Abstract
Neuropathic pain is a complex chronic condition characterized by a wide range of sensory, cognitive, and affective symptoms. A large percentage of neuropathic pain patients are also afflicted with depression and anxiety disorders – a pattern that is reliably replicated in animal models. Furthermore, clinical and preclinical studies indicate that chronic pain corresponds with adaptations in several brain networks involved in mood, motivation, and reward. Chronic stress is also a major risk factor for depression. We investigated whether chronic pain and stress affect similar mechanisms, and whether chronic pain can affect gene expression patterns known to be involved in depression. Using two mouse models for neuropathic pain and depression (spared nerve injury (SNI) and chronic unpredictable stress (CUS)) we performed next generation RNA-sequencing and pathway analysis to monitor changes in gene expression in the nucleus accumbens (NAc), the medial prefrontal cortex (mPFC), and the periaqueductal grey (PAG). These three brain regions are implicated in the pathophysiology of depression and in the modulation of pain. In addition to finding unique transcriptome profiles across the three brain regions,.we identified a substantial number of signaling pathway-associated genes with similar changes in expression in both SNI and CUS mice. Many of these genes have been implicated in depression, anxiety and chronic pain in patients. Our study provides a resource of the changes in gene expression induced by long-term neuropathic pain in three distinct brain regions and reveals molecular connections between pain and chronic stress.
Introduction
Chronic pain is a debilitating condition affecting over 100 million Americans (1), with more thanhalf showing comorbid depression and anxiety (2–4). Several recent studies using rodent models of chronic pain reveal adaptations in mood and stress-related brain regions with corresponding effects in nociceptive, motivation, anxiety, and depression related behaviors, as well as analgesic efficacy (5–11). Moreover, neuroimaging studies from chronic pain patients show altered activity in the nucleus accumbens (NAc) and the medial prefrontal cortex (mPFC) (12–14), two brain regions that are part of the brain reward center and have documented roles in the pathophysiology of depression (15). The NAc displays connections with multiple brain regions, including the ventral tegmental area, amygdala, thalamus, and the mPFC, and shows altered activity in murine models of stress-induced depression as well as upon noxious stimulation or relief from pain (5, 16–21). Notably, both chronic stress (22) and nerve injury (23) result in altered dendritic spine morphology in the mPFC, whilst optogenetic activation of mPFC-NAc projections modulates sensory and affective symptoms of neuropathic pain (6), and induces resilience to chronic social defeat stress (24). Accordingly, several studies report plasticity-related alterations in the PFC of rodents in models of neuropathic pain (21, 25–27).
The periaqueductal grey (PAG) plays a critical role in descending modulation of pain (28, 29), and mediates opioid analgesia (28, 30). Recent findings have also shown that chronic pain corresponds with greater functional connections between the ventromedial PFC and PAG in humans (31), and robust mPFC-PAG projections have been confirmed in rodents (32). Rodent models of chronic pain and stress induce plasticity within the PAG (33–35), and PAG activation modulates anxiety-like behaviors (36, 37).
Although chronic stress and chronic pain conditions have been shown to correspond with plasticity-related changes in the mPFC, NAc, and PAG, few studies have investigated network-wide cellular and molecular adaptations within these areas in response to long-term pain. Critically, several studies using rodent chronic pain models report that depression-like behaviors do not appear until at least six weeks after-injury (7, 38–40), and thus an understanding of the molecular adaptations in mood-related brain regions at these later time points is of paramount importance. Here, we used the murine spared nerve injury (SNI) model of neuropathic pain (41, 42), along with next generation RNA-sequencing (RNAseq) to monitor gene expression changes in mPFC, NAc, and PAG tissue of adult mice two and a half months after nerve injury. Our data indicate that distinct transcriptional profiles emerge across these brain regions in correspondence with long-term neuropathic pain, but show some overlapping effects in signaling pathways and biological functions. We demonstrate significant overlap between differentially expressed genes identified in this data set, and genes implicated in depression, anxiety, and pain from human postmortem or mutant mouse studies. Last, we confirm the correspondence of genes to depression by comparing the expression pattern in a distinct mouse model of depression, chronic unpredictable stress (CUS) (43).
Results
Spared nerve injury leads to nociceptive sensitization and enhances anxiety and depression related behaviors
We applied the spared nerve injury (SNI) model or sham surgery in adult (8 week old) male C57BL/6 mice (Fig 1A), which has been shown to produce typical symptoms associated with chronic pain and anxiety-like or depression-like behaviors (39, 41, 44). Mice exposed to SNI showed robust expression of mechanical allodynia, as indicated by decreases in mechanical thresholds in the Von Frey test that persisted for two months (Fig. 1B). In addition, 2 months after surgery, we used the elevated plus maze (EPM) and open field (OF) tests to assess anxiety-related exploratory behavior (45). SNI mice showed significantly less exploration of the open arms in the EPM (Sham: 12 ± 2.5 %, SNI: 4.2 ± 0.7 %; Fig 1C), and spent significantly less time in the center area of the OF compared to sham mice (Sham: 12.4 ± 2.3 %, SNI: 5.9 ± 1.7 %; Fig 1D). We next assessed if SNI affected depression-like behaviors through the sucrose consumption and forced swim tests (FST) (46). At the 9 week time point, SNI mice showed significantly reduced preference for sucrose consumption (Sham: 77 ± 1.4 %, SNI: 58 ± 3.9 Fig 1E), and also showed significantly longer duration of immobility in the FST (Sham: 128.14 ± 10.6 s, SNI: 167.29 ± 6.5 s; Fig 1F). Importantly, a 60 min assessment of locomotor activity showed no difference between the groups at 2 months (Fig. S1). These data show that SNI induces persistent nociceptive sensitization and long-term enhancements in anxiety and depression-like behaviors.
Fig 1. The SNI model of neuropathic pain induces mechanical allodynia and enhances anxiety- and depression-like behaviors. (A).
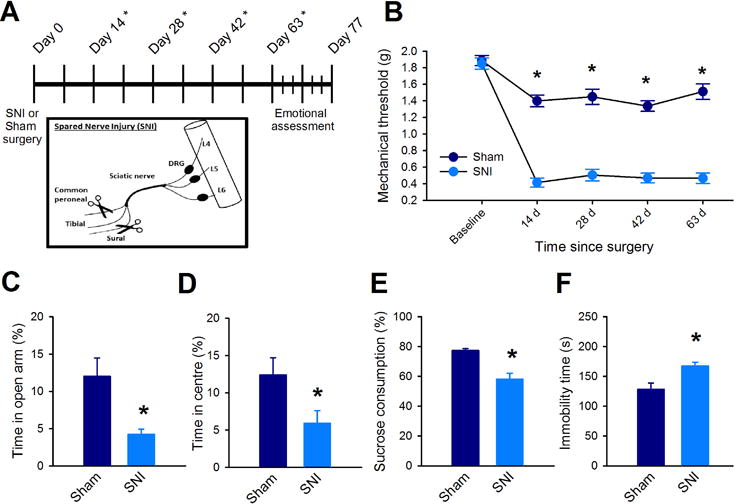
Top: experimental timeline; * denotes days where mechanical nociceptive thresholds were measured. Bottom: Spared nerve injury model (SNI). (B) Mechanical allodynia 50% mechanical thresholds were determined using Von Frey filaments before surgery (baseline) and every 2 weeks thereafter. (n = 16 mice per group, F(1,120) = 155, p < 0.001; two-way ANOVA and Holm-Sidak adjustment for multiple comparisons). (C to F) Anxiety-like behaviors assessed in an elevated plus maze (C; n = 9 mice per group, t(16) = 3.03, p = 0.008; unpaired t-test), in an open field test (D; n = 9 mice per group, t(16) = 2.24, p = 0.04; unpaired t-test), for sucrose consumption (E; n = 9 mice per group, t(16) = 4.62, p < 0.001; unpaired t-test), and in a forced swim test (F; n = 9, t(16) = −3.15, p = 0.004; unpaired t-test). Data in (B to F) are mean ± SEM, * p < 0.05.
Long-term neuropathic pain corresponds with distinct altered gene expression patterns in the mPFC, NAc, and PAG
We next investigated if long-term SNI was associated with changes in gene expression in brain regions known to be involved in anxiety and depression, and thus performed next generation RNA-sequencing (RNAseq) on mPFC, NAc, and PAG tissue collected from adult male mice 2.5 months after SNI or sham surgeries. Our RNAseq design maximized the trade-off between biological replicates and sequencing depth to obtain high statistical power (47), and sequenced 6 biological replicates per brain region, per group (Fig. S2). RNAseq read alignment and differential analysis were performed by using the Voom Limma and TopHat packages. We achieved an average mapping depth of 58.9 ± 20.8 M reads, with a mapping rate of 92 ± 0.33 %. We quantified gene expression levels in each brain region and compared levels from Sham and SNI groups to determine SNI-induced differentially expressed genes. Long-term SNI corresponded with robust changes in gene expression and distinct patterns of upregulated vs. downregulated genes (mPFC: upregulated: 179, downregulated: 204; NAc: upregulated: 135, downregulated: 691; PAG: upregulated: 6539, downregulated: 1225).
We next characterized whether differentially expressed genes showed patterns of co-expression across the three brain regions by employing the rank-rank hypergeometric overlap test (RRHO) (48, 49). This test characterizes the patterns and significance of overlap between gene expression profiles by comparing ranked lists of differentially expressed genes in the mPFC, NAc, and PAG. Although some co-regulation can be seen across all three brain regions, the NAc and PAG show a remarkably robust pattern of co-upregulated genes (Fig 2A). Accordingly, comparisons of the gene expression patterns between the NAc and PAG show stronger Spearman rank correlation coefficient values (rho = 0.547), compared to comparisons between the NAc and mPFC (rho = 0.332), and between the mPFC and PAG (rho = 0.195). Indeed 3594 genes were found to be upregulated by SNI in both the NAc and PAG brain regions of mice (Fig 2B). We examined the gene ontology of these specific genes, and found that several biological functions are implicated by these co-upregulated genes, including intracellular signaling cascade, regulation of biosynthesis, transcription, phosphorylation, and metabolic processes. Furthermore, histone deacetylases (HDACs) 4, 5, and 7 were found to be implicated in 40% of the biological processes represented by the co-upregulated genes. A protein-protein interaction map of the genes implicated in the regulation of biosynthesis showed that HDAC5 in particular interacted with a robust number of proteins coded by genes in this gene ontology group (Fig 2C), and several studies have linked its expression with stress-induced depression and antidepressant effects (7, 50, 51). In a separate group of mice, using real-time quantitative PCR, we confirmed that HDAC5 is upregulated in the NAc (Sham: 1.00 ± 0.14 FC, SNI: 3.98 ± 1.03 FC) and PAG (Sham: 0.98 ± 0.06 FC, SNI: 1.15 ± 0.04 FC) two and a half months after injury (Fig 2D).
Fig 2. The NAc and PAG show robust co-upregulation patterns of genes in correspondence with long term neuropathic pain. (A).
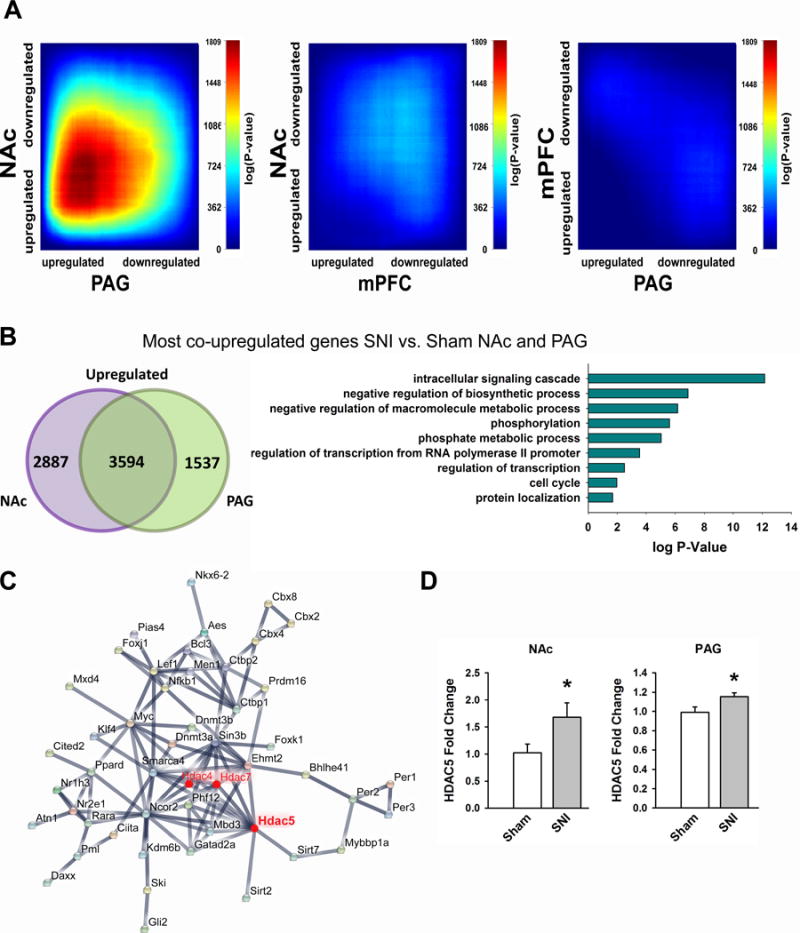
RRHO maps comparing the overlap of SNI-induced differentially expressed genes. Threshold free comparisons of up- and down- regulated genes are shown between NAc and PAG (left), NAc and mPFC (middle), and mPFC and PAG (right). (B) Venn diagram showing number of co-upregulated genes between NAc and PAG (left); enriched gene-ontology terms of co-upregulated genes (right). (C) Protein-protein interaction maps of enriched GO term negative-regulation of biosynthesis. (D) RT qPCR validation of SNI-induced HDAC5 upregulation in the NAc (n=6 mice per group, t(11) = −2.24, p = 0.047; unpaired t-test) and PAG (n = 16 mice per group, t(30)= −2.32, p = 0.027; unpaired t-test). Data are mean ± SEM are reported, * p < 0.05.
HDAC5 plays a critical role in neuropathic-pain states
Given the implications of HDAC5 in depression-related behaviors and antidepressant effects, we next sought to determine if a genetic mutant lacking the expression of HDAC5 (HDAC5 KO) showed altered behavioral effects to long-term SNI. We exposed HDAC5 KO and HDAC5 WT mice to SNI and found both groups displayed robust mechanical allodynia in response to SNI (Fig 3A), with no differences detected between HDAC5 KO and HDAC5 WT in either SNI or sham treated groups. In contrast, emotion related behavior differed significantly in response to long-term SNI, with HDAC5 KO mice displaying significantly reduced SNI-induced depression- and anxiety-like behaviors compared to HDAC5 WT mice. Nine weeks after surgery, sham treated HDAC5 WT and HDAC5 KO showed similar sucrose preference values (HDAC5 WT: 75 ± 3.4 %, HDAC5 KO: 72 ± 3.2 %), whereas SNI treated HDAC5 WT mice showed significantly lower sucrose preference values than HDAC5 KO mice (HDAC5 WT: 49 ± 5.9 %, HDAC5 KO: 63 ± 2.4 %; Fig 3B). In a similar manner, HDAC5 KO mice also showed resiliency to SNI-induced increases in anxiety-like behavior. In the EPM, although sham treated HDAC5 WT and HDAC5 KO mice showed equal levels of exploratory behavior in the open arm (HDAC5 WT: 13.41 ± 2.5 %, HDAC5 KO: 13.49 ± 0.8 %), long-term SNI reduced exploratory behavior only in HDAC5 WT mice, whereby HDAC5 KO mice showed significantly greater levels of exploratory time in the open arm compared to HDAC5 WT (HDAC5 WT: 5.03 ± 1.2 %, HDAC5 KO: 14.97 ± 4.1 %; Fig 3C). Likewise in the open field, SNI treated HDAC5 KO mice showed significantly more exploration in the center area compared to HDAC5 WT mice (HDAC5 WT: 5.11 ± 1.6 %, HDAC5 KO: 10.86 ± 1.9 %; Fig 3D), whereas groups exposed to sham surgery showed similar levels (HDAC5 WT: 8.56 ± 1.7 %, HDAC5 KO: 9.78 ± 1.5 %). Antidepressants are commonly prescribed for chronic pain symptoms and previous work indicates that HDAC5 modulates antidepressant effects in chronic stress models (50).Therefore, we next tested the possibility that mice lacking HDAC5 may respond differently to chronic treatment with duloxetine, a serotonin and norepinephrine reuptake inhibitor that has anti-allodynic properties (52). Three weeks after SNI surgery, HDAC5 WT and HDAC5 KO mice showed similar levels of mechanical allodynia as evidenced by very low 50% mechanical thresholds (HDAC5 WT: 0.37 ± 0.10 g, HDAC5 KO: 0.43 ± 0.09 g). We then commenced chronic duloxetine treatment (5mg/kg every 12h), and tested their nociceptive responses to Von Frey filaments every 5 days. Remarkably, HDAC5 KO mice showed a significantly faster and stronger anti-allodynic response to chronic duloxetine that was evident as early as after 10 days of treatment (Fig 3E). A two-way repeated measures ANOVA revealed a significant difference between groups. Together, these data indicate that HDAC5 modulates the emotional effects induced by long-term SNI, and is involved in the anti-allodynic effects of antidepressants.
