctiva and ocular skin, although enhanced apoptosis in IκBζ-deficient epithelial cells is involved in the onset of Sjögren’s syndrome-like autoimmune disease in IκBζ−/− mice24. It is not clear how the immune system is overactivated by depletion of IκBζ.
In this study, we demonstrated that Nfkbiz-deficient (Nfkbiz −/−) mice with atopic-like dermatitis have elevated serum IgE Abs and drastically increased numbers of IL-17A-secreting CD4+ T cells in skin. Microbiome analysis showed the predominant expansion of Staphylococcus sequences, particularly S. xylosus, and a significant decrease in the diversity of commensal bacteria. Antibiotics completely eliminated dermatitis symptoms, implying specific taxonomic alterations associated with the induction of hyper-immune responses in the skin. Of note, the inoculation of S. xylosus in the skin of Nfkbiz −/− and their heterozygous littermates (Nfkbiz +/−) resulted in the expansion of IL-17A production from CD4+ T cells.
Results
Nfkbiz−/− mice developed spontaneous skin inflammation
To clarify the mechanism underlying the development of dermatitis in Nfkbiz −/− mice, we first confirmed the disease progression in our facility. As reported by others23, we established that erosions and hair loss that were exhibited throughout the ocular region beginning about age 4 weeks in Nfkbiz −/− mice then extended to the whole body (Fig. 1A). In these mice, disease incidence and clinical score were also significantly increased from about age 4 weeks in Nfkbiz −/− mice (Fig. 1B). The levels of transepidermal water loss (TEWL), which show abnormal skin barrier function, and of serum IgE Ab, which has a positive correlation with the severity of atopic dermatitis and psoriasis27, 28, was higher in the Nfkbiz −/− mice than in Nfkbiz-sufficient (Nfkbiz +/−) mice (Fig. 1C). In addition, the levels of various pro-inflammatory cytokines (i.e., IL-1β, IFN-γ, TNF-α, IL-6, IL-17A, and IL-22) and chemokines (i.e., MCP-1, MIP-1α, RANTES, and KC) were significantly increased in the serum of Nfkbiz −/− mice when compared with Nfkbiz +/− mice (Fig. 1D). There were marked increases in the numbers of CD11b+ or CD207+ or CD11c+ cells and CD4+ or CD8α+ cells infiltrating into the epidermis and dermis of Nfkbiz −/− mice as determined by confocal microscopy analysis (Fig. 1E). We conducted an intense analysis and found no significant phenotype differences in the innate lymphoid cell subsets isolated from Nfkbiz +/− and Nfkbiz −/− mice (data not shown). Once we confirmed the development of dermatitis in Nfkbiz −/− mice, we performed further experiments.
Figure 1.
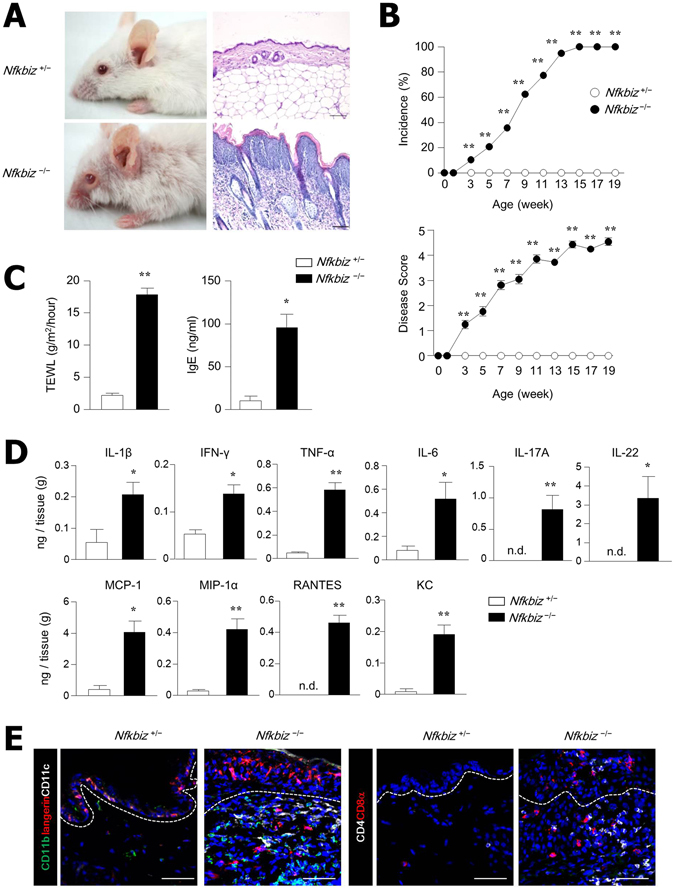
Spontaneous skin inflammation develops in Nfkbiz −/− mice. (A) Representative skin phenotype and histology of skin biopsy from Nfkbz +/− and Nfkbiz −/− mice at age 10 weeks. Scale bar = 50 µm. (B) Disease incidence and progression of dermatitis in Nfkbiz +/− and Nfkbiz −/− mice (n = 20). Dermatitis incidence (score ≥ 3) is shown as a percentage. (C) Trans-epidermal water loss (TEWL) and serum IgE antibody levels in Nfkbiz +/− and Nfkbiz −/− mice. (D) Cytokines and chemokine levels in a skin homogenate. (E) Confocal microscopy of skin biopsies from Nfkbiz +/− and Nfkbiz −/− mice. Scale bar = 50 μm. Data are representative of three independent experiments. All data are mean ± s.e.m. *p < 0.05, **p < 0.01.
IL-17A- and IL-22-secreting skin-resident T cells increased in Nfkbiz−/− mice
To determine the cell types involved in dermatitis in Nfkbiz −/− mice, we analyzed immune cells in the skin via flow cytometry. The skin-resident dendritic cells (DCs) include Langerhans cells (LCs) in the epidermis and DCs in the dermis (dDCs), which can be divided into various subsets29. Absolute numbers of MHCII+CD11c+ cells were highly elevated in the epidermis and dermis of Nfkbiz −/− mice in comparison with those in Nfkbiz +/− mice (Fig. S1A). Among four DC subsets in skin, the relative percentages of CD103+ dDCs (CD11b+/−CD207−CD103+) were increased while CD207+ dDCs (CD11b+CD207+CD103+) were decreased in Nfkbiz −/− mice when compared with Nfkbiz +/− mice (Fig. 2A). No significant changes of relative percentages were found in LCs (CD11b+CD207+CD103−) and CD11b+ dDCs (CD11b+CD207−CD103−) between Nfkbiz −/− and Nfkbiz +/− mice (Fig. 2A). We next addressed skin-resident T-cell subsets and found that γδhigh T cells with few CD4 and CD8 T cells were present in the skin of Nfkbiz +/− mice, but Nfkbiz −/− mice with AD had massive infiltration of CD4, CD8, and γδmid T cells (Fig. 2B). Interestingly, accumulated CD4, CD8, and γδmid skin-resident T cells obtained from Nfkbiz −/− mice predominantly produced IL-17A and IL-22 (Fig. 2C). When these findings are considered together, CD103+ dDCs and IL-17A- and IL-22-secreting skin-resident T cells may be associated with skin inflammation in Nfkbiz −/− mice.
