Abstract
Background
Wide acceptance of the colony-forming-unit (CFU) assay as a reliable potency test for stem cell products is hindered by poor inter-laboratory reproducibility. The goal of this study was to ascertain current laboratory practices for performing the CFU assay to identify practices that could be standardized to improve overall reproducibility.
Materials and Methods
A survey to evaluate current laboratory practices for performing CFU assays was designed and internationally distributed.
Results
A total of 105 individuals initiated the survey, of which 68% perform CFU assays. A majority specified that an automated rather than a manual cell count was performed on pre-diluted aliquots of stem cell products. Viability testing methods employed a variety of stains and when multiple sites used the same viability stain the methods differed. Cell phenotype used to prepare working cell suspensions for inoculating the CFU assay differed among sites. Most respondents scored CFU assays between 14–16 days of incubation, but culture plates were read with a variety of different microscopes. Of 57 respondents, 42% had not performed a validation study or established assay linearity. Only 63% of laboratories had criteria for determining if a plate was overgrown with colonies.
Conclusion
Survey results revealed inconsistent inter-laboratory practices for performing the CFU assay. Moreover, the relatively low number of centers with validated CFU assays raises concerns about assay accuracy and emphasizes a need for the establishment of central standards. The survey results shed light on a number of steps of the methodology that could be targeted for standardization across laboratories.
Keywords: Colony-Forming-Units, Hematopoietic, Hematopoietic Progenitor Cells, Potency Test, Hematopoietic
INTRODUCTION
Functional analysis of hematopoietic progenitor cell (HPC) products is critical for comparative selection of the highest quality stem cell product for a transplant recipient. Yet, the selection of stem cell products for transplantation is typically based primarily upon non-functional cellular parameters such as TNCs and cellular immunophenotypes (e.g. CD34+ cell counts). While these surrogate assays have demonstrated good inverse correlations with the period of clinically relevant cytopenia, the predictive value of these assays for hematopoietic engraftment may be reduced given that they do not provide functional information on the hematopoietic quality of the graft. As a result, the absence of functional hematopoietic information may result in the selection of low potency stem cell products that fail to engraft in a patient despite a unit having a high cell count with an acceptable phenotype.
The CFU assay is a hematopoietic functional assay, which is often used to measure the function or potency of hematopoietic progenitors present in stem cell products. However, poor inter-laboratory reproducibility of the CFU assay even among experienced laboratories precludes universal implementation of this assay (1, 2). As a consequence, the CFU assay fails to meet potency testing guidelines as set forth by the U.S. Federal Drug Administration (FDA) (3). These guidelines require that a potency assay be capable of predicting therapeutic outcome, establishing industry release criteria and defining product expiration. On the other hand, reasonably good intra-laboratory reproducibility for the CFU assay has resulted in some investigators reporting that there is a good correlation between numbers of CFU generating progenitors present in stem cell products and short-term hematopoietic reconstitution in autologous and allogeneic transplantation settings (4–9). Given that CFU assays performed at a single site can correlate with engraftment, it should be possible with stringent standardization of the method to improve inter-laboratory reproducibility so that results from different sites can be used to predict the in vivo efficacy of stem cell grafts for clinical applications.
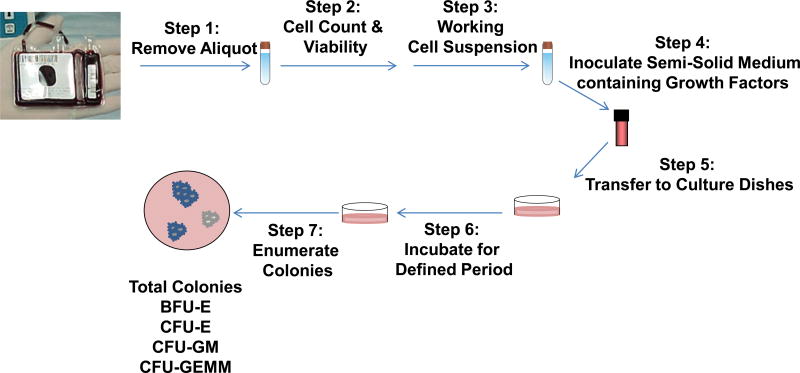
The CFU assay takes advantage of a hematopoietic progenitor’s ability to proliferate and differentiate to form a colony of cells committed to specific blood cell lineages. This in vitro assay is typically performed by removing an aliquot of cells from a stem cell product, preparing a working cell suspension, inoculating growth factor containing semi-solid medium with a desired cell concentration and transferring cells/methylcellulose into culture dishes (Fig 1.). The dishes are then placed in a humidified incubator for a defined period of time. At the end of the culture period the total number of colonies produced is counted microscopically and classified according to their morphological features as burst-forming unit-erythroid (BFU-E), colony-forming unit erythroid (CFU-E), CFUs containing granulocytes and macrophages (CFU-GM), and CFUs containing granulocytes, erythrocytes, macrophages, and megakaryocytes (CFU-GEMM). The type and number of the colonies obtained at the end of the culture period is driven by the amount and combination of growth factors present in the culture.
