Abstract
Triple negative breast cancer (TNBC) highly infiltrated with CD8+ tumor-infiltrating lymphocytes (TILs) has been associated with improved prognosis. This observation led us to hypothesize that CD8+ TIL could be utilized in autologous adoptive cell therapy for TNBC, although this concept has proven to be challenging, given the difficulty in expanding CD8+ TILs in solid cancers other than melanoma. To overcome this obstacle, we used an agonistic antibody (urelumab) to a TNFR family member, 4-1BB/CD137, which is expressed by recently activated CD8+ T cells. This approach was first utilized in melanoma and, in this study, led to advantageous growth of TILs for the majority of TNBC tumors tested. The agonistic antibody was only added in the initial setting of the culture and yet favored the propagation of CD8+ TILs from TNBC tumors. These expanded CD8+ TILs were capable of cytotoxic functions and were successfully utilized to demonstrate the presence of immunogenic mutations in autologous TNBC tumor tissue without recognition of the wild-type counterpart. Our findings open the way for a successful adoptive immunotherapy for TNBC.
Keywords: TIL, Triple negative breast cancer, 4-1BB/CD137, mutated peptide
INTRODUCTION
Despite the tremendous progress made in the past decades for early diagnosis and treatment of breast cancer, women affected by triple negative breast cancer (TNBC) have no treatment options beyond standard chemotherapy. TNBC is defined as such because of its lack of estrogen or progesterone receptors as well as the absence of gene amplification of the human epidermal growth factor receptor (HER)-2 (1). Infiltration of TNBC with CD8+ tumor-infiltrating lymphocytes (TIL) is associated with improved prognosis, suggesting that these infiltrating T cells could specifically recognize tumor antigens (Ag) and be utilized in autologous adoptive cell therapy (ACT) for TNBC (2). The potential of TIL therapy has now been shown in melanoma by multiple cancer centers (3–6). Because of the wide diversity of Ag-specific cells (encompassing both self and mutated) that TIL therapy offers, efforts are being made to transpose this success to other types of solid tumors, as found in gastric, cervical, and ovarian cancers (7–9). One of the major challenges faced in these new trials is the difficulty of expanding the CD8+ T cells from within the TIL population.
One approach to address this issue, which we first utilized in melanoma, is to manipulate a member of the TNFR family, 4-1BB/CD137, which is expressed on the cell surface of recently activated CD8+ T cells. 4-1BB provides a potent costimulatory signal for CD8+ T-cell activation/division and can prevent activation-induced cell death (AICD) of TILs (10). We can accelerate the propagation of Ag-enriched CD8+ TILs by stimulating 4-1BB with an agonistic 4-1BB monoclonal antibody (mAb) at the initiation of the tumor-fragment TIL cultures (11). A prosurvival and memory signature is associated with the early stimulation of 4-1BB, highly dependent on NF-κB activation and Ag-presentation by the dendritic cells within the tumor fragment (11). Based on the lessons learned in melanoma, we hypothesized that an agonistic anti-4-1BB mAb could favor CD8+ TIL propagation from TNBC tumors.
As demonstrated in metastatic melanoma and gastric cancer, ACT using TILs is an effective approach for therapy. TILs encompass a large and diverse amount of tumor-specific T cells that possess the ability to recognize mutation-driven neopeptides (12,13). In contrast to circulating T cells from the blood, TILs that recognize neoepitopes derived from tumor mutations are enriched at the tumor site (14). This enrichment makes TILs an essential screening tool used in the validation of neoantigens as targets for vaccination (15). It also makes them a rich resource for finding new T-cell receptors (TCRs) that recognize mutated peptides, to be later used in engineered TCR ACT. A major obstacle often encountered in solid cancers other than melanoma, like breast cancer for example, is the inefficient expansion of CD8+ TILs that would, in turn, be used to identify and target mutation-driven peptides. Here, we show that the addition of an agonistic 4-1BB mAb in the early phase of tumor-fragment cultures led to favorable propagation of CD8+ TILs from TNBC and can be used to demonstrate the presence of immunogenic mutations in TNBC tumor tissue. Although it was reported that breast cancer encompasses some non-basal mutations, breast cancer has a relatively low level of somatic mutations (16,17), making this technique a valuable approach.
