Abstract
Coenzyme Q (CoQ) is a lipid present in all cell membranes. One of the multiple metabolic functions of CoQ is to transport electrons in the reaction catalyzed by sulfide:quinone oxidoreductase (SQOR), the first enzyme of the oxidation pathway of sulfides (hydrogen sulfide, H2S). Early evidence of a defect in the metabolism of H2S in primary CoQ deficiency came from yeast studies in Schizosaccharomyces pombe strains defective for dps1 and ppt1 (homologs of PDSS1 and COQ2, respectively), which have H2S accumulation. Our recent studies in human skin fibroblasts and in murine models of primary CoQ deficiency show that, also in mammals, decreased CoQ levels cause impairment of H2S oxidation. Patient fibroblasts carrying different mutations in genes encoding proteins involved in CoQ biosynthesis show reduced SQOR activity and protein levels proportional to the levels of CoQ. In Pdss2kd/kd mice, kidney, the only organ clinically affected, shows reduced SQOR levels and downstream enzymes, accumulation of H2S, and glutathione depletion. Pdss2kd/kd mice have also low levels of thiosulfate in plasma and urine, and increased C4–C6 acylcarnitines in blood, due to inhibition of short-chain acyl-CoA dehydrogenase. Also in Coq9R239X mice, the symptomatic organ, cerebrum, shows accumulation of H2S, reduced SQOR, increase in thiosulfate sulfurtransferase and sulfite oxidase, and reduction in the levels of glutathione and glutathione enzymes, leading to alteration of the biosynthetic pathways of glutamate, serotonin, and catecholamines. Coq9R239X mice have also reduced blood pressure, possible consequence of H2S-induced vasorelaxation. Since liver is not clinically affected in Pdss2 and Coq9 mutant mice, the effects of the impairment of H2S oxidation in this organ were not investigated, despite its critical role in metabolism. In conclusion, in vitro and in vivo studies of CoQ deficient models provide evidence of tissue-specific H2S oxidation impairment, an additional pathomechanism that should be considered in the understanding and treatment of primary CoQ deficiency.
Keywords: coenzyme Q, CoQ, sulfides, H2S, sulfide:quinone oxidoreductase, SQOR
Introduction: sulfide metabolism and mitochondria
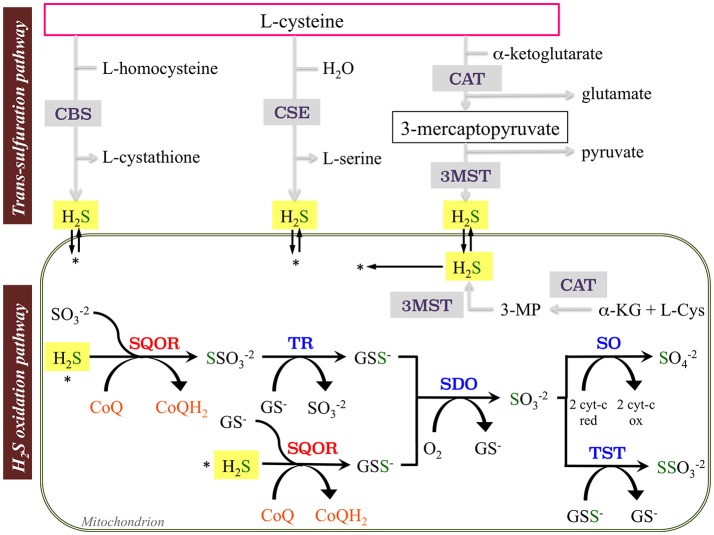
Sulfide metabolism in mammalian cells includes the trans-sulfuration (biosynthetic) and the hydrogen sulfide (H2S) oxidation (catabolic) pathways. H2S is produced endogenously by the desulfuration of cysteine or homocysteine by the cytoplasmic enzymes cystathionine β-synthase (CBS) and cystathionine γ-lyase (CSE, CTH). H2S is also produced in the reaction catalyzed by the cytosolic/mitochondrial enzyme 3-mercaptopyruvate sulfurtransferase (3-MST), which uses 3-mercaptopyruvate as substrate (Kabil and Banerjee, 2014; Figure 1). Involvement of the trans-sulfuration pathway in mitochondrial pathology has been recently demonstrated. In human dopaminergic neurons, the complex I inhibitor MPP+ induces activation of branches of the trans-sulfuration pathway, mediated by the transcription factor ATF4, to increase glutathione (GSH; Krug et al., 2014). ATF4-mediated activation of serine biosynthesis and trans-sulfuration pathway was also observed in HEK-293 cells and muscle with mitochondrial DNA depletion (Bao et al., 2016; Nikkanen et al., 2016).
Figure 1.
Schematic representation of sulfides (H2S) synthesis and oxidation pathways. The enzymes involve in the trans-sulfuration pathway are cystathionine β-synthase (CBS), cystathionine γ-lyase (CSE), and PLP-independent 3-mercaptopyruvate sulfurtransferase (3MST). The enzymes involved in the mitochondrial H2S oxidation pathway are sulfide:quinone oxidoreductase (SQOR), sulfur dioxygenase (SDO; also known as ETHE1 or persulfide dioxygenase), sulfite oxidase (SO), thiosulfate sulfurtransferase or rhodanese (TST), and thiosulfate reductase (TR) (Tables A1, A2).
