Abstract
Delineation and noise removal play a significant role in clinical quantification of PET images. Conventionally, these two tasks are considered independent, however, denoising can improve the performance of boundary delineation by enhancing SNR while preserving the structural continuity of local regions. On the other hand, we postulate that segmentation can help denoising process by constraining the smoothing criteria locally. Herein, we present a novel iterative approach for simultaneous PET image denoising and segmentation. The proposed algorithm uses generalized Anscombe transformation priori to non-local means based noise removal scheme and affinity propagation based delineation. For non-local means denoising, we propose a new regional means approach where we automatically and efficiently extract the appropriate subset of the image voxels by incorporating the class information from affinity propagation based segmentation. PET images after denoising are further utilized for refinement of the segmentation in an iterative manner. Qualitative and quantitative results demonstrate that the proposed framework successfully removes the noise from PET images while preserving the structures, and improves the segmentation accuracy.
Keywords: PET Denoising, PET Segmentation, Regional Means, Affinity Propagation, Generalized Anscombe Transformation
1 Introduction
Positron emission tomography (PET) reveals metabolic activities by detecting gamma photon pairs caused by positron annihilation from radiotracers localized to specific regions. It provides diagnostic and therapeutic interpretation for evaluation of many diseases. As compared with anatomical imaging techniques of computed tomography (CT) and magnetic resonance imaging, PET images are known by their high sensitivity and low spatial resolution. Moreover, PET images suffer from noise caused by random and scattered coincidences, and has low signal-to-noise ratios (SNRs).
Quantitative measurements of PET images such as mean and maximum standardized uptake value (SUVmax and SUVmean) and metabolic tumor volume (MTV) are strongly affected by the denoising and segmentation. For instance, noise distribution in PET images (non-Gaussian) [1] degrades the sensitivity of SUV-based quantitative metrics and affects the correct boundary of lesions. Meanwhile, object information through delineation can help promoting the denoising performance. However, joint interaction between denoising and delineation is often ignored by conventional approaches [2,3]. For denoising, the major challenge is to reduce noise while preserving the small structures under low resolution condition. Incorporating knowledge of small structures can address this challenge. Therefore, segmentation can improve the performance of denoising by introducing additional constraints for smoothing process. For segmentation, on the other hand, the challenge is to robustly extract the regions under low SNR. Hence, denoising can promote the accuracy of segmentation by increasing SNR as long as there is no resolution loss. In order to address this coupled problem, an efficient segmentation algorithm is required to estimate boundaries of metabolically active uptake regions; and an accurate denoising method is desirable to enhance SNR.
Current approaches in PET denoising focus on Gaussian smoothing, adaptive diffusion filtering [2], filtering in transformation domain with anatomical information [3], and soft thresholding method [1]. Since Gaussian smoothing and adaptive diffusion filtering rely on neighboring voxels, they are only able to catch local similarities. As PET images are low resolution, such methods are not optimal and cause local blurring and information loss. Incorporation of anatomical information can help defining the candidate structures, however, it may create artifacts due to the fact that anatomical-functional correspondence does not always hold for all locations. Soft thresholding methods have been shown to be effective in noise removal; however, accurately modeling noise distribution is a challenging problem. Recently, non-local means method [4] gained increasing attention for its effectiveness in noise reduction and structure preservation of low SNR images by considering global similarity measurement instead of intensity gradient alone.
Detailed noise characteristics of PET images are still unknown. A Poisson model is proposed recently [1] where noise is transformed to Gaussian using variance stabilizing transform (VST). In this study, we assume both multiplicative and additive noise component in a mixed Poisson-Gaussian model because pure Poisson modeling may be suboptimal either. Next, the generalized Anscombe transformation (GAT) with its optimal inversion [5] were utilized for variance stabilization.
To address all challenges described above, in this paper, we propose an iterative approach for simultaneous PET image denoising and segmentation by (1) stabilizing noise under mixed Poisson-Gaussian model with GAT, (2) estimating local uptake regions using affinity propagation (AP) clustering, (3) removing Gaussianized noise using regional means within the non-local means framework by incorporating class information from segmentation, and (4) applying the optimal inverse GAT (IGAT) to obtain denoised PET images. Steps (2) and (3) are utilized in an iterative manner to enhance the performance of each other and final segmentation result is the clustering result of (2) at convergence. The proposed method can be performed on either 2-D or 3-D, and the presented results are using 3-D algorithm. A flowchart for the presented scheme is shown in Fig. 1. In the next section, the proposed framework is presented in detail.
