Highlights
-
•
Complex glycans play multifaceted roles in interplay between virus and its host.
-
•
Host glycans serve co-receptors or specific primary receptors for viral entry.
-
•
Viral glycan – host lectin or antibody interaction favors either virus or host.
-
•
This review covers role of glycan–protein interactions in viral pathogenesis.
Abstract
The surfaces of host cells and viruses are decorated by complex glycans, which play multifaceted roles in the dynamic interplay between the virus and the host including viral entry into host cell, modulation of proteolytic cleavage of viral proteins, recognition and neutralization of virus by host immune system. These roles are mediated by specific multivalent interactions of glycans with their cognate proteins (generally termed as glycan-binding proteins or GBPs or lectins). The advances in tools and technologies to chemically synthesize and structurally characterize glycans and glycan–GBP interactions have offered several insights into the role of glycan–GBP interactions in viral pathogenesis and have presented opportunities to target these interactions for novel antiviral therapeutic or vaccine strategies. This review covers aspects of role of host cell surface glycan receptors and viral surface glycans in viral pathogenesis and offers perspectives on how to employ various analytical tools to target glycan–GBP interactions.
Current Opinion in Structural Biology 2016, 40:153–162
This review comes from a themed issue on Carbobydrate-protein interactions and glycosylation
Edited by Nagasuma R Chandra and Harry J Gilbert
For a complete overview see the Issue and the Editorial
Available online 25th October 2016
http://dx.doi.org/10.1016/j.sbi.2016.10.003
0959-440X/© 2016 Published by Elsevier Ltd.
Introduction
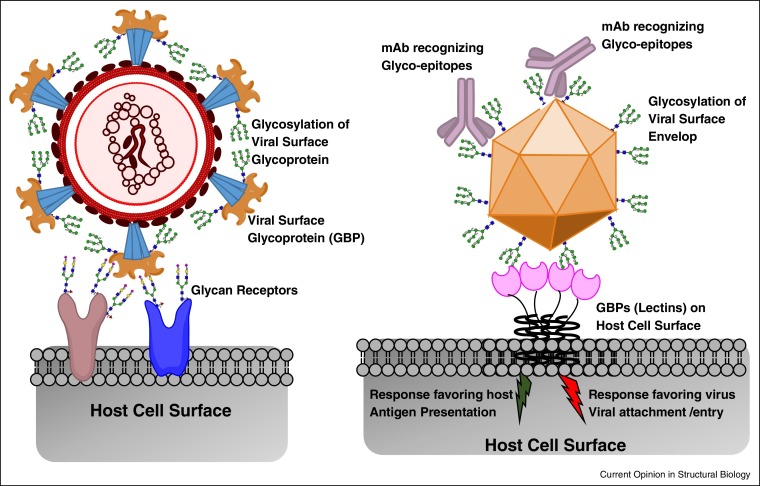
Complex glycans decorate surfaces of both host cells (and tissues) and viruses and play multifaceted roles in interactions between the viruses and host organisms that critically govern viral pathogenesis [1••, 2•, 3•, 4]. These complex glycans are attached N-linked or O-linked to proteins as a part of post-translational modifications or attached to lipids. The complexity and structural heterogeneity of the glycans displayed on host cell/tissue surface and on viral surface glycoproteins predominantly arises from the complex non-template driven biosynthetic machinery of the host cell involving several enzymes that show tissue-specific expression patterns [5, 6, 7]. The interplay between the virus and the host involves interactions between two surfaces decorated by complex glycans. In addition to the glycans, a critical component involved in this interplay is a set of proteins known as glycan-binding proteins (GBPs) or lectins (Figure 1 ).
Figure 1.
Schematic of complex glycans in the interplay between virus and host. Shown in the figure (on the left) is a schematic of a virus surface glycoprotein (such as influenza A virus hemagglutinin) that recognized glycans on the host cell surface as their primary receptors for viral attachment and entry. The viral surface protein is in itself glycosylated and depending on the site of glycan occupancy, the glycosylation would impact the binding of this protein to the host-cell glycan receptor. Shown on the right is a schematic of glycan on the surface of virus envelop proteins (such as dengue) interacting with GBP anchored on the host cell. This interaction could either be beneficial for the virus wherein it plays a role in viral attachment and entry into a cell capable of promoting the productive infection or it could be beneficial to the host wherein antigen presenting cells could uptake the virus and prime the host immune response.
The GBPs involved in virus–host interplay mediated by glycans are typically anchored on the surfaces of cells (for example C-type lectins, Siglecs, etc. [1••, 8] and viruses (envelop proteins or spike glycoproteins) [2•, 3•, 4]. The functional unit of GBP is often multimeric comprising of homo-oligomers of individual protein domains each having a glycan-binding site [1••]. Each glycan-binding site shows specific recognition to glycan motif or substructure of the complex glycan structure comprising of 2-5 sugars; however, the binding affinity for a given site to a glycan motif is typically low to moderate. The binding affinity and specificity are further enhanced through avidity and multivalency wherein multiple glycan motifs displayed on the surface bind to multiple glycan binding sites in the GBP oligomer unit [1••, 9].
The complex glycans displayed on host cell surfaces typically act as attachment factors, co-receptors or primary receptors that are specifically recognized by the viral surface glycoproteins. For example, complex glycans terminated by α2-3 or α2-6-linked sialic acid (N-acetyl neuraminic acid) act as receptors for several different viruses [10•]. Linear sulfated glycosaminoglycans such as heparan sulfate act as co-receptors for a variety of viruses [11] including dengue virus [12, 13], hepatitis C virus [14], and foot-and-mouth disease virus [15]. The predominant display of specific glycan motifs on surfaces of different cells and tissues contributes to the host restriction and cell/tissue tropism of the virus. As an example, the human upper respiratory epithelial surface predominantly display sialylated glycan receptors terminated by α2-6-linked sialic acid and these receptors are specifically recognized by hemagglutinin glycoprotein (HA) on surface of influenza A viruses that are known to infect and transmit via respiratory droplets in humans [4, 16, 17, 18]. On the other hand, when influenza A viruses that are commensal or epizootic in birds infect humans, they typically affect deep lung and other tissues that predominantly display sialylated glycan receptors terminated by α2-3-linked sialic acid and are unable to transmit efficiently via respiratory droplets.
The glycans displayed on viral surfaces are posttranslational modifications of envelop proteins in viruses such as flaviviruses including dengue and zika virus or of glycoprotein spikes as observed in influenza A virus, Ebola virus, etc. These glycans are added to the viral glycoproteins as a part of the host–cell glycan biosynthesis upon viral replication in the host. In most cases, glycosylation sites on viral surface proteins are highly conserved since the glycans at these sites critically maintain the stability of these proteins and the viral particles as a whole. In addition to maintaining the stability of the viral particles, these glycans also play key roles in mediating infection of host cells by certain viruses such as dengue and Ebola viruses through specific interactions with GBPs (such as C-type lectins) displayed on the host cell surface [1••, 19].
