Abstract
The advent of rapid and progressively more affordable sequencing and gene expression studies have spurred research on therapies for cancer targeted to specific gene alterations. With few exceptions, such as those cancers with either a paucity of mutations or major chromosomal rearrangements driving the neoplastic transformation, the approaches based on one mutational target-one drug have achieved only modest outcomes in cancer. Using the paradigm of aggressive breast cancers, we will show the mathematical explanation that predicts our failures and indicates a plausible way forward. An integrated network modeling approach to intracellular signaling, metabolism, and microenvironment interactions, coupled with the use of synthetic devices engineered to understand phenotypic heterogeneity of cancer lesions, may form the basis for selection of the next-generation of personalized therapies for cancer. Academia can play a larger role in bringing effective drugs to first-in-human trials in this context.
Tumor heterogeneity and the concept of actionability
It is well established that solid tumors are highly heterogeneous (1). Even for tumors detected relatively “early” at approximately 1 cm in size, the presence of nearly a billion cancer cells in such a lesion is the first harbinger of profound heterogeneity. With cancer cells mutating at a rate of approximately 1 every 10,000 to 100,000 cells, a lesion with a billion cells may already contain tens of thousands of clones. In fact, if the order in which mutations are acquired does not matter for the phenotype of the clones, there would be theoretically 2n distinct genomic clones to contend with during treatment; for a tumor that is found to contain 10 mutations, that number is already quite large, namely 1,024. If the order in which mutations occur in a cell matters in determining the phenotype, however, the number of distinct clones grows much faster with n than 2n; in fact, it is en!-1, where e is the Euler constant (2.718…) and n! is the expression 1x 2x3x..(n-1)xn. It follows that the sheer numbers of potentially distinct clones harbored by most solid tumors in principle would strongly mathematically argue against educated guesses regarding the actionability of one mutation over others when deciding on personalized treatments: yet, this is essentially the state of the art. At present, actionability is influenced less by tumor heterogeneity or phenotypic characteristics of individual clones than by the availability of drugs directed against specific mutated proteins. Several lines of current investigations attempt to establish the phenotypic potential for metastases of individual subpopulations of cells. This work is ongoing and relies on innovations that include microfluidics (2), artificial organ niches (3), and tissue engineering (4), as well as more traditional murine-based models (5). In all cases, however, the analyses involve just a few clones in each tumor, rather than hundreds or thousands of clones. As we make progress in all these fronts, we are enhancing the potential to realize the promise of personalized oncology.
Costs and risks associated with drug discovery
Drug discovery remains a high-risk and expensive endeavor. The average out-of-pocket cost of developing a drug now exceeds $1.5 billion (6) with an overall likelihood of approval (LOA) at 9.6% for a candidate entering the clinic (7). The average time from early-stage discovery to approval was 2 years in 1960. Increased oversight and regulation, in part due historically to the devastating teratogenic effects of thalidomide in Europe, led to sharply increased timelines for drug approval peaking at 12 to 15 years at present (8). Longer timelines, increased costs, and lower research and development budgets in pharmaceutical companies have led to a renewed focus on drug discovery in academia. Rather than lead in drug development, academia has traditionally led in the basic science supporting target identification and lately, increasingly, also in biomarker validation for those drugs with definitive targets. Although important, these tasks comprise still a small percentage of the overall effort entailed in bringing drugs to Phase I testing in the clinic, to first-in-human trials. The LOA of oncology drug candidates entering the clinic is 5.1%; the lowest among all therapeutic areas (7). Given the failure rate of industrial drugs in oncology, we pose that academia can contribute significantly to the discovery and testing of new drugs through specific directions of research and implementation that are amenable to the academic enterprise. Multifaceted partnerships within universities’ departments that can focus on different aspects of the drug development process as well as between academic institutions and companies stand to accelerate effective drug development in cancer and other chronic complex diseases.