Figure 2.
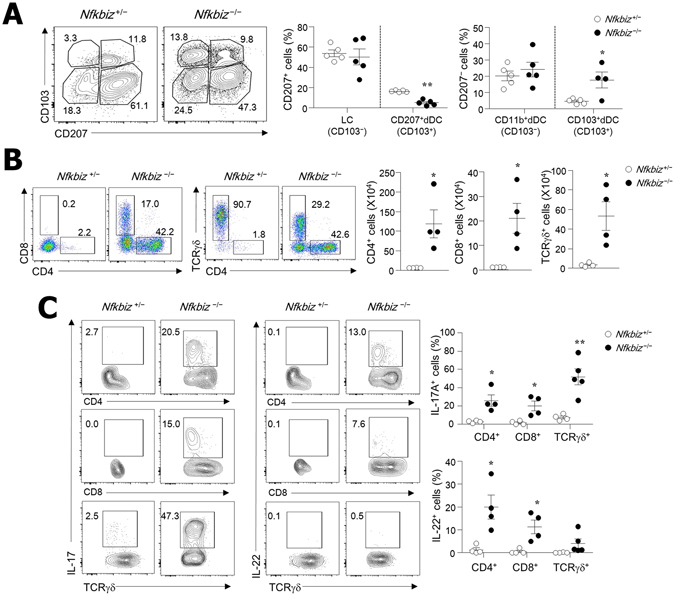
IL-17A- and IL-22-secreting T cells infiltrate into skin of Nfkbiz −/− mice. Flow cytometry analysis of (A) DC subsets in CD45+MHCII+CD11c+ cells and (B) T cells in skin of Nfkbiz +/− and Nfkbiz −/− mice. (C) IL-17A and IL-22 production from each cell subset after gating CD45-positive skin cells. Data are representative of three independent experiments. *p < 0.05, **p < 0.01.
Nfkbiz-deficient skin exhibited decreased diversity of the bacterial community and an increase in S. xylosus
Numerous studies have shown that dysbiosis of the skin microbiome is associated with development of AD and psoriasis in human patients and in animal models2, 3, 5. Hence, we next addressed the skin microbiota to determine whether the resident microbiota is also related to the development of dermatitis in Nfkbiz −/− mice. When quantity of bacterial 16S rRNA was assessed by PCR, there was a significantly higher bacterial burden in the skin homogenates of Nfkbiz −/− mice than from Nfkbiz +/− mice (Fig. 3A). Further pyrosequencing analysis using skin tissue homogenates revealed that Nfkbiz +/− and Nfkbiz −/− mice have segregated microbiota composition (Fig. 3B) and that Nfkbiz −/− mice have less diversity and species richness in than Nfkbiz +/− mice (Fig. 3C). The Nfkbiz −/− mice had decreased levels of Pseudomonas, Acinetobacter, Gemella, Ochrobactrum, and Rhodococcus genera (data not shown) but high abundance of S. xylosus (Fig. 3D). To further confirm the expansion of S. xylosus under disease conditions, we next cultured skin swabs from Nfkbiz +/− and Nfkbiz −/− mice on mannitol salt agar (MSA), a medium selective for Staphylococcus 30. Consistent with the pyrosequencing data, numerous yellow colonies on MSA, mostly identified as S. xylosus by 16S rRNA sequencing, were obtained from skin swab culture of Nfkbiz −/− mice (Fig. 3E). In addition, elevated numbers of Staphylococcus colonies were detected with increasing age (Fig. 3F), which correlated with the clinical score of Nfkbiz −/− mice (Fig. 1B). Overall, decreased bacterial diversity with the striking expansion of S. xylosus occurred in the skin of Nfkbiz −/− mice with dermatitis.
Figure 3.

Increased bacterial burden and decreased bacterial diversity in skin of Nfkbiz −/− mice. (A) Relative expression of 16 s rDNA universal bacteria in skin of Nfkbiz +/− and Nfkbiz −/− mice. (B) Principal coordinate analysis of unweighted and weighted UniFrac distances between samples from skin of Nfkbiz +/− and Nfkbiz −/− mice. (C) Shannon index, observed operational taxonomic units (OTUs), and phylogenetic diversity (PD) whole tree of Nfkbiz +/− and Nfkbiz −/− mice. (D) Composition of skin bacterial communities in Nfkbiz +/− and Nfkbiz −/− mice at phylum level (>1% of total 16S rRNA sequences) with additional speciation of Corynebacterium and Staphylococcus genera. Each bar represents relative abundance in one mouse. Results are representative of two experiments (n = 4–6 per group). (E and F) Quantification of Staphylococcus species from skin swabs of Nfkbiz +/− and Nfkbiz −/− mice performed using mannitol salt agar (MSA). Data are mean ± s.e.m. (A and F). *p < 0.05, **p < 0.01. n.d., not detected.
Antibiotic treatment ameliorated dermatitis in Nfkbiz−/− mice
To clarify the direct role of resident microbiota in controlling symbiosis in the skin, we next applied antibiotics to Nfkbiz −/− mice. Oral administration of antibiotics including cephalexin and enrofloxacin for 4 weeks significantly ameliorated the skin inflammation in Nfkbiz −/− mice, during which time the disease score of untreated mice worsened (Fig. 4A). To support this amelioration of skin inflammation, we found significantly lower levels of TEWL and IgE Abs in the antibiotic-treated Nfkbiz −/− mice than in the untreated Nfkbiz −/− mice (Fig. 4B). We also observed a decline in Staphylococcus species in the skin, indicating that the antibiotics effectively suppressed S. xylosus (Fig. 4B). The oral administration of antibiotics suppressed the levels of pro-inflammatory cytokines (i.e., IL-1β, TNF-α, IL-6, IL-17A, and IL-22) and chemokines (i.e., MCP-1, MIP-1α, RNATES, and KC) in the skin (Fig. 4C). In this regard, the numbers of infiltrated DCs and T cells were significantly diminished in the skin tissue of antibiotic-treated Nfkbiz −/− mice (Figs 4D and S1B).
Figure 4.