The complex glycans on the viral surface also play a key role in host immune response to counter the viral infection. While GBPs anchored on surfaces of host cells such as dendritic cells (DCs) play a role in viral entry, it also plays a dual role to enhance antigen presentation and processing for adaptive immune response [1••]. Sites of N-linked glycosylation are often positively selected during evolution of the virus in human host to increase glycans on the viral surface so as to present glycans that mimic self-antigens and mask the underlying protein epitope which in turn permits the virus to evade host immune response. In other cases, particularly with HIV, the clustering of glycosylation sites on the gp120 surface glycoprotein present novel glyco-eptiopes that do not mimic self-antigens and therefore lead to potent neutralization by antibodies that target these novel eptiopes across a broad spectrum of viral strains [20••]. This review covers some general concepts on role of host cell surface glycan receptors and viral surface glycans in viral pathogenesis. Some perspectives on developing the appropriate tools to define target glycan motifs on host cells or viral surface towards developing antiviral strategies is also presented.
Glycans on host surface mediating viral entry, infection and tropism
The viral surface glycoproteins have evolved to specifically recognize distinct glycan motifs on the host cell surface and these specific interactions are one of the many factors that contribute to virus entry into specific host cells and host tissue tropism. Depending on the virus, the host cell surface glycans act as general attachment factors, co-receptors or primary receptors that mediate viral infection and entry.
Acidic glycans such as linear heparan sulfate glycosaminoglycan polysaccharides or branched glycans terminated by sialic acid displayed on cell surfaces in many instances play a role in attracting viruses based on their negative charge and serve as initial attachment factors to the host cell. Specifically, heparan sulfate by virtue of its chain length and distribution of sulfate groups has been implicated to play key role as attachment factor or co-receptors mediating the initial attachment of several viruses including HIV [21, 22], enterovirus EV71 [23, 24], echovirus E5 [25], coxsackievirus A9 and B3 strains [26, 27], dengue virus [12, 13] and Ebola virus [28]. An established example of specific modification in heparan sulfate that mediates virus entry is that of 3-O sulfation of the glucosamine that plays a critical role in specific interactions with herpes simplex virus [29, 30, 31].
Sialylated glycan receptors play a more directed role in mediating binding, attachment and entry of viruses into host cells [10•]. It has been demonstrated that enterovirus EV-D68 binds sialylated glycan receptors along the canyon of the viral surface and that this binding event causes significant conformational change in the viral envelop that displaces the fatty acid (also known as pocket factor which maintains structural stability of the envelop) and prepares the virus for infection [32, 33]. Furthermore, it has been indicated that sialylated glycan receptors specifically in the context of O-linked glycans and glycolipids and not in the context of N-linked glycans mediate infection of EV-71 virus [34]. Picornaviruses including coxsackievirus A24 and EV-70 which cause severe conjunctivitis have been demonstrated to preferentially bind to α2-6-linked sialylated glycan receptors [35, 36, 37, 38, 39].
The influenza A virus is among the best studied viruses that are known to bind to host cell surface sialylated glycan receptors. The binding specificity of the viral surface hemagglutinin (HA) to sialylated glycan receptors on the host cell surface is one of many factors that critically govern adaptation of influenza to the human host. Avian virus HA binds with high specificity and affinity to glycans terminated by α2-3-linked sialic acid which are found in abundance in avian gut and lower respiratory tract of humans [40, 41, 42]. Human virus HAs possess characteristic glycan receptor binding properties; their HA predominantly binds with high affinity (or avidity) to glycan receptors terminated by α2-6-linked sialic acid, which are predominantly expressed in the upper respiratory epithelia of humans [40, 43, 44]. The human upper respiratory epithelium is the primary target site for infection of human-adapted viruses and is thought to be a prerequisite for efficient human-to-human transmission via respiratory droplets [4]. In addition to HA, influenza A virus surface also has an enzyme that cleaves sialic acid from both viral surface and host cell surface glycans to enhance productivity of infection, prevent self-aggregation of new virions that emerge from productive infection in a host cell [45, 46]. In fact one of the earliest antiviral therapeutic strategies based on host cell surface glycan receptors was the design of sialic acid analogues (oseltamivir, zanamivir, etc.) to specifically bind to and inactivate the influenza A virus NA [47].
In addition to acidic glycans, neutral glycans such as histo-blood group antigens present on bodily secretions and also epithelial surface of the human intestine play a key role in mediating initial infection and entry of most norovirus genotypes. The characteristic motif among these histo-blood group antigens is a fucose α1-2 linked to galactose which is specifically recognized by protruding domain (P) of the viral capsid [48, 49]. In fact, oligosaccharides in milk which also comprise of terminal α1-2 fucosyl glycan motif (similar to that in histo-blood group antigen) have been shown to have a potent inhibitory role in norovirus infection by serving as receptor decoys [50, 51].
Role of viral surface glycoproteins in pathogenesis
During the natural course of viral pathogenesis in different mammalian hosts including humans, the glycan biosynthetic machinery of the host cell also glycosylates the viral surface proteins. These viral surface glycans interact with the GBPs on the host cell. These interactions either favor the virus in terms of gaining entry to specific target cells that display the GBP or favor the host where the viral glycans are recognized by various circulating GBPs that mediate clearance of the virus or by anchored GBPs on antigen presenting cells that prime host immune response to target the viruses (see [1••] for a detailed review). The glycan motifs on the viral surface glycans can be classified into three broad categories: first, high mannose type; second, acidic (terminal sialylation) and three, neutral (not capped by sialic acid).
Some of the circulating GBPs in different hosts that help in viral clearance include collectins (recognize high mannose type and neutral glycans), surfactant proteins SP-A and SP-D (recognize high mannose and acidic glycans), mannose binding lectin, ficolins and galectins that show a broader diversity in recognition of high mannose, neutral and acidic glycans [1••, 8]. Langerin is a C-type lectin on surface of Langerhans cells (antigen presenting cells) that specifically recognize high-mannose type glycans on viral surface and plays a key role in priming the host immune response to recognize viral glycopeptides as antigens. A prototypic example of immune response targeted to viral surface glycans is that of HIV where numerous antibodies targeting the glyco-eptiopes achieve potent neutralization against a broad spectrum of viral strains [20••].
Among the GBPs on host cell surface that bind to viral surface glycans to facilitate viral entry and infection, the dendritic cell-specific ICAM-grabbing non-integrin (DC-SIGN) and liver/lymph node specific ICAM-grabbing non-integrin (L-SIGN) are the most prominent and play a key role in pathogenesis of numerous viruses including HIV, flaviviruses (dengue, zika), Ebola virus, measles virus, respiratory syncytial virus, coronavirus (SARS), etc. [1••, 52]. In fact it has been demonstrated that the dengue virus switches its specificity from DC-SIGN (high mannose and fucosylated N-linked glycans) which plays a key role in infection of the primary monocyte cells to L-SIGN (stricter binding to high mannose glycans) during subsequent secondary infection of other cells including hepatocytes [53•, 54, 55].