Successes in academic and nonprofit drug discovery
Successful examples of drug discovery in academia have typically relied on indications which receive little attention by pharmaceutical companies, such as orphan diseases. One noteworthy example of a model based on studying these diseases is the Drugs for Neglected Diseases initiative (DNDi). The DNDi has earned well-deserved notoriety as it has achieved approval for six treatments, and has a pipeline of 26 drugs, for an estimated value of $290 million (9). The relatively low cost of this portfolio, especially when compared to a typical large pharmaceutical company pipeline, indeed represents a small fraction of what the United States or European pharmaceutical industry would spend on a single drug. For DNDi this is due to a combination of several factors. The DNDi owns no laboratories of its own, relies on collaborators sharing their library of compounds for mutual benefit, and outsources the screening of libraries to universities. Clinical trials are typically small comprising dozens rather than hundreds or thousands of patients, with firm measurable endpoints often related to the treatment of an infection, and the initiative faces little competitive pressure. Many more diseases await effective cures and this mechanism appears very efficient to accomplish these goals.
While the open sharing of data and compounds may be a viable strategy in neglected diseases with low commercial pressure, low competition, and small markets, it is unlikely to be entirely applicable to broader goals in academic drug discovery. However, there are important features that warrant consideration in oncology. For instance, another area in which DNDi and drug discovery in academia have thrived is in repurposing existing drugs or compounds in the public domain for unexplored diseases. As preclinical research is not needed and the toxicity of existing drugs is generally known, costs are significantly lowered; moreover, as toxicity is less of a threat, repurposing drugs may prove attractive to certain investors or to academic entrepreneurs. For all these reasons, the LOA for repurposed drugs and combination therapies is 22.6% or 2.3-fold higher than for the average LOA. Thalidomide, is a salient example. Once pulled off the market for its teratogenic effects, thalidomide was repurposed for its anti-angiogenic effects and was approved in combination with dexamethasone for the treatment of multiple myeloma. A comprehensive study showed that approximately 15% of drugs approved in the decade before 2007 were originally discovered in academic laboratories (10). However, most of these approvals were likely derived for treatments for neglected diseases and through drug repurposing efforts. This is not necessarily a reflection of limits of academic drug discovery today, but rather represents a period in which most universities were ill-equipped to perform the multidisciplinary efforts required for early-stage and preclinical drug discovery.
In the last decade, several universities have launched multi-department drug discovery and development cores. A recent editorial calls for new relationships between such academic drug discovery centers and commercial partners, including drug companies that manufacture generics to control the increasing cost of drugs (11). However, even when relationships between academia and industry are successfully established, there remain significant limitations to academic drug discovery; notably, the relative scarcity of resources for initial investments in the research, as compared to the pharmaceutical industry. Thus, it is preferable to leverage the unique capabilities and skills of universities that make them distinct from drug companies rather than mimic a small-scale pharmaceutical company: in this endeavor, the research leading to and identifying and selecting appropriate targets is crucial.
Early-stage discovery
Two different approaches are typically used for developing molecularly targeted therapies for cancer — a target-based discovery approach and a phenotype-based approach.
A few cancers which have either a paucity of mutations or major chromosomal rearrangements driving transformation can be targeted using drugs that are extremely selective for the aberrant protein. A prototypical example is chronic myelogenous leukemia in which a chromosomal translocation leads to the fusion protein Bcr-Abl that primarily drives cell growth. The selective Bcr-Abl inhibitor imatinib shows robust activity in treating chronic myelogenous leukemia patients. A target-based drug discovery approach is especially suitable for these types of cancers where the oncogenic driver is a single molecular change which is known with certainty. In contrast, for cancers with multiple mutations and altered signaling pathways, the identification of actionable targets is exponentially more complex.
