Abstract
Investigation into the biological function of 5-benzylidene-4-oxazolidinones revealed dose-dependent inhibition of biofilm formation in Methicillin-resistant S. aureus (MRSA). This structurally unusual class of small molecules inhibit up to 89% of biofilm formation with IC50 values as low as 0.78 μM, and disperse pre-formed biofilms with IC50 values as low as 4.7 μM. Together, these results suggest that 4-oxazolidinones represent new chemotypes to enable the study of bacteral biofilms with small molecule chemical probes.
The growing prevalence of multi-drug resistant organisms represents an increasingly serious threat to human health1. Despite significant medical advances in the past three decades, distressingly few novel classes of antibiotics have been deployed in the clinic2. In that same span, antibacterial resistance has become endemic3. Gram-positive pathogen Methicillin-resistant Staphylococcus aureus (MRSA) has relevance to human health, especially in community- and hospital-acquired infections4. Multi-drug resistant strains of MRSA exhibit resistance to all know classes of antibiotics5, including last-resort antibiotics, linezolid6 and vancomycin7, further motivating the development of new pharmacophores.
A significant factor contributing to the pervasiveness of many bacterial infections is their ability to form biofilms8. A biofilm is a surface-associated community of bacteria encased in an extracellular polymeric matrix of polysaccharides, DNA and proteins, which offers a protective barrier from environmental threats, host immune response and antimicrobial agents9. The biofilm state constitutes a phenotypic shift from individual planktonic bacteria to a surface associated community of bacteria, often manifesting as contrasting gene expression profiles, cellular compositions and greatly reduced growth rates10. Biofilm formation can extend bacterial survivability on surfaces including indwelling medical devices, hospital equipment and clothing, in turn leading to higher rates of secondary infection11. Consequently, bacterial biofilms have been implicated in as many as 80% of human bacterial infections ranging from dental plaques to endocarditis12. In light of the significantly reduced efficacy of conventional antibiotic treatments against biofilms13, new classes of compounds are needed to combat biofilms and serve as small molecule chemical probes to study the fundamental properties of these complex bacterial communities14.
The search for novel scaffolds possessing anti-biofilm properties against MRSA has revealed several classes of bio-active structures4. Melander and co-workers have developed 2-aminoimidazole (2AI) containing small molecules15 based on the marine derived natural product oroidin16 (Figure 1) to inhibit biofilm formation and disperse preformed biofilms in a variety of Gram-positive and Gram-negative pathogens. These novel scaffolds have been shown to modulate biofilm formation across multiple species with no appreciable antimicrobial activity17. Small molecules that modulate biofilm formation without killing the bacteria are potentially attractive since the development of resistance may be delayed due to decreased selection pressure18.
Fig. 1.
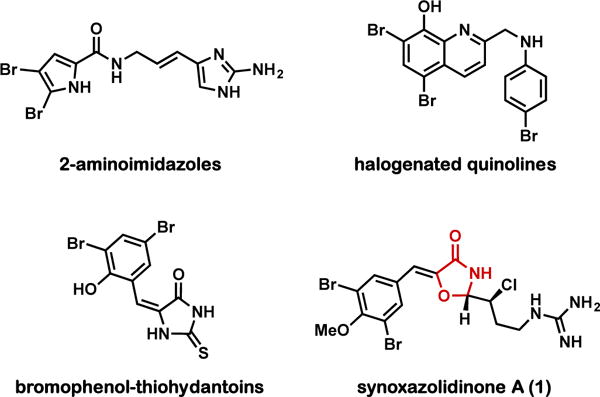
Synoxazolidinone A (1) and several representative classes of small molecules known to modulate MRSA biofilm formation
The recently reported halogenated quinoline class of anti-biofilm agents displays potent biofilm eradication activity19, an alternate mechanism which kills biofilm-associated cells rather than modulating gene expression and dispersing the bacteria20. Additionally, the cyclic dinucleuotide signalling pathway has become a promising target for antimicrobial development, as cyclic dinucleotides are important bacterial second messengers implicated in the modulation of a variety of bacterial growth phenotypes including biofilm formation, motility and virulence21–23. Accordingly, Sintim and co-workers have extensively characterized the c-di-AMP synthase inhibition activity of bromophenol-thoihydantoins and demonstrated their ability to modulate bacterial biofilms24.