In addition to their interactions with mammalian host GBPs, glycosylation of the viral surface proteins also impacts binding of these proteins to host cell glycan receptors as observed in the case of influenza A virus HA. Molecular dynamics simulation studies predicted that HA glycans may form interactions near the binding pocket to influence receptor binding [56]. Site-directed mutagenesis to knock-out glycosylation sites on HA or modifying structure of N-linked glycans on the virus by enzymatic treatment or transgenic cell lines have shown distinct changes in glycan-receptor binding specificity [57]. In the case of the 1918 pandemic H1N1 influenza viruses, a mutation that abrogates glycosylation at a single highly conserved site significantly impacted the ability of the virus to bind to α2-6 sialylated glycan receptors but not to α2-3 sialylated glycan receptors [58]. Site-specific loss-of-glycosylation mutations have also been shown to impact glycan receptor-binding specificity which in-turn governed virulence in mice [59, 60, 61]. Therefore, it is evident that the viral surface glycosylation governed by specific glycosylation sites and the structural heterogeneity of the glycans at each site in the relevant physiological context of virus interaction with the host cell has multidimensional roles in pathogenesis of the virus in the host.
Tools for defining glycan-specific targets in virus–host interactions
The dynamic interplay between virus and its mammalian host involving glycan–GBP interactions presents new opportunities to target these interactions so as to develop novel antiviral therapies. Based on what is known about role of complex glycans in viral–host interactions, the key question is how does one define a target in the right physiological context of these interactions? [43] Given that glycosylation biosynthesis machinery is complex and does not have a template, the results from in vitro experiments may be biased towards glycans made by a given cell line used in the experiment and may not be related to the actual physiological context of the virus–host cell interactions. For example, the glycosylation on gp120 protein on the surface of the virus is different from that of a recombinantly expressed protein in different cell lines [62•, 63, 64]. Therefore, framing this question would guide the integration of the datasets from the various tools developed to decode glycan structures and glycan–protein interactions. Some of the key tools and perspectives on how to integrate the tools from the standpoint of characterizing glycan–protein interaction targets is summarized in the following (schematic of the integration of tools shown in Figure 2 ).
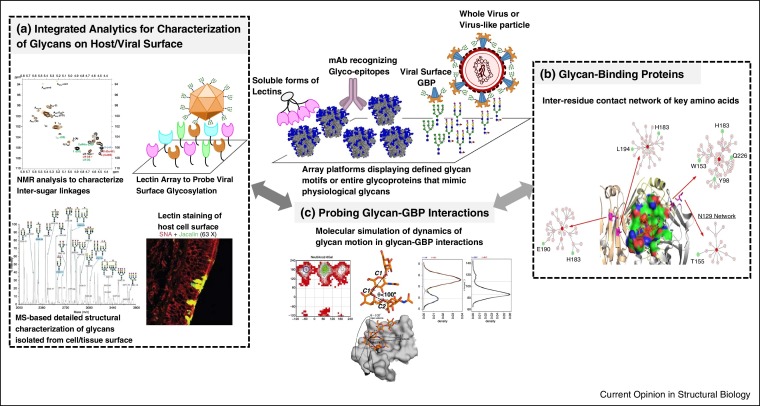
Figure 2.
Tools to define glycan-specific targets in viral–host interactions. (a) Shows snapshots of various analytical tools that either directly probe glycans in the appropriate physiological context (such as lectin-based staining and lectin array) or provide detailed characterization of glycans isolated from host or viral surface (using mass spectrometry and NMR based approaches. (b) Shows analysis of GBPs and their glycan-binding site using a network approach which not only takes into account key residues in glycan-binding but also those that are related to these residues through their inter-residue interaction network. (c) Shows a schematic of biochemical (top) and structural (bottom) tools to probe glycan–GBP interactions. As shown in the bottom, shape based definitions of the conformational space sampled by glycans relates the conformations defined by 7 different glyosidic torsion angle (for a tetrasaccharide) into a single parameter θ whose variation can be studied during molecular dynamics simulations.
Structural characterization of glycans
The characterization of glycan-specific targets in virus-host interactions can range from simple to very complex definitions. An example of a simple glycan-specific target is a single terminal sugar such as sialic acid as seen in the case of neuraminidase inhibitors or a defined oligosaccharide glycan motif such as tri or tetra-saccharide motif capped by α2-3 or α2-6-linked sialic acid that has specific recognition and high affinity binding by the viral surface glycoproteins. A highly complex description of glycan target would involve defining the glycan motif, the linkage of the glycan comprising this motif to specific glycosylation site on a specific protein and a geometrical arrangement of such glycosylation sites on the glycoprotein. An example of such a complex definition is the glyco-epitope surfaces on HIV gp120 targeted by potent broad spectrum neutralizing antibodies. Therefore the scope of the target definition would guide the development and integration of diverse analytical tools.
The GBPs or lectins that are known to recognize specific glycan motifs are valuable tools that can be used to assess the distribution of these motifs. Furthermore, the advances in chemical synthesis of glycans coupled with the ability to probe finer nuances of glycan motif recognition by various lectins using glycan array platforms has substantially improved the knowledge on the diversity of glycan motifs recognized by various GBPs [65•, 66]. These GBPs can be directly used to stain host cell or tissue surfaces and multiplexing the staining of more than one GBP can expand the knowledge of the glycan motifs distributed on these surfaces [4, 67, 68]. As an example co-staining of host tissues or cells with Sambucus nigra agglutinin I (SNA-I) which binds with high specificity to α2-6 sialylated glycans and Concanavalin A (ConA) which binds to trimannosyl core of N-linked glycans would give information on predominance of α2-6 sialylated glycans that are N-linked to the glycoproteins. These lectins can also be displayed in an array-like platform and samples such as whole viruses or virus-like particles comprising of the viral surface glycoprotein can be probed for their binding to such lectin-arrays [65•, 69, 70, 71]. This would again provide information on the prevalence of specific glycan motifs that are present as a part of the glycans on the viral surface glycoproteins. The advantages of using lectins is that it provides a read on the glycan motif in their physiological context either on the host tissue or cell or viral surface and do not require any additional processing that may change the structural diversity of the glycans before analysis. However, a limitation in this approach arises from the lack of a comprehensive panel of lectins that can completely decode the structural diversity of the glycans. As an example while lectins like SNA-I give an accurate picture of distribution of glycans capped by α2-6-linked sialic acid, the information on whether this motif is a part of highly branched N-linked glycans or a part of long oligosaccharide branch length cannot be deduced.
There are several analytical methods building on the use of different mass spectrometric methods such as MALDI-MS, LC–MS, tandem MS–MS, and other methods using NMR spectroscopy, and HPLC that have been developed over several decades for detailed structural characterization of complex glycans, glycopeptides and glycolipids. Each analytical method is unique but they provide overlapping information on the detailed nuances of glycan structure, occupancy of the glycosylation site, etc. Therefore integration of information from different analytical techniques would provide comprehensive information on the glycan structures displayed on host and viral surfaces. One limitation of this approach is the challenges in accessing and isolating glycans from their right physiological context before analysis. For example, the HA from influenza A virus subtypes H1N1 and H3N2 that have adapted well to the human host shows extensive staining of non-ciliated goblet cells in the upper respiratory epithelial surface. However it is challenging to obtain and grow these cells in vitro to extract the glycans from these cells for detailed structural analyses.