Fig. 1.
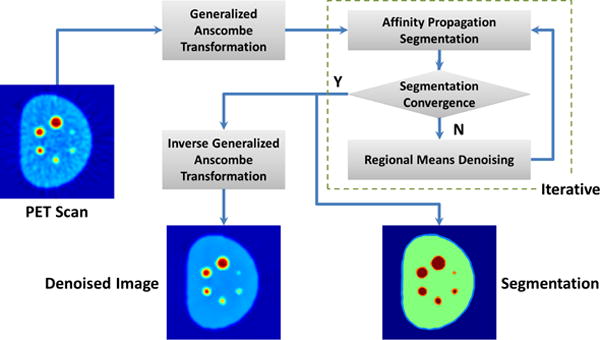
Flowchart of the iterative segmentation based denoising algorithm
2 Methods
In the proposed framework, we incorporate the region information from segmentation process for promoting the performance of denoising and the resulting image with higher SNR can enhance the performance of segmentation in an iterative manner.
Generalized Anscombe Transformation and Optimal Inverse
GAT is the general version of the classical Anscombe transformation designed for stabilizing the noise variance assuming a mixed Poisson-Gaussian model. According to the theorem, intensities are modeled as a scaled Poisson variable corrupted by additive Gaussian noise. Let p denote the Poisson variable with variance λ, α denote the scale parameter, and n denote the Gaussian noise with mean μ and standard deviation σ, we have the voxel intensity as
| (1) |
where p ∼ and . Noise stabilization can further be achieved by GAT as
| (2) |
so that the resulting y = GAT (x) has approximately unity variance. Once the PET images are transformed with GAT, Gaussianized noise can be removed with the proposed method, leading to a better delineation performance. Final step is the inverse GAT to transform the denoised PET images back to original image domain. Herein, we use the the exact unbiased inverse of the GAT [5] to recover the intensity information.
Affinity Propagation Based PET Image Segmentation
We propose an iterative way to tackle delineation of uptake regions in PET images by combining the power of a robust segmentation and structure preserving denoising algorithms. For a robust and accurate segmentation algorithm, we use affinity propagation (AP) algorithm [6] recently shown to be very effective for PET images [7]. Other PET segmentation techniques can instead be used for this purpose as long as highly accurate delienation results are guaranteed. Due to page limitation, we refer our readers the original AP paper in [7] that we follow in our study for the details of the novel similarity functions and delineation steps. Briefly, AP clustering is utilized to find optimal thresholding levels that separate the PET image into several regions.
Regional Means Denoising
AP clustering yields a robust estimation of uptake regions, although the group separation can be inaccurate locally due to intensity variation under low SNR. This information serves as a “pre-screening” for non-local means denoising, and hence we introduce the notion “regional means” for this technique. The key idea is to enhance SNR while preserve the true uptake regions, so that the enhanced image can help AP to generate a better segmentation result that in turn benefit the denoising in the next iteration. The basic formulation for non-local means [4] filtering is that for a point in image, its estimated value after filtering is the weighted average of all the points in the image instead of only its neighbors. The method can be performed on both 2-D and higher dimensional images. In the following, without loss of generality, we present a 3-D formulation for the proposed method with which the results were generated. The basic non-local means algorithm utilizes the redundant information of structural patterns within the image by considering the similarity between local patches with size N × N × N, which provides a more reliable reduction of noise than the conventional local intensity schemes, as it enables more robust comparison than conventional neighborhood filters. However, for computational purposes, the search of similar patches is usually restricted in a larger “search window” of M × M × M(M > N). Therefore, from implementation perspective, the information utilization is restricted. Since approximate region estimation can be efficiently performed by AP, it is possible to remove this computation restriction of search window. Instead of searching the entire image or restricted within local neighborhood, we propose the regional means scheme as follows.
