Abstract
Hepatocellular carcinoma (HCC) has one of the highest mortality rates among numerous types of cancer. It has been demonstrated that in hepatitis B (HBV)-associated HCC, the expression of chimeric fusion transcript HBx-long interspersed nuclear element-1 (LINE-1) initiated by HBV integration is correlated with hepatocarcinogenesis and poor patient survival rates. Furthermore, increased rates of LINE-1 hypomethylation have been detected in HCC tissues compared with adjacent tissues. This suggests that individual LINE-1 RNA (L1 RNA) serves an important role in the processes of hepatocarcinogenesis. The present study assessed the epigenic interaction between L1 RNA and polypyrimidine tract-binding protein-associated splicing factor (PSF) in the A549 human alveolar epithelial and 16HBE human bronchial epithelial cell lines. In addition, changes in the transcriptional regulatory activity of PSF on its target gene, proto-oncogene G antigen 6 (GAGE6), were investigated following overexpression of L1 RNA, as well as its impact on cell-proliferative capacity, carried out by plotting cell growth curves and 5-ethynyl-2′-deoxyuridine assay. It was observed that L1 RNA specifically bound to the RNA binding domain of PSF and released the GAGE6 promoter region from the DNA-binding domain of PSF. This increased the transcription of GAGE6 and led to the promotion of cell proliferation as well as colony formation. Furthermore, at least two binding sites specific for PSF were identified on L1 RNA. In conclusion, the transcriptional regulatory activity of L1 RNA may partially result in cell transformation, and endogenous L1 RNA may function as an important regulatory factor in the process of tumorigenesis.
Keywords: long interspersed nuclear element 1, polypyrimidine tract-binding protein-associated splicing factor, proliferation, tumor formation, hepatocellular carcinoma, GAGE6
Introduction
Hepatocellular carcinoma (HCC), which is one of the most common and fatal tumor types in the world, has a poor prognosis and is responsible for numerous cases of cancer-associated mortality (1). Hepatitis B virus (HBV) and HCV are generally considered to be risk factors for HCC and jointly account for its most common etiology (2). Furthermore, it has been suggested that HCC arising from HBV or HCV infection has an increased mortality rate, although the exact underlying mechanisms of virus-mediated hepatocarcinogenesis have remained elusive (3,4).
Polypyrimidine tract-binding protein-associated splicing factor (PSF) is an abundant ubiquitous nuclear protein that serves critical regulatory roles in vertebrates (5,6). One of its primary roles is as a component of spliceosomes and it is mediated by its RNA-binding domains (RBD) (7). In addition, PSF has been identified as a transcriptional regulator of the gene encoding for P450-linked side chain cleaving enzyme, which initiates the steroidogenic pathway, and binds to its regulatory region through its DNA-binding domain (DBD) (8,9). The RBD and DBD function independently and their novel mechanisms for reversible gene transcription mediated by PSF protein and noncoding RNA have been investigated. It has been demonstrated that their transcription is activated by the release of PSF protein from the repressed proto-oncogene G antigen 6 (GAGE6) by five PSF-binding noncoding RNA fragments (10). This indicates that the regulatory activity of noncoding RNA in tumorigenesis occurs through its binding to the RBD of PSF and the consequent release of the transcriptionally repressed proto-oncogene from its DBD.
Long interspersed nuclear element 1 (LINE-1; L1) is a repetitive element, which constitutes 17–25% of the human genome, and comprises a 5′-untranslated region (UTR), two open-reading frames and a 3′-UTR (11). The genome contains numerous copies of L1s, which have often been referred to as junk DNA sequences, as their function was unknown. However, it has been revealed that L1s serve important transcriptional regulatory functions, which is tuned by changes in their methylation status (12). Hypomethylation in the promoter region of L1 leads to transcriptional activation of the L1 element, causing transposition of the retro-element and chromosomal alteration (13). This indicates that physiological processes are regulated by the amount of L1 RNA expression; however, the exact mechanism has remained elusive.
It has been demonstrated that hypomethylation of L1 is associated with HBV infection, a larger tumor size and a more advanced disease stage (14). Pyrosequencing analysis of the methylation level of L1 revealed that increased hypomethylation of L1 is associated with an increased risk of developing HCC (15). Therefore, the present study aimed to assess the interaction between L1 RNA and PSF protein. Lau et al (16) have previously indicated that transcripts of the genome integration sites of viral-human fusion genes which function as hybrid RNAs with tumor-promoting properties. However, it has remained elusive whether L1 RNA affects the processes of tumorigenesis in HCC without HBV infection or of carcinogenesis in general, including lung carcinogenesis.
