SUMMARY
Many physiological functions of helicases are dependent on their ability to unwind nucleic acid duplexes in an ATP-dependent fashion. Determining the kinetic frameworks of these processes is critical to understand how these proteins function. We recently developed a fluorescence assay to monitor RNA duplex unwinding by DEAD-box helicase proteins in real time. Two fluorescently modified short reporter oligonucleotides are annealed to an unmodified loading strand of any length. One reporter is modified with cyanine 3 (Cy3), whereas the other is modified with a spectrally paired black hole quencher. Complete separation of the Cy3-modified reporter strand is detected as an increase in total fluorescence. This assay is compatible with reagentless biosensors to monitor ATPase activity so that the coupling between ATP hydrolysis and duplex unwinding can be determined. With the protocol described, obtaining data and analyzing results of unwinding and ATPase assays takes ~4 h.
Keywords: Helicase, RNA, DEAD box, ATPase
INTRODUCTION
Many aspects of DNA and RNA metabolism require remodeling of duplex regions by helicases. These enzymes can be classified into six superfamilies (SFs) that either form (SF3–6) or do not form (SF1–2) ring structures1,2. To date, all eukaryotic RNA helicases can be classified as either belonging to SF1 or SF21,3,4. The proposed mechanisms by which helicases perform their functions vary greatly. These include unwinding processively by translocating along nucleic acid duplexes, or by unwinding in a non-processive manner without translocation. Many physiological functions of helicases are dependent on these enzymes’ ability to unwind nucleic acid duplexes in an NTP-dependent fashion. The connection between energy expenditure and unwinding is best studied using quantitative in vitro measurements of both activities (i.e. NTP hydrolysis to NDP and nucleic acid unwinding) under identical conditions. Determining the kinetic frameworks of these coupled processes will help generate models that describe of how helicases remodel nucleic acid secondary structure.
Overview of fluorescence-based unwinding and ATP hydrolysis assays
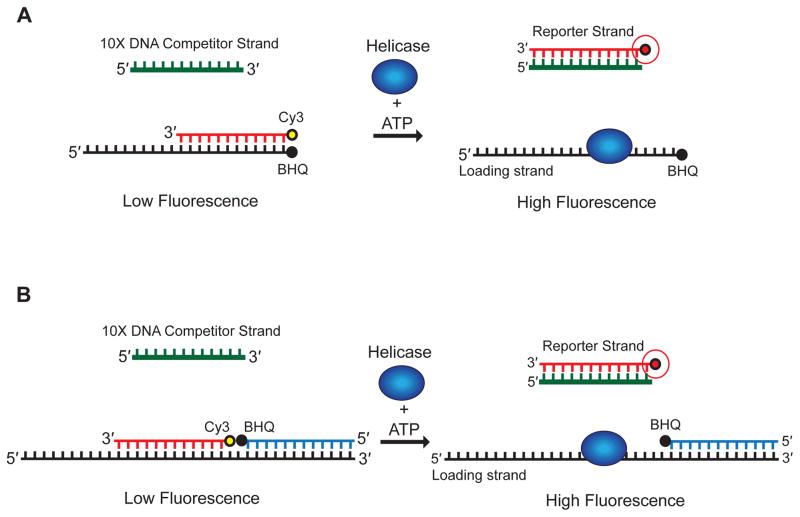
Recently, we and others developed a simple fluorescence quenching assay that uses two modified RNA strands to study duplex unwinding by RNA helicases (Fig. 1A)5,6. The assay monitors fluorescence resonance energy transfer (FRET) to detect the separation of a fluorophore-labeled reporter strand from a loading strand that is modified with a spectrally paired quencher dye. Complete strand separation is detected in real time as an increase in the total fluorescence of the reporter strand. The applicability of this approach is limited to relatively short RNA templates by the cost and reliability of RNA oligonucleotide synthesis and labeling. To overcome this limitation, we have adapted our assay to employ two short modified reporter strands designed to anneal to either natural or engineered binding sites on an unmodified RNA loading strand that is easily generated by in vitro transcription (Fig. 1B)7. Using this approach, we have analyzed duplex unwinding on much longer (~1 Kb) RNA substrates than those analyzed via the approach described in figure 1a.
Figure 1.
Representation of the two fluorescence-based approaches to probing RNA unwinding reactions described in the Introduction. (A) A reporter strand modified with Cy3 at its 5′ end (red) is annealed to a complementary loading strand (black) that is modified with a spectrally Cy3-paired BHQ at its 3′ end. The loading strand possesses a single-stranded 5′-end extension that can be varied in length. Annealing of the reporter strand to the loading strand results in the fluorescence signal from the Cy3 dye to be quenched. (B) A reporter strand modified with Cy3 at its 5′ end (red) is annealed to an unmodified complementary loading strand (black). A second reporter strand modified with a spectrally Cy3-paired BHQ on its 3′ end is annealed to the loading strand so that the BHQ and Cy3 dyes are positioned with a one nucleotide gap between them, and, once again, Cy3 fluorescence quenched as a result. The loading strand possesses a single stranded 5′-end extension that can be varied in length. In both assays, the addition of the helicase and ATP promotes reporter strand detachment, resulting in an increase in Cy3-associated fluorescence. A 10-fold molar excess of a DNA competitor strand (green) that is complementary to the reporter strand is added to each reaction to prevent reannealing after unwinding.
Both assays yield comparable results that can be used to reproducibly determine the kinetic parameters of duplex unwinding by DEAD-box helicase proteins. Importantly, ATP hydrolysis can be monitored in both of these unwinding assays using commercially available fluorescent biosensors that detect phosphate release8–10. The ability to measure both duplex unwinding and ATPase activities under identical conditions reveals the relative coupling efficiency of helicase proteins. Investigations of reaction conditions and the effects of accessory factors or drugs can easily be accommodated by these protocols in order to employ them to evaluate regulatory interactions and perform drug candidate screening. We have successfully used these methods to probe the mechanism of the DEAD-box helicase eIF4A, but they should be applicable to study any RNA helicase.
Applications of fluorescence unwinding assay
Fluorescence methods are widely applied to probe biochemical and kinetic mechanisms of duplex unwinding by helicases, both in bulk solution and single-molecule measurements11–13. Multiple fluorescent data can be combined so that information is obtained for different processes in real time, as described here for duplex unwinding and ATPase activities.
Our modified helicase assay offers some distinct advantages compared with an assay that use two modified RNA strands for studying unwinding by RNA helicases5,6. Using an unmodified RNA loading strand enables measurement of duplex unwinding within physiologically relevant RNAs without sequence length limitations. Altering the position of the fluorescent reporter strands offers a rapid and flexible way to investigate relationships between unwinding rates and distinct features of the RNA strand. By employing variously positioned reporters labeled with different fluorescent dyes, movements of helicases or entire complexes can be more precisely determined. One such application could be to monitor duplex unwinding within 5′ untranslated regions (5′UTRs) of mRNAs. By using a number of reporters, it should be possible to observe the progress of the scanning ribosome using either cell extracts or purified components.
Recently, a version of the FRET-based duplex unwinding assay that employs both fluorophore-modified loading and reporter strands (Fig. 1A) was successfully used by others to identify HCV helicase inhibitory activities in marine extracts5. Our improved version of this assay will enable screening to be carried out using more physiologically-relevant RNA substrates (Fig. 1B).
Comparisons with other unwinding and ATPase assays
A number of in vitro assays are available to determine helicase duplex unwinding and ATP hydrolysis and rates. Each assay has different advantages and disadvantages that should be carefully considered before deciding which to adopt. The most widely used analytical method to monitor unwinding is electrophoretic mobility shift assay (EMSA)14. Although by this approach reliable quantitative data are generated, the utility of EMSAs is limited by 1) temporal resolution; 2) the number of time points that can be conveniently analyzed; and 3) the relatively long amount of time (hours/overnight) it takes to obtain results. These limitations make EMSAs both less convenient than real-time assays and unsuitable for high-throughput screening.
By contrast, FRET-based assays have a number of advantages over traditional EMSAs. Firstly, they all report on duplex unwinding in real time using a single reaction5–7,15–18. This characteristic enables rapid data generation and avoids the need to remove a small fraction of the assay sample at different time points for gel-based analysis. Importantly, fluorescence assays are not limited by the frequency of time point acquisition, enabling data points to be collected on the second scale or faster. This feature can be particularly valuable when fitting data to single or double exponential rate equations. These assays therefore enable the collection of precise kinetic unwinding data within minutes of carrying out the experiment rather than the hours that are required for gel-based assays. In addition, fluorescence unwinding assays can be readily adapted for use in high-throughput assays to identify inhibitors and activators of helicase function.
The molecular beacon-based helicase assay (MBHA) is an elegant method to study RNA and DNA helicases. This assay monitors the dissociation of a hairpin-forming RNA or DNA oligonucleotide that is modified with a fluorescent moiety on one end and a quencher on the other15,17–20. Alternatively, the fluorophore and quencher can be split between two molecular beacons that are annealed side by side to an RNA loading strand21. The advantage of MBHA assays compared to our assay is that reannealing of the molecular beacon to the loading strand is not possible, thereby bypassing the need for a capture strand to measure unwinding rates. Although these assays have been employed very successfully to monitor unwinding by some DNA and RNA helicases, they are not effective for measuring DEAD-box RNA helicase function. DEAD-box helicases can load onto the single-stranded loop region of the dissociated molecular beacon hairpin and unwind the hairpin, making it difficult to distinguish the rate of unwinding of the molecular beacon from the loading strand from the rate of hairpin unwinding of the dissociated molecular beacon. To circumvent this problem, a DNA molecular beacon can be used, but this type of substrate is not physiologically relevant for DEAD-box helicases. To study DEAD-box protein function, the fluorescence unwinding assay that we describe here simply employs two relatively short modified fluorophore and quencher strands (19–24-nt long), that do not form hairpin structures when dissociated from the loading strand (Fig. 1B). These reporters can be annealed to unmodified loading strands of diverse sequences and lengths. Any desired loading strand is easily produced using in vitro transcription, enabling the rapid generation of a variety of targets7.
Limitations
All fluorescence methods also have intrinsic limitations. They do require expensive specialized equipment, such as a high-quality, sensitive fluorometer. Fluorescence intensity changes may vary with pH and temperature, limiting the use of these methods under particular assay conditions. In addition, calibrating a fluorescence intensity change to quantify the reaction progress can also be challenging, although it is relatively easy implementing the present protocol. Similarly to gel-based assays, only complete strand separation is detected in our assay, providing little information concerning unwinding intermediates. As an alternative approach, site-specific 2-aminopurine replacements of adenine can monitor intermediates in duplex unwinding. Loss of base pairing results in an increase in fluorescence that can be monitored in real time using standard or rapid mixing techniques22. Another concern is that the presence of fluorescent dyes can increase duplex stability, at least with DNA substrates23. When this tendency surfaces, it can make it difficult to accurately measure the effect of duplex stability on unwinding rates. This potential interference can be evaluated by comparing unwinding rates of modified and unmodified substrates using an alternative non-fluorescence assay such as EMSA. Fluorescent dyes can also interact with helicases, resulting in fluorescence changes not associated with unwinding24.