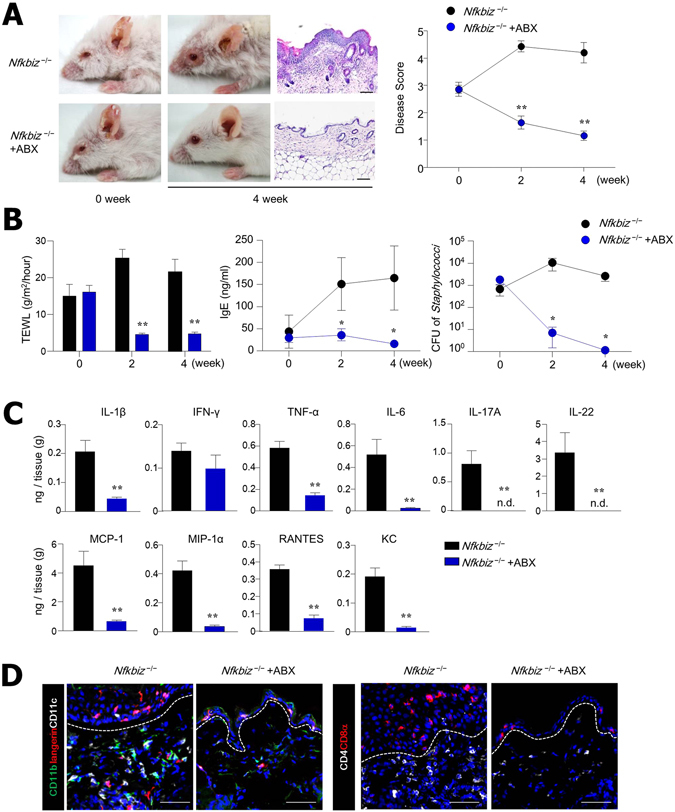
Antibiotic (ABX) treatment eliminates skin inflammation. (A) Representative skin phenotype and histology of skin biopsy from untreated (water alone) and ABX-treated Nfkbiz −/− mice at age 6 weeks. Scale bar = 50 μm. (B) TEWL, serum IgE levels, and colony-forming units (CFU) of Staphylococcus species. (C) Cytokine and chemokine levels in skin homogenate. (D) Confocal microscopy of skin biopsies. Scale bar = 50 μm. Data are representative of three independent experiments. All data are mean ± s.e.m. *p < 0.05, **p < 0.01.
We further explored DC and T-cell subsets associated with skin inflammation induced by resident bacteria. Among the DC subsets, only CD103+ dDC subsets were significantly reduced by antibiotic treatments (Fig. 5A). The oral administration of antibiotics diminished total recovered numbers of CD4, CD8, and γδT cells and the percentages of those cells (Fig. 5B). Interestingly, IL-17A-secreting CD4, CD8, and γδT cells were significantly reduced by antibiotic treatments (Fig. 5C). Consistent with the ELISA data shown in Fig. 4C, antibiotic treatments were not associated with IFN-γ secretion by CD4+ and CD8+ T cells (Fig. S2). Collectively, these data suggest that the dominant expansion of S. xylosus, which inhabits the skin of mammals as a commensal bacterium, is one factor that contributes to the onset of skin inflammation in the absence of the Nfkbiz gene. Moreover, IL-17A and IL-22 are strongly related to the expansion of S. xylosus.
Figure 5.

Infiltrated IL-17A- and IL-22-secreting T cells are reduced in skin of Nfkbiz −/− mice after ABX treatment. Flow cytometry analysis of (A) DCs in CD45+MHCII+CD11c+ cells and (B) T cells in CD45+ cells isolated from skin of water- and ABX-treated Nfkbiz −/− mice at age 6 weeks. (C) IL-17A and IL-22 production by CD45+ cells in skin. Data are representative of three independent experiments. *p < 0.05, **p < 0.01.
Antibiotic treatment reduced gene expression related to skin inflammation of Nfkbiz−/− mice
To investigate which genes are involved in the regulation of skin inflammation induced by resident bacteria in the absence of the Nfkbiz gene, we compared the gene expression profiles in the skin tissues of Nfkbiz +/− mice, Nfkbiz −/− mice, and antibiotic-treated Nfkbiz −/− mice. The expression levels of 694 and 368 genes were higher and lower (i.e., changed ≥ 2-fold) in Nfkbiz −/− mice than in Nfkbiz +/− mice (Fig. S3A) and in antibiotic-treated Nfkbiz −/− mice than in untreated Nfkbiz −/− mice (Fig. 6A), respectively. As expected, the gene expression levels of pro-inflammatory cytokines (i.e., IL-17A, IFN-γ, TNF, and IL-18) were higher in the skin of Nfkbiz −/− mice than in Nfkbiz +/− mice. The expression of the S100a9 gene, which is a marker of inflammation, was also significantly increased in the inflamed skin of Nfkbiz −/− mice (Fig. S3A). In contrast, the gene expression level of ectodysplasin A (Eda), which contributes to human and murine skin repair31, was lower in the skin of Nfkbiz −/− mice than in Nfkbiz +/− mice (Fig. S3A). Furthermore, IL-17A target genes (i.e., S100a9, S100a8, CAMP, and LCN2) were remarkably upregulated in the skin of Nfkbiz −/− mice with dermatitis (Fig. S3A). We next created and analyzed a hit gene network predicted by Ingenuity Pathway Analysis (IPA) software. Activation of the NF-kB signal pathway in the skin of Nfkbiz −/− mice was as predicted by the IPA software (Fig. S3B). Notably, the depletion of resident microbiota by antibiotic treatment resulted in the reduction of gene expression related to inflammation (i.e., S100a8, S100a9 and Tnfrsf9), but the activation of gene expression was associated with skin repair (i.e., Eda and Eda receptor) (Fig. 6A). Th17 cell-associated genes, (i.e., S100a8, CAMP, BATF, IL-23A, LCN2, and HIF-1α) highly expressed in Nfkbiz −/− mice with dermatitis, were downregulated after antibiotic treatments (Fig. 6A). By analysis with the IPA software, downregulation of the NF-kB signal pathway was predicted in the skin of Nfkbiz −/− mice in the absence of resident microbiota after antibiotic treatment, in comparison with that in untreated Nfkbiz −/− mice (Fig. 6B). Overall, these results support our conclusion that unusual expansion of the resident microbiota may be a key factor in the development of skin inflammation.
Figure 6.

ABX treatment reduces gene expression related to inflammation in skin of Nfkbiz −/− mice. (A) Microarray analysis of skin from water- and ABX-treated Nfkbiz −/− mice (n = 3 per group). Genes with significantly changed expression (fold change > 1.0 and p < 0.05) after ABX treatment clustered in a heat map. (B) A gene interaction network, associated with the amelioration of spontaneous dermatitis in Nfkbiz −/− mice, identified by Ingenuity Pathway Analysis software. Solid lines indicate direct interaction; dashed lines indicate indirect interaction.
