Abstract
Nonalcoholic fatty liver disease (NAFLD) is the most prevalent liver disease worldwide, the morbidity of which closely correlates with diversity of ethnicity, minority, family and location. Its histology spans from simple steatosis, to nonalcoholic steatohepatitis, which ultimately results in fibrosis, cirrhosis and hepatocellular carcinoma. The accelerating prevalence of NAFLD is due to an incremental incidence of metabolic syndrome that is distinguished by dyslipidemia, glucose impairment, obesity, excessive oxidative stress and adipocytokine impairment. Additionally, the pathogenesis of NAFLD is thought to be a multifactorial and complicated disease associated with lifestyle habits, nutritional factors and genetics. However, the pathogenesis and underlying mechanism in the development of NAFLD caused by genetics remains unclear. People have been increasingly emphasizing on the relationship between NAFLD and gene polymorphisms in recent years, with the aim of having a comprehensive elucidation of associated gene polymorphisms influencing the pathogenesis of the disease. In the current article, the authors attempted to critically summarize the most recently identified gene polymorphisms from the facets of glucose metabolism, fatty acid metabolism, oxidative stress and related cytokines in NAFLD that contribute to promoting the progression of the disease.
Keywords: liver, nonalcoholic fatty liver disease, nonalcoholic steatohepatitis, gene polymorphism
1. Introduction
Nonalcoholic fatty liver disease (NAFLD) is defined as a clinicopathological syndrome with the primary characteristic being hepatocellular macrovesicular steatosis, where pathology and other explicit factors give rise to hepatic injury, ranging from simple steatosis to nonalcoholic steatohepatitis (NASH), which has the potential to develop into cirrhosis, hepatocellular carcinoma (HCC) and end-stage liver disease (1). NAFLD is proven to be related to obesity, type 2 diabetes (T2DM), insulin resistance (IR), hyperlipidemia and gender. The onset and persistence of steatohepatitis results from the early predisposing factors of accumulating visceral adiposity and free fatty acids (FFAs). According to the ‘two hit hypothesis’ (2), the degree of obesity and presence of IR being the first hit promote the accumulation of fatty acids in liver, and increase the susceptibility of hepatic injury caused by the second hit. Active factors of the ‘two hit hypothesis’, for instance, inflammation, dysfunction of Kupffer cells, oxidative stress, mitochondrial dysfunction and a regulative disorder of adipocytokines, lead to a greater risk of serious liver disease. The primary factor in causing NAFLD is the presence of IR that is associated with overexpression of hypoadiponectinemia, hyperleptinemia and cytokines, all of which can intensify IR and promote the accumulation of fatty acids in liver. Excessive FFAs in hepatocytes are said to induce the occurrence of oxidative stress, and the release of inflammatory factors and adipogenesis-related factors result in the formation of necroinflammation, hepatofibrogenesis and cirrhosis.
Currently, NAFLD is gradually becoming more prevalent and affects ~20–30% of the general population worldwide. Most estimates indicate that 70% obese patients may have hepatic steatosis, among which, the risk of developing NASH accounts for 20%, and patients with NASH that progresses to cirrhosis is estimated to be ~20–25% (3). NASH is now accepted as the major underlying cause of cryptogenic cirrhosis, which suggests that genetic factors are involved in the development of NAFLD. As of yet, the genetic mechanism of NAFLD is not able to be comprehensively and thoroughly understood, but research on genetic variation influencing IR, lipid metabolism and oxidative stress are important issues of scientific interest to improve patients' management. The aims of the present review is to concentrate on the available knowledge of gene polymorphisms implicated in the development of NAFLD induced by a disorder in glucose metabolism and fatty acid metabolism, oxidative stress and related cytokines.
2. Pathogeneisis
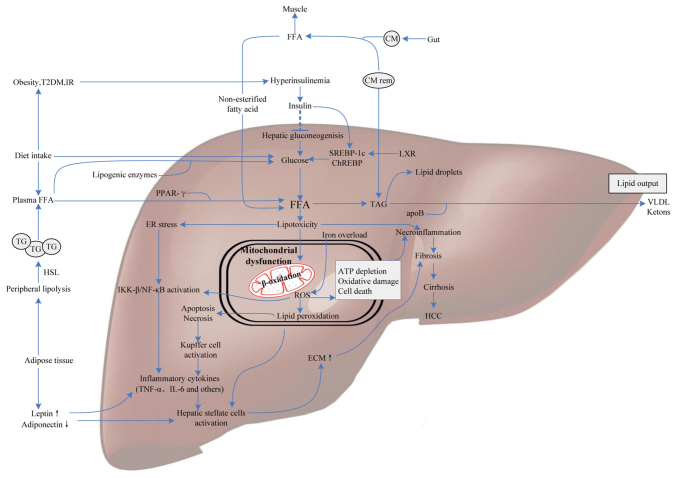
The molecular events lead to the occurrence of intrahepatic lipid accumulation followed by increased FFAs in the liver, reduced fatty acid oxidation, or elevated de novo synthesis of triglycerides (TG), whereby the very low-density lipoprotein (VLDL) synthetic rate is unable to keep up. FFAs derive from dietary fatty acids and lipolysis takes place in the peripheral, especially visceral, subcutaneous adipose tissue. Hepatic glucose and FFA uptake occurs via an insulin-independent manner. Insulin serves a crucial role in the pathogenesis of NAFLD by upregulating colony-stimulating factor expression. Almost all patients with NAFLD suffer from hepatic IR with a failure to suppress lipolysis as a result from a deficiency of sensitivity to insulin in adipose tissue. IR leads to lipid storage in hepatocytes for two major routes: One is hyperlipidemia and the other is hyperinsulinemia. The storage and release of FFAs from adipose tissue is mediated by hormones and cytokines, such as estrogen, cortisol, growth hormone, glucagon, insulin and insulin-like growing factor, which augment levels of fatty acids through altering energy metabolism in turn to enhance FFA uptake and synthesis in liver, ultimately resulting in lipid accumulation. FFA is an amphipathic molecule with high toxicity, and the reinforcement of peroxidation engendered by unsaturated fatty acids finally impairs hepatocytes. Lipid peroxidation free radicals and end-products like malondialdehyde are able to bring about abnormal fluidity and permeability of cell membrane, causing cell dysfunction, apoptosis or death. Excess FFAs conduce to hepatotoxicity in NAFLD/NASH, since FFA oxidation in hepatic mitochondria leads to oxidative stress. Following this, elevated levels of peroxides and free radicals, as a consequence of fatty acid oxidation, causes oxidative damage, endoplasmic reticulum stress and apoptosis (4). Once the absorption of FFAs increases, this is followed by a compensatory increase in mitochondrial β-oxidation, which will further enhance the generation of reactive oxygen species (ROS) that can tightly bind to membrane phospholipids to block the respiratory chain of electron transfer in mitochondria (5). Another location where ROS is generated is in microsomes. FFA is the inductive agent of P450-2E1, which occupies an important position in the cascade of ROS and lipid peroxidation generation (6). In microsomes, FFAs are easily oxidized and degraded into acyl-CoA. As the ligand combines with some enzymes of the fatty acid oxidation system in the liver, acyl-CoA controls gene-induction and promoting the synthesis of unwinding protein, which could prevent hepatocyte apoptosis. Patients carrying variant alleles of acyl-CoA dehydrogenases may result in the progression of steatohepatitis, because of aberrant fatty acid oxidation (7). In addition, the following undoubtedly contribute to lipid storage within the liver (5,8–10): Insufficient synthesis and inhibition of apolipoproteins; Inhibition of newly synthesized VLDL transportation from the endoplasmic reticulum to the Golgi complex; Dysfunction of the Golgi-endoplasmic reticulum-lysosome complex; Reduction of saccharification or secretory vesicles of newly synthesized VLDL; Interference with exocytosis of newly synthesized VLDL as a result from malfunction of secretory vesicles migrating to hepatocyte serosa; Chronic hunger, poor digestion and absorption, low protein diet leading to the deficiency of ‘anti-fatty liver factors’ (including methionine, choline and phosphatidylcholine), bringing about insufficient synthesis of apolipoproteins, in particular apoB and VLDL. It has been demonstrated that lipid metabolism disturbance is regulated insufficiently by vital transcription factors that are indispensable for lipogenesis, such as the activation of AMP-activated protein kinase (AMPK), Jun N-terminal kinase (JNK), protein kinase C and nuclear factor (NF)-κB (11). Other adipokines, such as resistin and leptin also serve as prospectively latent markers of NAFLD. These events cooperatively promote the susceptibility to NAFLD and its progression (Fig. 1).