The aim of the present study was to investigate whether, apart from the chimeric HBx-LINE-1 transcript, interactions between L1 RNA and PSF protein and the changes occurring in the regulatory activities of PSF have a role in lung carcinogenesis. The binding of L1 RNA to the RBD of PSF, the release of the DNA promoter region of GAGE6, and the effects on the proliferation of the A549 lung cancer and the 16HBE normal bronchial cell lines were assessed in order to identify the potential role of L1 RNA in these processes.
Materials and methods
Cell lines
The A549 human non-small cell lung carcinoma cell line, the 16HBE human bronchial epithelial cell line and the 293 human embryonic kidney (HEK293) cell line were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and all cells were obtained from Dr Xu Song (Department of Center for Functional Genomics and Bioinformatics, College of Life Science, Sichuan University, Chengdu, China). All cells were supplemented with 10% (v/v) fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin in an atmosphere containing 5% CO2 at 37°C.
Construction of plasmid encoding L1 RNA
The L1 complementary (c) DNA fragment was amplified by polymerase chain reaction (PCR) using the genome of A549 cells as a template and primers were designed to generate HindIII and XbaI restriction sites at the 5′ and 3′ ends of the amplified fragments, respectively. The primer sequences used were as follows: L1 forward, 5′-CCCAAGCTTGTGTTGTTGAGGATGTGAAG-3′ and reverse, 5′-TGCTCTAGATCCTGTGATTGATTTATTTTACTTA-3′ [Beijing Genomics Institute (BGI), Beijing, China]. The PCR mixture contained forward and reverse primers (0.2 µM), DNA template (1 ng/µl), dNTP, Taq polymerase and PCR buffer (primeSTAR; Takara Bio., Inc., Otsu, Japan). The PCR procedure consisted of 34 cycles of denaturation at 95°C for 10 sec, annealing at 55°C for 10 sec, and extension at 72°C for 30 sec, with initial denaturation of template DNA at 95°C for 5 min. The amplified L1 cDNA was subsequently digested with HindIII and XbaI (Takara Bio, Inc.) and cloned into multiple cloning sites of pcDNA3.1 (+) (Thermo Fisher Scientific, Inc.).
Stable transfection of cell lines
The A549-L1 and 16HBE-L1 cell lines were constructed by stably transfecting plasmid pcDNA3.1-L1 encoding the 380-nt L1 RNA fragment into A549 or 16HBE cells. The A549-vector or 16HBE-vector cell lines were constructed by transfecting the empty vector, pcDNA3.1, into A549 or 16HBE cells. All transfections were performed using the transfection reagent Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) in accordance with the manufacturer's instructions. After 2 days of incubation, stably transfected A549 or 16HBE cells were selected with 450 or 900 g/ml geneticin (G418; Sigma-Aldrich; Merck Millipore, Darmstadt, Germany), respectively. Following selection, cells were maintained with half-doses of G418. The expression levels of target genes were analyzed using reverse transcription-quantitative PCR (RT-qPCR).
RNA extraction and RT-qPCR
TRIzol reagent (Thermo Fisher Scientific, Inc.) was used to isolate total RNA from cells according to the manufacturer's instructions. A sample of 500 ng RNA was reverse transcribed into cDNA in a 10-µl reaction volume using M-MLV reverse transcriptase reagent (Vazyme Biotech Co., Ltd., Nanjing, China) and qPCR was performed using 0.25 µl cDNA and SsoFast EvaGreen™ Supermix (Bio-Rad Laboratories, Inc., Hercules, CA, USA) on a Bio-Rad CFX96 Real-Time PCR detection system (Bio-Rad Laboratories, Inc.). Data were analyzed using Bio-Rad CFX 2.1 Manager software (Bio-Rad Laboratories, Inc.) and GAPDH gene expression was used as an internal reference. Data was normalized using the 2−ΔΔCq method (17). The forward and reverse primers used for PCR were as follows: GAGE6 forward, 5′-GCCTCCTGAAGTGATTGGGCCTA-3′ and reverse, 5′-CAGGCGTTTTCACCTCCTCTGGA-3′; PSF forward, 5′-ATGTCTCGGGATCGGTTCCGGA-3′ and reverse, 5′-CCAACAAACAACCGACATCGCTG-3′; L1 forward, 5′-TGAAGTAAAGAAAACCCTTGCCT-3′ and reverse, 5′-TCTTGGTCATTGTGAATAGTGCT-3′; and GAPDH forward, 5′-ACCACAGTCCATGCCATCAC-3′ and reverse, 5′-TCCACCACCCTGTTGCTGTA-3′. All primers were supplied by BGI. The thermocycling conditions were 1 min at 95°C, followed by 40 cycles of 95°C for 10 sec, 60°C for 30 sec and 72°C for 20 sec.