Differentiation of Th17 cells in the skin after topical application of S. xylosus
The amelioration of skin inflammation resulting from the administration of antibiotics prompted us to investigate whether the topical application of S. xylosus induces skin inflammation in Nfkbiz −/− mice. We pretreated Nfkbiz +/− and Nfkbiz −/− mice with antibiotics for 2 weeks to reduce resident commensals and topically applied S. xylosus to the back skin for 2 weeks. Topical inoculation resulted in the persistent colonization of S. xylosus on the skin of both Nfkbiz +/− and Nfkbiz −/− mice after antibiotic treatment (Fig. S4). The frequency of LCs and dDCs in the skin was not altered by S. xylosus inoculation (data not shown). Of interest, the topical application of S. xylosus induced the accumulation of CD4+ T cells in the skin and markedly enhanced the production of IL-17A in the skin of Nfkbiz +/− and Nfkbiz −/− mice (Figs 7A and S5A). There were many more IL-17A-secreting CD4+ T cells in the skin of Nfkbiz −/− mice than in Nfkbiz +/− mice. The numbers of IL-22-secreting CD4+ T cells were only slightly increased by S. xylosus application, which did not reach statistical significance (Fig. 7A). As expected, there was no significant induction of IL-17A-secreting CD4+ T cells when heat-killed S. xylosus was applied topically onto Nfkbiz +/− and Nfkbiz −/− mice (Figs 7A and S5A). Furthermore, we observed significant increases of pro-inflammatory cytokines (i.e., IL-1β, IFN-γ, TNF-α, and IL-17A) and chemokines (i.e., MCP-1, MIP-1α, RANTES, and KC) in Nfkbiz −/− skin after inoculation of S. xylosus (Fig. 7B), which also elevated the levels of IL-1β, TNF-α, MCP-1, MIP-1α, and RANTES in the skin of Nfkbiz +/− mice (Fig. S5B). When these findings are considered together, a single commensal bacterial species (i.e., S. xylosus) in the skin has the potential to lead to the differentiation of IL-17A-secreting CD4+ T cells and to elicit various pro-inflammatory cytokines and chemokines, which may be involved in the onset of skin inflammation.
Figure 7.
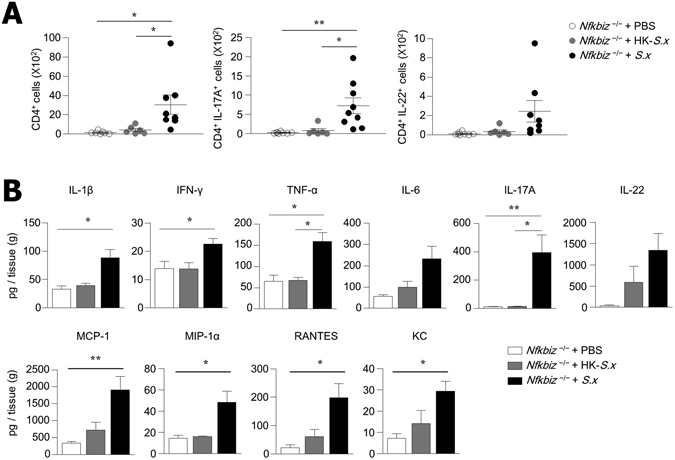
Topical application of S. xylosus leads to differentiation of Th17 cells in skin of Nfkbiz −/− mice. (A) Flow cytometry analysis of IL-17A and IL-22 production in CD4+ cells from skin of PBS, heat-killed S. xylosus (HK-S.x), or S. xylosus(S.x)-inoculated Nfkbiz −/− mice. (B) Cytokine and chemokine levels in skin homogenate. Data are representative of three independent experiments. Data are mean ± s.e.m. *p < 0.05, **p < 0.01.
Discussion
In this study, we showed that spontaneous dermatitis developed in Nfkbiz −/− mice along with expansion of IL-17A- and IL-22-secreting CD4+ T cells. Pyrosequencing analysis indicated significantly decreased bacterial diversity and drastically expanded pathobiont S. xylosus in the skin of Nfkbiz −/− mice. Antibiotic treatment relieved the dermatitis in Nfkbiz −/− mice, accompanied by decreased numbers of IL-17A- and IL-22-secreting CD4+ T cells. The introduction of S. xylosus to the skin of Nfkbiz −/− mice resulted in the predominant expansion of IL-17A-secreting CD4+ T cells. These results demonstrate that the expansion of pathobiont S. xylosus under Nfkbiz-deficient conditions may be a cause of skin dysbiosis and may be mediated by pathogenic IL-17A-secreting CD4+ T cells.
Previous studies described humans with AD and psoriasis that exhibited general pattern dysbiosis in the skin. These subjects had decreased bacterial diversity and altered predominant bacterial species2, 32. In this study, we found a similar dysbiotic patterns in Nfkbiz −/− mice with spontaneous dermatitis. Specifically, S. xylosus accounted for more than 80% of the resident bacteria in the skin of Nfkbiz −/− mice. S. xylosus is a commensal bacterium that inhabits the skin of mammals and occasionally humans33, and its ability to cause opportunistic infections has been reported10–12. We found a correlation between the expansion of S. xylosus and the severity of inflammation in Nfkbiz −/− mice, a finding that is supported by a recent study describing the dominant expansion of S. aureus among patients with more severe AD34. Moreover, the fact that antibiotics specific to S. xylosus resulted in the complete suppression of dermatitis35, 36 supports the notion that the dysbiosis provoked by the predominance of S. xylosus is closely related to skin inflammation.
AD has been considered an allergic Th2-mediated disease due to the high levels of IgE Ab37; however, others suggest that AD is more heterogeneous, given the contributions of other T cells such as Th17 and Th2216, 38, 39. Among several subsets of AD, intrinsic AD and Asian AD patients preferentially exhibit the activation of Th17 and Th22 cytokines40, 41. These patients show features typical of persons with psoriasis, such as parakeratosis and epidermal hyperplasia41. Although the numbers of Th17 and Th22 cells were found to be increased systemically and in the skin of AD patients, the roles of IL-17A and IL-22 in the pathogenesis of this disease remain unclear. In this context, another recent study addressed the possibility that IL-17A acts as an inducer for Th2-type immune responses in murine AD models42. On the other hand, IL-22 is known to promote keratinocyte proliferation. IL-23 produced by keratinocytes in response to endogenous TLR4 ligands polarized skin DCs to stimulate the IL-22 response43. In our study, we found that IL-17A and IL-22, more than any other cytokines, were significantly reduced in response to the administration of antibiotics (Fig. 5C) and that S. xylosus inoculation onto Nfkbiz −/− mice led to elevated IL-17A and IL-22 cytokines in skin homogenates (Fig. 7A). In this context, others demonstrated that the onset of skin inflammation in Nfkbiz −/− is independent of the Th2 response by showing that dermatitis still developed in Nfkbiz −/−Stat6−/− mice26. We further observed that IFN-γ remained elevated even after antibiotic treatment and improvement of skin inflammation (Figs 4C and S2). These lines of evidence suggest that Th17 and Th22 cells rather than Th1 cells may be directly involved in the development of skin inflammation in Nfkbiz −/− mice.