MATERIALS AND METHODS
Patient selection
Following informed consent, nine patients with triple-negative breast cancer were enrolled to this study as shown in Table 1. Eight patients underwent neoadjuvant chemotherapy and 6 of 8 patients underwent taxane- and anthracycline-based chemotherapy. Tissue from surgical resections was used to expand TILs under an IRB-approved protocol (PA12-0728) approved by the Institutional Review Board of The University of Texas MD Anderson Cancer Center.
Table 1.
| Case | Prior treatments | Age | Subtype | cStage | pStage | Grade | Method of isolation | Total number of TIL (IL-2) 106 | Total number of TIL (IL-2 + 4-1BB) 106 | TIL/fragment (IL-2) 106 | TIL/fragment (IL-2 + 4-1BB) 106 | Number of fragment (IL-2) | Number of fragment (IL-2 + 4-1BB) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Panitsumab, Abraxane, Carboplatin, FAC | 52 | IBC-TNBC | cT4dN3bM0 IIIc | ypT4bN3b | 2 | Enzymatic digestion | * NA | NA | ** NG | NG | NA | NA |
| 2 | PTX, FAC | 63 | IBC-TNBC | cT4dNxM1 IV | ypT1miN2a | 3 | Enzymatic digestion Mechanical disruption |
NA | NA | NG | NG | NA | NA |
| 3 | Capecitabine | 76 | IBC-TNBC | cT4dN2M0 IIIB | ypT4dN3 | 2 | Fragment | NA | NA | NG | NG | NA | NA |
| 4 | none | 71 | TNBC | cT1cN0M0 IA | ypT1aN0 | 2 | Fragment | 20.8 | 34.3 | 3.5 | 11.4 | 6 | 3 |
| 5 | Abraxane, FAC | 55 | TNBC | cT2N0M0 IIA | ypT1aN0 | 3 | Fragment | 16.6 | 34.6 | 4.2 | 17.3 | 4 | 2 |
| 6 | PTX, FAC | 57 | IBC-TNBC | cT4dN2M0 IIIB | ypT0N0 | 3 | Fragment | 16.9 | 520.3 | 1.2 | 37.2 | 14 | 14 |
| 7 | PTX, FAC | 34 | TNBC | cT2N1M0 IIB | ypT1cN1 | 3 | Fragment | 30.8 | 519.3 | 3.8 | 64.9 | 8 | 8 |
| 8 | PTX, FAC | 51 | TNBC | cT2N0M0 IIA | ypT1cN0 | 3 | Fragment | 111.1 | 160.1 | 9.3 | 13.3 | 12 | 12 |
| 9 | PTX, FAC | 79 | TNBC | cT3N1M0 IIIA | ypT2N0 | 3 | Fragment | 0.00629 | 5.8 | 0.000524 | 0.481 | 12 | 12 |
NA = None Available
NG = No Growth
Reagents
A fully human and purified IgG4 monoclonal antibody against human CD137, urelumab (663513; Lot 6A20377) was kindly provided by Bristol Myers Squib (BMS) through a Material Transfer Agreement. Human recombinant IL2 (proleukin) was generously provided by Prometheus Therapeutics and Diagnostics.
Isolation and expansion of TILs from human TNBC tumors
The tumor samples were either enzymatically digested followed by centrifugation over a step gradient of 75% and 100% Ficoll, with the TILs collected from above the 100% Ficoll layer, or cut into 3–5 mm2 fragments and placed in TIL culture media [TIL-CM, RPMI 1640 with Glutamax supplemented with 2 mM L-glutamine, 1 mM pyruvate, 1× of HEPES, 50 μM 2-mercaptoethanol, 1X pen-strep (Invitrogen) and 10% heat-inactivated human AB Serum (Sigma-Aldrich)] with IL2 (3000 IU/ml) in 24-well plates for a period of 4 weeks, as previously described (18). For the 4-1BB condition, both IL2 (3000 IU/ml) and 4-1BB agonist mAb (10ug/ml) were added in the culture plates on day 0. Half of the media was changed every 3 to 4 days with the addition of IL2 (3000 IU/ml) with each media change. Cells were split upon confluency but kept in one or multiple 24-well plates. TILs were expanded for 28 days prior to performing the described assays.
