Abstract
Background
Pseudomonas spp are important opportunistic and nosocomial pathogens. One such species is Pseudomonas monteilii (P. monteilii). It has been described as an environmental contaminant and potential pathogen. We identified this organism as the causative agent of an exacerbation of bronchiectasis and an environmental contaminant in our hospital on two separate occasions.
Case presentation
P. monteilii was the cause of an exacerbation of bronchiectasis in a 30-year-old HIV negative male. Patient presented with cough with sputum production and exertional dyspnea. The isolate was recovered from a sputum sample in significant counts and definitively identified by Matrix-Assisted Laser Desorption/Ionisation- Time of Flight Mass Spectrometry (MALDI-TOF MS). He was treated with piperacillin-tazobactam and recovered clinically and microbiologically. Another two isolates of the organism were contaminants from the hospital environment. The three isolates were susceptible to all tested antibiotics. Typing by Random amplification of polymorphic DNA (RAPD) found no clonal relationship between them.
Conclusions
Less common species of Pseudomonas need to be identified accurately. This organism is identified by commonly used phenotypic systems as P. putida which may have contributed to a lower reported prevalence. P. monteilii is a known environmental contaminant and must also be considered as a potential pathogen, particularly in patients with chronic lung disease.
Electronic supplementary material
The online version of this article (doi:10.1186/s12879-017-2600-9) contains supplementary material, which is available to authorized users.
Keywords: Pseudomonas monteilii, Bronchiectasis, MALDI-TOF MS, RAPD, Pseudomonas putida
Background
The genus Pseudomonas consists of non-fermenting gram negative bacilli which are opportunistic human pathogens, plant pathogens and environmental contaminants. The common human pathogens in this genus include the type species Pseudomonas aeruginosa, and Pseudomonas putida. Pseudomonas monteilii is a species closely related to P. putida. It was first isolated from clinical samples almost two decades ago [1], but has since been only rarely isolated from clinical specimens [2, 3]. Most isolates are environmental, including resistant strains from the hospital environment [4]. Though it is considered a coloniser and potential pathogen, its status as a human pathogen is unclear.
During surveillance among environmental and clinical specimens for the presence of P.aeruginosa, non-lactose fermenting, oxidase positive gram negative bacilli were subjected to PCR for identification. Two primers targeting different genes which have been used to identify P. aeruginosa were employed [5, 6]. Two isolates, one clinical and one environmental, tested positive for the O-Antigen acetylase gene but not for 16S rDNA v2 and v8. These isolates were identified by the Vitek 2 Compact system as P. putida and as P. monteilii by Matrix-Assisted Laser Desorption/Ionisation- Time of Flight Mass Spectrometry (MALDI-TOF MS) using the MALDI BioTyper software version 3.1 (Bruker Daltonik GmbH, Leipzig, Germany). All isolates showed a score value >2 indicating valid species identification as P monteilii. On re-identification of Pseudomonas spp isolates by MALDI-TOF MS, another environmental isolate presumptively identified as P. putida by the Vitek 2 Compact system was found to be P. monteilii. These isolates were not temporally or spatially related.
Case presentation
The clinical isolate was cultured from a sputum sample of a 30-year-old HIV negative male suffering from an exacerbation of bronchiectasis in September 2015. He was referred to our institute for evaluation of cough with sputum production and repeated episodes of sneezing and nasal discharge for the last 15 years and breathlessness for the past one year. His clinical course was characterised by progressive exertional dyspnoea along with wheezing. A week prior to presentation, he experienced low-grade intermittent fever along with chills and rigors, which prompted the referral. Based on his symptomatic and radiological profile, he had received anti-tuberculous therapy for nine months three years back without relief. He was never a smoker with no history of exposure to environmental smoke, biomass fuel smoke or toxic fumes. General physical examination revealed presence of digital clubbing. There was no evidence of pallor, cyanosis or lymphadenopathy. He was afebrile with a respiratory rate of 18 per minute and oxygen saturation of 94% on oxygen @ 2 L/min. On auscultation, vesicular breath sounds were audible bilaterally along with coarse creptations over all areas of the lung. The total leucocyte count was 17.9 × 103 cells/ mm3, with neutrophilic predominance. Spirometry was suggestive of severe restriction with no response to bronchodilators. Chest radiograph showed multiple ring like shadows in bilateral lower zones. High resolution computed tomography of the thorax revealed multiple dilated bronchi with classical signet ring sign and string of pearls appearance in bilateral lower lobes and right middle lobe, suggestive of cystic bronchiectasis.
