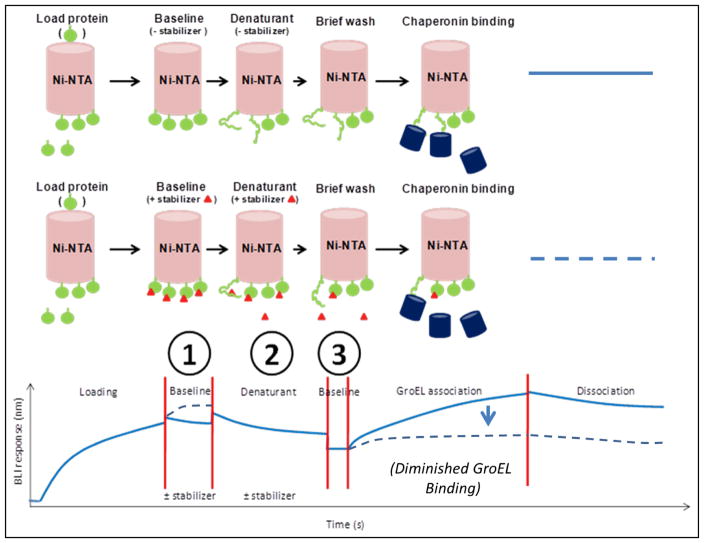
Figure 1.
Schematic illustration of a generic denaturation pulse platform for identification of kinetic protein stabilizers. Key steps include target protein loading onto biosensors followed by dipping into a buffer with or without a potential stabilizer (baseline), dipping into a chaotropic solution with or without a potential stabilizer, a wash step to remove denaturant (and compound if any) and subsequent GroEL association with protein species that expose hydrophobic residues. The following steps highlight the processes that can be varied, including 1) pretreatment time with a potential stabilizer to ensure adequate binding, 2) denaturation time to increase unfolded population and 3) wash time to remove denaturant and compound. In the presence of a stabilizer (red triangle) prior to and during the denaturant pulse phase, the GroEL binding amplitude and kinetics is diminished (dotted line). As a necessary control, profiles with any evidence of stabilizer dissociation in the absence of GroEL need to be subtracted from GroEL association phase.