Figure 1.
Molecular mediators involved in NAFLD pathogenesis. FFAs derive from dietary fatty acids and lipolysis in the periphery, and then are delivered to the liver by CM via the portal vein. Meanwhile, individuals with obesity, IR or T2DM influence hepatic gluconeogenesis and transcriptional activity of SREBP-1c and ChREBP by increasing the generation of glucose, ultimately leading to intrahepatic FFA accumulation. Excess FFAs present lipotoxicity with the ability to impair hepatocytes triggering necroinflammation and mitochondrial dysfunction. Some FFAs in the liver are converted into TAG and then exported out of liver in the form of VLDL, others are oxidized in hepatic mitochondria, generating a mass of ROS. Additionally, iron overload generating hydroxyl radicals through the Fenton reaction also contributes to oxidative stress. ROS can cause oxidative damage, endoplasmic reticulum stress and apoptosis leading to necroinflammation via IKK-β activation, NF-κB activation and Kupffer cell activation pathways. Subsequently, the release of inflammatory cytokines, like TNF-α and IL-6, which give rise to hepatofibrogenesis, and finally progress to cirrhosis and HCC. Therefore, molecular mediators involved in glucose metabolism, fatty acid metabolism and oxidative stress may contribute to the progression of NAFLD. NAFLD, nonalcoholic fatty liver disease; FFAs, free fatty acids; IR, insulin resistance; CM, chylomicron; T2DM, type 2 diabetes; SREBP-1c, sterol regulatory element binding protein-1c; ChREBP, carbohydrate-responsive element-binding protein; TAG, triacylglycerol; VLDL, very low-density lipoprotein; ROS, reactive oxygen species; IKK-β, IkB kinase-β; NF-κB, nuclear factor-κB; HCC, hepatocellular carcinoma; PPAR, peroxisome proliferator-activated receptor; ApoB, apolipoprotein B; TG, triglyceride; ECM, extracellular matrix; TNF-α, tumor necrosis factor-α; LXR, liver X receptor; HSL, hormone-sensitive lipase.
3. Gene polymorphisms in mediators of glucose metabolism
IR and aberration of glucose metabolism are a central part of the onset and development of NAFLD. The formation of IR in the liver is caused by some factors that do the same to the progression of NAFLD/NASH, and will be discussed below.
Insulin receptor substrate (IRS) and ectoenzyme nucleotide pyrophosphatase phosphodiesterase 1 (ENPP1)
Among the genes concerned with regulating IR, IRS-1 and ENPP1 directly interact with insulin receptor (INSR) signaling. Mechanically, insulin binds to the INSR with a high affinity for hepatocytes, resulting in INSR phosphorylation. The activation of IRS-1 and the downstream kinase AKT phosphorylates the transcription factor FOXO1 in turn to decrease glucose production and cell apoptosis. FOXO1 expression is observed to be increased and resistant to the inhibition of hyperinsulinemia in patients with NASH. ENPP1 is a membrane glycoprotein, and overexpression of ENPP1 induces the occurrence of IR and the disturbance of glucose metabolism (12). IR, as determined by the Lys121Gln single nucleotide polymorphism (SNP), is seen to function through different mechanisms, for instance, the alteration of appetite controlled by hypothalamus, and the reduced levels of high-density lipoprotein (HDL) (13). The IRS protein functions as a regulator in the insulin-like growth factor-1-activated signaling. The Gly972Arg SNP, as previously reported, is also associated with IR, which brings about more serious liver fibrosis and progression of NAFLD (14). Presumably, NAFLD patients carrying both ENPP1 and IRS-1 SNPs possess higher possibility of fibrogenesis than those with unitary ENPP1 or IRS-1 SNP (13,14). Thereby, the coincident presence of ENPP1 and IRS-1 SNPs in NAFLD patients synergistically triggers hepatic injury.
Adiponectin
Adiponectin is a specific plasma protein derived from brown adipose tissue that serves a crucial role in regulating glucose homeostasis. In liver tissue, adiponectin plays a part in reducing the generation of glucose and FFAs, the concentration of which circulating in plasma negatively associates with TG, and is downregulated in obese populations. In addition, adiponectin has capable of exerting insulin sensitivity effects and acts as an anti-inflammatory molecule. However, inflammation is able to impede the release of adiponectin, thus adipose tissue inflammation is taken for one of the prime mechanisms involved in decreasing plasma levels of adiponectin in obesity (15). Adiponectin predominantly activates AMPK and peroxisome proliferator-activated receptor (PPAR)-γ to improve insulin sensitivity, reduce the synthesis of fatty acids and enhance fatty acids oxidation for combating steatosis. Therefore, the levels of adiponectin attribute to the development of NAFLD/NASH, and the adiponectin gene has the ability of modulating individual differences. In a Japanese population, adiponectin gene variants, +45GT and +276GT locus of alleles were correlated to the risk of NAFLD. The frequency of the +45GG genotype was significantly more pervasive among NAFLD females, whose hepatofibrosis were more serious than that in control group (16). However, no association was observed between NAFLD and the genetic polymorphism of adiponectin at positions 11391, −11377, +45 and +276 in a Chinese population. Nonetheless, subjects carrying −11377 and +45 SNPs were associated with higher TG levels, body mass index (BMI) and lower plasma adiponectin levels (17). Another study suggested that +45TG and +276GT genetic variations were closely related to the predisposition to NAFLD in Chinese and Indian populations (18). The adiponectin rs266729 (−11377G/C) polymorphism may be a candidate gene associated with the susceptibility to NAFLD in a southeast Iranian population, but has not yet been confirmed in other ethnicities (19). Collectively, adiponectin gene polymorphisms have an intimate association with BMI, blood lipid levels, T2DM, obesity and hyperlipemia, which add to the risk of NAFLD.
Resistin
Resistin is a hormone secreted by fat cells, and is regarded as the link of adiposity to diabetes. The major target of resistin is the liver in vivo, and reduced or increased resistin levels respectively gives rise to improvement or worsening of hepatic IR through interfering with glycometabolism (20). The reduction of hepatic glucose in the absence of resistin is integral to the activation of the AMPK pathway. It was demonstrated that hepatic steatosis and VLDL secretion were decreased in resistin-deficient mice treated with high-fat diet, which suggested a function of resistin on steatosis induced (21). Resistin suppresses the effect of insulin on glucose absorption and glucose tolerance impairment, which was regulated by CCAAT/enhancer binding protein α and PPAR-γ in the phosphatidyl inositol 3-kinase and mitogen-activated protein kinase pathways (22). Therefore, SNPs may influence resistin gene expression. For adult women, the −420C/G SNP of the resistin gene was related with abdominal obesity and metabolic risk factors; this polymorphism in the promoter region captures only a small part of the total genetic contribution to the variation of circulating resistin (23). Another genotype of the resistin +229AA polymorphism was confirmed to be associated with higher risk of obesity and NAFLD incidence in T2DM patients (24). This evidence is consistent with the research in obese Egyptian patients (25). However, how resistin gene polymorphisms influence the signaling pathway mechanism of NAFLD has not been figured out clearly yet. Obviously, resistin as a proinflammatory factor is involved in the inflammatory cascade reaction and triggering ‘the second hit’.
Leptin and leptin receptor (LepRb)
Leptin is protein-related product encoded by an obesity gene and is involved in the modulation of body fat, insulin signaling and the immune system. Leptin acts through leptin receptors (LepRb), a member of the class-1 cytokine receptor family, originally demonstrated in hypothalamic neurons, to control satiety and maintain the body's constant energy balance. Leptin is a contributing factor of elevating intracellular fatty acids via promoting IR and altering insulin signaling in hepatocytes for hepatic steatosis. At a later stage, hepatic steatosis caused by leptin acting on proinflammatory response amplification evolves into steatohepatitis. However, both roles of leptin and LepRb in NAFLD are not completely clear. The LepRb gene polymorphism in humans has been reported to be in association with IR, T2DM, obesity and lipid metabolism, as well as the distribution of local body mass. The LepRb SNP (rs6700896) increased the risk for NAFLD as well as T2DM in an Egyptian population, and this enhanced leptin level was accompanied by a decrease of LepRb concentration (26). Furthermore, by mediating lipid metabolism and insulin sensitivity, the LepRb G2057A SNP is conducive to the onset of NAFLD (27). However, the LepRb Arg233Gln polymorphism has been indicated in relation to lower cholesterol, low-density lipoprotein (LDL) levels and fibrosis score in NAFLD (28). Patients carrying the SNPs of LepRb (rs1137100) with patatin-like phospholipase domain containing 3 (rs738409) have ~three-fold risk of suffering from NAFLD than those carrying either SNP, both genes are correlatively upregulated in the absence of excess lipids (28). Leptin inhibits steatosis via downregulating stearoyl-CoA desaturase-1 and sterol regulatory element binding protein (SREBP) 1c. Gene polymorphisms that code leptin or regulate leptin secretion and tissue sensitivity proteins may be potential genes in the development of NAFLD.