Fig 3. Genetic elimination of HDAC5 attenuates emotional but not nociceptive effects of long-term neuropathic pain, and enhances antidepressant efficacy. (A).
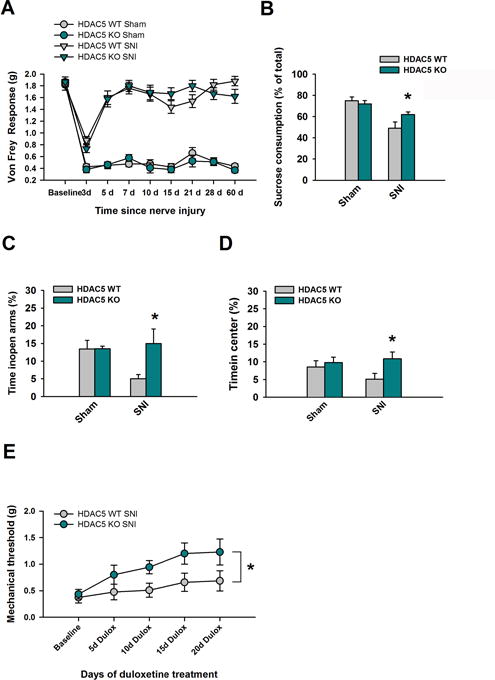
50% mechanical thresholds were compared between HDAC5 WT and HDAC5 KO mice before sham or SNI surgery (baseline) and up to 60 days thereafter. Measurements were made using Von Frey filaments. No significant difference was observed between groups at any time-point (F(1,324) =1.11, p = 0.355), and there was no interaction between genotype, typed of surgery, and time (F(8, 324) = 1.36, p = 0.214); n = 10 mice per group. SNI treated HDAC5 WT and HDAC5KO mice were monitored (B) in the sucrose preference test (n = 10 mice per group, t(18) = −2.2, p = 0.041; unpaired t-test). (C) in the EPM test (n = 10 mice per group, t(18) =−2.3, p = 0.033; unpaired t-test). (D) in the open field test (n = 10 mice per group, t(18) = −2.3, p = 0.035; unpaired t-test). (E) and in the Von Frey assay following chronic duloxetine (5 mg/kg every 12 hours; n = 7 mice per group, F(1,48) = 9.0; P = 0.01; two-way ANOVA and Holm-Sidak adjustment for multiple comparisons). Data are mean ± SEM, *p < 0.05
Gene expression patterns in the mPFC, NAc, and PAG that undergo robust fold change after long-term neuropathic pain
We next sought to identify the genes that undergo the most robust changes in expression as a consequence of chronic neuropathic pain, and applied a strict threshold to generate lists of genes that showed a differentially expressed fold change (FC) of 1.50 or greater and only included genes with p-values of 0.05 or less (table S1). With these cutoffs, hierarchical clustering revealed distinct overall patterns in gene expression profiles across the three brain regions (Fig 4A). The patterns of upregulated versus downregulated genes varied across all three brain regions (mPFC: upregulated genes: 36, downregulated: 53; NAc: upregulated genes: 13, downregulated genes: 110; PAG upregulated genes: 724, downregulated genes: 71; Fig 4B). We next used VennPlex (53) to group differentially expressed genes from all three brain regions to assess the commonalities and differences based on their corresponding expression values. Remarkably, little overlap in regulated genes was observed, with only 1 upregulated gene (Capn11) and 1 downregulated gene (Cyp2e1) in common across all three brain regions (Fig 4C).
Fig 4. Long-term neuropathic pain corresponds with robust and distinct transcriptome changes in the mPFC, NAc, and PAG. (A).
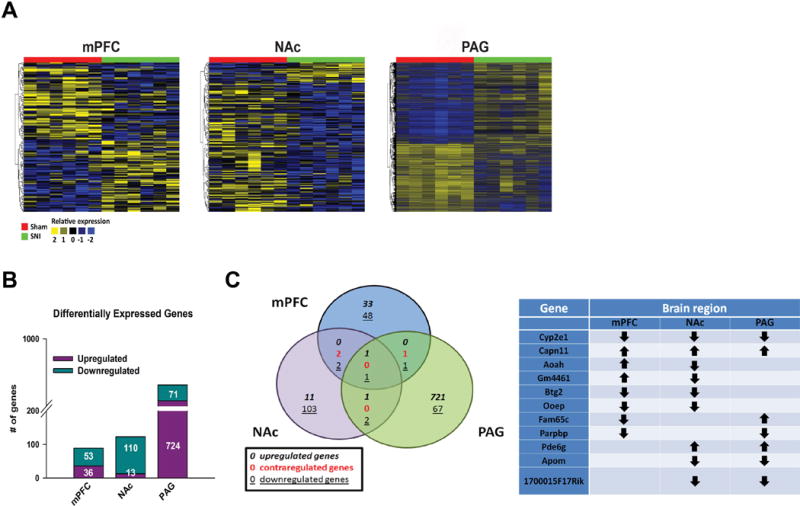
RNA sequencing experimental timeline and design comparing mPFC, NAc, and PAG from mice exposed to SNI or sham. 6 biological replicates were used per condition for each brain region, composed of 2 pooled samples/biological replicate. (B) Hierarchical cluster heat maps of the mPFC, NAc, and PAG showing relative expression of genes across sham and SNI samples. (C) Numbers of significantly upregulated or downregulated differentially expressed genes in SNI vs. Sham mice in mPFC, NAc, and PAG brain regions. Genes reported showed at least log2(1.50) fold change compared to sham. (D) Left: VennPlex assessment of gene expression overlaps in the mPFC, NAc, and PAG in SNI vs. sham. Right: Common differentially expressed genes across brain regions, data is expansion of Venn diagram on left. Down arrow = down-regulation; Up arrow = up-regulation.
Gene expression patterns in the mPFC, NAc, and PAG reflect common biological functions and signaling pathways
We next sought to characterize the SNI-induced changes in gene expression may have on biological networks and neural function. We used the Ingenuity Pathway Analysis (IPA), allowing us to compare our differentially expressed data set with over 1,480,000 experimentally observed interactions between molecules (54). Using the Fisher’s exact test, network scores are generated based on the number of molecules they contain, with higher scores representing lower probabilities of finding the observed genes by chance alone. The total number of network eligible differentially expressed genes from our dataset varied across the three brain regions, with the PAG displaying the largest amount (mPFC: 16, NAc: 21, and PAG: 238). Table 1 displays the top biological networks that contain more than four differentially expressed genes. As expected from the distinct pattern of differentially expressed genes observed, the mPFC, NAc, and PAG showed unique molecular networks. Nevertheless, the top network represented in all three brain regions involved a network implicated to affect behavior, and both the NAc and PAG had a significant overrepresentation of genes corresponding to neurological disease. We next performed IPA’s causal network analysis which identifies upstream molecules experimentally shown to control the expression of the differentially expressed genes from our dataset. This analysis recognizes regulators known to exert direct relationships as well as molecules through which a master regulator modulates gene expression. Activation states (activated or inhibited) are determined from z-scores that indicate if there is a significant match between the predicted and observed gene expression pattern, only z-scores of ≥2 or ≤ −2 are considered to be significant predictors of activity. As with the biological networks, distinct master and participating regulators were observed across the three brain regions, but only molecules from the mPFC and PAG showed relative expression levels that could be considered statistically significant (Table 2). We next used IPA to assess the overlap between differentially expressed genes in the mPFC, NAC, and PAG with genes sets known to be involved in specific signaling pathways. Despite the stringent cutoffs, a vast amount of signaling pathways were implicated in each brain region (mPFC: 78; NAc: 142; PAG: 189). Nineteen canonical signaling pathways showed significant levels of differentially expressed genes in at least two brain regions, including 5 pathways which were affected in all three brain regions: cAMP-mediated, G-protein coupled receptor, IGF-1, pregnenolone biosynthesis, and glucocorticoid receptor signaling pathways (Fig 5A). Thus, while SNI promotes unique adaptations in gene expression in the mPFC, NAc, and PAG, some of these adaptations appear to impact signal transduction networks that are similarly regulated in all three regions examined.
Table 1. Top biological networks corresponding to differentially expressed genes in the mPFC, NAc, and PAG.
Networks are composed of molecules and differentially expressed genes experimentally observed to interact with each other and scored relative to all molecules they are known to be connected to. Using a Fisher’s exact test, network scores are generated based on the number of molecules they contain, with higher scores representing lower probabilities of finding the observed genes in a randomly selected network by chance alone. Genes in bold indicate SNI-induced differentially expressed genes in mPFC, NAc, or PAG. Only networks that include more than one differentially expressed gene are presented.
| Brain region | Network | Molecules in network | Score | # of molecules in dataset | Top diseases and functions |
|---|---|---|---|---|---|
| mPFC | 1 | BATF2,BTG2,CALCB,CDH1,CREB1,CREM,CXCR4,CXCR5,CYR61, DRD1,DUSP1, EGR1,F2RL1, FOS, GCG, GLP1R, GNRH1, HTT, IFI16, IFNG, IL4, JUN, MAPK10, NGF, NOS2, NPAS4, NPY, PSEN1, RAC1,STAT1,SYN1,TAC1,TFAP2A,TP73,VEGFA | 21 | 11 | Behavior, Cell Death and Survival, Cellular Development |
| 2 | 5-hydroxytryptamine, APP, ASCL2, BCL2L1,BDNF, CARTPT, CNR1, corticosterone, CREB1, CRH, CRHR2, CXCR4, cyclic AMP, CYP2E1, dopamine, EGR1, EIF2AK2, ethanol, FOS, GABA, GRIA1, GRIN1, GRIN2A, GRIN2B, GSK3B, HDAC6, LEP, MAPT, NR3C1, PIK3R1, POMC, SLC1A7, SP1, TGFB1, TPH2 | 6 | 4 | Behavior, Nervous System Development and Function, Amino Acid Metabolism | |
|
| |||||
| NAc | 1 | ACPP, adenosine, Agtr1b,APP,beta-estradiol, BTG2, Ca2+, CCL11, CCL3L3, CCND1, CHRNA4, CHRNA6, CHRNB3, CYP2E1, E2F7, ENPP3, ethanol, FOS, IL6, L-dopa, MAPK8, MPZ, NPTX2, PIEZO2, POU1F1, POU2AF1, PTGDR, SDC1, SHISA3, SUMO2, TGFB1, TGM1, THBS1, TRPV4, ZFHX3 | 39 | 18 | Behavior, Neurological Disease, Nucleic Acid Metabolism |
|
| |||||
| PAG | 1 | ARHGEF19, C2CD2L, CASKIN2, FES, JPH3, KDM4B, L-dopa, MAPT, NAGLU, NPHP4, PALM, RGS14, ROMO1, SLC2A6, SPINK8, TMC6, TRIB3, ZNF76 | 20 | 16 | Neurological Disease, Psychological Disorders, Behavior |
| 2 | adenosine, ATN1, ATP13A2, CBFB, CC2D1A, CDKN1B, Cdkn1c, CYBA, DRD2, EGFR, FABP7, FBXW7, Fxyd2, IFI16, ITGB4, JUP, LINGO1, MAP3K10, MAPK8IP3, Mt3, NEUROD1, NKX6-2, NOTCH1, NTRK2, PITPNM1, POSTN, PTPN22, PVALB, SIM1, SOCS2, SOX10, SPP1, ST8SIA5, TCF3, Tmsb4x (includes others) | 16 | 18 | Nervous System Development and Function, Cell Death and Survival, Gene Expression | |
| 3 | AADAT,acetylcholine,ACHE,ARHGEF7,BAX,BBC3,CHAT,CYCS,EGL N2,EPAS1, FAM173A, FOXO3, GAPDH, GIT1, GRIK2, HIF1A, HSPB1, HTT, IGFBP5, ITGB1, KCNJ4, kynurenic acid, MAP2K4, MAP3K5, MAPK10, MYH14, PER1, PRKCD, SGK1, SLC18A3, SPHK1, TSPO, VDR, VIPR2, VPS18 | 13 | 16 | Neurological Disease, Psychological Disorders, Cell Death and Survival | |
| 4 | ADORA2A, ALDOC, ATP1A2, Atp5e, C1QA, C1QC, CKB, CNTFR, CSF1R, DSTN, Erdr1, FBL, GJA1, GJC2, HBA1/HBA2, Hbb, HES5, HPCA, JRK, MAP2K2, Mt2, MT-CO1, NAPB, NPAS1, PGAM1, PRDX6, PSEN1, RELN, Rplp1 (includes others), SOCS1, SOD1, STAB1, STAT4, TUBB2A, TUBB4A | 13 | 16 | Organismal Injury and Abnormalities, Neurological Disease, Cell- To-Cell Signaling and Interaction | |
| 5 | AATK, ABL1, AGAP2, Ap2b1, ASIC3, CDK5, CSNK1G2, DCTN1, DDAH2, DLG4, DLGAP3,DMWD, DOT1L, EIF4E, EIF4EBP1, FMR1, HOXD10, KCNA2, KIRREL2, MAOB, MAPK1, MTOR, PRKCH, PURA, RAPGEF3, RARA, RPS3, RPS26, RPS6KB1, RPTOR, SLC9A3R2, SND1, STX1B, VGF, ZFPM1 | 13 | 16 | Cell Morphology, Cellular Assembly and Organization, Cellular Development | |
| 6 | ABCA1, ADAM10, APOE, ARHGAP33, BAD, CREB1, CREM, CSRNP1, DFNB31, EPB41, GADD45G, GAL, GRIA3, GRIK2, GRM2, HIC1, HMGCR, IER2, INHBA, INS, IRS2, JUNB, L-glutamic acid, LDLR, melatonin, MPP1, MYO15A, NCDN, NFKBID, NR4A1, PPP1R13L, RFX1, SIRT1, SLC17A8, SLC1A1 | 13 | 16 | Neurological Disease, Behavior, Lipid Metabolism | |
| 7 | APEX1, ARPC1B, BAX, BCL2, BCL2L12, CCND1, CDKN1A, CDKN1B, CDKN2A, DDR1, FAU, GAPDH, IGF1, inosine, ITGB1, LRRN3, MGMT, MMP2, MYC, MYCN, PLEC, PTEN, RARA, RPL8, RPLP2, RPS15, RUSC2, SIM2, SLC2A1, SP1, TIMP2, TP53, VIM, ZBTB17, ZYX | 12 | 15 | Cell Cycle, Cancer, Organismal Injury and Abnormalities | |
| 8 | ACE, AGT, AGTR1, BAX, BCL2, BCL2L1, beta-estradiol, CDC42EP1, EPHA2, ESR1, ESR2, FKBP8, GNRH1, GRIN2C, HCRT, IGF2, IGFBP2, IGFBP6, KISS1, KLF2, LHB, MAP1S, MIF, NOS3, OPRK1, PPARG, PPARGC1A, PRL, progesterone, PSD, PTGDS, RABEP2, SFN, TJP1, VEGFA | 12 | 15 | Renal and Urological System Development and Function, Reproductive System Development and Function, Cardiovascular Disease | |
| 9 | BAIAP2, CASC5, CASK, CASKIN1, CCND1, CDC6, Crip2, DLG2, EPOR, FAM83D, FOSB, GDNF, GRIN2B, HDAC1, HTR1A, ITPKA,L- dopa, LIN7B, LMNA, LMNB2, LZTS3, MIB2, NT5E, NTRK3, PCLO, PHOX2B, SATB1, SHANK3, SNCA, SNCB, TH, TLE1, TPPP3, VAMP2, YIF1B | 12 | 15 | Cell Death and Survival, Behavior, Cell-To-Cell Signaling and Interaction | |
| 10 | acetylcholine, ACHE, ACTA1, AP2M1, APP, BACE1, DPYSL2, ELN, FBXO2, FGF2, FLNA, FZR1, GIPC1, GNA12, GNAS, GRIN1, ITGB1, MAP1LC3A, MAP2K4, MDK, Mt1, MT-CO1, NF1, NGFR, NRP1,PAWR, RAC1, RHOA, RTN4R, SLC5A7, SOD1, SPHK2, TFEB, TIMM23B, TOMM40 | 10 | 14 | Nervous System Development and Function, Cellular Assembly and Organization, Cellular Development | |
| 11 | acetic acid, ADRA1D, ATF3, CARTPT, CHODL, COL18A1, corticosterone, CRH, CRY1, CRY2, cyclic GMP, F2, FGF2, formaldehyde, FOS, FOXO1, FOXO3, GNAI2, GNAO1, GPRIN2, GRM2, GRM4, histamine, JUNB, LIF, MATK, nitric oxide, NPPC, OPRM1, PER1, PITX2, SOCS3, SPRY2, SRC, UCN | 10 | 14 | Amino Acid Metabolism, Small Molecule Biochemistry, Behavior | |
| 12 | ACTC1, ascorbic acid, BNC1, CARTPT, CNR1, COL5A1, CRABP1, CYP2E1, DAGLA, EPHA8, ethanol, FOXJ1, HOXC8, HOXC9, IRX6, ISL1, JAK3, LHX2, LILRB3, MAB21L1, mir-9,MYL1, NEUROD4, Nrgn, PAPPA2, PIRT, POU4F1, POU4F3, PPP1R1C, RBM24, REST, RTN4RL2, RUNX3, Speer1-ps1, ZNF335 | 10 | 14 | Embryonic Development, Organismal Development, Cellular Development | |
| 13 | BCKDK, BDNF, C5AR1, CALB2, CCL5, CD14, CSPG4, EIF2AK4, FOSB, FZD2, GAL3ST1, GJA1, GNAO1, GNB2, GRIP1, IFNG, LAMB2, LAMC1, MAP2K1, MBP, MOG, MYRF, PACSIN3, PICK1, PLP1, PNOC, PPARD, PPP1R1B, PRKCZ, RTN4, SERPINH1, SIRT1, TLR4, UNC5A, YWHAG | 9 | 13 | Nervous System Development and Function, Cellular Movement, Neurological Disease | |
| 14 | AMHR2, ARL4D, BCL3, CRLF2, GALK1, GRHPR, JAG2, KMT2D, MED1, NUDT14, TCOF1 | 9 | 8 | Auditory and Vestibular System Development and Function, Carbohydrate Metabolism, Cellular Development | |
| 15 | ACADS, ARHGEF1, ARX, ascorbic acid, CARTPT, cyclic AMP, DBH, DIO3, dopamine, DTNBP1, FLYWCH2, FST, GABRB3, GLO1, GPR37, GPR171, histamine, HRH3, INHA, INHBA, INHBB, KCNA1, MAOA, MAOB, melatonin, norepinephrine, PARK2, PARK7, PCSK4, PCSK1N, PI4K2A, PTPN5, SMAD7, SPTBN4, TNFRSF25 | 8 | 12 | Cell-To-Cell Signaling and Interaction, Drug Metabolism, Molecular Transport | |
| 16 | ARHGEF7, ATF3, AVP, BAX, BMP2, BMPR1B, cholesterol, CTNNB1, CXCL8, DUSP1, EMX1, FURIN, GAL, GDNF, GHRH, GHRL, GRIK2, HSPG2, ID3, IL1A, JUN, LZTS2, MAP3K11, MAPK10, MID1, NCOR2, NGF, NGFR, Npy4r, NR1H2, quinolinic acid, SCRIB, SHH, SREBF1, VEGFA | 7 | 11 | Cell Death and Survival, Tissue Morphology, Nervous System Development and Function | |
Table 2. Causal networks composed of.