Purification of His-tagged proteins
Construction of a prokaryotic expression vector carrying the intact structural gene of human PSF protein was amplified by PCR, inserted into the plasmid pET-28a and expressed in Escherichia coli (E. coli) BL21 cells (Vazyme Biotech Co., Ltd.). The DBD, RBD-1 and RBD-2 of the PSF protein gene were inserted into the plasmid pET-42a and expressed in E. coli BL21 cells (Vazyme Biotech Co., Ltd.). Cells were cultured in LB medium at 37°C with shaking at 200 rpm until the inoculum optical density at 600 nm reached 0.6, and then shaken overnight at 25°C and 100 rpm in the presence of 0.1 mM isopropyl β-D-1-thiogalactopyranoside. Proteins were purified using Ni-nitrilotriacetic acid agarose (Qiagen, Inc., Valencia, CA, USA).
Electrophoretic mobility shift assays (EMSA)
The RNA probe was transcribed in vitro using T7 RNA polymerase (Promega Corp., Madison, WI, USA) from the PCR product containing the T7 RNA polymerase promoter. The 61-bp DNA probe was the PSF binding motif in the GAGE6 promoter (18) and was synthesized with the following sequence: 5′-GCCTTCTGCAAAGAAGTCTTGCGCATCTTTTGTGAAGTTTATTTCTAGCTTTTTGATGCTG-3′, and was synthesized by BGI. The RNA or DNA probe was biotin-labeled using an RNA 3′ End Biotinylation kit (Pierce; Thermo Fisher Scientific, Inc.) or a Biotin 3′ End DNA labeling kit (Pierce; Thermo Fisher Scientific, Inc.), respectively. A total of 5 ng biotin-labeled RNA or DNA fragments were mixed with 100–500 ng recombinant PSF protein, DBD of PSF, RBD-1 of PSF, RBD-2 of PSF or the total protein of E. coli lysates. EMSA was performed using a LightShift Chemiluminescent EMSA kit (Thermo Fisher Scientific, Inc.) following the manufacturer's instructions.
Cell proliferation assay
A total of 5×103 cells per well were cultured in a 12-well plate at 37°C as attached monolayers in DMEM with 10% FBS. Each day, the cell numbers in three wells were counted with a hemocytometer following detachment with trypsin. Each test had two replicates (n=3).
For the 5-ethynyl-2′-deoxyuridine (EdU) assay, ~4×103 cells per well were grown in a 96-well dish and processed with the EdU labeling kit from RiboBio Co., Ltd. (Guangzhou, China).
Soft-agar colony assay
A total of 1×103 cells were suspended in 1 ml 0.3% BD Difco™ Agar (BD Biosciences, Franklin Lakes, NJ, USA) in DMEM with 10% FBS and seeded in each well of 6-well plates containing 1 ml of a solidified layer of 0.6% agar in the same medium. Each test had two replicates (n=3). Following 21 days of incubation, 100 µl 1.5 mM nitro blue tetrazolium in phosphate-buffered saline (PBS) was added to the dishes and after 4 h, the colonies were counted and images were captured with an Epson 4990 scanner.
Chromatin immunoprecipitation (ChIP) and RNA immunoprecipitation (RIP) assays
ChIP assays were performed using a ChIP assay kit (Upstate Biotechnology, Inc., Lake Placid, NY, USA) according to the manufacturer's protocol. Cells were cultured in 100-mm plates to 70–80% confluence, reversibly cross-linked with formaldehyde and immunoprecipitated with an anti-PSF antibody (P2860; Sigma-Aldrich; Merck Millipore). The GAGE6 promoter DNA fragments in PSF-DNA complexes were analyzed by qPCR using the following primers: GAGE6 forward, 5′-GCCTTCTGCAAAGAAGTCTTGCGC-3′ and reverse, 5′-ATGCGAATTCGAGGCTGAGGCAGACAAT-3′. Dihydrofolate reductase (DHFR) 5′ UTR DNA was used as a negative control and the primers used were as follows: Forward, 5′-CTGATGTCCAGGAGGAGAAAGG-3′ and reverse, 5′-AGCCCGACAATGTCAAGGACTG-3′. All primers were obtained from BGI. RIP assays were performed using an RIP-Assay kit (MBL International Co., Woburn, MA, USA) following the manufacturer's instructions. The pcDNA3.1-PSF (encoding human PSF protein) and pcDNA3.1-L1 plasmids mixed with Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) were transiently transformed into HEK293 cells. After 4 h of transfection, the cells were washed once with PBS and fresh medium was added. The cells were cultured for 48 h prior to testing. L1 RNA in PSF-RNA complexes was analyzed by extracting the RNA with TRIzol reagent and assaying by RT-qPCR (as described above). Rabbit IgG (provided in the kit) was compared with anti-PSF antibody to determine the fold enrichment of the target gene. DHFR 5′ UTR DNA was compared with GAGE6 DNA, and GAPDH mRNA with L1 RNA to assess the specific binding of PSF with GAGE6 DNA or L1 RNA.