4. Gene polymorphisms in mediators of lipid metabolism
The increased release of FFAs is closely relevant to the onset and development of NAFLD. Lipotoxicity have an impact on insulin signaling cascade, and then results in the accumulation of lipids, ultimately leading up to the unbalance of gene-mediated lipid metabolism.
Uncoupling protein 3 (UPC3)
UPC3 is a family of mitochondrial transporters known for uncoupling oxidative phosphorylation via proton leakage from the inner mitochondrial membrane, with primarily selective expression in skeletal muscle, as well as brown adipose tissue, and is implicated in modulating thermoregulation and energy metabolism (29). Thereby, the expression of UPC3 mRNA undeniably has a bearing on an increase in metabolic rate and a lower BMI. A number of polymorphisms have been identified in the UPC3 gene. Particularly, the −55C/T polymorphism is responsible for UPC3 mRNA levels, T2DM and weight gain (29). In obese Indonesian children, subjects with T/T genotypes of UPC3 were demonstrated to present a lower total energy expenditure than those with other genotypes (30). The level of serum LDL-cholesterol at baseline was substantiated in relation to the rs3781907, rs1726745, rs11235972 and rs1800849 variants of the UCP3 gene. Xu et al (31) demonstrated that the rs11235972GG genotype appears at a higher frequency in NAFLD, but no increased risk associating the rs1800849 variant with the development of NAFLD was confirmed. However, this remains controversial with respect to the impact of these polymorphisms on NAFLD susceptibility. Further additional larger studies should also be conducted concerned with allowing stratification for ethnicity and gene-gene interactions in order to distinctly elucidate the possible roles of UCP3 polymorphisms in NAFLD.
Microsomal triglyceride transfer protein (MTTP)
MTTP, as a pivotal enzyme involved in the incorporation of TG into VLDL for lipid export out of the liver, is localized to the endoplasmic reticulum of hepatocytes and enterocytes. The human MTTP gene is located on chromosome 4q24 and made up of 18 exons and 17 introns (32). Low hepatic expression of MTTP induced by MTTP gene polymorphisms may be related to the pathogenesis of NAFLD. Functional polymorphisms of the MTTP gene (promoter −493G/T, −164 T/C and Ile128Thr) are involved in lower LDL levels and are protected against other traits of metabolic syndrome, some studies indicated (33,34). Among these, lower transcription of MTTP was exhibited in a common SNP at position −493G/T, which predisposed to NASH by modulating lipoprotein metabolism (35). The MTTP-493G>T resulting from a G>T substitution in the intron region of NM_000253.2, which may reduce the expression of MTTP and lead to an inadequate formation of VLDL and chylomicron, resulting in the dysregulation of hepatic lipid metabolism. In a study involving women treated with a western type diet, higher fasting levels for plasma cholesterol (but not synthesis) and high-absorption status were demonstrated in TT homozygote women compared with G carriers (36). The presence of the G allele of MTTP −493G/T has an association with lower hepatic MTTP expression, which protects against steatosis in chronic hepatitis (37). Now, it is universally acknowledged that the MTTP −493G>T polymorphism is correlated with the risk of NAFLD. Thus, the MTTP −493G>T polymorphism in the future may be used as a valuable and practical biomarker for early diagnosis of NAFLD.
PPAR-α and PPAR-γ signaling
PPAR-α belongs to the nuclear hormone receptor superfamily and acts as a primary regulatory role on lipid metabolism. PPAR-α is able to activate fatty acid oxidation and lipid hydrolysis, as well normalize glucose and insulin levels to prevent hepatic lipid accumulation (38). Downregulation of PPAR-α is involved in NASH pathogenesis by lessening FFA catabolism (39). PPAR-α-agonist fibrates and fenofibrates are used to improve dyslipidemia and insulin sensitivity in patients. The state of PPAR-α deficiency is involved in the steatohepatitis pathogenesis. Several studies have demonstrated a correlation between the Val227Ala of PPAR-α and low serum lipids (40,41). It may have a protective role against the development of obesity and be implicated in the pathogenesis of NAFLD. The carriers of the Leu162Val SNP, which develops IR by attenuating oxidative stress, had a association with decreased hepatic lipolysis, dyslipidemia and T2DM, but not with liver damage in NAFLD (41).
PPAR-γ, as another transcription factor, also belongs to the nuclear hormone receptor superfamily, and is primarily known to mediate adipocyte differentiation. A number of genetic variants at several nucleotide loci of the PPAR-γ gene bring about conformational changes in protein structures and gene location. For disparate ethnic groups, a missense Pro12Ala substation in the PPARγ2 gene (rs1801282) was demonstrated to have a bearing on higher NAFLD risk, but was neither associated with liver damage, nor insulin sensitivity (42). The decreasing DNA-binding affinity caused by rs1801282 variant weakens transcriptional activation that reduces PPAR-γ activity and activates lipogenic enzymes, resulting in the exacerbation of disease (38,42). Nevertheless, the association between rs1801282 variants and NAFLD risk was thrown into doubt in a meta-analysis study (43). In a middle-aged and older Chinese population, smokers with the C/C genotype of the PPARγ2 gene polymorphism synergistically demonstrated a 3.75-fold higher risk of NAFLD than non-smokers with the C/G genotype by aggravating oxidative stress (44). Another evidence indicated that a higher susceptibility to NAFLD was associated with G/G genotype at rs10865710 loci, whereas no influence of rs7649970 (45). Gawrieh et al (46) first provided the evidence that populations were at risk-reduction for NAFLD with a haplotype containing both minor alleles of Pro12Ala and C1431T that histological features manifested as lobular inflammation and fibrosis, which were associated with NASH. It may be a promising for subjects with NASH carrying this haplotype for treating with PPARG agonists.
Notwithstanding genetic variants of PPARα/β may influence disease progression of these studies make recommendations, the implications of these SNPs on the risk of NAFLD remain ill-defined, and further elucidation is required for the functional role of these two PPAR genes on mechanism of NAFLD pathogenesis.
SREBPs and SREBP cleavage activating protein (SCAP)
SREBP serves a modulatory role in lipid metabolism. Three informs of SREBP has been identified that contain SREBP-1a, SREBP-1c and SREBP-2 in the human SREBP family, which are correlated with lipogenesis, cholesterol homeostasis and adipocyte development (47). SREBP-1a is capable of activating the genes relating to fatty acid and cholesterol synthesis. Compared with SREBP-1a, the SREBP-1c is a weaker transcriptional activator, the expression of SREBP-1c is negatively related to tumor necrosis factor (TNF)-α and IL-1β secretion that is involved in liver fibrosis progression. In response to oxidative stress, the activation of AMPK pathway inhibits de novo lipogenesis via restraining the transcriptional activity of SREBP-1c (48). Evidently, the development of NASH may be associatively dependent on the downregulation of SREBP-1c. Both SREBP-1a and SREBP-1c originate from a single gene referred to as SREBF-1. Other than SREBF-1 acting on fatty acid synthesis, TG synthesis and phospholipid synthesis, the activation of the SREBP-2 gene directly functions on maintaining cholesterol homeostasis. Recently, a potential biomarker for predicting the development of NAFLD in a Han Chinese population was recommended by determining the genotypes of the SREBP-2 rs2228314 G>C polymorphism, the variant gene was indicated to carry a risk of NAFLD (49). SCAP is implicated in delivering SREBPs from the endoplasmic reticulum to the Golgi complex. The activation of SREBPs is subsequently translocated into the nucleus to stimulate the expression of target genes for synthesis. In liver-specific SCAP knockout mice, the synthesis rate of fatty acids in liver was decreased by 70–80% as a result of the significant reduction of expression levels of SREBP-1, SREBP-2 and SREBP target genes. Interestingly, the SCAP rs2101247 A allele (AA and GA genotypes) suffered from the risk of NAFLD may decrease, which has been reported in a female metabolic syndrome population (50), but it is unclear how, and the potential mechanisms, for how the gender-specific distribution in genetic susceptibility to NAFLD development are caused by particular genotypes.