Figure 1. Basic steps for setting up the CFU assay.
Step 1) An aliquot of cells is removed from a stem cell product; Step 2) A pre-dilution cell count and viability test are performed; Step 3) A working cell suspension is made from the aliquot of cells removed from step 1; Step 4) Semi-solid medium containing growth factors is inoculated with a defined volume of the working cell suspension; Step 5) The semi-solid medium containing growth factors and cells are transferred to a culture vessel; Step 6) The culture vessel is placed in an incubator for a defined period of time; Step 7) At the end of the culture period, the colonies are enumerated and differentiated.
As a first step towards inter-laboratory standardization of the CFU assay, the Cell Therapy Team of the Biomedical Excellence for Safer Transfusion (BEST) Collaborative designed a survey to evaluate current practices among different laboratories to identify sources of variability that may contribute to assay variability. The survey focused on practices associated with performing the CFU assay on fresh samples and was distributed internationally through membership rosters of the AABB, International Society for Cell Therapy (ISCT) and European Bone Marrow Transplant (EBMT) societies. Results from the survey expose highly variable laboratory practices, which support a need for the establishment of inter-laboratory standards for the CFU assay. Using survey results, in this document we provide suggestions for areas of practices to be considered for standardization of the CFU assay and how to address some of them to improve inter-laboratory assay precision.
MATERIALS AND METHODS
The BEST Collaborative Cellular Therapy Team used SurveyMonkey (http://www.surveymonkey.com/) to assess current laboratory practices for the CFU assay when performed on fresh samples. No attempt was made to assess laboratory practices for performing the CFU assay on previously frozen and thawed samples. The survey was designed to ask questions about how laboratories perform the CFU assay when evaluating hematopoietic progenitor cells in apheresis (HPC-A), marrow (HPC-M), and umbilical cord blood (HPC-C) products. The survey was designed with primarily closed-ended questions that provided one answer or multiple responses to a fixed set of possible choices. Skip Logic, or conditional branching rules were also used to direct respondents through different questions based on their response to a previous questions. Prior to a wide distribution of the survey, the BEST membership pilot tested the survey to facilitate the removal of inconsistent questions.
The survey was widely distributed to members of the AABB, the ISCT and EBMT. Individuals who responded to an invitation to complete the survey were directed to a specific page on the BEST Collaborative website. Each participant was provided with a brief explanation of the survey’s purpose and a direct link to the survey. Participants were provided the option of providing contact information. Internet Protocol addresses were captured to allow tracking of results and to eliminate duplicate entries by participants at the same computer. After completion respondents were not allowed to re-enter the survey. Surveys were collected over a 5–6 month period and responses were collated into an Excel spreadsheet. Analysis of survey data was conducted after eliminating duplicate entries that were performed by a given institution or by different participants at the same computer.
RESULTS
A total of 105 individuals initiated the survey. Of the 89 participants (93 respondents), who provided their institutional affiliation, 56 resided in North America, 21 in Europe, 7 in Asia, 4 in Australia and 1 in New Zealand. Since not all participants responded to each question, data is summarized by providing the number of respondents for each question in parentheses.
Of the 105 respondents, 67.6% (71) performed CFU assays while 32.4% (34) did not. Sixty-seven respondents specified the type of hematopoietic progenitor cell (HPC) product their institution evaluated. Of these 67 respondents, 73%, 67% and 69% said they performed CFU assays on HPC-A, HPC-M, and HPC-C, respectively.
Pre-dilution Total Nucleated Cell Counts
The first series of questions were designed to establish the time point at which laboratories removed an aliquot of cells from a product of HPC stem cells and how pre-dilution cell counts and viabilities were performed. The more common practice was to remove an aliquot of cells before (75% of 67 respondents) rather than after the addition of DMSO to hematopoietic stem cell products (Fig. 1; step 1). Automated cell counts were performed by a majority of laboratories (74%) while 26% still performed manual cell counts (65 respondents) on aliquots of cells removed from final products (i.e. pre-dilution cell count) (Fig. 1; step 2). Forty-eight percent (30/65) performed replicate cell counts to obtain average counts. When replicate counts were performed, 60% said that the replicate count was done using an automated cell counter while 40% of laboratories did a manual cell count (30 respondents). Thus, less than half of the laboratories surveyed performed replicate cell counts and when replicate counts were performed they were more likely to be done using an automated cell counter.