For point u in GAT transformed image J = GAT (I), its intensity is J(u) and class label given by AP segmentation is L(u) with corresponding group size G(L(u)). Class labels are ordered consecutive natural number such that L(u) > L(v), if J(u) > J(v). In order to determine its similar patches over the image, we search the following regions Ω:
Local search window of size M × M × M,
Random sample min{M3, G(L(u))} points in the regions with class label L(u) (candidate region),
Random sample min{M3, G(L(u)−1)} points in the regions with class label L(u) − 1 (neighbor region I) if L(u) > 1,
Random sample min{M3, G(L(u) + 1)} points in the regions with class label L(u) + 1 (neighbor region II) if L(u) < maxu L(u).
Last step is to apply regional means to point u as
| (3) |
where the weights w(u, v) depend on the similarity between the two patches and centered at point u and v as
| (4) |
where represents the Euclidean distance between the intensity vectors from the two intensity vectors from the two patches. is the normalizing constant such that
| (5) |
so that the weighting parameter satisfies the weighting conditions of 0 ≤ w(u, v) ≤ 1 and Σvw(u,v)=1, parameter h determines the degree of filtering. In this way, the resulting Ω is not restricted to local areas anymore, but with the expense of increased computational complexity by the order of four. With the information from segmentation, our technique is able to cover sufficient points that can contribute to the denoising at point u.
3 Experiments and Results
In order to evaluate both denoising and segmentation performance, we used 20 PET-CT images on two different NEMA phantoms with different reconstruction parameters. The first phantom contains six spheres with diameters of 10, 13, 17, 22, 28, and 37 mm, background concentration is 0.44 uCi/ml, and hot sphere concentration is 1.75 uCi/ml. The spatial resolution is 128×128×47 with spacing 2.73×2.73×3.27 mm, and the image is in units of Bq/ml. The second phantom has five spheres with diameters of 4, 5, 6, 8, and 10 mm. The true activities are 32.2 mCi/ml in the spheres and 6.2mCi/ml in the background. The spatial resolution is 256×256×95 with spacing 0.95×0.95×1.90 mm, and the image is in units of mCi/ml. In addition to phantom images, 20 MRI-PET images relating to different diseases were collected with IRB approval from 20 patients. The spatial resolution is 172×172 in plane with slice number from 189–211, and spacing 4.17×4.17×2.00 mm. The denoising performance of the proposed iterative regional means method is compared with commonly used methods including Gaussian filtering, anisotropic diffusion, non-local means, and block matching [8] methods. All methods for comparison were performed over the image after GAT stabilization. The segmentation performance is compared with CT ground truth for phantom images, and manual reference for patient images.
Evaluation of Denoising
Using phantom images, the boundaries between each uptake region and background can be accurately defined from CT scans. For patient scans, 20 regions with lesions were manually determined from all subjects by experts. Statistics of noise reduction performance, including SNR and max/mean uptake value, were then computed for all uptake regions from several ROIs within each of them (the convention for PET image measurements). The ROIs were identical among original and filtered images for comparison.
Qualitative results at sample slices from both phantom and human images are shown in Fig. 2 with the original image (A), the corresponding CT/MRI (B), the filtering result of Gaussian filtering (C), anisotropic diffusion (D), non-local means (E), block matching (F), and the proposed method (G). The red arrows show noise reduction areas where other methods show limited success; while the yellow arrows point out the preserved fine details with the proposed method where other methods over-smooth and blur the small structures.
Fig. 2.
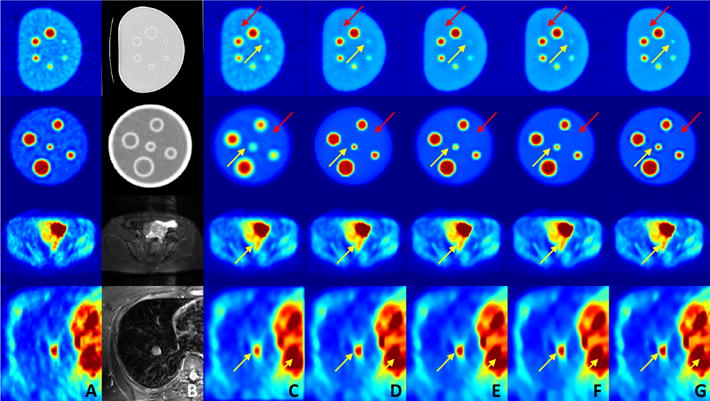
Qualitative results for denoising: the original image (A), the corresponding CT/MR image (B), the filtering result of Gaussian filtering (C), anisotropic diffusion (D), non-local means (E), block matching (F), and the proposed method (G). Yellow arrows point out the small uptake regions and red arrows shows the residual noise regions.