5. Gene polymorphisms in mediators of oxidative stress
The occurrence of oxidative stress induced by hepatonecrosis, inflammation and fibrosis, may be a factor of ‘the second hit hypothesis’ in the onset and development of NAFLD. Excessive oxidative stress derives from mitochondrial dysfunction and iron overload. The unceasingly increased ROS, and the product of oxidative stress, results in the rapid depletion of ATP, DNA injury, instability of protein and release of inflammatory factors, sequentially break cellular completeness of structure and function, which promotes the development of NAFLD (6,10).
Manganese superoxide dismutase (MnSOD)
MnSOD is nuclear encoded mitochondrial protein for scavenging ROS. The MnSOD activity protects against ROS by detoxifying superoxides into oxygen and hydrogen peroxide, the impairment of which contributes to the higher oxidative stress for NAFLD. In a Japanese population with NASH, a common variant of the MnSOD gene 1183T/T results in an amino acid substitution in the signaling sequence targeting the enzyme to the mitochondria, which weakens the capability of MnSOD through mitochondria, leading to the reduction of MnSOD and augmentation of oxygen radicals in mitochondria, ultimately resulting in hepatocellular damage (51). Another MnSOD polymorphism (C47T, rs4880) has been investigated as a possible susceptible factor in NASH and fibrosis, due to protein-import diminished into mitochondria (52). More interestingly, the concentration of MnSOD reduced in males with NAFLD, instead of in females, possibly because due to the differing sex hormone levels in different sexes (53). Thereby, the low level of MoSOD in males makes them more susceptible to increase oxidative stress and disease progression. In addition, the presence of the Ala16Vla-MnSOD allele with the G-463A myeloperoxidase variant may be attribute to an excess of hepatic iron accumulation and HCC development, along with the increase of ROS-related oxidative stress and DNA damage (54).
Cytochrome P4502E1 (CYP4502E1)
CYP4502E1 is an endoplasmic mono-oxygenase and it serves a role in NAFLD. The elicitors such as ethanol, diet and endogenously produced fatty acids, affect CYP4502E1 activation. As a result of incomplete transfer of electrons to molecular oxygen, CYP2450E1 releases superabundant ROS generation that leads to raised oxidative stress in the pathophysiology of NAFLD. With excessive CYP4502E1 activation in the perivenular hepatocytes, the area of hepatocyte injury was the greatest (55). There are six kinds of restriction fragment length polymorphism in CYP2450E1, and two point mutations of PstI/RsaI located in 5′-flanking region affect the expression of CYP2450E1 at the level of transcription. It was indicated that the variant of c2 allele makes the CYP2450E1 level increase, which functions by blocking tyrosine phosphorylation of IRS-1 and IRS-2, ultimately resulting in IR and an increased risk of HCC (56). The CYP4502E1*5B (RsaI, −1053C>T) variant is relevant to the improvement of its transcription and enzyme activity, which increase the generation of ROS in alcoholic cirrhotic patients, but not in NAFDL patients (57). Consequently, larger cohort studies should be explored in future research to verify and evaluate the effect of the CYP2E1 Pst I/Rsa I polymorphism-associated NAFLD risk.
Iron overload and hemochromatosis (HFE)
Among the candidate factors of oxidative stress, iron is a notable pro-oxidant and acts through the Fenton reaction to generate hydroxyl radicals (58). Excessive hepatic iron accumulation causes oxidative stress and promotes chronic liver damage in patients with genetic hemochromatosis associated with HFE variants, especially C282Y (rs1800562) and H63D (rs1799945). In mouse, iron-induced inflammatory activation may contribute to the necroinflammation, while the iron-mediated downregulation M2 pathway in macrophages has an impact on fibrogenesis in NASH (58), which supports for a mechanistic role for iron in NASH. HFE is characterized by the enhanced absorption of dietary iron that results in progressive deposition of metal in the liver. The HFE-knockout mice presented a deficiency of hepatic-intestinal iron and lipid signaling, which enabled them to be subjected to an accelerated progression of injury to fibrosis with diet-induced hepatic lipotoxicity. The mechanism is relevant to reduced hepcidin release leading to elevated iron absorption and deposition, which promotes hepatic IR by disability of glucose clearance and the facilitation of fibrogenesis by ROS generation of hepatic stellate cells (HSCs) (59). In general, HFE variants are common in white populations: The frequency of the C282Y homozygote, H63D homozygote, H63D/wild-type heterozygote in white populations respectively accounts for 0.44, 2.4 and 24%, while in Asians, this was ~0, 0–1 and 2.6–11%, respectively (60). In American patients with NAFLD, carriers of the C282Y variant, instead of H63D HFE variant, were associated with hepatocellular iron deposition and lower serum ALT and AST levels than wt/wt subjects, implying that NASH pathogenesis may be independent of mild hepatocellular iron deposition (61). However, the iron deposition, rather than the HFE mutation, is the key factor in the development of NAFLD, NASH and HCC (62). Also, β-globin variants are associated with both hepatic iron and fibrosis, and arising rates of iron accumulation are comparatively common in subjects with C282Y HFE hemochromatosis and the beta thalassemia trait. The presence of β-globin variants seems to be related to more severe liver disease. Therefore, routine blood analysis may be conducive to identify the risk of NASH progression in the carriers of the beta thalassemia trait.
6. Gene polymorphisms in mediators of liver fibrosis
There are countless factors associated with liver fibrosis, including the activation of HSCs and the synthesis and degradation of collagen. Fibrosis is a dynamic process of continuous extracellular matrix (ECM) remodeling. The genes encoding hepatofibrotic and fibrinolytic proteins are candidates for fibrotic progression of NAFLD.
The leakage of endotoxins from the gut, bacterial overgrowth and endotoxemia has been identified as a possible stimulus of NASH. Toll-like receptor 4 (TLR4), a lipopolysaccharide receptor, interacting with endotoxins, which leads to the release of a mass of proinflammatory mediators that induce hepatic injury and fibrogenesis (63,64). The importance of TLR4 signaling with Kupffer cells in the pathogenesis of steatohepatitis was verified in mice (65). TLR4 in Kupffer cells serve a crucial role in the genesis of NASH via ROS-dependent induction and X-box binding protein 1 activation. Dysregulation of TLR4 signaling as a result of SNPs, alters the ligand binding and balance between pro- and anti-inflammatory cytokines. It was reported that the rs4986790 (D299G) and rs4986791 (T399I) were related to IR, metabolic syndrome and T2DM (66). Nevertheless, a meta-analysis of 15,059 subjects drew a conclusion that Asp299Gly and Thr399Ile polymorphisms of the TLR4 gene makes no difference with increased risk of T2DM (67). In a small portion of the general population, Kiziltas et al (68) first revealed in humans that TLR4 signaling is pivotal for the pathogenesis of NASH, and the TLR4 heterozygous gene mutation (Asp299Gly) may have a preventive role against the genesis of NAFLD, which strongly suggests that TLR4 genotypes will be used as probable biomarkers in future clinical trials for developing antagonists and agonists of TLR4 to better manage patients with NAFLD.
Transforming growth factor (TGF)-β1 is another potent inducer of hepatic fibrogenesis. TGF-β1 mediates hepatic fibrosis via activating HSCs with the generation of ECM proteins. TGF-β1 also indirectly act through increasing connective tissue growth factors leading to liver fibrogenesis (69). In the human TGF-β1 locus, several SNPs have been described previously. Two commonly studied polymorphisms of the TGF-β1 gene are C-509T (rs1800469) and T-869C (rs1800470). The level of TGF-β1 expression resulted from allelic variants are significantly different (70). In the Egyptian population, the serum TGF-β1 level in hepatocirrhosis patients with the TGF-β1 −509TT genotype were remarkably increased compared with those with CT/CC genotypes, and the −509 T allele carriers were seven-fold more prone to develop liver cirrhosis than −509 C allele carriers (71), which is in agreement with the study in Italian populations (72). Additionally, the frequency of T allele is remarkably high in HCV patients, more frequently leading to progression of fibrosis (73). However, with respect to the T-869C polymorphism associated with HCC, there are some controversial results. The reasons for this large difference in risk in different studies is unknown.