The marine natural product synoxazolidinone A (1, Figure 1) possesses an unusual 5-benzylidene-4-oxazolidinone core decorated with a brominated aromatic moiety and chlorinated, guanidine-containing sidechain25. Recent efforts in our lab to develop new synthetic methods for the synthesis of these unusual heterocycles culminated in the first total synthesis of (±)-synoxazolidinone A (±-1)26. Enabled by these efforts, initial investigation into structure-activity-relationship (SAR) generated interest in synoxazolidinone analogues with a 2-dichloroalkyl sidechain. (±)-1 and several of these analogues are moderately active against Staphylococcus aureus and Methicillin-resistant Staphylococcus aureus and somewhat less active against Acinetobacter baumannii; however, no clear SAR could be assessed. Interestingly, 1 and a related natural product, synoxazolidinone C27, were recently shown to exhibit antifouling properties against a variety of marine bacteria28, prompting our investigation into the potential biological function of this heterocyclic scaffold outside of its antimicrobial activity.
To enable an expanded exploration into 2-dichloroalkyl-5-benzylidene-4-oxazolidinones, we prepared a panel of analogues using a modification of the method previously described in the total synthesis of (±)-1 (Figure 2. a, b)26. Initial challenges accessing the necessary α,α-dichloroimine starting materials were overcome by employing tert-butyl imines for the chlorination in carbon tetrachloride, according to an established literature procedure (Figure S2.)29. These α,α-dichloro tert-butylimines were then subjected to the acylation/cyclization conditions, furnishing the desired N-tert-butyl-5-benzylidene-4-oxazolidinones in modest yields (see SI for full details). Subsequent removal of the tert-butyl group using trifluoroacetic acid provided the title compounds in short order, employing a single chromatographic separation.
Fig. 2.
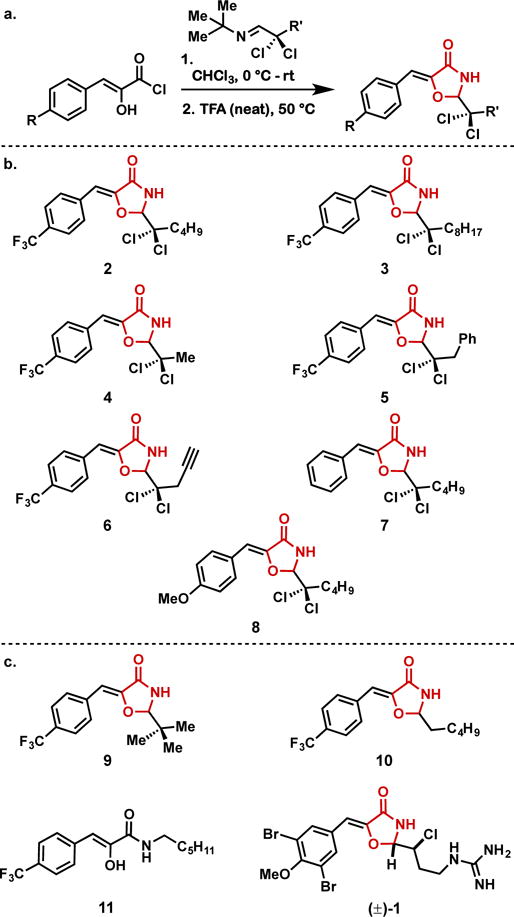
Compounds investigated for biofilm inhibition activity; a) Synthesis of 5-benzylidene-4-oxazolidinones; b) 2-dichloroalkyl-5-benzylidene-4-oxazolidinones; c) additional compounds investigated (see SI).