upstream molecules known to control the expression of genes showing differential expression in the mPFC and PAG. No statistically significant causal networks were identified for the NAc. Master regulators: predicted upstream molecules at the root of the causal network. Participating regulators: molecules through which a master regulator modulates gene expression in target genes. Data presented have p-value overlap of < 0.05, calculated using Fisher’s exact test.
| Brain region | Master Regulator | Molecule Type | Participating regulators | Predicted Activation State | Activation Z- score | Target molecules in dataset |
|---|---|---|---|---|---|---|
| mPFC | melatonin | Endogenous chemical | BAX,CREB1, JUN, melatonin | Activated | 2.236 | BTG2,CDH1,CYR61,DUSP1, NPAS4 |
| IFNG | cytokine | CREB1,IFNG, MAPK1,RAC1 | Inhibited | −2.121 | BATF2,BTG2,CDH1,CYR61 DUSP1,IFI16,NPAS4 | |
| IL15 | cytokine | CREB1,IL15, MAPK1, STAT1 | Inhibited | −2.449 | BATF2,BTG2,CYR61,DUSP1, IFI16,NPAS4 | |
| NTRK1 | kinase | CREB1,JUN, NTRK1 | Inhibited | −2.236 | BTG2,CDH1,CYR61,DUSP1, NPAS4 | |
| K+ | endogenous chemical | CREB1,JUN,K+ | Inhibited | −2.236 | BTG2,CDH1,CYR61,DUSP1, NPAS4 | |
| FGF2 | growth factor | CREB1,FGF2, RAC1, STAT1 | Inhibited | −2.646 | BTG2,CDH1,CYR61,DUSP1, NPAS4 | |
| NTRK2 | kinase | CREB1,NTRK2, RAC1,SRC | Inhibited | −2.236 | BTG2,CDH1,CYR61,DUSP1, NPAS4 | |
|
| ||||||
| PAG | inosine | endogenous chemical | inosine | Activated | 2.449 | ARPC1B,C1QA,C1QC, EIF4EBP1,Mt1,TSPO |
| CREM | transcription regulator | CREM | Activated | 2.236 | CSRNP1,IRS2,JUNB, NFKBID,PER1 | |
| PRKCH | kinase | MTOR,PRKCH, STAT3 | Activated | 2.121 | DCTN1,DDAH2,HBA1/HBA2,HES5, HSPB1,MBP,NOTCH1,SOCS3 | |
Fig 5. SNI-induced gene expression patterns indicated common signal transduction pathways are affected in the mPFC, NAc and PAG and show robust overlap with genes previously implicated in anxio-depressive states and pain. (A).
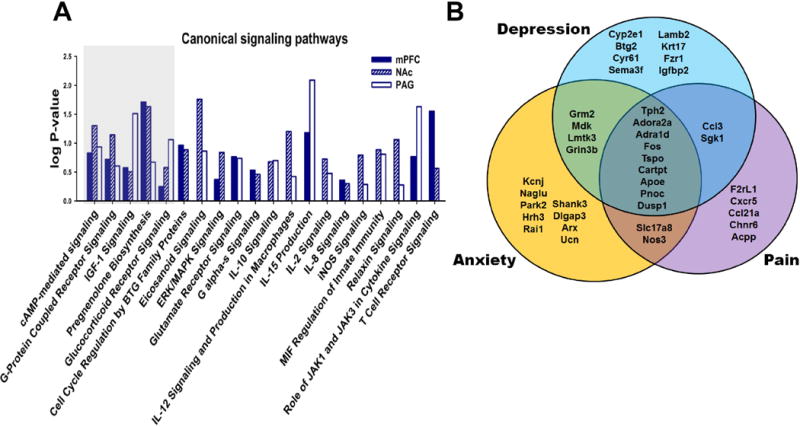
Comparison of IPA derived canonical signaling pathways affected by gene expression patterns in mPFC, NAc, and PAG samples from SNI vs. sham. (B) Overlap of differentially expressed genes from present RNAseq study (mPFC, NAc, and PAG combined) to genes previously implicated in depression, anxiety, and pain, mostly from post-mortem human brain samples or mutant KO mouse behavioral studies.
Our data, as well as several other studies demonstrate that SNI does not only produce long-term nociceptive sensitization, but also promotes anxiety and depression-like behaviors. We next took advantage of IPA’s extensive database to determine the extent of overlap between the differentially expressed genes in our study and genes identified in the literature as being implicated in pain, anxiety or depression. Remarkably, a large subset of differentially expressed genes across all three brain regions overlapped with IPA identified molecules implicated in the literature in depression, anxiety, and pain-like conditions (Fig 5B and Table 3). A total of 39 genes were identified, including 8 genes that had been implicated in depression; 9 in anxiety; 5 in pain; 4 in depression and anxiety; 2 in pain and depression; 2 in pain and anxiety; and 9 genes that have been implicated in all three conditions. However, taken together with our behavioral findings, our data indicates that all 39 of these genes correspond with neuropathic pain-induced depression and anxiety. Indeed, our findings extend beyond 39 genes and implicate 89 genes in the mPFC, 123 in the NAc, and 795 in the PAG. In order to validate the SNI-induced differential expression patterns of key genes, we next performed qPCR on a variety of differentially expressed genes implicated in the 19 common pathways and in the overlap with the literature (Fig 6A). Multiple genes that have been previously implicated in anxio-depressive and nociceptive states were validated, including Cyp2e1, Btg2, Cyr61, Tph2, Ccl3, Fos, Sgk1, Dusp1, and Slc17a8. In addition, several previously unreported genes were also validated, including Capn11, Oacyl, Mt1, and Vgf. As the protein product of Capn11 (calpain 11) belongs to a large family of calcium-dependent cysteine proteases (55), we also assessed the mRNA levels of the two most prototypical calpains, calpain 1 (Capn1) and calpain 2 (Capn2), which showed no SNI-induced regulation. As increased Capn11 expression was confirmed in all three brain regions, we next sought to determine if inhibition of calpain proteins can rescue SNI-induced mechanical allodynia, and enhanced anxiety and depression like behaviors. We exposed adult (8 week-old) male C57BL/6 mice to SNI and confirmed the presence of mechanical allodynia for 60 days, after which they were separated into two groups, one receiving systemic injections of the calpain inhibitor, MDL 28170 (3 mg/kg daily), and the other receiving daily saline injections (Fig 6B). We performed Von Frey assessments of mechanical thresholds one and two weeks after drug treatment, and found that MDL 28170 increased mechanical thresholds, indicating that it reduced mechanical allodynia. A two-way repeated measures ANOVA revealed a significant interaction between drug treatment and time, whereby only mice receiving MDL 28170 showed increased thresholds after two weeks of daily injections (Fig 6C). Compared with daily saline injections, mice receiving daily MDL28170 showed similar low levels of exploratory behavior in the EPM (Fig. 6D), and similar levels of immobility in the forced swim test (Fig 6E). These data indicate that chronic calpain inhibition can reduce SNI induced nociceptive sensitization after two months of neuropathic pain, but cannot rescue enhanced expression of anxiety and depression related behaviors.
Table 3. SNI-induced differentially expressed genes across all three brain regions that have been previously implicated in depression, anxiety, or pain.
Table lists selected studies where experimental observations indicate the involvement of specific genes. Genes included were derived from IPA analysis. Data is expansion of Venn diagram in Fig 5B. Arrows indicate direction of regulation of differentially expressed genes (for example, downward arrow indicates decrease in expression).
| Gene | Coded protein | Brain region in present study | Associated disorder | Reference |
|---|---|---|---|---|
| Cyp2e1 | cytochrome P450, family 2, subfamily e, polypeptide 1 | mPFC (↓), NAc (↓), & PAG (↓) | Depression | (122) |
|
|
||||
| Btg2 | B cell translocation gene 2, anti-proliferative | mPFC (↓), NAc (↓) | Depression | (103) |
|
|
||||
| Cyr61 | cysteine rich protein 61 | mPFC (↓) | Depression | (103) |
|
|
||||
| Tph2 | tryptophan hydroxylase 2 | mPFC (↓) | Depression, anxiety, and pain | (98, 99, 123) |
|
|
||||
| Dusp1 | mitogen-activated protein kinase phosphatase 1 | mPFC (↓) | Depression, anxiety, and pain | (102,103,124) |
|
|
||||
| Ccl3 | chemokine (C-C motif) ligand 3 | NAc (↑) | Depression and pain | (113, 125) |
|
|
||||
| Fos | FBJ osteosarcoma oncogene | NAc (↓) | Depression, anxiety, and pain | (103, 105, 126) |
|
|
||||
| Sgk1 | serum/glucocorticoid regulated kinase 1 | PAG (↑) | Depression and pain | (118, 119, 127) |
|
|
||||
| Adora2a | adenosine A2A receptor | PAG (↑) | Depression, anxiety, and pain | (128–130) |
|
|
||||
| Adra1d | adrenergic receptor alpha-1 | PAG (↑) | Depression, anxiety, and pain | (131–133) |
|
|
||||
| Grin3b | N-methyl-D-aspartate receptor subunit 3B | PAG (↑) | Depression and anxiety | (134) |
|
|
||||
| Tspo | translocator protein | PAG (↑) | Depression, anxiety, and pain | (135–137) |
|
|
||||
| Cartpt | cocaine- and amphetamine-regulated transcript protein | PAG (↓) | Depression, anxiety, and pain | (138–141) |
|
|
||||
| Apoe | apolipoprotein E | PAG (↑) | Depression, anxiety, and pain | (142–145) |
|
|
||||
| sema3f | semaphorin 3f | PAG (↑) | Depression | (146) |
|
|
||||
| Igfbp2 | insulin-like growth factor binding protein 2 | PAG (↑) | Depression | (146) |
|
|
||||
| Lamb2 | intermediate filament protein | PAG (↑) | Depression | (146) |
|
|
||||
| Krt17 | keratin 17 | PAG (↑) | Depression | (146) |
|
|
||||
| Mdk | midkine (neurite growth-promoting factor 2) | PAG (↑) | Depression and anxiety | (147) |
|
|
||||
| Fzr1 | fizzy/cell division cycle 20 related 1 | PAG (↑) | Depression | (146) |
|
|
||||
| Grm2 | mGluR2/mGluR7 | PAG (↑) | Depression and anxiety | (148) |
|
|
||||
| Kcnj11 | inwardly rectifying potassium channel, subfamily J, member 11 | PAG (↑) | Anxiety | (149) |
|
|
||||
| Naglu | alpha-N-acetylglucosaminidase | PAG (↑) | Anxiety | (150) |
|
|
||||
| Park2 | Parkin | PAG (↑) | Anxiety | (151) |
|
|
||||
| Hrh3 | histamine H3 receptor | PAG (↑) | Anxiety | (152) |
|
|
||||
| Shank3 | SH3 and multiple ankyrin repeat domains 3 | PAG (↑) | Anxiety | (153) |
|
|
||||
| Nos3 | nitric oxide synthase 3 (endothelial cell) | PAG (↑) | Anxiety and pain | (154, 155) |
|
|
||||
| Slc17A8 | solute carrier family 17 (vesicular glutamate transporter), member 8; VGLUT3 | PAG (↓) | Anxiety and pain | (120) |
|
|
||||
| Lmtk3 | lemur tyrosine kinase 3 | PAG (↑) | Depression and anxiety | (156) |
|
|
||||
| Pnoc | Prepronociceptin | PAG (↑) | Depression, anxiety, and pain | (157–160) |
|
|
||||
| Ucn | Urocortin | PAG (↑) | Anxiety | (161) |
|
|
||||
| Dlgap3 | discs, large (Drosophila) homolog-associated protein 3 | PAG (↑) | Anxiety | (162) |
|
|
||||
| Arx | aristaless-related homeobox | PAG (↓) | Anxiety | (163) |
|
|
||||
| F2rl1 | coagulation factor II (thrombin) receptor-like 1 | mPFC (↑) | Pain | (164) |
|
|
||||
| Cxcr5 | chemokine receptor 5 | mPFC (↑) | Pain | (165) |
|
|
||||
| Ccl21a | chemokine ligand 21A | mPFC (↑) | Pain | (166) |
|
|
||||
| Chnra6 | cholinergic receptor, nicotinic, alpha polypetide 6 | NAc (↓) | Pain | (167) |
|
|
||||
| Acpp | prostatic acid phosphatase | NAc (↓) | Pain | (168) |
Fig 6. qPCR validation of genes of interest and behavioral effects of chronic calpain inhibition (A).