7. Gene polymorphisms in mediators of related cytokines
TNF-α
TNF-α is a cytokine induced by macrophages and others cells (such as adipocytes and hepatocytes) with comprehensive functionality in metabolic regulation, inflammation and tumor apoptosis. Now, enhanced TNF-α expression has been demonstrated in NASH/NAFLD, and is therefore regarded as a risk biomarker of disease progression. TNF-α blocks IRS signaling and induces IR by triggering IkB kinase-β (IKK-β), the upstream activator of NF-κB, and other critical intracellular kinases, such as JNK activation. In animal models of genetic and diet-induced obesity, an improvement of NASH and IR were attained by various strategies targeting blockade of TNF-α activity. The TNF-α −238 promoter polymorphism was identified a higher prevalence in patients with NAFLD, and the −863 variant was in relation to the decrease of TNF-α concentration (74). Furthermore, lower LDL-cholesterol, BMI and susceptibility of NASH in Mexican and Chinese cohorts were observed in association with the −238 polymorphism. However, frequencies of variants −1031C and −863A were more prevalent in subjects with NASH in a different study (75). A meta-analysis evaluating polymorphisms in TNF-α confirmed the relationship of the −238 polymorphism with NAFLD, whereas the −308 polymorphism was not determined as a susceptible factor (76). Additionally, TNF-α G-308A, G-238A and C-863A polymorphisms are also related to HCC in Asians populations (77). It is still unclear whether, and certainly which, TNF-α polymorphism is relevant to the development of NASH. Further studies are in progress to focus on replicating results larger cohorts and different populations.
Interleukin (IL)-6
IL-6 is a cytokine involved controlling the balance between proinflammatory and anti-inflammatory pathways. The IL-6 gene is located on chromosome 7p21, and the −174 polymorphism of the IL-6 gene promoter is described to be in linkage disequilibrium with other commonly studied makers of inflammation. The G-174C of IL-6 gene polymorphism has been reported to relate to its transcription rate and the development of T2DM. In children with obesity, BMI, middle upper arm circumference, tricipital skin-fold thickness and serum albumin levels were demonstrated to have a correlation with the CC allele carriers of IL-6 gene (78). Moreover, the IL-6 −174C variant is more frequent in NASH individuals than that in cohorts with NAFLD. However, its possible role in NAFLD has not been fully studied. Currently, the presence of the GG genotype was observed to be in high frequency in HCV-related chronic hepatitis, liver cirrhosis and HCC (79). The variant predisposes to produce higher levels of IL-6 that contributes to the progression of HCC. In addition, the IL-6 receptor SNP (rs6684439) in Chinese patients with HBV is related to a lower risk of HCC (80), because of decreased levels of circulating sIL-6R.
8. Conclusions
NAFLD is the most common chronic liver disease that influences mortality. It is becoming a public health problem, especially the common progressive types of NAFLD, which ultimately progress to HCC. Patients with NAFLD frequently suffer from metabolic syndrome (81). The occurrence of NAFLD caused by many factors involving an overwhelmingly complex pathophysiological process and the joint action of multiple mechanisms. Ethnic background, gender, age and geographic location influence the results varied widely. The function of gene polymorphisms, which implicate IR, fatty acid metabolism, oxidative stress and hepatofibrogenesis, are reflected in every part of the pathogenesis of NAFLD. Therefore, obesity, impairment of glucose homeostasis and genetic influences are deemed as important pathogenic contributors in the advancing of NAFLD and NASH. The molecular mechanisms of oxidative stress, lipotoxicity and abnormal adipocytokine release caused by gene polymorphisms in NAFLD/NASH, have been gradually elucidated. It is of great significance for developing effective strategies in the future. However, due to the complexity and limitation of gene polymorphism research on NAFLD, most studies meet with the absence of statistical power, replication in various cohorts, well-matched samples and the presence of heterogeneities in disease stages, thus, researchers still do not completely understand the physiopathological mechanism of NAFLD caused by gene polymorphisms. This makes the research on NAFLD gene polymorphisms be filled with unknown and challenges. With the technological development and applications of disease gene location, the authors believe that a further breakthrough through studying NAFLD gene polymorphisms will be achieved to help identify an effective treatment for NAFLD.
Glossary
Key words
- NAFLD
nonalcoholic fatty liver disease
- NASH
nonalcoholic steatohepatitis
- HCC
hepatocellular carcinoma
- T2DM
type 2 diabetes
- IR
insulin resistance
- FFAs
free fatty acids
- TNF-α
tumor necrosis factor-α
- TG
triglyceride
- VLDL
very low-density lipoprotein
- ROS
reactive oxygen species
- AMPK
AMP protein kinase
- JNK
Jun N-terminal kinase
- NF-κB
nuclear factor-κB
- IRS
insulin receptor substrate
- ENPP1
ectoenzyme nucleotide pyrophosphate phosphodiesterase 1
- SNP
nucleotide polymorphism
- HDL
high-density lipoprotein
- SCD-1
stearoyl-CoA desaturase-1
- IGF-1
insulin-like growth factor-1
- PPAR-α/γ
peroxisome proliferator-activated receptor-α/γ
- BMI
body mass index
- ALT
alanine transaminase
- AST
aspartate aminotransferase
- PI3-K
phosphatidyl inositol 3-kinase
- MAPK
mitogen-activated protein kinase
- LepRb
leptin receptor
- LDL
low-density lipoprotein
- SREBP
sterol regulatory element binding protein
- UPC3
uncoupling protein 3
- MTTP
microsomal triglyceride transfer protein
- CM
chylomicron
- SCAP
SREBP cleavage activating protein
- MnSOD
manganese superoxide dismutase
- CYP4502E1
cytochrome P4502E1
- HFE
hemochromatosis
- TLR4
Toll-like receptor 4
- XBP-1
X-box binding protein 1
- TGF-β1
transforming growth factor-β1
- ECM
extracellular matrix
- IKK-β
IkB kinase-β
- IL-6
interleukin-6
References
- 1.Zhang X, Harmsen WS, Mettler TA, Kim WR, Roberts RO, Therneau TM, Roberts LR, Chaiteerakij R. Continuation of metformin use after a diagnosis of cirrhosis significantly improves survival of patients with diabetes. Hepatology. 2014;60:2008–2016. doi: 10.1002/hep.27199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Day CP, James OF. Steatohepatitis: A tale of two ‘hits’? Gastroenterology. 1998;114:842–845. doi: 10.1016/S0016-5085(98)70599-2. [DOI] [PubMed] [Google Scholar]
- 3.Anstee QM, Daly AK, Day CP. Genetics of alcoholic and nonalcoholic fatty liver disease. Semin Liver Dis. 2011;31:128–146. doi: 10.1055/s-0031-1276643. [DOI] [PubMed] [Google Scholar]
- 4.Lake AD, Novak P, Hardwick RN, Flores-Keown B, Zhao F, Klimecki WT, Cherrington NJ. The adaptive endoplasmic reticulum stress response to lipotoxicity in progressive human nonalcoholic fatty liver disease. Toxicol Sci. 2014;137:26–35. doi: 10.1093/toxsci/kft230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson N, Borlak J. Molecular mechanisms and therapeutic targets in steatosis and steatohepatitis. Pharmacol Rev. 2008;60:311–357. doi: 10.1124/pr.108.00001. [DOI] [PubMed] [Google Scholar]