The patient was admitted to the ward and a sputum sample was sent to the aerobic culture laboratory. Empirical treatment with intravenous infusion of piperacillin-tazobactam 4.5 gm QID and oral azithromycin 500 mg OD was started. On direct microscopic examination of sputum, the sample had 15-20 pus cells and 0-5 epithelial cells/ low power field. Plenty of gram negative bacilli were observed under the oil immersion objective. The sample was processed by semi-quantitative method using a calibrated loop after treatment with N-acetyl cysteine. It was cultured on sheep blood agar and MacConkey agar plates. The plates were incubated overnight at 37 ° C in ambient air and 5% CO2 for Blood agar plates. More than 105 cfu/ ml non-hemolytic, 2 mm in diameter, greyish, translucent, moist, low convex colonies on Blood agar and non-lactose fermenting colonies in MacConkey agar, were isolated which were identified as P. monteilii. Since significant counts of a single type of organism were isolated from a good quality sputum specimen, it was considered pathogenic. Antimicrobial susceptibility for piperacillin, cefepime, ceftazidime, meropenem, ciprofloxacin, levofloxacin, gentamicin and amikacin was tested using Kirby Bauer's disk diffusion method; for colistin E-test was used (MIC of 2 mcg/ml). Pseudomonas aeruginosa ATCC 27853 strain was used as control. The organism was found to be susceptible to all tested antimicrobials [7]. Treatment with piperacillin-tazobactam was continued. Surveillance samples - nasal and pharyngeal swabs, urine and stool specimens were collected on day 1 and day 8 of admission as part of a separate study. None of these samples grew pathogenic organisms. The patient’s symptoms settled, total leucocyte count returned to normal (9.4 × 103 cells/ mm3), and sputum culture was negative before he was discharged after eight days of hospital stay. A brief summary of his stay in the hospital has been depicted in the timeline (Additional file 1). On discharge, the patient was shifted to oral piperacillin-tazobactam, which was continued for a further 7 days. On follow up, P. monteilii has not been cultured from clinical samples from this patient since.
Surveillance cultures from the hospital environment in the month of September 2015 did not grow any P. monteilii . The two environmental isolates were cultured one each from a bed railing in the Intensive Care Unit in February 2016, and from a bedside table in the ward in March 2016. Both tested susceptible to all antimicrobials (colistin MIC 1.5 mcg/ml). We did not identify any related clinical isolates in that time period. Though the isolates had identical antibiograms, typing by Random amplification of polymorphic DNA (RAPD) (Fig.1) using the ERIC2 primer found 35–57% homology between the strains in UPGMA generated dice coefficients, indicating no clonal relationship. This was expected since the patient acquired the infection prior to admission and the isolates were recovered months apart. The discriminatory power of this primer is unknown for P. monteilii. However, RAPD being affected by differences in factors other than antimicrobial susceptibility, is a more sensitive method for identifying heterogeneity [8].
Fig. 1.