Duplex unwinding by helicases requires NTP hydrolysis. Understanding how NTP hydrolysis is coupled with duplex unwinding is important and requires quantitative assays of both NTP hydrolysis and unwinding. The requirement and stoichiometry of the hydrolysis step for unwinding may vary between different helicase mechanisms. For DEAD-box proteins, ATP hydrolysis is only required for the release of the single-stranded product during the recycling step25. A widely used approach to investigate ATP hydrolysis is to measure conversion rates of radioactively labeled ATP into ADP and inorganic phosphate (Pi). In this assay, the separation of these products can be monitored at different time points using thin layer chromatography26,27. Like an EMSA, this method does not require specialized instrumentation, but it is time-consuming, requiring hours/overnight to obtain results, and unsuitable for high-throughput assays. Alternatively, continuous time assays can be used where the ATP hydrolysis reaction is coupled to reactions that use NADH as a substrate28,29. Depletion of NADH can be monitored simply by the change in absorbance at 340 nm or in fluorescence (ex 340 nm/em 460 nm). The extents of these changes are proportional to the amount of ATP being hydrolyzed. The main limitation of coupled assays is that they generally require additional enzymatic and buffer components that might interfere with the helicase assay.
Reagentless biosensors have become a powerful method for measuring NTP hydrolysis in real time10. These biosensors are fluorescently modified proteins for which the intensity of fluorescence increases upon binding a target molecule. For instance, the amount of free phosphate can be monitored using E. coli phosphate-binding protein modified with a coumarin dye (PBP-MDCC)8,9,30. ATP hydrolysis in the nanomolar range can be sensitively measured in real time via phosphate release. This method requires the addition of only a single component, reducing the likelihood of assay incompatibility.
More recently, different alternative fluorescent biosensors have become available for measuring ATP hydrolysis in real time. These include ADP transcreener (Bellbrook labs), and ADP Quest (DiscoverRx). Although we have not used these reagents in our helicase assay, they should be compatible with our real time assay conditions.
Experimental Design
Nucleic acid substrate design and preparation
If the RNA substrate to be tested has less than 50 nucleotides, modified loading and reporter strands can be synthesized by a commercial supplier (Fig. 1A)6. The reporter RNA oligonucleotide, labeled with Cyanine 3 (Cy3) at its 5′-end, is then annealed to the loading RNA oligonucleotide labeled with a spectrally paired Black Hole Quencher (BHQ) at its 3′-end. We have successfully used a number of different duplex sequences with various stabilities that can be melted by eIF4A (Table 1)6. Increasing or decreasing the length of the reporter or loading strand increases or decreases the length of the 5′ overhang relative to the duplex region (Table 1). The length of the 3′ overhang can also be increased or decreased in a similar manner. The primary limitation of commercial RNA or DNA oligonucleotide synthesis is the loading strand length. This approach also limits the position where the reporter oligonucleotide can anneal on the loading strand, since the Cy3 and BHQ, which have to come in close proximity following formation of the double strand, are positioned at the 5′ and 3′ ends of the two nucleic acid strands. To bypass this limitation, we optimized a double-reporter system with a 5′-end Cy3 labeled reporter strand and an additional reporter strand with a 3′-end BHQ label designed to anneal at adjacent positions on an unlabeled loading strand (Fig. 1B)7. In this assay, we have successfully used the same 24-nucleotide Cy3 reporter from the single reporter assay (Table 2). For efficient quenching, we have used a BHQ-modified reporter strand that binds too strongly to the loading strand (calculated ΔG = −49.5 kcal/mol) to be released from it by eIF4A (Table 2). The stability of this inter-strand interaction ensures that only the rate of Cy3 strand dissociation is monitored in the assay. The positions of the reporters on the loading strand can also be easily adjusted. The unmodified loading strand is generated using published methods for in vitro transcription reactions with minor modifications31–33, which enable researchers to obtain easily strands of different length and sequence. Steps 1–18 of the Procedure describe an approach to generate large amounts of unmodified RNA for the in vitro helicase assays. This method is based on previously published protocols with minor modifications31,32,34.
Table 1.
Sequences and annealing data for fluorescently labeled RNA oligonucleotides used for a single-reporter RNA unwinding experiment.
| Single-stranded overhang (nt) |
Duplex Length (bp) |
Annealing ΔG (kcal/mol) |
Helicase substrate |
|---|---|---|---|
| 0 | 12 | −24.2 | 3′-CGUGGCAUUUCG-Cy3-5′ 5′-GCACCGUAAAGC-BHQ-3′ |
| 10 | 12 | −24.2 | 3′-CGUGGCAUUUCG-Cy3-5′ 5′-GAACACCAUGGCACCGUAAAGC-BHQ-3′ |
| 20 | 12 | −24.2 | 3′-CGUGGCAUUUCG-Cy3-5′ 5′-GAACAACAACAACAACCAUGGCACCGUAAAGC-BHQ-3′ |
| 30 | 12 | −24.2 | 3′-CGUGGCAUUUCG-Cy3-5′ 5′-GAACAACAACAACAACAACAACAACCAUGGCACCGUAAAGC-BHQ-3′ |
| 20 | 18 | −23.9 | 3′-CUUUUAGUUUUGAUUUUG-Cy3-5′ 5′-GAACAACAACAACAACCAUGGAAAAUCAAAACUAAAAC-BHQ-3′ |
| 20 | 24 | −24.7 | 3′-CUUUUUUAAUUUUUUAAUUUUUUG-Cy3-5′ 5′-GAACAACAACAACAACCAUGGAAAAAAUUAAAAAAUUAAAAAAC-BHQ-3′ |
| 20 | 24* | −29.3 | 3′-CUUUUUUCAUUUUUUAACUUUUUG-Cy3-5′ 5′-GAACAACAACAACAACCAUGGAAAAAAGUAAAAAAUUGAAAAAC-BHQ-3′ |
Table 2.
Sequences and annealing data for fluorescently labeled RNA oligonucleotides used for a double-reporter RNA unwinding experiment (nucleotide modified with Cy3 in red; nucleotide modified with BHQ in blue).
RNA Synthesis and Purification
The RNA loading strands are transcribed by recombinant T7 RNA polymerase from DNA oligonucleotides or PCR products. When using DNA oligonucleotides, a T7 RNA Polymerase promoter sequence is annealed to the single-stranded template oligonucleotide that carries a complementary T7 polymerase promoter sequence followed by the sequence to be transcribed31,32,34. Alternatively, PCR-amplified DNA sequences originating from either DNA oligonucleotides or plasmids can be used. For template strands with more than 100 nucleotides, we typically use a 1-ml PCR solution to obtain large quantities (50–100 μg) of DNA template for in vitro transcription. It is important to verify that the PCR reaction yields a single band of expected size by agarose gel electrophoresis to ensure homogeneous RNA loading strand preparation. Prior to in vitro transcription, it is essential to remove RNase contamination by phenol-chloroform extraction of the DNA template followed by ethanol precipitation.
To transcribe milligram-scale quantities of the RNA template strand, we carry out transcription reactions in a 1–5 ml total volume for 1 h at 37 °C, which gives optimal yields while minimizing product degradation. Following transcription, the RNA product can be efficiently gel purified, if it is typically less than 200 nucleotides in length. The gel-eluted RNA is then phenol-chloroform-extracted and ethanol-precipitated. Although gel purification of longer RNAs can be undertaken, it will result in a substantial loss of RNA. However, this approach is recommended when initially characterizing the activity of a long RNA strand in the helicase assay. Alternatively, RNAs with more than 200 nucleotides can generally be extracted with phenol-chloroform and ethanol-precipitated directly from the transcription reaction. Compared with the gel-purification approach, the main problem with this approach is that the loading strand is likely to be more heterogeneous in length and will still possess the contaminating DNA template. In addition, residual contaminating nucleotides, including ATP, may affect the activity of certain helicases. To minimize this contamination, we use ammonium acetate in the ethanol precipitation step to reduce precipitation of free nucleotides. Additional removal of free nucleotides is accomplished by passing the sample through Sephadex G25 or G50 spin columns, or proprietary desalting columns. With this methodology, we have had good success studying RNA remodeling on ~800-nucleotide-long loading strands.
DNA or RNA Duplex Annealing
Annealing reactions must be optimized for each set of substrates used in the unwinding assay. This often requires testing different magnesium and salt concentrations so that any structures in the loading strand folds correctly in addition to efficient annealing of the reporter strand to the loading strand. In the single reporter assay, the BHQ-modified loading strand and the Cy3-modified reporter strand are mixed in a 1:1 ratio. The ratio of the two strands can be varied by ~10% to optimize annealing efficiency, using the degree of fluorescence quenching as a gauge. However, the exact amount of duplex formation can only be verified by electrophoresis of the annealing reaction mixture on a non-denaturing gel22. In the double reporter system assay, Cy3-labeled reporter strand, BHQ-modified reporter and loading strand are mixed in 1:1:1 ratio, again varying the ratio to achieve efficient fluorescence quenching. The annealed complexes can be purified using native gel electrophoresis if it is essential to ensure that the exact stoichiometry is achieved14.