With a representative series of 2-dichloroalkyl-5-benzylidene-4-oxazolidinones in hand, we next investigated the biological activity of these compounds along with several related small molecules (Figure 2. C; synthesis of 10 and (±)-1 was previously reported26, see SI for synthesis and characterization of compounds 930 and 11). Table 1 provides minimum inhibitory concentrations (MICs) for these compounds against MRSA (ATCC BAA-44). 2 exhibited similar antibacterial activity to that of (±)-1, as we have previously reported with other strains of S. aureus. Antimicrobial activity was completely abolished with a change from an n-butyl side chain (2) to an n-octyl side chain (3). Shortening the side chain from n-butyl (2) to methyl (4) resulted in significantly reduced activity. Conversely, a change from an n-butyl side chain (2) to a benzyl side chain (5) resulted in a 2-fold enhancement of the MIC, from 12 μg/mL. to 6 μg/mL. Inclusion of a terminal alkyne in the side chain (6) resulted in somewhat reduced activity compared to 2. We postulated that the electron-withdrawing trifluoromethyl substituent on the aromatic ring was crucial to the antimicrobial activity of the 2-dichloroalkyl-5-benzylidene-4-oxazaolidinones, and this was supported by the significantly diminished activity of 7 and 8, both bearing an electron-rich aromatic ring. Further, a direct comparison of the antimicrobial activity of 4 to that of 9 reveals the importance of the dichloromethylene moiety for potency, whereby replacing the two chlorine atoms (4) with methyl groups (9) results in a 4-fold reduction in antimicrobial activity. In contrast, replacing the chlorine atoms (2) with hydrogen atoms (10) led to only a slight reduction in MIC, suggesting the benefit of the chlorine atoms is a result of the polar substituents rather than simply steric bulk. Finally, 11 exhibited no antimicrobial activity, illustrating the importance of the 4-oxazolidinone core.
Table 1.
Biological activity of 5-benzylidene-4-oxazolidinones against MRSA (ATCC BAA 44)
| Cpd | MICab (μg/mL) | % Biofilm Inhib. (5 μM)b | % Biofilm Inhib. (40 μM)b | % Biofilm Disp. (5 μM)b | % Biofilm Disp. (40 μM)b |
|---|---|---|---|---|---|
| 1 | 10 | −36 | 62 | −32 | 9.9 |
| 2 | 12 | 89 | NDc | 59 | 87 |
| 3 | >128 | 68 | 75 | 19 | 19 |
| 4 | 32 | 55 | NDc | 50 | NDc |
| 5 | 6 | 82 | NDc | 57 | 69 |
| 6 | 16 | 64 | NDc | 63 | NDc |
| 7 | 64 | 51 | NDc | 14 | 26 |
| 8 | >128 | 39 | 52 | 49 | 58 |
| 9 | 128 | 31 | 51 | 5.7 | 31 |
| 10 | 64 | 17 | 53 | NDc | NDc |
| 11 | >128 | 24 | 41 | −24 | −6.5 |
We next set out to investigate other biological functions of these 4-oxazolidinones and began exploring their activity against bacterial biofilms, given their known anti-fouling properties.28 Biofilm inhibition assays were carried out according to the established crystal violet staining protocol31. The results of these studies are summarized in Table 1. For these studies, we initially explored the percent inhibition of biofilm formation compared to an untreated control and further defined IC50 values for the most potent compounds.