Statistical analysis
All results are presented as the mean ± standard error of the mean. Student's t-test was used for comparisons between two groups. P<0.05 was considered to indicate a statistically significant difference. Data were analyzed using GraphPad Prism 5 software (GraphPad Software, Inc., San Diego, CA, USA).
Results
Binding ability of L1 RNA and PSF protein in vitro
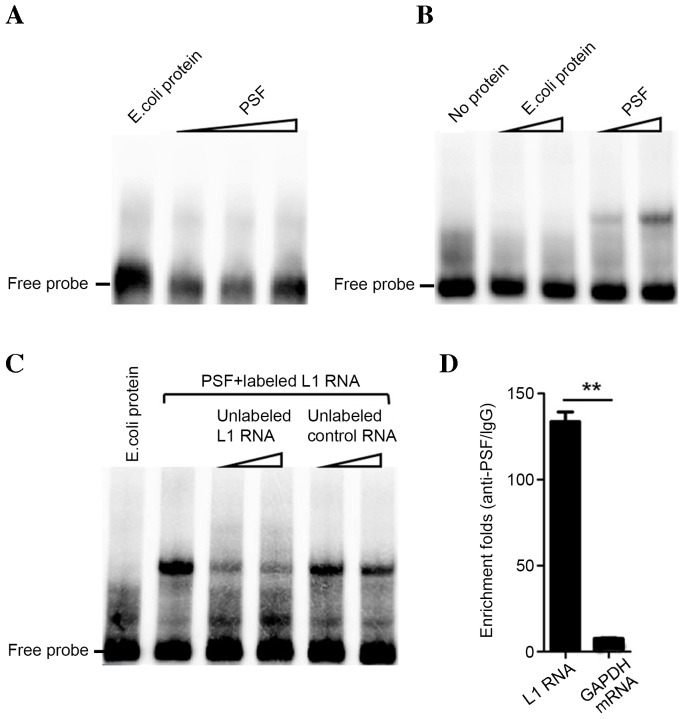
It has been suggested that HBV may cause HCC by integrating into L1 retrotransposons and lead to L1 transcription (16). Therefore, it was hypothesized that transcripts of retrotransposons L1 may also influence lung carcinogenesis. The present study focused on the role L1 RNA may serve in lung carcinogenesis without HBV infection, which is different from HBx-LINE-1 causing hepatocarcinogenesis (16). It has previously been demonstrated that L1PA16 RNA binds to PSF protein, which is part of the LINE retrotransposons family (10). Therefore, L1 RNA, although it shares no sequence similarity with L1PA16 RNA, may bind to PSF. The results of in vitro testing by EMSA indicated that L1 RNA binds to PSF protein, while RNA control pcDNA3.1 RNA exhibits no detectable binding (Fig. 1A and B). Addition of unlabeled L1 RNA probe reduced the binding band of L1/PSF, indicating its binding capacity and specificity to PSF (Fig. 1C). An in vitro RIP assay revealed that L1 RNA and PSF have a strong binding interaction. Compared with the enrichment fold of GAPDH mRNA (negative control), L1 RNA was significantly enriched by immunoprecipitation with anti-PSF antibody (P<0.01; Fig. 1D). Therefore, the results indicated that L1 RNA strongly and specifically binds to PSF in vitro.
Figure 1.
Binding of L1 RNA to PSF protein in vitro. Binding of (A) biotin-labeled negative RNA probe or (B) biotin-labeled L1 RNA to PSF protein or E. coli total protein measured by electrophoretic mobility shift assay. (C) Increasing amounts of competitive unlabeled L1 RNA or negative control RNA were added followed by incubation for 20 min. (D) RIP assay was performed to test the binding of L1 RNA and PSF protein by transient transfection of L1 RNA and PSF-coded plasmid for 48 h. The enrichment folds of immunoprecipitated L1 RNA were identified by comparing the products of the RIP assay using anti-PSF antibodies with those using normal rabbit IgG. **P<0.01. PSF, polypyrimidine tract-binding protein-associated splicing factor; IgG, immunoglobulin G; L1, long interspersed nuclear element 1; RIP, RNA immunoprecipitation; E. coli, Escherichia coli.