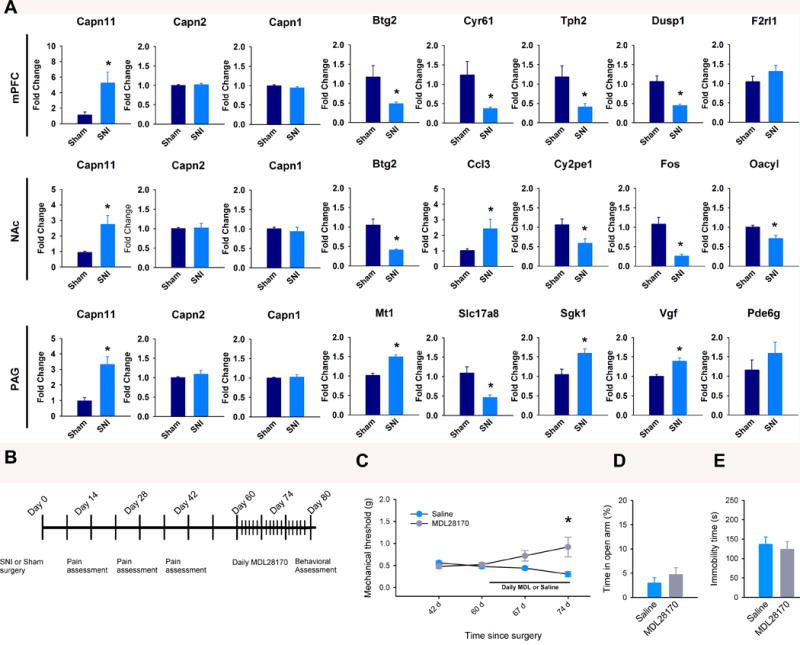
Expression of the indicated genes in each brain region. Top, mPFC: Capn11 (t(16) = −2.83, p = 0.012), Capn2 (t(10) = −0.02, p = 0.669), Capn1 (t(10) = 1.44, p = 0.168), Btg2 (t(10) = 2.35, p = 0.041), Cyr61 (t(10) = 2.48, p = 0.033), Tph2 (t(10) = 2.61, p = 0.037), Dusp1 (t (14) = 4.27, p < 0.001), F2rl1 (t(10) = −1.28, p = 0.23. Middle, NAc: Capn11(t(10) = −3.06, p = 0.012), Capn2 (t(10) = 0.61, p = 0.558), Capn1 (t(10) = −0.15, p = 0.883), Btg2 (t(10) = 4.14, p = 0.002), Ccl3 (t(10) = −2.26, p = 0.048), Cyp2e1 (t(12) = 2.54, p = 0.026), Fos (t (10) = 4.57, p = 0.001), Oacyl (t(10) = 3.27, p = 008). Bottom, PAG: Capn11 (t(12) = −4.15, p < 0.001), Capn2( t(10) = −0.91, p = 0.383), Capn1 (t(10) = −0.36, p = 0.727), Mt1 (t(10) = −6.14, p < 0.001), Slc17a8 (t(10) = 3.12, p = 0.011), Sgk1 (t(10) = −3.03, p = 0.013), Vgf (t (14) = − 4.15, p < 0.001), Pde6g (t(10) = −0.92, p= 0.380). Data are mean fold change ± SEM from n = 6–9 mice per group . values determined by unpaired t-tests. (B) Timeline of MDL 28170 or saline treatments and behavior assessment. (C) Mechanical thresholds after two weeks of treatment with MDL28170 (n = 5 mice per group; F(3,39) = 7.12, P = 0.001; two-way ANOVA and Holm-Sidak adjustment for multiple comparisons). (D and E) Percent open arm exploratory behavior in the EPM test (D) and immobility time in the forced swim test (E) in SNI mice treated with saline or MDL 28170 for two weeks, no significant effect found by unpaired t-tests. * p < 0.05.
Differentially expressed genes in the mPFC, NAc, and PAG under neuropathic pain states are also regulated by chronic unpredictable stress
To determine if SNI-induced differentially expressed genes are linked to anxio-depressive behaviors, we tested if chronic stress alters the expression of genes shown to be regulated by SNI. We used the chCUS model, which has been shown to induce depression-like behaviors in mice (56, 57). In our protocol, we did not use any stimuli that promote injury, inflammation or nociception (see methods; Fig 7A). Four weeks of CUS promoted depression-like states, as indicated by significantly increased latencies to eat in the novelty suppressed feeding (NSF) test (baseline: 73.8 ± 6.8 s; CUS: 134.4 ± 21.6 s; Fig 7B), and significantly reduced sucrose preference (baseline: 87.3 ± 0.6 %; CUS: 77.7 ± 1.7 %;; Fig 7C). NSF has been shown to reflect changes in anxiety and depression-like behavior (58). Remarkably, multiple genes were regulated by both CUS and SNI. Specifically, three genes that were regulated by SNI in the mPFC, Btg2, (B-cell translocation gene 2, anti-proliferative), Cyr61, (cysteine rich protein), and Dusp1, (dual specificity phosphatase 1), showed similar patterns of regulation in the mPFC following CUS (Btg2: 0.57 ± 0.09 fold change (FC); Cyr61: 0.46 ± 0.06 FC; Dusp1: 0.50 ±0.04; Fig 7D). In the NAc, Btg2, Capn11 (calpain 11), the cytokine Ccl3, also known as macrophage inflammatory protein 1-alpha, and FOS, (FBJ osteosarcoma oncogene) were also regulated in the same direction by SNI and CUS (Btg2: 0.74 ± 0.09 FC; Capn11: 2.71 ± 0.68 FC; Ccl3: 1.45 ± 0.08 FC; Fos: 0.53 ±0.13; Fig 5E). In the PAG, Sgk1 (serum glucocorticoid-regulated kinase), the vesicular glutamate transporter, Slc17a8, (solute carrier family 17 member 8, also known as Vglut3), and Capn11 were also similarly regulated by SNI and CUS (Sgk1: 1.38 ± 0.13 FC; Slc17a8: 0.37 ± 0.08 FC; Capn11: 1.28 ±0.05; Fig 7F). These data demonstrate that several genes in the mPFC, NAc, and PAG are regulated in a similar manner by neuropathic pain states and chronic unpredictable stress.
Fig 7. SNI and chronic stress cause similar gene expression patterns in the mPFC, NAc, and PAG. (A).
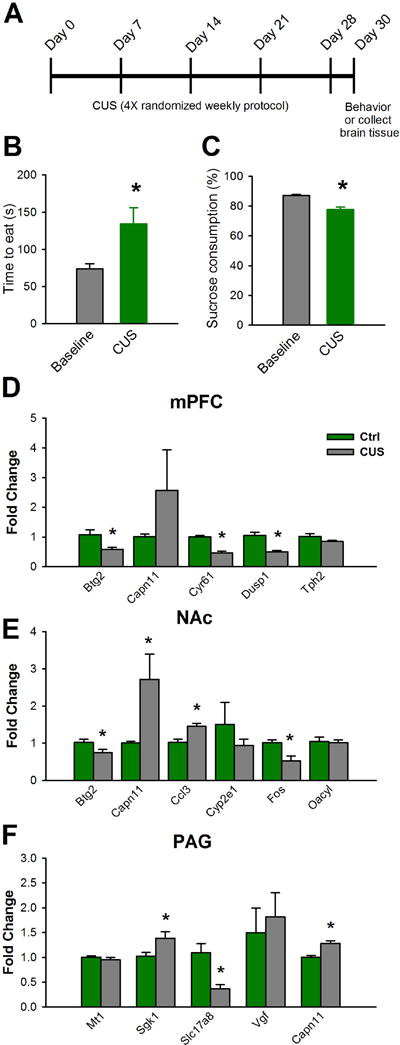
Chronic unpredictable stress (CUS) experimental timeline; CUS was administered for 4 consecutive weeks, after which behavior was assessed or animals were sacrificed and brains extracted for further analysis. (B) In the novelty suppressed feeding test, CUS increased latencies to eat (n = 11, t(20) = −2.7, p = 0.015; unpaired t-tests). (C) CUS decreased sucrose consumption rates (n = 11, t(20) = −5.3, p < 0.001; unpaired t-tests). (D) Genes regulated by CUS and SNI in the mPFC: Btg2 (n = 7, t(12) = 2.8, p = 0.017; unpaired t-tests), Cyr61 (n = 7 mice per group, t(12) = 7.3, p < 0.001; unpaired t-tests), Dusp1 (n = 6, t(10) = 4.4 p < 0.001; unpaired t-tests). (E) Genes regulated by CUS and SNI in the NAc: Btg2 (n = 7, t(12) = 2.8, p = 0.042; unpaired t-tests), Capn11: (n = 9, t(16) = −2.5, p = 0.024; unpaired t-tests), Ccl3 (n = 7, t(12) = −3.65, p = 0.003; unpaired t-tests), Fos (n = 7, t(12) = 3.3 p = 0.006; unpaired t-tests). (F) Genes regulated by CUS and SNI in the mouse PAG: Sgk1 (n = 7 mice per group, t(12) = −2.4, p = 0.034; unpaired t-tests), Slc17a8 (n = 7, t(12) = 3.6, p = 0.004; unpaired t-tests), Capn11 (n = 7, t(12) = − 4.5 p < 0.001; unpaired t-tests). Data are mean fold change ± SEM, *p < 0.05.
Discussion
Using next generation RNA sequencing, we present the first report of gene expression adaptations in three mood-associated brain regions in response to long-term neuropathic pain in mice. We show that SNI enhanced anxiety- and depression-like behaviors two months after nerve injury, which corresponded with distinct differentially expressed gene patterns, biological networks, and upstream regulators, in all three brain regions. Despite these differences, common biological functions and signaling pathways were identified. Our findings also indicate that neuropathic pain states promote several gene expression adaptations that are also observed in models of depression.
Our data indicate that HDAC5 is regulated by long term neuropathic pain states in the NAc and PAG. Using RRHO, we identified robust co-upregulation of multiple genes in the NAc and PAG in correspondence with long-term SNI. Through gene ontology analysis, we found that HDAC5, which encodes for an enzyme (HDAC5) known to promote the stability of compact chromatin forms, was implicated in several biological processes over-represented by co-upregulated genes in the NAc and PAG. We further found that genetic deletion of HDAC5 attenuates SNI-induced anxiety and depression-like phenotypes. Previous studies have documented the role of HDAC5 in modulation of antidepressant drug efficacy in models of stress or neuropathic pain (7,50). In line with these findings, our study suggests that HDAC5 is a negative modulator of SNRI actions and points to HDAC5 inhibitors as potential targets for the treatment of neuropathic pain. Unexpectedly, HDAC5 expression in the NAc and PAG was not captured by our subsequent analysis employing the stringent fold change cutoff of 1.50. It is worth noting however that within the PAG, HDAC5 expression was increased in the SNI group by a fold change of 1.48, just barely missing the 1.50 cutoff.
Despite stark differences in predicted upstream regulators, we observed that one gene (Capn11) was upregulated, and one gene (Cyp2e1) was downregulated in all three brain regions of mice exposed to SNI. Cyp2e1 is a cytochrome p450 family member linked to ethanol and nicotine intake (59–62) that is known to be regulated by inflammatory cytokines (63–65). Future studies will need to clarify the role of Cyp2e1 in reward mechanisms under chronic pain states, and its role in sensory and affective symptoms of pain conditions. Similarly to the network-wide Cyp2e1 reduction, all three brain regions showed upregulated Capn11 expression; this gene codes for the protein calpain 11, an intracellular, calcium-dependent cysteine protease (55). Calpain 11 is most similar to the classical calpains (1 and 2), containing all three typical domains in the large subunit. Cortical expression of Capn11 mRNA is observed in mice, mainly in astrocytes, but is also expressed in neurons and microglia (66). Calpain substrates in neurons include synaptic proteins such as membrane receptors, cytoskeletal proteins, postsynaptic density proteins, and intracellular mediators involved in synaptic function (67, 68), indicating that neuropathic pain-induced calpain activity may mediate various plasticity-related processes. Although nociception-induced changes in calpain 11 have not been investigated, calpain inhibitors that prevent the action of all calpains are used in most investigations, and findings include reports that calpains mediate AMPA receptor subtype editing (69) and glycine-mediated increases in dendritic spine density of cultured hippocampal neurons (70). Furthermore, our data showing that a non-selective calpain inhibitor has anti-allodynic effects is in accordance with two recent rat studies which have shown that 14 days of systemic calpain inhibition reduces cancer bone pain (71) and nociceptive sensitization induced by diabetic neuropathy (72).
Our findings also suggest that the patterns of differentially expressed genes induced by SNI are indicative of altered activity of various signal transduction pathways, including glucocorticoid receptor, cAMP, and GPCR mediated signaling. Glucocorticoids have been established to be involved in the effects of chronic stress in humans (73–75), and rodents (76–78). Our finding that SNI alters glucocorticoid signaling is also in agreement with studies showing that stress-induced increases in corticosterone potentiate SNI-induced allodynia in mice (44). Similarly, we report that SNI-induced differential expression of genes implicated in cAMP-mediated signaling, in accordance with studies linking perturbations of cAMP signalling pathways to depression-like (79) and chronic pain-like states (80–85). The pregnenolone biosynthesis signaling pathway was also affected in all three brain regions in response to SNI. Decreased neuro-steroid biosynthesis has been implicated as a possible contributor to the development of anxiety and depressive disorders (86, 87). Attention has turned to allopregnenolone (88) that acts as a potent positive allosteric modulator of GABAA receptors (89), has anxiolytic effects in rodents and promotes the efficacy of fluoxetine (90–92). Pregnenolone biosynthesis deficiencies resulting in altered GABAA modulation are supported by recent findings that GABAA signaling in the PAG is altered by persistent inflammation in rats (93).
Among the 19 pathways that showed altered activation in at least two brain regions, several have been previously implicated in anxiety or depression. For example, we demonstrate changes in IGF-1 signaling and several pro- and anti-inflammatory interleukin-mediated pathways, in accordance with observations that stressful experiences affect the function of inflammatory molecules (94). Moreover, alterations in inflammatory pathways promote changes in brain circuitry involved in mood, stress, and pain (44, 95–96).
Numerous neuroimaging studies show that in humans, pain is associated with activity in the NAc, mPFC and PAG (13,14, 28). These regions display robust reciprocal connections, (11), and form a network capable of exerting top-down regulation of nociceptive input (28,29). The PAG in particular plays a prominent role in decending moudlation of pain through its projections to the nucleus raphe magnus (28, 29), and can either excert facilitatory or inhibitory effects on spinal regions. Notably, mPFC neurons project to the PAG (32), and therefore gene expression changes in this brain region may affect PAG activity and the modulation of pain transmission. PFC projections to NAc have also been shown to modulate sensory and affective symptoms of neuropathic pain (6). In addition, the NAc is connected to several brain regions that modulate nociceptive perception and mood, icluding the PFC, the amygdala and the hippocampus (6,15,24,31,32). Gene expression changes in mood-related networks can impact the perception of nociceptive information as well as the responsiveness to antidepressant medications (7). We have shown that disruption of HDAC5 expression, which we found to be upregulated in the NAc and PAG in SNI mice, affects behavioral responses to SNRI treatment.
Various SNI-induced differentially expressed genes identified in our study have previously been reported to be implicated in depression, anxiety, and pain-like conditions. Tph2 was robustly reduced in the mPFC, and decreases in TPH2 abundance is being considered as a trait marker for depression (97–99). Likewise, we found that SNI caused a robust decrease of Dusp1 in the mPFC. DUSP1 controls the expression of several inflammation-induced genes, including IL-6, IL-10, Ccl3, Ccl4, and Cxcl2 (100). Overexpression of Dusp1 in the rat spinal cord prevents neuropathic pain (101), whereas mice with genetic deletion of Dusp1 are resilient to stress-induced depression (102), suggesting distinct roles for Dusp1 in spinal versus supraspinal sites. Some of the differentially expressed genes identified showed opposite expression patterns from those previously reported on depression and anxiety. For example, Btg2 mRNA is enhanced in the neocortex of mice in response to acute immobilization stress (103), whereas we found it to be decreased in the mPFC and NAc of SNI and CUS mice. Because the upregulation of Btg2 was observed in a model of acute stress, it remains to be investigated how chronic stress conditions affect the expression of this gene in mPFC and NAc. Similarly, we found SNI reduced the expression of Cyr61 and Dusp1 in the mPFC, but previous studies have linked increases in the expression of these genes in the neocortex and hippocampus to anxiety and depression (102–103). These differences may underlie brain region and cell type-specific effects of stress and nerve injury on Cyr61 and Dusp1 expression, or may suggest that these genes play distinct roles in neuropathic pain versus chronic stress states. Moreover, we observed decreases in Fos expression in the NAc of SNI and CUS mice, and although the antidepressants imipramine (104), and milnacipran (10) enhance c-Fos expression in the PFC of rats, homozygous FosKO mice show reduced anxiety-like behaviors (105). Nevertheless, these observations highlight the value of network-wide analysis, and the potential detrimental effects disruptions in normal gene expression (be it enhanced or decreased) may exert on normal emotional function.