Therefore, depending on the scope of glycan target definition it would be necessary to employ both lectin-based tools and analytical tools and integrate the information from applying these tools for an unbiased definition of the target. Such an approach was used to expand the target definition of sialylated glycan receptors for human adapted influenza A viruses going beyond just the terminal α2-6 sialic acid linkage to a long oligosaccharide motif capped by α2-6 sialic acid both in the context of N-linked glycans on the ciliated epithelial surface and non-ciliated goblet cell surface in respiratory tract of humans and established animal models such as ferrets [44, 68, 72].
Probing glycan–GBP interactions
A variety of biochemical methods have been used to characterize the specificity in the context of multivalent GBP–glycan interactions. Among the various tools, glycan array platforms are rapidly emerging as a popular tool to probe finer nuances of glycan structures that are recognized by various GBPs. Glycan platforms consist of hundreds of synthetic glycan motifs (typically present on N and O-linked glycoproteins and glycolipids) displayed on the surface of the array. Multiple types of arrays have been developed that utilize different strategies including the formation of neoglycolipids [73], neoglycoproteins [74], or the direct application of glycans to various surfaces [75, 76, 77, 78, 79, 80]. Studies have also begun to adapt these technologies towards the presentation of natural glycans by harvesting glycans from the cells or tissues and imprinting these on a glycan array format, thus allowing one to probe the glycan repertoire of a biological system [81].
Designing the experimental method and array platform to probe glycan–GBP interactions needs to be carefully done based on the scope of the analysis. In most cases the scope of the analysis is to obtain a primary screen of a whole virus or viral surface glycoprotein or mammalian GBP against the maximum diversity of glycan motifs that can be generated in a defined fashion synthetically. Following this primary screen, the second objective is to usually probe deeper into the quantitative differences in affinity and avidity between different viruses for specific glycan motifs. This would often involve doing a dose-dependent binding of the viral surface glycoprotein (or whole virus) to defined motifs in secondary assays such as surface plasmon resonance or isothermal titration calorimetry [77, 82]. It is important to understand the glycan target motif while designing the glycan arrays for probing nuances of glycan binding specificities. For example, if one were trying to understand the nuances in binding of antibodies that target glycan shield of HIV surface protein gp120-gp41, then using an array of just different high mannose and complex glycans may not fully capture the nuances in the antibody-binding given that the underlying peptide epitope would not be present in such a platform. This might lead to confounding interpretations on link between target glycan motif recognized by these antibodies and their neutralization potential in in vitro and in vivo assays. It is therefore be more appropriate to use the recently developed BG505 SOSIP.644 gp120-gp41 trimeric unit [83] expressed in different cell types (and also mutant forms that lack specific glycosylation sites) by presenting this trimeric unit and its variants on an platform to probe the nuances of glycan recognition by the glycan shield targeting antibodies.
Structural and molecular specificity of glycan–GBP interactions
At the three-dimensional structure and molecular level, the interactions between glycans and GBPs have been captured in numerous X-ray co-crystal structures of viral surface glycoprotein–glycan receptor complexes and antibody-glyco-eptiope complexes as in the case of HIV. Over the past several years, there has been a substantial progress in developing tools for accurate molecular simulation of glycan–protein interactions including improvement in force-fields to include glycan-specific parameters [84]. This progress has led to improvements in modeling the electron density of glycans to assign their coordinates in the co-crystal structures and also rectify errors in past assignments pertaining to incomplete ring closures, inaccurate ring and exocyclic torsion angles, etc. [85, 86•, 87]. The growing wealth of GBP-glycan co-crystal structures and development of molecular simulation tools have permitted successful predictions of amino acid changes that would alter glycan-binding specificity of the GBP. A couple of areas where this approach has been extensively applied is in prediction and validation of mutations in influenza A virus HA that would switch its binding specificity from α2-3 to α2-6 sialylated glycan receptors [4, 88, 89, 90] and in engineering antibodies that target known glycan epitopes to alter their epitope specificity [91, 92, 93].
More recent studies have described glycan-binding sites by mapping the three-dimensional structure into a two-dimensional network of non-bonded inter-residue interactions involving key residues that contact the glycan structure [88, 94]. This network approach has permitted obtaining insights into the molecular determinants of glycan receptor-binding specificity of influenza A virus HA going beyond just the residues that contact the glycans. This include role of site-specific glycosylation that impacts receptor-binding specificity [58, 59]. This network approach had also provided a framework to investigate amino acid changes required to switch glycan receptor-specificity (from α2-3 to α2-6 sialylated glycan receptors) of the H5N1 influenza HA in the context of additional changes in the glycan-binding site resulting from natural sequence evolution [88]. Importantly the network analyses studies transformed the notion of hallmark residues across different strains associated with glycan-binding specificity by demonstrating that each strain depending on its sequence evolution needed distinct set of amino acid changes to switch receptor specificity.
On the glycan side, the availability of co-crystal structures and molecular simulation tools has provided insights into the preferential conformations of the glycans sampled in the unbound and protein-bound states. In fact, the conformational space sampled by sialylated glycan receptors in binding site of avian and human influenza HA has been described using shape-based definitions such umbrella and cone-like topologies. These shape-based definition of the glycan conformational space simplifies the parameterization to capture the conformational sample going from a series of glyosidic torsion angles to a one or two parameters that capture the shape [44]. In addition to X-ray co-crystal structures, glycan–GBP complexes have been analyzed in solution using various NMR methods including STD-NMR. For example, NMR analyses of α2-3 and α2-6 sialylated glycan receptors bound to different HAs complimented the information on glycan–protein contacts from the X-ray co-crystal structures by providing additional information on dynamics of glycan motion in the free and protein-bound states [95, 96].
Unlike protein–protein interactions that cover a large surface area on both interacting proteins, protein–glycan interactions cover substantially lower surface area on the protein which is consistent with the low to moderate binding affinity of a given glycan-binding site to a given glycan-motif. Despite this affinity, there is specificity in the glycan-binding site — glycan contacts which is also governed by the dynamics of glycan motion in the binding site compared to the unbound state. As a result, mutations in the amino acid that directly contacts the glycan or in other amino acids in its network of inter-residue contacts are likely to significantly impact the glycan-binding properties of the GBP. Thus it is important to capture the network properties on the protein and dynamics of glycan motion while modeling the molecular interactions between glycans and GBPs so as to engineer these interactions in a predictable fashion.
Summary
Complex glycans play multidimensional roles in various stages of viral pathogenesis in a given host starting from viral attachment and entry to host immune response to counter the infection. The advances in development of tools to chemically synthesize glycans, sophisticated analytical methods for structural characterization of glycans, novel array platforms to simultaneously probe interactions between multitude of GBPs and glycan motifs have transformed our understanding of these multidimensional roles. However, given that the context of glycan–GBP interactions (in different hosts, within the host versus outside the host in in vitro assays) critically impacts the structural diversity of the glycans, there are gaps in bridging the definition of glycan-based targets to the biological functions in viral pathogenesis mediated by these interactions. Therefore one has to take an integrated approach by bridging different tools such as synthetic, analytical, array platforms, molecular modeling to fill these gaps and appropriately frame and validate target definition in these interactions. Such an approach would substantially augment ongoing efforts to in targeting glycan-GBP interactions to develop vaccine strategies as in the case of HIV or novel antiviral therapeutics.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgement
This work was funded in part by National Institutes of Health Merit Award (R37 GM057073-13).