Our results indicate that the topical association of S. xylosus elevates chemokine secretion in the skin (Fig. 7B). Others showed that MCP-1 and MIP-1α participate in both immediate and delayed skin reactions and trigger the recruitment of local T cells, macrophages, and granulocytes44. Transgenic mice overexpressing MCP-1 in the epidermis also exhibit a dramatic increase of MHCII+ cells in the dermis and show augmentation of the contact hypersensitivity reaction45. We speculate that the expansion of S. xylosus first stimulates chemokine secretion by keratinocytes and/or fibroblasts, which may be involved in the recruitment of innate immune cells in the skin that are indispensable in the onset of skin inflammation. In support of this possibility, the inoculation of S. xylosus also increased migratory DCs (CD11c+ MHCIIhi) in skin-draining lymph nodes (Fig. S6). Other studies indicate that MCP-1 is also responsible for the recruitment of Th17 cells46, while IL-17A stimulates fibroblasts to produce MCP-1 and RANTES47, 48. Overall, these results imply that the expansion of a single pathobiont such as S. xylosus provokes inflammation in local mucosal tissues through the induction of chemokines and Th17 and Th22 responses.
In Nfkbiz −/− mice with severe dermatitis, we found a marked increase in CD103+ dDCs, which are rare in normal skin (Fig. 2A). This subset was significantly reduced after antibiotic treatment (Fig. 5A). However, topical inoculation of S. xylosus was unable to drive the recruitment of CD103+ dDCs in the skin (data not shown). Others found that CD103+ DCs in non-lymphoid tissues are functionally specialized in the cross-presentation of antigens to CD8+ cells49. CD207+ dDCs, often referred to as CD103+ DCs in the skin, induce CD8+ T-cell and Th1-cell responses15, 50. However, there have been no reports of CD103+ dDCs in skin of Nfkbiz −/− mice with dermatitis, though its presence in steady-state skin-draining lymph nodes is known51. Further studies on CD103+ dDC function are needed to assess their role in Th17 differentiation during S. xylosus infection.
IκBζ, encoded by the Nfkbiz gene, is a transcription factor known to promote IL-17A production. For instance, IκBζ−/− mice show resistance to diseases with enhanced Th17 responses, including experimental autoimmune encephalomyelitis (EAE) and psoriasis19, 52. However, in this study, we observed robust Th17 responses as well as expression of IL-17A signature genes (i.e., S100a8, S100a9, CAMP, and LCN2) in the skin of Nfkbiz −/− mice with spontaneous dermatitis (Fig. S3). We assume that, unlike RORγt, IκBζ is not a master regulator for Th17 cells. It could thus function as a Th17 cell-specific transcription factor in cooperation with ROR nuclear factors. The overexpression of ROR nuclear factors drove IL-17A production in Nfkbiz −/− T cells19. In our study, we detected the upregulation of other transcription factors that contribute to Th17 differentiation, such as BATF, which remodels chromatin to enable the access of transcription factors to Th17-specific loci53; Hif-1α, which can directly increase RORγt expression and drive IL-17A expression with RORγt54; and IL-23A, which is essential for the induction of pathogenic Th17 cells55 in the skin of Nfkbiz −/− mice with dermatitis (Fig. S3). These results provide support for a molecular mechanism of an enhanced Th17 response in Nfkbiz −/− mice.
In conclusion, our findings demonstrate that S. xylosus is an important pathobiont that accelerates dermatitis in Nfkbiz-deficient mice. We also found that a single commensal bacterium can modulate innate and acquired immune responses in the skin. Although the commensal microbiota maintains a symbiotic relationship with the host immune system and plays a beneficial role in the host under steady-state conditions, it can provoke and trigger inflammation under Nfkbiz-deficient conditions. These results suggest that the Nfkbiz gene has a potential regulatory role in the microbiota-skin immunity axis.
Methods
Ethics statement
All animal experiments were approved by the Institutional Animal Care and Use Committee of Asan Biomedical Research Center (Approval No. PN 2014-12-063) and the studies were carried out in accordance with the approved guidelines and regulations. All experiments were performed under anesthesia with a mixture of ketamine (100 mg/kg) and xylazine (20 mg/kg), and all efforts were made to minimize suffering.
Mice
Nfkbiz −/− mice were provided by Prof. Shizuo Akira (IFReC, Osaka University, Japan). All mice were maintained under specific pathogen-free conditions in the animal facility at Asan Biomedical Research Center (Seoul, Korea), where they received sterilized food and water ad libitum.
Clinical score
Pathological phenotypes of mice were scored according to the criteria as described previously24, with slight modification: (1) swelling of the eyelid; (2) periocular skin erosion and eyes with discharge that were difficult to open; (3) inflammation reaching the skin of the ear and part of the skin on the body; (4) erosion and loss of hair in the whole body; and (5) severe erosion and loss of hair in the whole face and body.
Preparation of mononuclear cells from skin
Mouse trunk skin was shaved, peeled off, and placed epidermal side down in a Petri dish. Subcutaneous fat tissue and blood vessels were scrapped off with a Scalpel (Feather, Osaka, Japan). Skin tissue was then incubated with 0.25% trypsin (Gibco, Waltham, MA) at 37 °C for 2 h. Epidermal cells were separated from the dermis by mechanical separation and vigorously agitated in RPMI 1640 (Gibco) containing 10% FBS (Gibco) at 37 °C for 10 min. The dermis was then minced with scissors and further treated with 2.5 mg/ml collagenase IV (Sigma Aldrich, St. Louis, MO) and 0.2 mg/ml DNase (Roche) at 37 °C with stirring. After 1 h, digestion was stopped by the addition of 10 mM EDTA. Epidermal and dermal cell suspensions were filtered and enriched for immune cells using a 40–75% Percoll density gradient (GE Healthcare, Buckinghamshire, UK).
Flow cytometry analysis and antibodies
For cytokine analysis, cell suspensions were cultured in RPMI 1640 medium supplemented with 10% FBS and 1% penicillin and streptomycin (Gibco) with 50 ng/ml PMA (Sigma Aldrich) and 1 μg/ml ionomycin (Sigma Aldrich) in the presence of Brefeldin A (BD Biosciences, Franklin Lakes, NJ) overnight at 37 °C. Cells were stained with Live/Dead Cell Stain kit (Invitrogen, Carlsbad, CA) and surface markers after treatment with purified anti-mouse CD16/32 Ab (BD Biosciences). Fixation and permeabilization were performed with BD Cytofix/Cytoperm (BD Biosciences), followed by intracellular cytokine staining. The following Abs from BD Biosciences, eBioscience (Santa Clara, CA), or BioLegend (San Diego, CA) were used for analysis: anti-CD45 (30-F11), anti-CD4 (RM4–5), anti-CD8α (53–6.7), anti-TCRγδ (GL3), anti-IFN-γ (XMG1.2), anti-IL-4 (11B11), anti-IL-17A (TC11-18H10), anti-IL-22 (IL-22JOP), anti-MHC II (M5/114.15.2), anti-CD207 (4C7), anti-CD11b (M1/70), anti-CD11c (HL3), and anti-CD103 (2E7). Cell acquisition was performed on a BD Biosciences FACSCanto II flow cytometer and data were analyzed using FlowJo software (Tree Star, Ashland, OR).