Identification of the interacting domain of PSF protein and L1 RNA
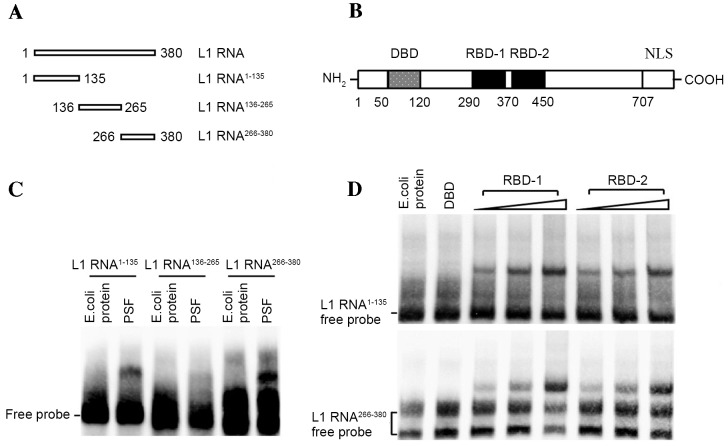
To biochemically analyze the interaction between PSF and L1 RNA, several independent RNA-protein interaction assays were performed. First, to define a region within L1 RNA sufficient for PSF binding, biotin-labeled RNAs corresponding to different regions of the L1 RNA were incubated with purified PSF protein and the reactions were analyzed by EMSA (Fig. 2A and B). Fragments 1 (L1 RNA1–135) and 3 (L1 RNA266–380) efficiently bound the intact PSF protein (Fig. 2C). By contrast, no interacting bands between fragment 2 (L1 RNA136–265) and PSF were observed. Two RBDs, referred to as RBD-1 and RBD-2, have been detected in PSF, which is responsible for binding non-coding RNA (18). The release of the target DNA fragment from the DBD of PSF by RNA binding is controlled by RBD-1 but not RBD-2. This means that it is critical to identify which RBD is bound by L1 RNA. For this, biotin-labeled L1 RNA fragments were incubated with increasing amounts of the respective RBDs (amino acids 290–370 of RBD-1 and amino acids 370–455 of RBD-2) of PSF for subsequent EMSAs. The results demonstrated that, with increasing amounts of RBD-1 or RBD-2 truncate, stronger bands of protein-RNA complex were observed, indicating the effective interactions (Fig. 2D).
Figure 2.
Identification of interacting domain of PSF protein and L1 RNA. (A) Sub-fragments of L1 RNA. (B) PSF structural domains. (C) Binding of PSF protein to three fragments of L1 RNA separately by EMSA. Total E. coli protein was used as the protein negative control. (D) Identification of the binding of the DBD, RBD-1 and RBD-2 of PSF to L1 RNA1–135 (upper panel) and L1 RNA266–380 (lower panel) by EMSA. Total E. coli protein was used as the control protein. PSF, polypyrimidine tract-binding protein-associated splicing factor; L1, long interspersed nuclear element 1; E. coli, Escherichia coli; EMSA, electrophoretic mobility shift assay; DBD, DNA binding domain; RBD, RNA binding domain; NLS, nuclear localization signal.
Binding of L1 RNA to PSF competitively inhibits PSF from binding to the downstream target gene GAGE6
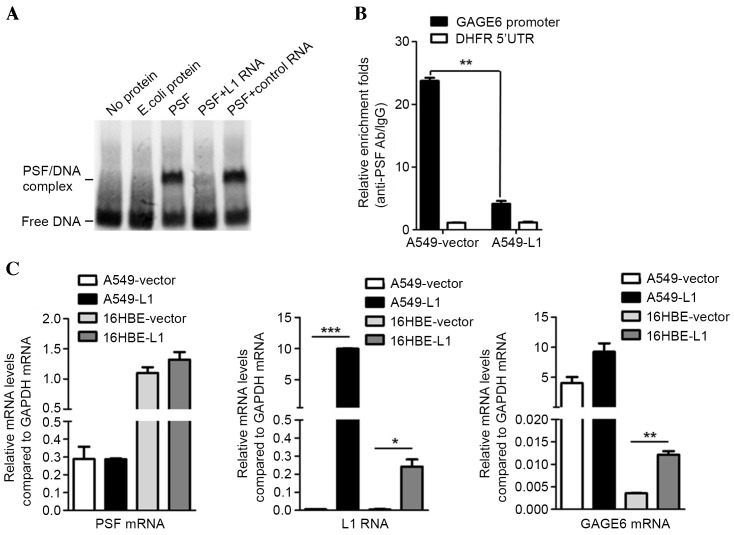
It has been suggested that PSF regulates mouse Rab23 expression by binding to the Rab23 promoter and repressing its transcriptional activity (19). However, certain types of non-coding RNA, including VL30-1, may bind to the PSF protein and suppress its ability to bind to the Rab23 response element (19). The effect of L1 RNA on the expression of the Rab23 human homolog GAGE6 was assessed by competitive EMSA. The unlabeled L1 RNA probe reduced the binding band of biotin-DNA/PSF, while it remained unaffected by the plasmid encoding control RNA (Fig. 3A). This result indicated that L1 RNA inhibits the binding of biotin-DNA with PSF protein in vitro. Subsequently, A549 cells were transfected with L1-encoding plasmid or with the empty pcDNA3.1 plasmid as a control and subjected to a CHIP assay. qPCR demonstrated that compared with the control, overexpression of L1 RNA reduced the binding of the PSF/GAGE6 promoter (P<0.01; Fig. 3B). These results demonstrated that L1 RNA exerts its function, at least in part, by binding to PSF and reversing PSF-mediated gene repression (Fig. 3C). Therefore, overexpression of L1 RNA leads to a release of PSF from the promoter region of GAGE6 and causes an increase in the expression of GAGE6 mRNA (P<0.05; Fig. 3C).
