Abstract
Due to tetracycline abuse, the safe bifidobacteria in the human gastrointestinal intestinal tract (GIT) may serve as a reservoir of tetracycline resistance genes. In the present investigation of 92 bifidobacterial strains originating from the human GIT, tetracycline resistance in 29 strains was mediated by the tet(W), tet(O) or tet(S) gene, and this is the first report of tet(O)- and tet(S)-mediated tetracycline resistance in bifidobacteria. Antibiotic resistance genes harbored by bifidobacteria are transferred from other bacteria. However, the characteristics of the spread and integration of tetracycline resistance genes into the human intestinal bifidobacteria chromosome are poorly understood. Here, conserved sequences were identified in bifidobacterial strains positive for tet(W), tet(O), or tet(S), including the tet(W), tet(O), or tet(S) and their partial flanking sequences, which exhibited identity with the sequences in multiple human intestinal pathogens, and genes encoding 23 S rRNA, an ATP transporter, a Cpp protein, and a membrane-spanning protein were flanking by the 1920-bp tet(W), 1920-bp tet(O), 1800-bp tet(O) and 252-bp tet(S) in bifidobacteria, respectively. These findings suggest that tetracycline resistance genes harbored by human intestinal bifidobacteria might initially be transferred from pathogens and that each kind of tetracycline resistance gene might tend to insert in the vicinity of specific bifidobacteria genes.
Introduction
There are up to 1013–1014 total bacteria in the human gastrointestinal intestinal tract (GIT)1, 2. Due to the abuse of tetracycline in the clinical and nonclinical treatment of various human infections3, the carriage of tetracycline resistance genes by bacteria in the human GIT has been an area of intense investigation4. Most studies have focused on the tetracycline resistance genes carried by clinical pathogens or opportunistic pathogens5 and have continuously detected new tetracycline resistance genes harbored by the intestinal pathogens, such as the tet(40) gene in the human intestinal firmicute bacterium6. However, because bifidobacteria are ingested as probiotics in the human GIT and have acquired a “generally regarded as safe” (GRAS) status7–9, so far, only tet(W)- and tet(M)-mediated tetracycline resistance have been detected in intestinal bifidobacteria of human origin10–13, and only tet(L)-, tet(O/W)-, tet(W/32/O)-, and tet(O/W/32/O/W/O)-mediated tetracycline resistance have been detected in intestinal bifidobacteria of pig origin14. Therefore, it remains unknown whether tetracycline resistance genes other than tet(W) and tet(M) can be detected in the bifidobacterial strains originating in the human GIT.
Antibiotic resistance (AR) genes within potentially mobile elements can spread horizontally across genera in the human GIT15. Comparative analysis of sequences flanking the same AR gene in one genus of bacteria can therefore further reveal the spread characteristics of the AR gene. However, although two tetracycline resistance genes [tet(W) and tet(M)] have been detected in human intestinal bifidobacteria10–13, only the sequences flanking the tet(W) gene in bifidobacteria have been analyzed10, 12. Scott previously found a conserved tet(W) gene sequence of 2154 bp in 10 gut bifidobacterial strains of 5 species12. Ammor analyzed the flanking sequences of the tet(W) genes in another six human intestinal bifidobacteria and found an orfY gene in the downstream flanking region of the tet(W) gene in one B. thermophilum strain and one B. longum strain and a transposase gene in the downstream flanking region of the tet(W) gene in two B. longum strains10. Based on these results, it is not possible to determine whether the tet(W) gene inserts into common sites in the chromosome of the human intestinal bifidobacteria or whether other tetracycline resistance genes may exhibit conservation in their integration into the human intestinal bifidobacteria chromosome.
As a result of the misuse and overuse of tetracycline, the traditionally safe bifidobacteria in the human GIT may serve as a reservoir of tetracycline resistance genes and increasingly become a threat to human health. Therefore, this study was performed to assess 92 bifidobacterial strains isolated from the feces of 14 healthy individuals, one type strain and seven commercial strains via phenotypically and genotypically screening the acquired tetracycline resistance profiles and to comparatively analyze the upstream and downstream sequences flanking the tetracycline resistance genes harbored by different strains.
Results
Tetracycline susceptibility profiles
The MIC values of tetracycline in the 100 bifidobacterial strains tested are presented in Tables 1 and 2. Twenty-nine bifidobacterial strains, including the seven Bifidobacterium longum strains shown in Table 1 and two Bifidobacterium bifidum strains, six Bifidobacterium pseudocatenulatum strains, 13 Bifidobacterium lactis strains and one Bifidobacterium breve strain shown in Table 2, exhibited strong tetracycline resistance [minimum inhibitory concentration (MIC) ≥256 μg/ml], with MIC values that higher than the breakpoint for Bifidobacterium defined by the European Food Safety Authority (EFSA) (MIC = 8 μg/ml)16.
Table 1.
MIC susceptibility profiles of tetracycline and the corresponding genotypes for 45 B. longum strains one B. infantis strain.