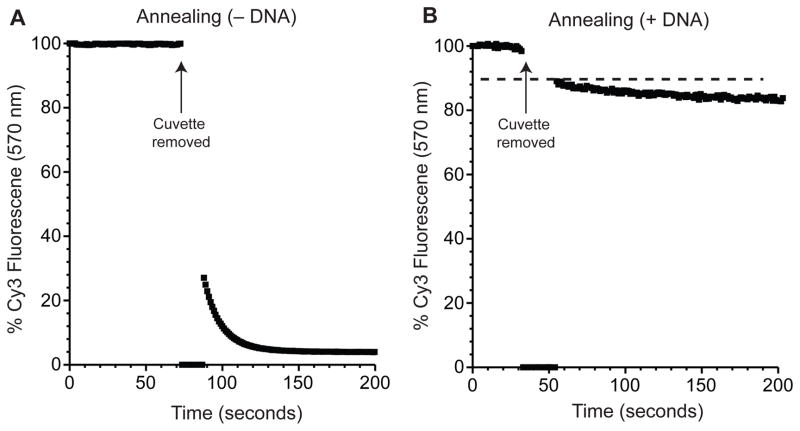
Strand annealing is carried out by denaturing the mix of the oligonucleotides at 80 °C in 1X reaction buffer. The reaction mixture is cooled slowly to room temperature (~25 °C) over about an hour before it is placed on ice for an additional 10 min. In our hands, annealing at high DNA or RNA concentrations (~500 nM) results in better initial fluorescence quenching (Fig. 2A). Following annealing and cooling to 4 °C, we add an excess amount of a competitor DNA strand, which is complementary to the Cy3-modified reporter strand (Table 3). This competitor prevents re-annealing of the reporter strand during the unwinding reaction, and should be used for accurate determination of the unwinding rate constant. The degree of reannealing in the presence of the competitor strand can be determined by adding the loading strand, reporter strand(s), and DNA competitor together and monitoring the degree of quenching upon reaching equilibrium in the cuvette. We use a 10-fold molar excess of competitor DNA to Cy3 reporter strand that inhibits ~90% of reannealing (Fig. 2B). After addition of the competitor, the reaction mixture is incubated on ice for another 10 min to ensure that any unbound Cy3-modified reporter strand is trapped by the DNA competitor before the unwinding reaction begins.
Figure 2.
Monitoring reporter strand annealing to the loading strand. Representative time courses of a Cy3-modified reporter strand annealing to a spectrally paired BHQ-modified loading strand using the single-reporter assay illustrated in Fig. 1A. The reporter strand (24 nt) is complementary to a loading strand (44 nt) and forms a duplex with a stability of −24.7 kcal mol−1 (Table 1). Each assay solution contains a 50 nM concentration of the reporter strand in the absence (A) or presence (B) of a preannealed 10-fold molar excess of a DNA competitor strand. Total Cy3 fluorescence (at 570 nm) of the reporter strand prior to the addition of the loading strand is determined and set as 100%. The cuvette is removed from the fluorometer as indicated and the loading strand to a final concentration of 50 nM is added and mixed by pipetting up and down. The cuvette is placed back in the fluorometer and the change in fluorescence is observed over time. The dashed line in panel (B) represents the theoretical 100% Cy3 fluorescence of the reporter strand following the 10% dilution that results from the addition of the loading strand solution.
Table 3.
Competitor DNA and Cy3-labeled RNA reporter sequences and resulting DNA/RNA hybrids.
| Competitor DNA/RNA hybrid products |
|---|
| 3′-CGUGGCAUUUCG-Cy3-5′ 5′-GCACCGTAAAGC-3′ |
| 3′-CUUUUAGUUUUGAUUUUG-Cy3-5′ 5′-GAAAATCAAAACTAAAAC-3′ |
| 3′-CUUUUUUAAUUUUUUAAUUUUUUG-Cy3-5′ 5′-GAAAAAATTAAAAAATTAAAAAAC-3′ |
| 3′-CUUUUUUCAUUUUUUAACUUUUUG-Cy3-5′ 5′-GAAAAAAGTAAAAAATTGAAAAAC-3′ |
One caveat when using the DNA competitor approach is that the resulting RNA/DNA hybrid may serve as a substrate for some RNA helicases, reducing the amount of helicase available for the unwinding reaction. Should the helicase bind to the RNA/DNA hybrid, this interaction will likely stimulate the helicase’s ATPase activity and further complicate the interpretation of coupling between ATP hydrolysis and duplex unwinding steps. This problem is not relevant to our studies of the human eIF4F complex, which does not unwind RNA/DNA hybrids35. A fluorescent DNA reporter strand could be used to solve this potential problem, since the resulting DNA/DNA duplex product is not a substrate for an RNA helicase. A hairpin-forming DNA molecular beacon may, however, be a more straightforward solution18,19,21.
Unwinding Reactions
After annealing, the double-stranded substrate can be directly used in the unwinding reaction. We carry out our unwinding assays at 25 °C, since the phosphate release assay is also carried out at this temperature10. First, it is essential to calibrate the maximum possible fluorescence signal of the RNA reporter strand (Fig. 3). A mock unwinding assay reaction mixture is prepared that contains all of the reaction components at the appropriate concentration and temperature in the absence of the BHQ-modified oligonucleotide (Cy3-modified oligonucleotide, loading oligonucleotide, ATP, competitor DNA, and proteins). The maximum fluorescence reading also serves as a useful guide to determine how much Cy3 photobleaching occurs over time.
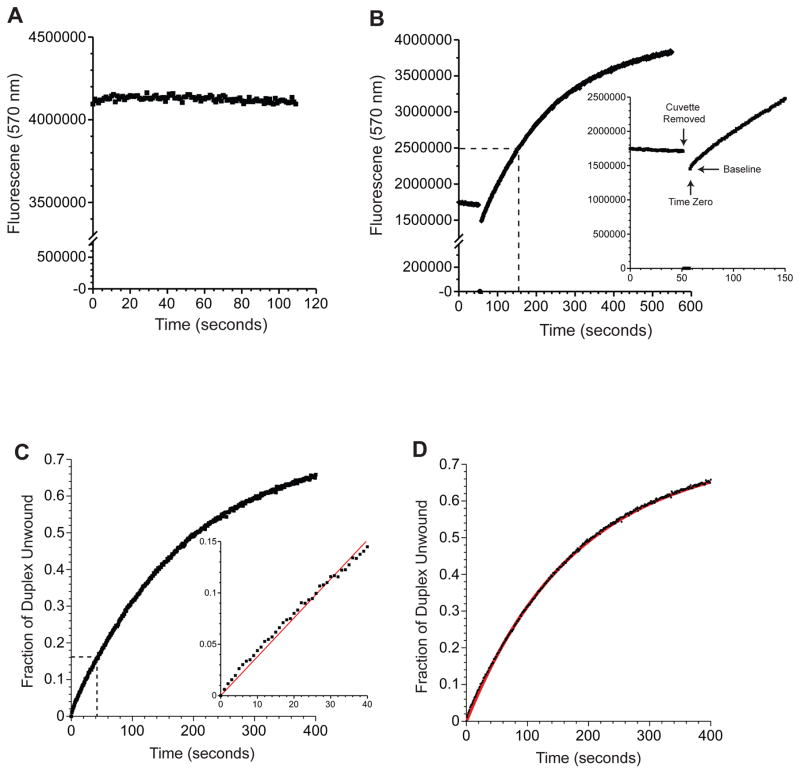
Figure 3.
Evaluating duplex melting by eIF4A and its accessory proteins using the double-reporter fluorescence unwinding assay. Data are generated using the 24-bp duplex substrate (Table 3) and a 1-μM concentration of eIF4E, eIF4A, eIF4G and eIF4B. (A) Maximum Cy3 fluorescence at 570 nm is shown for a control reaction mixture containing 50 nM Cy3 reporter, 50 nM loading strand, 500 nM DNA competitor, 2 mM ATP, eIF4A and accessory proteins (see above for concentrations) in reaction buffer. (B) Representative time course of Cy3 fluorescence before and after ATP addition. A zoomed-in view of the dashed region of the graph is shown in the inset. Total Cy3 fluoresence is recorded for an annealed duplex substrate (50 nM) in the presence of the BHQ reporter strand and proteins. The cuvette is removed after 50 s and ATP to a final concentration of 2 mM is added. The cuvette is placed back in the fluorometer and the change in Cy3 fluorescence is recorded. The removal of the cuvette, addition of ATP (time zero), and the baseline are indicated in the inset. (C) The fluorescence data reported in panel B are converted into the fraction of duplex unwound versus time after ATP addition, as described in the data analysis section. A zoomed-in view of the dashed region of the graph is shown in the inset. The initial rate of unwinding is determined by a linear fit (red line) of the first 40 s of the unwinding reaction. (D) The fluorescence unwinding time course shown in panel C (black line) is fitted to a double exponential rate equation (red line).
To measure unwinding activity, we combine our annealed RNA substrate (50 nM), the helicase, and any other accessory proteins in our 1X reaction buffer. The baseline fluorescence is monitored for at least 100 s to verify substrate annealing and the absence of extraneous RNase activity (Fig. 3B). The background fluorescence is generally 10–50% of the maximum fluorescence. For ATP-dependent helicases, the unwinding reaction can be initiated by adding a saturating amount of ATP. Alternatively, the reaction can be initiated by the addition of the helicase itself. In this case, the background fluorescence should be measured with all the components present except for the helicase. Our assay measures the total fluorescence as a function of time. When using eIF4A and its accessory proteins6,7, the unwinding profile reaches a maximum fluorescence within 6 to 8 min (Fig. 3B). Please note that, to ensure reproducibility, the unwinding assay should be repeated at least three times.
Monitoring ATPase Activity
The measurement of ATPase activity in the duplex unwinding reaction is a variation of the helicase assay described above, the only difference consisting in the addition of an extra protein component, the coumarin-modified phosphate binding protein (PBP-MDCC) (Fig. 4). Our protocol closely follows those previously described in detail by Martin Webb’s group8–10. Importantly, we have determined that the presence of this phosphate sensor in our unwinding assay does not change the rate of duplex unwinding by eIF4A. To be able to compare the rate of ATP hydrolysis with duplex unwinding, it is essential to carry out the unwinding assay and the ATPase assay under identical conditions. The only variation to the unwinding assay described above is that the ATPase assay is initiated by the addition of the helicase protein rather than ATP (which is instead present in the initial reaction mixture). As a control, the unwinding assay can be initiated by the addition of helicase to an ATP-containing reaction mixture to determine if any meaningful changes in unwinding rates are observed. Also for the ATPase activity assay, it is important to repeat each acquisition at least three times to ensure reproducibility.
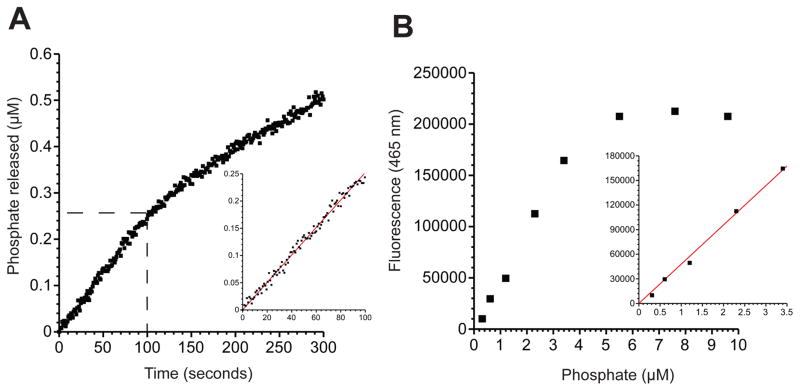
Figure 4.
