Abstract
Long noncoding RNAs (lncRNAs) are emerging as key players in various fundamental cellular biological processes, and many of them are likely to have functional roles in tumorigenesis. Maternally expressed gene 3 (MEG3) is an imprinted gene located at 14q32 that encodes an lncRNA, and the decreased MEG3 expression has been reported in multiple cancer tissues. However, nothing is known about the alteration and role of MEG3 in environmental carcinogen-induced lung tumorigenesis. Our present study, for the first time to the best of our knowledge, discovered that environmental carcinogen nickel exposure led to MEG3 downregulation, consequently initiating c-Jun-mediated PHLPP1 transcriptional inhibition and hypoxia-inducible factor-1α (HIF-1α) protein translation upregulation, in turn resulting in malignant transformation of human bronchial epithelial cells. Mechanistically, MEG3 downregulation was attributed to nickel-induced promoter hypermethylation via elevating DNMT3b expression, while PHLPP1 transcriptional inhibition was due to the decreasing interaction of MEG3 with its inhibitory transcription factor c-Jun. Moreover, HIF-1α protein translation was upregulated via activating the Akt/p70S6K/S6 axis resultant from PHLPP1 inhibition in nickel responses. Collectively, we uncover that nickel exposure results in DNMT3b induction and MEG3 promoter hypermethylation and expression inhibition, further reduces its binding to c-Jun and in turn increasing c-Jun inhibition of PHLPP1 transcription, leading to the Akt/p70S6K/S6 axis activation, and HIF-1α protein translation as well as malignant transformation of human bronchial epithelial cells. Our studies provide a significant insight into understanding the alteration and role of MEG3 in nickel-induced lung tumorigenesis.
Keywords: LncRNA MEG3, DNMT3b, PHLPP1, HIF-1α translation, Nickel
Introduction
Lung cancer is the leading cause of cancer mortality around the world with an estimated 89,320 and 73,190 deaths predicted to occur in men and women, respectively, in 2016 in the United States1. Although, lung cancer cases are attributable in part to smoking tobacco, environmental pollution is more likely cause nonsmoking-related lung cancer2. Therefore, understanding the environmental factors that predispose an individual to nonsmoking-related lung cancer is of prime importance. Nickel is one of the most common environmental factors and is considered to be a group 1 human carcinogen by the International Agency Research on Cancer (IARC) in 19903. The carcinogenic action of nickel compounds is thought to involve oxidative stress, epigenetic effects, and the regulation of gene expression by activation of certain transcription factors related to corresponding signal transduction pathways4. Hypoxia-inducible factor-1 (HIF-1) is a heterodimeric transcription factor that plays a key role in cellular adaptations to hypoxia by controlling the expression of a series of genes involved in angiogenesis, oxygen transport, and glucose metabolism5. HIF-1 consists of HIF-1α and HIF-1β (also known as aryl hydrocarbon nuclear receptor translocator, ARNT). Both HIF-1α and HIF-1β are required for formation of the HIF-1 heterodimer, whereas HIF-1α is a key regulatory subunit and is responsible for HIF-1 transcriptional function6. Our previous studies indicate that nickel exposure results in HIF-1α protein accumulation7–10, consequently contributing to cell transformation9. However, the molecular mechanisms underlying the HIF-1α protein accumulation upon nickel exposure are far from fully understood although it is well-known that the inhibition of HIF-1α protein degradation is involved.
Long noncoding RNAs (lncRNAs) are a heterogeneous group of non-coding transcripts longer than 200 nucleotides. Abnormal expression of many lncRNAs has been reported to be involved in cancer predisposition, development, and progression11. Maternally expressed gene 3 (MEG3) is an lncRNA with a length of ~1.6 kb nucleotides and is reciprocally imprinted with the paternally expressed gene DLK1 constituting an imprinting domain on human chromosome 14q32 12. In humans, MEG3 is expressed in many normal tissues13, while the loss of MEG3 expression has been found in various types of tumor tissues and cell lines, which are associated with tumor development and progression14. However, the role of MEG3 in environmental carcinogen-induced lung cancer remains largely unknown.
Epigenetic mechanisms have been well known for their contribution to the carcinogenic characteristics of nickel 15, and multiple epigenetic mechanisms have been identified that mediate gene silencing following nickel exposure 16. Nevertheless, to the best of our knowledge, there is no report on the implication of lncRNA as an upstream regulator in nickel-induced lung tumorigenesis. In the present study, we discovered that nickel exposure led to MEG3 downregulation through its promoter hypermethylation, and MEG3 downregulation resulted in PHLPP1 transcriptional inhibition, consequently leading to HIF-1α protein translation elevation, and in turn promoting malignant transformation of human bronchial epithelial cells.
Results
Nickel-induced MEG3 downregulation contributed to the malignant transformation of Beas2B cells
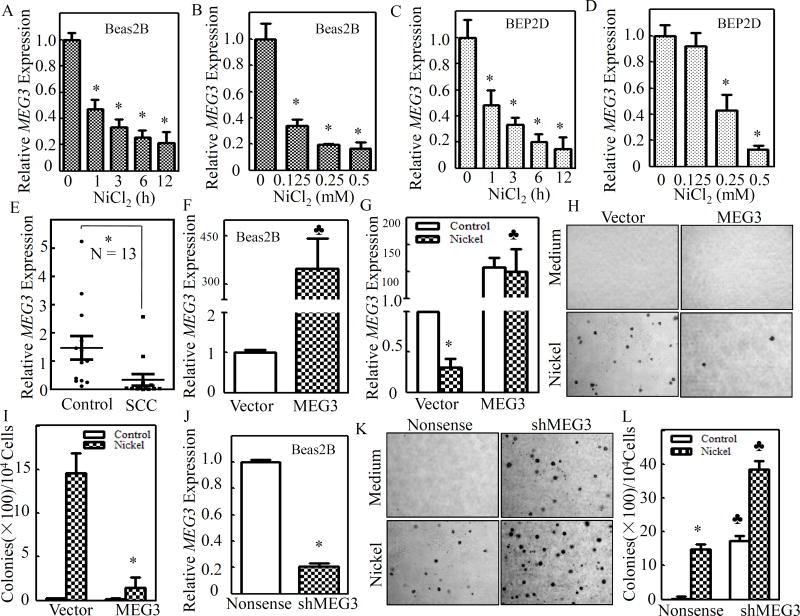
Although MEG3 downregulation has been observed in various human tumors, including non-small cell lung cancer (NSCLC)17, the impact of environmental carcinogens on MEG3 expression and their potential contribution to lung tumorigenesis have never been explored. To test this, human bronchial epithelial cell line Beas2B cells were exposed to different doses of NiCl2 over varying time periods, and MEG3 expression was assessed by quantitative PCR. As shown in Figures 1A & 1B, exposure to NiCl2 caused a time- and dose-dependent downregulation of MEG3 expression in Beas2B cells. The human bronchial epithelial cell line BEP2D is an established clonal population of HPV-18-immortalized human bronchial epithelial cells, and provides another model to study the molecular pathogenesis of lung cancer18. In the present study, as shown in Figures 1C& D, downregulation of MEG3 expression was also observed in BEP2D cells in a time- and dose-dependent fashion. These results demonstrate that nickel exposure results in attenuation of MEG3 expression in human bronchial epithelial cells.
Figure 1. Nickel-induced MEG3 downregulation contributed to malignant transformation of Beas2B cells.
(A – D) Beas2B or BEP2D cells (2×105) were seeded into each well of 6-well plates and cultured till the cell density reached 80–90%. The cells were then exposed to 0.5 mM of NiCl2 for the indicated time periods (A & C) or to various concentrations of NiCl2 for 12 h (B & D). (E) Thirteen pairs of human primary lung squamous cell carcinomas and their adjacent normal lung tissues were obtained from the First Affiliated Hospital of Wenzhou Medical University. The relative levels of MEG3 were determined by qPCR using gene specific primers as described in the section of “Materials and Methods”, and gapdh was measured as an internal control. (F) MEG3 overexpression was identified in Beas2B(MEG3) in comparison to Beas2B(Vector) cells. The spade (♣) indicates a significant increase in comparison to Beas2B(Vector) cells (P< 0.05). (G) Nickel exposure resulted in downregulation of endogenous MEG3, but not ectopically expressed MEG3. The asterisk (*) indicates a significant decrease in comparison to medium control (P< 0.05). The spade (♣) indicates a significant increase in comparison to Beas2B(Vector) (P< 0.05). (H&I) Beas2B(MEG3) and Beas2B(Vector) were repeatedly exposed to 0.5 mM of NiCl2 for 3 months and the nickel treated cells were subjected to anchorage-independent assay in soft agar (H). The number of colonies were scored and presented as colonies per 104 seeded cells (I). The asterisk (*) indicates a significant decrease in comparison to Beas2B(Vector) (P< 0.05). (J) shMEG3 knockdown plasmid was stably transfected into Beas2B cells and the stable transfectants were identified. The asterisk (*) indicates a significant decrease in comparison to Beas2B(Nonsense) (P< 0.05). (K&L) Beas2B(shMEG3) and Beas2B(Nonsense) cells were repeatedly exposed to 0.5 mM of NiCl2 for 3 months and nickel treated cells were subjected to anchorage-independent growth assay (K). The number of colonies were scored and presented as colonies per 104 seeded cells (L). The asterisk (*) indicates a significant decrease in comparison to medium control (P< 0.05;left). The spade (♣) indicates a significant increase in comparison to Beas2B(Nonsense) cells (P< 0.05).