16s rRNA gene pyrosequencing analysis
Genomic DNA was extracted from skin tissue using the NucleoSpin Soil kit (Macherey-Nagel, Düren, Germany). PCR amplification was performed using primers targeting the segment from the V1 to V3 regions of the 16S rRNA gene with extracted gDNA. For bacterial amplifications, we used barcoded primers of 9 F (5′-CCTATCCCCTGTGTGCCTTGGCAGTC-TCAG-AC-AGAGTTTGATCMTGGCTCAG-3′; the underlined sequence indicates the target region primer) and 541 R (5′-CCATCTCATCCCTGCGTGTCTCCGAC-TCAG-X-AC-ATTACCGCGGCTGCTGG-3′; X indicates the unique barcode for each subject). The amplifications were performed under the following conditions: initial denaturation at 95 °C for 5 min, followed by 30 cycles of denaturation at 95 °C for 30 sec, primer annealing at 55 °C for 30 sec, and extension at 72 °C for 30 sec, with final elongation at 72 °C for 5 min. The amplified products were purified with the QIAquick PCR purification kit (Qiagen, Valencia, CA). Equal concentrations of purified products were pooled together and short fragments (non-target products) were removed with an AMPure bead kit (Agencourt Bioscience, Beverly, MA). The quality and product size were assessed on a Bioanalyzer 2100 (Agilent, Palo Alto, CA) using a DNA 7500 chip. Mixed amplicon sequencing was performed by emulsion PCR and then deposited on picotiter plates. The sequencing was carried out at Chunlab (Seoul, Republic of Korea) on a GS Junior Sequencing System (Roche, Branford, CT), in accordance with the manufacturer’s instructions.
Microarray analysis
RNA from skin tissues was isolated with TRIzol (Invitrogen) reagent. RNA labeling and hybridization were performed by using the Agilent one-color microarray-based gene expression analysis kit (Agilent Technology, Santa Clara, CA). In brief, total RNA (200 ng) from each sample was linearly amplified and labeled with Cy3-dCTP. The labeled cRNAs were purified by RNAeasy mini kit (Qiagen). Each labeled cRNA (600 ng) was fragmented by adding blocking agent and fragmentation buffer, and then heated at 60 °C for 30 min. Finally GE hybridization buffer was added to dilute the labeled cRNA. Hybridization solution was dispensed into the gasket slide and assembled with the Agilent SurePrint G3 Mouse GE 8X60 K kit. The slides were incubated for 17 h at 65 °C in an Agilent hybridization oven and then washed. The hybridized array was immediately scanned with an Agilent Microarray Scanner D.
IPA (Ingenuity Pathway Analysis)
IPA (Ingenuity Systems, http://www.ingenuity.com) was used for gene network analysis, which was carried out to identify the most significant gene sets associated with disease process and molecular and cellular functions in significantly altered genes (described in the Figs 6 and S3). The significance of over-represented gene sets was estimated by the right-tailed Fisher’s exact test. Gene network analysis was carried out by using a global molecular network developed from information contained in the Ingenuity Knowledge Base. Identified gene networks were ranked according to scores provided by IPA.
Antibiotic treatment
Mice were treated with 0.5 mg/ml cephalexin and enrofloxacin (Sigma Aldrich) in drinking water.
Bacterial isolation and inoculation onto mice
To isolate S. xylosus from skin, swabs from the shaved back skin of mice (1 cm2 or the infection area) were placed in PBS and cultured on MSA (BD Biosciences). The colonies from the plates were sequenced by Macrogen (Seoul, Republic of Korea). S. xylosus was cultured in mannitol salt broth (BD Biosciences) at 37 °C overnight in a shaking incubator. Bacteria were enumerated by assessing the colony-forming units (CFUs) using a standard plate count method and by measuring optical density at 600 nm. For the topical inoculation of bacteria, a bacterial suspension (109 CFUs) in PBS was topically applied to the shaved back skin of mice using a sterile cotton swab. Bacteria were applied every other day.
Histological analysis and immunofluorescence microscopy
After sacrifice, mouse back skin was fixed in 4% paraformaldehyde, embedded in paraffin, and stained with hematoxylin-eosin. Microscopic images were obtained (BX53; Olympus Optical, Tokyo, Japan). For immunofluorescence microscopy, skin tissues were frozen, embedded in OCT compound, sectioned, and fixed with −20 °C acetone for 5 min. Slides were blocked with PBS containing 1% BSA for 1 h at room temperature and stained with anti-CD8α (53–6.7), anti-CD4 (RM4-5), anti-CD11b (M1/70), anti-CD207 (4C7), and anti-CD11c (HL3) Abs (BD Biosciences) overnight at 4 °C. Tissues were washed and stained with 4′,6-diamidino-2-phenylindole (DAPI; BD Pharmingen) for 2 min at room temperature, followed by mounting with PermaFluor mountant (Thermo, Waltham, MA). Images were captured on an LSM 710 confocal microscope (Carl Zeiss, Oberkochen, Germany).
IgE and cytokine analysis
Serum total IgE Ab level was determined using mouse IgE ELISA kit (BD Biosciences). Supernatant was collected after skin tissues were weighed and homogenized with Tris-EDTA buffer (10 mM Tris-HCl and 1 mM EDTA, pH 7.4, 0.05% sodium azide, 1% Tween-80, protease inhibitor cocktail) and centrifuged at 11,000 × g for 10 min at 4 °C. Cytokines and chemokines from tissue homogenates were measured by a ProcartaPlex Multiplex immunoassay kit (eBioscience), in accordance with the manufacturer’s instructions.
Trans-epidermal water loss
TEWL was measured by VapoMeter SWL4001JT (Delfin, Kuopio, Finland).
Statistics
GraphPad Prism software (GraphPad, La Jolla, CA) was used for statistical analysis. Significant differences between two groups were analyzed with two-tailed unpaired t-test. Multiple groups were analyzed by two-way ANOVA followed by Bonferroni’s post hoc test (*p < 0.05; **p < 0.01).