Determining the rate of phosphate release during duplex unwinding. (A) A representative time course of phosphate release by eIF4A in the presence of eIF4G682-1105 on a 12-bp duplex with a 20-nt overhang (Table 1). The amount of phosphate released (in μM) versus time is shown. The zero time point is taken as the time of helicase solution addition. These data are obtained after conversion of the total fluorescence of PBP-MDCC at 465 nm data versus time, as described in the data analysis section. A zoomed-in view of the dashed region of the graph is shown in the inset. The initial rate of phosphate release is determined by a linear fit (red line) of the first 100 s of the unwinding reaction. (B) Titration of phosphate to PBP-MDCC. Each reaction contains 10 μM PBP-MDCC in reaction buffer incubated at 25 °C. Various dilutions of phosphate are added and the total fluorescence at 465 nm is recorded. The linear response of the PBP-MDCC to phosphate binding is identified as the linear region of the titrations (inset; 0–3.5 μM). The slope of the linear fit of the fluorescence versus phosphate is used to calibrate the phosphate release assay.
It is essential to calibrate the fluorescent signal generated by phosphate binding to the PBP-MDCC protein. To this end, dilutions of inorganic phosphate (Pi) are generated and the total fluorescence of the PBP-MDCC protein for each concentration of Pi is recorded (Fig. 4B). The change in fluorescence per micromole of Pi is simply calculated from the slope of the linear portion of the dilution range. The total change in fluorescence during the unwinding reaction can then be compared to the linear fit of the Pi calibration to determine the rate of Pi produced. Importantly, a large excess of PBP-MDCC (~10 μM) must be used in the reaction so that the protein itself does not limit the rate of Pi binding.
To measure the ATPase activity during unwinding, the reaction is incubated with all the components (minus helicase) at 25 °C for 100 s, or until the fluorescence signal has stabilized. A large increase in fluorescence generally indicates that one or more of the components is contaminated with Pi. If it is not possible to identify and remove these contaminants, a phosphate mop can be used to reduce the background10. In a separate control reaction, the helicase should also be tested for contamination by Pi by monitoring total fluorescence in the absence of ATP. The ATPase reaction is initiated by the addition of the helicase to the cuvette. Total coumarin fluorescence emission is then recorded over time until the fluorescence reading reaches a plateau. This generally occurs in the same time frame as observed with the unwinding reactions.
Data Analysis
To quantify the unwinding activity, the fraction of duplex unwound at each time point must be calculated. This quantification is made possible by calibrating the assay using the maximum fluorescence and the baseline fluorescence immediately after addition of ATP. The baseline fluorescence is subtracted from the values for each time point and from the maximum fluorescence (Fig 3B and 3C). A control reaction should be initially carried out adding buffer rather than ATP solution to verify that the expected baseline for a particular substrate is obtained (Fig. 3B). Performing this control experiment is particularly important if duplex unwinding is rapid upon ATP addition. We also find that fluorescence intensity can vary slightly depending on the exact cuvette position in the holder of the fluorometer. Care should therefore be taken to obtain reproducible cuvette placement in the fluorometer holder. The fraction of duplex unwound for each time point is obtained by simply dividing the baseline-corrected fluorescence values at each time point by the corrected maximum fluorescence value (Fig. 3C):
The fraction of duplex unwound versus time can be fit to single or double exponential equations to obtain rate constants for this process (Fig. 3D). It is worth mentioning that we sometimes discount the first 10 s after the addition of ATP because the data obtained during this time is sometimes not reliable due to variations in mixing time.
Alternately, it is possible to measure the initial rate of unwinding from the slope of the initial linear part of the unwinding curve as fraction of duplex unwound per minute. The fraction of duplex unwound per minute for different concentrations of helicase can then be plotted against the protein concentration and fit to the Hill equation:
Where: fraction/minute is the initial rate of duplex unwinding at any given protein concentration [x]; A is the maximum initial rate of unwinding; Kd is the apparent dissociation constant of the protein–substrate interaction; and n is the Hill Coefficient. It is important to repeat this fit with a reaction that uses half the concentration of substrate to confirm the apparent dissociation constants.
MATERIALS
CRITICAL: All aqueous solutions should be prepared using Milli-Q or equivalent RNase-free filtered water.
REAGENTS
Milli-Q filtered water (Milli-Q water purification system, Millipore, cat. no. Z00Q0V0WW)
7-methyl guanosine (7-MEG, Sigma-Aldrich, cat. no. M0627)
Purine nucleoside phosphorylase (PNPase, Sigma-Aldrich, cat. no. N8264)
Tris base (Fisher Scientific, cat. no. BP152-5)
Acetic Acid (Fisher Scientific, cat. no. A38S-212) CAUTION; Flammable liquid and vapor. Causes severe burns.
Hydrochloric Acid (Fisher Scientific, cat. no. A114C-212) CAUTION; Corrosive. Causes severe burns.
Sodium Hydroxide (Fisher Scientific, cat. no. S318-500) CAUTION; Causes severe burns.
Potassium Hydroxide (Fisher Scientific, cat. no. P250-500) CAUTION; Causes severe burns.
HEPES (Fisher Scientific, cat. no. BP310)
Magnesium acetate (Fisher Scientific, cat. no. BP215)
Potassium chloride (Fisher Scientific, cat. no. P217)
Ammonium acetate (Sigma-Aldrich, cat. no. A1542)
DTT (Gold Biotechnology, cat. no DTT50)
Glycerol (Fisher Scientific, cat. no. G33-4)
Potassium phosphate monobasic (Fisher Scientific, cat. no. P285-3)
Nitric acid (Sigma-Aldrich, cat. no. 438073) CAUTION; Highly corrosive.
RNA oligonucleotides (Integrated DNA technology-IDT). Sequences for fluorescently modified reporter oligonucleotides that we have successfully used are given in Table 1 and Table 2. Oligonucleotides are commercially synthesized so that they are modified with either Cy3 at the 5′-end or a spectrally paired BHQ at the 3′-end. CRITICAL: Purchase oligonucleotides that have been HPLC-purified.
DNA oligonucleotides (Integrated DNA technology-IDT). Sequences for DNA competitor oligonucleotides are given in Table 3. Please note that we have not found it necessary to gel-purify DNA oligonucleotides, but they can be, if required.
Unlabeled RNA loading strand. Prepare according to directions in Steps 1–18.
MgCl2 (Sigma-Aldrich, cat. no. M8266)
Triton X-100 (Sigma-Aldrich, cat. no. T8787)
Bromophenol blue (Bio-Rad, cat. no.161-0404)
Xylene Cyanole (Bio-Rad, cat. no. 161-0423)
Formamide (Fisher-Scientific, cat. no. BP227)
Ethanol (VWR, cat. no. 89125-170). CAUTION; Ethanol is highly flammable and volatile.
Chloroform (Fisher Scientific, cat. no. BP1145-1). CAUTION; Harmful and volatile.
Phenol/choloroform/isoamyl alcohol (25:24:1) (Ambion, cat. no. 15593031). CAUTION; Harmful and volatile.
Spermidine (Sigma-Aldrich, cat. no. S2626)
ATP (Sigma-Aldrich, cat. no. A7699)
CTP (Sigma-Aldrich, cat. no. C1506)
GTP (Sigma-Aldrich, cat. no. G8877)
UTP (Sigma-Aldrich, cat. no. U6875)
Urea Electrophoresis Grade (Sigma-Aldrich, cat. no. U6504)
Acrylamide/Bis-acrylamide (40% w/v; 29:1 ratio) (Fisher Scientific, cat. no. BP14081)
Ammonium persulfate (Bio-Rad, cat. no. 161-0700)
TEMED (Sigma-Aldrich, cat. no. T9281). CAUTION; Flammable and toxic.
Boric acid (Fisher Scientific, cat. no. A74-1)
EDTA (Sigma-Aldrich, cat. no. EDS-500G). CAUTION; Causes serious eye irritation.
Amicon Ultra-15 Filter Unit (Millipore, cat. no. EDS-500)
DNA template (Annealed oligonucleotides or PCR product, see Procedure)
T7 DNA oligonucleotide (5′-TAATACGACTCACTATAG-3′; custom synthesized from IDT oligo)
Purified PBP-MDCC (Expression plasmid a kind gift from Martin Webb (MRC, UK). Available for purchase from Invitrogen, cat. no. D10253)
Purified T7 RNA Polymerase (1 mg/ml)36
EQUIPMENT
50-μl quartz submicrocell (Starna Cells, cat. no. 16.50F-Q-10/Z8.5)
Spectrofluorometer (Horiba Scientific, Fluorolog-3)
Cooling thermostat (Lauda-Brinkmann, Lauda-Alpha RA8)
RNAse-free 1.5-ml microcentrifuge tubes (Eppendorf, cat. no. 022363204)
Filter pipette tips 10 μl (Fisher Scientific, cat. no. 02-682-240)
Filter pipette tips 200 μl (Fisher Scientific, cat. no. 02-682-231)
Filter pipette tips 1000 μl (Fisher Scientific, cat. no. 02-682-244)
Dry bath incubator (Fisher Scientific, cat. no. 11-718Q)
High-voltage electrophoresis power supply (Bio-Rad, 164-5056)
Refrigerated tabletop microcentrifuge (Eppendorf, cat. no. 022620667)
Vortex mixer (Fisher Scientific, cat. no. 02-215-365)
DeltaGraph software (Redrock software)
Origins software (Horiba Scientific)
Nanodrop (Fisher Scientific, cat. no. 13-400-411)
Dry Block Heater (VWR, cat. no. 12621-104)
Rotating mixer (Fisher Scientific, cat. no. 22-363-152)
Dissecting forceps (Fisher Scientific, cat. no. 1381239)
Analytical vertical electrophoresis unit (C.B.S. Scientific, cat. no. ASG-250)
Preparative vertical electrophoresis unit (C.B.S. Scientific, cat. no. LSU-400-20)
Razor blade (VWR, cat. no. 55411-050)
Saran wrap (VWR, cat. no. 89136-664)
Sharpie Pen (VWR, cat. no. 500034-938)
Serological 10 ml Pipet (Fisher Scientific, cat. no. 13-675-20)
Eppendorf Tubes Safe-Lock tubes 1.5 ml (Fisher Scientific, cat. no. 05-402-25)
Conical tubes 50 ml (Fisher Scientific, cat. no. 12-565-270)
Sterile bottle top filters (Fisher Scientific, cat. no. 09-741-07)
Conical tube filter top 50ml (Corning, cat. no. 430320)
Power supply (Bio-Rad, cat. no. 164-5056)
Eppendorf Refrigerated Microcentrifuge (Fisher Scientific, cat. no. 02-262-0603)
Precoated TLC glass plates (EMD, cat. no. 5715-7)
Handheld UV lamp (UVP, cat. no. 95-0017-09) CAUTION Use appropriate eye protection when using UV light.