Flow Cytometric Analysis of TILs
Expanded TILs were stained in FACS Wash Buffer (Dulbecco’s phosphate-buffered saline 1X with 1% bovine serum albumin) for 30 min using fluorochrome-conjugated monoclonal antibodies for CD3, CD4, CD8 and Caspase 3 (BD Bioscience). Stained cells were acquired using the BD FACS Canto™ II and analyzed using FlowJo software (Tree star). Dead cells were excluded using an AQUA live/dead staining (Invitrogen). For recognition of mutated antigens, intracellular staining was performed for IFNγ (BD Biosciences), preceded by surface staining for CD3, CD4, and CD8. The following gating strategy was used; FSC-A vs SSC-A followed by doublet exclusion. IFNγ expression was gated on live, CD3+CD4−CD8+ cells.
Cytotoxic T-cell Assay
TILs expanded for 28 days with or without 4-1BB agonistic mAb were assayed for cytotoxic ability using a flow cytometry–based assay that measures caspase-3 cleavage using Fc receptor–positive P815 mastocytoma cells coated with CD3 mAb in a redirected CTL assay. The target cells were labeled with DDAO-SE (Molecule Probes-Invitrogen) for 15 min at 37°C. TILs and target cells were incubated at different effector-to-target ratios for 3h at 37°C before harvesting and staining for cleaved caspase 3 in the target cells (19).
Mutation calling and peptide predictions
The Broad Institute’s Mutect algorithm was used to generate mutation calls from raw whole exome sequencing data (20). Class I neopeptides were predicted as described in Snyder et al. (21). All possible 8, 9, 10, 11, and 12-mer mutated and wild-type peptides were generated based on the mutation call variant descriptions. The netMHC algorithm was used to predict peptide - HLA (HLA-A, -B, -C) binding affinities (22). A peptide was considered to bind to an HLA if the predicted binding affinity was below 500 nM. DNA derived from the patient’s PBMCs was used for germline expression. Patient HLA types were determined from the exome sequencing data using the Broad Institute’s Athlates software (23).
IFNγ ELISPOT assay
IFNγ ELISPOT (Enzyme-linked immunospot) assay was performed using Millipore MultiScreen-HA 96-well filter plates (Millipore Cat No. MAHAS4510). Plates were coated with 5 ug/ml anti-IFNγ mAb in pH 9.5 carbonate buffer (Sigma) overnight at 4°C and blocked with PBS, 2% BSA (Sigma Cat No. A8577). Rapidly expanded (REP) TILs [previously described (18)] were thawed and rested overnight with IL2 at 37°C. The following day, cells were washed and rested in media without IL2 for 6 h at 37°C. The cells were plated in triplicates at a concentration of 1 × 106 cells/well (BC7) or 5 × 105 cells/well (BC9). For BC7, 1 × 106 cells/well were stimulated due to the expected low level of neo-epitope reactivity. The patient peptides and the control peptides were added at a concentration of 10 μM. Plates were incubated for 16 hours at 37°C prior to incubation with biotin-labeled anti-IFNγ mAb (Mabtech #3420-6-250) for 1h at RT. After treatment with a 1:5000 dilution of extravidin-alkaline phosphatase (Sigma) for 1.5h at RT, plates were developed with filtered 5-bromo-4-chloro-3-indolyl-phosphate in conjunction with nitro blue tetrazolium (BCIP/NBT substrate; Sigma). Spots were counted on an ImmunoSpot ELISPOT reader (CTL Immunospot Reader, software version 5.1.36). The HIV-gag peptide was used as a negative control as well as the TILs incubated only with media (TILs alone). Cells treated with PMA and ionomycin (Sigma) were used as positive controls in all experiments. Peptide recognition was consider positive when the spot were three times higher than the negative control (peptide).
Statistical Analysis
GraphPad prism version 6.0 (GraphPad Software) was used for graphing and statistical analysis. Statistical analysis for comparison of 2 groups was done using the Wilcoxon signed rank test (paired datasets).