At least four enzymes participate to the catabolism of H2S in the mitochondria and sequentially perform the oxidation of the sulfide into a sulfate ion (Figure 1). The first one, sulfide:quinone oxidoreductase (SQOR), transfers sulfane sulfur atoms from H2S to free sulfites, and generates thiosulfate. During this reaction, electrons are shuttled from sulfide to the mitochondrial electron transport chain by reduction of ubiquinone (CoQ) to ubiquinol (CoQH2; Jackson et al., 2012). Then, a sulfur dioxygenase (SDO or ETHE1) converts the product of this reaction, GSH persulfide (GSSH) to sulfite, releasing GSH. Sulfite can then be oxidized to sulfate by sulfite oxidase (SO); alternatively, the thiosulfate sulfurtransferase or rhodanese (TST) converts sulfide to thiosulfate via addition of a persulfide. The sulfane sulfur from thiosulfate can be remobilized by another sulfurtransferase called thiosulfate reductase (TR) and sulfate can be secreted into the blood and eliminated through the urine (Muller et al., 2004; Hildebrandt and Grieshaber, 2008; Figure 1).
The interplay of catabolism and the upstream biosynthesis pathways probably contributes to the regulation of H2S levels, as suggested by recent studies in patients and an animal model of Crohn's disease (Mottawea et al., 2016). Mottawea and colleagues showed that patients with Crohn's disease have an increase in the intestinal H2S microbial producers with a parallel decrease in the enzymes of the H2S oxidation pathway (Mottawea et al., 2016). Consistently, administration of an H2S scavenger in mice with Crohn's disease mitigated their colitis, revealing the importance of H2S oxidation pathway in inflammatory bowel disease (Mottawea et al., 2016).
Hydrogen sulfide, together with nitric oxide and carbon monoxide, is a gas modulator involved in numerous physiological functions such as cell proliferation, angiogenesis, cardioprotection, neural development, prevention of oxidative stress, and apoptosis (Bouillaud and Blachier, 2011). Several lines of evidence also indicate that at concentrations of 1–10 μM, H2S is utilized by SQOR to maintain mitochondrial electron transport and to produce ATP in mammalian cells (Modis et al., 2013). In physiological conditions, the oxidation of H2S appears to contribute marginally to the cellular oxygen consumption and mitochondrial ATP synthesis, due to the major utilization of reducing equivalents through complex I. However, cells in sulfide-rich environments as colonocytes allow SQOR functioning at its maximal rate independently of the presence of other substrates and cellular ATP demand (Lagoutte et al., 2010). Albeit, if accumulated (>10 μM), H2S becomes toxic, causing cytochrome c oxidase (COX, complex IV) deficiency, by inhibiting heme a (Di Meo et al., 2011), and dicarboxylic aciduria, through inhibition of the enzymatic activity of short-chain acyl CoA dehydrogenase (SCAD; Pedersen et al., 2003).
Therefore, H2S seems to have a consistent and congruent biphasic effect in the mitochondrial respiratory chain: at over-physiological concentrations it is a COX inhibitor while at physiological concentrations it serves as mitochondrial substrate equivalent to Krebs cycle—derived electron donors—such as, NADH or FADH2.
Hydrogen sulfide also participates in the relaxation of blood vessels by opening ATP-sensitive K+ channels in vascular smooth muscle (Yang et al., 2008), in inflammatory modulation (Yang et al., 2013) and in the production of reactive oxygen species (ROS; Eghbal et al., 2004). Accumulation of H2S in the nervous system induces increase in the concentration of serotonin and a decrease in GABA, aspartate, norepinephrine, and glutamate (Skrajny et al., 1992; Roth et al., 1995). One mechanism of action of H2S is through modification of cysteine residues of target proteins by S-sulfhydration (sulfhydration, persulfhydation). Oxidative post-translational modifications of Cys residues in proteins are important for regulation of different cell functions. S-sulfhydration usually affects proteins function exerting opposite effects of nitrosylation, therefore enhancing their function (Mustafa et al., 2009; Paul and Snyder, 2012). For example, S-sulfhydration has been shown to regulate ATP5A1 (a subunit of the mitochondrial ATP synthase; Modis et al., 2016), to increase transcriptional activity of the Krupper-like factor 5 (KLF5; Meng et al., 2016), and to induce Nrf2 dissociation from Keap1, thus enhancing Nrf2 nuclear translocation (Xie et al., 2016).
Yeast models: the first evidence of H2S accumulation in CoQ deficiency
In fission yeast, the enzyme sulfite reductase is responsible for the synthesis of sulfide from sulfite (Vande Weghe and Ow, 1999). Sulfide is necessary for the biosynthesis of cysteine and methionine; cysteine is synthetized by cysteine synthase from O-acetylserine and sulfide (Fujita and Takegawa, 2004), while homocysteine is synthetized by homocysteine synthase from O-acetylhomoserine and sulfide (Brzywczy et al., 2002; Fujita et al., 2006). Sulfide is oxidized by sulfide-quinone oxidoreductase encoded by hmt2, which was originally identified in a mutant highly sensitive to Cd2+ (Vande Weghe and Ow, 2001).