Viability testing
Forty-two laboratories (65%) indicated that they used trypan blue, whereas 23 (26%) of the participants used 7-aminoactinomycin D (7-AAD) (65 total respondents) (Fig. 1;Step 2). Other viability stains used by participants included acridine orange/propidium idodide (AO/PI), AO/ethidium bromide (AO/EB), and erythrosin B. A majority of laboratories (72%) indicated that cells were incubated for a defined period when using either trypan blue or 7-AAD (Table 1). Incubation times ranged from 1–10 min with trypan blue and 5–20 min with 7-AAD (Table 1). No defined incubation times were given for AO/PI, AO/EB and erythrosine B. All viability assays performed with 7-AAD were analyzed using flow cytometry; in contrast 86% of viability assays performed with trypan blue were read microscopically. Three out of 65 laboratories indicated that flow cytometry was used to evaluate cells stained with trypan blue.
Table I.
Practices for Performing Viability Testing
| Stain | *Total | Defined Incubation Period for Cells and Viability Stain |
*Incubation Period (min) | Stain |
*Method used to Read Viability |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Yes | No | N/A | 1 | 2 | 5 | 10 | 15 | 20 | 2–3 | 2–5 | N/A | Microscope | Fow Cytometry |
N/A | |||
| Trypan Blue | 42 | 29 | 12 | 1 | 15 | 2 | 8 | 2 | 0 | 0 | 1 | 1 | Trypan Blue | 36 | 3 | 3 | |
| 7-AAD | 20 | 16 | 3 | 1 | 2 | 6 | 1 | 6 | 1 | 7-AAD | 17 | 3 | |||||
| AO/PI | 1 | 1 | AO/PI | 1 | |||||||||||||
| AO/EB | 1 | 1 | AO/EB | 1 | |||||||||||||
| Erythrosin | 1 | 1 | Erythrosin | 1 | |||||||||||||
Number of Respondents
N/A=Respondents did not answer question
Working Cell Suspension Preparation
Seventy-one percent of respondents (62 individuals) indicated that the aliquot that was used for performing a pre-dilution cell count and viability test was also the one used to prepare a working cell suspension to set-up the CFU assay. To prepare a working cell suspension for inoculating the semi-solid medium for CFU plating, 81% of laboratories (62 respondents) said the aliquot came from the final stem cell product.
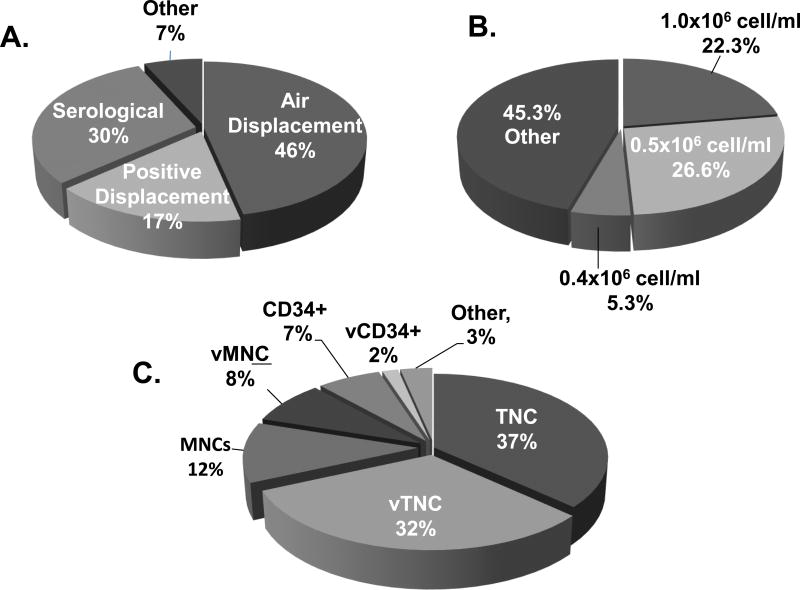
The preparation of the working cell suspension varied considerably from laboratory to laboratory (Fig. 2A). First, a significant variety of pipetting devices was used. Air displacement pipets were the most commonly used devices followed by serological and positive displacement pipettes (Fig. 2A) After making the working cell suspension, 39% of laboratories performed an additional cell count on the final dilution to verify the accuracy of the dilution (51 respondents). Second, the working cell concentrations were another source of variability. Forty-five percent of respondents used working cell concentrations unique to their institution (Fig. 2B). Finally, the calculations to determine the concentration of working cell suspensions were based on different cellular phenotypes that included TNC, viable TNCs, total MNCs, total viable MNCs, CD34+ and viable CD34+ (vCD34+) cells (Fig. 2C).