Quantitative analysis confirms the qualitative observation that the proposed method effectively removed the noise from PET image and preserved the fine details of small regions. As shown in Fig. 3 A/D, ROIs were divided into high uptake regions (blue) and low uptake regions (red). Low uptake background regions are not influenced by partial volume effect; thus, provide reliable estimation of overall Signal-to-Noise Ratio (SNR). Further, relative contrast (RC) were calculated for high uptake regions and compared with low uptake regions, measuring object-to-background contrast relative to the noise in each region.
| (6) |
where , , , and denote the mean and standard deviation of high / low uptake regions. For denoising methods to be effective, two most commonly used markers for PET images, SUVmax and SUVmean, should not be changed significantly before and after denoising. Fig. 3 C/E illustrates an intensity profile along a sample line within the PET image Fig. 3 B/D, as compared with the block matching method (blue), which is the state-of-the-art for denoising, the proposed algorithm (red) successfully preserved the intensity level for different objects and minimized the noise from the original image (black). Table 1 presents the quantitative results of SNR, RC, max and mean value reduction rate (RR) of the ROIs as well as the ratio of uptake values as compared with phantom ground truth for object and background regions - object ratio (OR), and background ratio (BR). Our experimental results show that the proposed method outperforms other methods in all measurements for the task of PET image denoising. Note that for human subjects (H), OR and BR are not available because the true local uptake value is not known unlike in phantom case (P).
Fig. 3.
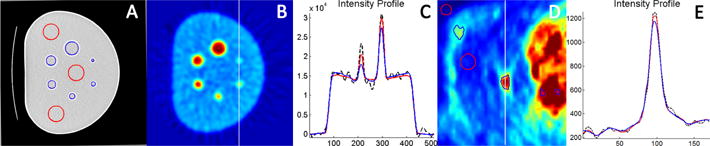
(A, D) ROI distribution in high uptake regions (blue) and low uptake regions (red), (B, D) original image showing sample line location, and (C, E) intensity profile on the sample line for original image (black), block matching method (blue), and proposed method (red).
Table 1.
Statistics of denoising performance for different methods. P: phantom; H: human.
| SNR | RC | Max RR | Mean RR | OR | BR | |||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| P | H | P | H | P | H | P | H | P | P | |
|
| ||||||||||
| Original | 11.73 | 7.08 | 11.32 | 34.14 | 0% | 0% | 0% | 0% | 59.28% | 85.02% |
| Gaussian | 15.62 | 7.01 | 12.88 | 27.76 | 11.08% | 9.27% | 9.55% | 5.51% | 53.99% | 85.21% |
| Diffusion | 21.05 | 7.25 | 16.38 | 33.98 | 13.20% | 7.20% | 7.37% | 3.37% | 55.99% | 85.72% |
| Non-Local Means | 21.88 | 7.69 | 13.70 | 37.67 | 13.27% | 7.04% | 11.03% | 3.61% | 54.41% | 85.89% |
| Block Matching | 20.36 | 9.92 | 14.02 | 39.97 | 7.32% | 6.96% | 6.87% | 3.59% | 56.27% | 85.41% |
| Proposed | 35.55 | 11.82 | 19.15 | 52.40 | 2.97% | 3.55% | 2.61% | 1.29% | 58.24% | 86.26% |
Evaluation of Segmentation
Fig. 4 illustrates the segmentation result given by AP over the phantom image and denoised results. The regions (B) defined by CT image (A) are used as ground truth for assessing segmentation performance. As shown, initial segmentation result (C) can include redundant thresholding levels due to the noise, while the final result recovers the information by enhancing the image (D). Other de-noising techniques are also able to reduce the noise, however, local structures are not preserved (E). Dice similarity coefficient (DSC), and Hausdorff distance (HD) are calculated for further evaluation, and the result are 92.75% for DSC and 3.14 mm for HD (pixel size 2.73 × 2.73 mm).