We next evaluated MEG3 expression in lung tissues from patients with lung squamous cell carcinoma (SCC). MEG3 expression in lung SCC was profoundly downregulated in comparison to those of the corresponding adjacent non-tumor lung tissues (n=13, p<0.05) (Figure 1E & Supplemental Table S1). It was noted that MEG3 expression in adenocarcinoma was also inhibited (n=17, p<0.05) (Supplemental Figure S1 & Table S1). We made a comparison of the MEG3 level in Beas2B and BEP2D cells with the normal lung tissue samples. There was no significant difference of the MEG3 level between the normal lung tissues and Beas2B cells or BEP2D cells (t=1.472, p=0.087; t=1.365, p=0.099, respectively). Moreover, the MEG3 levels in Beas2B or BEP2D cells with 0.5 mM nickel treatment for 6h also showed no significant difference in comparison to the lung cancer samples (t=1.037, p=0.136; t=1.047, p=0.139, respectively). Collectively, our results demonstrate that MEG3 downregulation was not only observed in nickel-exposed human bronchial epithelial cells, but also exhibited in human lung cancer tissues.
To determine whether MEG3 downregulation plays a role in nickel-induced cell transformation, MEG3 overexpression plasmid was stably transfected into Beas2B cells with G418 antibiotic selection. MEG3 was overexpressed over 300-fold in stable transfectant Beas2B(MEG3) in comparison to the scramble control transfectant Beas2B(Vector) (Figure 1F). It is significant to note that ectopically expressed MEG3 was not downregulated by nickel exposure, while endogenous MEG3 was attenuated in Beas2B(Vector) cells due to nickel exposure (Figure 1G). We then repeatedly exposed the Beas2B(MEG3) and Beas2B(Vector) cells to nickel for 3 months, and the anchorage-independent growth capability of nickel-treated cells was evaluated in soft agar. As shown in Figures 1H & 1I, the MEG3 overexpression led to significant inhibition of Beas2B cell transformation by nickel in comparison to the scramble control transfectant under the same experimental conditions, suggesting that MEG3 provides an inhibitory effect on cell transformation of human bronchial epithelial cells induced by nickel exposure. To further test this notion, the short hairpin RNA (shRNA) knockdown endogenous MEG3 expression was employed. As shown in Figure 1J, the stably expressed shMEG3 profoundly impaired MEG3 expression in Beas2B cells. Stable knockdown of MEG3 expression alone resulted in a remarkably spontaneous cell transformation of Beas2B cells, and also significantly promoted nickel-induced transformation of Beas2B cells (Figures 1K & 1L). The regulatory effect of MEG3 on cell growth was also consistently observed in monolayer growth assay with above gain- or loss-expression of MEG3 in Beas2B cell transfectants, including the parental Beas2B, Beas2B(vector), Beas2B(MEG3), Beas2B(nonsense), and Beas2B(shMEG3)(Supplemental Figure S2). Those results strongly demonstrate that MEG3 downregulation contributes to malignant transformation of human bronchial epithelial cells due to nickel exposure.
MEG3 promoted HIF-1α protein translation following nickel exposure
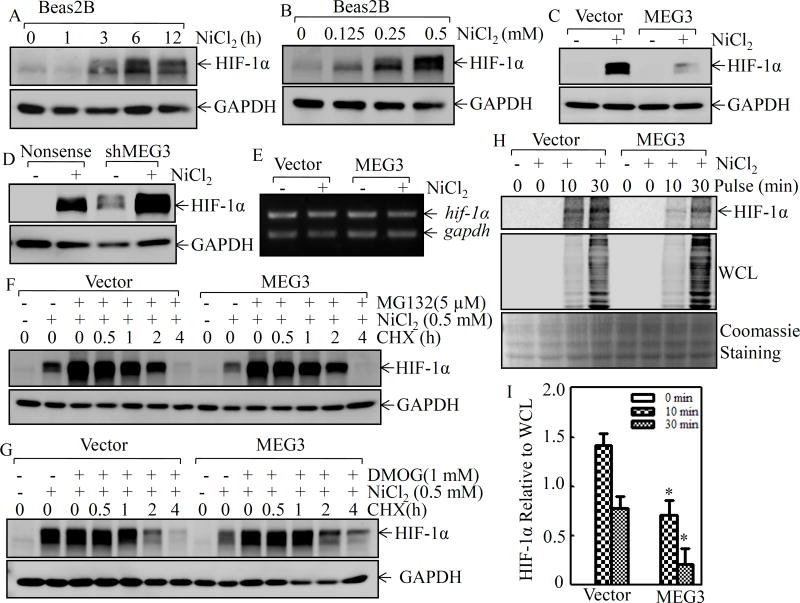
Our previous studies have shown that nickel exposure elevates HIF-1α protein accumulation in various cells, and HIF-1α is involved in cell transformation in anchorage-independent growth assay9. As expected, nickel exposure accumulated HIF-1α protein in a time- and dose-dependent manner in Beas2B cells (Figures 2A & 2B). To evaluate whether MEG3 is involved in nickel-induced HIF-1α protein accumulation, HIF-1α protein level in Beas2B(MEG3) was compared with that in Beas2B(Vector) cells following nickel exposure. As shown in Figure 2C, nickel-induced HIF-1α protein was barely observable in Beas2B(MEG3) following nickel exposure, whereas Beas2B(Vector) showed a marked induction of HIF-1α protein under the same experimental conditions. Moreover, stable knockdown of MEG3 expression alone resulted in an increase in the basal level of HIF-1α protein expression, and also significantly promoted nickel-induced HIF-1α protein abundance (Figure 2D). Our results reveal that MEG3 is an important player in the regulation of HIF-1α protein abundance in human bronchial epithelial cells following nickel exposure.
Figure 2. MEG3 inhibited HIF-1α protein translation in Beas2B cells.
(A & B) Beas2B cells were exposed to 0.5 mM of NiCl2 for the indicated time periods(A) or to various doses of NiCl2 for 6 h(B). The extracts were subjected to Western blotting to determine HIF-1α protein expression. GAPDH was used as a protein loading control. (C & D) Beas2b(MEG3) vs. Beas2B(Vector) (C) or Beas2B(shMEG3) vs. Beas2B(Nonsense) cells (D) were exposed to 0.5 mM of NiCl2 for 6 h. The cell extracts were subjected to Western blotting for determination of HIF-1α protein expression. (E) Beas2B(MEG3) and Beas2B(Vector) cells were exposed to 0.5 mM of NiCl2 for 6 h and the cells were then used to extract total RNA using Trizol reagent. RT-PCR was carried out to detect hif-1α mRNA expression. gapdh was used a loading control. (F & G) Beas2B(MEG3) and Beas2B(Vector) cells were pre-treated with NiCl2 plus MG132 (F) or NiCl2 plus DMOG (G) for 6 h. The cells were then used to determine of HIF-1α protein degradation rates in the presence of cycloheximide (CHX; 50µg/mL) for different time periods. (H & I) Beas2B(MEG3) and Beas2B(Vector) cells were exposed to 0.5 mM of NiCl2 for 6 h. The newly synthesized HIF-1α protein was monitored by pulse assay using [35S]-labeled methionine-cysteine. WCL stands for whole cell lysate and Coomassie blue staining was used for protein loading control (H). The content of the newly synthesized HIF-1α protein was normalized to the total newly synthesized protein (I).
HIF-1α expression is well known for its regulation at the transcriptional and protein degradation levels due to hypoxia or the hypoxia mimetic agents5. However, our results from RT-PCR showed no observable difference at hif-1α mRNA levels between Beas2B(MEG3) and Beas2B(Vector) cells following nickel exposure (Figure 2E), suggesting that MEG3-mediated HIF-1α abundance upon nickel exposure might occur at the post-transcriptional level. Following this, the HIF-1α protein degradation rate was then evaluated. As shown in Figure 2F, after treatment of cells with nickel plus the proteasome inhibitor MG132 for 6 h, the cell culture medium was replaced with the medium containing cycloheximide (CHX) to prevent de novo new protein synthesis, and both Beas2B(Vector) and Beas2B(MEG3) were used to determine HIF-1α protein degradation rates. The results showed that two stable transfectants have very similar HIF-1α protein degradation rates (Figure 2F), excluding MEG3 regulates HIF-1α protein expression at the protein degradation level. This notion was consistently supported by the results obtained from the observation of HIF-1α protein degradation in both Beas2B(Vector) and Beas2B(MEG3) treated with dimethyloxaloylglycine (DMOG), an inhibitor of prolyl hydroxylase (PHD) (Figure 2G). Thus, we next determined the possibility of MEG3 regulation of HIF-1α protein translation using [35S]-labeled methionine and cysteine in pulse analysis. As shown in Figures 2H & 2I, the incorporation of [35S]-labeled methionine and cysteine into newly synthesized HIF-1α protein was inhibited in Beas2B(MEG3) cells, although it was gradually increased along with the incubation time periods in both Beas2B(MEG3) and Beas2B(Vector) cells. Our results demonstrate that MEG3 downregulation was crucial for HIF-1α protein translation following nickel exposure in human bronchial epithelial cells.