Here we found that SNI causes anxio-depressive symptoms and differentially expressed genes in the mPFC, NAc, and PAG similar to mice exposed to CUS, a distinct stress model validated to induce depression in mice. Most of these genes have been observed to be involved in plasticity related processes, and their altered expression may reflect adapted brain states in chronic pain and mood disorders. Within the mPFC, Btg2 expression was reduced, and previous findings have shown Btg2 involvement in neurite outgrowth (106) and histone H4 arginine methylation and acetylation processes (107). Similarly, Cyr61 expression was also reduced in SNI and CUS groups of mice, and its activity is critical for dendritic growth in hippocampal neurons (108). Within the NAc, Ccl3 was enhanced in SNI and CUS groups, and its protein product, the pro-inflammatory cytokine, Ccl3, is known to enhance inflammatory responses within the CNS and increases Ca2+ signaling in cultured neurons (109–110). Notably, spinal cord injury leads to Ccl3 upregulation in the hippocampus (111), and its expression is positively correlated with depression (112–113). NAc samples of SNI and CUS mice also showed similar decreases in Fos, and the link between activity-dependent Fos expression, synaptic plasticity, and learning and memory have been well documented (114–117). Likewise, multiple differentially expressed genes in the PAG of SNI and CUS mice have been linked with anxiety and depression. The expression of Sgk1, upregulated in both SNI and CUS mice, is upregulated in the corpus callosum (118), and hippocampus (119) by chronic stress, and mRNA amounts of Sgk1 are increased in peripheral blood samples from patients with major depression (119). Similarly, genetic deletion of Slc17a8 (Vglut3), the expression of which was reduced in the PAG of both SNI and CUS groups in our study, enhances anxiety-like behaviors in adult mice that correspond with decreases in 5-HT1A-mediated neurotransmission in the raphe nuclei (120). Some SNI-induced gene expression patterns were not observed in the CUS model, however, including changes in the expression of Tph2 and Capn11 in the mPFC, Oacyl and Cyp2e1 in the NAc, and Mt1 and Vgf in the PAG. These differences may rely on the models or time points selected, or on the influences of input from nociceptive neurons in the examined brain regions. Recent work characterizing long-term neuropathic pain-induced methylation changes in the PFC and peripheral T cells of rats showed that SNI-induced gene expression patterns correlate with the severity of nociceptive sensitization (25). Future work should determine whether similar correlations can be observed with anxio-depressive symptom severity, and whether gene expression patterns can be predictive of anxio-depressive symptom development in models of neuropathic pain.
An understanding of the genes and signal transduction networks regulated by long-term neuropathic pain in mood-regulating networks will help determine if drugs targeting nociceptive transmission and perception also affect long-term adaptations that lead to depression, or if adjunct medications need to be used. Several types of antidepressants, such as tricyclic antidepressants and selective serotonin-norepinephrine reuptake inhibitors target both spinal pathways and mood-regulating networks, and are among the top prescribed medications for neuropathic pain (121). However, full efficacy is only reached by a subset of patients, and their use is limited by the slow onset of action and severe adverse effects. Future studies must investigate cell-type specific adaptations in gene expression following long-term neuropathic pain, and should include several other regions modulating cognition, motivation and responses to stress. These studies are expected to provide important information on the genes and networks that need to be targeted in order to efficiently treat neuropathic pain.
Materials and Methods
Animals
We used adult (8 – 9 weeks) male C57Bl/6 mice purchased from Jackson Laboratories, and HDAC5 knockout or wildtype controls bred on a C57Bl/6 background and maintained in a colony at our facility. Mice were group housed (3 per cage), and were kept on a 12h light/dark cycle, with food and water available ad libitum. All procedures were performed in accordance to the guidelines of the Animal Care Committees at Icahn Medical School at Mount Sinai, and approved by IACUC.
Spared Nerve Injury model
The spared nerve injury (SNI) model of neuropathic pain was performed under Avertine (2,2,2-tribromoethanol, Sigma-Aldrich) general anesthesia, as previously described (7, 41). Briefly, skin and muscle incisions were made on the left hind leg at mid-thigh level, revealing the sciatic nerve and its three branches. The common peroneal and sural nerves were carefully ligated with 6.0 silk suture (Ethicon, Johnson & Johnson Intl.), transected, and a 1–2mm sections of each of these nerves were removed. The tibial nerve was left intact. Skin was then closed and stitched with silk 4.0 sutures (Ethicon, Johnson & Johnson Intl). Sham operated mice were exposed to the same procedure but all nerves were left intact.
Drugs
Duloxetine (Tocris) was dissolved in phosphate buffered saline (PBS) to a concentration of 0.5 mg/ml. MDL 28170 (Tocris) was first dissolved in DMSO and then diluted prior to injection to the desired concentration, so that the injected solution contained 5% DMSO. Control saline injections included equal amounts of DMSO. All injections were administered intraperitoneally (i.p.), alternating sides between each injection.
Behavioral Analysis
Mechanical allodynia was measured through the up-down method (44) using Von Frey filaments (Stoelting, IL) with ascending forces expressed in grams (0.04–2.0 g). Each filament was applied five times in a row against the medial plantar surface of the hind paw. Prolonged hindpaw withdrawal, rapid fanning, or licking induced by the filament was defined as a positive nociceptive response. The lowest gram force eliciting positive responses in three out of five repetitive stimuli was defined as the 50 % withdrawal threshold. Mice were habituated to the Von Frey apparatus for 1 hour on test days prior to analysis.
In the forced swim test, mice were placed in individual glass cylinders (46-cm height × 18-cm diameter) containing water at room temperature (25 ± 1°C) at a depth of 15 cm, and were allowed to freely swim in the container for 6 min; sessions were videotaped for offline analysis. Immobility measurements were determined as time spent immobile with the exception of a single paw flick to remain afloat. Analysis was performed by a blind observer.
To assess sucrose consumption, mice were single housed with sucrose (1%) or water available for a period of 24 hours. Sucrose consumption was determined as total sucrose consumed divided by the total water and sucrose consumption, multiplied by 100. Bottles were rotated at 3, 6, and 12 hours to prevent side bias. Measurements were taken by a blind observer.
Elevated plus maze (EPM) and Open Field (OF) tests were performed under red light conditions, and mice were acclimatized to the room for 1 hour before testing. The EPM consisted of two open arms and two closed arms situated opposite to each other (each measuring 12 × 50 cm). The number of entries and time spent in each arm was recorded and analyzed using Noldus EthoVision 10.1 software. For OF test, mice were allowed to freely explore the open field arena (50 × 50 × 20 cm) for 5 min. Time spent in the center vs. border regions of the arena were recorded and analyzed using Noldus EthoVision 10.1. Analysis was performed by a blind observer.
Novelty Suppressed Feeding (NSF)
NSF was performed as reported previously with minor modifications (45). Briefly, mice were food deprived for 24 hours prior and habituated to the testing room for 1 hour. Under red light conditions, mice were placed in a plastic chamber (50 × 50 × 20 cm) with wood chip bedding. A single pellet of regular food chow was placed in the center of the chamber. Mice were placed in the corner of the box, and latency to eat was measured, with a cutoff time of 10 min.
Chronic Unpredictable Stress Model
CUS was performed as published previously (46) with minor modifications. Briefly, groups of male C57BL/6 mice (8-week old) were either exposed to 4 weeks of CUS, or were housed under the same conditions but left undisturbed (control). All mice were given access to food and water ad libitum. The CUS protocol was based on a weekly schedule of the following stressors: overnight illumination (light on during nighttime), cage tilting (45° angle), bedding removal, wet bedding, restraint (2 hrs, restraint by attaching tail on cage wall), and removal of food for 12h. Two different stressors were applied per day, each one during the light or dark phase cycle. The sequence of stressors applied was designed to maximize unpredictability.
RNA-Sequencing Experiments
RNA-sequencing was performed on polyadenylated fractions of isolated RNA. Briefly, two groups of mice (SNI and Sham) with six biological replicates per group were used. Bilateral punches were taken from the mPFC, NAc, and PAG, and punches were pooled from two mice per sample, so that there was a total of twelve animals per treatment group. Total RNA was isolated from pooled bilateral punches of two animals per replicate (six biological replicates per treatment group) using TRIzol (Life Technologies) according to manufacturer’s instructions. RNA was purified with RNeasy Micro Kit 50 (cat. 74004, Qiagen) according to manufacturer’s instructions, and an Agilent 2100 Bioanalyzer confirmed that the RNA integrity numbers were >9.0. Poly-A–containing mRNA was purified by using poly-T oligo-attached magnetic beads, and the mRNA-seq library was prepared from each pooled RNA sample by using the Illumina TruSeq RNA Sample Preparation Kit v2, according to the manufacturer’s protocol. RNA-seq was performed on the Illumina HiSeq2000 machine at the Mount Sinai School of Medicine’s Genomic Core facility. Fifty bp, paired end short reads were aligned to the mouse genome (mm10) using TopHat2. Uniquely aligned reads were counted against the Gencode vM2 annotation using HTSeq. Differential analysis was performed using the voom-limma R library, determining differentially expressed genes using a cutoff of p < 0.05. GO analysis was performed by using the functional annotation and clustering software tool available through the online Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Resources (Version 6.7; https://david.ncifcrf.gov/). Only terms with . < 0.05 are reported. To identify potential protein interactions between the products of the differentially expressed genes in our results, we used the STRING (Version 9.1) software and database. Venn diagrams were generated by using VennPlex Version 1.0.0.2 (NIH). Ingenuity Pathway Analysis (IPA) was used to identify genes involved in depression, and to determine potential activated pathways, upstream regulators, and biological functions (7).
Rank Rank Hypogeometric Overlap (RRHO)
To evaluate global gene expression profiles, the Rank Rank Hypergeometric Overlap test (RRHO) test (48, 49) was performed between two sets of brain region specific differential gene expression comparisons. RRHO employs a threshold-free approach and ranks differentially expressed genes to determine the statistical significance of the number of overlapping genes, providing hypergeometric P-values for the enrichment of overlapping genes for all possible rank pairs for both brain regions. The direction signed log10-transformed hypergeometric P-values were corrected for multiple comparisons according to the Benjamini and Yekutieli method (48).
Hierarchical Clustering Heatmaps
Variance stabilized expression values were computed using DESeq R package. To visualize relative expression in each brain region separately, the variance stabilized values were z-scored for each differentially expressed gene. Hierarchical clustering was performed on these genes using Euclidean distance and plotted.
Quantitative Real-Time PCR
qPCR was performed as reported previously (7). Total RNA was isolated from frozen bilateral punches of single animals (NAc and PAG), or pooled two per sample (mPFC) using TRIzol (Life Technologies) according to manufacturer’s instructions, and was purified using chloroform (Sigma-Aldrich), precipitated with isopropanol (Sigma-Aldrich) and 75% Ethanol (Sigma-Aldrich), and eluted in RNAse-free water (Life Technologies). RNA was reverse transcribed into cDNA using qScript cDNA Synthesis Kit (Quanta Biosciences). Primers were designed to amplify regions of 50 – 250 bp (table S2) located in the genes of interest selected from IPA analysis. All reactions run in triplicate and assessed using the ΔΔCt method and normalized to glyceraldehyde-3-phosphate dehydrogenase (Gapdh).
Data Analysis
Results are presented as mean ± SEM. Statistical analyses were performed using unpaired or paired student t-tests, or with two-way repeated measures ANOVA, with post-hoc adjustments for multiple comparisons using Holm-Sidak method. For analysis of sham and SNI HDAC5 WT and HDAC5 KO von Frey measurements, we used a 3-Way ANOVA. In all cases, the significance threshold was set at P < 0.05.
Supplementary Material
Figure S1: Locomotor behavior of sham and SNI mice 2 months after surgery.
Figure S2: Column dendrogram for all three brain regions.
Table S1: Differentially expressed genes in the mPFC, NAc, and PAG of SNI mice.
Table S2: Primers used for qPCR experiments.
Acknowledgments
Funding: This work has been supported by NIDA PPG-POIDAO8227 (VZ), NINDS NS086444 (VZ, LS) and the Banting Postdoctoral Fellowships Program (GD).
Footnotes
Author contributions: GD, VM, SG, IP, EL, LS, and VZ participated in research design and data analysis. GD, VM, SG, CA, performed the experiments. GD and VZ wrote the manuscript.
Competing interests: The authors declare that they have no financial interests or potential conflicts of interest.
Data and materials availability: RNAseq data has been uploaded to the public repository GEO (https://www.ncbi.nlm.nih.gov/geo/info/seq.html), accession numbers GSM2422758 - GSM2422793.