RESULTS AND DISCUSSION
Addition of agonistic 4-1BB mAb increases yield of TNBC TILs and expands CD8+ T cells
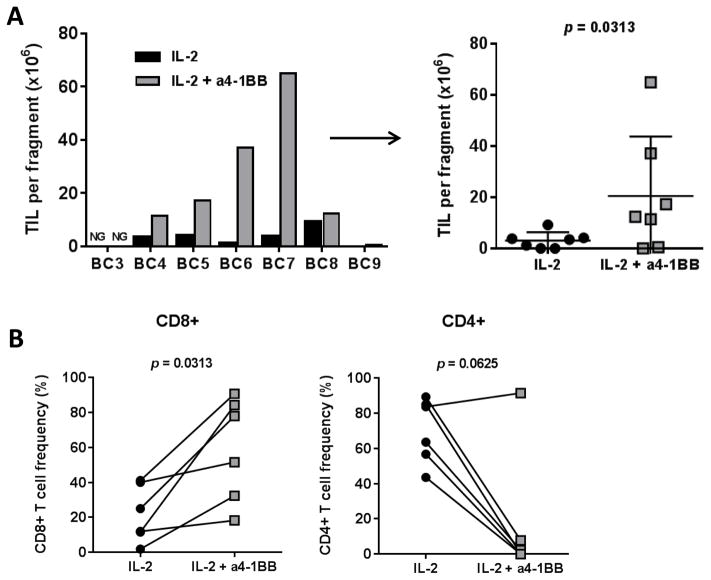
We first evaluated the ability of the agonistic anti-4-1BB mAb to improve the global propagation of TILs in TNBC when added in the early phase of the tumor fragment culture. TILs expanded in the presence of anti-4-1BB mAb in combination with high dose IL2 yielded statistically significantly higher numbers of CD3+ T cells per fragment than those with IL2 alone (mean of 20.5×106 vs 3.1 ×106, p = 0.0313; Table 1 and Fig. 1A). A single dose of the anti-4-1BB mAB on day 0 of a 28-day culture was sufficient to generate a higher number of TILs with a significant increase in the percentage of CD8+ T cells within the CD3+ TIL population (P = 0.0313) and a proportionate decrease in the CD4+ T-cell population in all patients except one (Fig. 1B). These results show that, similar to what we reported in melanoma, stimulation of 4-1BB using an agonistic antibody favors expansion of the CD8+ T-cell population from TNBC tissue.
Figure 1. Addition of agonistic anti-4-1BB antibody allows for the generation of CD8+ TILs from TNBC tumor fragments.
(A) The total number of TILs expanded from 7 TNBC patients, reported as per single-fragment growth, because diverse number of fragments were put in culture depending on the tumor sample size obtained post-surgery. The absolute number is shown as determined by trypan blue exclusion. Growth per patient (left) and per culture condition (right) is shown. (B) The percentage of CD8+ TILs (left) and CD4+ TILs (right) obtained after 28 days of culture with IL2 alone or with IL2 + anti-4-1BB from 6 independent lines.
4-1BB agonistic antibody increased the cytotoxicity of CD8+ TIL
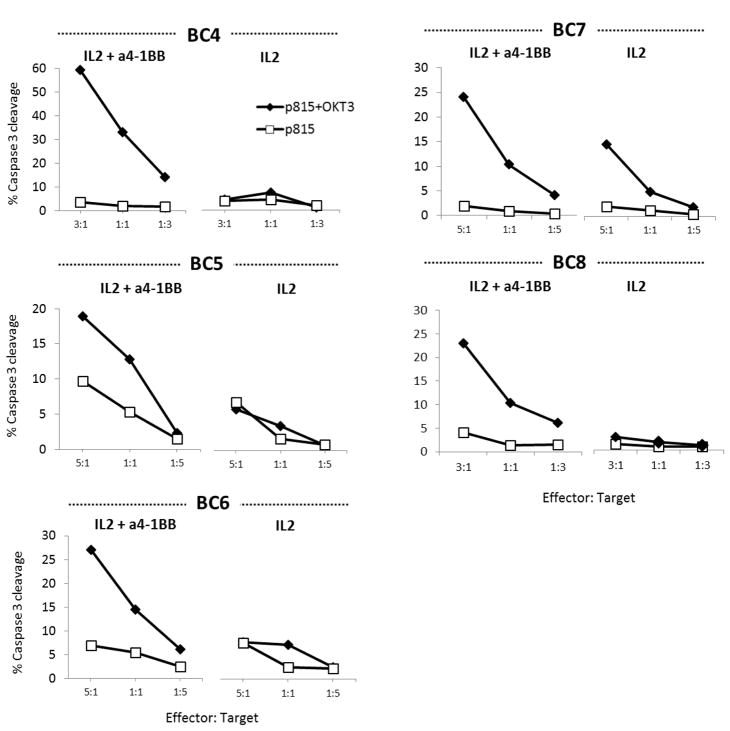
Melanoma TILs propagated with anti-4-1BB have an enhanced cytotoxic capacity, so we hypothesized that this may also be true with TNBC TILs. Due to our lack of success in the generation of autologous or HLA-matched tumor cell lines, CTL activity of both post-expansion TIL products (expanded for 28 days with or without anti-4-1BB mAB) were assessed using a flow cytometry-based redirected killing assay. This assay depicts CTL activity by cleavage of caspase-3 in p815 cells (DDAO-SE labeled) loaded with the CD3 mAb, OKT3. Five independent TNBC TIL lines exposed to an agonist stimulation of 4-1BB in culture showed greater cytotoxic capacity compared to their IL2-alone counterpart (Fig. 2). Although this is not a direct assay for antitumor recognition, these results support the enhanced cytotoxic capacity favored by expansion with 4-1BB mAb costimulation. The fact that the addition of 4-1BB mAb also increases the expansion of CD8+ T cells in the TIL product must be taken into account. However, this costimulation provides an opportunity to unveil potential antitumor reactivity that, in an IL2 alone setting, could go undetected.