Akt/p70S6K/S6 pathway played an important role in MEG3 regulation of HIF-lα protein translation
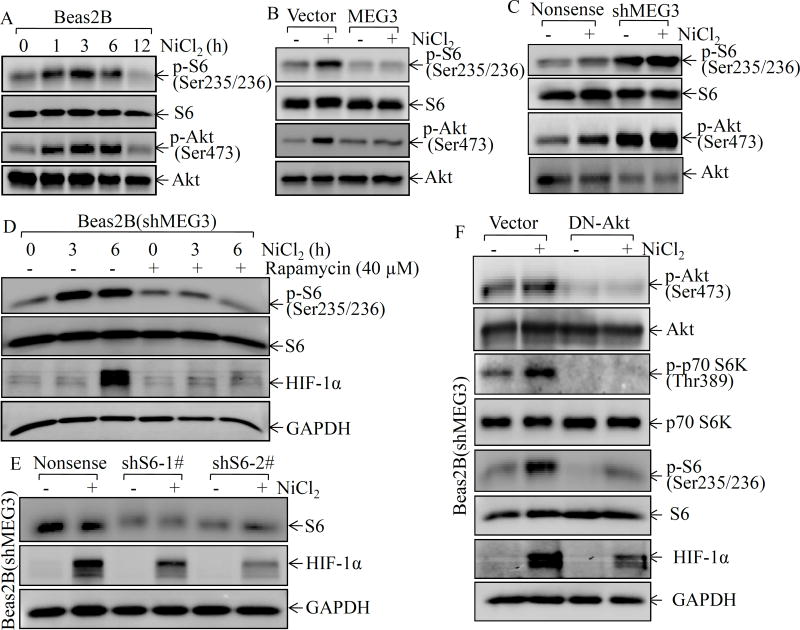
Ribosome biogenesis is a fundamental process that provides cells with the molecular factories for cellular protein production19. The ribosome is a complex molecular machine that is composed of a small 40S and a large 60S subunit. Phosphorylation at Ser235/236 of the 40S ribosomal protein S6 plays an important role in the regulation of protein translation20. Thus, the phosphorylation at Ser235/236 of S6 was evaluated following nickel exposure in Beas2B cells. As shown in Figure 3A, nickel exposure led to an increase in S6 phosphorylations at Ser235/236 in the early time phase (1–6 h after exposure). To examine whether MEG3 was implicated in modulation of S6 phosphorylations at Ser235/236 following nickel exposure, we compared the S6 phosphorylation at Ser235/236 between Beas2B(MEG3) cells and Beas2B(Vector) cells. The results indicated that S6 phosphorylations at Ser235/236 was completely inhibited in Beas2B(MEG3) cells in comparison to the Beas2B(Vector) cells (Figure 3B). The findings were further validated in MEG3 knockdown transfectants, which showed an increase in S6 phosphorylation at Ser235/236 as compared with Beas2B(Nonsense) cells following nickel exposure (Figure 3C). The mammalian target rapamycin (mTOR) is the upstream kinase mediating S6 phosphorylations at Ser235/23621. To test the role of S6 phosphorylation in HIF-1α protein translation, rapamycin, a specific inhibitor of mTOR, was employed. As shown in Figure 3D, pretreatment of cells with rapamycin blocked S6 phosphorylation at Ser235/236 and HIF-1α protein accumulation following nickel exposure. Moreover, knockdown of S6 expression by shRNA also inhibited HIF-1α accumulation following nickel exposure (Figure 3E), strongly supporting our notion that MEG3 inhibited HIF-1α translation via attenuation of the S6 phosphorylation at Ser235/236.
Figure 3. MEG3 inhibited nickel-induced HIF-1α protein expression by targeting Akt/p70S6K/S6 ribosomal pathway.
(A) Beas2B cells were exposed to 0.5 mM of NiCl2 for the indicated time periods. The cell extracts were subjected to Western blotting to determine p-S6 phosphorylation at Ser235/236, S6, Akt phosphorylation at Ser473, and Akt. (B & C) Beas2B(MEG3) vs. Beas2B(Vector) or Beas2B(shMEG3) vs. Beas2B(Nonsense) cells were exposed to 0.5 mM of NiCl2 for 6 h. The cell extracts were subjected to Western blotting to determinate p-S6 phosphorylation at Ser235/236, S6, Akt phosphorylation at Ser473, and Akt. (D) Beas2B(shMEG3) cells were pretreated with rapamycin (40µM) for 30min, the cells were then exposed to 0.5 mM of NiCl2 for the indicated time periods. The cell extracts were subjected to Western blotting to determinate p-S6 phosphorylation at Ser235/236, S6, HIF-lα, and GAPDH protein expression. (E) shRNA specific targeting S6 and its scramble control were transiently transfected into Beas2B cells and the transfectants were exposed to 0.5 mM of NiCl2 for 6 h. The cell extracts were subjected to Western blotting to determinate S6, HIF-lα, and GAPDH protein expression. (F) DN-AKT plasmid and its scramble control were transiently transfected into Beas2B(shMEG3) cells. The transfectants were exposed to 0.5 mM of NiCl2 for 6 h and then were extracted to determinate Akt phosphorylation at Ser473, Akt, p70S6K phosphorylation at Thr389, p70S6K, S6 phosphorylation at Ser235/236, S6, HIF-1α, and GAPDH by Western blot.
Our previous studies have demonstrated that nickel exposure leads to activation of phosphatidylinositol 3-kinase (PI-3K), Akt, and p70S6 kinase (p70S6k)7. Akt activation has been found to be implicated in ribosomal S6 kinase (S6K1)/S6-dependent p53 protein translation in arsenite-treated cells22. Thus, Akt phosphorylation at Ser473 was evaluated in both MEG3 overexpressed and knockdown Beas2B cells in comparison to the corresponding vector control transfectants following nickel exposure. The results showed that the increase of Akt phosphorylation at Ser473 was consistent with the increase of S6 phosphorylation at Ser235/236 following nickel exposure in Beas2B cells (Figure 3A). Moreover, the increase of Akt phosphorylation at Ser473 by nickel was completely abolished by ectopic overexpression of MEG3 and remarkably enhanced by knockdown of MEG3 (Figures 3B & 3C). To test whether Akt activation mediated S6 phosphorylation at Ser235/236 and HIF-1α protein translation following nickel exposure, we transiently transfected the dominant-negative mutant Akt (DN-Akt, AktT308A/S473A) into Beas2B(shMEG3) cells, and the transient transfectant Beas2B(shMEG3/DN-Akt) and its vector control transfectant Beas2B(shMEG3/Vector) were evaluated. Overexpression of DN-Akt blocked nickel-induced Akt phosphorylation at Ser473, p70S6K phosphorylation at Thr389, and S6 phosphorylation at Ser235/236, as well as HIF-1α protein accumulation following nickel exposure (Figure 3F). Our results strongly indicate that the Akt/p70S6k/S6 pathway plays an important role in MEG3 regulation of HIF-1α protein translation.