References
- 1••.Van Breedam W., Pohlmann S., Favoreel H.W., de Groot R.J., Nauwynck H.J. Bitter-sweet symphony: glycan–lectin interactions in virus biology. FEMS Microbiol Rev. 2014;38:598–632. doi: 10.1111/1574-6976.12052. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is a review of outstanding interest given that it captures all the known mammalian host lectins known to play key roles in viral pathogensis through their interactions with viral surface glycosylation.
- 2•.Stroh L.J., Stehle T. Glycan engagement by viruses: receptor switches and specificity. Annu Rev Virol. 2014;1:285–306. doi: 10.1146/annurev-virology-031413-085417. [DOI] [PubMed] [Google Scholar]; This is a review of special interest given that it captures knowledge on virus–host systems where specificity of interaction with glycan receptor plays a key role in host tropism and viral pathogenesis.
- 3•.Smith D.F., Cummings R.D. Investigating virus–glycan interactions using glycan microarrays. Curr Opin Virol. 2014;7C:79–87. doi: 10.1016/j.coviro.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; This review covers analyses of viruses and viral proteins using glycan array platforms and the information that is obtained from such analyses.
- 4.Raman R., Tharakaraman K., Shriver Z., Jayaraman A., Sasisekharan V., Sasisekharan R. Glycan receptor specificity as a useful tool for characterization and surveillance of influenza A virus. Trends Microbiol. 2014:10. doi: 10.1016/j.tim.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Varki A., Cummings R.D., Esko J.D., Freeze H.H., Hart G.W., Etzler M.E. edn 2. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2008. Essentials of glycobiology. [Google Scholar]
- 6.Taylor M.E., Drickamer K. Oxford University Press; Oxford; New York: 2003. Introduction to glycobiology. [Google Scholar]
- 7.Lowe J.B., Marth J.D. A genetic approach to Mammalian glycan function. Annu Rev Biochem. 2003;72:643–691. doi: 10.1146/annurev.biochem.72.121801.161809. [DOI] [PubMed] [Google Scholar]
- 8.Mason C.P., Tarr A.W. Human lectins and their roles in viral infections. Molecules. 2015;20:2229–2271. doi: 10.3390/molecules20022229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins B.E., Paulson J.C. Cell surface biology mediated by low affinity multivalent protein–glycan interactions. Curr Opin Chem Biol. 2004;8:617–625. doi: 10.1016/j.cbpa.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 10•.Stencel-Baerenwald J.E., Reiss K., Reiter D.M., Stehle T., Dermody T.S. The sweet spot: defining virus–sialic acid interactions. Nat Rev Microbiol. 2014;12:739–749. doi: 10.1038/nrmicro3346. [DOI] [PMC free article] [PubMed] [Google Scholar]; This review focuses specifically on role of sialylated glycans as receptors for viral infection, entry and pathogenesis.
- 11.Wadstrom T., Ljungh A. Glycosaminoglycan-binding microbial proteins in tissue adhesion and invasion: key events in microbial pathogenicity. J Med Microbiol. 1999;48:223–233. doi: 10.1099/00222615-48-3-223. [DOI] [PubMed] [Google Scholar]
- 12.Artpradit C., Robinson L.N., Gavrilov B.K., Rurak T.T., Ruchirawat M., Sasisekharan R. Recognition of heparan sulfate by clinical strains of dengue virus serotype 1 using recombinant subviral particles. Virus Res. 2013;176:69–77. doi: 10.1016/j.virusres.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y., Maguire T., Hileman R.E., Fromm J.R., Esko J.D., Linhardt R.J., Marks R.M. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate [see comments] Nat Med. 1997;3:866–871. doi: 10.1038/nm0897-866. [DOI] [PubMed] [Google Scholar]
- 14.Baumert T.F., Meredith L., Ni Y., Felmlee D.J., McKeating J.A., Urban S. Entry of hepatitis B and C viruses – recent progress and future impact. Curr Opin Virol. 2014;4:58–65. doi: 10.1016/j.coviro.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 15.Fry E.E., Lea S.M., Jackson T., Newman J.W., Ellard F.M., Blakemore W.E., Abu-Ghazaleh R., Samuel A., King A.M., Stuart D.I. The structure and function of a foot-and-mouth disease virus-oligosaccharide receptor complex. Embo J. 1999;18:543–554. doi: 10.1093/emboj/18.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ge S., Wang Z. An overview of influenza A virus receptors. Crit Rev Microbiol. 2011;37:157–165. doi: 10.3109/1040841X.2010.536523. [DOI] [PubMed] [Google Scholar]
- 17.Malik Peiris J.S. Avian influenza viruses in humans. Rev Sci Tech. 2009;28:161–173. doi: 10.20506/rst.28.1.1871. [DOI] [PubMed] [Google Scholar]
- 18.Neumann G., Kawaoka Y. Host range restriction and pathogenicity in the context of influenza pandemic. Emerg Infect Dis. 2006;12:881–886. doi: 10.3201/eid1206.051336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pipirou Z., Powlesland A.S., Steffen I., Pohlmann S., Taylor M.E., Drickamer K. Mouse LSECtin as a model for a human Ebola virus receptor. Glycobiology. 2011;21:806–812. doi: 10.1093/glycob/cwr008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20••.Doores K.J. The HIV glycan shield as a target for broadly neutralizing antibodies. FEBS J. 2015;282:4679–4691. doi: 10.1111/febs.13530. [DOI] [PMC free article] [PubMed] [Google Scholar]; This comprenehsive reivew on glycosylation of HIV envelop and their roles as glycoeptiopes for potent neutralizing antibodies is of outstanding interest.
