Abstract
Transcription factor DksA contains a four-Cys Zn2 +-finger motif thought to be responsible for structural integrity and the relative disposition of its domains. Pseudomonas aeruginosa encodes an additional DksA paralog (DksA2) that is expressed selectively under Zn2+ limitation. Although DksA2 does not bind Zn2+, it complements the Escherichia coli dksA deletion and has similar effects on transcription in vitro. In this study, structural and biochemical analyses reveal that DksA2 has a similar fold, domain structure and RNA polymerase binding properties to those of the E. coli DksA despite the lack of the stabilizing metal ion.
Keywords: DksA, DksA2, RNA polymerase, Zinc, Cysteine
1. Introduction
Zn2+ is an essential cofactor for many proteins [1] and its homeostasis is essential for bacterial cell survival, a need often exploited by mammalian cells which resist bacterial invasion by either depleting [2] or increasing [3] Zn2+ in infected areas. One of the strategies utilized by bacteria to maintain sufficient Zn2+ levels is to use Zn2+-containing proteins as a reservoir. For example, YtiA, a Zn-independent paralog of the ribosomal protein L31, is known to mobilize Zn2+ from the ribosomes under Zn2+ limitation [4].
A bacterial stress regulator DksA belongs to a family of DNA-independent transcription factors. Escherichia coli (EC) DksA consists of a coiled-coil (CC) domain, which approaches the RNA polymerase (RNAP) active site through the secondary channel, and a globular domain, most of which lacks secondary structure and binds to the β′ rim helices (RH) outside of the channel (Fig. 1). The fold of the globular domain and relative orientation of the two domains appear to be maintained by a Zn2+ ion chelated to four cysteines [5]. We recently reported that some bacteria also encode DksA2, a functional Zn-independent DksA paralog that is expressed under Zn2+ starvation [6]. Similar to the DksA role in bacterial virulence [7–9], a Zn-independent paralog could confer a selective advantage in evading the immune response.
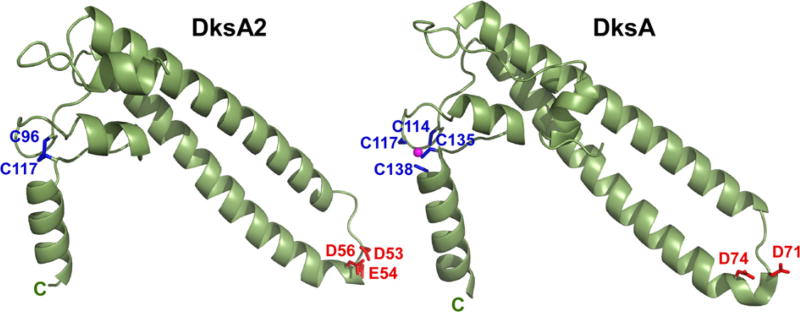
Fig. 1.
Structures of (A) PA DksA2 and (B) EC DksA (1TJL) with the key residues shown as sticks. The Zn2+ ion is shown as a sphere, the C-termini are marked.
In this work we report the structure of DksA2 from Pseudomonas aeruginosa (PA), which, to our knowledge, this is the first reported structure of a Zn-independent paralog. The structure shows remarkable preservation of the domain organization of the DksA-fold and illustrates an evolutionary strategy to stabilize a protein scaffold without the normally required metal ion. Our analysis highlights the preservation of an appropriate inter-domain interface which is essential for maintenance of the global DksA2 structure; even small perturbations at this interface impair protein folding and function.
2. Methods
2.1. Reagents
All general reagents and oligonucleotides were obtained from Sigma–Aldrich (St. Louis, MO), NTPs from GE Healthcare (Piscataway, NJ, USA), ApC from Iba (Goettingen, Germany), PCR reagents from Gene Choice (Frederick, MD, USA), restriction and modification enzymes from New England Biolabs (Ipswich, MA, USA), and [32P]-NTPs from Perkin–Elmer (Waltham, MA, USA). Plasmids used in this work are listed in Supplementary Table S1.