Electronic supplementary material
Acknowledgements
We thank Prof. Jin-Woo Bae and Dr. Min-Soo Kim for their advice and evaluation of the metagenomic analysis. This work was supported by the National Research Foundation of Korea (NRF-2017R1A2B3002132) and the Korean Healthcare Technology R&D Project, Ministry for Health, Welfare and Family Affairs of Korea (HI13C0016).
Author Contributions
Y.K. performed all experiments and data analysis and wrote the manuscript; Y.L. and J.Y. helped to design experiments, analyze data, and review the manuscript; S.L. helped in data collection and analysis; Y.P. assisted in data analysis, interpretation and manuscript review; M.K. contributed to the conception and design of the study and supervised the study.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-05740-z
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Belkaid Y, Segre JA. Dialogue between skin microbiota and immunity. Science. 2014;346:954–959. doi: 10.1126/science.1260144. [DOI] [PubMed] [Google Scholar]
- 2.Kong HH, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012;22:850–859. doi: 10.1101/gr.131029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marcus E, Demmler D, Rudolph A, Fischer M. Bacterial colonization of psoriasis plaques. Is it relevant? Dermatol Reports. 2011;3:e14. doi: 10.4081/dr.2011.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freeman, A. F. & Holland, S. M. The hyper-IgE syndromes. Immunol Allergy Clin North Am28, 277–291 viii (2008). [DOI] [PMC free article] [PubMed]
- 5.Kobayashi T, et al. Dysbiosis and Staphylococcus aureus colonization drives inflammation in atopic dermatitis. Immunity. 2015;42:756–766. doi: 10.1016/j.immuni.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dordet-Frisoni E, et al. Genomic diversity in Staphylococcus xylosus. Appl Environ Microbiol. 2007;73:7199–7209. doi: 10.1128/AEM.01629-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giordano N, et al. Erythema nodosum associated with Staphylococcus xylosus septicemia. J Microbiol Immunol Infect. 2016;49:134–137. doi: 10.1016/j.jmii.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Tselenis-Kotsowilis AD, Koliomichalis MP, Papavassiliou JT. Acute pyelonephritis caused by Staphylococcus xylosus. J Clin Microbiol. 1982;16:593–594. doi: 10.1128/jcm.16.3.593-594.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makki AR, et al. Phenotypic and genotypic characterization of coagulase negative staphylococci (CoNS) other than Staphylococcus epidermidis isolated from ocular infections. Invest Ophthalmol Vis Sci. 2011;52:9018–9022. doi: 10.1167/iovs.11-7777. [DOI] [PubMed] [Google Scholar]
- 10.Won YS, et al. Identification of Staphylococcus xylosus isolated from C57BL/6J-Nos2(tm1Lau) mice with dermatitis. Microbiol Immunol. 2002;46:629–632. doi: 10.1111/j.1348-0421.2002.tb02744.x. [DOI] [PubMed] [Google Scholar]
- 11.Russo M, Invernizzi A, Gobbi A, Radaelli E. Diffuse scaling dermatitis in an athymic nude mouse. Vet Pathol. 2013;50:722–726. doi: 10.1177/0300985812463408. [DOI] [PubMed] [Google Scholar]
- 12.Gozalo AS, et al. Spontaneous Staphylococcus xylosus infection in mice deficient in NADPH oxidase and comparison with other laboratory mouse strains. J Am Assoc Lab Anim Sci. 2010;49:480–486. [PMC free article] [PubMed] [Google Scholar]
- 13.Belkaid Y, Tamoutounour S. The influence of skin microorganisms on cutaneous immunity. Nat Rev Immunol. 2016;16:353–366. doi: 10.1038/nri.2016.48. [DOI] [PubMed] [Google Scholar]
- 14.Naik S, et al. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337:1115–1119. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naik S, et al. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature. 2015;520:104–108. doi: 10.1038/nature14052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eyerich K, et al. IL-17 in atopic eczema: linking allergen-specific adaptive and microbial-triggered innate immune response. J Allergy Clin Immunol. 2009;123:59–66 e54. doi: 10.1016/j.jaci.2008.10.031. [DOI] [PubMed] [Google Scholar]
- 17.Niebuhr M, et al. Staphylococcal alpha-toxin is a strong inducer of interleukin-17 in humans. Infect Immun. 2011;79:1615–1622. doi: 10.1128/IAI.00958-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reginald K, et al. Immunoglobulin E antibody reactivity to bacterial antigens in atopic dermatitis patients. Clin Exp Allergy. 2011;41:357–369. doi: 10.1111/j.1365-2222.2010.03655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okamoto K, et al. IκBζ regulates Th17 development by cooperating with ROR nuclear receptors. Nature. 2010;464:1381–1385. doi: 10.1038/nature08922. [DOI] [PubMed] [Google Scholar]
- 20.Karlsen JR, Borregaard N, Cowland JB. Induction of neutrophil gelatinase-associated lipocalin expression by co-stimulation with interleukin-17 and tumor necrosis factor-a is controlled by IκBζ but neither by C/EBP-b nor C/EBP-d. J Biol Chem. 2010;285:14088–14100. doi: 10.1074/jbc.M109.017129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MaruYama T, et al. Control of IFN-g production and regulatory function by the inducible nuclear protein IκBζ in T cells. J Leukoc Biol. 2015;98:385–393. doi: 10.1189/jlb.2A0814-384R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamazaki S, et al. Gene-specific requirement of a nuclear protein, IκBζ, for promoter association of inflammatory transcription regulators. J Biol Chem. 2008;283:32404–32411. doi: 10.1074/jbc.M802148200. [DOI] [PubMed] [Google Scholar]
- 23.Yamamoto M, et al. Regulation of Toll/IL-1-receptor-mediated gene expression by the inducible nuclear protein IκBζ. Nature. 2004;430:218–222. doi: 10.1038/nature02738. [DOI] [PubMed] [Google Scholar]
- 24.Okuma A, et al. Enhanced apoptosis by disruption of the STAT3- IκBζ signaling pathway in epithelial cells induces Sjogren’s syndrome-like autoimmune disease. Immunity. 2013;38:450–460. doi: 10.1016/j.immuni.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 25.Motoyama M, et al. Positive and negative regulation of NF-kB-mediated transcription by IκBζ, an inducible nuclear protein. J Biol Chem. 2005;280:7444–7451. doi: 10.1074/jbc.M412738200. [DOI] [PubMed] [Google Scholar]
- 26.Ueta M, et al. Stat6-independent tissue inflammation occurs selectively on the ocular surface and perioral skin of IκBζ−/− mice. Invest Ophthalmol Vis Sci. 2008;49:3387–3394. doi: 10.1167/iovs.08-1691. [DOI] [PubMed] [Google Scholar]