- 6.Naik A, Belič A, Zanger UM, Rozman D. Molecular interactions between NAFLD and xenobiotic metabolism. Front Genet. 2013;4:2. doi: 10.3389/fgene.2013.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang D, Liu ZX, Choi CS, Tian L, Kibbey R, Dong J, Cline GW, Wood PA, Shulman GI. Mitochondrial dysfunction due to long-chain Acyl-CoA dehydrogenase deficiency causes hepatic steatosis and hepatic insulin resistance. Proc Natl Acad Sci USA. 2007;104:17075–17080. doi: 10.1073/pnas.0707060104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duvnjak M, Lerotić I, Barsić N, Tomasić V, Jukić L Virović, Velagić V. Pathogenesis and management issues for non-alcoholic fatty liver disease. World J Gastroenterol. 2007;13:4539–4550. doi: 10.3748/wjg.v13.i34.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bradbury MW, Berk PD. Lipid metabolism in hepatic steatosis. Clin Liver Dis. 2004;8(xi):639–671. doi: 10.1016/j.cld.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 10.de Almeida IT, Cortez-Pinto H, Fidalgo G, Rodrigues D, Camilo ME. Plasma total and free fatty acids composition in human non-alcoholic steatohepatitis. Clin Nutr. 2002;21:219–223. doi: 10.1054/clnu.2001.0529. [DOI] [PubMed] [Google Scholar]
- 11.Malaguarnera L, Di Rosa M, Zambito AM, dell'Ombra N, Nicoletti F, Malaguarnera M. Chitotriosidase gene expression in Kupffer cells from patients with non-alcoholic fatty liver disease. Gut. 2006;55:1313–1320. doi: 10.1136/gut.2005.075697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kadowaki T, Yamauchi T, Kubota N, Hara K, Ueki K, Tobe K. Adiponectin and adiponectin receptors in insulin resistance, diabetes, and the metabolic syndrome. J Clin Invest. 2006;116:1784–1792. doi: 10.1172/JCI29126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Müssig K, Heni M, Thamer C, Kantartzis K, Machicao F, Stefan N, Fritsche A, Häring HU, Staiger H. The ENPP1 K121Q polymorphism determines individual susceptibility to the insulin-sensitising effect of lifestyle intervention. Diabetologia. 2010;53:504–509. doi: 10.1007/s00125-009-1612-4. [DOI] [PubMed] [Google Scholar]
- 14.Dongiovanni P, Valenti L, Rametta R, Daly AK, Nobili V, Mozzi E, Leathart JB, Pietrobattista A, Burt AD, Maggioni M, et al. Genetic variants regulating insulin receptor signalling are associated with the severity of liver damage in patients with non-alcoholic fatty liver disease. Gut. 2010;59:267–273. doi: 10.1136/gut.2009.190801. [DOI] [PubMed] [Google Scholar]
- 15.Parola M, Marra F. Adipokines and redox signaling: Impact on fatty liver disease. Antioxid Redox Signal. 2011;15:461–483. doi: 10.1089/ars.2010.3848. [DOI] [PubMed] [Google Scholar]
- 16.Tokushige K, Hashimoto E, Noto H, Yatsuji S, Taniai M, Torii N, Shiratori K. Influence of adiponectin gene polymorphisms in Japanese patients with non-alcoholic fatty liver disease. J Gastroenterol. 2009;44:976–982. doi: 10.1007/s00535-009-0085-z. [DOI] [PubMed] [Google Scholar]
- 17.Wong VW, Wong GL, Tsang SW, Hui AY, Chan AW, Choi PC, So WY, Tse AM, Chan FK, Sung JJ, et al. Genetic polymorphisms of adiponectin and tumor necrosis factor-alpha and nonalcoholic fatty liver disease in Chinese people. J Gastroenterol Hepatol. 2008;23:914–921. doi: 10.1111/j.1440-1746.2008.05344.x. [DOI] [PubMed] [Google Scholar]
- 18.Wang BF, Wang Y, Ao R, Tong J, Wang BY. AdipoQ T45 G and G276 T polymorphisms and susceptibility to nonalcoholic fatty liver disease among Asian populations: A meta-analysis and meta-regression. J Clin Lab Anal. 2016;30:47–57. doi: 10.1002/jcla.21814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hashemi M, Bojd H Hanafi, Nasab E Eskandari, Bahari A, Hashemzehi NA, Shafieipour S, Narouie B, Taheri M, Ghavami S. Association of adiponectin rs1501299 and rs266729 gene polymorphisms with nonalcoholic fatty liver disease. Hepat Mon. 2013;13:e9527. doi: 10.5812/hepatmon.9527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muse ED, Obici S, Bhanot S, Monia BP, McKay RA, Rajala MW, Scherer PE, Rossetti L. Role of resistin in diet-induced hepatic insulin resistance. J Clin Invest. 2004;114:232–239. doi: 10.1172/JCI200421270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singhal NS, Patel RT, Qi Y, Lee YS, Ahima RS. Loss of resistin ameliorates hyperlipidemia and hepatic steatosis in leptin-deficient mice. Am J Physiol Endocrinol Metab. 2008;295:E331–E338. doi: 10.1152/ajpendo.00577.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomaru T, Steger DJ, Lefterova MI, Schupp M, Lazar MA. Adipocyte-specific expression of murine resistin is mediated by synergism between peroxisome proliferator-activated receptor gamma and CCAAT/enhancer-binding proteins. J Biol Chem. 2009;284:6116–6125. doi: 10.1074/jbc.M808407200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar S, Gupta V, Srivastava N, Gupta V, Mishra S, Mishra S, Shankar M Natu, Roy U, Chandra A, Negi MP, et al. Resistin 420C/G gene polymorphism on circulating resistin, metabolic risk factors and insulin resistance in adult women. Immunol Lett. 2014;162(2 Pt B):287–291. doi: 10.1016/j.imlet.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 24.Zhang LY, Jin YJ, Jin QS, Lin LY, Zhang DD, Kong LL. Association between resistin +299A/A genotype and nonalcoholic fatty liver disease in Chinese patients with type 2 diabetes mellitus. Gene. 2013;529:340–344. doi: 10.1016/j.gene.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 25.El-Shal AS, Pasha HF, Rashad NM. Association of resistin gene polymorphisms with insulin resistance in Egyptian obese patients. Gene. 2013;515:233–238. doi: 10.1016/j.gene.2012.09.136. [DOI] [PubMed] [Google Scholar]
- 26.Lu H, Sun J, Sun L, Shu X, Xu Y, Xie D. Polymorphism of human leptin receptor gene is associated with type 2 diabetic patients complicated with non-alcoholic fatty liver disease in China. J Gastroenterol Hepatol. 2009;24:228–232. doi: 10.1111/j.1440-1746.2008.05544.x. [DOI] [PubMed] [Google Scholar]
- 27.Swellam M, Hamdy N. Association of nonalcoholic fatty liver disease with a single nucleotide polymorphism on the gene encoding leptin receptor. IUBMB Life. 2012;64:180–186. doi: 10.1002/iub.597. [DOI] [PubMed] [Google Scholar]
- 28.Zain SM, Mohamed Z, Mahadeva S, Cheah PL, Rampal S, Chin KF, Mahfudz AS, Basu RC, Tan HL, Mohamed R. Impact of leptin receptor gene variants on risk of non-alcoholic fatty liver disease and its interaction with adiponutrin gene. J Gastroenterol Hepatol. 2013;28:873–879. doi: 10.1111/jgh.12104. [DOI] [PubMed] [Google Scholar]
- 29.Brondani LA, Assmann TS, de Souza BM, Bouças AP, Canani LH, Crispim D. Meta-analysis reveals the association of common variants in the uncoupling protein (UCP) 1–3 genes with body mass index variability. PLoS One. 2014;9:e96411. doi: 10.1371/journal.pone.0096411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mexitalia M, Yamauchi T, Utari A, Sjarif DR, Subagio HW, Soemantri A, Ishida T. The role of uncoupling protein 2 and 3 genes polymorphism and energy expenditure in obese Indonesian children. J Pediatr Endocrinol Metab. 2013;26:441–447. doi: 10.1515/jpem-2012-0311. [DOI] [PubMed] [Google Scholar]
- 31.Xu YP, Liang L, Wang CL, Fu JF, Liu PN, Lv LQ, Zhu YM. Association between UCP3 gene polymorphisms and nonalcoholic fatty liver disease in Chinese children. World J Gastroenterol. 2013;19:5897–5903. doi: 10.3748/wjg.v19.i35.5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hashemi M, Hoseini H, Yaghmaei P, Moazeni-Roodi A, Bahari A, Hashemzehi N, Shafieipour S. Association of polymorphisms in glutamate-cysteine ligase catalytic subunit and microsomal triglyceride transfer protein genes with nonalcoholic fatty liver disease. DNA Cell Biol. 2011;30:569–575. doi: 10.1089/dna.2010.1162. [DOI] [PubMed] [Google Scholar]