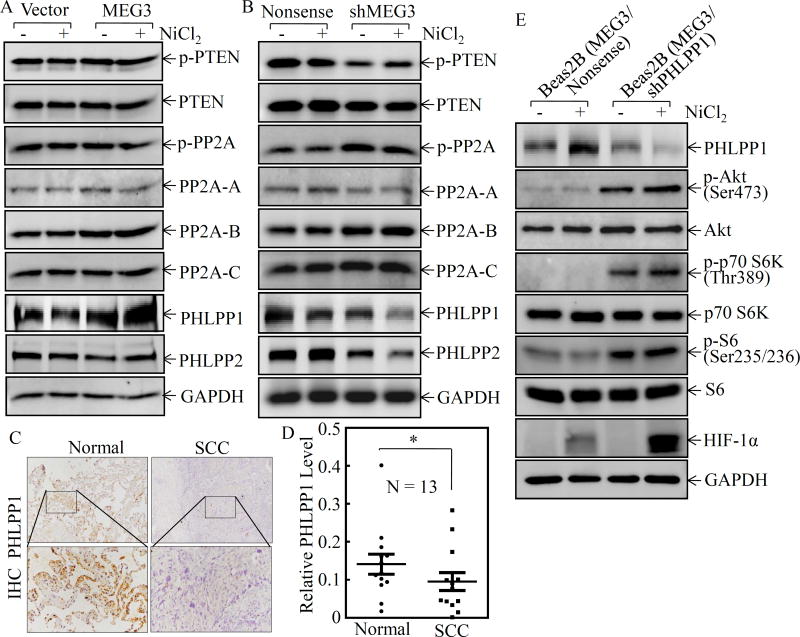
PHLPP1 was responsible for MEG3-mediated inhibition of Akt/p70S6K/S6 pathway activation
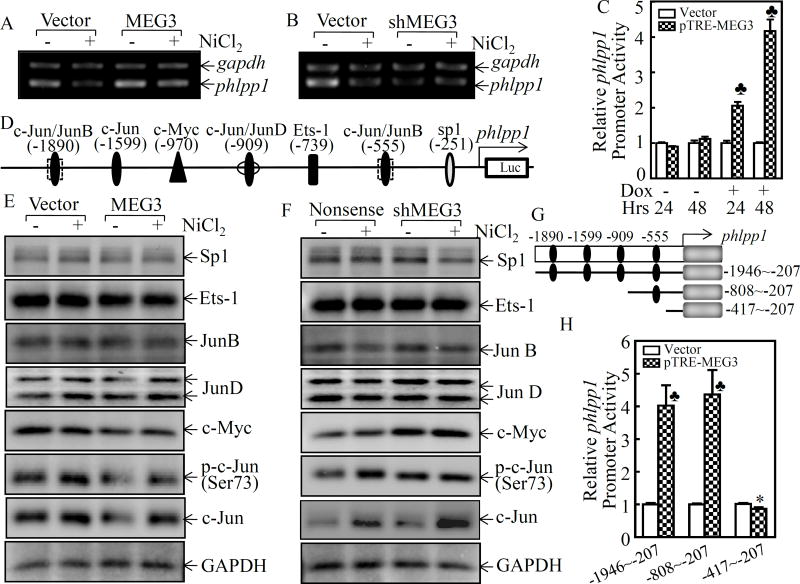
The phosphatase and tensin homolog (PTEN) is a tumor suppressor that negatively regulates the PI-3K/Akt pathway by dephosphorylation of the lipid second message, phosphatidylinositol-3,4,5-trisphosphate, a product of phosphatidylinositol 3-kinase (PI-3K), by which PTEN inhibits Akt activation 23. PH domain leucine-rich repeat protein phosphatase (PHLPP) is a protein phosphatase, that is comprised of PHLPP1 and PHLPP2 two members, which are able to specifically dephosphorylate the hydrophobic motif of Akt (Ser473 in Akt)24. Protein phosphatase 2A (PP2A) has also been reported to be a negative regulator of the Akt pathway 25. To identify the downstream effector of MEG3 that mediated the Akt/p70S6K/S6 pathway activation, we compared the expression of PTEN, PHLPP1, PHLPP2, and PP2A in Beas2B(MEG3) vs. Beas2B(Vector) and Beas2B(shMEG3) vs. Beas2B(Nonsense) cells following nickel exposure. As shown in Figure 4A, overexpression of MEG3 specifically reversed nickel downregulation of PHLPP1 protein expression in comparison to Beas2B(Vector). Although knockdown of MEG3 also changed the protein levels of p-PTEN, p-PP2A, PP2A-B, and PHLPP2, but those change could not be reversed by overexpression of MEG3 in Beas2B(MEG3) cells (Figures 4A & 4B). Therefore, we anticipate that PHLPP1 might be the potential target of MEG3 in Beas2B cells. It is highly significant to note that PHLPP1 downregulation was also observed in lung cancer tissues (Figures 4C & 4D). Moreover, there was a highly significant positive correlation between MEG3 and PHLPP1 (ρ=0.846, P=0.000) (Supplemental Figure S3A). These results reveal that PHLPP1 might be an MEG3 downstream phosphatase responsible for dephosphorylation of Akt following nickel exposure. To this end, we used shRNA to knockdown of PHLPP1 in Beas2B(MEG3) cells. As shown in Figure 4E, knockdown of PHLPP1 led to marked increases in the phosphorylation of Akt at Ser473, p70S6K at Thr389 and S6 at Ser235/236, as well as induction of HIF-1α protein. It was noted that MEG3 expression level in constitutive stable Beas2B(MEG3) transfectants is over 300-fold. To avoid constitutive over 300-fold MEG3 expression impacting on intrinsic cell function, we constructed an inducible expression plasmid pTRE-MEG3, which was also stably transfected into Beas2B cells. The stable transfectant, Beas2B(pTRE-MEG3), showed around 30-fold MEG3 expression upon treatment with 10 µg/ml Dox in comparison to the vector control transfectants under the same experimental conditions (Supplemental Figure S4). The effect of MEG3 on the expression of HIF-1α and PHLPP1, as well as the phosphorylation of c-Jun at Ser73, Akt at Ser473, p70 S6K at Thr389 and S6 at Ser235/236, has been validated in the inducible MEG3 stable transfectants in the absence or presence of Dox (Supplemental Figure S5). Collectively, our results demonstrate that PHLPP1 is an MEG3 downstream phosphatase in regulation of phosphorylations of Akt at Ser473, p70S6K at Thr389, and S6 at Ser235/236, as well as expression of HIF-1α protein following nickel exposure.
Figure 4. PHLPP1 contributed to the activation of Akt/p70S6K/S6 ribosomal pathway and induction of HIF1α protein.
(A & B) Beas2B(MEG3) vs. Beas2B(Vector) or Beas2B(shMEG3) vs. Beas2B(Nonsense) cells were exposed to 0.5 mM of NiCl2 for 6 h. The cell extracts were subjected to Western blotting for determining PHLPP1, PHLPP2, p-PP2A, PP2A-A, PP2A-B, PP2A-C, p-PTEN, PTEN, and GAPDH protein expression. (C&D) PHLPP1 expression in human lung squamous cell carcinomas was evaluated by IHC staining (C) and analyzed by calculating the integrated optical density per stained area (IOD/area) (D). (E) shRNA targeting PHLPP1 and its scramble control plasmids were transiently transfected into Beas2B(MEG3) cells and the transfectants were exposed to 0.5 mM of NiCl2 for 6 h. The cells were then extracted to determinate PHLPP1, Akt phosphorylation at Ser473, Akt, p70S6K phosphorylation at Thr389, p70S6K, S6 phosphorylation at Ser235/236, S6, HIF-lα, and GAPDH by Western blotting.
MEG3 regulated PHLPP1 transcription via interaction with transcription factor c-Jun
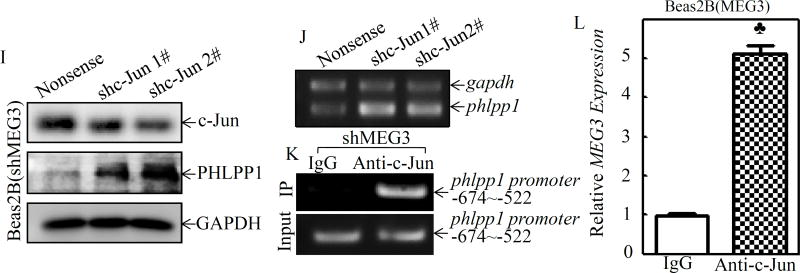
To evaluate the molecular mechanisms underlying MEG3 downregulation of PHLPP1 expression, we detected the phlpp1 mRNA levels in Beas2B(MEG3) vs. Beas2B(Vector) and Beas2B(shMEG3) vs. Beas2B(Nonsense) cells following nickel exposure. As shown in Figure 5A, overexpression of MEG3 was able to increase the basal level of phlpp1 mRNA expression and that it also brought phlpp1 mRNA in nickel-treated Beas2B(MEG3) up to the basal level observed in Beas2B(Vector) cells. Consistently, knockdown of MEG3 resulted in a remarkably reducing the phlpp1 mRNA expression (Figure 5B). Moreover, overexpression of MEG3 also increased the promoter transcriptional activity (Figure 5C), indicating MEG3 upregulates PHLPP1 expression at the transcriptional level. To test the possibility of transcriptional regulation, bioinformatics software was used to analyze the potential transcription factor binding sites in phlpp1 promoter region. The results showed that there were multiple potential transcription factor binding sites, including c-Jun, Jun B, c-Myc, JunD, Sp1, and Ets-1, in the phlpp1 promoter region (Figure 5D). Thus, we then focused on identification of the potential transcription factors contribution to phlpp1 mRNA expression by comparing the effect of MEG3 on the transcription factors expression and their activation following nickel exposure. As the results shown in Figure 5E, overexpression of MEG3 inhibited expression of c-Jun and c-Myc, while it did not show a remarkable effect on other transcription factors, including Jun B, Jun D, Sp1, or Ets-1. The effect of MEG3 on the expression of c-Jun and c-Myc was further validated in Beas2B(shMEG3) cells(Figure 5F). These results are also consistent with our most recently findings that nickel exposure leads to a marked nuclear translocation of c-Jun and c-Myc26. Therefore, we proposed that c-Jun and c-Myc might be the transcription factors for negative regulation of phlpp1 promoter transactivation through direct binding. To test this possibility, a series of phlpp1 promoter-driven luciferase reporters containing incrementally deleted sequences in the −1946/−207 region (numbers relative to the transcription start site), as indicated in Figure 5G, were stably transfected into Beas2B(pTRE-MEG3) and Beas2B(Vector) cells. The results showed that the promoter transcriptional activity was completely attenuated in the transfectant with the −417/−207 luciferase reporter in comparison to the transactivation observed in the transfectants with other luciferase reporters (Figure 5H), suggesting that the transcription factor binding site at −555 (c-Jun) is the critical one responsible for phlpp1 transcription. To test this possibility, c-Jun was specifically knockdown in Beas2B(shMEG3) cells, as expected, knockdown of c-Jun promoted PHLPP1phlpp1 mRNA and protein expression (Figures 5I & 5J). To provide experimental evidence showing c-Jun directly binding to phlpp1 promoter, ChIP assay was performed. As shown in Figure 5K, the −808/−417 region of phlpp1 promoter that contains tentative c-Jun binding site was specifically presented in the immunoprecipitated complex using anti-c-Jun antibody in ChIP assay. To demonstrate the inhibition of c-Jun transactivation activity by its interaction with MEG3, an RNA immunoprecipitation assay was further performed. The results indicated that the immunoprecipitated complex pulled down by anti-c-Jun from Beas2B(MEG3) cells contained MEG3 transcript (Figure 5L), strongly demonstrating that MEG3 positively regulates phlpp1 mRNA transcription by interacting with c-Jun and disrupting c-Jun binding to phlpp1 promoter.
Figure 5. MEG3 interacted with transcription factor c-Jun, consequently regulating PHLPP1 promoter transcription.