- 21.Connell B.J., Lortat-Jacob H. Human immunodeficiency virus and heparan sulfate: from attachment to entry inhibition. Front Immunol. 2013;4:385. doi: 10.3389/fimmu.2013.00385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilen C.B., Tilton J.C., Doms R.W. HIV: cell binding and entry. Cold Spring Harb Perspect Med. 2012:2. doi: 10.1101/cshperspect.a006866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamayoshi S., Fujii K., Koike S. Receptors for enterovirus 71. Emerg Microbes Infect. 2014;3:e53. doi: 10.1038/emi.2014.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan C.W., Poh C.L., Sam I.C., Chan Y.F. Enterovirus 71 uses cell surface heparan sulfate glycosaminoglycan as an attachment receptor. J Virol. 2013;87:611–620. doi: 10.1128/JVI.02226-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Israelsson S., Gullberg M., Jonsson N., Roivainen M., Edman K., Lindberg A.M. Studies of Echovirus 5 interactions with the cell surface: heparan sulfate mediates attachment to the host cell. Virus Res. 2010;151:170–176. doi: 10.1016/j.virusres.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 26.Merilahti P., Karelehto E., Susi P. Role of heparan sulfate in cellular infection of integrin-binding Coxsackievirus A9 and human parechovirus 1 isolates. PLOS ONE. 2016;11:e0147168. doi: 10.1371/journal.pone.0147168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pourianfar H.R., Kirk K., Grollo L. Initial evidence on differences among Enterovirus 71, Coxsackievirus A16 and Coxsackievirus B4 in binding to cell surface heparan sulphate. Virusdisease. 2014;25:277–284. doi: 10.1007/s13337-013-0172-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Hearn A., Wang M., Cheng H., Lear-Rooney C.M., Koning K., Rumschlag-Booms E., Varhegyi E., Olinger G., Rong L. Role of EXT1 and glycosaminoglycans in the early stage of filovirus entry. J Virol. 2015;89:5441–5449. doi: 10.1128/JVI.03689-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tiwari V., Tarbutton M.S., Shukla D. Diversity of heparan sulfate and HSV entry: basic understanding and treatment strategies. Molecules. 2015;20:2707–2727. doi: 10.3390/molecules20022707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thacker B.E., Xu D., Lawrence R., Esko J.D. Heparan sulfate 3-O-sulfation: a rare modification in search of a function. Matrix Biol. 2014;35:60–72. doi: 10.1016/j.matbio.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J., Shriver Z., Pope R.M., Thorp S.C., Duncan M.B., Copeland R.J., Raska C.S., Yoshida K., Eisenberg R.J., Cohen G. Characterization of a heparan sulfate octasaccharide that binds to herpes simplex virus type 1 glycoprotein d. J Biol Chem. 2002;277:33456–33467. doi: 10.1074/jbc.M202034200. [DOI] [PubMed] [Google Scholar]
- 32.Baggen J., Thibaut H.J., Staring J., Jae L.T., Liu Y., Guo H., Slager J.J., de Bruin J.W., van Vliet A.L., Blomen V.A. Enterovirus D68 receptor requirements unveiled by haploid genetics. Proc Natl Acad Sci U S A. 2016;113:1399–1404. doi: 10.1073/pnas.1524498113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y., Sheng J., Baggen J., Meng G., Xiao C., Thibaut H.J., van Kuppeveld F.J., Rossmann M.G. Sialic acid-dependent cell entry of human enterovirus D68. Nat Commun. 2015;6:8865. doi: 10.1038/ncomms9865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang B., Chuang H., Yang K.D. Sialylated glycans as receptor and inhibitor of enterovirus 71 infection to DLD-1 intestinal cells. Virol J. 2009;6:141. doi: 10.1186/1743-422X-6-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zocher G., Mistry N., Frank M., Hahnlein-Schick I., Ekstrom J.O., Arnberg N., Stehle T. A sialic acid binding site in a human picornavirus. PLoS Pathog. 2014;10:e1004401. doi: 10.1371/journal.ppat.1004401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mistry N., Inoue H., Jamshidi F., Storm R.J., Oberste M.S., Arnberg N. Coxsackievirus A24 variant uses sialic acid-containing O-linked glycoconjugates as cellular receptors on human ocular cells. J Virol. 2011;85:11283–11290. doi: 10.1128/JVI.05597-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nilsson E.C., Jamshidi F., Johansson S.M., Oberste M.S., Arnberg N. Sialic acid is a cellular receptor for coxsackievirus A24 variant, an emerging virus with pandemic potential. J Virol. 2008;82:3061–3068. doi: 10.1128/JVI.02470-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nokhbeh M.R., Hazra S., Alexander D.A., Khan A., McAllister M., Suuronen E.J., Griffith M., Dimock K. Enterovirus 70 binds to different glycoconjugates containing alpha2,3-linked sialic acid on different cell lines. J Virol. 2005;79:7087–7094. doi: 10.1128/JVI.79.11.7087-7094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alexander D.A., Dimock K. Sialic acid functions in enterovirus 70 binding and infection. J Virol. 2002;76:11265–11272. doi: 10.1128/JVI.76.22.11265-11272.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Riel D., Munster V.J., de Wit E., Rimmelzwaan G.F., Fouchier R.A., Osterhaus A.D., Kuiken T. Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. Am J Pathol. 2007;171:1215–1223. doi: 10.2353/ajpath.2007.070248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong S.S., Yuen K.Y. Avian influenza virus infections in humans. Chest. 2006;129:156–168. doi: 10.1378/chest.129.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shinya K., Ebina M., Yamada S., Ono M., Kasai N., Kawaoka Y. Avian flu: influenza virus receptors in the human airway. Nature. 2006;440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 43.Shriver Z., Raman R., Viswanathan K., Sasisekharan R. Context-specific target definition in influenza a virus hemagglutinin–glycan receptor interactions. Chem Biol. 2009;16:803–814. doi: 10.1016/j.chembiol.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chandrasekaran A., Srinivasan A., Raman R., Viswanathan K., Raguram S., Tumpey T.M., Sasisekharan V., Sasisekharan R. Glycan topology determines human adaptation of avian H5N1 virus hemagglutinin. Nat Biotechnol. 2008;26:107–113. doi: 10.1038/nbt1375. [DOI] [PubMed] [Google Scholar]
- 45.Yen H.L., Liang C.H., Wu C.Y., Forrest H.L., Ferguson A., Choy K.T., Jones J., Wong D.D., Cheung P.P., Hsu C.H. Hemagglutinin-neuraminidase balance confers respiratory-droplet transmissibility of the pandemic H1N1 influenza virus in ferrets. Proc Natl Acad Sci U S A. 2011;108:14264–14269. doi: 10.1073/pnas.1111000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fanning T.G., Reid A.H., Taubenberger J.K. Influenza A virus neuraminidase: regions of the protein potentially involved in virus–host interactions. Virology. 2000;276:417–423. doi: 10.1006/viro.2000.0578. [DOI] [PubMed] [Google Scholar]
- 47.Saladino R., Barontini M., Crucianelli M., Nencioni L., Sgarbanti R., Palamara A.T. Current advances in anti-influenza therapy. Curr Med Chem. 2010;17:2101–2140. doi: 10.2174/092986710791299957. [DOI] [PubMed] [Google Scholar]
- 48.Schroten H., Hanisch F.G., Hansman G.S. Human norovirus interactions with histo-blood group antigens and human milk oligosaccharides. J Virol. 2016;90:5855–5859. doi: 10.1128/JVI.00317-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tan M., Jiang X. Histo-blood group antigens: a common niche for norovirus and rotavirus. Expert Rev Mol Med. 2014;16:e5. doi: 10.1017/erm.2014.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Etzold S., Bode L. Glycan-dependent viral infection in infants and the role of human milk oligosaccharides. Curr Opin Virol. 2014;7:101–107. doi: 10.1016/j.coviro.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 51.Weichert S., Koromyslova A., Singh B.K., Hansman S., Jennewein S., Schroten H., Hansman G.S. Structural basis for norovirus inhibition by human milk oligosaccharides. J Virol. 2016;90:4843–4848. doi: 10.1128/JVI.03223-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang F., Ren S., Zuo Y. DC-SIGN, DC-SIGNR and LSECtin: C-type lectins for infection. Int Rev Immunol. 2014;33:54–66. doi: 10.3109/08830185.2013.834897. [DOI] [PubMed] [Google Scholar]
- 53•.Idris F., Muharram S.H., Diah S. Glycosylation of dengue virus glycoproteins and their interactions with carbohydrate receptors: possible targets for antiviral therapy. Arch Virol. 2016;161:1751–1760. doi: 10.1007/s00705-016-2855-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; This review talks about the known glycoforms on dengue virus envelop proteins and their role in viral entry and transmission.