| Species | Strain | Origin | MIC (μg/ml) | Tetracycline resistance genes | |||
|---|---|---|---|---|---|---|---|
| tet(W) | tet(O) | tet(S) | The other 10 genes | ||||
| B. infantis | Pronova BI211a | Human | <0.016 | − | − | − | − |
| B. longum | Pronova BL88-Onllya | Human | <0.016 | − | − | − | − |
| A33 | Child feces | <0.016 | − | − | − | − | |
| A42 | Child feces | <0.016 | − | − | − | − | |
| W11 | Adult feces | <0.016 | − | − | − | − | |
| W12 | Adult feces | <0.016 | − | − | − | − | |
| W14 | Adult feces | <0.016 | − | − | − | − | |
| W210 | Adult feces | <0.016 | − | − | − | − | |
| W22 | Adult feces | <0.016 | − | − | − | − | |
| N34 | Adult feces | <0.016 | − | − | − | − | |
| N45 | Adult feces | <0.016 | − | − | − | − | |
| N51 | Adult feces | <0.016 | − | − | − | − | |
| Y27 | Adult feces | <0.016 | − | − | − | − | |
| Y35 | Adult feces | <0.016 | − | − | − | − | |
| Z21 | Child feces | <0.016 | − | − | − | − | |
| Z31 | Child feces | <0.016 | − | − | − | − | |
| D41 | Child feces | <0.016 | − | − | − | − | |
| D510 | Child feces | <0.016 | − | − | − | − | |
| D512 | Child feces | <0.016 | − | − | − | − | |
| D514 | Child feces | <0.016 | − | − | − | − | |
| X41 | Child feces | <0.016 | − | − | − | − | |
| H1 | Child feces | <0.016 | − | − | − | − | |
| H32 | Child feces | <0.016 | − | − | − | − | |
| L2 | Adult feces | <0.016 | − | − | − | − | |
| L8 | Adult feces | <0.016 | − | − | − | − | |
| N7 | Adult feces | <0.016 | − | − | − | − | |
| W211 | Adult feces | <0.016 | − | − | − | − | |
| W21 | Adult feces | <0.016 | − | − | − | − | |
| W24 | Adult feces | <0.016 | − | − | − | − | |
| W29 | Adult feces | <0.016 | − | − | − | − | |
| W212 | Adult feces | <0.016 | − | − | − | − | |
| W41 | Adult feces | <0.016 | − | − | − | − | |
| a44 | Child feces | <0.016 | − | − | − | − | |
| A31 | Child feces | <0.016 | − | − | − | − | |
| A44 | Child feces | <0.016 | − | − | − | − | |
| A45 | Child feces | <0.016 | − | − | − | − | |
| A47 | Child feces | <0.016 | − | − | − | − | |
| F7 | Adult feces | <0.016 | − | − | − | − | |
| Y2 | Adult feces | <0.016 | − | − | − | − | |
| H21 | Child feces | ≥256 | − | + | − | − | |
| H34 | Child feces | ≥256 | − | + | − | − | |
| F313 | Adult feces | ≥256 | − | + | − | − | |
| F21 | Adult feces | ≥256 | + | − | − | − | |
| X33 | Child feces | ≥256 | + | − | − | − | |
| Y33 | Adult feces | ≥256 | − | + | − | − | |
| Z1 | Child feces | ≥256 | − | + | − | − | |
aCommercial strain obtained from the Shanghai Jiao Da Onlly Co. (Shanghai, PR China).
Table 2.
MIC susceptibility profiles of tetracycline and the corresponding genotypes for 2 B. adolescentis strains, 3 B. bifidum strains, 12 B. pseudocatenulatum strains, 18 B. breve strains and 19 B. lactis strains.
| Species | Strain | Origin | MIC (μg/ml) | Tetracycline resistance genes | |||
|---|---|---|---|---|---|---|---|
| tet(W) | tet(O) | tet(S) | The other 10 genes | ||||
| B. adolescentis | W25 | Adult feces | <0.016 | − | − | − | − |
| W42 | Adult feces | <0.016 | − | − | − | − | |
| B. bifidum | Pronova BB47a | Human | <0.016 | − | − | − | − |
| Y24 | Adult feces | ≥256 | + | − | − | − | |
| Y21 | Adult feces | ≥256 | + | − | − | − | |
| B. pseudocatenulatum | L37 | Adult feces | <0.016 | − | − | − | − |
| W13 | Adult feces | <0.016 | − | − | − | − | |
| W28 | Adult feces | <0.016 | − | − | − | − | |
| N2 | Adult feces | <0.016 | − | − | − | − | |
| A35 | Child feces | <0.016 | − | − | − | − | |
| D52 | Child feces | <0.016 | − | − | − | − | |
| J56 | Adult feces | ≥256 | + | − | − | − | |
| H23 | Child feces | ≥256 | + | − | − | − | |
| Z25 | Child feces | ≥256 | + | − | − | − | |
| a39 | Child feces | ≥256 | + | − | − | − | |
| Y1 | Adult feces | ≥256 | − | + | − | ||
| F312 | Adult feces | ≥256 | − | − | + | − | |
| B. breve | ATCC 15700b | Human | <0.016 | − | − | − | − |
| Pronova BB8a | Human | <0.016 | − | − | − | − | |
| BBW | Child feces | <0.016 | − | − | − | − | |
| BBM | Child feces | <0.016 | − | − | − | − | |
| BB2 | Child feces | <0.016 | − | − | − | − | |
| BB | Child feces | <0.016 | − | − | − | − | |
| N1 | Adult feces | <0.016 | − | − | − | − | |
| N24 | Adult feces | <0.016 | − | − | − | − | |
| L211 | Adult feces | <0.016 | − | − | − | − | |
| W46 | Adult feces | <0.016 | − | − | − | − | |
| SQS3-56 | Child feces | <0.016 | − | − | − | − | |
| SQS3-64 | Child feces | <0.016 | − | − | − | − | |
| SQS5-51 | Child feces | <0.016 | − | − | − | − | |
| SQS5-52 | Child feces | <0.016 | − | − | − | − | |
| A34 | Child feces | <0.016 | − | − | − | − | |
| a313 | Child feces | <0.016 | − | − | − | − | |
| a37 | Child feces | <0.016 | − | − | − | − | |
| A27 | Child feces | ≥256 | − | − | + | − | |
| B. lactis | Pronova BL99a | Human | <0.016 | − | − | − | − |
| Pronova BL25a | Human | <0.016 | − | − | − | − | |
| Pronova BI516a | Human | <0.016 | − | − | − | − | |
| J316 | Adult feces | <0.016 | − | − | − | − | |
| F5 | Adult feces | <0.016 | − | − | − | − | |
| F18 | Adult feces | <0.016 | − | − | − | − | |
| F9 | Adult feces | ≥256 | + | − | − | − | |
| F10 | Adult feces | ≥256 | + | − | − | − | |
| F11 | Adult feces | ≥256 | + | − | − | − | |
| F12 | Adult feces | ≥256 | + | − | − | − | |
| J310 | Adult feces | ≥256 | + | − | − | − | |
| J311 | Adult feces | ≥256 | + | − | − | − | |
| J317 | Adult feces | ≥256 | + | − | − | − | |
| L35 | Adult feces | ≥256 | + | − | − | − | |
| L36 | Adult feces | ≥256 | + | − | − | − | |
| L38 | Adult feces | ≥256 | + | − | − | − | |
| L310 | Adult feces | ≥256 | + | − | − | − | |
| L311 | Adult feces | ≥256 | + | − | − | − | |
| L312 | Adult feces | ≥256 | + | − | − | − | |
aCommercial strain obtained from the Shanghai Jiao Da Onlly Co. (Shanghai, PR China).
bType strain.