References and Notes
- 1.C. Committee on Advancing Pain Research, and Education. Relieving Pain in America: A Blueprint for Transforming Prevention, Care, Education, and Research. Institute of Medicine (US); Washington, DC: 2011. [Google Scholar]
- 2.Lerman SF, Rudich Z, Brill S, Shalev H, Shahar G. Longitudinal associations between depression, anxiety, pain, and pain-related disability in chronic pain patients. Psychosomatic medicine. 2015;77:333–341. doi: 10.1097/PSY.0000000000000158. [DOI] [PubMed] [Google Scholar]
- 3.McWilliams LA, Goodwin RD, Cox BJ. Depression and anxiety associated with three pain conditions: results from a nationally representative sample. Pain. 2004;111:77–83. doi: 10.1016/j.pain.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 4.Bair MJ, Wu J, Damush TM, Sutherland JM, Kroenke K. Association of depression and anxiety alone and in combination with chronic musculoskeletal pain in primary care patients. Psychosomatic medicine. 2008;70:890–897. doi: 10.1097/PSY.0b013e318185c510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor AM, Castonguay A, Taylor AJ, Murphy NP, Ghogha A, Cook C, Xue L, Olmstead MC, De Koninck Y, Evans CJ, Cahill CM. Microglia disrupt mesolimbic reward circuitry in chronic pain. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2015;35:8442–8450. doi: 10.1523/JNEUROSCI.4036-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee M, Manders TR, Eberle SE, Su C, D’Amour J, Yang R, Lin HY, Deisseroth K, Froemke RC, Wang J. Activation of corticostriatal circuitry relieves chronic neuropathic pain. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2015;35:5247–5259. doi: 10.1523/JNEUROSCI.3494-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitsi V, Terzi D, Purushothaman I, Manouras L, Gaspari S, Neve RL, Stratinaki M, Feng J, Shen L, Zachariou V. RGS9-2–controlled adaptations in the striatum determine the onset of action and efficacy of antidepressants in neuropathic pain states. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E5088–5097. doi: 10.1073/pnas.1504283112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwartz N, Temkin P, Jurado S, Lim BK, Heifets BD, Polepalli JS, Malenka RC. Chronic pain. Decreased motivation during chronic pain requires long-term depression in the nucleus accumbens. Science. 2014;345:535–542. doi: 10.1126/science.1253994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goffer Y, Xu D, Eberle SE, D’Amour J, Lee M, Tukey D, Froemke RC, Ziff EB, Wang J. Calcium-permeable AMPA receptors in the nucleus accumbens regulate depression-like behaviors in the chronic neuropathic pain state. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:19034–19044. doi: 10.1523/JNEUROSCI.2454-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takeda R, Watanabe Y, Ikeda T, Abe H, Ebihara K, Matsuo H, Nonaka H, Hashiguchi H, Nishimori T, Ishida Y. Analgesic effect of milnacipran is associated with c-Fos expression in the anterior cingulate cortex in the rat neuropathic pain model. Neuroscience research. 2009;64:380–384. doi: 10.1016/j.neures.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 11.Zhuo M. Neural Mechanisms Underlying Anxiety-Chronic Pain Interactions. Trends in neurosciences. 2016 doi: 10.1016/j.tins.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 12.Hashmi JA, Baliki MN, Huang L, Baria AT, Torbey S, Hermann KM, Schnitzer TJ, Apkarian AV. Shape shifting pain: chronification of back pain shifts brain representation from nociceptive to emotional circuits. Brain : a journal of neurology. 2013;136:2751–2768. doi: 10.1093/brain/awt211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baliki MN, Geha PY, Fields HL, Apkarian AV. Predicting value of pain and analgesia: nucleus accumbens response to noxious stimuli changes in the presence of chronic pain. Neuron. 2010;66:149–160. doi: 10.1016/j.neuron.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moayedi M, Weissman-Fogel I, Crawley AP, Goldberg MB, Freeman BV, Tenenbaum HC, Davis KD. Contribution of chronic pain and neuroticism to abnormal forebrain gray matter in patients with temporomandibular disorder. NeuroImage. 2011;55:277–286. doi: 10.1016/j.neuroimage.2010.12.013. [DOI] [PubMed] [Google Scholar]
- 15.Russo SJ, Nestler EJ. The brain reward circuitry in mood disorders. Nature reviews Neuroscience. 2013;14:609–625. doi: 10.1038/nrn3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vialou V, Robison AJ, Laplant QC, Covington HE, 3rd, Dietz DM, Ohnishi YN, Mouzon E, Rush AJ, 3rd, Watts EL, Wallace DL, Iniguez SD, Ohnishi YH, Steiner MA, Warren BL, Krishnan V, Bolanos CA, Neve RL, Ghose S, Berton O, Tamminga CA, Nestler EJ. DeltaFosB in brain reward circuits mediates resilience to stress and antidepressant responses. Nature neuroscience. 2010;13:745–752. doi: 10.1038/nn.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim BK, Huang KW, Grueter BA, Rothwell PE, Malenka RC. Anhedonia requires MC4R-mediated synaptic adaptations in nucleus accumbens. Nature. 2012;487:183–189. doi: 10.1038/nature11160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaudhury D, Walsh JJ, Friedman AK, Juarez B, Ku SM, Koo JW, Ferguson D, Tsai HC, Pomeranz L, Christoffel DJ, Nectow AR, Ekstrand M, Domingos A, Mazei-Robison MS, Mouzon E, Lobo MK, Neve RL, Friedman JM, Russo SJ, Deisseroth K, Nestler EJ, Han MH. Rapid regulation of depression-related behaviours by control of midbrain dopamine neurons. Nature. 2013;493:532–536. doi: 10.1038/nature11713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kupchik YM, Brown RM, Heinsbroek JA, Lobo MK, Schwartz DJ, Kalivas PW. Coding the direct/indirect pathways by D1 and D2 receptors is not valid for accumbens projections. Nature neuroscience. 2015;18:1230–1232. doi: 10.1038/nn.4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacAskill AF, Cassel JM, Carter AG. Cocaine exposure reorganizes cell type- and input-specific connectivity in the nucleus accumbens. Nature neuroscience. 2014;17:1198–1207. doi: 10.1038/nn.3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Navratilova E, Xie JY, Meske D, Qu C, Morimura K, Okun A, Arakawa N, Ossipov M, Fields HL, Porreca F. Endogenous opioid activity in the anterior cingulate cortex is required for relief of pain. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2015;35:7264–7271. doi: 10.1523/JNEUROSCI.3862-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liston C, Miller MM, Goldwater DS, Radley JJ, Rocher AB, Hof PR, Morrison JH, McEwen BS. Stress-induced alterations in prefrontal cortical dendritic morphology predict selective impairments in perceptual attentional set-shifting. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2006;26:7870–7874. doi: 10.1523/JNEUROSCI.1184-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Metz AE, Yau HJ, Centeno MV, Apkarian AV, Martina M. Morphological and functional reorganization of rat medial prefrontal cortex in neuropathic pain. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2423–2428. doi: 10.1073/pnas.0809897106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bagot RC, Parise EM, Pena CJ, Zhang HX, Maze I, Chaudhury D, Persaud B, Cachope R, Bolanos-Guzman CA, Cheer JF, Deisseroth K, Han MH, Nestler EJ. Ventral hippocampal afferents to the nucleus accumbens regulate susceptibility to depression. Nature communications. 2015;6:7062. doi: 10.1038/ncomms8062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Massart R, Dymov S, Millecamps M, Suderman M, Gregoire S, Koenigs K, Alvarado S, Tajerian M, Stone LS, Szyf M. Overlapping signatures of chronic pain in the DNA methylation landscape of prefrontal cortex and peripheral T cells. Scientific reports. 2016;6:19615. doi: 10.1038/srep19615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu H, Wu LJ, Wang H, Zhang X, Vadakkan KI, Kim SS, Steenland HW, Zhuo M. Presynaptic and postsynaptic amplifications of neuropathic pain in the anterior cingulate cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28:7445–7453. doi: 10.1523/JNEUROSCI.1812-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alvarado S, Tajerian M, Millecamps M, Suderman M, Stone LS, Szyf M. Peripheral nerve injury is accompanied by chronic transcriptome-wide changes in the mouse prefrontal cortex. Molecular pain. 2013;9:21. doi: 10.1186/1744-8069-9-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fields HL. Pain modulation: expectation, opioid analgesia and virtual pain. Progress in brain research. 2000;122:245–253. doi: 10.1016/s0079-6123(08)62143-3. [DOI] [PubMed] [Google Scholar]
- 29.Porreca F, Ossipov MH, Gebhart GF. Chronic pain and medullary descending facilitation. Trends in neurosciences. 2002;25:319–325. doi: 10.1016/s0166-2236(02)02157-4. [DOI] [PubMed] [Google Scholar]
- 30.Basbaum AI, Bautista DM, Scherrer G, Julius D. Cellular and molecular mechanisms of pain. Cell. 2009;139:267–284. doi: 10.1016/j.cell.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu R, Gollub RL, Spaeth R, Napadow V, Wasan A, Kong J. Disrupted functional connectivity of the periaqueductal gray in chronic low back pain. NeuroImage Clinical. 2014;6:100–108. doi: 10.1016/j.nicl.2014.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Floyd NS, Price JL, Ferry AT, Keay KA, Bandler R. Orbitomedial prefrontal cortical projections to distinct longitudinal columns of the periaqueductal gray in the rat. The Journal of comparative neurology. 2000;422:556–578. doi: 10.1002/1096-9861(20000710)422:4<556::aid-cne6>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 33.Du L, Wang SJ, Cui J, He WJ, Ruan HZ. The role of HCN channels within the periaqueductal gray in neuropathic pain. Brain research. 2013;1500:36–44. doi: 10.1016/j.brainres.2013.01.035. [DOI] [PubMed] [Google Scholar]
- 34.Hu J, Wang Z, Guo YY, Zhang XN, Xu ZH, Liu SB, Guo HJ, Yang Q, Zhang FX, Sun XL, Zhao MG. A role of periaqueductal grey NR2B-containing NMDA receptor in mediating persistent inflammatory pain. Molecular pain. 2009;5:71. doi: 10.1186/1744-8069-5-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berton O, Covington HE, 3rd, Ebner K, Tsankova NM, Carle TL, Ulery P, Bhonsle A, Barrot M, Krishnan V, Singewald GM, Singewald N, Birnbaum S, Neve RL, Nestler EJ. Induction of deltaFosB in the periaqueductal gray by stress promotes active coping responses. Neuron. 2007;55:289–300. doi: 10.1016/j.neuron.2007.06.033. [DOI] [PubMed] [Google Scholar]
- 36.Graeff FG. Serotonin, the periaqueductal gray and panic. Neuroscience & Biobehavioral Reviews. 2004;28:239–259. doi: 10.1016/j.neubiorev.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 37.Roncon CM, Biesdorf C, Santana RG, Zangrossi H, Jr, Graeff FG, Audi EA. The panicolytic-like effect of fluoxetine in the elevated T-maze is mediated by serotonin-induced activation of endogenous opioids in the dorsal periaqueductal grey. Journal of psychopharmacology. 2012;26:525–531. doi: 10.1177/0269881111434619. [DOI] [PubMed] [Google Scholar]
- 38.Yalcin I, Bohren Y, Waltisperger E, Sage-Ciocca D, Yin JC, Freund-Mercier MJ, Barrot M. A time-dependent history of mood disorders in a murine model of neuropathic pain. Biol Psychiatry. 2011;70:946–953. doi: 10.1016/j.biopsych.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 39.Terzi D, Gaspari S, Manouras L, Descalzi G, Mitsi V, Zachariou V. RGS9–2 modulates sensory and mood related symptoms of neuropathic pain. Neurobiology of learning and memory. 2014;115:43–48. doi: 10.1016/j.nlm.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goncalves L, Silva R, Pinto-Ribeiro F, Pego JM, Bessa JM, Pertovaara A, Sousa N, Almeida A. Neuropathic pain is associated with depressive behaviour and induces neuroplasticity in the amygdala of the rat. Experimental neurology. 2008;213:48–56. doi: 10.1016/j.expneurol.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 41.Shields SD, Eckert WA, 3rd, Basbaum AI. Spared nerve injury model of neuropathic pain in the mouse: a behavioral and anatomic analysis. The journal of pain : official journal of the American Pain Society. 2003;4:465–470. doi: 10.1067/s1526-5900(03)00781-8. [DOI] [PubMed] [Google Scholar]
- 42.Stratinaki M, Varidaki A, Mitsi V, Ghose S, Magida J, Dias C, Russo SJ, Vialou V, Caldarone BJ, Tamminga CA, Nestler EJ, Zachariou V. Regulator of G protein signaling 4 [corrected] is a crucial modulator of antidepressant drug action in depression and neuropathic pain models. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:8254–8259. doi: 10.1073/pnas.1214696110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willner P. Validity, reliability and utility of the chronic mild stress model of depression: a 10-year review and evaluation. Psychopharmacology. 1997;134:319–329. doi: 10.1007/s002130050456. [DOI] [PubMed] [Google Scholar]
- 44.Norman GJ, Karelina K, Zhang N, Walton JC, Morris JS, Devries AC. Stress and IL-1beta contribute to the development of depressive-like behavior following peripheral nerve injury. Molecular psychiatry. 2010;15:404–414. doi: 10.1038/mp.2009.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holter SM, Einicke J, Sperling B, Zimprich A, Garrett L, Fuchs H, Gailus-Durner V, Angelis M Hrabe de, Wurst W. Tests for Anxiety-Related Behavior in Mice. Current protocols in mouse biology. 2015;5:291–309. doi: 10.1002/9780470942390.mo150010. [DOI] [PubMed] [Google Scholar]
- 46.Berton O, Nestler EJ. New approaches to antidepressant drug discovery: beyond monoamines. Nature reviews Neuroscience. 2006;7:137–151. doi: 10.1038/nrn1846. [DOI] [PubMed] [Google Scholar]
- 47.Liu Y, Zhou J, White KP. RNA-seq differential expression studies: more sequence or more replication? Bioinformatics. 2014;30:301–304. doi: 10.1093/bioinformatics/btt688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Plaisier SB, Taschereau R, Wong JA, Graeber TG. Rank-rank hypergeometric overlap: identification of statistically significant overlap between gene-expression signatures. Nucleic Acids Res. 2010;38:e169. doi: 10.1093/nar/gkq636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bagot RC, Cates HM, Purushothaman I, Lorsch ZS, Walker DM, Wang J, Huang X, Schluter OM, Maze I, Pena CJ, Heller EA, Issler O, Wang M, Song WM, Stein JL, Liu X, Doyle MA, Scobie KN, Sun HS, Neve RL, Geschwind D, Dong Y, Shen L, Zhang B, Nestler EJ. Circuit-wide Transcriptional Profiling Reveals Brain Region-Specific Gene Networks Regulating Depression Susceptibility. Neuron. 2016;90:969–983. doi: 10.1016/j.neuron.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nature neuroscience. 2006;9:519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- 51.Covington HE, 3rd, Maze I, LaPlant QC, Vialou VF, Ohnishi YN, Berton O, Fass DM, Renthal W, Rush AJ, 3rd, Wu EY, Ghose S, Krishnan V, Russo SJ, Tamminga C, Haggarty SJ, Nestler EJ. Antidepressant actions of histone deacetylase inhibitors. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29:11451–11460. doi: 10.1523/JNEUROSCI.1758-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kremer M, Salvat E, Muller A, Yalcin I, Barrot M. Antidepressants and gabapentinoids in neuropathic pain: Mechanistic insights. Neuroscience. 2016 doi: 10.1016/j.neuroscience.2016.06.057. [DOI] [PubMed] [Google Scholar]
- 53.Cai H, Chen H, Yi T, Daimon CM, Boyle JP, Peers C, Maudsley S, Martin B. VennPlex–a novel Venn diagram program for comparing and visualizing datasets with differentially regulated datapoints. PloS one. 2013;8:e53388. doi: 10.1371/journal.pone.0053388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kramer A, Green J, Pollard J, Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu HY, Lynch DR. Calpain and synaptic function. Molecular neurobiology. 2006;33:215–236. doi: 10.1385/MN:33:3:215. [DOI] [PubMed] [Google Scholar]
- 56.Willner P, Towell A, Sampson D, Sophokleous S, Muscat R. Reduction of sucrose preference by chronic unpredictable mild stress, and its restoration by a tricyclic antidepressant. Psychopharmacology. 1987;93:358–364. doi: 10.1007/BF00187257. [DOI] [PubMed] [Google Scholar]
- 57.Papp M, Moryl E, Willner P. Pharmacological validation of the chronic mild stress model of depression. European journal of pharmacology. 1996;296:129–136. doi: 10.1016/0014-2999(95)00697-4. [DOI] [PubMed] [Google Scholar]
- 58.Santarelli L, Saxe M, Gross C, Surget A, Battaglia F, Dulawa S, Weisstaub N, Lee J, Duman R, Arancio O, Belzung C, Hen R. Requirement of hippocampal neurogenesis for the behavioral effects of antidepressants. Science. 2003;301:805–809. doi: 10.1126/science.1083328. [DOI] [PubMed] [Google Scholar]