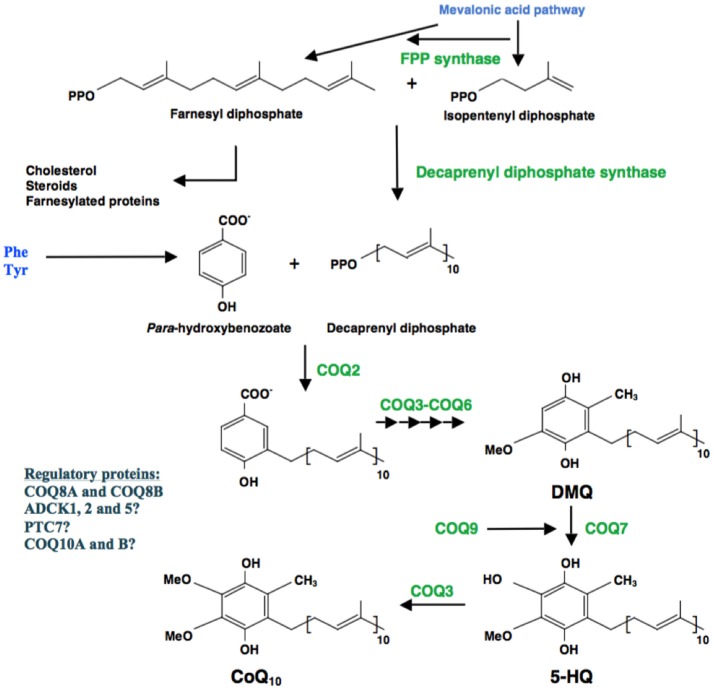
In 2000, Uchida and colleagues characterized a strain of Schizosaccharomyces pombe with a defect in its PHB polyprenyltransferase gene, ppt1 (COQ2 homolog), encoding the second enzyme of the CoQ biosynthetic pathway (Figure 2), unable to produce CoQ, and accumulating H2S (Uchida et al., 2000). This observation suggested that if cells lack CoQ, SQOR cannot function and thus the cell accumulates H2S. The same phenotype was subsequently shown to be present in S. pombe strains disrupted for all the individual coq genes (Figure 2; Zhang et al., 2008; Hayashi et al., 2014). Moreover, in all tested strains grown in both, rich and minimum media, sulfide levels were lowered by addition of cysteine, suggesting that cysteine controls the production of sulfide.
Figure 2.
Schematic representation of CoQ biosynthesis. Coenzyme Q10 (CoQ10) is the predominant form of CoQ in humans and is synthesized in the mitochondrial inner membrane. CoQ10 is composed of a benzoquinone ring, derived from tyrosine or phenylalanine, and an isoprenoid side chain, synthetized in multiple steps by the enzyme decaprenyl diphosphate synthase. PHB-polyprenyl transferase (COQ2) is responsible for the condensation of decaprenyl diphosphate and para-hydroxybenzoate (PHB). The benzoate ring is then modified by at least six enzymes, which catalyze methylation, decarboxylation, and hydroxylation reactions to synthesize CoQ10 (Tables A1, A2).
Mammalian studies In vitro: the H2S oxidation pathway is impaired in human CoQ deficiency, proportionally to the degree of CoQ deficiency and independently of the molecular defect
In mammals, CoQ is a lipid-soluble present in all cell membranes and is involved in multiple metabolic functions. One of these functions is to shuttle electrons in the first reaction of the H2S oxidation pathway, catalyzed by SQOR (Figure 1). Our studies in human fibroblasts confirm that low levels of CoQ cause decrease of SQOR protein levels, proportionally to the degree of CoQ deficiency (Luna-Sanchez et al., 2017; Ziosi et al., 2017). We showed that fibroblasts carrying mutations in different CoQ biosynthetic genes—PDSS2, COQ2, COQ4, COQ8A, and COQ9—have decrease of SQOR driven-respiration, and SQOR steady state protein levels, proportional to the severity of CoQ deficiency (Luna-Sanchez et al., 2017; Ziosi et al., 2017). The defects observed in COQ mutant fibroblasts are rescued by CoQ supplementation (Luna-Sanchez et al., 2017; Ziosi et al., 2017). Moreover, pharmacological inhibition of CoQ biosynthesis via 4-NB in wild-type fibroblasts and by COQ8A/ADCK3 depletion in HeLa cells partially recapitulate the COQ mutant cells phenotype, indicating that they are caused by CoQ deficiency (Ziosi et al., 2017). The levels of residual SQOR protein and the availability of catabolites seem to regulate the pathway downstream, since in patients and 4-NB treated fibroblasts, and in COQ8A depleted HeLa cells, SQOR protein levels are partially reduced, and the other enzymes of the pathway are increased; on the contrary, severe SQOR depletion in Hela cells shuts down the pathway (Ziosi et al., 2017).
The mRNA levels of the enzymes of the H2S oxidation pathway differ among CoQ deficient cell lines, suggesting that in CoQ deficiency, SQOR levels are regulated by CoQ amount at translational level; if the residual CoQ is very low, SQOR protein is degraded because unstable. However, the striking effects of CoQ synthesis inhibition and of CoQ supplementation on SQOR mRNA levels suggest that changes in CoQ levels modulate gene expression (Ziosi et al., 2017). There are previous evidences that CoQ affects biological processes, as lipid metabolism, inflammation, and cell signaling, through regulation of gene expression, mediated by its antioxidant function (Schmelzer et al., 2008; Fischer et al., 2016).