Detection of tetracycline resistance genes
As Tables 1 and 2 show, each of the 29 tetracycline-resistant bifidobacterial strains possessed one tetracycline resistance determinant [tet(W), or tet(O), or tet(S) gene], and none of the 13 tetracycline resistance determinants tested were detected in the 71 tetracycline-sensitive bifidobacterial strains. The occurrence of the tet(W), tet(O), and tet(S) genes among the 100 bifidobacterial strains of the seven Bifidobacterium species tested are further summarized in Table 3.
Table 3.
Tetracycline resistance and occurrence of tetracycline resistance genes among 100 bifidobacterial strains of seven species.
| Species | Total strain number | Tetracycline resistant strains | tet(W) | tet(O) | tet(S) |
|---|---|---|---|---|---|
| B. adolescentis | 2 | 0 | − | − | − |
| B. infantis | 1 | 0 | − | − | − |
| B. longum | 45 | 7 | 2 | 5 | − |
| B. lactis | 19 | 13 | 13 | − | − |
| B. pseudocatenulatum | 12 | 6 | 4 | 1 | 1 |
| B. breve | 18 | 1 | − | − | 1 |
| B. bifidum | 3 | 2 | 2 | − | − |
| Total | 100 | 29 | 21 | 6 | 2 |
In the 21 tet(W)-positive strains, including 2 B. longum subsp. longum strains, 13 B. animalis subsp. lactis strains, 4 B. pseudocatenulatum strains, and 2 B. bifidum strains, tet(W) exhibited an identical DNA sequence of 1560 bp, which encoded a protein consisting of 520 amino acids that displayed 100% identity with the ribosomal protection protein tetW previously identified in Bifidobacterium animalis subsp. lactis strain IPLAIC4 (GenBank accession number GU361625.1).
In the 6 tet(O)-positive strains including 5 B. longum subsp. longum strains and one B. pseudocatenulatum strain, tet(O) exhibited an identical DNA sequence of 1457 bp, which encoded a protein consisting of 458 amino acids that displayed 100% identity with the ribosomal protection protein tetO previously identified in Streptococcus suis BM407 (GenBank FM252032.1).
In the two tet(S)-positive strains, B. pseudocatenulatum strain F312 and B. breve strain A27, tet(S) exhibited an identical DNA sequence of 210 bp, which encoded a protein consisting of 70 amino acids that displayed 100% identity with the ribosomal protection protein tetS previously identified in Lactococcus lactis subsp. lactis strain ILIBB-JZK (GenBank KF278750.1).
The complete sequence lengths of the tet(W), tet(S), and tet(O) genes were further confirmed by determining the sequences flanking the tet(W), tet(O), and tet(S) genes (see section “Sequence conservation of the tet(W), tet(O), tet(S) genes and their flanking regions”).
Sequence conservation of the tet(W), tet(O), tet(S) genes and their flanking regions
The nucleotide sequences of the 1560-bp tet(W), 1457-bp tet(O), and 210-bp tet(S) genes and their flanking sequences were compared in different bifidobacterial strains (Figs 1, 2, 3).
Figure 1.
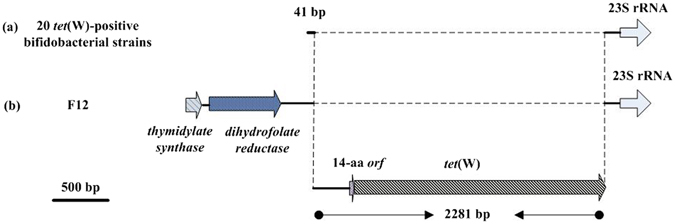
Genetic organization of the regions upstream and downstream of tet(W) in the 21 tet(W)-positive bifidobacterial strains. (a) 20 tet(W)-positive bifidobacterial strains. (b) The tet(W)-positive B. animalis subsp. lactis strain F12.
Figure 2.
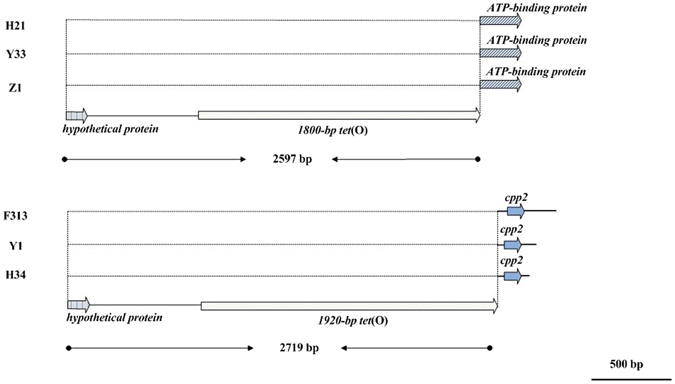
Genetic organization of the regions upstream and downstream of tet(O) in the 6 tet(O)-positive bifidobacterial strains.
Figure 3.
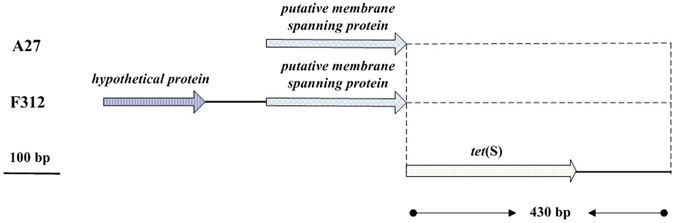
Genetic organization of the regions upstream and downstream of tet(S) in the 2 tet(S)-positive bifidobacterial strains.
The 21 tet(W)-positive bifidobacterial strains shared a core DNA region of 2281 bp, including a sequence of 298 bp, an upstream flanking sequence of 45 bp encoding an 14-amino-acid tet(W)-regulatory peptide, and the complete sequence of the 1920-bp tet(W) gene (Fig. 1). The 2281-bp sequence showed 99-100% nucleotide identity with the sequence previously identified in Corynebacterium diphtheria strain BH8 (GenBank CP003209.1), Streptococcus suis strain GZ1 (GenBank CP000837.1), and Arcanobacterium pyogenes strain OX4 (GenBank DQ517519.1).