Figure 2. Enhanced cytolytic function of TNBC TILs generated with anti-4-1BB.
TNBC TILs grown with IL2 alone or with IL2 + anti-4-1BB for 28 days from 5 independent lines were cocultured with target P815 (DDAO-SE labeled) cells pulsed with anti-CD3 (OKT3, diamonds). P815 cells not pulsed with OKT3 were used as a negative control (squares). Cells were stained for active caspase-3 by flow cytometry as a measure of tumor killing by TIL effectors. Gating was performed on DDAO-SE–positive target cells to determine the level of active caspase-3 detection. The graphs show the percentage of caspase-3 positive tumor targets at decreasing TIL: tumor ratios. Each experiment was independently performed per patient.
BC7 and BC9 T cells from TILs recognize class I–restricted mutated peptides
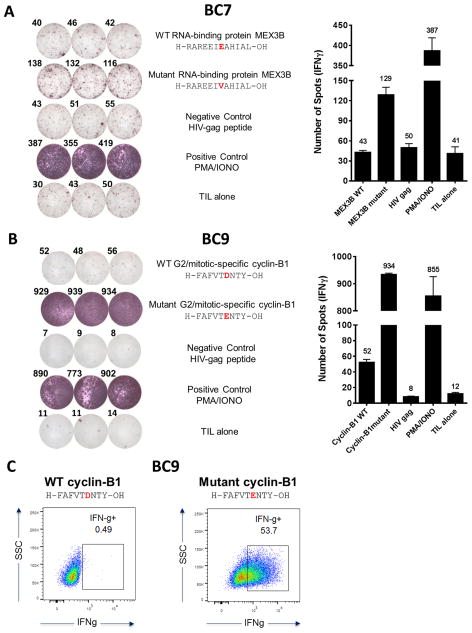
Given our ability to access a larger number of TNBC CD8+ TILs, obtained by expansion following an initial anti-4-1BB stimulation, we explored the potential presence of neo-Ag–specific TILs in triple-negative breast cancer. In melanoma, TILs are a great source of CD8+ T cells that can recognize tumor-associated mutated peptides (12). It is unclear whether the same is true in breast cancer, so we decided to explore this avenue using our 4-1BB stimulated TNBC TILs. Due to the inability to expand CD8+ TILs from IL2-alone cultures, we were unable to test for reactivity differences between the two culture conditions. Thus, only TILs expanded with anti–4-1BB were tested for neoepitope reactivity. Whole exome from DNA extracted from formalin-fixed paraformaldehyde embedded tumor tissue were sequenced from patients BC7 and BC9. Mutation calls were made using the Broad Institute’s Mutect algorithm. Class I neopeptides were predicted according to the patient HLA-A, B, and C loci. Patient BC7 loci were A*01:01, A*30:01, B*08:01, B*13:02, C*07:01, C*06:02, (Supplementary Table S1) and patient BC9 loci were A*33:03, A*68:01, B*18:01, B*58:01, C*12:03 and C*03:02 (Supplementary Table S2). All possible 8, 9, 10, 11, and 12-mer mutated and wild-type peptides were generated based on the mutation call variant descriptions. A peptide was considered to bind to an HLA if the predicted binding affinity was below 500 nM. The peptides used in this study were selected based on this affinity. Overall, we identified 73 and 92 missense mutations from BC7 and BC9, respectively. Given the low number of predicted mutations, we decided to directly screen the TILs expanded with anti–4-1BB for reactivity. Recognition against one mutated peptide was found in TNBC TILs from patient BC7 (Fig. 3A). The IFNγ ELISPOT assay showed recognition of the mutated version of the RNA-binding protein MEX3B, but not to its wild-type counterpart (means of 129 vs 41 spots, respectively) (Fig. 3A). Wild-type MEX3B recognition was at the same level as the negative control HIV-gag peptide. A blast of both the mutant and wild-type sequences identified the 12-mer peptide, residues 216 to 227. The wild-type sequence was identified as H-RAREEIEAHIAL-OH and the mutated sequence was identified as H-RAREEIVAHIAL-OH. The mutated version of MEX3B was predicted to bind to HLA-A*30:01 with an affinity that ranged from 16–216 nM, based upon the prediction algorithm and the peptide length (Supplementary Table S1). The mutation from “E” to “V” at position 7 appears to be sufficient to generate an immunogenic neoantigen that was recognized by BC7 CD8+ TIL.