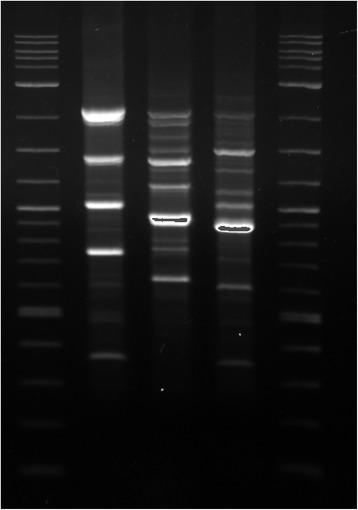
RAPD banding pattern of Pseudomonas monteilii isolates. Legend: Lane 1- 2 Log ladder marker, Lane 2- Isolate from sputum specimen, Lane 3-Isolate from bed railing, ICU, Lane 4- Isolate from bedside table, Ward, Lane 5- 2 Log ladder marker
Discussion and conclusions
Accurate identification of less common Pseudomonas spp is important. In this case, P. monteilii was identified by the Vitek 2 Compact system as P. putida, which has been previously noted [4]. The prevalence of this organism probably appears to be low due to its absence from the Vitek 2 database. The few studies which have identified it have used either 16S rRNA sequencing (primers 27f and 1525r) or MALDI-TOF MS based identification [2, 9, 10].
Pseudomonas aeruginosa is associated with severe disease and rapid decline in lung function in bronchiectasis. Infection once established proves difficult to cure, leading to chronic colonization of the airways. Other species of Pseudomonas though less virulent, are also opportunistic pathogens and may cause colonization of the airways. P. monteilii is an uncommon but increasingly recognized organism in the hospital environment. In the domestic environment, it has been found to be the most common Pseudomonas species isolated and has been cultured most frequently from soil, garbage and drains [10]. It has also been cultured from clinical specimens such as bronchial aspirates, urine, stool, bile and blood though evidence of infection was not collected [1–3]. It was identified as a colonizer in pharyngeal and rectal swabs collected from Syrian refugees, including a meropenem resistant isolate [11]. This organism is also a potential MBL reservoir, with environmental strains having been found to harbour bla VIM-13 and bla VIM-2 genes [3–12].
Our observations indicate that P. monteilii is also an opportunistic pathogen, probably under-reported due to difficulty in identification. P. monteilii, a known environmental contaminant must also be considered as a potential pathogen, particularly in patients with chronic lung disease. It has long been suspected of being potentially pathogenic to humans, and concrete evidence is likely to build with rapid and easy MALDI-TOF MS based identification.
Additional files
Stating all the details of the case report including history, diagnosis and treatment. (DOCX 17 kb)
Acknowledgements
We would like to thank Dr. S. K. Jha for his assistance with the interpretation of clinical and radiological information.
Funding
This research did not receive any specific grant from funding agencies in public, commercial, or not-for-profit sectors.
Availability of data and materials
All data generated or analysed during this study are included in the submitted manuscript.
Abbreviations
- MALDI-TOF MS
Matrix-assisted laser desorption/ionisation- time of flight mass spectrometry
- MBL
Metallo-β-lactamase
- MIC
Minimum inhibitory concentration
- RAPD
Random amplification of polymorphic DNA
- UPGMA
Unweighted pair group method with arithmetic mean
Author’s contributions
MS conceptualized the study, generated dendrograms, analysed the RAPD results and edited the manuscript, Aditi collected the samples, processed the samples and performed the laboratory tests, wrote the manuscript, KB Performed VITEK & MALDI-TOF. All authors read and approved the final manuscript
Ethics approval and consent to participate
The study was ethically approved by the Institutional Ethics Committee, Vallabhbhai Patel Chest Institute, University of Delhi. Written consent to participate was taken from the patient.
Consent for publication
Written consent to publish was obtained from the patient.
Competing interests
The authors declare that they have no competing interests
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s12879-017-2600-9) contains supplementary material, which is available to authorized users.
Contributor Information
Aditi, Email: aditi.lh@gmail.com.
Malini Shariff, Phone: +91-11-27402424, Email: malini.shariff@gmail.com.
Kiran Beri, Email: kiranberi1@gmail.com.