REAGENT SETUP
Buffers
When using the helicase assay with RNA substrates, ensure that all buffers are RNase-free by using Milli-Q filtered water or equivalent to prepare them. We also recommend filtering buffers through a 0.2 μm bottle top filter before use.
Tris-acetate (1 M; pH 7.5) buffer is adjusted to pH to 7.5 with acetic acid and stored at room temperature for 1 year.
Tris-HCl (1 M; pH 8) buffer is adjusted to pH to 8 with concentrated hydrochloric acid and stored at room temperature for 1 year.
HEPES (1 M; pH 7.5) buffer is adjusted to pH to 7.5 with potassium hydroxide and stored at room temperature for 1 year.
Ammonium acetate (3 M; pH 5.2) is adjusted to pH to 5.2 with acetic acid and stored at room temperature for 1 year.
EDTA (0.5 M; pH 8.0) is adjusted to pH 8.0 with sodium hydroxide and stored at room temperature for 1 year.
Reaction buffer 10X
Mix together the following components at the mentioned final concentration: 200 mM Tris-acetate (pH 7.5), 20 mM magnesium acetate, 1 M potassium chloride and 2 mM DTT. We generally prepare this buffer without DTT added, filter sterilize it, and store it at room temperature indefinitely. Fresh DTT is added to a small volume of 10X buffer (~1 ml) for use on the day of the experiment. Small aliquots of the 10X buffer containing DTT can be stored at −20 °C for several months, but care should be taken not to freeze-thaw these aliquots multiple times.
NTP stocks (ATP, CTP, GTP, UTP)
Individual nucleotide triphosphate stocks are prepared at a 100 mM concentration. Each stock is prepared using Milli-Q filtered water and carefully adjusted to pH 7.5 using 10 M sodium hydroxide. Each NTP stock can be stored at −20 °C for ~2 years.
ATP solution
Prepare a 20 mM Mg-ATP stock in 1X reaction buffer: mix 100 μl of 40 mM ATP (ATP stock is pH 7.5) in Milli-Q filtered H2O with 40 mM of magnesium acetate. Prepare fresh with RNase-free reagents and keep on ice before use. Alternatively, aliquot (~50 μl) and store at −20 °C for up to 1 year. CRITICAL: Avoid repeated freeze-thaw cycles.
Purified proteins
We typically store purified protein components (e.g. eIF4A6,7) at −80 °C in 20 mM HEPES (pH 7.5), 200 mM potassium chloride, 10% (vol/vol) glycerol, and 1 mM DTT. We typically store our proteins at a concentration of at least 10 μM for up to 2 years at −80 °C7.
Inorganic phosphate (Pi) standards
Prepare 10, 20, 30, 40, 50 and 100 μM potassium phosphate monobasic solutions in 1X reaction buffer and store for 1 year at −20 °C. These solutions will be used to generate a calibration curve (see Procedure).
Oligonucleotides
Suspend all synthesized lyophilized oligonucleotides (received from commercial supplier IDT) to a 100-μM concentration in 10 mM Tris-HCl, pH 8 at −20 °C. Prepare multiple small aliquots (~50 μl) to avoid multiple freeze thawing cycles of fluorescently labeled oligonucleotides. Fluorescent oligonucleotides should be stored in the dark at −20 °C for ~2 years.
Loading strand
Transcribe an unlabeled RNA loading strand from oligonucleotides or PCR products using previously published protocols with minor modifications (see Procedure)31,32,34. Store the purified loading strand RNA at −80 °C in 10 mM Tris-HCl, pH 8 for ~2 years.
10X transcription buffer
Prepare a solution containing 400 mM Tris HCl pH 8.1, 0.1% (vol/vol) Triton X-100, 300 mM MgCl2, 10 mM spermidine, and 50 mM DTT. CRITICAL Store the 10X transcription reaction buffer at 4 °C for upto 1 year, and add spermidine and DTT fresh prior to transcription.
5X Tris-borate-EDTA (TBE 5X) buffer
Prepare a solution containing 450 mM Trizma base, 450 mM boric acid, 10 mM EDTA (pH 8). This buffer can be stored at room temperature for 6 months.
Ammonium persulfate solution
Prepare a 10% (wt/vol) solution of ammonium persulfate and pass it through a 0.22-μm filter. Store at 4 °C for 1 month.
Gel markers
Prepare a 2X loading dye marker by mixing 0.025% (wt/vol) bromophenol blue and 0.025% (wt/vol) xylene cyanol with 95% formamide.
Annealing buffer
Prepare a buffer containing the following components: 20 mM Tris-acetate pH 7.5, 2 mM magnesium acetate, 0.2 mM DTT, and 100 mM potassium chloride.
Denaturing acrylamide gel
| Component | Percentage | ||||
|---|---|---|---|---|---|
|
| |||||
| 16% | 12% | 10% | 8% | 6% | |
| Acrylamide, Bis-acrylamide 40% wt/vol (29:1) | 24 ml | 18 ml | 15 ml | 12 ml | 9 ml |
| TBE (5X) | 12 ml | 12 ml | 12 ml | 12 ml | 12 ml |
| Urea | 30 g | 30 g | 30 g | 30 g | 30 g |
| H2O | – | 6 ml | 9 ml | 12 ml | 15 ml |
| 10% (wt/vol) ammonium persulfate | 400 μl | 400 μl | 400 μl | 400 μl | 400 μl |
| TEMED | 60 μl | 60 μl | 60 μl | 60 μl | 60 μl |
|
| |||||
| Total Volume | 60 ml | 60 ml | 60 ml | 60 ml | 60 ml |
EQUIPMENT SETUP
Cuvette
Clean and store the cuvette in 5 M nitric acid according to manufacturer’s guidelines. Rinse the cuvette thoroughly with RNase-free water prior to use. Blot with kimwipes until water residue is eliminated.
Fluorometer
Turn on fluorometer as outlined in the manufacturer’s guidelines and do it about 30 min prior to start of experiments to allow for lamp warm-up. Excitation and emission wavelengths will depend on the Fluorophore-Quencher pair. The following settings are for the Cy3 fluorophore (excitation λ = 550 nm and emission λ = 570 nm):
| Excitation λ | 550 nm |
| Entrance slit width | 6 nm |
| Exit slit width | 6 nm |
|
| |
| Emission λ | 570 nm |
| Entrance slit width | 6 nm |
| Exit slit width | 6 nm |
|
| |
| Integration time | 0.1 s |
| Time increment | 1.0 s |
For the phosphate release assay, fluorescence measurements are made using excitation and emission wavelengths of 430 nm and 465 nm, respectively. To reduce signal-to-noise ratio, optimize entrance and exit slit width and integration time for your particular machine. Time increments will depend on the kinetics of your unwinding reactions and can be varied accordingly.
Dry bath heating block
Set the block temperature to 80°C.
PRODEDURE
CRITICAL: Proper techniques must be used to avoid RNase contamination when preparing and working with RNA substrates. The use of gloves and RNase-free reagents and consumables is essential. We strongly recommend using filter-tip pipette tips for all work with RNA. Fluorescently modified oligonucleotides are sensitive to light and should be stored appropriately to avoid light damage.
Preparation of DNA template TIMING 1.5 h
-
1
Generat the DNA template for transcription by using either oligonucleotides or PCR products. Oligonucleotides are designed to include a T7 promoter sequence that must be located in a duplex region, as described previously7,31,32,34. We recommend implementing the oligonucleotide option if the template to be obtained is relatively short, up to ~40 base pairs. For longer templates, PCR can be used to amplify a defined sequence of plasmid DNA. A T7 promoter can also be added during PCR, if one is not already present in the plasmid sequence32. We use a standard published protocol for generating templates by PCR reactions37. Reactions typically include 10–500 ng of plasmid template; 200 μM dNTPs; ~0.2 μM forward and reverse primers; and 5 units/μl PFU DNA polymerase. PCR reactions are carried out for 25–30 cycles.
PAUSE POINT. PCR reactions can be stored at −20 °C indefinitely.
-
2
Add an equal volume of phenol:chloroform/isoamyl alcohol (25:24:1) to the solution of template DNA. We recommend starting with between 100 μl and 200 μl volume of DNA template solution in a 1.5-ml tube. Please note that we have not found it necessary to incubate the tube on ice during the phenol/chloroform extraction procedure.
-
3
Vortex the 1.5-ml tube for 15 s and then centrifuge at 20,000 g for 2 min to separate the phases. This centrifugation can be carried out at room temperature (~25 °C) or at 4 °C with equal success.
-
4
Remove the bottom phase carefully (phenol:chloroform) and discard it into the laboratory phenol waste. We recommend leaving a small amount (~10 μl) of the bottom phase so that no amount of upper phase is mistakenly discarded.
-
5
Repeat steps 2–4 a further two times so that three phenol:chloroform extractions are carried out in total.
-
6
Add to the tube an equal volume of chloroform.
-
7
Vortex the tube for 15 s and then centrifuge at 20,000 g for 2 min to separate the phases. This centrifugation can be carried out at room temperature or at 4 °C.
-
8
Isolate the top aqueous layer (containing the DNA template) and transfer it to a new tube. Be careful not to remove any of the lower phase at this stage. Add 1/10 volume of 3 M ammonium acetate (pH 5.2), and then 2.5 volumes of ice-cold 100% ethanol.
-
9
Vortex the tube containing the template DNA solution from step 8 for 15 s and place it on dry ice for 30 min or at −80 °C for 1 h.
PAUSE POINT The precipitation may be left overnight on dry ice or at −80 °C.
-
10
Centrifuge the tube from step 9 at 20,000 g for 30 min at 4 °C in a microfuge. Carefully remove all the liquid from the tube and leave the tube open to dry on the bench for 10 min. We recommend placing a kimwipe over the tube to prevent any dust from entering the tube.
-
11
Resuspend the DNA dried at the bottom of the tube in an appropriate volume of 10 mM Tris (pH 8) so that the final DNA concentration is ~100 μM.
PAUSE POINT. The purified DNA solution can be stored at −20 °C indefinitely.
DNA Template Annealing TIMING 15 min
CRITICAL: We have found that the annealing reaction is not required if the DNA template has been obtained via PCR, so if the DNA template was prepared via PCR, we advise readers to skip steps 12–14 and go directly to step 15
-
12
Prepare 50-μl of an annealing solution containing 20 μM T7 DNA oligonucleotide and 20 μM template DNA oligonucleotide in a 1:1 ratio in annealing buffer. This annealing reaction volume is sufficient for 5 ml of transcription reaction and can be prepared in a 1.5 ml Eppendorf tube.