Figure 3. CD8+ TILs from patients BC7 and BC9 recognize class I–restricted mutated peptides.
(A) IFNγ ELISPOT showing BC7 TIL recognition of the mutant version (H-RAREEIVAHIAL-OH) of RNA-binding protein MEX3B (mutation E→V at position 7) but not the wild-type form nor the negative control HIV-gag. The quantification of BC7 ELISPOT reactivity against mutant MEX3B is shown as number of spots/106 TILs. A TIL-alone control is also included; n = 2 independent experiments (B) IFNγ ELISPOT demonstrating BC9 TIL recognition of the mutant version (H-FAFVTENTY-OH) of G2/mitotic-specific cyclin-B1 (mutation D→E at position 6) as compared to the wild-type form. Quantification of BC9 ELISPOT reactivity against mutant cyclin-B1 is shown as number of spots/5×105 TIL; n = 3 independent experiments. For A) and B) the average number of spot is shown per condition as a label over the bar graphs. (C) IFNγ intracellular staining of BC9 TILs stimulated with either WT or mutant cyclin-B1 showing that mutated G2/mitotic-specific cyclin-B1–specific CD8+ T cells constituted over 50% of BC9 TILs. IFNγ expression is gated on live, CD3+CD4−CD8+ TILs.
An exceptional recognition of a mutated peptide was also identified using an IFNγ ELISPOT for patient BC9 (Fig. 3B). The blast of both the mutant and wild-type forms identified the peptide as being the G2/mitotic-specific cyclin-B1 (CCNB1), residues 269 to 277. The mutated version of cyclin-B1 was predicted to bind to three of the patient’s HLA alleles: HLA-A*68:01, HLA-B*58:01, and HLA-C*12:03 (Supplementary Table S2). The predicted affinity of this neoepitope ranged from 3–479 nM based upon the prediction algorithm and peptide length. The wild-type sequence of cyclin-B1 is H-FAFVTDNTY-OH, which compared to the negative control (mean of 52 vs 8 spots respectively), was weakly recognized by BC9 CD8+ TILs, but not to the extent of the mutated peptide (mean of 934 spots) (Fig. 3B). The mutated sequence, H-FAFVTENTY-OH, which was generated from a mutation from “D” to “E” at position 6, was again sufficient to generate a neoantigen that was recognized by a large portion of BC9 CD8+ T TILs, perhaps because this mutated peptide could bind to, and be presented by, multiple alleles of this patient (Fig. 3B, Supplementary Table S2). When the flow cytometry intracellular staining for IFNγ ICS was also taken into account (Fig. 3C), we concluded that over 50% of the BC9 CD8+ T cells from the TIL product were specific for the mutated form of G2/mitotic-specific cyclin-B1, but not the wild-type form.