- 27.Paparo SB, Guaragna MA, Albanesi M. High IgE levels in patients affected by psoriasis: review of the literature and personal observations. Clin Ter. 2014;165:91–93. doi: 10.7471/CT.2014.1682. [DOI] [PubMed] [Google Scholar]
- 28.Laske N, Niggemann B. Does the severity of atopic dermatitis correlate with serum IgE levels? Pediatr Allergy Immunol. 2004;15:86–88. doi: 10.1046/j.0905-6157.2003.00106.x. [DOI] [PubMed] [Google Scholar]
- 29.Henri S, et al. Disentangling the complexity of the skin dendritic cell network. Immunol Cell Biol. 2010;88:366–375. doi: 10.1038/icb.2010.34. [DOI] [PubMed] [Google Scholar]
- 30.Shittu A, Lin J, Morrison D, Kolawole D. Identification and molecular characterization of mannitol salt positive, coagulase-negative staphylococci from nasal samples of medical personnel and students. J Med Microbiol. 2006;55:317–324. doi: 10.1099/jmm.0.46072-0. [DOI] [PubMed] [Google Scholar]
- 31.Garcin CL, et al. Ectodysplasin A pathway contributes to human and murine skin repair. J Invest Dermatol. 2016;136:1022–1030. doi: 10.1016/j.jid.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alekseyenko AV, et al. Community differentiation of the cutaneous microbiota in psoriasis. Microbiome. 2013;1:31. doi: 10.1186/2049-2618-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagase N, et al. Isolation and species distribution of Staphylococci from animal and human skin. J Vet Med Sci. 2002;64:245–250. doi: 10.1292/jvms.64.245. [DOI] [PubMed] [Google Scholar]
- 34.Totte JE, et al. Prevalence and odds of Staphylococcus aureus carriage in atopic dermatitis: a systematic review and meta-analysis. Br J Dermatol. 2016;175:687–695. doi: 10.1111/bjd.14566. [DOI] [PubMed] [Google Scholar]
- 35.Bochniarz M, Wawron W, Szczubial M. Production of slime by coagulase-negative staphylococci (CNS) isolated from clinical and subclinical mastitis in cows. Pol J Vet Sci. 2014;17:447–452. doi: 10.2478/pjvs-2014-0064. [DOI] [PubMed] [Google Scholar]
- 36.Zamankhan Malayeri H, Jamshidi S, Zahraei Salehi T. Identification and antimicrobial susceptibility patterns of bacteria causing otitis externa in dogs. Vet Res Commun. 2010;34:435–444. doi: 10.1007/s11259-010-9417-y. [DOI] [PubMed] [Google Scholar]
- 37.Brandt, E. B. & Sivaprasad, U. Th2 cytokines and atopic dermatitis. J Clin Cell Immunol2 (2011). [DOI] [PMC free article] [PubMed]
- 38.Gittler JK, et al. Progressive activation of Th2/Th22 cytokines and selective epidermal proteins characterizes acute and chronic atopic dermatitis. J Allergy Clin Immunol. 2012;130:1344–1354. doi: 10.1016/j.jaci.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koga C, et al. Possible pathogenic role of Th17 cells for atopic dermatitis. J Invest Dermatol. 2008;128:2625–2630. doi: 10.1038/jid.2008.111. [DOI] [PubMed] [Google Scholar]
- 40.Suarez-Farinas M, et al. Intrinsic atopic dermatitis shows similar Th2 and higher Th17 immune activation compared with extrinsic atopic dermatitis. J Allergy Clin Immunol. 2013;132:361–370. doi: 10.1016/j.jaci.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Noda S, et al. The Asian atopic dermatitis phenotype combines features of atopic dermatitis and psoriasis with increased Th17 polarization. J Allergy Clin Immunol. 2015;136:1254–1264. doi: 10.1016/j.jaci.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 42.Nakajima S, et al. IL-17A as an inducer for Th2 immune responses in murine atopic dermatitis models. J Invest Dermatol. 2014;134:2122–2130. doi: 10.1038/jid.2014.51. [DOI] [PubMed] [Google Scholar]
- 43.Yoon J, et al. IL-23 induced in keratinocytes by endogenous TLR4 ligands polarizes dendritic cells to drive IL-22 responses to skin immunization. J Exp Med. 2016;213:2147–2166. doi: 10.1084/jem.20150376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaga M, et al. Skin reactivity and local cell recruitment in human atopic and nonatopic subjects by CCL2/MCP-1 and CCL3/MIP-1a. Allergy. 2008;63:703–711. doi: 10.1111/j.1398-9995.2007.01578.x. [DOI] [PubMed] [Google Scholar]
- 45.Nakamura K, Williams IR, Kupper TS. Keratinocyte-derived monocyte chemoattractant protein 1 (MCP-1): analysis in a transgenic model demonstrates MCP-1 can recruit dendritic and Langerhans cells to skin. J Invest Dermatol. 1995;105:635–643. doi: 10.1111/1523-1747.ep12324061. [DOI] [PubMed] [Google Scholar]
- 46.Pelletier M, et al. Evidence for a cross-talk between human neutrophils and Th17 cells. Blood. 2010;115:335–343. doi: 10.1182/blood-2009-04-216085. [DOI] [PubMed] [Google Scholar]
- 47.Brembilla NC, et al. Th17 cells favor inflammatory responses while inhibiting type I collagen deposition by dermal fibroblasts: differential effects in healthy and systemic sclerosis fibroblasts. Arthritis Res Ther. 2013;15:R151. doi: 10.1186/ar4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fang S, et al. IL-17A promotes RANTES expression, but not IL-16, in orbital fibroblasts via CD40-CD40L combination in thyroid-associated ophthalmopathy. Invest Ophthalmol Vis Sci. 2016;57:6123–6133. doi: 10.1167/iovs.16-20199. [DOI] [PubMed] [Google Scholar]
- 49.Ginhoux F, et al. The origin and development of nonlymphoid tissue CD103+ DCs. J Exp Med. 2009;206:3115–3130. doi: 10.1084/jem.20091756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Igyarto BZ, et al. Skin-resident murine dendritic cell subsets promote distinct and opposing antigen-specific T helper cell responses. Immunity. 2011;35:260–272. doi: 10.1016/j.immuni.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Henri S, et al. CD207+CD103+ dermal dendritic cells cross-present keratinocyte-derived antigens irrespective of the presence of Langerhans cells. J Exp Med. 2010;207:189–206. doi: 10.1084/jem.20091964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johansen C, et al. IκBζ is a key driver in the development of psoriasis. Proc Natl Acad Sci USA. 2015;112:E5825–5833. doi: 10.1073/pnas.1509971112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ciofani M, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dang EV, et al. Control of Th17/Treg balance by hypoxia-inducible factor 1. Cell. 2011;146:772–784. doi: 10.1016/j.cell.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee Y, et al. Induction and molecular signature of pathogenic Th17 cells. Nat Immunol. 2012;13:991–999. doi: 10.1038/ni.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