Of the 6 tet(O)-positive bifidobacterial strains, three B. longum strains (H21, Y33 and Z1) shared a core DNA region of 2597 bp; however, an additional two B. longum strains, H34 and F313, and one B. pseudocatenulatum strain, Y1, shared a core DNA region of 2719 bp (Fig. 2). The conserved 2597-bp or 2719-bp sequences in the 6 tet(O)-positive bifidobacterial strains contained a sequence of 156 bp encoding a hypothetical protein, an upstream flanking sequence of 643 bp, and an 1800-bp or 1920-bp tet(O) gene, and exhibited 99-100% nucleotide identity with the 2597-bp or 2719-bp sequences previously identified in Campylobacter coli strain 6461 (GenBank JQ613156.1), Streptococcus pyogenes strain ICESp2905 (GenBank FR691055.1), and Streptococcus suis strain NSUI002 (GenBank CP011419.1).
The 2 tet(S)-positive bifidobacterial strains shared a core DNA region of 430 bp, including the 252-bp tet(S) gene and a downstream flanking sequence of 178 bp, which exhibited 99-100% identity with the sequences previously identified in Listeria monocytogenes strain LM78 (GenBank JX865374.1), Streptococcus suis strain G52 (GenBank JQ762256.1), and Enterococcus faecium strain E241 (GenBank JN980096.1).
Analysis of ORFs in regions flanking the tet(W), tet(O), and tet(S) genes
In the 21 tet(W)-positive bifidobacterial strains shown in Fig. 1, a 23SrRNA gene was found 97 bp downstream of the tet(W) gene and showed 100% nucleotide identity with the sequence previously identified in Bifidobacterium animalis strain A6 (GenBank CP010433.1). Additionally, another two open reading frames (ORFs), including a 140-bp sequence encoding thymidylate synthase and a 648-bp sequence encoding dihydrofolate reductase, were found upstream of the tet(W) gene in one B. animalis subsp. lactis strain, F12, which exhibited 98–100% nucleotide identity with the sequence previously identified in Bifidobacterium pseudocatenulatum DSM 20438 (GenBank AP012330.1) and Bifidobacterium kashiwanohense PV20-2 (GenBank CP007456.1).
In the 6 tet(O)-positive bifidobacterial strains shown in Fig. 2, a 198-bp ORF encoding an ABC transporter was found downstream of the 1800-bp tet(O) gene in B. longum strains H21, Y33 and Z1, and a 99-bp cpp2 gene was found downstream of the 1920-bp tet(O) gene in B. longum strains H34 and F313 and B. pseudocatenulatum strain Y1.
In the 2 tet(S)-positive bifidobacterial strains (B. pseudocatenulatum strain F312 and B. breve strain A27), a 270-bp ORF encoding a putative membrane-spanning protein was found in the adjacent upstream region flanking the tet(S) gene. Additionally, in B. pseudocatenulatum strain F312, another 186-bp ORF encoding a hypothetical protein was found 400 bp upstream of the tet(S) gene.
Mobility of the tet(W), tet(O), and tet(S) genes
Filter matings of the 21 tet(W)-positive bifidobacterial strains, the six tet(O)-positive bifidobacterial strains, and the two tet(S)-positive bifidobacterial strains with Enterococcus faecalis StF-EFM failed in laboratory conditions.
Discussion
In our previous investigation of a collection of 92 bifidobacterial strains originating from the human GIT, the macrolide, lincosamide, and streptogramin (MLS) resistance gene erm(X) was detected in 30 bifidobacterial strains. This study further investigated the tetracycline-resistant phenotype and genotype of these 92 strains and found that 29 bifidobacterial strains exhibited tetracycline resistance. Notably, nine bifidobacterial strains, including B. longum strains F313 and F21, B. pseudocatenulatum strains J56, H23, Z25, a39, Y1, and F312, and B. bifidum strain Y21, simultaneously exhibited MLS and tetracycline resistance. Bifidobacteria have been regarded as traditional safe probiotics in the human GIT7, 8, and only tet(W)- and tet(M)-mediated tetracycline resistance had been reported in human intestinal bifidobacteria10–13. However in the present study, acquired tetracycline resistance in the 29 bifidobacterial strains was mediated by tet(W), tet(O) or tet(S), and this study provides the first report of tet(O)- and tet(S)-mediated tetracycline resistance in bifidobacteria. The finding of two new tetracycline resistance genes [tet(O) and tet(S)] in bifidobacteria suggest that the selective pressure of intensive tetracycline use has caused human intestinal bifidobacteria to acquire more tetracycline resistance genes to survive and eventually become a reservoir of tetracycline resistance genes as previously speculated by many researchers17–19.
It has been generally considered that the AR resistance genes carried by bifidobacteria are transferred from other bacteria in the human GIT via a number of complex mechanisms15, 20. Previously, it was reported that the tetracycline resistance gene tet(W) in 10 human intestinal bifidobacterial strains of 5 species had a conserved sequence of 2154 bp10. In the present study, the tetracycline resistance gene tet(W) in 21 human intestinal bifidobacterial strains of 4 species had a conserved sequence of 2281 bp that included the previously reported 2154 bp sequence, while the 1800-bp tet(O) gene in three human intestinal B. longum strains had a conserved sequence of 2599 bp, the 1920-bp tet(O) gene in another three human intestinal bifidobacterial strains of two species had a conserved sequence of 2719 bp, and the tet(S) gene in two human intestinal bifidobacterial strains of 2 species had a conserved sequence of 430 bp. All of these conserved sequences contained the sequence of the tetracycline resistance gene [tet(W), tet(O) or tet(S)] and its partial flanking sequence, which showed 98–100% nucleotide identity with the sequence previously identified in multiple human intestinal pathogens (Arcanobacterium, Streptococcus, Corynebacterium, Campylobacter, Listeria, etc.). Not unexpectedly, with the widespread use of tetracycline in the treatment of various human bacterial infections, pathogens are indeed more likely to harbor and retain AR genes and retain them than other bacteria in the human GIT3, 21. Therefore, our results indicate that different tetracycline resistance genes acquired by human intestinal bifidobacteria might initially be transferred from intestinal pathogens.