Given the lack of accessibility to autologous tumor lines or preserved tumor lysate, we were unable to directly demonstrate natural presentation (and processing) by the tumor cells of the mutated peptide for both patients. Typically, the use of PBMCs, autologous or HLA-matched APCs, transfected with the mutated protein can give a good indication of the immunogenicity of a particular peptide; however, in this study we instead assessed the pre-existing antitumor T-cell pool present in the tumor for recall responses to the cognate peptides predicted from WES. This approach circumvents the limitations introduced through differential antigen presentation by normal cells versus tumors (12). In addition, pre-existing TIL populations could respond to epitopes only cross-presented by APCs, which cannot be recapitulated in vitro. Our approach allowed us to amplify CD8+ TILs to assess a pre-existing antitumor immune response and to identify immunogenic, mutated peptides in a cancer with a low mutational incidence. In the two cases presented here, few mutated peptides were predicted from WES sequencing of the TNBC tumor tissue (73 and 92, respectively), therefore leading to a manageable number of peptides to test for immunogenicity. The use of mass spectrometry following HLA-elution from the tumor as well as the tandem, mini-gene approach to narrow the scope of antigens to be screened for reactivity by TILs was not necessary for this study (12,24,25). Moreover, mass spectrometry of eluted peptides is limited by its very nature in that highly abundant peptides are the most likely to be detected, while missing low frequency neoantigens that may be highly immunogenic. Overall, TNBC provided an advantage over highly mutated solid cancers, such as non-small cell lung cancer or metastatic melanoma, in terms of the number of neoantigens predicted. Indeed it is possible that even more immunogenic peptides may be present in TNBC, as our approach did not take into account the conformational changes that can alter the peptide-MHC complex, resulting in the expression of immunogenic peptides with low affinity HLA-binding (26). Further understanding this class of mutations within solid tumors with low mutation burdens would be an interesting avenue of future exploration.
Our approach enabled the identification of TILs specifically directed against tumor mutations in breast cancer and, more specifically, triple-negative breast cancer. This was rendered possible by a specific costimulation of the breast cancer TILs that, in turn, favored the expansion of CD8+ T cells from the tumor fragments. Although the immunogenic mutations identified here might fall into the category of “unique” antigens, this study certainly provides a strong argument in favor of utilizing immunotherapy in patients with TNBC and provides the basis for future large-scale studies of TNBC TIL.
Supplementary Material
Acknowledgments
Supported by the Cancer Center Support Grant CA016672 (to MD Anderson Cancer Center), by the Morgan Welch Inflammatory Breast Cancer Research Program, and State of Texas Rare and Aggressive Breast Cancer Research Program Grant. M.H. was supported by My Oncology Dream Award from Japan Cancer Society and Oncology Education Promotion Foundation. Neoantigen experiments performed by the Immunotherapy Platform at MDACC. We thank Bristol Myers Squibb (Marie Jure-Kunkel and Stacey Goldberg) for urelumab (BMS-663513), Li Li and Wendy Woodward for their help in collecting samples, Jie Willey and Huiming Sun for help in consenting patients, and Orenthial J. Fulbright for critical reading of the manuscript.
Abbreviations
- TIL
Tumor-infiltrating lymphocytes
- TNBC
Triple negative breast cancer
Footnotes
Conflict of Interest: The authors declare no potential conflicts of interest
References
- 1.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Loi S, Sirtaine N, Piette F, Salgado R, Viale G, Van Eenoo F, et al. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02–98. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2013;31:860–7. doi: 10.1200/JCO.2011.41.0902. [DOI] [PubMed] [Google Scholar]
- 3.Rosenberg SA, Yang JC, Sherry RM, Kammula US, Hughes MS, Phan GQ, et al. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin Cancer Res. 2011;17:4550–7. doi: 10.1158/1078-0432.CCR-11-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Besser MJ, Shapira-Frommer R, Treves AJ, Zippel D, Itzhaki O, Hershkovitz L, et al. Clinical responses in a phase II study using adoptive transfer of short-term cultured tumor infiltration lymphocytes in metastatic melanoma patients. Clin Cancer Res. 2010;16:2646–55. doi: 10.1158/1078-0432.CCR-10-0041. [DOI] [PubMed] [Google Scholar]