Figure 2. Techniques for Preparing working cell suspensions for inoculating CFU semi-solid medium (60 respondents).
(A) Percent of respondents who use the indicated type of pipet devices to prepare a working cell suspension. (B) Percent of survey participants that use the indicated cell concentrations to inoculate the CFU assay. (C) Percent of laboratories that use the specified cell immunophenotypes to calculate the concentration of the working cell suspension.
Methylcellulose-based medium Inoculation
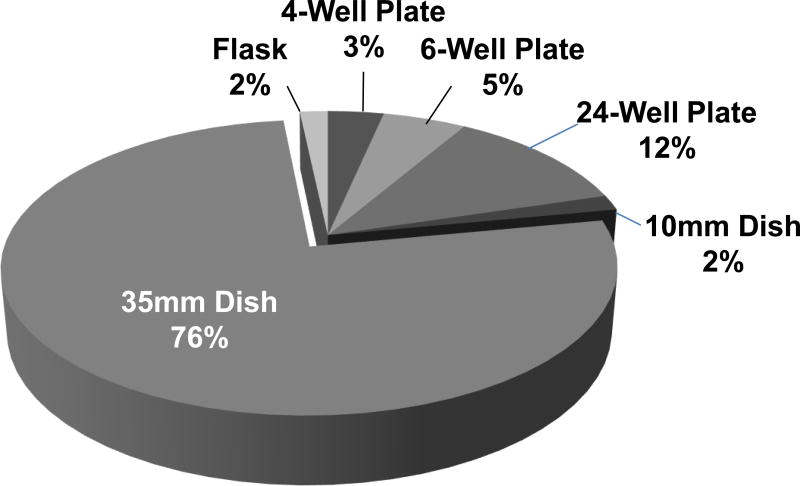
A majority of survey participants (90%) indicated that they purchased methylcellulose based medium for the CFU assay. Five percent said they use medium made in-house and the remaining 5% indicated that they did not use methylcellulose-based medium (60 respondents). Batch preparations (i.e. methycellulose+cytokines+cells) sufficient to perform replicate plating’s were made by 76% of laboratories. The numbers of replicate plates per assay ranged from 1–4. The most popular size of the culture plate was 35mm followed by 24-well culture plates (Fig. 3). Other types of culture plates reported included: 4-well and 6-well plates, 10 mm dishes and flasks (Fig. 3).
Figure 3. Types Culture Vessels used for CFU Plating (59 respondents).
To transfer methylcellulose containing cytokines and cells to culture plates/dishes, 61% of laboratories use a syringe and 37% a pipet (59 respondents). For laboratories using syringes, 81% used a blunt-ended needle to transfer medium to a culture plate (36 respondents). For the 21 laboratories that use a pipet, 38.1%, 9.5%, and 47.6% employ an air-displacement, positive displacement or serological pipet, respectively. One of 59 laboratories indicated that they use a 16 gauge blunt end cannula. One laboratory did not specify the type of pipet.
The final total number of cells plated per dish/well varied from site to site. For laboratories using 35 mm dishes (45 of 57 respondents), the range of cells plated was 10,000–200,000 for HPC-A, 25,000–100,000 for HPC-M and 5,000–100,000 for HPC-CB. One laboratory indicated that the number of cells plated was dependent on the percentage of viable CD34+ cells. For laboratories using 24 well plates, three of four laboratories indicated they plated 100,000 and 50,000 cells for both HPC-A, HPC-M. Laboratories (59.6%) tended to plate CFU based on viable cell counts (57 respondents). After plating cells, 26.3% of individuals indicated that they performed a cell count on the remaining working cell suspension to verify an accurate dilution (57 respondents).
Culture plates were placed in incubators at 37°C, in 5% CO2 at 95% humidity by the vast majority of respondents. Ninety-eight percent indicated that cultures plates were maintained at a temperature of 37°C with 1 laboratory indicating culture plates were maintained at 22 °C (57 respondents). Ninety-three percent said they used 5% CO2/air mixture and the remaining 7% of respondents indicated that they used 7% CO2/air 6% CO2+10%O2, 5.5% CO2 or 5%CO2+5O2. Most laboratories placed culture dishes within larger dishes along with a water dish to help maintain humidity. Eighty-eight percent of labs indicated that they used 95% humidity while one lab indicated a range of 50–100% humidity; another lab said that they were changing to 85% humidity; and two labs did not know the percent humidity of the incubator.