Because bifidobacteria rarely harbor plasmids, it is generally believed that the acquired AR genes tend to be integrated into the chromosome of bifidobacteria22, 23. However, the integration characteristics of the tetracycline resistance genes in the chromosome of human intestinal bifidobacteria are poorly understood. Previously, only one report had investigated the insertion site of the tetracycline resistance gene tet(W) in six intestinal bifidobacterial strains, showing that the tet(W) gene was flanked downstream by an orfY gene in one B. thermophilum strain and one B. longum strain and by a transposase gene in two B. longum strains12. In the present study, the tetracycline resistance gene tet(W) was flanked downstream by a 23 S rRNA gene in 21 bifidobacterial strains, while the tet(S) was flanked upstream by a gene encoding a membrane-spanning protein in two bifidobacterial strains. In addition, in the six tet(O)-positive bifidobacterial strains, the tet(O) gene exhibited two different lengths, 1801 bp and 1920 bp; the 1800-bp tet(O) gene was flanked downstream by a gene encoding an ATP transporter, and the 1920-bp tet(O) gene was flanked downstream by a gene encoding a Cpp2 protein. Moreover, these genes flanking the tet(W), tet(O) or tet(S) in the bifidobacterial strains in this study only exhibited 98–100% nucleotide identity with these sequences previously identified in Bifidobacterium. Hence, our results provide evidence for revealing the insertion regularity of different tetracycline resistance genes into the chromosome of human intestinal bifidobacteria, and we speculate that each kind of acquired tetracycline resistance gene might tend to insert into the vicinity of specific genes in bifidobacteria. In Gram-positive anaerobes other than bifidobacteria, a few researchers had also investigated the integration characteristic of the acquired tetracycline resistance genes tet(W) and tet(S). However, no similar genes was found flanking the tetracycline resistance genes tet(W) in the two Lactobacillus reuteri strains24 and no similar genes were found flanking the tetracycline resistance genes tet(S) in the six Streptococcus dysgalactiae subsp. equisimilis strains25. Thus, unlike in bifidobacteria, the tetracycline resistance genes tet(W) and tet(S) in the other Gram-positive anaerobes might exhibit random insertion sites, which remains to be further studied.
Commercially used bifidobacterial strains are commonly screened from the healthy human GIT26, 27. However, it had been verified that one B. longum strain F8 isolated from the healthy human GIT could transfer the tetracycline resistance gene tet(W) to Butyrivibrio adolescentis strain L2-322912. Thus, considering that the AR genes harbored by bifidobacterial strains could have the potential risk of transfer to pathogenic bacteria in the human GIT and become a treat to human healthy28, 29, the EFSA recommended that bacterial strains for commercial use should not harbor any transferable AR genes16. Over the past few years, only tet(W)- and tet(M)-mediated tetracycline resistance had been detected in human intestinal bifidobacteria10–13; thus, human intestinal bifidobacterial strains lacking the tet(W) and tet(M) genes would be considered as relatively safe. However, this study detected two new tetracycline resistance genes, tet(O) and tet(S), in human intestinal bifidobacteria in addition to tet(W) and further investigated the potential transferability of tet(W), tet(O) and tet(S) in bifidobacteria via filter mating experiments. Although no transfer of tet(W), tet(O) or tet(S) was observed via filter mating, this does not confirm that the tet(W), tet(O) or tet(S) in these bifidobacterial strains could not be transferred in the human GIT, since the actual transfer process of AR genes that occurs in the GIT usually occurs over a much longer period of time15. Therefore, the presence of the tetracycline resistance genes tet(O) and tet(S) should also be considered in the safety assessment of human intestinal bifidobacterial strains prior to commercial use.
In summary, this study has provided additional genetic knowledge regarding acquired tetracycline resistance in bifidobacteria isolated from the healthy human GIT. The detection of two new tetracycline resistance genes [tet(O) and tet(S)] in human bifidobacteria indicates that human intestinal bifidobacteria have begun to harbor more AR genes, and that the screening of bifidobacterial strains from the healthy human GIT for commercial use faces additional challenges.
Methods
Ethical Statement
Ethics approval for this study was obtained within the framework of the National Basic Research Program of China (973 Program) (No. 2012CB720802). Final approval was obtained from the Research Ethics Committee of Shanghai Jiaotong University, China. The methods were carried out in accordance with the approved guidelines. The written informed consent was obtained from all participants or their legal guardians in the study.
Bacterial strains and growth conditions
One hundred individual bifidobacterial strains belonging to seven species were investigated in the present study: of these, one was a type strain, seven were commercial strains, and 92 were isolated from the feces of 14 healthy individuals (Tables 1 and 2). The first letter in the names of the 92 strains, “J”, “L”, “F”, “W”, “N”, “Y”, “A”, “Z”, “D”, “X”, “H”, “a”, “B”, or “S”, indicates the origin among the 14 individuals. The number of strains of each species in the 100 tested strains was as follows: Bifidobacterium longum, 45; Bifidobacterium breve, 18; Bifidobacterium lactis 19; Bifidobacterium pseudocatenulatum, 12; Bifidobacterium bifidum, 3; Bifidobacterium adolescentis, 2; Bifidobacterium infantis, 1.
All of the strains were cultured in de Man Rogosa Sharpe (MRS) medium supplemented with 0.05% (w/v) L-cysteine (MRSC). Incubations were performed at 37 °C for 12–48 h under anaerobic conditions (AnaeroGenTM, Oxoid Ltd, Basingstoke, UK).