2.2. Proteins
DksA [5], DksA2 [6] and RNAP [10] were purified as described previously. Proteins were dialyzed against buffers containing 100 mM NaCl and 0.1 mM EDTA (with or without 5 mM DTT or β-mercaptoethanol, BME); 20 mM Na-phosphate, pH 7, was used for CD analysis, 20 mM Tris–HCl, pH 6.9, was used for all other assays. DksA2 (15 µM) was oxidized in Tris–HCl, pH 7.9, 50 mM NaCl at room temperature with 3.75 mM diamide (30 min) or 0.1% hydrogen peroxide (1 h).
2.3. Thiol modification
Modification of free cysteines in vitro was done using 15–200 µMof DksA2 and 250 mMof [PEG]12-maleimide (Thermo Fisher, Waltham, MA) in 20 mM Na-phosphate buffer, pH 7.5, 50 mM NaCl for 2 h at room temperature. For in vivo modification, E. coli cultures induced for 2 h with 0.3 mM IPTG were incubated for 30 min on ice with 10 mMof N-ethylmaleimide (NEM) or iodoacetamide (IAM), as described in [11]. Cells were then harvested and proteins were purified as described above.
2.4. X-ray crystallography and NMR analysis
Crystallization of DksA2 was carried out by vapor diffusion in hanging drops. The X-ray diffraction data were collected at the sector 21-ID of the Advanced Synchrotron Source at the Argonne National Laboratories. The details of crystallization, data collection and crystal structure determination, as well as the description of the NMR analysis, are given in Supplementary Methods. The structure and structure factor amplitudes were deposited into the Protein Data Bank under accession number 4IJJ. Calculation of solvent accessible surface area (ASA) of the globular domain was carried out with Surface Racer [12] and the ASA of this peptide in the unfolded state was estimated as previously reported [13].
3. Results
3.1. PA DksA2 structure is similar to that of EC DksA
We showed that EC DksA and PA DksA2 function similarly in vivo and in vitro, despite the absence of the Zn-finger in the latter [6]. In addition, modest similarity between DksA and DksA2 (34% identity in overall sequence, 18% in their N-terminal halves) prompted us to characterize DksA2 structurally. We overexpressed and purified PA DksA2 from E. coli and determined its crystal structure. The crystallographic data and refinement statistics are given in Supplementary Table S2.
The crystals contain three molecules of DksA2 per asymmetric unit. This crystallographic trimer is stabilized by three specifically bound sulfate ions from the crystallization solution, with one sulfate coordinating two adjacent DksA2 molecules (Supplementary Fig. S1). The structure of DksA2 is generally similar to that of EC DksA (Fig. 1), but there are several minor differences in the lengths of helices and their orientations (Supplementary Fig. S2), as well as in conformations of the loops of the globular region and the CC hairpin. In the most divergent N-terminal region, these differences include a shorter loop after the N-terminal helix in DksA2 and, consequently, a different orientation of this helix relative to the rest of the protein.
Notably, instead of a bound Zn2+ ion, the two Cys residues of DksA2 (Cys96 and Cys117) are in the immediate proximity of each other consistent with the presence of a disulfide bond, likely stabilizing this region of the protein. Remarkably, the fold of the very small globular domain of DksA2 is very similar to that of DksA despite the absence of Zn2+ coordination in DksA2. In the 64-residue globular domain of DksA2, only about 3700 Å2 of solvent accessible surface area is buried upon folding of an extended peptide, indicative of a very small hydrophobic core. Such small globular domains (in the absence of structural metals) are very marginally stable [14].