- 5.Radvanyi LG, Bernatchez C, Zhang M, Fox PS, Miller P, Chacon J, et al. Specific lymphocyte subsets predict response to adoptive cell therapy using expanded autologous tumor-infiltrating lymphocytes in metastatic melanoma patients. Clin Cancer Res. 2012;18:6758–70. doi: 10.1158/1078-0432.CCR-12-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersen R, Donia M, Ellebaek E, Borch TH, Kongsted P, Iversen TZ, et al. Long-Lasting Complete Responses in Patients with Metastatic Melanoma after Adoptive Cell Therapy with Tumor-Infiltrating Lymphocytes and an Attenuated IL2 Regimen. Clin Cancer Res. 2016;22:3734–45. doi: 10.1158/1078-0432.CCR-15-1879. [DOI] [PubMed] [Google Scholar]
- 7.Tran E, Ahmadzadeh M, Lu YC, Gros A, Turcotte S, Robbins PF, et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science. 2015;350:1387–90. doi: 10.1126/science.aad1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stevanovic S, Draper LM, Langhan MM, Campbell TE, Kwong ML, Wunderlich JR, et al. Complete regression of metastatic cervical cancer after treatment with human papillomavirus-targeted tumor-infiltrating T cells. J Clin Oncol. 2015;33:1543–50. doi: 10.1200/JCO.2014.58.9093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santoiemma PP, Powell DJ., Jr Tumor infiltrating lymphocytes in ovarian cancer. Cancer biology & therapy. 2015;16:807–20. doi: 10.1080/15384047.2015.1040960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernandez-Chacon JA, Li Y, Wu RC, Bernatchez C, Wang Y, Weber JS, et al. Costimulation through the CD137/4–1BB pathway protects human melanoma tumor-infiltrating lymphocytes from activation-induced cell death and enhances antitumor effector function. J Immunother. 2011;34:236–50. doi: 10.1097/CJI.0b013e318209e7ec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chacon JA, Sarnaik AA, Chen JQ, Creasy C, Kale C, Robinson J, et al. Manipulating the tumor microenvironment ex vivo for enhanced expansion of tumor-infiltrating lymphocytes for adoptive cell therapy. Clin Cancer Res. 2015;21:611–21. doi: 10.1158/1078-0432.CCR-14-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robbins PF, Lu YC, El-Gamil M, Li YF, Gross C, Gartner J, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013;19:747–52. doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tran E, Turcotte S, Gros A, Robbins PF, Lu YC, Dudley ME, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344:641–5. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schumacher TN, Hacohen N. Neoantigens encoded in the cancer genome. Current opinion in immunology. 2016;41:98–103. doi: 10.1016/j.coi.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 15.van Buuren MM, Calis JJ, Schumacher TN. High sensitivity of cancer exome-based CD8 T cell neo-antigen identification. Oncoimmunology. 2014;3:e28836. doi: 10.4161/onci.28836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–9. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forget MA, Malu S, Liu H, Toth C, Maiti S, Kale C, et al. Activation and propagation of tumor-infiltrating lymphocytes on clinical-grade designer artificial antigen-presenting cells for adoptive immunotherapy of melanoma. J Immunother. 2014;37:448–60. doi: 10.1097/CJI.0000000000000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He L, Hakimi J, Salha D, Miron I, Dunn P, Radvanyi L. A sensitive flow cytometry-based cytotoxic T-lymphocyte assay through detection of cleaved caspase 3 in target cells. J Immunol Methods. 2005;304:43–59. doi: 10.1016/j.jim.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 20.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–9. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Snyder A, Chan TA. Immunogenic peptide discovery in cancer genomes. Curr Opin Genet Dev. 2015;30:7–16. doi: 10.1016/j.gde.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nielsen M, Lundegaard C, Worning P, Lauemoller SL, Lamberth K, Buus S, et al. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003;12:1007–17. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu C, Yang X, Duffy B, Mohanakumar T, Mitra RD, Zody MC, et al. ATHLATES: accurate typing of human leukocyte antigen through exome sequencing. Nucleic Acids Res. 2013;41:e142. doi: 10.1093/nar/gkt481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalaora S, Barnea E, Merhavi-Shoham E, Qutob N, Teer JK, Shimony N, et al. Use of HLA peptidomics and whole exome sequencing to identify human immunogenic neo-antigens. Oncotarget. 2016;7:5110–7. doi: 10.18632/oncotarget.6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yadav M, Jhunjhunwala S, Phung QT, Lupardus P, Tanguay J, Bumbaca S, et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature. 2014;515:572–6. doi: 10.1038/nature14001. [DOI] [PubMed] [Google Scholar]
- 26.Duan F, Duitama J, Al Seesi S, Ayres CM, Corcelli SA, Pawashe AP, et al. Genomic and bioinformatic profiling of mutational neoepitopes reveals new rules to predict anticancer immunogenicity. J Exp Med. 2014;211:2231–48. doi: 10.1084/jem.20141308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.