CFU readout
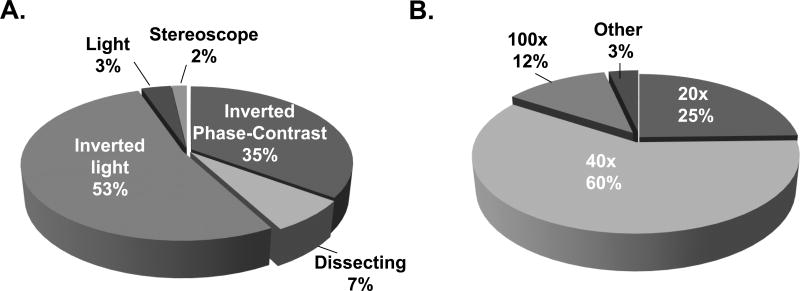
Culture periods for the CFU assay were predominately 14–16 days independent of whether the assay was used to measure colony formation in HPC-A, HPC-M or HPC-C Products Table 2. To assist in the enumeration and differentiation of colonies, 84% of respondents (57) use a microscopic grid. The most popular type of microscope and the magnification used to read colonies was an inverted light microscope and optical magnification of 400× (10× eyepiece and 40× objective lens (Fig. 4). Colonies were most commonly scored and reported for CFU-GM (Table 3). To calculate the total number of colonies present in a product, some laboratories (73%) used the post-dilution cell count and others (27%) used the pre-dilution cell count (15 respondents).
Table 2.
Incubation Periods Reported for CFU Assay
| Cells per Dish/Well | <7 Days |
7–9 Days |
10–13 Days |
14–16 Days |
Other | N/A | Response Count |
|---|---|---|---|---|---|---|---|
| HPC, Apheresis | 0.0% (0) | 1.8% (1) | 16.4% (9) | 63.6% (35) | 0.0% (0) | 18.2% (10) | 55 |
| HPC, Marrow | 0.0% (0) | 3.8% (2) | 15.4% (8) | 53.8% (28) | 0.0% (0) | 26.9% (14) | 52 |
| HPC, Cord Blood | 0.0% (0) | 3.8% (2) | 11.3% (6) | 58.5% (31) | 0.0% (0) | 26.4% (14) | 53 |
Figure 4. Microscopic readout for CFU.
(A) Magnification used to read colonies; (B) Type of microscope used to read colonies.
Table 3.
Colony Phenotypes by Sites
| Colony Type | Yes | No | Response Count |
|---|---|---|---|
| BFU-E | 78.3% (36) | 21.7% (10) | 46 |
| CFU-GEMM | 64.3% (27) | 35.7% (15) | 42 |
| CFU-GM | 94.2% (49) | 5.8% (3) | 52 |
| Total Colony | 61.9% (26) | 38.1% (16) | 42 |
| Other | *3 |
One site indicated they report CFU-E; a second BFU-E & CFU-GM; a third site that they do not report data to an outside institution.
Validation and Proficiency Testing
Validation studies to test the linearity of tissue culture plates for determining at which cell dose a CFU culture plate/dish reached a saturation plateau were performed by 58% of respondents (54 respondents). Only 37% had established criteria for determining whether a culture plate/dish was overgrown and the remaining 63% did not (57 respondents). Overlapping colonies was given by 61% of 21 respondents as the primary criteria for determining whether a CFU plate was overgrown. Other criteria given to define if a culture plate was overgrown included: 1) when colonies exceeded a maximum number per plate (e.g. >120, >150, 150–200, and >300 colonies per plate); 2) the number of colonies were too numerous to count; and 3) when phenol red in the methylcellulose turned yellow indicating that a change in the pH of the medium had occurred. Interestingly, if a CFU plate was determined to be overgrown, only 12.3% of labs repeated the assay by adding fewer cells per plate, 49.1% said that they did not repeat the test and the remaining 38.6% indicated that it was essentially not applicable to them (57 respondents).
A majority (98%) of sites indicated that they participated in a proficiency testing program (57 respondents). The most commonly used proficiency testing program was from StemCell Technologies (59.6%) followed by the College of American Pathologists (36.8%). One laboratory indicated that their proficiency testing program involved the use of a third independent party to verify scoring. While another laboratory said that they planned to participate in a program in the future. The average number of individuals trained to perform a CFU assay per laboratory was 4.1 (range 1–10). Third party formal training was attended by staff from 47.4% of reporting laboratories (57 respondents).