Sulfide accumulation in COQ mutant fibroblasts leads to increased protein S-sulfhydration, particularly targeting cytoplasmic ribosomal proteins and proteins involved in cell redox status. Further studies are needed to assess whether S-sulfhydration affects these proteins function.
Mammalian In vivo studies: the connection of the disruption in sulfide metabolism and the decreased levels of glutathione as a possible pathogenic mechanism in CoQ deficiency
We investigated the tissue-specific effects of CoQ deficiency on H2S oxidation in three mouse models with different phenotype associated to CoQ deficiency: Pdss2kd/kd mice, which carry a spontaneous mutation in Pdss2, which encodes the subunit 2 of polyprenyl-diphosphate synthase, the first enzyme of CoQ biosynthesis (Peng et al., 2004; Saiki et al., 2005), and two knock-in mice harboring mutation in Coq9 (Garcia-Corzo et al., 2013; Luna-Sanchez et al., 2015), which encodes COQ9, a protein that interact with COQ7, the enzyme responsible for the hydroxylation of demethoxyubiquinone to 5-hydroxyquinone (Figure 2; Garcia-Corzo et al., 2013). Adult Pdss2kd/kd mice develop nephrotic syndrome, and subsequently kidney failure (Madaio et al., 2005; Peng et al., 2008). The Coq9R239X knock-in mice have 10–15% of residual CoQ levels in cerebrum and kidney, and 10–20% in muscle, and manifest fatal mitochondrial encephalopathy, while the Coq9Q95X mice have 40–50% of residual CoQ in cerebrum and kidney, but 10–20% in muscle, and manifest late-onset mild mitochondrial myopathy (Garcia-Corzo et al., 2013; Luna-Sanchez et al., 2015). Lohman and colleagues previously reported that steady-state levels of SQOR were reduced in heart and kidney of Coq9R239X mice (Lohman et al., 2014).
We observed that the protein levels of SQOR in the three mouse models studied, correlate with the level of CoQ deficiency, and affect the downstream enzymes of the H2S oxidation pathway (Luna-Sanchez et al., 2017; Ziosi et al., 2017). In kidney of Pdss2kd/kd mice, which has only 15% residual CoQ, severely reduced SQOR protein levels were associated with down-regulation of all the downstream enzymes of the pathway, gluthatione (GSH) and thiosulfates reduction and mild accumulation of H2S, all indicative of a shut-down of the oxidation pathway. This alteration of the H2S oxidation pathway was also observed by severe SQOR knock down in HeLa cells, suggesting that the levels of SQOR regulate the enzymes of the down-stream pathway. In brain of Pdss2kd/kd mice, which has ~30% residual CoQ concentrations and does not show any clinical phenotype, SQOR protein levels were slightly increased in mutant mice, and the downstream H2S oxidation pathway was normal (Ziosi et al., 2017).
Also in brain, kidneys and muscle of Coq9R239X and Coq9Q95X mice, SQOR protein levels and SQOR activity correlate with the severity of CoQ deficiency. Two months of ubiquinol-10 supplementation in Coq9R239X mice increased muscle and kidney SQOR, proportionally to the increase of CoQ level, indicating that indeed CoQ deficiency causes the decrease of SQOR (Luna-Sanchez et al., 2017). As a consequence of the reduced SQOR levels, in Coq9R239X mice, TST activity was increased in cerebrum and kidneys and SO levels were increased in brain. Administration of the H2S donor GYY4137 did not increase TST levels in wild-type mice, suggesting that the increase in TST activity is not a direct consequence of increased levels of H2S, but possibly of increased protein sulfhydration, caused by H2S accumulation. The function of proteins that can be regulated by this post-translational modification would be affected, and the expression of enzymes potentially involved in the removal of persulfide groups, such as, sulfurtransferases, might be induced.
GSH, the major non-protein thiol in cells, was decreased in affected organs of Pdss2kd/kd and Coq9R239X mice. In kidney of Pdss2kd/kd GSH depletion may be caused by reactive sulfur and oxygen radical produced by H2S autoxidation (Truong et al., 2006), or by down-regulation of synthesis of GSH, to balance the increase of GSH caused by decrease of TST.
In cerebrum of Coq9R239X mice, GSH depletion may be due to a decrease in the levels of glutamate, one of the three amino acids components of GSH, with the parallel increase in N-acetylglutamate, or to a reduction of its precursors, as suggested by the decreased cerebral levels of L-glutamate, an essential aminoacid for GSH biosynthesis, or to a reduction of the levels and activity of the enzymes GPx4 and GRd, which utilize GSH. These enzymes were indeed decreased, consistently with the GSH levels. This may be critical for the increase of oxidative damage previously observed in affected organs of Pdss2kd/kd and Coq9 mutant mice (Garcia-Corzo et al., 2013; Quinzii et al., 2013), as well as in CoQ deficient human fibroblasts, where ROS and oxidative stress levels correlate with cell viability (Lopez et al., 2010; Quinzii et al., 2010, 2012).
Since we showed that SQOR depleted Hepa1c1c7 cells have GSH levels comparable to controls (Luna-Sanchez et al., 2017), it is possible that tissue-specific abnormalities of H2S metabolism contribute to oxidative stress in CoQ deficiency through alteration of the GSH system. Nevertheless, we cannot exclude other factors that may be causing the low levels of GSH in CoQ deficiency.