- 54.Dejnirattisai W., Webb A.I., Chan V., Jumnainsong A., Davidson A., Mongkolsapaya J., Screaton G. Lectin switching during dengue virus infection. J Infect Dis. 2011;203:1775–1783. doi: 10.1093/infdis/jir173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hacker K., White L., de Silva A.M. N-linked glycans on dengue viruses grown in mammalian and insect cells. J Gen Virol. 2009;90:2097–2106. doi: 10.1099/vir.0.012120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kasson P.M., Pande V.S. Structural basis for influence of viral glycans on ligand binding by influenza hemagglutinin. Biophys J. 2008;95:L48–L50. doi: 10.1529/biophysj.108.141507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang C.C., Chen J.R., Tseng Y.C., Hsu C.H., Hung Y.F., Chen S.W., Chen C.M., Khoo K.H., Cheng T.J., Cheng Y.S. Glycans on influenza hemagglutinin affect receptor binding and immune response. Proc Natl Acad Sci U S A. 2009;106:18137–18142. doi: 10.1073/pnas.0909696106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jayaraman A., Koh X., Li J., Raman R., Viswanathan K., Shriver Z., Sasisekharan R. Glycosylation at Asn91 of H1N1 haemagglutinin affects binding to glycan receptors. Biochem J. 2012;444:429–435. doi: 10.1042/BJ20112101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun X., Jayaraman A., Maniprasad P., Raman R., Houser K.V., Pappas C., Zeng H., Sasisekharan R., Katz J.M., Tumpey T.M. N-linked glycosylation of the hemagglutinin protein influences virulence and antigenicity of the 1918 pandemic and seasonal H1N1 influenza A viruses. J Virol. 2013;87:8756–8766. doi: 10.1128/JVI.00593-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tate M.D., Brooks A.G., Reading P.C. Specific sites of N-linked glycosylation on the hemagglutinin of H1N1 subtype influenza A virus determine sensitivity to inhibitors of the innate immune system and virulence in mice. J Immunol. 2011;187:1884–1894. doi: 10.4049/jimmunol.1100295. [DOI] [PubMed] [Google Scholar]
- 61.Reading P.C., Pickett D.L., Tate M.D., Whitney P.G., Job E.R., Brooks A.G. Loss of a single N-linked glycan from the hemagglutinin of influenza virus is associated with resistance to collectins and increased virulence in mice. Respir Res. 2009;10:117. doi: 10.1186/1465-9921-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62•.Pritchard L.K., Harvey D.J., Bonomelli C., Crispin M., Doores K.J. Cell- and protein-directed glycosylation of native cleaved HIV-1 envelope. J Virol. 2015;89:8932–8944. doi: 10.1128/JVI.01190-15. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study using a combination of analytical techniques to analyze glycans derived from gp120 on viruses, pseudo viruses and recombinantly expressed in different cell lines such as PBMCs and HEK 293T cells to determine which glycoforms mimic the native gp120 glycans on virion surface to assess the production of this viral antigen for vaccine strategies.
- 63.Raska M., Takahashi K., Czernekova L., Zachova K., Hall S., Moldoveanu Z., Elliott M.C., Wilson L., Brown R., Jancova D. Glycosylation patterns of HIV-1 gp120 depend on the type of expressing cells and affect antibody recognition. J Biol Chem. 2010;285:20860–20869. doi: 10.1074/jbc.M109.085472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Doores K.J., Bonomelli C., Harvey D.J., Vasiljevic S., Dwek R.A., Burton D.R., Crispin M., Scanlan C.N. Envelope glycans of immunodeficiency virions are almost entirely oligomannose antigens. Proc Natl Acad Sci U S A. 2010;107:13800–13805. doi: 10.1073/pnas.1006498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65•.Wang L., Cummings R.D., Smith D.F., Huflejt M., Campbell C.T., Gildersleeve J.C., Gerlach J.Q., Kilcoyne M., Joshi L., Serna S. Cross-platform comparison of glycan microarray formats. Glycobiology. 2014;24:507–517. doi: 10.1093/glycob/cwu019. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper describes analysis of mammalian and viral lectins on different glycan mirocarray formats to assess the comparability and validation of information on glycan binding specificity generated on the same analyte across platforms.