Figure 3.
Effects of L1 RNA binding to PSF on GAGE6. (A) Binding of L1 RNA releases GAGE6 promoter DNA from PSF. (B) Release of GAGE6 promoter region from PSF by overexpression of L1 RNA demonstrated by chromatin immunoprecipitation. (C) Expression levels of PSF mRNA, L1 RNA and GAGE6 mRNA in A549 and 16HBE cells transfected with L1 RNA overexpression or empty vector measured using reverse-transcription quantitative polymerase chain reaction. *P<0.05, **P<0.01, ***P<0.001. GAGE6, G antigen 6; 5′UTR, 5′ untranslated region; Ab, antibody; IgG, immunoglobulin G; E. coli, Escherichia coli; L1, long interspersed nuclear element 1; PSF, polypyrimidine tract-binding protein-associated splicing factor; DHFR, dihydrofolate reductase.
Effects of L1 RNA overexpression on the proliferation and colony formation of A549 and 16HBE cells
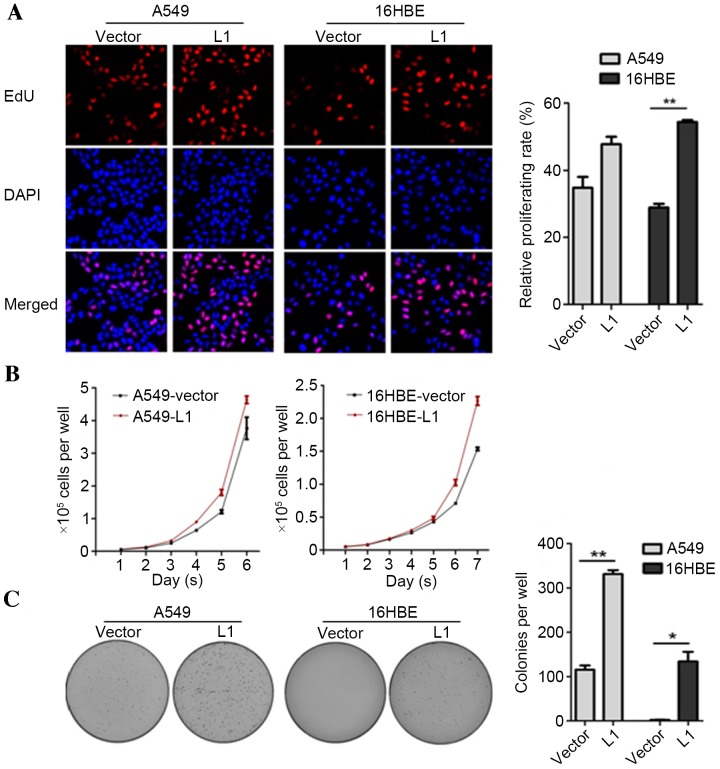
As a previous study has indicated the regulatory activities of GAGE6 in cell proliferation and tumor formation (18), the effects of L1 RNA on cell proliferation and colony formation were investigated in the present study. An EdU incorporation assay was employed to detect whether overexpression of L1 RNA may affect the number of proliferating cells. The results demonstrated that the number of EdU-positive cells in the A549-L1 RNA and 16HBE-L1 RNA groups were increased compared with the A549-vector and 16HBE-vector groups, respectively (P<0.01; Fig. 4A). Consistent with the EdU staining results, cell proliferation and colony formation rates were significantly higher in A549-L1 RNA and 16HBE-L1 RNA cells compared with the control groups (P<0.05; Fig. 4B and C). Taken together, the results demonstrated that overexpression of L1 RNA promoted cell proliferation and colony formation.
Figure 4.
Effect of L1 RNA on cell proliferation and tumorigenesis in A549 and 16HBE cell lines. (A) EdU assays measuring cell proliferation. Magnification, ×200. Red, EdU-stained nuclei of proliferating cells; blue, all nuclei counterstained with Hoechst. (B) Cell growth curves over 6–7 days. (C) Colony formation rates in 0.3% soft agar. *P<0.05, **P<0.01. L1, long interspersed nuclear element 1; DAPI, 4′,6-diamidino-2-phenylindole; EdU, 5-ethynyl-2′-deoxyuridine.
Discussion
The results of the present study expand on a previous study demonstrating that the chimeric HBx-LINE-1 functions as a hybrid RNA and promotes tumorigenesis in HCC (16). A lower hypomethylation level of L1 and higher transcriptional level of L1 RNA has been detected in HBV-transfected tumor samples compared with adjacent tissues (20). The present study described a mechanism of gene regulation involving formation of a complex between L1 RNA and the regulatory protein PSF, accounting for the effect of L1 RNA on cell proliferation.