Initial results from biofilm inhibition assays (MRSA ATCC BAA-44) with (±)-1 revealed no biofilm inhibition activity up to 20 μM, followed by moderate inhibition at 40 and 80 μM. The observed activity did not appear to be dose-dependent. Instead 1 displayed no activity until ~30–40 μM when biofilm and planktonic bacterial growth was greatly inhibited (the MIC of 1 is ~20 μM). In contrast, compounds 2, 3 and 5 exhibited dose-dependent biofilm inhibition at concentrations well below their MIC. Additionally, 2 inhibited 89% and 5 inhibited 82% of biofilm formation at 5 μM providing IC50 values of 0.78 μM and 1.2 μM respectively (dose-response curves are provided in the SI). It is important to highlight that the range of concentrations tested directly correlated with the activity observed and the MIC of the compounds to avoid testing at concentrations significantly above the threshold where bacterial growth was greatly impacted. We next explored the impact of the relative size of the substituent on the aminal carbon of the 4-oxazolidinone and found small to medium aliphatic substituents more advantageous.
Compound 4, for example, exhibited a reduction in activity compared to 2 as was also observed for alkyne 6 to a lesser extent. In addition to the alkyl chain length, the electron deficient aromatic ring also seems to play a critical role in the biofilm inhibition potency (as was observed in the antimicrobial activity), whereby replacing the 4-trifluoromethyl substituent with a hydrogen atom reduced the activity significantly (7) and a methoxy substituent (8) resulted in < 50% biofilm inhibition at 5 μM. The dichloromethylene moiety also plays a significant role in the observed activity. Replacing the two chlorine atoms (4) with methyl groups (9) reduced the activity by 3-fold, reinforcing the necessity of the electronegative chlorine atoms. Similarly, substituting the two chlorine atoms (2) with hydrogen atoms (10) resulted in a 5-fold reduction in biofilm inhibition activity. Finally, secondary α-oxoamide 11 exhibited only modest biofilm activity, even at 40 μM, affirming the necessity of the 4-oxazolidinone for biofilm modulating properties.
We also evaluated the activity of this panel of 4-oxazolidinones against pre-formed biofilms using an established crystal violet staining protocol15 (see SI for full experimental details). Percent inhibition values are shown in Table 1. Compound 2 resulted in 59% dispersion of pre-formed biofilm at 5 μM and 87% dispersion at 40 μM (IC50 = 4.7 μM), while 5 displayed 57% dispersion at 5 μM, although a maximum dispersion of 69% was observed at 40 μM.
The antibacterial and anti-biofilm properties of 2 and 5 were also evaluated against three additional MRSA strains (Table 2, ATCC BAA 43 300, 1685, and 1770). Both 2 and 5 exhibited modest biofilm inhibition activity against MRSA strains BAA 43 300, 1685 and 1770 as compared to strain BAA-44. The efficacy of 2 was somewhat reduced against strain 43 300 and 1770. 5 exhibited slightly lower activity against strains 43 300 and 1685, although virtually no activity against strain 1770. On the other hand, biofilm dispersion efficacy of 2 was significantly reduced against MRSA strain BAA 43 300 and 1685, whereas 5 exhibited no dose-dependent dispersion activity against strain 43 300. Further studies are required to evaluate the efficacy of these 4-oxazolidinones on a larger panel of Gram-positive pathogens.
Table 2.
Antibiofilm Activity of 5B4Os Against Relevant MRSA strains
| MRSA Strain | ||||
|---|---|---|---|---|
| Cpd | 43 300 | 1685 | 1770 | |
| 2 | MIC (μg/mL) | 8 | 16 | 16 |
| % Biofilm Inhib. (5/40 μM) | 59/54 | 45/66 | 49/66 | |
| % Biofilm Disp. (5/40 μM) | 23/63 | 24/61 | 52/46 | |
| 5 | MIC (μg/mL) | 8 | 8 | 8 |
| % Biofilm Inhib. (5/40 μM) | 41/63 | 19/17 | 3/−16 | |
| % Biofilm Disp. (5/40 μM) | 37/ND | 15/ND | 29/ND | |
Analysis of the growth curves of 2 and 5 (See SI for full experimental details) reveals substantial inhibition of growth in the first 8 hours by 2 at 1 μM and 5 μM. Although the final cellular density of the culture was similar to that of the untreated control, the delayed growth could account, in part, for the observed biofilm inhibition activity. On the other hand, growth curve of 5 showed modest deviation from the untreated control at 1 μM and moderate growth inhibition in the first 8 hours at 5 μM, indicating minimal inhibition of the growth of planktonic bacteria by this 4-oxazolidinone at optimal biofilm inhibition concentrations. It is worth noting that antimicrobial activity alone does not provide anti-biofilm activity, particularly in the case of dispersion where clinically employed antibiotics possess 10 – 1000× decreased efficacy, highlighting a unique aspect of the small molecules reported herein. Finally, lysis of red blood cells by compounds 2, 3, and 5 was evaluated. 2 and 5 lysed < 1% of red blood cells at 100 μM, while 3 lysed < 1% of red blood cells at 20 μM and 7% of red blood cells at 100 μM.