The primary mechanism of H2S toxicity is the inhibition of mitochondrial complex IV (CIV; Nicholls and Kim, 1982), and mutations in the gene encoding the SDO ETHE1 causes accumulation of H2S in critical tissues, including colonic mucosa, liver, muscle, and brain, leading to inhibition of short-chain CoA dehydrogenase (SCAD) and CIV activities (Tiranti et al., 2004, 2009). Pdss2kd/kd mice show increased blood levels of C4-C6 acylcarnitines, indicative of a defect of short-chain fatty acids oxidation caused by SCAD inhibition, however, surprisingly, we did not find CIV deficiency in the affected tissues of the Pdss2kd/kd and Coq9 mutant mice (Luna-Sanchez et al., 2017; Ziosi et al., 2017). It is possible that in CoQ deficiency H2S levels are not high enough to suppress CIV activity, since patients with ethylmalonic aciduria present with a much more severe phenotype associated with CIV deficiency (Tiranti et al., 2009). However, since the ethylmalonic aciduria mouse model shows normal CIV activity and level in kidney and liver, despite the high thiosulfate and H2S concentrations, we cannot exclude the presence of tissue-specific alternative metabolic pathways for H2S detoxification, or different buffering mechanisms (Tiranti et al., 2009).
Conclusions
Several evidences in vitro and in vivo show that CoQ deficiency causes dis-regulation of the H2S oxidation pathway and accumulation of H2S that may affect multiple physiological processes, possibly through modification of protein S-sulfhydration.
Impairment of H2S oxidation may contribute to oxidative stress in CoQ deficiency or may play a synergistic role with oxidative stress in the pathogenesis of tissue-specificity in CoQ deficiency. The role of H2S metabolism defects in CoQ deficiency deserves further investigation since it may have therapeutic implications.
Author contributions
CQ and LL: Study concept and design, acquisition of data, analysis and interpretation, critical revision of the manuscript for important intellectual content; ML, MZ, AH, and GK: Acquisition, analysis and interpretation of data, writing of the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Appendix
Table A1.
Abbreviations used for enzymes and other proteins.
| Abbreviation | Name |
|---|---|
| SQOR | Sulfide:quinone oxidoreductase |
| PDSS1 | Subunit 1 of polyprenyl-diphosphate synthase |
| PDSS2 | Subunit 2 of polyprenyl-diphosphate synthase |
| COQ2 | PHB-polyprenyl transferase |
| CBS | Cystathionine b-synthase |
| CSE | Cystathionine g-lyase |
| CAT | Cysteine aminotransferase |
| 3-MST | 3-Mercaptopyruvate sulfurtransferase |
| TST (TR) | Thiosulfate sulfurtransferase |
| ETHE1 (SDO) | Ethylmalonic encephalopathy protein 1 (Sulfur dioxygenase) |
| SUOX (SO) | Sulfide oxidase |
| COX | Cytochrome c oxidase |
| SCAD | Acyl CoA dehydrogenase |
| ATP5A1 | Subunit A1 of the mitochondrial ATP synthase |
| KLF5 | Krupper-like factor 5 |
| Nrf2 | Nuclear factor (erythroid-derived 2)-like 2 |
| Keap1 | Kelch like-ECH-associated protein 1 |
| COQ8A/ADCK3 | Atypical kinase COQ8A |
| COQ9 | Ubiquinone biosynthesis protein COQ9 |
| COQ7 | 5-Demethoxyubiquinone hydroxylase |
| COQ3 | Ubiquinone biosynthesis O-methyltransferase |
| COQ4 | Ubiquinone biosynthesis protein COQ4 |
| COQ5 | 2-Methoxy-6-polyprenyl-1,4-benzoquinol methylase |
| COQ6 | Ubiquinone biosynthesis monooxygenase COQ6 |
Table A2.
Abbreviations used for substrates and products in enzymatic reactions.
| Abbreviation | Name |
|---|---|
| CoQ | Coenzyme Q (ubiquinone) |
| H2S | Hydrogen sulfide |
| GSH | Reduced gluthatione |
| GSSG | Oxidized gluthatione |
| CoQH2 | Reduced coenzyme Q (ubiquinol) |
| Sulfite | |
| Thiosulfate | |
| Sulfate | |
| ROS | Reactive oxygen species |
| 2 cyt-c red | Cytochrome c reduced |
| 2 cyt-c ox | Cytochrome c oxidized |
| PHB | Para-hydroxybenzoate |
| 4-NB | 4-Nitrobenzoate |
| DMQ | Demetoxyubiquinone |
| 5-HQ | 5-Hidroxyquinone |
| L-Cys | L-Cysteine |
| α-KG | α-Ketoglutarate |
| 3-MP | 3-Mercaptopyruvate |
| FPP | Farnesyl Diphosphate |
| Phe | Phenilalanine |
| Tyr | Tyrosine |
| GPx4 | Glutathione peroxidase |
| GRd | Glutathione reductase |
Footnotes
Funding. This work was supported by NIH P01 HD080642-01 (CQ and LL). CQ receives funding also by the Muscle Dystrophy Association (MDA) and the Department of Defense (DOD). LL is supported also by grants from Ministerio de Economía y Competitividad, Spain, and the ERDF (RYC-2011-07643 and SAF2015-65786-R) and from the call “todos somos raros, todos somos únicos.” ML is a postdoctoral fellow from Fundación Ramón Areces. AH is supported by the FPU Program from the Ministerio de Educación, Cultura y Deporte, Spain.