-
13
Incubate the reaction mixture at 80 °C for 2 minutes to cause the two DNA strands to denature, and then allow it to cool to room temperature in an Eppendorf tube rack (for ~5 min).
-
14
Place the reaction mixture on ice for 10 min to ensure optimal annealing.
Transcription TIMING 1 h
-
15
(OPTIONAL) The scale of the transcription reaction will depend on how much RNA is required. Before proceeding with the preparative scale reaction detailed in steps 16 and 17, test a new template in a relatively small reaction (~20 μl) to ensure that the template is free of RNases and can be transcribed successfully. This is achieved by appropriately scaling the volumes provided in the in-text table shown in step 16. Analyze the products of the reaction by separating them on an analytical denaturing acrylamide gel (see below in step 18).
-
16
For preparative scale reactions of short RNAs (<200 nt) typically prepare a 5-ml transcription reaction as described in the in-text table below. This large volume is appropriate when the final RNA product will be purified using a denaturing gel. For longer RNA constructs (>200 nt), we have had good success using a smaller, 1-ml transcription reaction mixture followed directly by phenol:chloroform extraction (see step 18).
Component and Initial Concentration Volume (μl) Final Concentration 10X Transcription reaction buffer 500 1X 100 mM ATP 400 8 mM 100 mM GTP 400 8 mM 100 mM CTP 400 8 mM 100 mM UTP 400 8 mM 1 M MgCl2 150 30 mM 1 mg/ml T7 Polymerase 2000 0.4 mg/ml 20 μM Annealed oligos 50 200 nM Milli-Q filtered water 700 -
Total 5000 If using a PCR-obtained DNA template, we recommend using a 50 μg/ml concentration of the DNA template solution (which will not have gone through the annealing procedure of steps 12–14) in the transcription reaction.
-
17
Incubate the transcription reaction at 37 °C for 1 h.
CRITICAL STEP: Avoid incubating for more than 1 h, as long incubation times typically lead to an increase in the amount of RNA products that are shorter than desired.
PAUSE POINT This reaction mixture can be stored at −20 °C overnight or for weeks at −80 °C.
RNA PURIFICATION TIMING 1.5–4 h
-
18
Following transcription, purify short RNAs (<200 nt) produced according to option A (steps i-ix) or long RNAs (>200 nt) according to option B.
A. Denaturing gel RNA purification
Use 30 cm x 20 cm gel plates with a 6-mm spacer to purify a 5-ml transcription reaction. This size gel requires ~250-ml gel solution. The gel percentage will depend on the size of the RNA product and can be chosen according to published data for migration of nucleotides on denaturing gels38. A single large well across the gel is used to load the transcription reaction, with a second smaller well used to load 50 μl of RNA gel marker (bromophenol blue and xylene cyanol).
Pre-run the gel for 1 h in 0.5X TBE running buffer at ~40 W constant power in the cold room (4 °C) to ensure a constant gel temperature and running buffer equilibration. Mix the transcription reaction with an equal volume of formamide and heat the resulting solution to 70 °C for 3 min. Place then the sample on ice for 5 min, before loading it onto the gel. Make sure to flush out the gel lane(s) thoroughly with running buffer using a syringe prior to loading the transcription reaction.
Run the gel electrophoresis experiment at ~40 W for roughly 3 h, increasing or decreasing this duration depending on the size of the RNA product. Use the gel markers’ rate of migration through the gel as a rough guide of the size-based corresponding rate of the RNA bands38.
Remove the gel plates from the apparatus and place them flat on the bench. We recommend using a fresh piece of bench coat to place the gel on to avoid possible contamination. Carefully separate the gel plates and lay a piece of clean saran wrap over the gel. Turn the gel over so that it is resting on the saran wrap and remove the other gel plate. Place another piece of saran wrap over the gel.
Visualize the RNA product by UV shadowing. Place a TLC plate underneath the saran wrap underneath the gel, and the nucleic acids, if present, will appear as dark bands when illuminated from above by a handheld UV lamp. Use a sharpie pen to outline the RNA band on the saran wrap.
Use a sterile razor blade (passed through a flame using forceps) to cut around the RNA band. Transfer the gel piece to a 50-ml conical tube using clean, RNase-free forceps. Use a sterile 10-ml serological pipet to crush the gel into small (~2-mm) sized pieces. Accomplish this task by repeatedly pushing the gel against the tube wall with the pipet. Add Milli-Q filtered water to fill the tube to the 50-ml mark and place the tube on a rotating mixer. Activate the mixer and keep the tube on it overnight at 4 °C.
Recover the RNA in solution by centrifuging the 50-ml conical tube for 3 min at 1000 g and at 4 °C. Filter the supernatant into a new 50-ml conical tube using a 0.22-μm conical tube filter top. Resuspend the gel pieces in ~20 ml of Milli-Q filtered water and vortex for 30 s. Centrifuge the conical tube for 3 min at 1000 g and at 4 °C. Filter the supernatant as described above and pool it with the first supernatant.
Concentrate the supernatant down to ~2 ml using a 15-ml Amicon Ultra filtration unit with a molecular weight cutoff that is appropriate for the size of the RNA product.
-
Aliquot the concentrated RNA solution into 1.5-ml Eppendorf safe lock tubes so that each tube contains 500 μl of RNA solution. Phenol:chloroform-extract the RNA in each tube as detailed in steps 2–11 above. Following ethanol precipitation, resuspend each pellet in 100 μl of 10 mM Tris (pH 8). Determine the concentration of the RNA using a NanoDrop spectrophotometer, or equivalent, and subdivide the solution in ~ 50 μl aliquots. Freeze each aliquot and store it at −80 °C.
PAUSE POINT RNA purified in this way can be stored at −80 °C for at least 6 months without significant degradation.
The integrity of the purified RNA should be checked by loading ~5 μg of the purified RNA onto an analytical denaturing gel and visualizing by UV shadowing or ethidium bromide staining.
B. RNA purification by phenol:chloroform extraction
Perform phenol:chloroform extraction of the RNA from the transcription reaction mixture from step 17 as described in steps 2–11 above,
Duplex Annealing. Timing: 1–1.5 h
-
19
Prepare a 1-μM working stock solution of the Cy3-labeled reporter strand diluted in 1X reaction buffer. CRITICAL STEP This concentration is employed so that the same stock can be directly used to prepare the helicase assay reaction and the reaction needed to measure the maximum fluorescence (Step 29).
-
20
Prepare a 10-μM working stock solution of BHQ-labeled reporter strand in 1X reaction buffer.
-
21
Prepare a 10-μM working stock of unlabeled loading strand in 1X reaction buffer.
-
22
Mix the three RNA strand solutions to obtain a final 500-nM working concentration for each strand:
Component Volume (μl) Final Concentration 1 μM Cy3-labeled RNA 200 500 nM 10 μM BHQ-labeled RNA 20 500 nM 10 μM Loading RNA 20 500 nM 1X Reaction buffer 160 1X
Final Volume 400 -
23
Remove a prewarmed 80-°C heating block from the dry bath and place it on the lab bench. Place the annealing reaction mixture prepared in step 22 into the 80-°C heating block and let it cool to room temperature (~1 h). CRITICAL STEP Cover the block containing the annealing reaction mixture with foil to avoid photobleaching by light. ?TROUBLESHOOTING
-
24
Spin the tube at 10,000 g for 5 s in a microfuge at room temperature and place on ice for 10 min.
-
25
Transfer 250 μl of the solution from step 24 to a new RNase-free 1.5-ml Eppendorf tube and add 12.5 μl of a 100-μM solution of competitor DNA and 237.5 μl of 1X reaction buffer. Mix and incubate on ice for 10 min to sequester any free Cy3-labeled RNA. ?TROUBLESHOOTING
PAUSE POINT. The annealed RNA can be stored in the dark on ice for ~3 hours.
Unwinding Assay. TIMING 10–15 min per assay
-
26
Dilute all protein stocks to 10 μM into 1X reaction buffer. ?TROUBLESHOOTING
-
27
Combine the components shown in the in-text table below in a 1.5-ml Eppendorf tube. CRITICAL STEP Use the same aliquot of Cy3-labeled RNA that was used for the annealing reaction.
Component and Concentration Volume (μl) Final Concentration 1 μM Cy3-labeled RNA 3.75 50 nM 1 μM Loading RNA 3.75 50 nM 5 μM DNA competitor 3.75 500 nM 10 μM eIF4A (Helicase) 7.5 1 μM 20 mM ATP 7.5 2 mM 1x Reaction buffer 48.75 1x
Total 75 -
28
Transfer 70 μl of this mix to the 50-μl quartz cuvette and measure the fluorescence at the plateau, which should be reached after ~100 s. Record the raw fluorescence units as the maximum fluorescence. Repeat steps 27 and 28 two more times to obtain an average fluorescence reading (Fig.3A). CRITICAL STEP Avoid the formation of bubbles when transferring the sample to the cuvette. Bubbles will dramatically influence the fluorescence reading. ?TROUBLESHOOTING.
-
29
Combine the annealed RNA substrate solution from step 25 and the solution of proteins prepared in step 23 in 1X reaction buffer in a 1.5-ml RNase-free Eppendorf tube, as outlined in the in-text table below. CRITICAL STEP: Do not add the ATP at this stage.
Component and Concentration Volume (μl) Final Concentration 250 nM annealed substrate 15 50 nM 10 μM helicase 7.5 1 μM 1x Reaction buffer 45 1x
Total 67.5 -
30
Transfer 63 μl of the solution prepared in step 29 to the 50-μl quartz cuvette. Measure Cy3 fluorescence with RNA substrate and proteins after allowing the reaction to equilibrate to 25 °C (or to the reaction temperature of choice) for ~100 s. ?TROUBLESHOOTING
-
31
Remove the cuvette from spectrophotometer and add to it 7 μl of 20 mM ATP to initiate the reaction. Tap the cuvette three times to mix and place back into the chamber of the fluorometer. Please note that it is also possible to add the ATP directly to the cuvette without removing it from the machine. In this case, the sample must be mixed well by pipetting. We have obtained similar results with either approach.
-
32
Record total fluorescence (duplex unwinding activity) until saturation is reached, which typically takes 3–10 min, depending on the efficiency of the helicase (Fig. 3B). CRITICAL STEP: Repeat steps 29–32 two more times. ?TROUBLESHOOTING
ATPase Assay TIMING 30 min
-
33
Combine annealed RNA substrate solution prepared in step 25 and accessory protein components if used (minus eIF4A/helicase) in a 1.5-ml RNase-free Eppendorf tube, as outlined in the in-text table below. CRITICAL STEP: Do not add the helicase at this stage.