3.2. DksA2 interacts with RNAP similarly to DksA
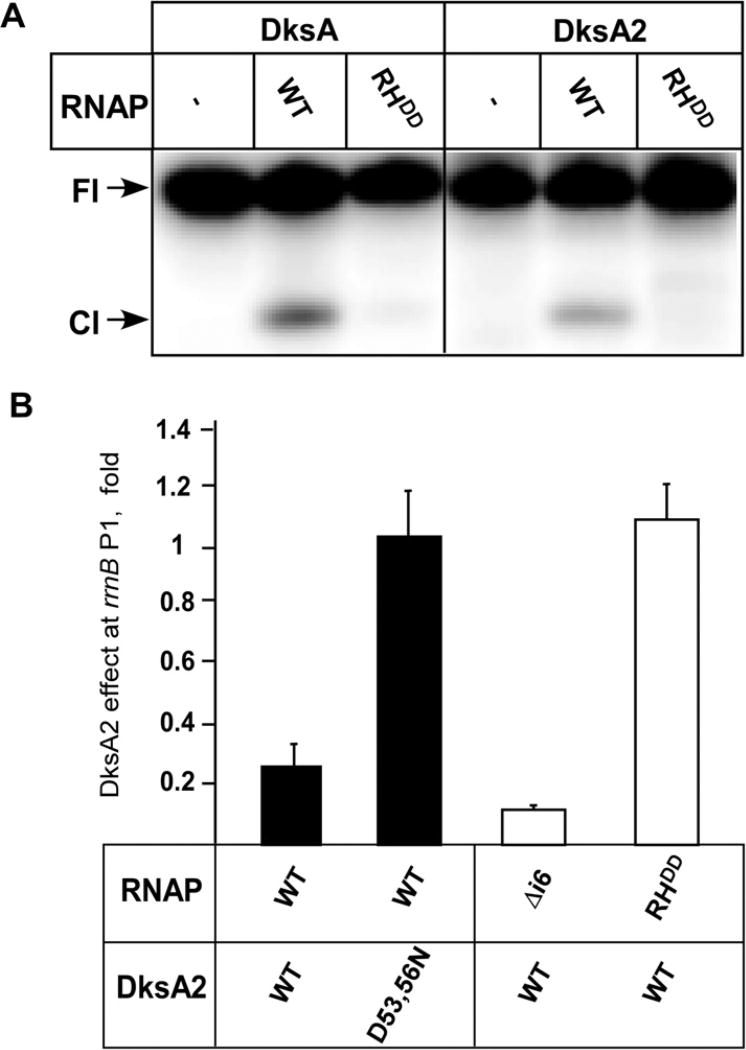
To examine whether, like EC DksA, DksA2 binds in close proximity to the RNAP active site located at the base of the secondary channel, we used Fe2+-mediated cleavage. In this assay, ▪OH radicals generated by Fe2+ ion bound in the RNAP active site cleave nearby protein or nucleic acid regions [5,15,16]. We found that the end-labeled DksA2 was cleaved similarly to DksA, and only in the presence of RNAP (Fig. 2A), indicating that DksA2 also binds near the enzyme’s active site.
Fig. 2.
DksA2 and DksA bind to RNAP similarly. (A) Binding through the secondary channel was assayed using localized Fe2+ cleavage as described in [16] using 20 nM [32P]-labeled DksA or DksA2 and 400 nM E. coli RNAP (WT or the RHDD variant). (B) Effects of mutations in DksA2 and RNAP on transcription from the rrnB P1 promoter. Assays were performed in three repeats as described in [6] with 5 µMDksA2, 30 nM RNAP, and 10 nM template; ApC, UTP and [α32P]-GTP were used as substrates. Gels were quantified by phosphorimaging (ImageQuant software, Molecular Dynamics).
Next, we tested the effect of a double substitution (β′672D, 673D) at the tip of the RH domain (RHDD) on DksA2 activity; these substitutions abolished EC DksA binding to RNAP [17]. We measured synthesis of a 4-nt RNA product from the rrnB P1 promoter, which is inhibited by both DksA and DksA2 [18]. We found that DksA2 was unable to act on and bind to RHDD RNAP (Fig. 2), suggesting that the RH serve as a binding site for DksA2, as they do for DksA.
We recently proposed that an i6 insertion in the β′ trigger loop hinders EC DksA access into the secondary channel; consistently, deletion of i6 augments DksA effects on transcription [19]. In support for a common mode of interactions with RNAP, inhibition of transcription from rrnB P1 by DksA2 was increased in the absence of i6 (Fig. 2B).
Finally, all characterized secondary channel regulators have at least one essential acidic residue at the tip of their CC domains [5,15].We found that, similarly to all other members of this family, substitution of CC tip residues Asp53 and Asp56 of DksA2 for Asn abolished DksA2 activity (Fig. 2B).