(A & B) Beas2B(MEG3) vs. Beas2B(Vector) or Beas2B(shMEG3) vs. Beas2B(Nonsense) cells were exposed to 0.5 mM of NiCl2 for 6 h. The cells were then used to extract total RNA using Trizol reagent. RT-PCR was carried out to detect phlpp1 mRNA expression and gapdh was used as a loading control. (C) the full long phlpp1 promoter-driven luciferase reporter constructs co-transfected with TK into Beas2B(pTRE-MEG3) and Beas2B(Vector), respectively. (D) The potential transcriptional factor binding sites in PHLPP1 promoter region (−2000~+0) were analyzed using the TRANSFAC 8.3 engine online. (E & F) Beas2B(MEG3) vs. Beas2B(Vector) or Beas2B(shMEG3) vs. Beas2B(Nonsense) cells were exposed to 0.5 mM of NiCl2 for 6 h. The cell extracts were subjected to Western blotting to determinate p-c-Jun(Ser73), c-Jun, sp1, Ets-1, JunB, JunD, and c-Myc. GAPDH was used as a protein loading control. (G) A schematic illustration of the construction of phlpp1 promoter-driven luciferase reporter constructs. (H) The various phlpp1 promoter-driven luciferase reporter constructs together with TK were stably transfected into Beas2B(pTRE-MEG3) and Beas2B(Vector), respectively. The transfectants were extracted to evaluate the luciferase activity after 48 h and the results are presented as relative phlpp1 promoter activity by normalization of luciferase activity to Renilla luciferase activity. The spade (♣) indicates a significant increase in comparison to the vector control group (P< 0.05) and the asterisk (*) indicates a significant decrease in comparison to other phlpp1 promoter-driven luciferase reporters (P< 0.05). (I) shRNA targeting c-Jun and its scramble control plasmid was transiently transfected into Beas2B(shMEG3) cells and the transfectants were extracted to determinate c-Jun, PHLPP1, and GAPDH by Western blot. (J) The cell transfectants were used to extract total RNA and phlpp1 expression was evaluated by using RT-PCR. (K) ChIP assays were performed to determine c-Jun binding to the phlpp1 promoter in Beas2B(shMEG3) cells, as described in the section of “Materials and Methods”. (L) RNA-IP was carried out to evaluate specific interaction of MEG3 with c-Jun. The spade (♣) indicates a significant increase in comparison to IgG control group (P< 0.05).
DNMT3b-mediated promoter hypermethylation contributed to MEG3 downregulation and HIF-1α protein upregulation following nickel exposure
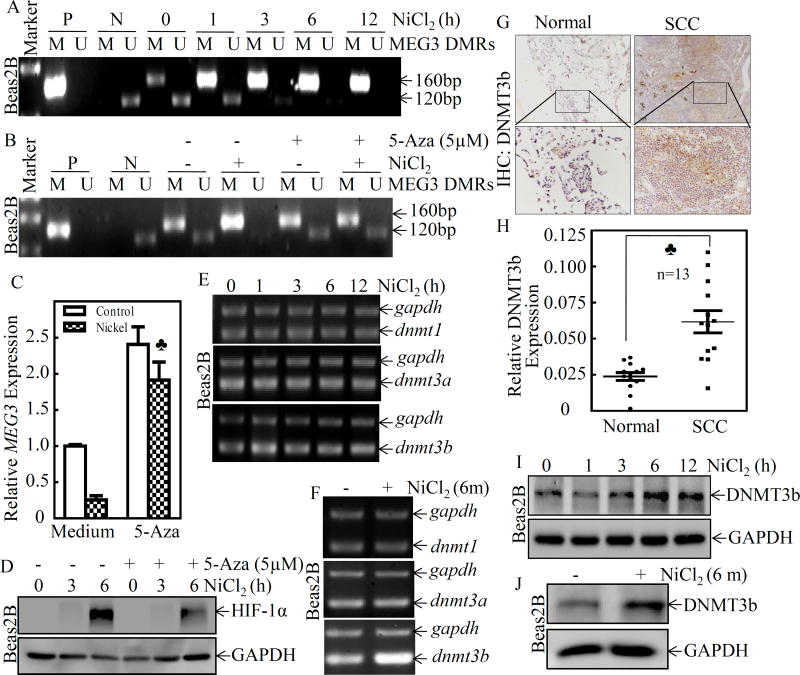
It has been reported that the MEG3 promoter hypermethylation is an important mechanism associated with the loss of MEG3 expression in clinically nonfunctioning pituitary tumors27. To investigate whether the downregulation of MEG3 expression in nickel exposure is due to promoter hypermethylation, we treated Beas2B cells with NiCl2 over various time periods, and assessed MEG3 promoter methylation at the differentially methylated region (DMR) using Methylation-Specific PCR (MS-PCR). The extent of the DMR of human MEG3 was shown to span approximately 4 kb of genomic sequence that encompasses the putative promoter region and includes two consensus CTCF (CCCTC binding factor) binding sites that also exhibit differential methylation28. In the present study, we referenced the studies from Murphy et al. 28, which have designed two independent primer sets to amplify the methylated and unmethylated DMR. The region analyzed by MS-PCR is from nt 64,450 to nt 66,150 of accession no. AL117190, spans ~1,700 bp, which correspond to sequences within the CpG-rich regions upstream of MEG3. As shown in Figure 6A, nickel treatment caused a time-dependent upregulation of methylated DNA (M), produced a 160-bp band, accompanied with the downregulation of unmethylated DNA (U), produced a 120-bp band, in Beas2B cells. To further explore the role of promoter hypermethylation in MEG3 downregulation, the DNA methyltransferase inhibitor, 5-aza-2’-deoxycytidine, was used to inhibit DNA methylation in genomic DNA. The results showed that the treatment of cells with 5-aza-2’-deoxycytidine inhibited nickel-induced methylation of MEG3 promoter (Figure 6B) and reversed the inhibition of nickel-induced MEG3 downregulation that was observed in Beas2B cells treated with nickel only (Figure 6C). Consistently, 5-aza-2’-deoxycytidine treatment also attenuated HIF-1α protein abundance due to nickel exposure in Beas2B cells (Figure 6D). These results reveal that nickel exposure led to MEG3 promoter hypermethylation contributes to HIF-1α protein expression in human bronchial epithelial cells.
Figure 6. DNMT3b mediated promoter hypermethylation contributed to MEG3 downregulation and HIF-1α protein expression following nickel exposure.
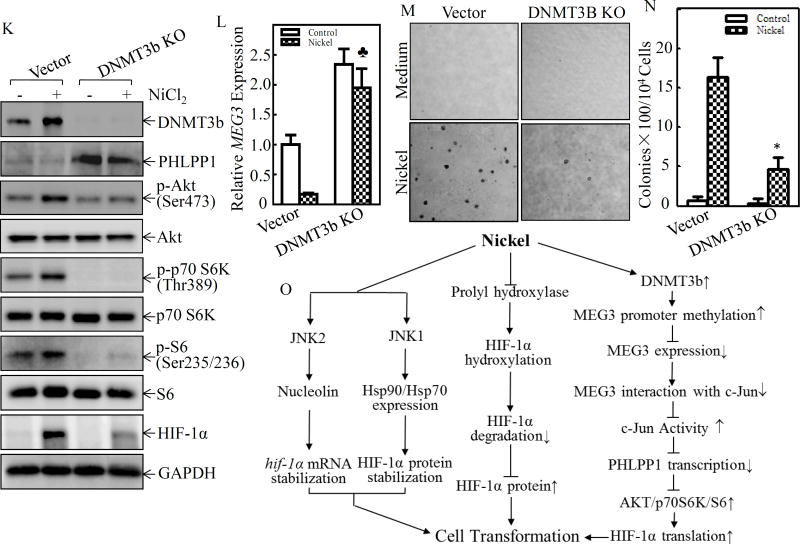
(A) Beas2B cells were exposed to 0.5 mM of NiCl2 for the indicated time periods. The methylation status of the MEG3 promoter in the differentially methylated region (DMR) was determined using the methylation-specific PCR (MS-PCR) assay. A primer set was used to evaluate the methylated (M) and unmethylated (U) copies of the MEG3 DMR gene. Methylated control was used as the positive control (P), while unmethylated control was used as the negative control (N) A 160 bp PCR product represents the methylated state and a 120 bp PCR product stands for the unmethylated allele. (B–D) Beas2B cells were pretreated with 5-aza-2’-deoxycytidine (5µM) for 72h, then exposed to 0.5 mM of NiCl2 for 6h. The cells were extracted to determinate the methylation status of the MEG3 promoter DMR by using the MS-PCR (B). The relative levels of MEG3 were determined by qPCR using gene specific primers and gapdh was used as an internal control. The spade (♣) indicates a significant increase in comparison to the medium control (P< 0.05) (C). The cells were also extracted for determination of HIF-1α and GAPDH protein expression by Western Blot (D). (E & F) Beas2B cells were exposed to 0.5 mM of NiCl2 for the indicated time periods and total RNA was extracted to detect dnmt1, dnmt3a, and dnmt3b expression using RT-PCR. (G & H) Immunohistochemically staining was carried out to evaluate DNMT3b expression in human lung squamous cell carcinomas and protein expression levels were analyzed by calculating the integrated optical density per stained area (IOD/area). (I & J) Beas2B cells were exposed to 0.5 mM of NiCl2 for the indicated time periods and the cells were extracted to determinate the DNMT3b and GAPDH protein expression by Western Blot. (K) DNMT3B CRISP knockout cell, Beas2B(DNMT3B KO), and its scramble control transfectant, Beas2B(Vector) cells, were exposed to 0.5 mM of NiCl2 for 6 h. The cell protein extracts were subjected to Western blot to determinate protein expression, as indicated (K), mRNA extracts were subjected to determinate MEG3 by qPCR (L). (M & N) Beas2B(DNMT3B KO) and Beas2B(Vector) cells and were repeatedly exposed to 0.5 mM of NiCl2 for 3 months. The cells were than subjected to determinate the anchorage-independent growth capability in soft agar (M). The number of colonies were scored and presented as colonies per 104 seeded cells (N). The asterisk (*) indicate a significant decrease in comparison to Beas2B(Vector) cells (P< 0.05). (O) A schematic illustration of the role and molecular mechanism underlying MEG3 in promotion of HIF-1α protein translation and malignant transformation of human bronchial epithelial cells following nickel exposure.