Transplant outcome
Finally, sites were asked whether they observed a clinical correlation between engraftment and the results they obtained from the CFU assay. Of 57 respondents who answered this question, 35% indicated that they saw a significant negative correlation with the length of peripheral blood cytopenias and their institutional CFU assay data. Nineteen percent indicated that they did not see any significant correlation and the remaining 46% of sites indicated that they were unable to determine a correlation between engraftment and the CFU assay.
DISCUSSION
Results from this study highlight that current inter-laboratory practices for setting-up the CFU assay are highly variable at multiple steps of the procedure. As a consequence, there are several technical aspects of the CFU assay that could be standardized. Based on best laboratory practices and/or previously published data, a list of recommendations for variables to be considered for standardization is presented in table 4. For example, a majority of laboratories (74%) performed automated cell counts, but a substantial number of laboratories (26%) indicated that they perform manual counts. Evidence is available that manual counts are less well controlled and are associated with more sources of error (i.e. preparing a dilution to charge a hemacytometer, manual counting, calculation errors) that lead to lower accuracy and precision than automated counts (10). This is confirmed by proficiency testing studies that show automated methods have better inter-laboratory reproducibility than manual counts (1, 11). Subsequently, one step to improve the standardization of the CFU assay would be for all laboratories to adopt well-controlled automated cell counts and eliminate the use of manual counts to perform cell counts on pre-dilution samples.
Table 4.
Prospects for Standardization
|
|
|
|
|
Another step of the CFU assay that could be standardized or possibly eliminated is that of viability testing. Answers regarding current laboratory practices for performing viability counts indicate that not only are different stains used among laboratories for conducting a viability assay, but even when the same viability stain is used by different laboratories the procedure is not the same among centers. Of the laboratories using the same dye, they reported the use of different incubation periods for staining cells and different techniques for assessing viable and non-viable cells (microscopically vs. flow cytometry). Interestingly, despite reports that trypan blue is inferior to other vital stains (12–14), the results of this study show that trypan blue is the most commonly used stain (65% of respondents) while alternative stains such as AO/PI, AO/EB, or erythrosin B are used by only a small number of sites. Given the variability associated with viability testing (i.e. trypan blue, AO/EB, AO/PI and 7-AAD) (15), we suspect that practices to integrate cell viability with cell counts contributes to CFU assay inter-laboratory testing variability. Based on the fact that the CFU assay inherently assesses cell viability (i.e. dead cells do not form colonies), one can argue that performing a viability assessment is redundant and unnecessary and only introduces error into the final calculation for determining CFU counts.
Different strategies are used to calculate the concentration of the working cell suspension for inoculating the semi-solid medium for plating CFU. For some participants, TNC, total MNC, and CD34+ cell counts are used, while other participants couple these parameters with a cell viability determination to prepare a working cell suspension that is used to plate viable TNCs, total viable MNCs and viable CD34+ (vCD34+) cells. Current practices of using different cellular phenotypes among laboratories to plate cells make it difficult to compare and interpret inter-laboratory results. Likewise, practices by some sites to not use the same aliquot to prepare the working cell suspension that is used to determine cell counts is a laboratory practice that may contribute to inter-laboratory variability. The importance of accurate cell counts and a standardized strategy to prepare a working cell suspension to set-up the CFU assay cannot be underestimated, as it will ultimately be used to determine the total number of colony forming progenitors in a product. Given the importance of performing accurate cell counts and the preparation of a working cell suspension, it would be prudent to verify the cell concentration of the working cell suspension with an automated cell counter.
Answers to questions pertaining to the inoculation of culture medium (i.e. transfer of cells into semi-solid medium), how semi-solid culture medium containing cells is transferred to culture plates and how culture plates are incubated and enumerated for colonies are also informative. Answers related to these steps of the CFU protocol provide insight into other areas of the assay that may represent additional sources of assay variability and potential target areas for standardization (Table 4). In the case of transferring semi-solid medium inoculated with cells, a majority (61%) of laboratories use a syringe while there are some that use air-displacement pipets (8 respondents) and serological pipets (10 respondents) to transfer methylcellulose medium (total respondents 59). This is in light of available information demonstrating that the transfer of a viscous medium (i.e. methylcellulose) is best achieved using a positive displacement pipet (16) and a recommendation that transfer of methylcellulose into culture plates should occur with a syringe (17). A majority of laboratories indicated that they purchase culture media containing cytokines; other laboratories said that they use an in-house formulation. Acknowledging that different cytokine combinations and cytokine concentrations can affect colony growth and differentiation (18), it is important that all sites performing CFU assays to assess a stem cell product for clinical application use the same concentration and combination of cytokines. Enumeration of colonies using different types of microscopes and the fact that not all sites report colony numbers in an agreed upon fashion also make it difficult to compare results. In particular, some sites report CFU-GM while other sites report total CFU. Finally, with 42% of sites not having established the linear range of their culture plates/dishes, this raises concerns about the accuracy of results being reported and underscores the need for the establishment of assay standards.