Antimicrobial susceptibility
The MIC values of tetracycline in these 100 bifidobacterial strains were determined using Etest strips (bioMérieux, Marcy-l’Étoile, France), according to the manufacturer’s recommendations. Prior to the assay, the strains were anaerobically cultured in MRSC medium at 37 °C for 24 h. An inoculum was then suspended in MRSC broth to achieve the turbidity of a 1.0 McFarland standard (3 × 108 cells/ml) and was subsequently uniformly applied to an agar plate with a sterile cotton swab in three directions. After drying for 20 or 30 min, tetracycline Etest strips with antimicrobial gradients ranging from 0.016 to 256 μg/ml were placed on the agar plates. The MIC values were visually defined as the lowest tetracycline concentration at which no growth was observed with the Etest strip after aerobic incubation at 37 °C for 48 h. The interpretation of the tetracycline susceptibility status of these strains was based on the tetracycline breakpoint for Bifidobacterium (MIC = 8 μg/ml) defined by the EFSA16. Each assay was repeated three times in duplicate.
PCR amplification and sequencing
Genomic DNA from the 100 bifidobacterial strains was extracted according to the method of Ausubel and colleagues30. The primers used to amplify five ribosomal protection genes [tet(M), tet(O), tet(S), tet(W), and tet(T)] and eight efflux genes [tet(A), tet(B), tet(C), tet(D), tet(E), tet(G), tet(K), and tet(L)] are listed in Table 4. The primers used to detect tet(M), tet(T), tet(A), tet(B), tet(C), tet(D), tet(E), tet(G), tet(K), and tet(L) were chosen as previously described31–33, while three sets of primers (tetW_F and tetW_R, tetO_F and tetO_R, and tetS_F and tetS_R) were designed to detect the tet(W), tet(O), and tet(S) genes based on the tet(W) sequence of Bifidobacterium animalis subsp. lactis CNCM I-2494 (GenBank CP002915.1), the tet(O) sequence of Streptococcus suis BM407 (GenBank FM252032.1), and the tet(S) sequence of Lactococcus lactis subsp. lactis strain ILIBB-JZK (GenBank KF278750.1), respectively. PCR assay was performed with TaKaRa Ex Taq DNA polymerase using the component concentration recommended by the provider (TaKaRa, Dalian, China). PCR products were separated by electrophoresis on a 1.0% agarose gel and visualized by ethidium bromide staining. All positive amplicons were purified by a PCR purification spin kit (Qiagen, Germany) and subsequently sequenced by the BGI Company (Shanghai, China). The obtained sequences were compared with those in GenBank.
Table 4.
Primers used in the present study.
| Name | Sequence (5′-3′) | Target | Reference |
|---|---|---|---|
| tetM_F | ACAGAAAGCTTATTATATAAC | tet(M) | 32 |
| tetM_R | TGGCGTGTCTATGATGTTCAC | ||
| tetO_F | AACTTAGGCATTCTGGCTCAC | tet(O) | This study |
| tetO_R | CTATGGACAACCCGACAGAAG | ||
| tetS_F | TAGATACTCCTGGACACAT | tet(S) | This study |
| tetS_R | ATGAGAATGACCTCGTTAC | ||
| tetW_F | CGGATTGTGGCATTTGT | tet(W) | This study |
| tetW_R | GCATAGAGGGTGAAGGAG | ||
| tetT_F | AAGGTTTATTATATAAAAGTG | tet(T) | 34 |
| tetT_R | AGGTGTATCTATGATATTTAC | ||
| tetA_F | GTAATTCTGAGCACTGTCGC | tet(A) | 32 |
| tetA_R | CTGCCTGGACAACATTGCTT | ||
| tetB_F | AAAACTTATTATATTATAGTG | tet(B) | 34 |
| tetB_R | TGGAGTATCAATAATATTCAC | ||
| tetC_F | TCTAACAATGCGCTCATCGT | tet(C) | 32 |
| tetC_R | CGTTGAAGGCTCTCAAGGGC | ||
| tetD_F | ATTACACTGCTGGACGCGAT | tet(D) | 32 |
| tetD_R | CTGATCAGCAGACAGATTGC | ||
| tetE_F | GTGATGATGGCACTGGTCAT | tet(E) | 32 |
| tetE_R | CTCTGCTGTACATCGCTCTT | ||
| tetG_F | TTTCGGATTCTTACGGTC | tet(G) | 32 |
| tetG_R | TCCTGCGATAGAGCTTAGA | ||
| tetK_F | TTATGGTGGTTGTAGCTAGAAA | tet(K) | 33 |
| tetK_R | AAAGGGTTAGAAACTCTTGAAA | ||
| tetL_F | GTMGTTGCGCGCTATATTCC | tet(L) | 33 |
| tetL_R | GTGAAMGRWAGCCCACCTAA | ||
| tetW_U_SP1 | GGAGGTTGTTTCCGCTTTGCTG | Upstream region of tet(W) | This study |
| tetW_U_SP2 | GGTAAAGGAACCCACCGTCATT | ||
| tetW_U_SP3 | TCTGTTACACCACTCCCGCTTG | ||
| tetW_D_SP1 | CATCTGTGCCACTGGAAGGAAG | Downstream region of tet(W) | This study |
| tetW_D_SP2 | TCCGTCCTCGTTGTCCCTTTTT | ||
| tetW_D_SP3 | AAGGTCGTCTTTCCAGCGTCTA | ||
| tetO_U_SP1 | GCAAATCAATCCCCTCTTGGTCA | Upstream region of tet(O) | This study |
| tetO_U_SP2 | GTCTGTGCCTGTATGCCATCCTTT | ||
| tetO_U_SP3 | CCACTGAAAAGATGTCACTGCTGT | ||
| tetO_D1_SP1 | CGATACAGCCTGCTCTGGTGAT | Downstream region of the1457-bp tet(O) | This study |
| tetO_D1_SP2 | CTCCCTATGCTCCAAACAACGA | ||
| tetO_D1_SP3 | TATTGCTTGGGGCACTTACAGA | ||
| tetO_D2_SP1 | TTTCTGGGCTTCTGTCGGGTTGTC | Downstream region of the 1800-bp tet(O) | This study |
| tetO_D2_SP2 | AAATGCGGTTATGGAGGGGGTTCT | ||
| tetO_D2_SP3 | GCAGGGACAGAACTATTAGAGCCA | ||
| tetS_U_SP1 | GATAGCGGTACAACGAAAACGGTA | Upstream region of tet(S) | This study |
| tetS_U_SP2 | TTTGGAACGCCAGAGAGGTATT | ||
| tetS_U_SP3 | CTGGACATGGATTTTTGGCAG | ||
| tetS_D_SP1 | TGCCAAAATCCATGGTCCAGG | Downstream region of tet(S) | This study |
| tetS_D_SP2 | CGGTCTGAATAGTAATACCTGTGTGG | ||
| tetS_D_SP3 | CCGTTTTGGTTGTACCGCTATC |
Genome walking
Nested PCR was conducted to amplify the flanking sequences of the tet(W) genes in 21 bifidobacterial strains, the tet(O) genes in 6 bifidobacterial strains, and the tet(S) genes in two bifidobacterial strains using a Genome Walking Kit (TaKaRa, Dalian, China), following the manufacturer’s recommendations. The nested PCR assays were performed in three steps using the same AP primer and three reverse SP primers (SP1, SP2, and SP3) designed under the conditions suggested by the kit instructions. The SP primers groups (SP1, SP2, and SP3) are listed in Table 3 and were designed to amplify the upstream and downstream sequences flanking the tet(W), tet(S), and tet(O) genes. In particular, two groups of SP primers were designed to amplify the downstream flanking sequences of the 1457-bp and 1800-bp tet(O) genes. All positive amplicons obtained in the third cycle of nested PCR were purified by a PCR purification spin kit (Qiagen, Germany) and subsequently sequenced by the BGI Company (Shanghai, China).