3.3. DksA2 cysteines can form a disulfide bond
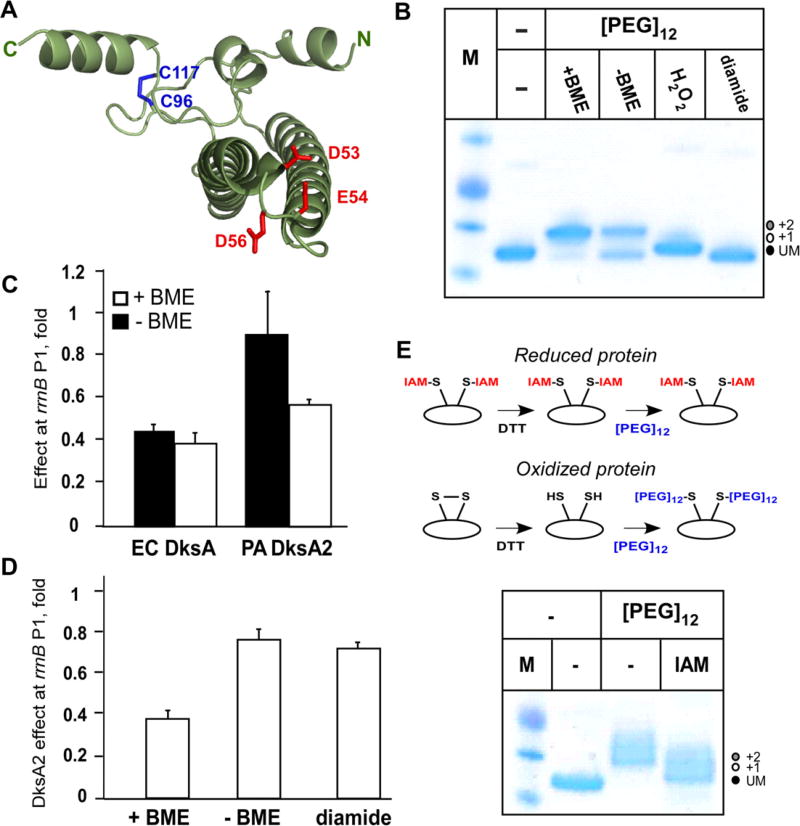
In DksA, the Zn-finger is formed by four cysteines, two from the region lacking the secondary structure and two from the C-terminal helix (Fig. 1). Thus, the Zn-finger has been proposed to dictate the orientation between these two structural elements and possibly the overall stability. In DksA2, the two remaining cysteines are juxtaposed (Fig. 3A), suggesting that a disulfide bond could play an analogous role. To test this idea, we used [PEG]12-maleimide, which binds covalently to free thiols and consequently increases the protein size by 710 Da per free cysteine. Fig. 3B shows that reduced (during dialysis in the presence of 2 mM BME) DksA2 reacted almost completely with [PEG]12-maleimide, whereas only ~60% of protein was modified in the absence of BME, suggesting partial oxidation. Treatment with hydrogen peroxide, which oxidizes free thiols predominantly into sulfonyls, or diamide, which induces the formation of disulfide bonds, protected DksA2 from modification. These results indicate that Cys96 and Cys117 can form a covalent bond.
Fig. 3.
A disulfide bond in DksA2. (A) A bridge between Cys96 and Cys117 observed in the structure. (B) Probing the thiols with [PEG]12-maleimide. Labeling was done for 2 h at room temperature and samples were analyzed on a 12% SDS–PAGE gel. (C) DksA2 activity is modestly increased in buffers containing BME. Transcription from the rrnB P1 promoter was assayed as described in Fig. 2; the fold effect corresponds to a fraction of overall transcription in the absence of DksA. (D) Oxidation has a modest inhibitory effect on DksA2 activity. (E) DksA2 cysteines are mostly in a reduced form in the cell. Cell were treated with IAM prior to lysis. DksA2 was purified and treated with [PEG]12, as above.
3.4. The disulfide bond plays a minor role in DksA2 activity in vivo and in vitro
If the disulfide bond is necessary to maintain the correct orientation between its domains, reducing conditions would be expected to decrease DksA2 activity. We tested this prediction by measuring DksA2 activity at the rrnB P1 promoter in the presence or absence of BME; EC DksA was used as a control to ensure that removing BME does not affect the transcription complex. Surprisingly, DksA2 activity was modestly increased in the presence of BME (Fig. 3C). Treating DksA2 with diamide or dialyzing DksA2 in non-reducing conditions had a similar modest inhibitory effect (Fig. 3D). We conclude that the disulfide bond is not essential, and may be even inhibitory, for DksA2 function; we cannot exclude the possibility that inhibition under the oxidizing conditions is due to modification of other DksA2 residues.