The results of this survey represent a cross-sectional analysis of the current practices of clinical laboratories in the United States, Europe and Asia and provide a number of areas of action for intervention. Having identified procedural differences among laboratories raises the question as to whether it is reasonable for an ad hoc international committee of experts in the field to develop a comprehensive set of international guidelines for performing a CFU assay. Especially given that reasonably good intra-laboratory CFU reproducibility allows some investigators to report that there is a correlation between numbers of CFU generating progenitors present in stem cell products and short-term hematopoietic reconstitution (4). In the end, for the CFU assay to meet the requirements of a true potency assay for accuracy, precision, specificity, linearity, and robustness as set forth by regulatory agencies (FDA and EU), a standardization of procedural steps and integration of standards and controls that are currently not available to run alongside test samples are necessary.
Acknowledgments
Support: This work was supported in part by the BEST collaborative group and the Puget Sound Blood Center.
We would like to thank members of the Cell Therapy Team of the Biomedical Excellence for Safer Transfusion (BEST) Collaborative Group.
ABBREVIATIONS
- CFU
colony-forming-unit
- TNC
total nucleated cell count
- MNCs
mononuclear cells
- HPC
hematopoietic progenitor cell
- FDA
US Food and Drug Administration
- BFU-E
burst-forming unit-erythroid containing
- CFU-E
colony-forming unit erythroid containing
- CFU-GM
CFUs containing granulocytes and macrophages
- CFU-GEMM
colony-forming-unit granulocytes, erythroid, macrophages, and megakaryocytes
- BEST
Biomedical Excellence for Safer Transfusion
- ISCT
International Society for Cell Therapy
- EBMT
European Bone Marrow Transplant
- HPC-A
hematopoietic progenitor cell-apheresis
- HPC-M
hematopoietic progenitor cell-marrow
- HPC-C
hematopoietic progenitor cell-umbilical cord blood
- 7-AAD
7-aminoactinomycin D
- AO/PI
acridine orange/propidium idodide
- AO/EB
AO/ethidium bromide
- vCD34+
viable CD34+
Footnotes
All authors have no conflict of interest.
References
- 1.Spellman S, Hurley CK, Brady C, Phillips-Johnson L, Chow R, Laughlin M, et al. Guidelines for the development and validation of new potency assays for the evaluation of umbilical cord blood. Cytotherapy. 2011;13(7):848–55. doi: 10.3109/14653249.2011.571249. Epub 2011/04/01. [DOI] [PubMed] [Google Scholar]
- 2.Serke S, Arseniev L, Watts M, Fritsch G, Ingles-Esteve J, Johnsen HE, et al. Imprecision of counting CFU-GM colonies and CD34-expressing cells. Bone marrow transplantation. 1997;20(1):57–61. doi: 10.1038/sj.bmt.1700830. Epub 1997/07/01. [DOI] [PubMed] [Google Scholar]
- 3.Center for Biologics Evaluation and Research R, MD. FDA Guidance for Industry. US Department of Health and Human Services, Food and Drug Adminstration; [January 2011]. Potency Tests for Cellular and Gene Therapy Products. http;//wwwfdagov/BiologicsBloodVaccines/GuidanceComplianceRegulatoryInformation/Guidances/defaulthtm. [Google Scholar]
- 4.Page KM, Zhang L, Mendizabal A, Wease S, Carter S, Gentry T, et al. Total colony-forming units are a strong, independent predictor of neutrophil and platelet engraftment after unrelated umbilical cord blood transplantation: a single-center analysis of 435 cord blood transplants. Biology of blood and marrow transplantation : journal of the American Society for Blood and Marrow Transplantation. 2011;17(9):1362–74. doi: 10.1016/j.bbmt.2011.01.011. Epub 2011/02/01. [DOI] [PubMed] [Google Scholar]