DNA methyltransferases (DNMTs) are enzymes that are responsible for the transfer of a methyl group from the universal methyl donor, S-adenosyl-L-methionine (SAM), to the 5-position of cytosine residues in DNA29. To investigate the effect of DNMTs on the hypermethylation of MEG3 differentially methylated regions (DMRs), we first evaluated the potential effect of short and long-term nickel exposure on the mRNA expression of DNMTs, including dnmt1, dnmt3a and dnmt3b. The results indicated that nickel exposure specifically induced mRNA expression of dnmt3b, but not dnmt1 and dnmt3a, following treatment of Beas2B cells with nickel for either 12 hours or 6 months (Figures 6E & 6F). The upregulation of DNMT3b protein has also been observed in human lung cancer tissues (Figures 6G & 6H) and nickel-exposed Beas2B cells (Figures 6I & 6J). Moreover, data analyses showed a highly negative correlation between DNMT3b expression and MEG3 level (ρ=−0.725, P=0.005), whereas no significant correlation was observed between PHLPP1 and DNMT3b (ρ=−0.523, P= 0.067) (Supplemental Figures S3B & S3C), revealing that DNMT3b might be directly regulating MEG3 expression, but not PHLPP1, following nickel exposure. To explore this relationship between DNMT3b and MEG3, specific sgRNA targeting DNMT3b was used to stably knockout dnmt3b in Beas2B cells. As shown in Figures 6K & 6L, DNMT3b knockout reversed nickel-induced MEG3 downregulation, increased in PHLPP1 expression and blocked Akt phosphorylation at Ser473, p70S6K phosphorylation at Thr389, S6 phosphorylation at Ser235/236, and HIF-1α protein induction in Beas2B cells due to nickel exposure. Consistently, DNMT3b knockout also abolished malignant transformation of human bronchial epithelial cells due to nickel exposure (Figures 6M& 6N). Taken together, our results strongly demonstrate that DNMT3b upregulation contributes to nickel-induced MEG3 promoter hypermethylation, which leads to inhibition of MEG3 expression, consequently resulting in c-Jun activation, phlpp1 transcriptional reduction, and in turn promoting HIF-1α protein translation via activation of the Akt/p70S6K/S6 axis in Beas2B cells as summarized in Figure 6O.
Discussion
MEG3, as a tumor suppressor, is expressed in multiple normal tissues of human internal organs13. The loss of MEG3 expression has been thought to be a primary feature of human cancers, such as liver cancer30, gastric cancer31, colorectal cancer32, glioma33, cervical cancer34, bladder cancer35, prostate cancer36, and lung cancer17. However, whether MEG3 is reducible upon environmental carcinogen exposure, what are the upstream regulators that lead to MEG3 expression and the downstream effectors mediating tumor suppressive function, as well as how MEG3 contributes to the tumorigenicity of environmental carcinogen has never been explored. In the present study, we found that nickel was able to inhibit MEG3 expression in human bronchial epithelial cells. Since squamous cell carcinoma is the major histological type in lung cancers of nickel-refinery workers, we have compared MEG3 expression in lung cancer tissues with their adjacent non-tumor lung tissues. The results consistently show the downregulation of MEG3 expression in squamous cell carcinomas. We have further demonstrated that downregulation of MEG3 is an important player for nickel-induced the transformation of human bronchial epithelial cells. Thus, our present studies not only, for the first time, discover the downregulation of MEG3 expression due to nickel exposure, they also demonstrate the contribution of MEG3 downregulation to malignant transformation of human bronchial epithelial cell following nickel exposure.
Hypoxia inducible factor-1 (HIF-1) is a master transcription factor that is critical for the regulation of the hypoxic response in eukaryotic cells. Deregulation of HIF-1 activity is mainly associated with the expression of HIF-1α subunit, a key regulatory subunit responsible for HIF-1 transcriptional function37. Oxygen-dependent hydroxylation of the HIF-1α subunit by prolyl hydroxylase domain (PHD) proteins signals their polyubiquitination and proteasomal degradation, and plays a critical role in regulating HIF-1α abundance and oxygen homeostasis38. Some environmental factors, including nickel, can inhibit PHD activity and cause HIF-1α stabilization39. Moreover, our published studies show that HIF-1α mRNA and protein stabilization can also be regulated by JNK210 and JNK19, respectively. However, translational regulation of HIF-1α, particularly under environmental carcinogen such as nickel, has never been explored. Our current studies demonstrate that MEG3 play a crucial role in nickel-induced HIF-1α protein translation. We showed that overexpression of MEG3 inhibited HIF-1α protein abundance, while knockdown of MEG3 increased HIF-1α protein expression by nickel exposure. In contrast to the protein abundance, HIF-1α mRNA levels and protein degradation rates were comparable between Beas2B(MEG3) vs. Beas2B(Vector) following nickel exposure, excluding the possibility that MEG3 regulates HIF-1α abundance at levels of transcription, mRNA stability or protein degradation although they are implicated in HIF-1α protein induction by nickel. The results obtained using [35S]-labeled methionine and cysteine for new protein synthesis clearly reveal that MEG3 is able to inhibit HIF-1α protein translation. Ribosomal protein S6 is a well-known mediator of protein translation, phosphorylation of S6 promotes the recruitment of the 40S ribosome to the mRNA and therefore has a paramount effect on protein translation40. Our results indicated that overexpression of MEG3 inhibited nickel-induced S6 phosphorylation at Ser235/Ser236, whereas knockdown of MEG3 significantly increased S6 phosphorylation at Ser235/Ser236. We further found that MEG3 downregulation resulted in S6 activation, and in turn directly promoting protein translation as demonstrated by the results obtained from comparison of S6 phosphorylation at Ser235/Ser236 in Beas2B(MEG3) vs. Beas2B(Vector) and Beas2B(shMEG3) vs. Beas2B(Nonsense), and utilization of mTOR inhibitor rapamycin and the shRNA specific targeting S6. By tracking the upstream pathways responsible for phosphorylation of S6, we defined MEG3 inhibition of Akt phosphorylation at Ser473, which is an upstream regulator directly mediating p70S6K/S6 phosphorylation and activation. Precise control of the balance of protein phosphorylation and dephosphorylation that is catalyzed by protein kinases and phosphatases is essential for cellular homeostasis. PTEN41, PP2A25 and PHLPP24 are three ubiquitously expressed phosphatases that have been reported to dephosphorylate many critical cellular molecules, such as Akt. Our further mechanistic elucidation indicates that PHLPP1, but not PP2A and PTEN, is an MEG3 downstream effector responsible for its inhibition of Akt phosphorylation and HIF-1α protein translation upon nickel exposure. Those results may be extent to human, since we found a strong positive correlation between MEG3 and PHLPP1 expression in lung cancer tissues.
c-Jun has been reported to play an important role in regulation of cellular proliferation, transformation and death42 and it promotes cellular survival by negatively regulating the expression of the tumor-suppressor PTEN, resulting in the concomitant activation of the Akt survival pathway43. Here we show that MEG3 regulates phlpp1 transcription through its direct interaction with the c-Jun and inhibition of c-Jun activity, while c-Jun is a negative transcription factor for phlpp1 promoter transcription. We found that c-Jun negatively regulates phlpp1 transcription by binding directly to the phppp1 promoter, and that MEG3 is able to bind to the c-Jun transcription factor and decrease c-Jun activity in the inhibition of phlpp1 promoter transcription. Thus, a highly specific and stable triplex structure formed by MEG3, together with the c-Jun and the phlpp1 promoter, results in MEG3 promotion of phlpp1 transcription.
Silencing of genes by aberrant promoter hypermethylation is one of the mechanisms leading to tumor suppressor downregulation and cancer initiation and progression44. It has been reported that hypermethylation of the MEG3 regulatory region has also been associated with the loss of MEG3 expression in human tumors cell lines, such as pituitary tumors27, gliomas45 and hepatoma46. In the present study, we found that environmental carcinogen nickel resulted in the hypermethylation of MEG3 regulatory region in normal human bronchial epithelial cells, and that treatment of cells with DNA methylation inhibitor 5-aza-2-deoxycytidine (5-Aza) increased in MEG3 expression, strongly indicating that the promoter hypermethylation mediates downregulation of MEG3 transcription due to nickel exposure. Methylation of mammalian genomic DNA is catalyzed by DNMTs47. In mammals, there are 3 major DNMTs: DNMT1, DNMT3a, and DNMT3b. DNMT1 is a maintenance DNMT, while DNMT3a and 3b are de novo DNMTs48. Our further elucidation shows that nickel exposure upregulated DNMT3b expression, and that knockout of DNMT3b promoted MEG3 and PHLPP1 expression, accompanied by inhibition of Akt/p70/S6 pathway activation, HIF-1α accumulation, and malignant transformation of Beas2B cells. Collectively, our studies demonstrate the crucial role of DNMT3b induction in the mediation of promoter hypermethylation and downregulation of MEG3, as well as their contribution to nickel-induced lung tumorigenicity. Those results may also be extent to human, since we found a strong negative correlation between DNMT3b and MEG3 expression in lung cancer tissues.