Filter mating experiments
The potential transferability of the tet(W) genes from 21 bifidobacterial strains, the tet(O) genes from 6 bifidobacterial strains, and the tet(S) genes from two bifidobacterial strains (donors) to Enterococcus faecalis StF-EFM (recipient) was investigated by filter mating experiments, following the method of Gevers and colleagues34. Briefly, the donor and recipient cells were grown to mid-exponential phase in MRSC medium prior to assay, and 1 ml of donor and 1 ml of recipient culture were mixed. Subsequently, the mixture (2 ml) was dispensed onto a sterile filter (0.45 μm; MF-Millipore membrane filter, HAWP 02500, Millipore) that was then anaerobically incubated on non-selective BHI agar (Oxoid) at 37 °C for 24 h. The cells were collected from the filters by centrifugation and resuspended in 1 ml of PBS. The transconjugants were aerobically detected on Pfizer Enterococcus Selective (PSE) agar supplemented with tetracycline (16 μg/ml), since only Enterococcus faecalis StF-EFM (recipient) can grow on PSE agar under aerobic conditions. Transfer frequencies were defined as the number of transconjugant colonies per recipient colony formed after the mating period.
Nucleotide sequence accession numbers
The nucleotide sequences of the regions flanking the tet(W) gene in 21 bifidobacterial strains were submitted to the GenBank database under accession numbers KY682293-KY682303, KY689744-KY689752, and KY697297. The nucleotide sequences of the regions flanking the tet(O) gene in 6 bifidobacterial strains were submitted to the GenBank database under accession numbers KY697298-KY697303. The nucleotide sequences of the regions flanking the tet(S) gene in the 2 bifidobacterial strains were submitted in the GenBank database under accession numbers KY818315 and KY818316.
Data Availability
The datasets generated during the current study are included in this article and are available from the corresponding author on reasonable request.
Acknowledgements
This work was supported by Shanghai Industry-University Joint Research Program [grant number HU CXY-2016-010].
Author Contributions
N.W. and H.Y. designed experiments. N.W., X.H., M.Z and X.L. performed the experiments. N.W., X.H. and H.Y. analyzed the data. N.W. and H.Y. wrote the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Butel MJ. Probiotics, gut microbiota and health. Med Mal Infect. 2014;44:1–8. doi: 10.1016/j.medmal.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Sanchez, B. et al. Probiotics, gut microbiota, and their influence on host health and disease. Mol Nutr Food Res61, doi:10.1002/mnfr.201600240 (2017). [DOI] [PubMed]
- 3.Grossman TH. Tetracycline Antibiotics and Resistance. Cold Spring Harb Perspect Med. 2016;6:a025387. doi: 10.1101/cshperspect.a025387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salyers AA, Gupta A, Wang Y. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 2004;12:412–416. doi: 10.1016/j.tim.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Rolain JM. Food and human gut as reservoirs of transferable antibiotic resistance encoding genes. Front Microbiol. 2013;4:173. doi: 10.3389/fmicb.2013.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kazimierczak KA, et al. A new tetracycline efflux gene, tet(40), is located in tandem with tet(O/32/O) in a human gut firmicute bacterium and in metagenomic library clones. Antimicrob Agents Chemother. 2008;52:4001–4009. doi: 10.1128/AAC.00308-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matto J, et al. Genetic heterogeneity and functional properties of intestinal bifidobacteria. J Appl Microbiol. 2004;97:459–470. doi: 10.1111/j.1365-2672.2004.02340.x. [DOI] [PubMed] [Google Scholar]
- 8.Tojo R, et al. Intestinal microbiota in health and disease: role of bifidobacteria in gut homeostasis. World J Gastroenterol. 2014;20:15163–15176. doi: 10.3748/wjg.v20.i41.15163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Babinska I, Rotkiewicz T, Otrocka-Domagala I. The effect of Lactobacillus acidophilus and Bifidobacterium spp. administration on the morphology of the gastrointestinal tract, liver and pancreas in piglets. Pol J Vet Sci. 2005;8:29–35. [PubMed] [Google Scholar]
- 10.Ammor MS, Florez AB, Alvarez-Martin P, Margolles A, Mayo B. Analysis of tetracycline resistance tet(W) genes and their flanking sequences in intestinal Bifidobacterium species. J Antimicrob Chemoth. 2008;62:688–693. doi: 10.1093/jac/dkn280. [DOI] [PubMed] [Google Scholar]
- 11.Florez AB, Ammor MS, Alvarez-Martin P, Margolles A, Mayo B. Molecular analysis of tet(W) gene-mediated tetracycline resistance in dominant intestinal Bifidobacterium species from healthy humans. Appl Environ Microbiol. 2006;72:7377–7379. doi: 10.1128/AEM.00486-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kazimierczak KA, Flint HJ, Scott KP. Comparative analysis of sequences flanking tet(W) resistance genes in multiple species of gut bacteria. Antimicrob Agents Chemother. 2006;50:2632–2639. doi: 10.1128/AAC.01587-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aires J, Thouverez M, Doucet-Populaire F, Butel MJ. Consecutive human bifidobacteria isolates and acquired tet genes. Int J Antimicrob Agents. 2009;33:291–293. doi: 10.1016/j.ijantimicag.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 14.van Hoek AHAM, et al. Mosaic tetracycline resistance genes and their flanking regions in Bifidobacterium thermophilum and Lactobacillus johnsonii. Antimicrob Agents Ch. 2008;52:248–252. doi: 10.1128/AAC.00714-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huddleston JR. Horizontal gene transfer in the human gastrointestinal tract: potential spread of antibiotic resistance genes. Infect Drug Resist. 2014;7:167–176. doi: 10.2147/IDR.S48820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.European Food Safety Authority. Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. The EFSA Journal10, 2740, doi:10.2903/j.efsa.2012.2740 (2012).