- 5.Prasad VK, Mendizabal A, Parikh SH, Szabolcs P, Driscoll TA, Page K, et al. Unrelated donor umbilical cord blood transplantation for inherited metabolic disorders in 159 pediatric patients from a single center: influence of cellular composition of the graft on transplantation outcomes. Blood. 2008;112(7):2979–89. doi: 10.1182/blood-2008-03-140830. Epub 2008/07/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scaradavou A, Smith KM, Hawke R, Schaible A, Abboud M, Kernan NA, et al. Cord blood units with low CD34+ cell viability have a low probability of engraftment after double unit transplantation. Biology of blood and marrow transplantation : journal of the American Society for Blood and Marrow Transplantation. 2010;16(4):500–8. doi: 10.1016/j.bbmt.2009.11.013. Epub 2009/11/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jagannath S, Vesole DH, Glenn L, Crowley J, Barlogie B. Low-risk intensive therapy for multiple myeloma with combined autologous bone marrow and blood stem cell support. Blood. 1992;80(7):1666–72. Epub 1992/10/01. [PubMed] [Google Scholar]
- 8.Tricot G, Jagannath S, Vesole D, Nelson J, Tindle S, Miller L, et al. Peripheral blood stem cell transplants for multiple myeloma: identification of favorable variables for rapid engraftment in 225 patients. Blood. 1995;85(2):588–96. Epub 1995/01/15. [PubMed] [Google Scholar]
- 9.Cancelas JA, Querol S, Canals C, Picon M, Azqueta C, Sola C, et al. Peripheral blood CD34+ cell immunomagnetic selection in breast cancer patients: effect on hematopoietic progenitor content and hematologic recovery after high-dose chemotherapy and autotransplantation. Transfusion. 1998;38(11–12):1063–70. doi: 10.1046/j.1537-2995.1998.38111299056318.x. Epub 1998/12/05. [DOI] [PubMed] [Google Scholar]
- 10.Read EJ, Carter CS. Enumeration of cells in bone marrow and peripheral blood stem cell collections: technical issues and prospects for standardization. Journal of hematotherapy. 1992;1(2):175–82. doi: 10.1089/scd.1.1992.1.175. Epub 1992/01/01. [DOI] [PubMed] [Google Scholar]
- 11.Moroff G, Eichler H, Brand A, Kekomaki R, Kurtz J, Letowska M, et al. Multiple-laboratory comparison of in vitro assays utilized to characterize hematopoietic cells in cord blood. Transfusion. 2006;46(4):507–15. doi: 10.1111/j.1537-2995.2006.00758.x. Epub 2006/04/06. [DOI] [PubMed] [Google Scholar]
- 12.Altman SA, Randers L, Rao G. Comparison of trypan blue dye exclusion and fluorometric assays for mammalian cell viability determinations. Biotechnology progress. 1993;9(6):671–4. doi: 10.1021/bp00024a017. Epub 1993/11/01. [DOI] [PubMed] [Google Scholar]
- 13.Krause AW, Carley WW, Webb WW. Fluorescent erythrosin B is preferable to trypan blue as a vital exclusion dye for mammalian cells in monolayer culture. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 1984;32(10):1084–90. doi: 10.1177/32.10.6090533. Epub 1984/10/01. [DOI] [PubMed] [Google Scholar]
- 14.Mascotti K, McCullough J, Burger SR. HPC viability measurement: trypan blue versus acridine orange and propidium iodide. Transfusion. 2000;40(6):693–6. doi: 10.1046/j.1537-2995.2000.40060693.x. Epub 2000/06/24. [DOI] [PubMed] [Google Scholar]
- 15.Brand A, Eichler H, Szczepiorkowski ZM, Hess JR, Kekomaki R, McKenna DH, et al. Viability does not necessarily reflect the hematopoietic progenitor cell potency of a cord blood unit: results of an interlaboratory exercise. Transfusion. 2008;48(3):546–9. doi: 10.1111/j.1537-2995.2007.01568.x. Epub 2007/12/11. [DOI] [PubMed] [Google Scholar]
- 16.Gast U, Hartmann I. Dispensing of highly viscous liquids. Eppendorf-Application Technical Report. 2009:1–9. [Google Scholar]
- 17.Technical Manual Stem Cell Technologies version 3.1.0: Human colony-forming cell (CFC) assays using methocult. 2009 [Google Scholar]
- 18.Dexter TM. Haemopoietic growth factors. British medical bulletin. 1989;45(2):337–49. doi: 10.1093/oxfordjournals.bmb.a072326. Epub 1989/04/01. [DOI] [PubMed] [Google Scholar]