In summary, our results define a novel effect of nickel on MEG3 reduction and the upstream epigenetic regulator DNMT3b leading to this reduction, as well as the mechanisms underlying its interaction with downstream effector c-Jun, consequently resulting in attenuation of PHLPP1 expression, Akt/p70S60K/S6 activation, HIF-1α protein translation as well as malignant transformation of human bronchial epithelial cells. These findings demonstrate the association and function of MEG3 in nickel-induced lung tumorigenesis and provide novel insights into understanding of the multiple levels of HIF-1α protein regulation in nickel responses.
Materials and Methods
Cell lines, Plasmids, Antibodies, and Other reagents
These details are described in Supplemental Information.
Patients and Tumor Sample Preparation
Thirteen pairs of lung squamous cell carcinoma, seventeen pairs of lung adenocarcinoma and adjacent non-tumor tissue specimens were obtained from surgical specimens at The First Affiliated Hospital of Wenzhou Medical University (Wenzhou, China) after informed consent. Lung squamous cell carcinoma was diagnosed histopathologically and clinical characteristics of the patients included in this study were presented in Supplementary materials. Adjacent non-tumor tissue specimens were taken from a standard distance (3 cm) from the margin of resected neoplastic tissues of patients with tumors who ensured surgical lung ablation. All these specimens were snap-frozen in liquid nitrogen after excision. This study was compliant with the Declaration of Helsinki Guidelines and was approved by the Medical Ethics Committee of Wenzhou Medical University. Informed consent was obtained from each participant and the procedures were carried out in accordance with the approved guidelines.
Cell Transformation and Anchorage-independent Growth Assay
Cell transformation was performed using anchorage-independent growth in soft agar as described in our previous publication9.
Luciferase Reporter Assay
Luciferase reporter assays were performed as described in our previous publication49.
[35S]-labeled Methionine-Cysteine Pulse Assay
The [35S]-labeled methionine-cysteine pulse assay was performed as described in our previous publication9. [35S]-labeled HIF-1α protein was detected with the Phosphor Imager (Molecular Dynamics, Kent City, MI, USA) and analyzed by calculating the integrated optical density per band area (IOD/area) using the Alpha Innotech SP image system (Alpha Innotech Corporation, USA).
DNA Extraction, Bisulfite DNA Modification, and Methylation-Specific PCR
Genomic DNA from Beas2B cells was extracted using DNeasy Blood & Tissue Kit (Qiagen, Gaithersburg, MD, USA) according to the manufacturer’s guidelines. Sodium bisulfite modification of DNA and subsequent purification was performed according to the manufacturer’s guidelines for sodium bisulfite conversion of unmethylated cytosine in DNA using EpiTect Bisulfite kit (Qiagen, Gaithersburg, MD, USA).
Bisulfite-treated genomic DNA was subjected to an optimized methylation-specific PCR, and was performed as previously described28.
Semiquantitative and quantitative RT-PCR
Semiquantitative or quantitative RT-PCR was performed to examine the expression level of mRNA and lncRNA MEG3, respectively, as previously described49. The primers used in this study are listed in Supplemental Table S2.
Immunohistochemistry (IHC) Staining
For immunohistochemistry staining, we respectively used antibodies specific against PHLPP1 and DNMT3b, and the protein expression levels were analyzed as previously described50.
Western Blotting
Western Blotting was performed as previously described10.
Chromatin Immunoprecipitation (ChIP)
Chromatin immunoprecipitation (ChIP) assays were performed using the EZ-ChIP chromatin immunoprecipitation kit (Millipore, Billerica, MA, USA) according to the manufacturer’s guidelines. Immunoselections of cross-linked protein-DNA were performed using anti-c-Jun antibody together agarose beads A/G, at 4 °C for overnight. The anti-rabbit IgG was used as a negative control. The purified DNAs were analyzed by PCR, the forward and reverse primers were: 5’- GGT GTG ACG GGT GTG TAT CT −3’ and 5’- GTA TTT TAC CCG CAC TCC AGC −3’ for human phlpp1 promoter.
RNA Immunoprecipitation (RIP)
RNA Immunoprecipitation was performed as previously described10. The RNAs in the buffer were extracted by TriZol reagent. qPCR was performed to detect the MEG3 presented in the immune-complex.
Statistical Analysis
All of the statistical analyses were performed by SPSS 17.0 software (SPSS Inc., Chicago, IL, USA). The student’s t-test was used to determine the significance of differences between two groups. The MEG3 expression in lung cancer tissues was compared with that of the corresponding adjacent non-tumor lung tissues using the paired samples t-test. P value with less than 0.05 was considered to be statistically significant.
Acknowledgments
We greatly appreciated Dr. Shau-Ping Lin (Institute of Biotechnology, National Taiwan University, Taipei, Taiwan) for his generous gifts about shRNA constructs against MEG3 and control vector. This work was supported partially by grants from NIH/NCI CA112557, CA165980 and CA177665, NIH/NIEHS ES000260, National Natural Science Foundation of China (81441090, 81229002, 81372946, 81673141 and 81371730), the Specialized Research Fund for the Doctoral Program of Higher Education (20133420110001), the Key Project of Science and Technology Innovation Team of Zhejiang Province (2013TD10), and the Grant for Scientific Research of BSKY (XJ201418) from Anhui Medical University. We also thanks Ms. Nedda Tichi for her critically reading of the manuscript.
Footnotes
Conflicts of interest: The authors declare no conflict of interest.
Authors’ Contributions
Conception and design: C. Zhou, Q. Zhu, C. Huang
Acquisition of data (acquired and managed patients, provided facilities, etc.): C. Zhou, H. Huang, J. Li, Q. Xie, J. Zhu, Y. Li, D. Zhang, Y. Liu, and J. Wang
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): C. Zhou, H. Huang, and C. Huang
Writing, review, and revision of the manuscript: C. Zhou and C. Huang
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): C. Zhou, J. Li, Q. Zhu, and C. Huang
Study supervision: C. Huang
Supplemental Information
Supplemental Material is available online
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2016. Ca-Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Field RW, Withers BL. Occupational and environmental causes of lung cancer. Clin Chest Med. 2012;33:681–703. doi: 10.1016/j.ccm.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kasprzak KS, Sunderman FW, Jr, Salnikow K. Nickel carcinogenesis. Mutat Res. 2003;533:67–97. doi: 10.1016/j.mrfmmm.2003.08.021. [DOI] [PubMed] [Google Scholar]
- 4.Lu H, Shi X, Costa M, Huang C. Carcinogenic effect of nickel compounds. Mol Cell Biochem. 2005;279:45–67. doi: 10.1007/s11010-005-8215-2. [DOI] [PubMed] [Google Scholar]
- 5.Ke Q, Costa M. Hypoxia-inducible factor-1 (HIF-1) Mol Pharmacol. 2006;70:1469–1480. doi: 10.1124/mol.106.027029. [DOI] [PubMed] [Google Scholar]
- 6.Lee JW, Bae SH, Jeong JW, Kim SH, Kim KW. Hypoxia-inducible factor (HIF-1)alpha: its protein stability and biological functions. Exp Mol Med. 2004;36:1–12. doi: 10.1038/emm.2004.1. [DOI] [PubMed] [Google Scholar]
- 7.Li J, Davidson G, Huang Y, Jiang BH, Shi X, Costa M, et al. Nickel compounds act through phosphatidylinositol-3-kinase/Akt-dependent, p70(S6k)-independent pathway to induce hypoxia inducible factor transactivation and Cap43 expression in mouse epidermal Cl41 cells. Cancer research. 2004;64:94–101. doi: 10.1158/0008-5472.can-03-0737. [DOI] [PubMed] [Google Scholar]
- 8.Ouyang W, Zhang D, Li J, Verma UN, Costa M, Huang C. Soluble and insoluble nickel compounds exert a differential inhibitory effect on cell growth through IKKalpha-dependent cyclin D1 down-regulation. J Cell Physiol. 2009;218:205–214. doi: 10.1002/jcp.21590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang D, Li J, Costa M, Gao J, Huang C. JNK1 mediates degradation HIF-1alpha by a VHL-independent mechanism that involves the chaperones Hsp90/Hsp70. Cancer research. 2010;70:813–823. doi: 10.1158/0008-5472.CAN-09-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang D, Li J, Zhang M, Gao G, Zuo Z, Yu Y, et al. The requirement of c-Jun N-terminal kinase 2 in regulation of hypoxia-inducing factor-1alpha mRNA stability. The Journal of biological chemistry. 2012;287:34361–34371. doi: 10.1074/jbc.M112.365882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 12.Miyoshi N, Wagatsuma H, Wakana S, Shiroishi T, Nomura M, Aisaka K, et al. Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome 14q. Genes Cells. 2000;5:211–220. doi: 10.1046/j.1365-2443.2000.00320.x. [DOI] [PubMed] [Google Scholar]