References
- 1.Elomari M, Coroler L, Verhille S, Izard D, Leclerc H. Pseudomonas monteilii sp. nov., isolated from clinical specimens. Int J Syst Bacteriol. 1997;47:846–52. [DOI] [PubMed]
- 2.Ferreira L, Sanchez-Juanes F, Porras-Guerra I, Garcia-Garcia M, Garcia-Sanchez J, Gonzalez-Buitrago J, et al. Microorganisms direct identification from blood culture by matrix- assisted laser desorption/ionization time-of-flight mass spectrometry. Clin Microbiol Infect. 2011;2010:546–51. [DOI] [PubMed]
- 3.Ocampo-Sosa AA, Guzman-Gomez LP, Fernandez-Martinez M, Roman E, Rodriguez C, Marco F, et al. Isolation of VIM-2-producing Pseudomonas monteilii clinical strains disseminated in a tertiary hospital in northern Spain. Antimicrob Agents Chemother. 2015;59:1334–6. [DOI] [PMC free article] [PubMed]
- 4.Bogaerts P, Bouchahrouf W, Lissoir B, Denis O, Glupczynski Y. IMP-13-producing Pseudomonas monteilii recovered in a hospital environment. J Antimicrob Chemother. 2011;66:2434–5. [DOI] [PubMed]
- 5.Choi HJ, Kim MH, Cho MS, Kim BK, Kim JY, Kim C, et al. Improved PCR for identification of Pseudomonas aeruginosa. Appl Microbiol Biotechnol. 2013;97:3643–51. [DOI] [PubMed]
- 6.Spilker T, Coenye T, Vandamme P, Lipuma J, Lipuma JJ. PCR-based assay for differentiation of Pseudomonas aeruginosa from other Pseudomonas species recovered from cystic fibrosis patients. J Clin Microbiol. 2004;42:2074–9. [DOI] [PMC free article] [PubMed]
- 7.Abou-Dobara MI, Deyab MA, Elsawy EM, Mohamed HH. Antibiotic susceptibility and genotype patterns of Escherichia coli, Klebsiella pneumoniae and Pseudomonas aeruginosa isolated from urinary tract infected patients. Pol J Microbiol. 2010;59:207–12. http://www.ncbi.nlm.nih.gov/pubmed/21033585. Accessed 2 Feb 2017. [PubMed]
- 8.Mellmann A, Cloud J, Maier T, Keckevoet U, Ramminger I, Iwen P, et al. Evaluation of matrix-assisted laser desorption ionization-time-of-flight mass spectrometry in comparison to 16S rRNA gene sequencing for species identification of nonfermenting bacteria. J Clin Microbiol. 2008;46:1946–54. [DOI] [PMC free article] [PubMed]
- 9.de Abreu PM, Farias PG, Paiva GS, Almeida AM, Morais PV. Persistence of microbial communities including Pseudomonas aeruginosa in a hospital environment: a potential health hazard. BMC Microbiol. 2014;14:118. doi:10.1186/1471-2180-14-118. [DOI] [PMC free article] [PubMed]
- 10.Remold SK, Brown CK, Farris JE, Hundley TC, Perpich JA, Purdy ME. Differential habitat use and niche partitioning by Pseudomonas species in human homes. Microb Ecol. 2011;62:505–17. doi:10.1007/s00248-011-9844-5. [DOI] [PubMed]
- 11.Angeletti S, Ceccarelli G, Vita S, Dicuonzo G, Lopalco M, Dedej E, et al. Unusual microorganisms and antimicrobial resistances in a group of Syrian migrants: Sentinel surveillance data from an asylum seekers centre in Italy. Travel Med Infect Dis. 2016;14:115–22. doi:10.1016/j.tmaid.2016.03.005. [DOI] [PubMed]
- 12.Scotta C, Juan C, Cabot G, Oliver A, Lalucat J, Bennasar A, et al. Environmental microbiota represents a natural reservoir for dissemination of clinically relevant metallo-β-lactamases. Antimicrob Agents Chemother. 2011;55:5376–9. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Stating all the details of the case report including history, diagnosis and treatment. (DOCX 17 kb)
Data Availability Statement
All data generated or analysed during this study are included in the submitted manuscript.