Component and Concentration Volume (μl) Final Concentration 250 nM annealed substrate 15 50 nM 100 μM PBP-MDCC 7.5 10 μM 1X Reaction buffer 45 1X
Total 67.5 -
34
Transfer 63 μl of the solution prepared in step 33 to the 50-μl quartz cuvette. Record PBP-MDCC fluorescence for a total of ~100 s as you allow the reaction mixture to equilibrate to room temperature (or the reaction temperature of choice). ?TROUBLESHOOTING
-
35
Remove the cuvette from spectrophotometer and add to it 7 μl of 10 μM helicase (eIF4A in our case) to initiate the reaction. Tap the cuvette three times to mix and place back into the chamber. It is also possible to add the helicase directly to the cuvette without removing it from the machine. We have obtained similar results with either approach.
-
36
Record the total fluorescence (ATPase activity) until a plateau is reached, which typically occurs 3–10 min, depending on the efficiency of the helicase (Fig. 4A). ?TROUBLESHOOTING
-
37
To determine the ATPase activity in the absence of RNA substrate, repeat steps 33–36 in the absence of RNA.
Generating a Pi Calibration curve TIMING 30 min
-
38
Add 10 μl of 100 μM PBP-MDCC and 90 μl of 1X reaction buffer to a RNase-free 1.5-ml Eppendorf tube. Mix and transfer the contents to a 50-μl quartz cuvette.
-
39
Incubate the solution in the cuvette for ~100 s and record the total fluorescence intensity at excitation and emission wavelengths of 430 nm and 465 nm, respectively. Consider this fluorescence value as the 0 μM Pi (Fig. 4B).
-
40
Take separate measurements of solutions with final concentrations of 1, 2, 3, 4, 5, and 10 μM Pi. For this purpose, add 10 μl of 10 μM, 20 μM, 30 μM, 40 μM, 50 μM, and 100 μM Pi, respectively, to 10 μl of 100 μM PBP-MDCC, and 80 μl of 1X reaction buffer, for a total volume of 100 μl (Fig. 4B).
Data analysis TIMING ~1 h
-
41
Calibrate the unwinding reaction using the maximum total fluorescence (step 28) and the baseline total fluorescence when unwinding is initiated (Step 31). Due to a dilution effect, the baseline fluorescence value measured immediately after addition of ATP should be ~10% lower to that measured prior to the addition of ATP. ?TROUBLESHOOTING
-
42
Subtract the baseline fluorescence value from the maximum fluorescence value.
-
43
Subtract the baseline fluorescence value from each fluorescence data point determined during the unwinding reaction.
-
44
Convert the fluorescence values into the fraction of quenched duplex unwound. This goal is achieved by simply dividing the fluorescence values (subtracted baseline) by the maximum fluorescence (subtracted baseline).
-
45
Use the time point at which the ATP is added as zero time. Subtract this time from all subsequent times to calibrate the time course. ?TROUBLESHOOTING
-
46
Plot the fraction of unwound duplex versus time using preferably a graphing program that enables to perform nonlinear regression analysis. We have used Deltagraph, Kaleidagraph, and Graphpad Prism with good results.
-
47Determine the initial rate of unwinding by measuring the fraction of duplex unwound during the initial linear portion of the unwinding reaction, which usually consists of the first 30 to 60 s of the unwinding reaction. Use a linear regression to fit the data and record the initial fraction of unwound duplex per minute:
Where:
y = the unwound duplex fraction
m = the slope of the line
x = the time point
b = the x-axis intercept (usually zero)
-
48(OPTIONAL) The data can also be fitted to an exponential equation to determine the rate constants of the duplex unwinding reaction. Whether a single or double exponential equation fits the data will depend on the specific helicase being used. In our hands, duplex unwinding by eIF4A produces rather complicated unwinding data that can only be fit with a double exponential equation. If the same holds true for the researcher’s helicase of choice, fit the data according to the following equation:
Where:
RNAss represents the fraction of single-stranded RNA released as a function of time.
A1 is the total unwinding amplitude of the rapid unwinding phase
k1 is the unwinding rate constant of the rapid unwinding phase
A2 is the total unwinding amplitude of the slow unwinding phase
k2 is the unwinding rate constant of the slow unwinding phase
-
49
To analyze the ATPase data, calibrate the change in fluorescence of PBP-MDCC in the same way described for the helicase assay (Steps 41–43).
-
50
Estimate the amount of ATP hydrolyzed per duplex unwound by dividing the initial rate of duplex unwinding by the initial rate of ATP hydrolysis.
?TROUBLESHOOTING
Step 23
Generating an annealed substrate by mixing equimolar amounts of each strand is not 100% efficient, and it is likely to result in some unannealed RNA strands. We have not found it necessary to purify the annealed RNA substrate, but this could be carried out using native gel purification. In either the single or double reporter systems, a suitable level of fluorescence quenching must be verified. We typically observe between 50% and 90% quenching efficiency in our assays. When using the double reporter system, we have found that the degree of quenching can depend on how extensively structured the loading RNA strand is in regions not involved in the interaction with the reporter strands. The quenching efficiency is easily determined by monitoring the total fluorescence before and after the annealing reaction. If a very low quenching efficiency (~50%) is observed, the annealing buffer and cooling procedure should be optimized for the RNA in question. If changes in the annealing buffer and conditions do not improve quenching efficiency, changing the positions and/or sequences of the reporter strands should be considered.
Step 25
The fluorophores are sensitive to light, and we have found their fluorescence intensity will diminish substantially after a few hours. To minimize this problem, keep all annealed stocks on ice and in the dark at all times, which is achieved by simply covering the ice bucket with a lid. Please note that we have not observed any substantial strand exchange from the loading strand between the reporter and the DNA competitor over the course of a few hours at 4 °C.
Step 26
Proteins may precipitate from solution as the day progresses. We have found that some purified proteins are relatively unstable after a number of hours in solution in the 1X reaction buffer. Check for protein precipitation throughout the day to ensure protein solubility and activity in the assay. Make new protein stocks in 1X reaction buffer if the proteins lose activity throughout the day.
Step 28
The experiment may suffer from a lack of reproducibility. We find that assay components such as the loading RNA strand, DNA competitor, ATP, the helicase of interest, and other proteins can alter the maximum fluorescence of the Cy3 dye. We therefore measure the maximum fluorescence in the presence of all components used in the unwinding reaction to avoid calibration errors. For reproducibility, we incubate the reaction components for at least 100 s, or until the maximum fluorescence reading has stabilized. A control reaction should also be implemented in which a non-hydrolyzable ATP analog is used instead of ATP to determine whether helicase binding to the duplex alters total fluorescence.
Step 30
A rapid increase in total fluorescence during this stage typically reflects RNase contamination of one or more of the components or the cuvette. The cuvette should be thoroughly washed in nitric acid and rinsed extensively with filtered water before attempting to set up another assay. Each component will need to be checked for RNase activity if the same rapid increase in fluorescence occurs after cleaning the cuvette. Prior to ATP addition, a less dramatic increase in fluorescence may indicate ATP-independent strand exchange on the loading strand between the reporter and the competitor DNA.
Step 32
Should a time course longer than ~10 min be needed to observe unwinding, constant excitation of the fluorophore will likely result in destruction of the fluorophore by photobleaching. To avoid this problem, it may be necessary to record time points less frequently so that the fluorophore is not constantly excited.
Step 34
Extraneous Pi contamination may be observed if the background coumarin fluorescence increases more than ~10% upon addition of the assay components. A phosphate mop (0.01 U/ml PNPase and 200 μM 7-MEG) can be added to the reaction to reduce the level of any contaminating Pi in the purified proteins and other components used. We have not observed any need for this precaution in our particular assays due to the low levels of contaminating Pi in our components.
Step 36
It is likely that the helicase will keep hydrolyzing ATP even after the duplex has been unwound. The reasons for this observation may include binding of the helicase to single-stranded RNA or to the DNA/RNA hybrid. The amount of DNA competitor strand used to prevent reannealing may also change the rate of ATP hydrolysis. Control experiments should therefore be carried out to determine the extent to which the presence and amount of different substrates stimulate the ATPase activity of the helicase being tested.
Step 41
The decrease in fluorescence baseline after addition of ATP may be larger than the dilution-associated 10%. This bigger-than-expected drop in fluorescence may occur if the helicase activity is rapid or if the cuvette is not in perfect alignment before and/or after ATP addition. If the change in baseline fluorescence level following ATP addition is substantially different from what is expected, an alternative method of ATP addition may be necessary. Using a rapid mixing technique, such as stopped-flow, may be necessary if manual pipetting is not reliable.
Step 45
The zero time point can be difficult to establish if the reaction is not properly mixed. Using a rapid mixing technique such as stopped-flow may be necessary if manual pipetting is not reliable.
TIMING
Steps 19–25, annealing of duplex substrate: 1–1.5 h
Steps 26–28, measuring maximum fluorescence: 15 min
Steps 29–32, measuring duplex unwinding: 15 min
Steps 33–37, measuring ATPase activity: 30 min
Steps 38–40, generating Pi calibration curve: 30 min
Steps 41–50, data analysis: 1 h
ANTICIPATED RESULTS
A representative data set from an unwinding assay is shown in Figure 3. The data shown is generated using fluorometer settings described with a 50 nM-concentration of substrate in a 50-μl cuvette. These data are generated using human eIF4A in the presence of eIF4G, eIF4E and eIF4B. In our hands, this helicase complex is the most active, and it unwinds roughly 70% of the duplex (24bp; Table 1) within 10 min at 25 °C. For this particular substrate, we observe a ~60% fluorescence quenching efficiency following annealing (Fig. 3A and 3B). This level of quenching clearly provides a good starting signal to be able to visualize unwinding. The fluorescence reading prior to ATP addition indicates that no RNase activity is associated with the components used (Fig. 3B). The baseline recorded upon ATP addition is within 5% of the expected baseline that would be obtained from the 10% volume dilution associated with the addition of ATP. The baseline fluorescence reading (~1.5×106) is then subtracted from the maximum fluorescence reading (Fig. 3A) and all of the unwinding data points (Fig. 3B). The zero of the X-axis is adjusted to reflect the time after ATP addition and the resulting calibrated unwinding curve is generated (Fig. 3C). The initial rate of unwinding is determined from the linear fit of the first 40 s of the unwinding reaction (Fig. 3C insert). The rate constants of the reaction are then obtained from fitting the data with a double exponential rate equation (Fig. 3D).