The mere correlation of altered protein levels or mutations with neoplastic transformation is not sufficient for validation as a target. A valid target must be responsible for part of the cancer phenotype, and distinguish the cancer from other cancer subtypes, other cancers, and normal nontransformed cells. Furthermore, the putative target must be tractable to inhibition by a small molecule or antibody. Thus, several known important targets such as mutant Kras, mutant p53, and Myc, are currently not considered to be easily “druggable.” If a target is identified and validated, a robust assay needs to be developed for the selected target, and screening a large library of compounds can yield hits which can be developed further using pharmacophore models based on structure-activity relationships. Alternatively, if the X-ray crystal structure of a target is available, in silico screening of the target can be performed or an available hit can be further developed using structure-based drug design.
Our group has a long-standing interest in exploring therapeutic options for triple-negative breast cancer (TNBC). TNBCs are defined by the lack of hormone receptors (estrogen receptor-negative [ER−] and progesterone receptor-negative [PR−]) and lack overexpression of HER2/neu, and can be further divided into several subtypes. TNBCs as a group are especially lethal with high metastatic potential; currently, there are no targeted therapies for TNBC. Thus, clinicians and patients rely entirely on cytotoxic chemotherapy despite high rates of recurrence of more than 40% at 5 years. A recent study published by our group confirmed an association between TNBC and patients of West African ancestry. The frequency among African American patients who are in fact largely of West African ancestry was observed to be intermediate between Caucasian Americans and West Africans, mirroring the genetic admixture observed following the trans-Atlantic slave trade (12). Further studies conducted using genomic data from patient-derived xenografts (PDX) of TNBC patients of diverse ancestries to identify molecular markers and putative targets are currently in progress.
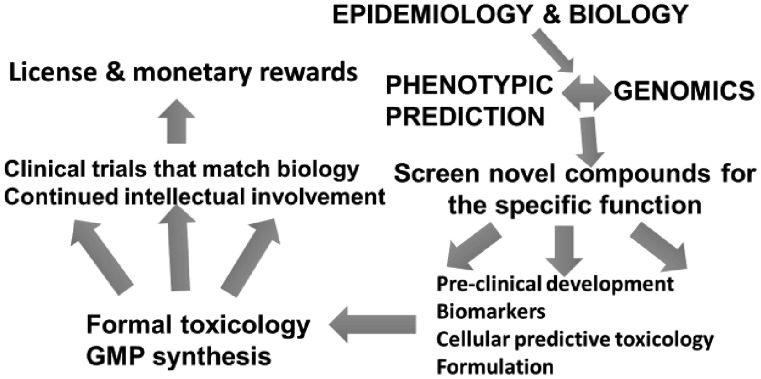
In the alternate strategy termed phenotype-based drug discovery, a library of compounds can be screened against various cell types to identify hits that modulate a specific response. This strategy is target-agnostic and the selected compounds may inhibit one or multiple targets concurrently which, in the case of oncology, bears the advantage of assessing antitumor effects in cells at an early stage. However, the trajectory from a first hit to a highly potent inhibitor is challenging, especially in the absence of biological information about the target. Such information may become vital at a later stage in the preclinical development of a drug to predict toxicities and also to identify biomarkers associated with clinical response. To conduct the steps outlined in Figure 1, many leading academic centers have developed specialized service cores.
Fig. 1.
Academic pipeline of drug development.
Most academic drug discovery cores are relatively recent and thus, it is difficult to fully judge their success in lowering costs and bringing new treatments to the forefront, especially when early-stage discovery is involved. The outcome milestones have not been reached in most cases. As a result, they constitute a significant long-term financial investment for universities, with as yet uncertain monetary return.
The traditional viewpoint for early-stage drug discovery in academia has been to pursue targets that are deemed too high-risk by industry due to their long time-frames, or those that face little commercial pressure (such as those involved in neglected diseases). In contrast, academic work on target classes with a large volume of prior work and competition from the pharmaceutical industry, such as kinases and G protein coupled receptors, tends to be approached with caution. However, even within these target classes, certain niches can be identified for successful academic drug discovery. Of the 518 kinases encoded in the human genome, small-molecule inhibitors have been approved for fewer than 20 kinases that were the intended primary target (13). Thus, a clear majority of the kinome remains untapped, despite several compounds active at the nanomolar level already available as starting points for drug discovery in the published literature.