- 66.Oyelaran O., Gildersleeve J.C. Glycan arrays: recent advances and future challenges. Curr Opin Chem Biol. 2009;13:406–413. doi: 10.1016/j.cbpa.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lakdawala S.S., Jayaraman A., Halpin R.A., Lamirande E.W., Shih A.R., Stockwell T.B., Lin X., Simenauer A., Hanson C.T., Vogel L. The soft palate is an important site of adaptation for transmissible influenza viruses. Nature. 2015;526:122–125. doi: 10.1038/nature15379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jayaraman A., Chandrasekaran A., Viswanathan K., Raman R., Fox J.G., Sasisekharan R. Decoding the distribution of glycan receptors for human-adapted influenza A viruses in ferret respiratory tract. PLoS ONE. 2012;7:e27517. doi: 10.1371/journal.pone.0027517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Krishnamoorthy L., Mahal L.K. Glycomic analysis: an array of technologies. ACS Chem Biol. 2009;4:715–732. doi: 10.1021/cb900103n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hsu K.L., Mahal L.K. A lectin microarray approach for the rapid analysis of bacterial glycans. Nat Protoc. 2006;1:543–549. doi: 10.1038/nprot.2006.76. [DOI] [PubMed] [Google Scholar]
- 71.Kanoelani T., Pilobello L.K.D.S.L.K.M. Development of a lectin microarray for the rapid analysis of protein glycopatterns. Chembiochem. 2005;6:985–989. doi: 10.1002/cbic.200400403. [DOI] [PubMed] [Google Scholar]
- 72.Srinivasan A., Viswanathan K., Raman R., Chandrasekaran A., Raguram S., Tumpey T.M., Sasisekharan V., Sasisekharan R. Quantitative biochemical rationale for differences in transmissibility of 1918 pandemic influenza A viruses. Proc Natl Acad Sci U S A. 2008;105:2800–2805. doi: 10.1073/pnas.0711963105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fukui S., Feizi T., Galustian C., Lawson A.M., Chai W. Oligosaccharide microarrays for high-throughput detection and specificity assignments of carbohydrate-protein interactions. Nat Biotechnol. 2002;20:1011–1017. doi: 10.1038/nbt735. [DOI] [PubMed] [Google Scholar]
- 74.Gildersleeve J.C., Oyelaran O., Simpson J.T., Allred B. Improved procedure for direct coupling of carbohydrates to proteins via reductive amination. Bioconjug Chem. 2008;19:1485–1490. doi: 10.1021/bc800153t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Blixt O., Westerlind U. Arraying the post-translational glycoproteome (PTG) Curr Opin Chem Biol. 2014;18:62–69. doi: 10.1016/j.cbpa.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 76.Grun C.H., van Vliet S.J., Schiphorst W.E., Bank C.M., Meyer S., van Die I., van Kooyk Y. One-step biotinylation procedure for carbohydrates to study carbohydrate–protein interactions. Anal Biochem. 2006;354:54–63. doi: 10.1016/j.ab.2006.03.055. [DOI] [PubMed] [Google Scholar]
- 77.Karamanska R., Clarke J., Blixt O., Macrae J.I., Zhang J.Q., Crocker P.R., Laurent N., Wright A., Flitsch S.L., Russell D.A. Surface plasmon resonance imaging for real-time, label-free analysis of protein interactions with carbohydrate microarrays. Glycoconj J. 2008;25:69–74. doi: 10.1007/s10719-007-9047-y. [DOI] [PubMed] [Google Scholar]
- 78.Liang P.H., Wu C.Y., Greenberg W.A., Wong C.H. Glycan arrays: biological and medical applications. Curr Opin Chem Biol. 2008;12:86–92. doi: 10.1016/j.cbpa.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mercey E., Sadir R., Maillart E., Roget A., Baleux F., Lortat-Jacob H., Livache T. Polypyrrole oligosaccharide array and surface plasmon resonance imaging for the measurement of glycosaminoglycan binding interactions. Anal Chem. 2008;80:3476–3482. doi: 10.1021/ac800226k. [DOI] [PubMed] [Google Scholar]
- 80.Xia B., Kawar Z.S., Ju T., Alvarez R.A., Sachdev G.P., Cummings R.D. Versatile fluorescent derivatization of glycans for glycomic analysis. Nat Methods. 2005;2:845–850. doi: 10.1038/nmeth808. [DOI] [PubMed] [Google Scholar]
- 81.Song X., Heimburg-Molinaro J., Cummings R.D., Smith D.F. Chemistry of natural glycan microarrays. Curr Opin Chem Biol. 2014;18:70–77. doi: 10.1016/j.cbpa.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dam T.K., Gerken T.A., Brewer C.F. Thermodynamics of multivalent carbohydrate-lectin cross-linking interactions: importance of entropy in the bind and jump mechanism. Biochemistry. 2009;48:3822–3827. doi: 10.1021/bi9002919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sanders R.W., Derking R., Cupo A., Julien J.P., Yasmeen A., de Val N., Kim H.J., Blattner C., de la Pena A.T., Korzun J. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kirschner K.N., Yongye A.B., Tschampel S.M., Gonzalez-Outeirino J., Daniels C.R., Foley B.L., Woods R.J. GLYCAM06: a generalizable biomolecular force field. Carbohydrates. J Comput Chem. 2008;29:622–655. doi: 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nivedha A.K., Thieker D.F., Makeneni S., Hu H., Woods R.J. Vina-Carb: improving glycosidic angles during carbohydrate docking. J Chem Theory Comput. 2016;12:892–901. doi: 10.1021/acs.jctc.5b00834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86•.Emsley P., Brunger A.T., Lutteke T. Tools to assist determination and validation of carbohydrate 3D structure data. Methods Mol Biol. 2015;1273:229–240. doi: 10.1007/978-1-4939-2343-4_17. [DOI] [PubMed] [Google Scholar]; Detailed protocols on validation of three-dimensional data on carbohydrate structures in databases such as PDB is covered in this paper.
- 87.Lutteke T., von der Lieth C.W. Data mining the PDB for glyco-related data. Methods Mol Biol. 2009;534:293–310. doi: 10.1007/978-1-59745-022-5_21. [DOI] [PubMed] [Google Scholar]
- 88.Tharakaraman K., Raman R., Viswanathan K., Stebbins N.W., Jayaraman A., Krishnan A., Sasisekharan V., Sasisekharan R. Structural determinants for naturally evolving H5N1 hemagglutinin to switch its receptor specificity. Cell. 2013;153:1475–1485. doi: 10.1016/j.cell.2013.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Imai M., Watanabe T., Hatta M., Das S.C., Ozawa M., Shinya K., Zhong G., Hanson A., Katsura H., Watanabe S. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012;486:420–428. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yamada S., Suzuki Y., Suzuki T., Le M.Q., Nidom C.A., Sakai-Tagawa Y., Muramoto Y., Ito M., Kiso M., Horimoto T. Haemagglutinin mutations responsible for the binding of H5N1 influenza A viruses to human-type receptors. Nature. 2006;444:378–382. doi: 10.1038/nature05264. [DOI] [PubMed] [Google Scholar]
- 91.Lak P., Makeneni S., Woods R.J., Lowary T.L. Specificity of furanoside-protein recognition through antibody engineering and molecular modeling. Chemistry. 2015;21:1138–1148. doi: 10.1002/chem.201405259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tessier M.B., Grant O.C., Heimburg-Molinaro J., Smith D., Jadey S., Gulick A.M., Glushka J., Deutscher S.L., Rittenhouse-Olson K., Woods R.J. Computational screening of the human TF-glycome provides a structural definition for the specificity of anti-tumor antibody JAA-F11. PLoS ONE. 2013;8:e54874. doi: 10.1371/journal.pone.0054874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pathiaseril A., Woods R.J. Relative energies of binding for antibody-carbohydrate-antigen complexes computed from free-energy simulations. J Am Chem Soc. 2000;122:331–338. doi: 10.1021/ja9914994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Soundararajan V., Zheng S., Patel N., Warnock K., Raman R., Wilson I.A., Raguram S., Sasisekharan V., Sasisekharan R. Networks link antigenic and receptor-binding sites of influenza hemagglutinin: mechanistic insight into fitter strain propagation. Sci Rep. 2011;1:200. doi: 10.1038/srep00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Elli S., Macchi E., Rudd T.R., Raman R., Sassaki G., Viswanathan K., Yates E.A., Shriver Z., Naggi A., Torri G. Insights into the human glycan receptor conformation of 1918 pandemic hemagglutinin-glycan complexes derived from nuclear magnetic resonance and molecular dynamics studies. Biochemistry. 2014;53:4122–4135. doi: 10.1021/bi500338r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sassaki G.L., Elli S., Rudd T.R., Macchi E., Yates E.A., Naggi A., Shriver Z., Raman R., Sasisekharan R., Torri G. Human (alpha2→6) and avian (alpha2→3) sialylated receptors of influenza A virus show distinct conformations and dynamics in solution. Biochemistry. 2013;52:7217–7230. doi: 10.1021/bi400677n. [DOI] [PMC free article] [PubMed] [Google Scholar]