References
- Bao X. R., Ong S. E., Goldberger O., Peng J., Sharma R., Thompson D. A., et al. (2016). Mitochondrial dysfunction remodels one-carbon metabolism in human cells. Elife 5:e10575. 10.7554/elife.10575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouillaud F., Blachier F. (2011). Mitochondria and sulfide: a very old story of poisoning, feeding, and signaling? Antioxid. Redox Signal. 15, 379–391. 10.1089/ars.2010.3678 [DOI] [PubMed] [Google Scholar]
- Brzywczy J., Sienko M., Kucharska A., Paszewski A. (2002). Sulphur amino acid synthesis in Schizosaccharomyces pombe represents a specific variant of sulphur metabolism in fungi. Yeast 19, 29–35. 10.1002/yea.798 [DOI] [PubMed] [Google Scholar]
- Di Meo I., Fagiolari G., Prelle A., Viscomi C., Zeviani M., Tiranti V. (2011). Chronic exposure to sulfide causes accelerated degradation of cytochrome c oxidase in ethylmalonic encephalopathy. Antioxid. Redox Signal. 15, 353–362. 10.1089/ars.2010.3520 [DOI] [PubMed] [Google Scholar]
- Eghbal M. A., Pennefather P. S., O'Brien P. J. (2004). H2S cytotoxicity mechanism involves reactive oxygen species formation and mitochondrial depolarisation. Toxicology 203, 69–76. 10.1016/j.tox.2004.05.020 [DOI] [PubMed] [Google Scholar]
- Fischer A., Onur S., Niklowitz P., Menke T., Laudes M., Doring F. (2016). Coenzyme Q10 redox state predicts the concentration of c-reactive protein in a large caucasian cohort. Biofactors 42, 268–276. 10.1002/biof.1269 [DOI] [PubMed] [Google Scholar]
- Fujita Y., Takegawa K. (2004). Characterization of two genes encoding putative cysteine synthase required for cysteine biosynthesis in Schizosaccharomyces pombe. Biosci. Biotechnol. Biochem. 68, 306–311. 10.1271/bbb.68.306 [DOI] [PubMed] [Google Scholar]
- Fujita Y., Ukena E., Iefuji H., Giga-Hama Y., Takegawa K. (2006). Homocysteine accumulation causes a defect in purine biosynthesis: further characterization of Schizosaccharomyces pombe methionine auxotrophs. Microbiology 152(Pt. 2), 397–404. 10.1099/mic.0.28398-0 [DOI] [PubMed] [Google Scholar]
- Garcia-Corzo L., Luna-Sanchez M., Doerrier C., Garcia J. A., Guaras A., Acin-Perez R., et al. (2013). Dysfunctional Coq9 protein causes predominant encephalomyopathy associated with CoQ deficiency. Hum. Mol. Genet. 22, 1233–1248. 10.1093/hmg/dds530 [DOI] [PubMed] [Google Scholar]
- Hayashi K., Ogiyama Y., Yokomi K., Nakagawa T., Kaino T., Kawamukai M. (2014). Functional conservation of coenzyme Q biosynthetic genes among yeasts, plants, and humans. PLoS ONE 9:e99038. 10.1371/journal.pone.0099038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt T. M., Grieshaber M. K. (2008). Three enzymatic activities catalyze the oxidation of sulfide to thiosulfate in mammalian and invertebrate mitochondria. FEBS J. 275, 3352–3361. 10.1111/j.1742-4658.2008.06482.x [DOI] [PubMed] [Google Scholar]
- Jackson M. R., Melideo S. L., Jorns M. S. (2012). Human sulfide:quinone oxidoreductase catalyzes the first step in hydrogen sulfide metabolism and produces a sulfane sulfur metabolite. Biochemistry 51, 6804–6815. 10.1021/bi300778t [DOI] [PubMed] [Google Scholar]
- Kabil O., Banerjee R. (2014). Enzymology of H2S biogenesis, decay and signaling. Antioxid. Redox Signal. 20, 770–782. 10.1089/ars.2013.5339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug A. K., Gutbier S., Zhao L., Poltl D., Kullmann C., Ivanova V., et al. (2014). Transcriptional and metabolic adaptation of human neurons to the mitochondrial toxicant MPP(+). Cell Death Dis. 5:e1222. 10.1038/cddis.2014.166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagoutte E., Mimoun S., Andriamihaja M., Chaumontet C., Blachier F., Bouillaud F. (2010). Oxidation of hydrogen sulfide remains a priority in mammalian cells and causes reverse electron transfer in colonocytes. Biochim. Biophys. Acta 1797, 1500–1511. 10.1016/j.bbabio.2010.04.004 [DOI] [PubMed] [Google Scholar]