We have used an approach that could be considered a hybrid of the two strategies discussed above in identifying candidate compounds for the treatment of TNBC. c-Src is an important nonreceptor tyrosine kinase that is highly overexpressed in TNBC. c-Src overexpression has been shown to play a role in proliferation, migration, and invasion of TNBC cell lines and previous findings have validated c-Src as an attractive target in TNBC. Despite data showing that the genetic knockdown of c-Src is an effective anti-cancer strategy, a profound discrepancy was noted between preclinical findings and clinical outcomes of pharmacologic anti-c-Src agents, a common occurrence in oncology. We hypothesized that one major reason for this discrepancy would be that the existing c-Src inhibitors did not inhibit all the functions of c-Src and in particular, they did not prevent the binding to many molecular partners (14).
Existing approved c-Src inhibitors, such as dasatinib and bosutinib, bind the active conformation of the kinase and have not been effective in the clinic. We reasoned that targeting the inactive conformation of the kinase could inhibit the catalytic and non-catalytic functions of c-Src, thus bringing about changes such as those observed when using genetic techniques such as siRNA knockdowns. Using structure-based drug design we developed a potent small molecule inhibitor of c-Src, termed UM-164, which inhibits the inactive conformation (15). We proceeded to comprehensively compare the effect of c-Src inhibition using dasatinib and UM-164 in vitro and in vivo. UM-164 consistently outperformed dasatinib in a diverse collection of TNBC cell lines and displayed highly potent inhibition of tumor growth in xenograft models with limited in vivo toxicity. Notably, through kinome-wide profiling of dasatinib and UM-164 in conjunction with in vitro studies, we could identify the p38 kinases as important targets of UM-164 (but not of dasatinib), contributing to UM-164’s excellent in vivo efficacy (14). Ongoing work in our laboratory is focused on synthesizing and testing analogs of UM-164 for optimization of its physiochemical and pharmacokinetic properties for its ultimate use in the clinic. A complementary approach we are currently undertaking involves screening of a library of well-characterized small-molecule kinase inhibitors with known selectivity spectrums against a wide variety of TNBC cell lines and PDXs. We intend to use computational methods to identifying novel kinase targets for TNBC and the hits could serve as leads for inhibitor development. As of now, we are continuing to guide the trajectory of our compounds, essentially following the path outlined in Figure 1, an unusual occurrence in academia.
Preclinical Development
Increasing the potency of a hit from a micromolar compound to a low nanomolar compound may yield a robust lead compound that must be validated using multiple assays. Further, significant synthetic effort is required for the optimization of a lead for parameters such as solubility, selectivity, pharmacokinetics, and metabolism. Toxicity studies in rodent and non-rodent animals, using a compound manufactured under good laboratory practice (GLP) procedures are required for filing an investigational new drug application with the US Food and Drug Administration. Despite the seemingly linear path in preclinical development, an iterative/cyclic path involving synthesis, optimization, and biological testing is more common.