Even though DksA2 is likely located in the cytoplasm, which is reducing in E. coli [20] and P. aeruginosa [21], the close proximity of the cysteines suggests that they may form a bridge in vivo. To test this prediction, we performed thiol alkylation of free cysteines in the cell. The method, reviewed by Hansen and Winther [22] and illustrated in Fig. 3E, is composed of four major steps: (i) irreversibly blocking free thiols with a membrane-permeable alkylating agent; (ii) washing the alkylating agent and purifying the protein; (iii) reducing any preexisting disulfide bonds by dialysis into BME-containing buffer; and (iv) detecting free thiols by treatment with [PEG]12-maleimide. Only those cysteines that were protected from alkylation in the cell, but are exposed after purification and treatment with BME, will react with [PEG]12-maleimide. We found that treating the cells with IAM (Fig. 3D) or NEM (Supplementary Fig. S3) significantly reduced modification by [PEG]12-maleimide after purification. This result indicates that Cys96 and Cys117 residues do not form a stable disulfide bond but cannot rule out a transient bridge in dynamic equilibrium with free thiols in vivo. We cannot exclude a possibility that overexpressing DksA2 may change its oxidative state, but we note that high levels of DksA2 are required to compensate for ΔdksA phenotypes in vivo [6].
3.5. Substitutions of DksA2 cysteines destabilize the tertiary structure
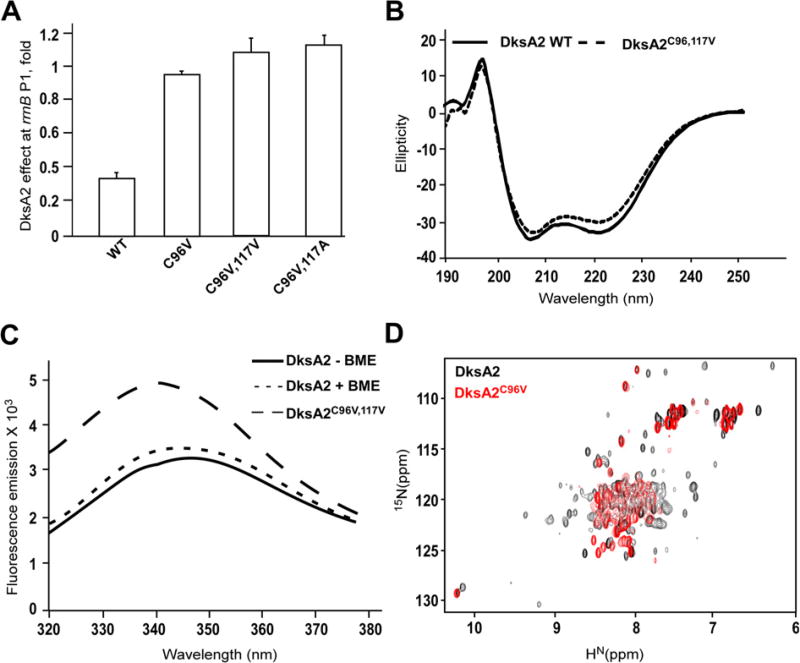
Since formation (or reduction) of a disulfide bond did not significantly affect DksA2 activity, we reasoned that substitutions of Cys96 and Cys117 for Val should have a similarly modest effect. Surprisingly, we found that even a single C96V substitution largely abolished DksA2 inhibitory activity at rrnB P1 promoter in vitro, whereas double substitutions C96V/C117V and C96V/C117A eliminated it (Fig. 4A). The latter result is particularly striking, since a Val–Ala pair is almost isosteric and similarly non-polar to a Cys–Cys pair [23], and suggests that the domain interface is very sensitive even to minor perturbations of geometry and/or polarity.
Fig. 4.