L1 is biologically significant not only as a retrotransposon that can function as a genetic modifier, but also as a transcript which is associated with tumorigenesis (13). Furthermore, the transcription of L1 is epigenetically controlled by hypomethylation. L1 elements contain a 5′-C-phosphate-G-3′ island in their 5′-UTR, which is usually heavily methylated in normal somatic cells (21). The importance of hypomethylation in this region has been highlighted in various types of human cancer, including HCC, chronic myeloid leukemia, bladder cancer, gastrointestinal stromal tumors, gastric cancer and ovarian cancer (12). Transcription of L1 induced by hypomethylation significantly correlates with the degree of malignancy, particularly in colorectal cancer (21–23). This suggests that L1 hypomethylation may serve a more important role in human cancer than the hypomethylation-induced inhibition of transcription of specific tumor suppressor genes.
PSF is a multifunctional protein, containing two RBD and one DBD. Originally, PSF was identified as a component of spliceosomes and observed to serve a transcriptional regulatory role on GAGE6, which functions as an oncogene (7,24). Repression of GAGE6 expression involves PSF binding to its regulatory region. The complex formed with L1 RNA inhibits and dissociates the binding of PSF to the GAGE6 regulatory region, allowing transcription to proceed (24). Complex formation with the RBDs on PSF and the resulting regulation of gene transcription was mapped to two fragments in L1 RNA. Binding of these two fragments to the RBDs of PSF may cause allosteric modification, resulting in weaker affinity of the DBD for the GAGE6 DNA motif. It was observed that increasing the concentration of L1 RNA increased its inhibitory effect on PSF/DNA binding affinity. L1 RNA is transcribed from endogenous genomic retrotransposons and the level varies depending on the cell type and the level of global hypomethylation (25). Thus, the key parameters that determine whether GAGE6 expression is repressed by PSF or induced by L1 RNA are the type of cell and the global hypomethylation status. It has been determined that human L1 retrotransposons are a major source of endogenous mutagens (26). The present study demonstrated the individual oncogenic activity of L1 RNA, which potentially specifies a functionally distinct class of long non-coding RNA-like transcripts.
In conclusion, the present study described a mechanism controlling cell proliferation and tumorigenesis in human cells, which involves the activation of oncogene GAGE6 transcription by L1 RNA through binding to PSF protein and its simultaneous release from the GAGE6 promoter region. In addition, overexpression of L1 exerted strong regulatory effects on the proliferation and colony formation of lung cancer and normal lung cells without HBV infection. Therefore, endogenous L1 RNA may potentially serve an important regulatory role in tumorigenesis.
Acknowledgements
The authors would like to thank Mr. Huimin Shi (Molecular Biology Laboratory, The Food and Drug Inspection Institute, Chengdu, China) for editing the draft manuscript. The current study was supported by the Sichuan Provincial Science and Technology Supporting Program (grant no. 2016FZ0093).
References
- 1.Guo XL, Li D, Hu F, Song JR, Zhang SS, Deng WJ, Sun K, Zhao QD, Xie XQ, Song YJ, et al. Targeting autophagy potentiates chemotherapy-induced apoptosis and proliferation inhibition in hepatocarcinoma cells. Cancer Lett. 2012;320:171–179. doi: 10.1016/j.canlet.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 2.Ng J, Wu J. Hepatitis B- and hepatitis C-related hepatocellular carcinomas in the United States: similarities and differences. Hepat Mon. 2012;12:e7635. doi: 10.5812/hepatmon.7635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGivern DR, Lemon SM. Virus-specific mechanisms of carcinogenesis in hepatitis C virus associated liver cancer. Oncogene. 2011;30:1969–1983. doi: 10.1038/onc.2010.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsai WL, Chung RT. Viral hepatocarcinogenesis. Oncogene. 2010;29:2309–2324. doi: 10.1038/onc.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kowalska E, Ripperger JA, Muheim C, Maier B, Kurihara Y, Fox AH, Kramer A, Brown SA. Distinct roles of DBHS family members in the circadian transcriptional feedback loop. Mol Cell Biol. 2012;32:4585–4594. doi: 10.1128/MCB.00334-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lowery LA, Rubin J, Sive H. Whitesnake/sfpq is required for cell survival and neuronal development in the zebrafish. Dev Dyn. 2007;236:1347–1357. doi: 10.1002/dvdy.21132. [DOI] [PubMed] [Google Scholar]