Lead optimization is one of the two predominant preclinical expenses in drug discovery and entails the synthesis of several analogs on a large scale (16), the other being formal toxicology with GLP product. In the 1950s, a period in which drugs entered the market at a rate significantly higher than today, 1 in 1,000 compounds synthesized would yield a successful drug. Today, 10,000 compounds synthesized yield 200 compounds entering clinical trials, with the ultimate approval of less than 10 drugs, of which typically only 1 provides a return on the investment (8). The low yield is despite the significant advances made in synthesis, purification, and characterization of organic compounds and the overall increase in throughput in the past 50 years. Adequate access to instrumentation and chemicals is typically not a problem in large research universities. However, a single postdoctoral fellow at an academic drug discovery center can be expected to synthesize 50 to 100 compounds of low-to-average complexity every year. Thus, drug discovery cores staffed with several postdoctoral fellows would still be able to synthesize compounds at a number far below what would be required to convert a weakly potent, high-micromolar hit from an assay into a low-nanomolar compound with acceptable physiochemical and pharmacokinetic properties (16). This underscores the importance of choosing the right target for discovery efforts from the inception of the trajectory. Consistent with the lower resources of academia as compared to industry, successful drug discovery in academia has often resulted from careful observations and insights using a smaller number of analogs, more focused on addressing a specialized structural goal.
The lead optimization of compounds using our strategy is far more limited and thus affordable, and the structural nature of kinase inhibitors enables good pharmacokinetic properties. Furthermore, the large increase in robust in vitro assay techniques in the last decade enables screening for selectivity and known problematic off-target effects at an early stage, with a concomitant reduction in costs and increase in feasibility.
Ultimately, a significant reduction in the cost of developing a drug can only be brought about by improving the overall LOA of a compound. This necessitates understanding the disease biology thoroughly, and using predictive models that have been shown to be successful. Although the overall LOA of oncology treatments is 5.1%, a split of phase transitions provides a clearer picture of areas where improvement is necessary. The success of transition of oncology candidates from phase I is 62.8%, from phase II is 24.6%, and from phase III is 40.1%. In comparison, the success of transition for drugs for all indications is 63.2%, 30.7%, and 58.1%, respectively (7). Thus, significantly fewer oncology drugs transition from phase II as compared to the average for all other diseases and the observation is even more pronounced for phase III transitions. This difference could be emblematic of the problems with current treatments in cancer, where patients are selected without an adequate understanding of tumor heterogeneity and tumor evolution. The same study investigated the differences in approval rates when patient selection criteria included or excluded the use of biomarkers. The use of biomarkers increased the overall LOA for drugs of all indications by 3-fold as compared to when biomarkers were not used in patient selection. The phase transitions for trials in which biomarkers were included versus those in which they were excluded are as follows: phase I transition (76.7% versus 63%), phase II transition (46.7% versus 28.8%), and phase III transition (76.5% versus 55%) (7), clearly justifying the care and intellectual engagement required upfront in drug development. Rushing a compound to the clinic without robust biomarkers is not only risky, but mathematically highly unlikely to succeed, considering typical solid tumor heterogeneity described earlier.
Understanding tumor heterogeneity and identifying biomarkers at an early stage can be pivotal in clinical success. A lack of confidence in established cell lines to adequately recapitulate tumor heterogeneity has led us and others to increasingly adopt PDXs as early preclinical models. Despite all of these efforts, persistence and the willingness to abandon a drug after several years of work is not necessarily ingrained in academic culture where perseverance in one’s objectives is considered a paramount value in scientific pursuits.
Future Directions and Conclusions
We have described challenges to drug development and have described the possible approaches, trajectories, and resources that academics can leverage to develop drugs into first-in-human trials, retaining at least partial intellectual control of clinical trial design and biomarker companion studies. In the era of personalized therapies, more so called “n of 1” trial are being envisioned. However, we contend that without extensive basic mechanistic science behind them, they are just as likely to fail as traditional trials with non-molecularly guided eligibility. By the year 2020, the global burden of cancer is expected to account for the largest disability adjusted lost years, tied for first place with mental illnesses, by the year 2030. The university communities, richly creative and sustainable teams of multidisciplinary scientists, stand to make a remarkable difference in the quality of human life around the world.
ACKNOWLEDGMENTS
This work was supported in part by the Breast Cancer Research Foundation (SDM), the Metavivor Foundation (SDM), the University of Michigan Comprehensive Cancer Center, the MTRAC initiative at the University of Michigan, and anonymous donors.