- 59.Howard LA, Miksys S, Hoffmann E, Mash D, Tyndale RF. Brain CYP2E1 is induced by nicotine and ethanol in rat and is higher in smokers and alcoholics. British journal of pharmacology. 2003;138:1376–1386. doi: 10.1038/sj.bjp.0705146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanchez-Catalan MJ, Hipolito L, Guerri C, Granero L, Polache A. Distribution and differential induction of CYP2E1 by ethanol and acetone in the mesocorticolimbic system of rat. Alcohol and alcoholism. 2008;43:401–407. doi: 10.1093/alcalc/agn012. [DOI] [PubMed] [Google Scholar]
- 61.Hipolito L, Sanchez-Catalan MJ, Polache A, Granero L. Induction of brain CYP2E1 changes the effects of ethanol on dopamine release in nucleus accumbens shell. Drug and alcohol dependence. 2009;100:83–90. doi: 10.1016/j.drugalcdep.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 62.Ledesma JC, Miquel M, Pascual M, Guerri C, Aragon CM. Induction of brain cytochrome P450 2E1 boosts the locomotor-stimulating effects of ethanol in mice. Neuropharmacology. 2014;85:36–44. doi: 10.1016/j.neuropharm.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 63.Projean D, Dautrey S, Vu HK, Groblewski T, Brazier JL, Ducharme J. Selective downregulation of hepatic cytochrome P450 expression and activity in a rat model of inflammatory pain. Pharmaceutical research. 2005;22:62–70. doi: 10.1007/s11095-004-9010-6. [DOI] [PubMed] [Google Scholar]
- 64.Hakkola J, Hu Y, Ingelman-Sundberg M. Mechanisms of down-regulation of CYP2E1 expression by inflammatory cytokines in rat hepatoma cells. The Journal of pharmacology and experimental therapeutics. 2003;304:1048–1054. doi: 10.1124/jpet.102.041582. [DOI] [PubMed] [Google Scholar]
- 65.Abdel-Razzak Z, Loyer P, Fautrel A, Gautier JC, Corcos L, Turlin B, Beaune P, Guillouzo A. Cytokines down-regulate expression of major cytochrome P-450 enzymes in adult human hepatocytes in primary culture. Molecular pharmacology. 1993;44:707–715. [PubMed] [Google Scholar]
- 66.Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, Deng S, Liddelow SA, Zhang C, Daneman R, Maniatis T, Barres BA, Wu JQ. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goll DE, Thompson VF, Li H, Wei W, Cong J. The calpain system. Physiological reviews. 2003;83:731–801. doi: 10.1152/physrev.00029.2002. [DOI] [PubMed] [Google Scholar]
- 68.Vinade L, Petersen JD, Do K, Dosemeci A, Reese TS. Activation of calpain may alter the postsynaptic density structure and modulate anchoring of NMDA receptors. Synapse. 2001;40:302–309. doi: 10.1002/syn.1053. [DOI] [PubMed] [Google Scholar]
- 69.Mahajan SS, Thai KH, Chen K, Ziff E. Exposure of neurons to excitotoxic levels of glutamate induces cleavage of the RNA editing enzyme, adenosine deaminase acting on RNA 2 and loss of GLUR2 editing. Neuroscience. 2011;189:305–315. doi: 10.1016/j.neuroscience.2011.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dore K, Labrecque S, Tardif C, De Koninck P. FRET-FLIM investigation of PSD95-NMDA receptor interaction in dendritic spines; control by calpain, CaMKII and Src family kinase. PloS one. 2014;9:e112170. doi: 10.1371/journal.pone.0112170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu JY, Jiang Y, Liu W, Huang YG. Calpain inhibitor reduces cancer-induced bone pain possibly through inhibition of osteoclastogenesis in rat cancer-induced bone pain model. Chinese medical journal. 2015;128:1102–1107. doi: 10.4103/0366-6999.155109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kharatmal SB, Singh JN, Sharma SS. Calpain inhibitor, MDL 28170 confer electrophysiological, nociceptive and biochemical improvement in diabetic neuropathy. Neuropharmacology. 2015;97:113–121. doi: 10.1016/j.neuropharm.2015.05.040. [DOI] [PubMed] [Google Scholar]
- 73.Jia P, Kao CF, Kuo PH, Zhao Z. A comprehensive network and pathway analysis of candidate genes in major depressive disorder. BMC systems biology. 2011;5(Suppl 3):S12. doi: 10.1186/1752-0509-5-S3-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bradley RG, Binder EB, Epstein MP, Tang Y, Nair HP, Liu W, Gillespie CF, Berg T, Evces M, Newport DJ, Stowe ZN, Heim CM, Nemeroff CB, Schwartz A, Cubells JF, Ressler KJ. Influence of child abuse on adult depression: moderation by the corticotropin-releasing hormone receptor gene. Archives of general psychiatry. 2008;65:190–200. doi: 10.1001/archgenpsychiatry.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wingenfeld K, Wolf OT. Effects of cortisol on cognition in major depressive disorder, posttraumatic stress disorder and borderline personality disorder - 2014 Curt Richter Award Winner. Psychoneuroendocrinology. 2015;51:282–295. doi: 10.1016/j.psyneuen.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 76.Niwa M, Jaaro-Peled H, Tankou S, Seshadri S, Hikida T, Matsumoto Y, Cascella NG, Kano S, Ozaki N, Nabeshima T, Sawa A. Adolescent stress-induced epigenetic control of dopaminergic neurons via glucocorticoids. Science. 2013;339:335–339. doi: 10.1126/science.1226931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhao Y, Ma R, Shen J, Su H, Xing D, Du L. A mouse model of depression induced by repeated corticosterone injections. European journal of pharmacology. 2008;581:113–120. doi: 10.1016/j.ejphar.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 78.Kalynchuk LE, Gregus A, Boudreau D, Perrot-Sinal TS. Corticosterone increases depression-like behavior, with some effects on predator odor-induced defensive behavior, in male and female rats. Behavioral neuroscience. 2004;118:1365–1377. doi: 10.1037/0735-7044.118.6.1365. [DOI] [PubMed] [Google Scholar]
- 79.Carlezon WA, Jr, Duman RS, Nestler EJ. The many faces of CREB. Trends in neurosciences. 2005;28:436–445. doi: 10.1016/j.tins.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 80.Wei F, Qiu CS, Kim SJ, Muglia L, Maas JW, Pineda VV, Xu HM, Chen ZF, Storm DR, Muglia LJ, Zhuo M. Genetic elimination of behavioral sensitization in mice lacking calmodulin-stimulated adenylyl cyclases. Neuron. 2002;36:713–726. doi: 10.1016/s0896-6273(02)01019-x. [DOI] [PubMed] [Google Scholar]
- 81.Wang H, Xu H, Wu LJ, Kim SS, Chen T, Koga K, Descalzi G, Gong B, Vadakkan KI, Zhang X, Kaang BK, Zhuo M. Identification of an adenylyl cyclase inhibitor for treating neuropathic and inflammatory pain. Science translational medicine. 2011;3:65ra63. doi: 10.1126/scitranslmed.3001269. [DOI] [PubMed] [Google Scholar]
- 82.Descalzi G, Fukushima H, Suzuki A, Kida S, Zhuo M. Genetic enhancement of neuropathic and inflammatory pain by forebrain upregulation of CREB-mediated transcription. Molecular pain. 2012;8:90. doi: 10.1186/1744-8069-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Barrot M, Olivier JD, Perrotti LI, DiLeone RJ, Berton O, Eisch AJ, Impey S, Storm DR, Neve RL, Yin JC, Zachariou V, Nestler EJ. CREB activity in the nucleus accumbens shell controls gating of behavioral responses to emotional stimuli. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:11435–11440. doi: 10.1073/pnas.172091899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Muschamp JW, Veer A Van’t, Parsegian A, Gallo MS, Chen M, Neve RL, Meloni EG, Carlezon WA., Jr Activation of CREB in the nucleus accumbens shell produces anhedonia and resistance to extinction of fear in rats. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:3095–3103. doi: 10.1523/JNEUROSCI.5973-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.O’Donnell JM, Zhang HT. Antidepressant effects of inhibitors of cAMP phosphodiesterase (PDE4) Trends in pharmacological sciences. 2004;25:158–163. doi: 10.1016/j.tips.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 86.Girdler SS, Klatzkin R. Neurosteroids in the context of stress: implications for depressive disorders. Pharmacology & therapeutics. 2007;116:125–139. doi: 10.1016/j.pharmthera.2007.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Romeo E, Strohle A, Spalletta G, di Michele F, Hermann B, Holsboer F, Pasini A, Rupprecht R. Effects of antidepressant treatment on neuroactive steroids in major depression. The American journal of psychiatry. 1998;155:910–913. doi: 10.1176/ajp.155.7.910. [DOI] [PubMed] [Google Scholar]
- 88.Marx CE, Keefe RS, Buchanan RW, Hamer RM, Kilts JD, Bradford DW, Strauss JL, Naylor JC, Payne VM, Lieberman JA, Savitz AJ, Leimone LA, Dunn L, Porcu P, Morrow AL, Shampine LJ. Proof-of-concept trial with the neurosteroid pregnenolone targeting cognitive and negative symptoms in schizophrenia. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2009;34:1885–1903. doi: 10.1038/npp.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Paul SM, Purdy RH. Neuroactive steroids. The FASEB Journal. 1992;6:2311–2322. [PubMed] [Google Scholar]
- 90.Holubova K, Nekovarova T, Pistovcakova J, Sulcova A, Stuchlik A, Vales K. Pregnanolone Glutamate, a Novel Use-Dependent NMDA Receptor Inhibitor, Exerts Antidepressant-Like Properties in Animal Models. Frontiers in behavioral neuroscience. 2014;8:130. doi: 10.3389/fnbeh.2014.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pinna G, Costa E, Guidotti A. Fluoxetine and norfluoxetine stereospecifically and selectively increase brain neurosteroid content at doses that are inactive on 5-HT reuptake. Psychopharmacology. 2006;186:362–372. doi: 10.1007/s00213-005-0213-2. [DOI] [PubMed] [Google Scholar]
- 92.Sripada RK, Marx CE, King AP, Rampton JC, Ho SS, Liberzon I. Allopregnanolone Elevations Following Pregnenolone Administration Are Associated with Enhanced Activation of Emotion Regulation Neurocircuits. Biological Psychiatry. 2013;73:1045–1053. doi: 10.1016/j.biopsych.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tonsfeldt KJ, Suchland KL, Beeson KA, Lowe JD, Li MH, Ingram SL. Sex Differences in GABAA Signaling in the Periaqueductal Gray Induced by Persistent Inflammation. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2016;36:1669–1681. doi: 10.1523/JNEUROSCI.1928-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hodes GE, Kana V, Menard C, Merad M, Russo SJ. Neuroimmune mechanisms of depression. Nature neuroscience. 2015;18:1386–1393. doi: 10.1038/nn.4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dantzer R, O’Connor JC, Freund GG, Johnson RW, Kelley KW. From inflammation to sickness and depression: when the immune system subjugates the brain. Nature reviews Neuroscience. 2008;9:46–56. doi: 10.1038/nrn2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Maletic V, Raison CL. Neurobiology of depression, fibromyalgia and neuropathic pain. Frontiers in bioscience. 2009;14:5291–5338. doi: 10.2741/3598. [DOI] [PubMed] [Google Scholar]
- 97.Moreno FA, Erickson RP, Garriock HA, Gelernter J, Mintz J, Oas-Terpstra J, Davies MA, Delgado PL. Association Study of Genotype by Depressive Response during Tryptophan Depletion in Subjects Recovered from Major Depression. Molecular neuropsychiatry. 2015;1:165–174. doi: 10.1159/000439114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zill P, Baghai TC, Zwanzger P, Schule C, Eser D, Rupprecht R, Moller HJ, Bondy B, Ackenheil M. SNP and haplotype analysis of a novel tryptophan hydroxylase isoform (TPH2) gene provide evidence for association with major depression. Molecular psychiatry. 2004;9:1030–1036. doi: 10.1038/sj.mp.4001525. [DOI] [PubMed] [Google Scholar]
- 99.Zhou Z, Roy A, Lipsky R, Kuchipudi K, Zhu G, Taubman J, Enoch MA, Virkkunen M, Goldman D. Haplotype-based linkage of tryptophan hydroxylase 2 to suicide attempt, major depression, and cerebrospinal fluid 5-hydroxyindoleacetic acid in 4 populations. Archives of general psychiatry. 2005;62:1109–1118. doi: 10.1001/archpsyc.62.10.1109. [DOI] [PubMed] [Google Scholar]
- 100.Hammer M, Mages J, Dietrich H, Servatius A, Howells N, Cato AC, Lang R. Dual specificity phosphatase 1 (DUSP1) regulates a subset of LPS-induced genes and protects mice from lethal endotoxin shock. The Journal of experimental medicine. 2006;203:15–20. doi: 10.1084/jem.20051753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ndong C, Landry RP, DeLeo JA, Romero-Sandoval EA. Mitogen activated protein kinase phosphatase-1 prevents the development of tactile sensitivity in a rodent model of neuropathic pain. Molecular pain. 2012;8:34. doi: 10.1186/1744-8069-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Duric V, Banasr M, Licznerski P, Schmidt HD, Stockmeier CA, Simen AA, Newton SS, Duman RS. A negative regulator of MAP kinase causes depressive behavior. Nature medicine. 2010;16:1328–1332. doi: 10.1038/nm.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kurumaji A, Ito T, Ishii S, Nishikawa T. Effects of FG7142 and immobilization stress on the gene expression in the neocortex of mice. Neuroscience research. 2008;62:155–159. doi: 10.1016/j.neures.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 104.Li B, Suemaru K, Kitamura Y, Gomita Y, Araki H, Cui R. Imipramine-induced c-Fos expression in the medial prefrontal cortex is decreased in the ACTH-treated rats. Journal of biochemical and molecular toxicology. 2013;27:486–491. doi: 10.1002/jbt.21510. [DOI] [PubMed] [Google Scholar]
- 105.Johnson RS, Spiegelman BM, Papaioannou V. Pleiotropic effects of a null mutation in the c-fos proto-oncogene. Cell. 1992;71:577–586. doi: 10.1016/0092-8674(92)90592-z. [DOI] [PubMed] [Google Scholar]
- 106.Miyata S, Mori Y, Tohyama M. PRMT1 and Btg2 regulates neurite outgrowth of Neuro2a cells. Neuroscience letters. 2008;445:162–165. doi: 10.1016/j.neulet.2008.08.065. [DOI] [PubMed] [Google Scholar]
- 107.Passeri D, Marcucci A, Rizzo G, Billi M, Panigada M, Leonardi L, Tirone F, Grignani F. Btg2 enhances retinoic acid-induced differentiation by modulating histone H4 methylation and acetylation. Molecular and cellular biology. 2006;26:5023–5032. doi: 10.1128/MCB.01360-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Malik AR, Urbanska M, Gozdz A, Swiech LJ, Nagalski A, Perycz M, Blazejczyk M, Jaworski J. Cyr61, a matricellular protein, is needed for dendritic arborization of hippocampal neurons. The Journal of biological chemistry. 2013;288:8544–8559. doi: 10.1074/jbc.M112.411629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Soares DM, Ott D, Melo MC, Souza GE, Roth J. Chemokine ligand (CCL)-3 promotes an integrated febrile response when injected within pre-optic area (POA) of rats and induces calcium signaling in cells of POA microcultures but not TNF-alpha or IL-6 synthesis. Brain, behavior, and immunity. 2013;34:120–129. doi: 10.1016/j.bbi.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 110.Chui R, Dorovini-Zis K. Regulation of CCL2 and CCL3 expression in human brain endothelial cells by cytokines and lipopolysaccharide. Journal of neuroinflammation. 2010;7:1. doi: 10.1186/1742-2094-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Knerlich-Lukoschus F, Noack M, von der Ropp-Brenner B, Lucius R, Mehdorn HM, Held-Feindt J. Spinal cord injuries induce changes in CB1 cannabinoid receptor and C-C chemokine expression in brain areas underlying circuitry of chronic pain conditions. Journal of neurotrauma. 2011;28:619–634. doi: 10.1089/neu.2010.1652. [DOI] [PubMed] [Google Scholar]
- 112.Lindqvist D, Hall S, Surova Y, Nielsen HM, Janelidze S, Brundin L, Hansson O. Cerebrospinal fluid inflammatory markers in Parkinson’s disease–associations with depression, fatigue, and cognitive impairment. Brain, behavior, and immunity. 2013;33:183–189. doi: 10.1016/j.bbi.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 113.Merendino RA, Di Pasquale G, De Luca F, Di Pasquale L, Ferlazzo E, Martino G, Palumbo MC, Morabito F, Gangemi S. Involvement of fractalkine and macrophage inflammatory protein-1 alpha in moderate-severe depression. Mediators of inflammation. 2004;13:205–207. doi: 10.1080/09511920410001713484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Greenberg ME, Ziff EB. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984;311:433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- 115.Frankland PW, Bontempi B, Talton LE, Kaczmarek L, Silva AJ. The involvement of the anterior cingulate cortex in remote contextual fear memory. Science. 2004;304:881–883. doi: 10.1126/science.1094804. [DOI] [PubMed] [Google Scholar]
- 116.Barth AL, Gerkin RC, Dean KL. Alteration of neuronal firing properties after in vivo experience in a FosGFP transgenic mouse. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2004;24:6466–6475. doi: 10.1523/JNEUROSCI.4737-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nestler EJ. FosB: a transcriptional regulator of stress and antidepressant responses. European journal of pharmacology. 2015;753:66–72. doi: 10.1016/j.ejphar.2014.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Miyata S, Yoshikawa K, Taniguchi M, Ishikawa T, Tanaka T, Shimizu S, Tohyama M. Sgk1 regulates desmoglein 1 expression levels in oligodendrocytes in the mouse corpus callosum after chronic stress exposure. Biochemical and biophysical research communications. 2015;464:76–82. doi: 10.1016/j.bbrc.2015.05.109. [DOI] [PubMed] [Google Scholar]