In summary, we have identified 2-dichloroalkyl-5-benzylidene-4-oxazolidinones as modulators of MRSA biofilms. We have synthesized a series of simplified analogues and assessed their biofilm inhibition and dispersal activity. Through structure-activity analyses, we have determined 1) electron-withdrawing substituents on the benzylidene moiety are important for antibiofilm potency, 2) the 4-oxazolidinone core is required for biological activity and 3) the dichloromethylene functionality improves biofilm activity. Ongoing efforts in our laboratory are focused on understanding the mechanisms by which these small molecules elicit their activity as well as investigation of biofilm inhibitory properties of this new class of compounds across an expanded set of bacterial species.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (Grant R01GM110154)‡
‡The authors would like to thank Prof. Christian Melander and Dr. Roberta Melander (NC State) for enlightening discussions on bacterial biofilms and assistance with the biofilm assay protocols. Mass spectrometry data was obtained from the NC State Mass Spectrometry Facility.
Footnotes
Electronic Supplementary Information (ESI) available: Detailed experimental procedures, analytical data for all previously unreported compounds, NMR spectra and all relevant biological data. See DOI: 10.1039/x0xx00000x
References
- 1.Bush K, Courvalin P, Dantas G, Davies J, Eisenstein B, Huovinen P, Jacoby GA, Kishony R, Kreiswirth BN, Kutter E, Lerner SA, Levy S, Lewis K, Lomovskaya O, Miller JH, Mobashery S, Piddock LJV, Projan S, Thomas CM, Tomasz A, Tulkens PM, Walsh TR, Watson JD, Witkowski J, Witte W, Wright G, Yeh P, Zgurskaya HI. Nat Rev Micro. 2011;9:894–896. doi: 10.1038/nrmicro2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clatworthy AE, Pierson E, Hung DT. Nat Chem Biol. 2007;3:541–548. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- 3.Hampton T. JAMA. 2013;310:1661–1663. doi: 10.1001/jama.2013.280695. [DOI] [PubMed] [Google Scholar]
- 4.M DJ, Jr, Hergenrother PJ. Curr Med Chem. 2006;13:2163–2177. doi: 10.2174/092986706777935212. [DOI] [PubMed] [Google Scholar]
- 5.Schito GC. Clin Microbiol Infect. 2006;12(Supplement 1):3–8. doi: 10.1111/j.1469-0691.2006.01343.x. [DOI] [PubMed] [Google Scholar]
- 6.Gu B, Kelesidis T, Tsiodras S, Hindler J, Humphries RM. J Antimicrob Chemother. 2012 doi: 10.1093/jac/dks354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sieradzki K, Tomasz A. J Bacteriol. 1997;179:2557–2566. doi: 10.1128/jb.179.8.2557-2566.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costerton JW, Stewart PS, Greenberg EP. Science. 1999;284:1318. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 9.Donlan RM, Costerton JW. Clin Microbiol Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith RS, Iglewski BH. Curr Opin Microbiol. 2003;6:56–60. doi: 10.1016/s1369-5274(03)00008-0. [DOI] [PubMed] [Google Scholar]