- 17.Ammor MS, et al. Molecular characterization of intrinsic and acquired antibiotic resistance in lactic acid bacteria and bifidobacteria. J Mol Microbiol Biotechnol. 2008;14:6–15. doi: 10.1159/000106077. [DOI] [PubMed] [Google Scholar]
- 18.Duranti, S. et al. Prevalence of Antibiotic Resistance Genes among Human Gut-Derived Bifidobacteria. Appl Environ Microbiol83, doi:10.1128/AEM.02894-16 (2017). [DOI] [PMC free article] [PubMed]
- 19.de Vries LE, et al. The gut as reservoir of antibiotic resistance: microbial diversity of tetracycline resistance in mother and infant. PLoS One. 2011;6:e21644. doi: 10.1371/journal.pone.0021644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang FX, Mao DQ, Luo Y, Wang Q, Mu QH. [Horizontal transfer of antibiotic resistance genes in the environment] Ying Yong Sheng Tai Xue Bao. 2013;24:2993–3002. [PubMed] [Google Scholar]
- 21.Scott KP, Melville CM, Barbosa TM, Flint HJ. Occurrence of the new tetracycline resistance gene tet(W) in bacteria from the human gut. Antimicrob Agents Chemother. 2000;44:775–777. doi: 10.1128/AAC.44.3.775-777.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma P, Tomar SK, Goswami P, Sangwan V, Singh R. Antibiotic resistance among commercially available probiotics. Food Res Int. 2014;57:176–195. doi: 10.1016/j.foodres.2014.01.025. [DOI] [Google Scholar]
- 23.Duranti, S. et al. Prevalence of Antibiotic Resistance Genes among Human Gut- Derived Bifidobacteria. Appl Environ Microb83, doi:UNSP e02894-1610.1128/AEM.02894–16 (2017). [DOI] [PMC free article] [PubMed]
- 24.Egervarn M, Roos S, Lindmark H. Identification and characterization of antibiotic resistance genes in Lactobacillus reuteri and Lactobacillus plantarum. J Appl Microbiol. 2009;107:1658–1668. doi: 10.1111/j.1365-2672.2009.04352.x. [DOI] [PubMed] [Google Scholar]
- 25.Liu LC, et al. Identification of tet(S) gene area in tetracycline-resistant Streptococcus dysgalactiae subsp. equisimilis clinical isolates. J Antimicrob Chemother. 2008;61:453–455. doi: 10.1093/jac/dkm500. [DOI] [PubMed] [Google Scholar]
- 26.Rivas P, Troncoso M, Figueroa G. Probiotic potencial of bifidobacterium spp isolated from healthy infants. Pediatric Research. 2006;60:639–639. doi: 10.1203/00006450-200611000-00046. [DOI] [Google Scholar]
- 27.Ruiz-Moyano S, Martin A, Benito MJ, Nevado FP. & Cordoba, M. D. Screening of lactic acid bacteria and bifidobacteria for potential probiotic use in Iberian dry fermented sausages. Meat Science. 2008;80:715–721. doi: 10.1016/j.meatsci.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 28.Ouoba LI, Lei V, Jensen LB. Resistance of potential probiotic lactic acid bacteria and bifidobacteria of African and European origin to antimicrobials: determination and transferability of the resistance genes to other bacteria. Int J Food Microbiol. 2008;121:217–224. doi: 10.1016/j.ijfoodmicro.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 29.van Reenen CA, Dicks LM. Horizontal gene transfer amongst probiotic lactic acid bacteria and other intestinal microbiota: what are the possibilities? A review. Arch Microbiol. 2011;193:157–168. doi: 10.1007/s00203-010-0668-3. [DOI] [PubMed] [Google Scholar]
- 30.Vincent D, Roy D, Mondou F, Dery C. Characterization of bifidobacteria by random DNA amplification. Int J Food Microbiol. 1998;43:185–193. doi: 10.1016/S0168-1605(98)00109-3. [DOI] [PubMed] [Google Scholar]
- 31.Guardabassi L, Dijkshoorn L, Collard JM, Olsen JE, Dalsgaard A. Distribution and in-vitro transfer of tetracycline resistance determinants in clinical and aquatic Acinetobacter strains. J Med Microbiol. 2000;49:929–936. doi: 10.1099/0022-1317-49-10-929. [DOI] [PubMed] [Google Scholar]
- 32.Thumu SC, Halami PM. Presence of erythromycin and tetracycline resistance genes in lactic acid bacteria from fermented foods of Indian origin. Antonie Van Leeuwenhoek. 2012;102:541–551. doi: 10.1007/s10482-012-9749-4. [DOI] [PubMed] [Google Scholar]
- 33.Gueimonde M, Salminen S, Isolauri E. Presence of specific antibiotic (tet) resistance genes in infant faecal microbiota. Fems Immunol Med Mic. 2006;48:21–25. doi: 10.1111/j.1574-695X.2006.00112.x. [DOI] [PubMed] [Google Scholar]
- 34.Gevers D, Huys G, Swings J. In vitro conjugal transfer of tetracycline resistance from Lactobacillus isolates to other Gram-positive bacteria. FEMS Microbiol Lett. 2003;225:125–130. doi: 10.1016/S0378-1097(03)00505-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during the current study are included in this article and are available from the corresponding author on reasonable request.