- 119.Anacker C, Cattaneo A, Musaelyan K, Zunszain PA, Horowitz M, Molteni R, Luoni A, Calabrese F, Tansey K, Gennarelli M, Thuret S, Price J, Uher R, Riva MA, Pariante CM. Role for the kinase SGK1 in stress, depression, and glucocorticoid effects on hippocampal neurogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:8708–8713. doi: 10.1073/pnas.1300886110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Amilhon B, Lepicard E, Renoir T, Mongeau R, Popa D, Poirel O, Miot S, Gras C, Gardier AM, Gallego J, Hamon M, Lanfumey L, Gasnier B, Giros B, El Mestikawy S. VGLUT3 (vesicular glutamate transporter type 3) contribution to the regulation of serotonergic transmission and anxiety. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:2198–2210. doi: 10.1523/JNEUROSCI.5196-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Attal N, Bouhassira D. Pharmacotherapy of neuropathic pain: which drugs, which treatment algorithms? Pain. 2015;156(Suppl 1):S104–114. doi: 10.1097/01.j.pain.0000460358.01998.15. [DOI] [PubMed] [Google Scholar]
- 122.Gurnot C, Martin-Subero I, Mah SM, Weikum W, Goodman SJ, Brain U, Werker JF, Kobor MS, Esteller M, Oberlander TF, Hensch TK. Prenatal antidepressant exposure associated with CYP2E1 DNA methylation change in neonates. Epigenetics. 2015;10:361–372. doi: 10.1080/15592294.2015.1026031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cai YQ, Wang W, Hou YY, Pan ZZ. Optogenetic activation of brainstem serotonergic neurons induces persistent pain sensitization. Molecular pain. 2014;10:70. doi: 10.1186/1744-8069-10-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ni HD, Yao M, Huang B, Xu LS, Zheng Y, Chu YX, Wang HQ, Liu MJ, Xu SJ, Li HB. Glial activation in the periaqueductal gray promotes descending facilitation of neuropathic pain through the p38 MAPK signaling pathway. Journal of neuroscience research. 2016;94:50–61. doi: 10.1002/jnr.23672. [DOI] [PubMed] [Google Scholar]
- 125.Delaleu N, Immervoll H, Cornelius J, Jonsson R. Biomarker profiles in serum and saliva of experimental Sjogren’s syndrome: associations with specific autoimmune manifestations. Arthritis research & therapy. 2008;10:R22. doi: 10.1186/ar2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li XY, Ko HG, Chen T, Descalzi G, Koga K, Wang H, Kim SS, Shang Y, Kwak C, Park SW, Shim J, Lee K, Collingridge GL, Kaang BK, Zhuo M. Alleviating neuropathic pain hypersensitivity by inhibiting PKMzeta in the anterior cingulate cortex. Science. 2010;330:1400–1404. doi: 10.1126/science.1191792. [DOI] [PubMed] [Google Scholar]
- 127.Geranton SM, Morenilla-Palao C, Hunt SP. A role for transcriptional repressor methyl-CpG-binding protein 2 and plasticity-related gene serum- and glucocorticoid-inducible kinase 1 in the induction of inflammatory pain states. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27:6163–6173. doi: 10.1523/JNEUROSCI.1306-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jacobson KA, Gao ZG. Adenosine receptors as therapeutic targets. Nature reviews Drug discovery. 2006;5:247–264. doi: 10.1038/nrd1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lahdesmaki J, Sallinen J, MacDonald E, Scheinin M. Alpha2A-adrenoceptors are important modulators of the effects of D-amphetamine on startle reactivity and brain monoamines. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2004;29:1282–1293. doi: 10.1038/sj.npp.1300428. [DOI] [PubMed] [Google Scholar]
- 130.Ledent C, Vaugeois JM, Schiffmann SN, Pedrazzini T, El Yacoubi M, Vanderhaeghen JJ, Costentin J, Heath JK, Vassart G, Parmentier M. Aggressiveness, hypoalgesia and high blood pressure in mice lacking the adenosine A2a receptor. Nature. 1997;388:674–678. doi: 10.1038/41771. [DOI] [PubMed] [Google Scholar]
- 131.Mishima K, Tanoue A, Tsuda M, Hasebe N, Fukue Y, Egashira N, Takano Y, Kamiya HO, Tsujimoto G, Iwasaki K, Fujiwara M. Characteristics of behavioral abnormalities in alpha1d-adrenoceptors deficient mice. Behavioural brain research. 2004;152:365–373. doi: 10.1016/j.bbr.2003.10.038. [DOI] [PubMed] [Google Scholar]
- 132.Harasawa I, Honda K, Tanoue A, Shinoura H, Ishida Y, Okamura H, Murao N, Tsujimoto G, Higa K, Kamiya HO, Takano Y. Responses to noxious stimuli in mice lacking alpha(1d)-adrenergic receptors. Neuroreport. 2003;14:1857–1860. doi: 10.1097/00001756-200310060-00020. [DOI] [PubMed] [Google Scholar]
- 133.Dale E, Bang-Andersen B, Sanchez C. Emerging mechanisms and treatments for depression beyond SSRIs and SNRIs. Biochemical pharmacology. 2015;95:81–97. doi: 10.1016/j.bcp.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 134.Niemann S, Kanki H, Fukui Y, Takao K, Fukaya M, Hynynen MN, Churchill MJ, Shefner JM, Bronson RT, Brown RH, Jr, Watanabe M, Miyakawa T, Itohara S, Hayashi Y. Genetic ablation of NMDA receptor subunit NR3B in mouse reveals motoneuronal and nonmotoneuronal phenotypes. The European journal of neuroscience. 2007;26:1407–1420. doi: 10.1111/j.1460-9568.2007.05774.x. [DOI] [PubMed] [Google Scholar]
- 135.Wang DS, Tian Z, Guo YY, Guo HL, Kang WB, Li S, Den YT, Li XB, Feng B, Feng D, Zhao JN, Liu G, Zhao MG. Anxiolytic-like effects of translocator protein (TSPO) ligand ZBD-2 in an animal model of chronic pain. Molecular pain. 2015;11:16. doi: 10.1186/s12990-015-0013-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rupprecht R, Rammes G, Eser D, Baghai TC, Schule C, Nothdurfter C, Troxler T, Gentsch C, Kalkman HO, Chaperon F, Uzunov V, McAllister KH, Bertaina-Anglade V, La Rochelle CD, Tuerck D, Floesser A, Kiese B, Schumacher M, Landgraf R, Holsboer F, Kucher K. Translocator protein (18 kD) as target for anxiolytics without benzodiazepine-like side effects. Science. 2009;325:490–493. doi: 10.1126/science.1175055. [DOI] [PubMed] [Google Scholar]
- 137.Rocca P, Beoni AM, Eva C, Ferrero P, Zanalda E, Ravizza L. Peripheral benzodiazepine receptor messenger RNA is decreased in lymphocytes of generalized anxiety disorder patients. Biol Psychiatry. 1998;43:767–773. doi: 10.1016/s0006-3223(97)00279-5. [DOI] [PubMed] [Google Scholar]
- 138.Chaki S, Kawashima N, Suzuki Y, Shimazaki T, Okuyama S. Cocaine- and amphetamine-regulated transcript peptide produces anxiety-like behavior in rodents. European journal of pharmacology. 2003;464:49–54. doi: 10.1016/s0014-2999(03)01368-2. [DOI] [PubMed] [Google Scholar]
- 139.Dominguez G, del Giudice EM, Kuhar MJ. CART peptide levels are altered by a mutation associated with obesity at codon 34. Molecular psychiatry. 2004;9:1065–1066. doi: 10.1038/sj.mp.4001578. [DOI] [PubMed] [Google Scholar]
- 140.Vicentic A, Jones DC. The CART (cocaine- and amphetamine-regulated transcript) system in appetite and drug addiction. The Journal of pharmacology and experimental therapeutics. 2007;320:499–506. doi: 10.1124/jpet.105.091512. [DOI] [PubMed] [Google Scholar]
- 141.Damaj MI, Hunter RG, Martin BR, Kuhar MJ. Intrathecal CART (55–102) enhances the spinal analgesic actions of morphine in mice. Brain research. 2004;1024:146–149. doi: 10.1016/j.brainres.2004.07.058. [DOI] [PubMed] [Google Scholar]
- 142.Feng F, Lu SS, Hu CY, Gong FF, Qian ZZ, Yang HY, Wu YL, Zhao YY, Bi P, Sun YH. Association between apolipoprotein E gene polymorphism and depression. Journal of clinical neuroscience : official journal of the Neurosurgical Society of Australasia. 2015;22:1232–1238. doi: 10.1016/j.jocn.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 143.Robertson J, Curley J, Kaye J, Quinn J, Pfankuch T, Raber J. apoE isoforms and measures of anxiety in probable AD patients and Apoe−/− mice. Neurobiology of aging. 2005;26:637–643. doi: 10.1016/j.neurobiolaging.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 144.Raber J, Akana SF, Bhatnagar S, Dallman MF, Wong D, Mucke L. Hypothalamic-pituitary-adrenal dysfunction in Apoe(−/−) mice: possible role in behavioral and metabolic alterations. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2000;20:2064–2071. doi: 10.1523/JNEUROSCI.20-05-02064.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Fullerton SM, Strittmatter WJ, Matthew WD. Peripheral sensory nerve defects in apolipoprotein E knockout mice. Experimental neurology. 1998;153:156–163. doi: 10.1006/exnr.1998.6872. [DOI] [PubMed] [Google Scholar]
- 146.Tochigi M, Iwamoto K, Bundo M, Sasaki T, Kato N, Kato T. Gene expression profiling of major depression and suicide in the prefrontal cortex of postmortem brains. Neuroscience research. 2008;60:184–191. doi: 10.1016/j.neures.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 147.Nakamura E, Kadomatsu K, Yuasa S, Muramatsu H, Mamiya T, Nabeshima T, Fan QW, Ishiguro K, Igakura T, Matsubara S, Kaname T, Horiba M, Saito H, Muramatsu T. Disruption of the midkine gene (Mdk) resulted in altered expression of a calcium binding protein in the hippocampus of infant mice and their abnormal behaviour. Genes to cells : devoted to molecular & cellular mechanisms. 1998;3:811–822. doi: 10.1046/j.1365-2443.1998.00231.x. [DOI] [PubMed] [Google Scholar]
- 148.Cryan JF, Kelly PH, Neijt HC, Sansig G, Flor PJ, van Der Putten H. Antidepressant and anxiolytic-like effects in mice lacking the group III metabotropic glutamate receptor mGluR7. The European journal of neuroscience. 2003;17:2409–2417. doi: 10.1046/j.1460-9568.2003.02667.x. [DOI] [PubMed] [Google Scholar]
- 149.Deacon RM, Brook RC, Meyer D, Haeckel O, Ashcroft FM, Miki T, Seino S, Liss B. Behavioral phenotyping of mice lacking the K ATP channel subunit Kir6.2. Physiology & behavior. 2006;87:723–733. doi: 10.1016/j.physbeh.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 150.Li HH, Yu WH, Rozengurt N, Zhao HZ, Lyons KM, Anagnostaras S, Fanselow MS, Suzuki K, Vanier MT, Neufeld EF. Mouse model of Sanfilippo syndrome type B produced by targeted disruption of the gene encoding alpha-N-acetylglucosaminidase. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:14505–14510. doi: 10.1073/pnas.96.25.14505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhu XR, Maskri L, Herold C, Bader V, Stichel CC, Gunturkun O, Lubbert H. Non-motor behavioural impairments in parkin-deficient mice. The European journal of neuroscience. 2007;26:1902–1911. doi: 10.1111/j.1460-9568.2007.05812.x. [DOI] [PubMed] [Google Scholar]
- 152.Rizk A, Curley J, Robertson J, Raber J. Anxiety and cognition in histamine H3 receptor−/− mice. The European journal of neuroscience. 2004;19:1992–1996. doi: 10.1111/j.1460-9568.2004.03251.x. [DOI] [PubMed] [Google Scholar]
- 153.Drapeau E, Dorr NP, Elder GA, Buxbaum JD. Absence of strong strain effects in behavioral analyses of Shank3-deficient mice. Disease models & mechanisms. 2014;7:667–681. doi: 10.1242/dmm.013821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Frisch C, Dere E, Silva MA, Godecke A, Schrader J, Huston JP. Superior water maze performance and increase in fear-related behavior in the endothelial nitric oxide synthase-deficient mouse together with monoamine changes in cerebellum and ventral striatum. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2000;20:6694–6700. doi: 10.1523/JNEUROSCI.20-17-06694.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Boettger MK, Uceyler N, Zelenka M, Schmitt A, Reif A, Chen Y, Sommer C. Differences in inflammatory pain in nNOS-, iNOS- and eNOS-deficient mice. European journal of pain. 2007;11:810–818. doi: 10.1016/j.ejpain.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 156.Inoue T, Hoshina N, Nakazawa T, Kiyama Y, Kobayashi S, Abe T, Yamamoto T, Manabe T, Yamamoto T. LMTK3 deficiency causes pronounced locomotor hyperactivity and impairs endocytic trafficking. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2014;34:5927–5937. doi: 10.1523/JNEUROSCI.1621-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gavioli EC, Vaughan CW, Marzola G, Guerrini R, Mitchell VA, Zucchini S, De Lima TC, Rae GA, Salvadori S, Regoli D, Calo G. Antidepressant-like effects of the nociceptin/orphanin FQ receptor antagonist UFP-101: new evidence from rats and mice. Naunyn-Schmiedeberg’s archives of pharmacology. 2004;369:547–553. doi: 10.1007/s00210-004-0939-0. [DOI] [PubMed] [Google Scholar]
- 158.Lutz PE, Zhou Y, Labbe A, Mechawar N, Turecki G. Decreased expression of nociceptin/orphanin FQ in the dorsal anterior cingulate cortex of suicides. European neuropsychopharmacology : the journal of the European College of Neuropsychopharmacology. 2015;25:2008–2014. doi: 10.1016/j.euroneuro.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Jenck F, Moreau JL, Martin JR, Kilpatrick GJ, Reinscheid RK, Monsma FJ, Jr, Nothacker HP, Civelli O. Orphanin FQ acts as an anxiolytic to attenuate behavioral responses to stress. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:14854–14858. doi: 10.1073/pnas.94.26.14854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Okuda-Ashitaka E, Minami T, Matsumura S, Takeshima H, Reinscheid RK, Civelli O, Ito S. The opioid peptide nociceptin/orphanin FQ mediates prostaglandin E2-induced allodynia, tactile pain associated with nerve injury. The European journal of neuroscience. 2006;23:995–1004. doi: 10.1111/j.1460-9568.2006.04623.x. [DOI] [PubMed] [Google Scholar]
- 161.Vetter DE, Li C, Zhao L, Contarino A, Liberman MC, Smith GW, Marchuk Y, Koob GF, Heinemann SF, Vale W, Lee KF. Urocortin-deficient mice show hearing impairment and increased anxiety-like behavior. Nature genetics. 2002;31:363–369. doi: 10.1038/ng914. [DOI] [PubMed] [Google Scholar]
- 162.Welch JM, Lu J, Rodriguiz RM, Trotta NC, Peca J, Ding JD, Feliciano C, Chen M, Adams JP, Luo J, Dudek SM, Weinberg RJ, Calakos N, Wetsel WC, Feng G. Cortico-striatal synaptic defects and OCD-like behaviours in Sapap3-mutant mice. Nature. 2007;448:894–900. doi: 10.1038/nature06104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Price MG, Yoo JW, Burgess DL, Deng F, Hrachovy RA, Frost JD, Jr, Noebels JL. A triplet repeat expansion genetic mouse model of infantile spasms syndrome, Arx(GCG)10+7 with interneuronopathy, spasms in infancy, persistent seizures, and adult cognitive and behavioral impairment. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29:8752–8763. doi: 10.1523/JNEUROSCI.0915-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Vergnolle N, Bunnett NW, Sharkey KA, Brussee V, Compton SJ, Grady EF, Cirino G, Gerard N, Basbaum AI, Andrade-Gordon P, Hollenberg MD, Wallace JL. Proteinase-activated receptor-2 and hyperalgesia: A novel pain pathway. Nature medicine. 2001;7:821–826. doi: 10.1038/89945. [DOI] [PubMed] [Google Scholar]
- 165.Jiang BC, Cao DL, Zhang X, Zhang ZJ, He LN, Li CH, Zhang WW, Wu XB, Berta T, Ji RR, Gao YJ. CXCL13 drives spinal astrocyte activation and neuropathic pain via CXCR5. The Journal of clinical investigation. 2016;126:745–761. doi: 10.1172/JCI81950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zhao P, Waxman SG, Hains BC. Modulation of thalamic nociceptive processing after spinal cord injury through remote activation of thalamic microglia by cysteine cysteine chemokine ligand 21. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27:8893–8902. doi: 10.1523/JNEUROSCI.2209-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wieskopf JS, Mathur J, Limapichat W, Post MR, Al-Qazzaz M, Sorge RE, Martin LJ, Zaykin DV, Smith SB, Freitas K, Austin JS, Dai F, Zhang J, Marcovitz J, Tuttle AH, Slepian PM, Clarke S, Drenan RM, Janes J, Sharari S Al, Segall SK, Aasvang EK, Lai W, Bittner R, Richards CI, Slade GD, Kehlet H, Walker J, Maskos U, Changeux JP, Devor M, Maixner W, Diatchenko L, Belfer I, Dougherty DA, Su AI, Lummis SC, Damaj M Imad, Lester HA, Patapoutian A, Mogil JS. The nicotinic alpha6 subunit gene determines variability in chronic pain sensitivity via cross-inhibition of P2X2/3 receptors. Science translational medicine. 2015;7:287ra272. doi: 10.1126/scitranslmed.3009986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zylka MJ, Sowa NA, Taylor-Blake B, Twomey MA, Herrala A, Voikar V, Vihko P. Prostatic acid phosphatase is an ectonucleotidase and suppresses pain by generating adenosine. Neuron. 2008;60:111–122. doi: 10.1016/j.neuron.2008.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Locomotor behavior of sham and SNI mice 2 months after surgery.
Figure S2: Column dendrogram for all three brain regions.
Table S1: Differentially expressed genes in the mPFC, NAc, and PAG of SNI mice.
Table S2: Primers used for qPCR experiments.