- 11.Flemming HC, Wingender J. Nat Rev Micro. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 12.Davies D. Nat Rev Drug Discov. 2003;2:114–122. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- 13.Rasmussen TB, Givskov M. Int J Med Microbiol. 2006;296:149–161. doi: 10.1016/j.ijmm.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Lyon GJ, Muir TW. Chem Biol. 2003;10:1007–1021. doi: 10.1016/j.chembiol.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 15.Richards JJ, Ballard TE, Melander C. Org Biomol Chem. 2008;6:1356–1363. doi: 10.1039/b719082d. [DOI] [PubMed] [Google Scholar]
- 16.Zidar N, Montalvão S, Hodnik Ž, Nawrot AD, Žula A, Ilaš J, Kikelj D, Tammela P, Mašič PL. Mar Drugs. 2014;12 doi: 10.3390/md12020940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richards JJ, Reyes S, Stowe SD, Tucker AT, Ballard TE, Mathies LD, Cavanagh J, Melander C. J Med Chem. 2009;52:4582–4585. doi: 10.1021/jm900378s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sintim HO, Smith JAI, Wang J, Nakayama S, Yan L. Future Med Chem. 2010;2:1005–1035. doi: 10.4155/fmc.10.185. [DOI] [PubMed] [Google Scholar]
- 19.Basak A, Abouelhassan Y, Huigens RW., III Org Biomol Chem. 2015;13:10290–10294. doi: 10.1039/c5ob01883h. [DOI] [PubMed] [Google Scholar]
- 20.Fletcher MH, Jennings MC, Wuest WM. Tetrahedron. 2014;70:6373–6383. [Google Scholar]
- 21.Huynh TN, Luo S, Pensinger D, Sauer JD, Tong L, Woodward JJ. Proc Nat Acad Sci. 2015;112:E747–E756. doi: 10.1073/pnas.1416485112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson JW, Sudarsan N, Furukawa K, Weinberg Z, Wang JX, Breaker RR. Nat Chem Biol. 2013;9:834–839. doi: 10.1038/nchembio.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rabin N, Zheng Y, Opoku-Temeng C, Du Y, Bonsu E, Sintim HO. Future Med Chem. 2015;7:493–512. doi: 10.4155/fmc.15.6. [DOI] [PubMed] [Google Scholar]
- 24.Zheng Y, Zhou J, Cooper SM, Jr, Opoku-Temeng C, De Brito AM, Sintim HO. Tetrahedron. 2016;72:3554–3558. [Google Scholar]
- 25.Tadesse M, Strøm MB, Svenson J, Jaspars M, Milne BF, Tørfoss V, Andersen JH, Hansen E, Stensvåg K, Haug T. Org Lett. 2010;12:4752–4755. doi: 10.1021/ol101707u. [DOI] [PubMed] [Google Scholar]
- 26.Shymanska NV, An IH, Pierce JG. Angew Chem Int Ed. 2014;53:5401–5404. doi: 10.1002/anie.201402310. [DOI] [PubMed] [Google Scholar]
- 27.Tadesse M, Svenson J, Jaspars M, Strøm MB, Abdelrahman MH, Andersen JH, Hansen E, Kristiansen PE, Stensvåg K, Haug T. Tet Lett. 2011;52:1804–1806. [Google Scholar]
- 28.Trepos R, Cervin G, Hellio C, Pavia H, Stensen W, Stensvåg K, Svendsen J-S, Haug T, Svenson J. J Nat Prod. 2014;77:2105–2113. doi: 10.1021/np5005032. [DOI] [PubMed] [Google Scholar]
- 29.Kimpe ND, Verhé R, Buyck LD, Schamp N. Synth Commun. 1975;5:269–274. [Google Scholar]
- 30.Shymanska NV, An IH, Guevara-Zuluaga S, Pierce JG. Bioorg Med Chem Lett. 2015;25:4887–4889. doi: 10.1016/j.bmcl.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 31.Rogers SA, Melander C. Angew Chem Int Ed. 2008;47:5229–5231. doi: 10.1002/anie.200800862. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