Representative data is shown for an ATPase assay using the PBP-MDCC biosensor in Figure 4. These data are generated from an assay using eIF4A, eIF4G682-1105 and a 50 nM concentration of a 12-bp duplex substrate6. Using the calibration method described in the Procedure, the units of the data shown here (Fig. 4A) have already been converted from total fluorescence intensity to the amount of phosphate released. Importantly, the calibration curve of fluorescence intensity versus phosphate concentration reveals the linear range of phosphate binding by the PBP-MDCC protein (10 μM) that is added to the reaction (Fig. 4B). The linear range of the PBP-MDCC protein is reliable up to ~3.5 μM phosphate, which is 5-fold greater than the amount of phosphate released after 300 s of reaction. A control experiment in which twice the amount of PBP-MDCC is used in the reaction can be carried out to ensure that the PBP-MDCC is not limiting for the rate of phosphate binding. The initial rate of phosphate release is easily determined from a linear fit of the first 100 s of the reaction (Fig. 4A). The amount of ATP hydrolyzed per duplex unwound can then be estimated by dividing the initial rate of duplex unwinding by the initial rate of ATP hydrolysis. However, one should be cautious when interpreting the results of this kind of analysis, since it can only provide a maximum possible amount of ATP that is hydrolyzed per duplex unwound6,10. Nevertheless, this analysis can be very useful when comparing how different helicase accessory proteins regulate coupling between ATP hydrolysis and duplex unwinding6. It is also worth noting that it is also possible to use a dual wavelength fluorometer to measure the unwinding reaction and the ATPase activity in the same reaction. This powerful approach will enable researchers to record these activities simultaneously in the same reaction mixture.
Acknowledgments
We thank Behzad Rad and Stephen Kowalczykowski and members of the Kowalczykowski laboratory for expert help and advice with developing this protocol. We also thank John Hershey for his many insightful comments throughout the protocol development. We gratefully acknowledge Martin Webb for his generous gift of PBP-MDCC and Angelie Do and Enkhee Tuvshintogs for expert technical assistance. This work was supported by the US National Institutes of Health (NIH) through an NIHR01 grant (R01GM092927; to C.S.F), an NIH training grant (T32 GM007377; to K.F.) and an American Heart Association and Myocarditis Foundation predoctoral fellowship (B.C.A.).
Footnotes
AUTHOR CONTRIBUTIONS
A.R.O., K.F., B.C.A., E.P.B. and C.S.F. designed the study. A.R.O., K.F., and B.C.A. performed the experiments. A.R.O., K.F., B.C.A., E.P.B. and C.S.F. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Current opinion in structural biology. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annual review of biochemistry. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 3.Jankowsky A, Guenther UP, Jankowsky E. The RNA helicase database. Nucleic acids research. 2011;39:D338–341. doi: 10.1093/nar/gkq1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jankowsky E. RNA helicases at work: binding and rearranging. Trends in biochemical sciences. 2011;36:19–29. doi: 10.1016/j.tibs.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tani H, et al. Real-time monitoring of RNA helicase activity using fluorescence resonance energy transfer in vitro. Biochemical and biophysical research communications. 2010;393:131–136. doi: 10.1016/j.bbrc.2010.01.100. [DOI] [PubMed] [Google Scholar]
- 6.Ozes AR, Feoktistova K, Avanzino BC, Fraser CS. Duplex unwinding and ATPase activities of the DEAD-box helicase eIF4A are coupled by eIF4G and eIF4B. Journal of molecular biology. 2011;412:674–687. doi: 10.1016/j.jmb.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feoktistova K, Tuvshintogs E, Do A, Fraser CS. Human eIF4E promotes mRNA restructuring by stimulating eIF4A helicase activity. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:13339–13344. doi: 10.1073/pnas.1303781110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brune M, Hunter JL, Corrie JE, Webb MR. Direct, real-time measurement of rapid inorganic phosphate release using a novel fluorescent probe and its application to actomyosin subfragment 1 ATPase. Biochemistry. 1994;33:8262–8271. doi: 10.1021/bi00193a013. [DOI] [PubMed] [Google Scholar]
- 9.Brune M, et al. Mechanism of inorganic phosphate interaction with phosphate binding protein from Escherichia coli. Biochemistry. 1998;37:10370–10380. doi: 10.1021/bi9804277. [DOI] [PubMed] [Google Scholar]
- 10.Toseland CP, Webb MR. Fluorescence tools to measure helicase activity in real time. Methods. 2010;51:259–268. doi: 10.1016/j.ymeth.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 11.Yodh JG, Schlierf M, Ha T. Insight into helicase mechanism and function revealed through single-molecule approaches. Quarterly reviews of biophysics. 2010;43:185–217. doi: 10.1017/S0033583510000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hilario J, Kowalczykowski SC. Visualizing protein-DNA interactions at the single-molecule level. Current opinion in chemical biology. 2010;14:15–22. doi: 10.1016/j.cbpa.2009.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer CJ, Tomko EJ, Wu CG, Lohman TM. Fluorescence methods to study DNA translocation and unwinding kinetics by nucleic acid motors. Methods in molecular biology. 2012;875:85–104. doi: 10.1007/978-1-61779-806-1_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jankowsky E, Putnam A. Duplex unwinding with DEAD-box proteins. Methods in molecular biology. 2010;587:245–264. doi: 10.1007/978-1-60327-355-8_18. [DOI] [PubMed] [Google Scholar]
- 15.Bjornson KP, Amaratunga M, Moore KJ, Lohman TM. Single-turnover kinetics of helicase-catalyzed DNA unwinding monitored continuously by fluorescence energy transfer. Biochemistry. 1994;33:14306–14316. doi: 10.1021/bi00251a044. [DOI] [PubMed] [Google Scholar]
- 16.Zhang XD, Dou SX, Xie P, Wang PY, Xi XG. RecQ helicase-catalyzed DNA unwinding detected by fluorescence resonance energy transfer. Acta biochimica et biophysica Sinica. 2005;37:593–600. doi: 10.1111/j.1745-7270.2005.00084.x. [DOI] [PubMed] [Google Scholar]
- 17.Houston P, Kodadek T. Spectrophotometric assay for enzyme-mediated unwinding of double-stranded DNA. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:5471–5474. doi: 10.1073/pnas.91.12.5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belon CA, Frick DN. Monitoring helicase activity with molecular beacons. BioTechniques. 2008;45:433–440. 442. doi: 10.2144/000112834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aydin C, Mukherjee S, Hanson AM, Frick DN, Schiffer CA. The interdomain interface in bifunctional enzyme protein 3/4A (NS3/4A) regulates protease and helicase activities. Protein science : a publication of the Protein Society. 2013;22:1786–1798. doi: 10.1002/pro.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nature biotechnology. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 21.Hanson AM, Hernandez JJ, Shadrick WR, Frick DN. Identification and analysis of inhibitors targeting the hepatitis C virus NS3 helicase. Methods in enzymology. 2012;511:463–483. doi: 10.1016/B978-0-12-396546-2.00021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Putnam A, Jankowsky E. Analysis of duplex unwinding by RNA helicases using stopped-flow fluorescence spectroscopy. Methods in enzymology. 2012;511:1–27. doi: 10.1016/B978-0-12-396546-2.00001-2. [DOI] [PubMed] [Google Scholar]
- 23.Moreira BG, You Y, Behlke MA, Owczarzy R. Effects of fluorescent dyes, quenchers, and dangling ends on DNA duplex stability. Biochemical and biophysical research communications. 2005;327:473–484. doi: 10.1016/j.bbrc.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 24.Anderson BJ, Larkin C, Guja K, Schildbach JF. Using fluorophore-labeled oligonucleotides to measure affinities of protein-DNA interactions. Methods in enzymology. 2008;450:253–272. doi: 10.1016/S0076-6879(08)03412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu F, Putnam A, Jankowsky E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20209–20214. doi: 10.1073/pnas.0811115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajagopal V, Lorsch JR. ATP and GTP hydrolysis assays (TLC) Methods in enzymology. 2013;533:325–334. doi: 10.1016/B978-0-12-420067-8.00025-8. [DOI] [PubMed] [Google Scholar]
- 27.Young C, Karbstein K. Analysis of cofactor effects on RNA helicases. Methods in enzymology. 2012;511:213–237. doi: 10.1016/B978-0-12-396546-2.00010-3. [DOI] [PubMed] [Google Scholar]
- 28.Renosto F, Seubert PA, Segel IH. Adenosine 5′-phosphosulfate kinase from Penicillium chrysogenum. Purification and kinetic characterization. The Journal of biological chemistry. 1984;259:2113–2123. [PubMed] [Google Scholar]
- 29.Ingerman E, Nunnari J. A continuous, regenerative coupled GTPase assay for dynamin-related proteins. Methods in enzymology. 2005;404:611–619. doi: 10.1016/S0076-6879(05)04053-X. [DOI] [PubMed] [Google Scholar]
- 30.Shutes A, Der CJ. Real-time in vitro measurement of intrinsic and Ras GAP-mediated GTP hydrolysis. Methods in enzymology. 2006;407:9–22. doi: 10.1016/S0076-6879(05)07002-3. [DOI] [PubMed] [Google Scholar]
- 31.Pleiss JA, Derrick ML, Uhlenbeck OC. T7 RNA polymerase produces 5′ end heterogeneity during in vitro transcription from certain templates. Rna. 1998;4:1313–1317. doi: 10.1017/s135583829800106x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic acids research. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vicens Q, Gooding AR, Duarte LF, Batey RT. Preparation of group I introns for biochemical studies and crystallization assays by native affinity purification. PloS one. 2009;4:e6740. doi: 10.1371/journal.pone.0006740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nilsen TW, Rio DC, Ares M., Jr High-yield synthesis of RNA using T7 RNA polymerase and plasmid DNA or oligonucleotide templates. Cold Spring Harbor protocols. 2013;2013 doi: 10.1101/pdb.prot078535. [DOI] [PubMed] [Google Scholar]
- 35.Rogers GW, Jr, Richter NJ, Lima WF, Merrick WC. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. The Journal of biological chemistry. 2001;276:30914–30922. doi: 10.1074/jbc.M100157200. [DOI] [PubMed] [Google Scholar]
- 36.Rio DC. Expression and purification of active recombinant T7 RNA polymerase from E. coli. Cold Spring Harbor protocols. 2013;2013 doi: 10.1101/pdb.prot078527. [DOI] [PubMed] [Google Scholar]
- 37.Green MR, Sambrook J, Sambrook J. Molecular cloning : a laboratory manual. 4. Cold Spring Harbor Laboratory Press; 2012. [Google Scholar]
- 38.Sambrook J, Maniatis T, Fritsch EF. Molecular cloning : a laboratory manual. 2. Cold Spring Harbor Laboratory; 1989. [Google Scholar]