Footnotes
Potentials Conflicts of Interest: None disclosed.
REFERENCES
- 1.Alizadeh AA, Aranda V, Bardelli A, et al. Toward understanding and exploiting tumor heterogeneity. Nat Med. 2015;21:846–53. doi: 10.1038/nm.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen W, Allen SG, Reka AK, et al. Nanoroughened adhesion-based capture of circulating tumor cells with heterogeneous expression and metastatic characteristics. BMC Cancer. 2016;16:614. doi: 10.1186/s12885-016-2638-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen SG, Chen YC, Madden JM, et al. Macrophages enhance migration in inflammatory breast cancer cells via RhoC GTPase signaling. Sci Rep. 2016;6:39190. doi: 10.1038/srep39190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen YC, Allen SG, Ingram PN, et al. Single-cell migration chip for chemotaxis-based microfluidic selection of heterogeneous cell populations. Sci Rep. 2015;5:9980. doi: 10.1038/srep09980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plaks V, Kong N, Werb Z. The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells? Cell Stem Cell. 2015;16:225–38. doi: 10.1016/j.stem.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DiMasi JA, Grabowski HG, Hansen RW. Innovation in the pharmaceutical industry: new estimates of R&D costs. J Health Econ. 2016;47:20–33. doi: 10.1016/j.jhealeco.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 7.Clinical Development Success Rates 2006–2015. 2016. Available at: https://www.bio.org/sites/default/files/Clinical Development Success Rates 2006-2015 - BIO, Biomedtracker, Amplion 2016.pdf. Accessed March 30, 2017. [Google Scholar]
- 8.Mochley-Rosen DGK. A Practical Guide to Drug Development in Academia. New York, New York: Springer International Publishing; 2014. [Google Scholar]
- 9.Maxmen A. Busting the billion-dollar myth: how to slash the cost of drug development. Nature. 2016;536:388–90. doi: 10.1038/536388a. [DOI] [PubMed] [Google Scholar]
- 10.Kneller R. The importance of new companies for drug discovery: origins of a decade of new drugs. Nat Rev Drug Discov. 2010;9:867–82. doi: 10.1038/nrd3251. [DOI] [PubMed] [Google Scholar]
- 11.Workman P, Draetta GF, Schellens JH, Bernards R. How much longer will we put up with $100,000 cancer drugs? Cell. 2017;168:579–83. doi: 10.1016/j.cell.2017.01.034. [DOI] [PubMed] [Google Scholar]
- 12.Jiagge E, Jibril AS, Chitale D, et al. Comparative analysis of breast cancer phenotypes in African American, White American, and West versus East African patients: correlation between African ancestry and triple-negative breast cancer. Ann Surg Oncol. 2016;23:3843–9. doi: 10.1245/s10434-016-5420-z. [DOI] [PubMed] [Google Scholar]
- 13.Wu P, Nielsen TE, Clausen MH. Small-molecule kinase inhibitors: an analysis of FDA-approved drugs. Drug Discov Today. 2016;21:5–10. doi: 10.1016/j.drudis.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 14.Gilani RA, Phadke S, Bao LW, et al. UM-164: a potent c-Src/p38 linase inhibitor with in vivo activity against triple-negative breast cancer. Clin Cancer Res. 2016;22:5087–96. doi: 10.1158/1078-0432.CCR-15-2158. [DOI] [PubMed] [Google Scholar]
- 15.Kwarcinski FE, Brandvold KR, Phadke S, et al. Conformation-selective analogues of dasatinib reveal insight into kinase inhibitor binding and selectivity. ACS Chem Biol. 2016;11:1296–304. doi: 10.1021/acschembio.5b01018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jorgensen WL. Challenges for academic drug discovery. Angew Chem Int Ed Engl. 2012;51:11680–4. doi: 10.1002/anie.201204625. [DOI] [PubMed] [Google Scholar]