Effects of substitutions of DksA2 cysteines. (A) Substitutions of Cys96 and Cys117 abolish DksA2 activity at rrnB P1. (B) Substitutions of two Cys residues have minimal effect on DksA2 secondary structure as monitored by circular dichroism, featuring strong helical signatures at 208 and 222 nm. (C) Intrinsic protein fluorescence emission of DksA2WT and DksA2C96V,117V (200 nM) in 20 mM HEPES, pH 7.5, 50 mM NaCl. Excitation was done at 285 nm and emission scan was done between 320 and 380 nm with a 5 nm slit size using F7000 Fluorimeter (Hitachi). (D) Overlay of two-dimensional 15N–1H HSQC spectra of DksA2 (black) and DksA2C96V (red). Loss of dispersed resonances in the DksAC96V spectrum suggests destabilization of the globular domain.
To examine possible changes in the secondary structure, we compared circular dichroism (CD) spectra of the wild-type (WT) DksA2 and the DksA2C96V,117V variant. As expected from the high proportion of helical structure in DksA2, the CD spectra are dominated by helices (i.e., strong minima at 208 and 222 nm). Notably, we did not observe significant changes between the two proteins (Fig. 4B).
To inspect differences in the tertiary structure, we used the intrinsic protein emission assay [24] which relies on the difference in emission of tryptophan residues in different environments. DksA2 contains a Trp residue in each domain and exhibits identical emission spectra with a peak at 345 nm both in the absence and in the presence of BME (Fig. 4C), arguing against disulfide bond-induced structural changes. In contrast, a significant increase in overall fluorescence and an emission peak shift to 340 nm in DksA2C96V,117V spectrum suggests conformational changes induced by substitutions.
To gain further insights into structural changes induced by a Cys substitution, we compared two-dimensional 1H–15N correlated NMR spectra of DksA2 and DksA2C96V. Although the DksA2 spectrum exhibits a high degree of resonance overlap, as expected from its coiled-coil structure, a number of dispersed resonances can be clearly observed, including two distinct resonances in the expected region for tryptophan indole Hε1. In contrast, the DksA2C96V spectrum is characterized by broadened peaks and very poor chemical shift dispersion; moreover, the signal for one of the tryptophan signals is no longer observed, indicating either overlap or excessive broadening (Fig. 4D). These features are consistent with the notion that the tertiary structure of the globular domain in DksAC96V is highly destabilized compared to the wild-type protein.
Structural similarity between DksA and DksA2 raises an interesting question: can EC DksA lose its Zn-finger and still function in a similar way to DksA2? To answer this question, we substituted two Cys residues for their counterparts in DksA2. This EC DksAC96T,138A variant was poorly soluble in the cell and was completely inactive at the rrnB P1 promoter in vitro (Supplementary Fig. S4), consistent with other reports on the importance of the Zn-finger for DksA activity [25].
4. Discussion
In this work we report the structure of P. aeruginosa DksA2, a paralog of the stress regulator DksA. This is the first available structure of a Zn-independent paralog (which are widespread in nature) that illustrates an alternative strategy for maintaining a protein fold in the absence of a “structural” metal. The structure and subsequent biochemical analysis indicate that despite the lack of two of the four Cys residues required for the Zn-finger motif and the consequent absence of a bound Zn2+, Dksa2 fold and its binding mode to RNAP are strikingly similar to that of EC DksA. The two cysteines of DksA2 are in close proximity to each other in the structure, suggesting that DksA2 could use a disulfide bond instead of a Zn-finger to maintain its structure.
Our analysis indicates that while this disulfide bond can form in vitro, it may only be formed transiently (if at all) in vivo. Irreversible formation of this disulfide bond in vitro has modest effects on DksA2 function, which may indicate that changes in the redox state of the cytoplasm may modulate the activity of DksA2. Notably, a chemical modification of the Cys thiols (likely to sulfonyl groups) or their substitutions for conserved residues abolish DksA2 activity and have dramatic effects on its tertiary structure. This suggests that maintaining the close distance between the sulfur atoms, even if transiently, is essential for the maintenance of the structural integrity of DksA2 and hence, for its function. This could explain why these two Cys residues are conserved among DksA2 – our results show that even the least perturbing substitutions are detrimental. We propose that the two cysteines are a critical part of a larger network of interactions that helps maintain the overall structure and the orientation between the globular and CC domain in the absence of a Zn-finger.