- Lohman D. C., Forouhar F., Beebe E. T., Stefely M. S., Minogue C. E., Ulbrich A., et al. (2014). Mitochondrial COQ9 is a lipid-binding protein that associates with COQ7 to enable coenzyme Q biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 111, E4697–E4705. 10.1073/pnas.1413128111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez L. C., Quinzii C. M., Area E., Naini A., Rahman S., Schuelke M., et al. (2010). Treatment of CoQ(10) deficient fibroblasts with ubiquinone, CoQ analogs, and vitamin C: time- and compound-dependent effects. PLoS ONE 5:e11897. 10.1371/journal.pone.0011897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna-Sanchez M., Diaz-Casado E., Barca E., Tejada M. A., Montilla-Garcia A., Cobos E. J., et al. (2015). The clinical heterogeneity of coenzyme Q10 deficiency results from genotypic differences in the Coq9 gene. EMBO Mol. Med. 7, 670–687. 10.15252/emmm.201404632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna-Sanchez M., Hidalgo-Gutierrez A., Hildebrandt T. M., Chaves-Serrano J., Barriocanal-Casado E., Santos-Fandila A., et al. (2017). CoQ deficiency causes disruption of mitochondrial sulfide oxidation, a new pathomechanism associated with this syndrome. EMBO Mol. Med. 9, 78–95. 10.15252/emmm.201606345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madaio M. P., Ahima R. S., Meade R., Rader D. J., Mendoza A., Peng M., et al. (2005). Glomerular and tubular epithelial defects in kd/kd mice lead to progressive renal failure. Am. J. Nephrol. 25, 604–610. 10.1159/000089709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng G., Xiao Y., Ma Y., Tang X., Xie L., Liu J., et al. (2016). Hydrogen sulfide regulates kruppel-like factor 5 transcription activity via specificity protein 1 S-sulfhydration at Cys664 to prevent myocardial hypertrophy. J. Am. Heart Assoc. 5:e004160. 10.1161/JAHA.116.004160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modis K., Coletta C., Erdelyi K., Papapetropoulos A., Szabo C. (2013). Intramitochondrial hydrogen sulfide production by 3-mercaptopyruvate sulfurtransferase maintains mitochondrial electron flow and supports cellular bioenergetics. FASEB J. 27, 601–611. 10.1096/fj.12-216507 [DOI] [PubMed] [Google Scholar]
- Modis K., Ju Y., Ahmad A., Untereiner A. A., Altaany Z., Wu L., et al. (2016). S-Sulfhydration of ATP synthase by hydrogen sulfide stimulates mitochondrial bioenergetics. Pharmacol. Res. 113(Pt. A), 116–124. 10.1016/j.phrs.2016.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottawea W., Chiang C. K., Muhlbauer M., Starr A. E., Butcher J., Abujamel T., et al. (2016). Altered intestinal microbiota-host mitochondria crosstalk in new onset Crohn's disease. Nat. Commun. 7:13419. 10.1038/ncomms13419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller F. H., Bandeiras T. M., Urich T., Teixeira M., Gomes C. M., Kletzin A. (2004). Coupling of the pathway of sulphur oxidation to dioxygen reduction: characterization of a novel membrane-bound thiosulphate:quinone oxidoreductase. Mol. Microbiol. 53, 1147–1160. 10.1111/j.1365-2958.2004.04193.x [DOI] [PubMed] [Google Scholar]
- Mustafa A. K., Gadalla M. M., Sen N., Kim S., Mu W., Gazi S. K., et al. (2009). H2S signals through protein S-sulfhydration. Sci. Signal. 2:ra72. 10.1126/scisignal.2000464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls P., Kim J. K. (1982). Sulphide as an inhibitor and electron donor for the cytochrome c oxidase system. Can. J. Biochem. 60, 613–623. 10.1139/o82-076 [DOI] [PubMed] [Google Scholar]
- Nikkanen J., Forsstrom S., Euro L., Paetau I., Kohnz R. A., Wang L., et al. (2016). Mitochondrial DNA replication defects disturb cellular dNTP pools and remodel one-carbon metabolism. Cell Metab. 23, 635–648. 10.1016/j.cmet.2016.01.019 [DOI] [PubMed] [Google Scholar]
- Paul B. D., Snyder S. H. (2012). H(2)S signalling through protein sulfhydration and beyond. Nat. Rev. Mol. Cell Biol. 13, 499–507. 10.1038/nrm3391 [DOI] [PubMed] [Google Scholar]
- Pedersen C. B., Bross P., Winter V. S., Corydon T. J., Bolund L., Bartlett K., et al. (2003). Misfolding, degradation, and aggregation of variant proteins. The molecular pathogenesis of short chain acyl-CoA dehydrogenase (SCAD) deficiency. J. Biol. Chem. 278, 47449–47458. 10.1074/jbc.m309514200 [DOI] [PubMed] [Google Scholar]
- Peng M., Falk M. J., Haase V. H., King R., Polyak E., Selak M., et al. (2008). Primary coenzyme Q deficiency in Pdss2 mutant mice causes isolated renal disease. PLoS Genet. 4:e1000061. 10.1371/journal.pgen.1000061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M., Jarett L., Meade R., Madaio M. P., Hancock W. W., George A. L., Jr., et al. (2004). Mutant prenyltransferase-like mitochondrial protein (PLMP) and mitochondrial abnormalities in kd/kd mice. Kidney Int. 66, 20–28. 10.1111/j.1523-1755.2004.00702.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinzii C. M., Garone C., Emmanuele V., Tadesse S., Krishna S., Dorado B., et al. (2013). Tissue-specific oxidative stress and loss of mitochondria in CoQ-deficient Pdss2 mutant mice. FASEB J. 27, 612–621. 