- 7.Patton JG, Porro EB, Galceran J, Tempst P, Nadal-Ginard B. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 1993;7:393–406. doi: 10.1101/gad.7.3.393. [DOI] [PubMed] [Google Scholar]
- 8.Urban RJ, Bodenburg Y, Kurosky A, Wood TG, Gasic S. Polypyrimidine tract-binding protein-associated splicing factor is a negative regulator of transcriptional activity of the porcine P450scc insulin-like growth factor response element. Mol Endocrinol. 2000;14:774–782. doi: 10.1210/mend.14.6.0485. [DOI] [PubMed] [Google Scholar]
- 9.Urban RJ, Bodenburg YH, Wood TG. NH2 terminus of PTB-associated splicing factor binds to the porcine P450scc IGF-I response element. Am J Physiol Endocrinol Metab. 2002;283:E423–E427. doi: 10.1152/ajpendo.00057.2002. [DOI] [PubMed] [Google Scholar]
- 10.Li L, Feng TT, Lian Y, Zhang G, Garen A, Song X. Role of human noncoding RNAs in the control of tumorigenesis; Proc Natl Acad Sci USA; 2009; pp. 12956–12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kazazian HH., Jr Mobile elements: Drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 12.Aporntewan C, Phokaew C, Piriyapongsa J, Ngamphiw C, Ittiwut C, Tongsima S, Mutirangura A. Hypomethylation of intragenic LINE-1 represses transcription in cancer cells through AGO2. PLoS One. 2011;6:e17934. doi: 10.1371/journal.pone.0017934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chowdhury S, Cleves MA, MacLeod SL, James SJ, Zhao W, Hobbs CA. Maternal DNA hypomethylation and congenital heart defects. Birth Defects Res A Clin Mol Teratol. 2011;91:69–76. doi: 10.1002/bdra.20761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tangkijvanich P, Hourpai N, Rattanatanyong P, Wisedopas N, Mahachai V, Mutirangura A. Serum LINE-1 hypomethylation as a potential prognostic marker for hepatocellular carcinoma. Clin Chim Acta. 2007;379:127–133. doi: 10.1016/j.cca.2006.12.029. [DOI] [PubMed] [Google Scholar]
- 15.Wu HC, Wang Q, Yang HI, Tsai WY, Chen CJ, Santella RM. Global DNA methylation levels in white blood cells as a biomarker for hepatocellular carcinoma risk: A nested case-control study. Carcinogenesis. 2012;33:1340–1345. doi: 10.1093/carcin/bgs160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lau CC, Sun T, Ching AK, He M, Li JW, Wong AM, Co NN, Chan AW, Li PS, Lung RW, et al. Viral-human chimeric transcript predisposes risk to liver cancer development and progression. Cancer Cell. 2014;25:335–349. doi: 10.1016/j.ccr.2014.01.030. [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Song X, Sun Y, Garen A. Roles of PSF protein and VL30 RNA in reversible gene regulation; Proc Natl Acad Sci USA; 2005; pp. 12189–12193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang G, Cui Y, Zhang GF, Garen A, Song X. Regulation of proto-oncogene transcription, cell proliferation, and tumorigenesis in mice by PSF protein and a VL30 noncoding RNA; Proc Natl Acad Sci USA; 2009; pp. 16794–16798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang YJ, Wu HC, Yazici H, Yu MW, Lee PH, Santella RM. Global hypomethylation in hepatocellular carcinoma and its relationship to aflatoxin B(1) exposure. World J Hepatol. 2012;4:169–175. doi: 10.4254/wjh.v4.i5.169.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogino S, Kawasaki T, Nosho K, Ohnishi M, Suemoto Y, Kirkner GJ, Fuchs CS. LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Int J Cancer. 2008;122:2767–2773. doi: 10.1002/ijc.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goel A, Xicola RM, Nguyen TP, Doyle BJ, Sohn VR, Bandipalliam P, Reyes J, Cordero C, Balaguer F, Castells A, et al. Aberrant DNA methylation in hereditary nonpolyposis colorectal cancer without mismatch repair deficiency. Gastroenterology. 2010;138:1854–1862. doi: 10.1053/j.gastro.2010.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sunami E, De Maat M, Vu A, Turner RR, Hoon DS. LINE-1 hypomethylation during primary colon cancer progression. PLoS One. 2011;6:e18884. doi: 10.1371/journal.pone.0018884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eynde B, Peeters O, De Backer O, Gaugler B, Lucas S, Boon T. A new family of genes coding for an antigen recognized by autologous cytolytic T lymphocytes on a human melanoma. J Exp Med. 1995;182:689–698. doi: 10.1084/jem.182.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trivedi M, Shah J, Hodgson N, Byun HM, Deth R. Morphine induces redox-based changes in global DNA methylation and retrotransposon transcription by inhibition of excitatory amino acid transporter type 3-mediated cysteine uptake. Mol Pharmacol. 2014;85:747–757. doi: 10.1124/mol.114.091728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burns KH, Boeke JD. Human transposon tectonics. Cell. 2012;149:740–752. doi: 10.1016/j.cell.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]