We show that while the formation of a long-lived disulfide bond per se is not required for DksA2 activity in vitro, substitutions of cysteines compromise its structure. Our data suggest that Cys96 and Cys117 make key contributions to the compact domain interface in DksA2 since even a nearly isosteric VA substitution, predicted to have the smallest effect on the geometry of a Cys/Cys pair based on studies of a model protein ROP [23], abolished DksA2 activity (Fig. 4A). Alternatively, formation of a transient disulfide bond may be required during the initial protein folding but is dispensable thereafter, as has been demonstrated for several proteins [20,26,27]. We evaluated this possibility by refolding DksA2 in the presence and absence of DTT. The refolded protein exhibited a comparable activity and 1H–15N HSQC spectrum to those of DksA2WT irrespective of the presence or absence of DTT (Supplementary Fig. S5), arguing against a key role of the disulfide bond in folding.
Altogether, our analysis demonstrates that DksA2 serves as a backup during Zn2+ starvation – it maintains the necessary quaternary structure in the absence of Zn2+ and functions similarly to DksA. Similar structural adaptation is likely to be found in paralogs of other Zn-finger proteins.
Supplementary Material
Acknowledgments
We thank Tom Magliery for comments on the manuscript and stimulating discussions, the staff of sector LS-CAT of the Advanced Photon Source for their assistance with the data collection, and Ben Blowers for assistance with optimization of crystallization conditions. This work was supported by grants from the National Science Foundation (MCB-0949569; to I.A.) and National Institutes of Health (R01 GM077234; to M.P.F.) and by University of Michigan College of Pharmacy start-up funds to O.V.T.
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.febslet.2013.01.073.
References
- 1.Blencowe DK, Morby AP. Zn(II) metabolism in prokaryotes. FEMS Microbiol. Rev. 2003;27:291–311. doi: 10.1016/S0168-6445(03)00041-X. [DOI] [PubMed] [Google Scholar]
- 2.Kehl-Fie TE, Skaar EP. Nutritional immunity beyond iron: a role for manganese and zinc. Curr. Opin. Chem. Biol. 2010;14:218–224. doi: 10.1016/j.cbpa.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Botella H, Peyron P, Levillain F, Poincloux R, Poquet Y, Brandli I, Wang C, Tailleux L, Tilleul S, Charriere GM, et al. Mycobacterial P-1-Type ATPases mediate resistance to zinc poisoning in human macrophages. Cell Host Microbe. 2011;10:248–259. doi: 10.1016/j.chom.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gabriel SE, Helmann JD. Contributions of Zur-controlled ribosomal proteins to growth under zinc starvation conditions. J. Bacteriol. 2009;191:6116–6122. doi: 10.1128/JB.00802-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perederina A, Svetlov V, Vassylyeva MN, Tahirov TH, Yokoyama S, Artsimovitch I, Vassylyev DG. Regulation through the secondary channel-structural framework for ppGpp-DksA synergism during transcription. Cell. 2004;118:297–309. doi: 10.1016/j.cell.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 6.Blaby-Haas CE, Furman R, Rodionov DA, Artsimovitch I, de Crecy-Lagard V. Role of a Zn-independent DksA in Zn homeostasis and stringent response. Mol. Microbiol. 2011;79:700–715. doi: 10.1111/j.1365-2958.2010.07475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma AK, Payne SM. Induction of expression of hfq by DksA is essential for Shigella flexneri virulence. Mol. Microbiol. 2006;62:469–479. doi: 10.1111/j.1365-2958.2006.05376.x. [DOI] [PubMed] [Google Scholar]
- 8.Webb C, Moreno M, Wilmes-Riesenberg M, Curtiss R, Foster JW. Effects of DksA and CIpP protease on sigma S production and virulence in Salmonella typhimurium. Mol. Microbiol. 1999;34:112–123. doi: 10.1046/j.1365-2958.1999.01581.x. [DOI] [PubMed] [Google Scholar]