10.1096/fj.12-209361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinzii C. M., Lopez L. C., Gilkerson R. W., Dorado B., Coku J., Naini A. B., et al. (2010). Reactive oxygen species, oxidative stress, and cell death correlate with level of CoQ10 deficiency. FASEB J. 24, 3733–3743. 10.1096/fj.09-152728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinzii C. M., Tadesse S., Naini A., Hirano M. (2012). Effects of inhibiting CoQ10 biosynthesis with 4-nitrobenzoate in human fibroblasts. PLoS ONE 7:e30606. 10.1371/journal.pone.0030606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S. H., Skrajny B., Reiffenstein R. J. (1995). Alteration of the morphology and neurochemistry of the developing mammalian nervous system by hydrogen sulphide. Clin. Exp. Pharmacol. Physiol. 22, 379–380. 10.1111/j.1440-1681.1995.tb02024.x [DOI] [PubMed] [Google Scholar]
- Saiki R., Nagata A., Kainou T., Matsuda H., Kawamukai M. (2005). Characterization of solanesyl and decaprenyl diphosphate synthases in mice and humans. FEBS J. 272, 5606–5622. 10.1111/j.1742-4658.2005.04956.x [DOI] [PubMed] [Google Scholar]
- Schmelzer C., Lindner I., Rimbach G., Niklowitz P., Menke T., Doring F. (2008). Functions of coenzyme Q10 in inflammation and gene expression. Biofactors 32, 179–183. 10.1002/biof.5520320121 [DOI] [PubMed] [Google Scholar]
- Skrajny B., Hannah R. S., Roth S. H. (1992). Low concentrations of hydrogen sulphide alter monoamine levels in the developing rat central nervous system. Can. J. Physiol. Pharmacol. 70, 1515–1518. 10.1139/y92-215 [DOI] [PubMed] [Google Scholar]
- Tiranti V., D'Adamo P., Briem E., Ferrari G., Mineri R., Lamantea E., et al. (2004). Ethylmalonic encephalopathy is caused by mutations in ETHE1, a gene encoding a mitochondrial matrix protein. Am. J. Hum. Genet. 74, 239–252. 10.1086/381653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiranti V., Viscomi C., Hildebrandt T., Di Meo I., Mineri R., Tiveron C., et al. (2009). Loss of ETHE1, a mitochondrial dioxygenase, causes fatal sulfide toxicity in ethylmalonic encephalopathy. Nat. Med. 15, 200–205. 10.1038/nm.1907 [DOI] [PubMed] [Google Scholar]
- Truong D. H., Eghbal M. A., Hindmarsh W., Roth S. H., O'Brien P. J. (2006). Molecular mechanisms of hydrogen sulfide toxicity. Drug Metab. Rev. 38, 733–744. 10.1080/03602530600959607 [DOI] [PubMed] [Google Scholar]
- Uchida N., Suzuki K., Saiki R., Kainou T., Tanaka K., Matsuda H., et al. (2000). Phenotypes of fission yeast defective in ubiquinone production due to disruption of the gene for p-hydroxybenzoate polyprenyl diphosphate transferase. J. Bacteriol. 182, 6933–6939. 10.1128/JB.182.24.6933-6939.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vande Weghe J. G., Ow D. W. (1999). A fission yeast gene for mitochondrial sulfide oxidation. J. Biol. Chem. 274, 13250–13257. 10.1074/jbc.274.19.13250 [DOI] [PubMed] [Google Scholar]
- Vande Weghe J. G., Ow D. W. (2001). Accumulation of metal-binding peptides in fission yeast requires hmt2+. Mol. Microbiol. 42, 29–36. 10.1046/j.1365-2958.2001.02624.x [DOI] [PubMed] [Google Scholar]
- Xie L., Gu Y., Wen M., Zhao S., Wang W., Ma Y., et al. (2016). Hydrogen sulfide induces keap1 S-sulfhydration and suppresses diabetes-accelerated atherosclerosis via Nrf2 activation. Diabetes 65, 3171–3184. 10.2337/db16-0020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Wu L., Jiang B., Yang W., Qi J., Cao K., et al. (2008). H2S as a physiologic vasorelaxant: hypertension in mice with deletion of cystathionine gamma-lyase. Science 322, 587–590. 10.1126/science.1162667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Wang T., Yang J., Rao K., Zhan Y., Chen R. B., et al. (2013). S-allyl cysteine restores erectile function through inhibition of reactive oxygen species generation in diabetic rats. Andrology 1, 487–494. 10.1111/j.2047-2927.2012.00060.x [DOI] [PubMed] [Google Scholar]
- Zhang M., Wakitani S., Hayashi K., Miki R., Kawamukai M. (2008). High production of sulfide in coenzyme Q deficient fission yeast. Biofactors 32, 91–98. 10.1002/biof.5520320111 [DOI] [PubMed] [Google Scholar]
- Ziosi M., Di Meo I., Kleiner G., Gao X. H., Barca E., Sanchez-Quintero M. J., et al. (2017). Coenzyme Q deficiency causes impairment of the sulfide oxidation pathway. EMBO Mol. Med. 9, 96–111. 10.15252/emmm.201606356 [DOI] [PMC free article] [PubMed] [Google Scholar]