- 33.Tian Y, Li H, Wang S, Yan J, Chen Z, Li Z, Feng H, Zhou H, Ouyang D. Association of L-FABP T94A and MTP I128T polymorphisms with hyperlipidemia in Chinese subjects. Lipids. 2015;50:275–282. doi: 10.1007/s11745-015-3990-3. [DOI] [PubMed] [Google Scholar]
- 34.Peng XE, Wu YL, Lu QQ, Hu ZJ, Lin X. MTTP polymorphisms and susceptibility to non-alcoholic fatty liver disease in a Han Chinese population. Liver Int. 2014;34:118–128. doi: 10.1111/liv.12220. [DOI] [PubMed] [Google Scholar]
- 35.Gambino R, Cassader M, Pagano G, Durazzo M, Musso G. Polymorphism in microsomal triglyceride transfer protein: A link between liver disease and atherogenic postprandial lipid profile in NASH? Hepatology. 2007;45:1097–1107. doi: 10.1002/hep.21631. [DOI] [PubMed] [Google Scholar]
- 36.Wolff E, Vergnes MF, Defoort C, Planells R, Portugal H, Nicolay A, Lairon D. Cholesterol absorption status and fasting plasma cholesterol are modulated by the microsomal triacylglycerol transfer protein −493 G/T polymorphism and the usual diet in women. Genes Nutr. 2011;6:71–79. doi: 10.1007/s12263-010-0174-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siqueira ER, Oliveira CP, Correa-Giannella ML, Stefano JT, Cavaleiro AM, Fortes MA, Muniz MT, Silva FS, Pereira LM, Carrilho FJ. MTP −493G/T gene polymorphism is associated with steatosis in hepatitis C-infected patients. Braz J Med Biol Res. 2012;45:72–77. doi: 10.1590/S0100-879X2011007500160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dongiovanni P, Rametta R, Fracanzani AL, Benedan L, Borroni V, Maggioni P, Maggioni M, Fargion S, Valenti L. Lack of association between peroxisome proliferator-activated receptors alpha and gamma2 polymorphisms and progressive liver damage in patients with non-alcoholic fatty liver disease: Α case control study. BMC Gastroenterol. 2010;10:102. doi: 10.1186/1471-230X-10-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stienstra R, Saudale F, Duval C, Keshtkar S, Groener JE, van Rooijen N, Staels B, Kersten S, Müller M. Kupffer cells promote hepatic steatosis via interleukin-1beta-dependent suppression of peroxisome proliferator-activated receptor alpha activity. Hepatology. 2010;51:511–522. doi: 10.1002/hep.23337. [DOI] [PubMed] [Google Scholar]
- 40.Chen S, Li Y, Li S, Yu C. A Val227Ala substitution in the peroxisome proliferator activated receptor alpha (PPAR alpha) gene associated with non-alcoholic fatty liver disease and decreased waist circumference and waist-to-hip ratio. J Gastroenterol Hepatol. 2008;23:1415–1418. doi: 10.1111/j.1440-1746.2008.05523.x. [DOI] [PubMed] [Google Scholar]
- 41.Naito H, Yamanoshita O, Kamijima M, Katoh T, Matsunaga T, Lee CH, Kim H, Aoyama T, Gonzalez FJ, Nakajima T. Association of V227A PPARalpha polymorphism with altered serum biochemistry and alcohol drinking in Japanese men. Pharmacogenet Genomics. 2006;16:569–577. doi: 10.1097/01.fpc.0000220565.90466.79. [DOI] [PubMed] [Google Scholar]
- 42.Domenici FA, Brochado MJ, Martinelli AL, Zucoloto S, da Cunha SF, Vannucchi H. Peroxisome proliferator-activated receptors alpha and gamma2 polymorphisms in nonalcoholic fatty liver disease: A study in Brazilian patients. Gene. 2013;529:326–331. doi: 10.1016/j.gene.2013.06.091. [DOI] [PubMed] [Google Scholar]
- 43.Sahebkar A. Does PPARγ2 gene Pro12Ala polymorphism affect nonalcoholic fatty liver disease risk? Evidence from a meta-analysis. DNA Cell Biol. 2013;32:188–198. doi: 10.1089/dna.2012.1947. [DOI] [PubMed] [Google Scholar]
- 44.Yang Z, Wen J, Li Q, Tao X, Ye Z, He M, Zhang W, Huang Y, Chen L, Ling C, et al. PPARG gene Pro12Ala variant contributes to the development of non-alcoholic fatty liver in middle-aged and older Chinese population. Mol Cell Endocrinol. 2012;348:255–259. doi: 10.1016/j.mce.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 45.Cao CY, Li YY, Zhou YJ, Nie YQ, Wan YJ. The C-681G polymorphism of the PPAR-γ gene is associated with susceptibility to non-alcoholic fatty liver disease. Tohoku J Exp Med. 2012;227:253–262. doi: 10.1620/tjem.227.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gawrieh S, Marion MC, Komorowski R, Wallace J, Charlton M, Kissebah A, Langefeld CD, Olivier M. Genetic variation in the peroxisome proliferator activated receptor-gamma gene is associated with histologically advanced NAFLD. Dig Dis Sci. 2012;57:952–957. doi: 10.1007/s10620-011-1994-2. [DOI] [PubMed] [Google Scholar]
- 47.Bien CM, Espenshade PJ. Sterol regulatory element binding proteins in fungi: Hypoxic transcription factors linked to pathogenesis. Eukaryot Cell. 2010;9:352–359. doi: 10.1128/EC.00358-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nagaya T, Tanaka N, Suzuki T, Sano K, Horiuchi A, Komatsu M, Nakajima T, Nishizawa T, Joshita S, Umemura T, et al. Down-regulation of SREBP-1c is associated with the development of burned-out NASH. J Hepatol. 2010;53:724–731. doi: 10.1016/j.jhep.2010.04.033. [DOI] [PubMed] [Google Scholar]
- 49.Wang Y, Tong J, Chang B, Wang BF, Zhang D, Wang BY. Relationship of SREBP-2 rs2228314 G>C polymorphism with nonalcoholic fatty liver disease in a Han Chinese population. Genet Test Mol Biomarkers. 2014;18:653–657. doi: 10.1089/gtmb.2014.0116. [DOI] [PubMed] [Google Scholar]
- 50.Sun S, Wang M, Song H, Wu T, Wei H, He S, Ding Z, Ji G. SCAP gene polymorphisms decrease the risk of nonalcoholic fatty liver disease in females with metabolic syndrome. J Genet. 2013;92:565–570. doi: 10.1007/s12041-013-0280-9. [DOI] [PubMed] [Google Scholar]
- 51.Younossi ZM, Gorreta F, Ong JP, Schlauch K, Del Giacco L, Elariny H, van Meter A, Younoszai A, Goodman Z, Baranova A, et al. Hepatic gene expression in patients with obesity-related non-alcoholic steatohepatitis. Liver Int. 2005;25:760–771. doi: 10.1111/j.1478-3231.2005.01117.x. [DOI] [PubMed] [Google Scholar]
- 52.Al-Serri A, Anstee QM, Valenti L, Nobili V, Leathart JB, Dongiovanni P, Patch J, Fracanzani A, Fargion S, Day CP, et al. The SOD2 C47T polymorphism influences NAFLD fibrosis severity: Evidence from case-control and intra-familial allele association studies. J Hepatol. 2012;56:448–454. doi: 10.1016/j.jhep.2011.05.029. [DOI] [PubMed] [Google Scholar]
- 53.Krautbauer S, Eisinger K, Lupke M, Wanninger J, Ruemmele P, Hader Y, Weiss TS, Buechler C. Manganese superoxide dismutase is reduced in the liver of male but not female humans and rodents with non-alcoholic fatty liver disease. Exp Mol Pathol. 2013;95:330–335. doi: 10.1016/j.yexmp.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 54.Nahon P, Sutton A, Rufat P, Ziol M, Akouche H, Laguillier C, Charnaux N, Ganne-Carrié N, Grando-Lemaire V, N'Kontchou G, et al. Myeloperoxidase and superoxide dismutase 2 polymorphisms comodulate the risk of hepatocellular carcinoma and death in alcoholic cirrhosis. Hepatology. 2009;50:1484–1493. doi: 10.1002/hep.23187. [DOI] [PubMed] [Google Scholar]
- 55.Kathirvel E, Chen P, Morgan K, French SW, Morgan TR. Oxidative stress and regulation of anti-oxidant enzymes in cytochrome P4502E1 transgenic mouse model of non-alcoholic fatty liver. J Gastroenterol Hepatol. 2010;25:1136–1143. doi: 10.1111/j.1440-1746.2009.06196.x. [DOI] [PubMed] [Google Scholar]
- 56.Liu W, Tian F, Dai L, Chai Y. Cytochrome P450 2E1 gene polymorphism and alcohol drinking on the risk of hepatocellular carcinoma: A meta-analysis. Mol Biol Rep. 2014;41:7645–7650. doi: 10.1007/s11033-014-3655-y. [DOI] [PubMed] [Google Scholar]