- 13.Zhou Y, Zhang X, Klibanski A. MEG3 noncoding RNA: a tumor suppressor. J Mol Endocrinol. 2012;48:R45–R53. doi: 10.1530/JME-12-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benetatos L, Vartholomatos G, Hatzimichael E. MEG3 imprinted gene contribution in tumorigenesis. Int J Cancer. 2011;129:773–779. doi: 10.1002/ijc.26052. [DOI] [PubMed] [Google Scholar]
- 15.Salnikow K, Zhitkovich A. Genetic and epigenetic mechanisms in metal carcinogenesis and cocarcinogenesis: nickel, arsenic, and chromium. Chem Res Toxicol. 2008;21:28–44. doi: 10.1021/tx700198a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hou L, Zhang X, Wang D, Baccarelli A. Environmental chemical exposures and human epigenetics. Int J Epidemiol. 2012;41:79–105. doi: 10.1093/ije/dyr154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu KH, Li W, Liu XH, Sun M, Zhang ML, Wu WQ, et al. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer. 2013;13:461. doi: 10.1186/1471-2407-13-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willey JC, Broussoud A, Sleemi A, Bennett WP, Cerutti P, Harris CC. Immortalization of normal human bronchial epithelial cells by human papillomaviruses 16 or 18. Cancer research. 1991;51:5370–5377. [PubMed] [Google Scholar]
- 19.Kressler D, Hurt E, Bassler J. Driving ribosome assembly. Biochim Biophys Acta. 2010;1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 20.Ruvinsky I, Sharon N, Lerer T, Cohen H, Stolovich-Rain M, Nir T, et al. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev. 2005;19:2199–2211. doi: 10.1101/gad.351605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimobayashi M, Hall MN. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat Rev Mol Cell Biol. 2014;15:155–162. doi: 10.1038/nrm3757. [DOI] [PubMed] [Google Scholar]
- 22.Yu Y, Zhang D, Huang H, Li J, Zhang M, Wan Y, et al. NF-kappaB1 p50 promotes p53 protein translation through miR-190 downregulation of PHLPP1. Oncogene. 2014;33:996–1005. doi: 10.1038/onc.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, Ouyang W, Li J, Wei L, Ma Q, Zhang Z, et al. Loss of tumor suppressor p53 decreases PTEN expression and enhances signaling pathways leading to activation of activator protein 1 and nuclear factor kappaB induced by UV radiation. Cancer research. 2005;65:6601–6611. doi: 10.1158/0008-5472.CAN-04-4184. [DOI] [PubMed] [Google Scholar]
- 24.Newton AC, Trotman LC. Turning off AKT: PHLPP as a drug target. Annu Rev Pharmacol Toxicol. 2014;54:537–558. doi: 10.1146/annurev-pharmtox-011112-140338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seshacharyulu P, Pandey P, Datta K, Batra SK. Phosphatase: PP2A structural importance, regulation and its aberrant expression in cancer. Cancer Lett. 2013;335:9–18. doi: 10.1016/j.canlet.2013.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang H, Zhu J, Li Y, Zhang L, Gu J, Xie Q, et al. Upregulation of SQSTM1/p62 contributes to nickel-induced malignant transformation of human bronchial epithelial cells. Autophagy. 2016;12:1687–1703. doi: 10.1080/15548627.2016.1196313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao J, Dahle D, Zhou Y, Zhang X, Klibanski A. Hypermethylation of the promoter region is associated with the loss of MEG3 gene expression in human pituitary tumors. J Clin Endocrinol Metab. 2005;90:2179–2186. doi: 10.1210/jc.2004-1848. [DOI] [PubMed] [Google Scholar]
- 28.Murphy SK, Wylie AA, Coveler KJ, Cotter PD, Papenhausen PR, Sutton VR, et al. Epigenetic detection of human chromosome 14 uniparental disomy. Human mutation. 2003;22:92–97. doi: 10.1002/humu.10237. [DOI] [PubMed] [Google Scholar]
- 29.Jin B, Robertson KD. DNA methyltransferases, DNA damage repair, and cancer. Adv Exp Med Biol. 2013;754:3–29. doi: 10.1007/978-1-4419-9967-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anwar SL, Krech T, Hasemeier B, Schipper E, Schweitzer N, Vogel A, et al. Loss of imprinting and allelic switching at the DLK1-MEG3 locus in human hepatocellular carcinoma. PLoS One. 2012;7:e49462. doi: 10.1371/journal.pone.0049462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun M, Xia R, Jin F, Xu T, Liu Z, De W, et al. Downregulated long noncoding RNA MEG3 is associated with poor prognosis and promotes cell proliferation in gastric cancer. Tumour Biol. 2014;35:1065–1073. doi: 10.1007/s13277-013-1142-z. [DOI] [PubMed] [Google Scholar]
- 32.Yin DD, Liu ZJ, Zhang E, Kong R, Zhang ZH, Guo RH. Decreased expression of long noncoding RNA MEG3 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer. Tumour Biol. 2015;36:4851–4859. doi: 10.1007/s13277-015-3139-2. [DOI] [PubMed] [Google Scholar]
- 33.Wang P, Ren Z, Sun P. Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation. J Cell Biochem. 2012;113:1868–1874. doi: 10.1002/jcb.24055. [DOI] [PubMed] [Google Scholar]
- 34.Zhang J, Yao T, Wang Y, Yu J, Liu Y, Lin Z. Long noncoding RNA MEG3 is downregulated in cervical cancer and affects cell proliferation and apoptosis by regulating miR-21. Cancer Biol Ther. 2016;17:104–113. doi: 10.1080/15384047.2015.1108496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ying L, Huang Y, Chen H, Wang Y, Xia L, Chen Y, et al. Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer. Mol Biosyst. 2013;9:407–411. doi: 10.1039/c2mb25386k. [DOI] [PubMed] [Google Scholar]
- 36.Luo G, Wang M, Wu X, Tao D, Xiao X, Wang L, et al. Long Non-Coding RNA MEG3 Inhibits Cell Proliferation and Induces Apoptosis in Prostate Cancer. Cell Physiol Biochem. 2015;37:2209–2220. doi: 10.1159/000438577. [DOI] [PubMed] [Google Scholar]
- 37.Weidemann A, Johnson RS. Biology of HIF-1alpha. Cell Death Differ. 2008;15:621–627. doi: 10.1038/cdd.2008.12. [DOI] [PubMed] [Google Scholar]
- 38.Fong GH, Takeda K. Role and regulation of prolyl hydroxylase domain proteins. Cell Death Differ. 2008;15:635–641. doi: 10.1038/cdd.2008.10. [DOI] [PubMed] [Google Scholar]
- 39.Salnikow K, Donald SP, Bruick RK, Zhitkovich A, Phang JM, Kasprzak KS. Depletion of intracellular ascorbate by the carcinogenic metals nickel and cobalt results in the induction of hypoxic stress. The Journal of biological chemistry. 2004;279:40337–40344. doi: 10.1074/jbc.M403057200. [DOI] [PubMed] [Google Scholar]
- 40.Magnuson B, Ekim B, Fingar DC. Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem J. 2012;441:1–21. doi: 10.1042/BJ20110892. [DOI] [PubMed] [Google Scholar]
- 41.Georgescu MM. PTEN Tumor Suppressor Network in PI3K-Akt Pathway Control. Genes Cancer. 2010;1:1170–1177. doi: 10.1177/1947601911407325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4:E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 43.Hettinger K, Vikhanskaya F, Poh MK, Lee MK, de Belle I, Zhang JT, et al. c-Jun promotes cellular survival by suppression of PTEN. Cell Death Differ. 2007;14:218–229. doi: 10.1038/sj.cdd.4401946. [DOI] [PubMed] [Google Scholar]
- 44.Belinsky SA. Gene-promoter hypermethylation as a biomarker in lung cancer. Nat Rev Cancer. 2004;4:707–717. doi: 10.1038/nrc1432. [DOI] [PubMed] [Google Scholar]
- 45.Li J, Bian EB, He XJ, Ma CC, Zong G, Wang HL, et al. Epigenetic repression of long non-coding RNA MEG3 mediated by DNMT1 represses the p53 pathway in gliomas. Int J Oncol. 2016;48:723–733. doi: 10.3892/ijo.2015.3285. [DOI] [PubMed] [Google Scholar]
- 46.Liu LX, Deng W, Zhou XT, Chen RP, Xiang MQ, Guo YT, et al. The mechanism of adenosine-mediated activation of lncRNA MEG3 and its antitumor effects in human hepatoma cells. Int J Oncol. 2016;48:421–429. doi: 10.3892/ijo.2015.3248. [DOI] [PubMed] [Google Scholar]
- 47.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 48.Subramaniam D, Thombre R, Dhar A, Anant S. DNA methyltransferases: a novel target for prevention and therapy. Front Oncol. 2014;4:80. doi: 10.3389/fonc.2014.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu Z, Zeng X, Xu J, Xu D, Li J, Jin H, et al. Isorhapontigenin suppresses growth of patient-derived glioblastoma spheres through regulating miR-145/SOX2/cyclin D1 axis. Neuro-oncology. 2016;18:830–839. doi: 10.1093/neuonc/nov298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang H, Pan X, Jin H, Li Y, Zhang L, Yang C, et al. PHLPP2 Downregulation Contributes to Lung Carcinogenesis Following B[a]P/B[a]PDE Exposure. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21:3783–3793. doi: 10.1158/1078-0432.CCR-14-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]