Fig. 4.
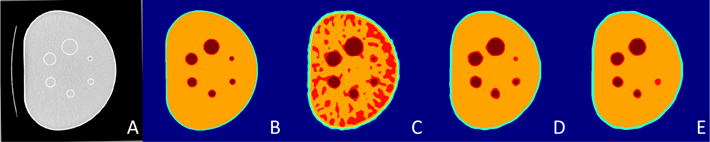
Segmentation results: (A) corresponding CT image, (B) ground truth from CT image, (C) initial segmentation result, (D) final segmentation result given by proposed method, (E) final segmentation result from image denoised by block matching method.
4 Discussion and Conclusion
In this study, we presented an effective tool for PET image denoising and segmentation. The proposed algorithm adopts affinity propagation for estimating different uptake regions, this information is further incorporated within a regional means denoising technique to enhance the SNR, which in turn helps promoting the accuracy of segmentation algorithm in an iterative manner. We also utilized generalized Anscombe transformation and its optimal inverse before and after the denoising-segmentation procedure, in order to Gaussianize the noise in PET images under mixed Poisson-Gaussian model. Experimental results demonstrated that the proposed framework effectively removes the noise from PET images while the structures are preserved, especially for small uptake regions.
Footnotes
This research is supported by CIDI, the intramural research program of the National Institute of Allergy and Infectious Diseases (NIAID) and the National Institute of Biomedical Imaging and Bioengineering (NIBIB).
References
- 1.Bagci U, Mollura DJ. Denoising PET images using singular value thresholding and stein’s unbiased risk estimate. In: Mori K, Sakuma I, Sato Y, Barillot C, Navab N, editors. MIC-CAI 2013, Part III. LNCS. Vol. 8151. Springer; Heidelberg: 2013. pp. 115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tauber C, Stute S, Chau M, Spiteri P, Chalon S, Guilloteau D, Buvat I. Spatiotemporal diffusion of dynamic PET images. Physics in Medicine and Biology. 2011;56(20):65–83. doi: 10.1088/0031-9155/56/20/004. [DOI] [PubMed] [Google Scholar]
- 3.Turkheimer FE, Boussion N, Anderson AN, Pavese N, Piccini P, Visvikis D. PET image denoising using a synergistic multiresolution analysis of structural (MRI/CT) and functional datasets. Journal of Nuclear Medicine. 2008;49(4):657–666. doi: 10.2967/jnumed.107.041871. [DOI] [PubMed] [Google Scholar]
- 4.Buades A, Coll B, Morel JM. A non-local algorithm for image denoising. IEEE Computer Society Conference on Computer Vision and Pattern Recognition. 2005;2:60–65. [Google Scholar]
- 5.Makitalo M, Foi A. Optimal inversion of the generalized Anscombe transformation for Poisson-Gaussian noise. IEEE Transactions on Image Processing. 2013;22(1):91–103. doi: 10.1109/TIP.2012.2202675. [DOI] [PubMed] [Google Scholar]
- 6.Frey BJ, Dueck D. Clustering by passing messages between data points. Science. 2007;315:972–976. doi: 10.1126/science.1136800. [DOI] [PubMed] [Google Scholar]
- 7.Foster B, Bagci U, Xu Z, Dey B, Luna B, Bishai W, Jain S, Mollura D. Segmentation of PET images for computer-aided functional quantification of tuberculosis in small animal models. IEEE Transactions on Biomedical Engineering. 2014;61(3):711–724. doi: 10.1109/TBME.2013.2288258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dabov K, Foi A, Katkovnik V, Egiazarian K. Image denoising by sparse 3-D transform-domain collaborative filtering. IEEE Transactions on Image Processing. 2007;16(8):2080–2095. doi: 10.1109/tip.2007.901238. [DOI] [PubMed] [Google Scholar]