- 57.Khan AJ, Ruwali M, Choudhuri G, Mathur N, Husain Q, Parmar D. Polymorphism in cytochrome P450 2E1 and interaction with other genetic risk factors and susceptibility to alcoholic liver cirrhosis. Mutat Res. 2009;664:55–63. doi: 10.1016/j.mrfmmm.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 58.Handa P, Morgan-Stevenson V, Maliken BD, Nelson JE, Washington S, Westerman M, Yeh MM, Kowdley KV. Iron overload results in hepatic oxidative stress, immune cell activation, and hepatocellular ballooning injury, leading to nonalcoholic steatohepatitis in genetically obese mice. Am J Physiol Gastrointest Liver Physiol. 2016;310:G117–G127. doi: 10.1152/ajpgi.00246.2015. [DOI] [PubMed] [Google Scholar]
- 59.Nelson JE, Brunt EM, Kowdley KV. Nonalcoholic Steatohepatitis Clinical Research Network: Lower serum hepcidin and greater parenchymal iron in nonalcoholic fatty liver disease patients with C282Y HFE mutations. Hepatology. 2012;56:1730–1740. doi: 10.1002/hep.25856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee SH, Jeong SH, Lee D, Lee JH, Hwang SH, Cho YA, Park YS, Hwang JH, Kim JW, Kim N, et al. An epidemiologic study on the incidence and significance of HFE mutations in a Korean cohort with nonalcoholic fatty liver disease. J Clin Gastroenterol. 2010;44:e154–e161. doi: 10.1097/MCG.0b013e3181d347d9. [DOI] [PubMed] [Google Scholar]
- 61.Nelson JE, Yeh MM, Kowdley KV. Relationship of C282Y HFE mutations, hepatic iron deposition and histologic features in patients with nonalcoholic fatty liver disease. Gastroenterology. 2011;140:S938–S939. [Google Scholar]
- 62.Valenti L, Fracanzani AL, Bugianesi E, Dongiovanni P, Galmozzi E, Vanni E, Canavesi E, Lattuada E, Roviaro G, Marchesini G, et al. HFE genotype, parenchymal iron accumulation, and liver fibrosis in patients with nonalcoholic fatty liver disease. Gastroenterology. 2010;138:905–912. doi: 10.1053/j.gastro.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 63.Rivera CA, Adegboyega P, van Rooijen N, Tagalicud A, Allman M, Wallace M. Toll-like receptor-4 signaling and Kupffer cells play pivotal roles in the pathogenesis of non-alcoholic steatohepatitis. J Hepatol. 2007;47:571–579. doi: 10.1016/j.jhep.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nagaya T, Tanaka N, Kimura T, Komatsu M, Tanaka E. Enhanced expression of toll-like receptor 4 and MyD88 is associated with disease progression in human nonalcoholic fatty liver disease. Gastroenterology. 2011;140:S–978. [Google Scholar]
- 65.Thuy S, Ladurner R, Volynets V, Wagner S, Strahl S, Königsrainer A, Maier KP, Bischoff SC, Bergheim I. Nonalcoholic fatty liver disease in humans is associated with increased plasma endotoxin and plasminogen activator inhibitor 1 concentrations and with fructose intake. J Nutr. 2008;138:1452–1455. doi: 10.1093/jn/138.8.1452. [DOI] [PubMed] [Google Scholar]
- 66.Peng D, Jiang F, Zhang R, Tang S, Chen M, Yan J, Sun X, Luo Y, Hu C, Jia W. Association of Toll-like Receptor 4 Gene polymorphisms with susceptibility to type 2 diabetes mellitus in the Chinese population. J Diabetes. 2015;7:485–492. doi: 10.1111/1753-0407.12206. [DOI] [PubMed] [Google Scholar]
- 67.Yin YW, Wang Q, Sun QQ, Hu AM, Liu HL. Toll-like receptor 4 gene Asp299Gly and Thr399Ile polymorphisms in type 2 diabetes mellitus: A meta-analysis of 15,059 subjects. Diabetes Res Clin Pract. 2015;107:338–347. doi: 10.1016/j.diabres.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 68.Kiziltas S, Ata P, Colak Y, Mesçi B, Senates E, Enc F, Ulasoglu C, Tuncer I, Oguz A. TLR4 gene polymorphism in patients with nonalcoholic fatty liver disease in comparison to healthy controls. Metab Syndr Relat Disord. 2014;12:165–170. doi: 10.1089/met.2013.0120. [DOI] [PubMed] [Google Scholar]
- 69.Tache D, Bogdan F, Pisoschi C, Baniţă M, Stănciulescu C, Fusaru AM, Comănescu V. Evidence for the involvement of TGF-β1-CTGF axis in liver fibrogenesis secondary to hepatic viral infection. Rom J Morphol Embryol. 2011;52:409–412. [PubMed] [Google Scholar]
- 70.Xiang TX, Cheng N, Li XN, Wu XP. Association between transforming growth factor-β1 polymorphisms and hepatocellular cancer risk: A meta-analysis. Hepatol Res. 2012;42:583–590. doi: 10.1111/j.1872-034X.2011.00958.x. [DOI] [PubMed] [Google Scholar]
- 71.Mohy A, Fouad A. Role of transforming growth factor-β1 in serum and − 509 C>T promoter gene polymorphism in development of liver cirrhosis in Egyptian patients. Meta Gene. 2014;2:631–637. doi: 10.1016/j.mgene.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Falleti E, Fabris C, Toniutto P, Fontanini E, Cussigh A, Bitetto D, Fornasiere E, Avellini C, Minisini R, Pirisi M. TGF-β1 genotypes in cirrhosis: Relationship with the occurrence of liver cancer. Cytokine. 2008;44:256–261. doi: 10.1016/j.cyto.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 73.Dai CY, Chuang WL, Lee LP, Pan WC, Huang JF, Hsieh MY, Hou NJ, Lin ZY, Chen SC, Hsieh MY, et al. Association between transforming growth factor-beta 1 polymorphism and virologic characteristics of chronic hepatitis C. Transl Res. 2008;152:151–156. doi: 10.1016/j.trsl.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 74.Valenti L, Fracanzani AL, Dongiovanni P, Santorelli G, Branchi A, Taioli E, Fiorelli G, Fargion S. Tumor necrosis factor alpha promoter polymorphisms and insulin resistance in nonalcoholic fatty liver disease. Gastroenterology. 2002;122:274–280. doi: 10.1053/gast.2002.31065. [DOI] [PubMed] [Google Scholar]
- 75.Tokushige K, Takakura M, Tsuchiya-Matsushita N, Taniai M, Hashimoto E, Shiratori K. Influence of TNF gene polymorphisms in Japanese patients with NASH and simple steatosis. J Hepatol. 2007;46:1104–1110. doi: 10.1016/j.jhep.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 76.Wang JK, Feng ZW, Li YC, Li QY, Tao XY. Association of tumor necrosis factor-α gene promoter polymorphism at sites −308 and −238 with non-alcoholic fatty liver disease: A meta-analysis. J Gastroenterol Hepatol. 2012;27:670–676. doi: 10.1111/j.1440-1746.2011.06978.x. [DOI] [PubMed] [Google Scholar]
- 77.Wei Y, Liu F, Li B, Chen X, Ma Y, Yan L, Wen T, Xu M, Wang W, Yang J. Polymorphisms of tumor necrosis factor-alpha and hepatocellular carcinoma risk: A HuGE systematic review and meta-analysis. Dig Dis Sci. 2011;56:2227–2236. doi: 10.1007/s10620-011-1617-y. [DOI] [PubMed] [Google Scholar]
- 78.Marginean O, Banescu C, Duicu C, Marginean M Oana, Marginean C. The role of interleukin-6 gene 572G/C polymorphism in child obesity. Clin Nutr. 2014;33:S117. doi: 10.1016/S0261-5614(14)50303-2. [DOI] [Google Scholar]
- 79.Giannitrapani L, Soresi M, Balasus D, Licata A, Montalto G. Genetic association of interleukin-6 polymorphism (−174 G/C) with chronic liver diseases and hepatocellular carcinoma. World J Gastroenterol. 2013;19:2449–2455. doi: 10.3748/wjg.v19.i16.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Deng Y, Li M, Wang J, Xie L, Li T, He Y, Lu Q, Li R, Tan A, Qin X, et al. Susceptibility to hepatocellular carcinoma in the Chinese population-associations with interleukin-6 receptor polymorphism. Tumour Biol. 2014;35:6383–6388. doi: 10.1007/s13277-014-1863-7. [DOI] [PubMed] [Google Scholar]
- 81.Bedogni G, Miglioli L, Masutti F, Tiribelli C, Marchesini G, Bellentani S. Prevalence of and risk factors for nonalcoholic fatty liver disease: The Dionysos nutrition and liver study. Hepatology. 2005;42:44–52. doi: 10.1002/hep.20734. [DOI] [PubMed] [Google Scholar]