- 9.Yun H, Jeon B, Barton YW, Plummer P, Zhang Q, Ryu S. Role of the DksA-like protein in the pathogenesis and diverse metabolic activity of campylobacter jejuni. J. Bacteriol. 2008;190:4512–4520. doi: 10.1128/JB.00105-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vassylyeva MN, Svetlov V, Dearborn AD, Klyuyev S, Artsimovitch I, Vassylyev DG. The carboxy-terminal coiled-coil of the RNA polymerase beta ‘-subunit is the main binding site for Gre factors. EMBO Rep. 2007;8:1038–1043. doi: 10.1038/sj.embor.7401079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bardwell JCA, Mcgovern K, Beckwith J. Identification of a protein required for disulfide bond formation in vivo. Cell. 1991;67:581–589. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- 12.Tsodikov OV, Record MT, Jr, Sergeev YV. Novel computer program for fast exact calculation of accessible and molecular surface areas and average surface curvature. J. Comput. Chem. 2002;23:600–609. doi: 10.1002/jcc.10061. [DOI] [PubMed] [Google Scholar]
- 13.Creamer TP, Srinivasan R, Rose GD. Modeling unfolded states of proteins and peptides II. Backbone solvent accessibility. Biochemistry. 1997;36:2832–2835. doi: 10.1021/bi962819o. [DOI] [PubMed] [Google Scholar]
- 14.De Sancho D, Doshi U, Munoz V. Protein folding rates and stability: how much is there beyond size? J. Am. Chem. Soc. 2009;131:2074–2075. doi: 10.1021/ja808843h. [DOI] [PubMed] [Google Scholar]
- 15.Lee JH, Lennon CW, Ross W, Gourse RL. Role of the coiled-coil tip of Escherichia coli Dksa in promoter control. J. Mol. Biol. 2012;416:503–517. doi: 10.1016/j.jmb.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lennon CW, Gaal T, Ross W, Gourse RL. Escherichia coli DksA binds to free RNA polymerase with higher affinity than to RNA polymerase in an open complex. J. Bacteriol. 2009;191:5854–5858. doi: 10.1128/JB.00621-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furman R, Sevostyanova A, Artsimovitch I. Transcription initiation factor DksA has diverse effects on RNA chain elongation. Nucleic Acids Res. 2012;40:3392–3402. doi: 10.1093/nar/gkr1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004;118:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 19.Furman R, Tsodikov OV, Wolf YI, Artsimovitch I. An insertion in the catalytic trigger loop gates the secondary channel of RNA polymerase. J. Mol. Biol. 2013;425:82–93. doi: 10.1016/j.jmb.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Messens J, Collet JF. Pathways of disulfide bond formation in Escherichia coli. Int. J. Biochem. Cell B. 2006;38:1050–1062. doi: 10.1016/j.biocel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 21.Price-Whelan A, Dietrich LEP, Newman DK. Pyocyanin alters redox Homeostasis and carbon flux through central metabolic pathways in Pseudomonas aeruginosa PA14. J. Bacteriol. 2007;189:6372–6381. doi: 10.1128/JB.00505-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen RE, Winther JR. An introduction to methods for analyzing thiols and disulfides: reactions, reagents, and practical considerations. Anal. Biochem. 2009;394:147–158. doi: 10.1016/j.ab.2009.07.051. [DOI] [PubMed] [Google Scholar]
- 23.Hari SB, Byeon C, Lavinder JJ, Magliery TJ. Cysteine-free rop: a four-helix bundle core mutant has wild-type stability and structure but dramatically different unfolding kinetics. Protein Sci. 2010;19:670–679. doi: 10.1002/pro.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beechem JM, Brand L. Time-resolved fluorescence of proteins. Annu. Rev. Biochem. 1985;54:43–71. doi: 10.1146/annurev.bi.54.070185.000355. [DOI] [PubMed] [Google Scholar]
- 25.Perron K, Comte R, van Delden C. DksA represses ribosomal gene transcription in Pseudomonas aeruginosa by interacting with RNA polymerase on ribosomal promoters. Mol. Microbiol. 2005;56:1087–1102. doi: 10.1111/j.1365-2958.2005.04597.x. [DOI] [PubMed] [Google Scholar]
- 26.Narhi LO, Hua QX, Arakawa T, Fox GM, Tsai L, Rosenfeld R, Holst P, Miller JA, Weiss MA. Role of native disulfide bonds in the structure and activity of insulin-like growth factor-I – genetic models of protein-folding intermediates. Biochemistry. 1993;32:5214–5221. doi: 10.1021/bi00070a033. [DOI] [PubMed] [Google Scholar]
- 27.Danek BL, Robinson AS. P22 tailspike trimer assembly is govemed by interchain redox associations. BBA-Proteins Proteomics. 2004;1700:105–116. doi: 10.1016/j.bbapap.2004.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.