Abstract
Providing temporally-regulated glial cell line-derived neurotrophic factor (GDNF) to injured nerve can promote robust axon regeneration. However, it is poorly understood why providing highly elevated levels of GDNF to nerve can lead to axon entrapment in the zone containing elevated GDNF. This limited understanding represents an obstacle to the translation of GDNF therapies to treat nerve injuries clinically. Here, we investigated how transgenic Schwann cells (SCs) overexpressing GDNF-IRES-DsRed impact nerve regeneration. Cultured primary SCs were transduced with lentiviruses (GDNF-overexpressing transgenic SCs), one of which provides the capability to express high levels of GDNF and regulate temporal GDNF expression. These SC groups were transplanted into a cellular nerve allografts (ANAs) bridging a 14mm rat sciatic nerve defect. GDNF-overexpressing transgenic SCs expressing GDNF for as little as 1 week decreased axon regeneration across ANAs and caused extensive extracellular matrix (ECM) remodeling. To determine whether additional gene expression changes beyond GDNF transgene expression occurred in GDNF-overexpressing transgenic SCs, microarray analysis of GDNF-overexpressing transgenic SCs compared to untreated SCs was performed. Microarray analysis revealed a set of common genes regulated in transgenic SC groups expressing high levels of GDNF compared to untreated SCs. A co-culture model of GDNF-overexpressing transgenic SCs with fibroblasts (FBs) revealed differential FB ECM-related gene expression compared to untreated SCs. These data suggest a component of axon entrapment is independent of GDNF's impact on axons.
Keywords: a cellular nerve allograft, candy store effect, gene therapy, glial cell line-derived neurotrophic factor, nerve regeneration, peripheral nerve
Introduction
Despite improved surgical techniques, recovery is usually incomplete after peripheral nerve injury and reconstruction (Mackinnon 2015). Common to all reconstructive efforts (end-to-end repair, nerve grafting, and nerve transfers) is a time-dependent element effecting nerve regeneration, where the growth supportive environment for axons in both the nerve and muscle declines over time (Fu and Gordon 1995a; Fu and Gordon 1995b; Gordon et al. 2011; Kobayashi et al. 1997). A substantial component of the time-dependent, growth-promoting environment within the nerve is provided via the endogenous expression of growth factors by support cells, such as Schwann cells (SCs) (Boyd and Gordon 2003b; Fu and Gordon 1997; Hoke et al. 2000).
Glial cell line-derived neurotrophic factor (GDNF) is one of these factors and is a potent component of the normal regenerative process after peripheral nerve injury. GDNF expression is transiently upregulated by SCs in the distal nerve following injury (Naveilhan et al. 1997; Trupp et al. 1995). Both sensory and motor neurons express receptors for GDNF (Ret/GFRα1) (Naveilhan et al. 1997), where GDNF signaling promotes axon outgrowth and neuronal survival (Bennett et al. 1998; Gavazzi et al. 1999; Leclere et al. 2007; Matheson et al. 1997; Trupp et al. 1995; Tucker et al. 2006). In addition, SCs also express receptors for GDNF (NCAM/GFRα1) (Iwase et al. 2005), where GDNF signaling activates pathways in SCs implicated in cell migration, differentiation, and growth factor production (Ellerbroek et al. 2003; Grimm et al. 1998; Iwase et al. 2005; Jesuraj et al. 2014; Kim et al. 1997; Kinameri and Matsuoka 2003; Klemke et al. 1997; Lang et al. 1996; Marquardt and Sakiyama-Elbert 2015; Meintanis et al. 2001; Morgan et al. 1991; Verity et al. 1998). However, the duration of endogenous GDNF expression following nerve injury is quite short (∼2 weeks) (Boyd and Gordon 2003a; Hoke et al. 2002) and is often less time than necessary for regenerating axons to reach end organ targets, as axon growth rates are ∼1-3 mm/day in humans (Hoke 2011). To overcome these limitations, exogenous GDNF can be administered directly to the nerve to improve regeneration when endogenous GDNF expression has declined (Boyd and Gordon 2003a; Wood et al. 2013a; Wood et al. 2013b).
While a variety of approaches including drug delivery systems and gene therapy have been developed to deliver growth factors locally to injured nerve (de Winter et al. 2013; Hoyng et al. 2015; Hoyng et al. 2011; Johnson et al. 2013; Marquardt and Sakiyama-Elbert 2013), finding the proper approach for GDNF delivery to improve nerve regeneration is still a major challenge. This issue arises in part due to an unusual phenomenon that occurs when supra-physiological levels of GDNF accumulate in nerve preceding regenerating axons. As axons grow through this area of supra-physiological GDNF levels, the axons become entrapped within the area, where few axons are able to extend beyond the zone of “high GDNF”; therefore, the resulting outcome is poor. Axon entrapment or the “candy store” effect is theorized to result from GDNF's chemoattractant properties for axons (Eggers et al. 2008; Tannemaat et al. 2008), but the mechanisms are poorly understood.
While much effort has focused on how GDNF levels impact neurons and their axons, GDNF impacts SC myelination in vivo (Eggers et al. 2013; Hoke et al. 2003) and their capabilities to support axon growth in neuronal culture (Marquardt and Sakiyama-Elbert 2015). In these studies, we investigated how elevated GDNF levels via lentiviral transduction and activation from transgenic SCs can impact nerve regeneration as well as the transduced SC gene expression. We hypothesized that GDNF transgenic SCs impact the SC's innate functions and may thus be a contributing factor to axon entrapment.
Materials and methods
All materials are from Sigma Aldrich (St. Louis, MO) unless otherwise specified. All surgical procedures were performed in strict accordance with the National Institutes of Health guidelines and were approved by Washington University's Institutional Animal Care and Use Committee (IACUC).
Isolation and expansion of primary SCs
SC cultures were prepared using previously described methods (Kaewkhaw et al. 2012; Morrissey et al. 1991; Wu-Fienberg et al. 2014). Briefly, the sciatic nerve was harvested from adult male Lewis rats (Charles River Laboratories, Wilmington, MA) using aseptic technique. The nerve was washed 2-3 times with growth medium: Dulbecco's Modified Eagle's Medium (DMEM; Invitrogen, Carlsbad, CA) or DMEM-D-valine (PAA Laboratories, Piscataway, NJ), containing 10% fetal bovine serum (FBS) supplemented with 100 U/mL penicillin, 100 μg/mL streptomycin, 0.25 μg/mL amphotericin B, 20 μg/mL bovine pituitary extract (FBS), and 5 μM forskolin. The nerve was subsequently incubated with growth medium for 7 days. These explants were then incubated overnight at 37°C in SC culture medium: DMEM, 10% FBS, 1% antibiotic antimitotic (ABAM; Invitrogen), which was supplemented with 1.25 U/mL dispase (PeproTech, Rocky Hill, NJ) and 0.05% collagenase type IV (PeproTech). Tissue was dissociated then centrifuged at 400xg for 6 minutes to obtain a pellet. The pellet was washed in DMEM/10% FBS, then seeded on tissue culture plates coated with poly-L-lysine (pLL) in SC culture medium. After 6 days, fibroblasts were complement-killed using a 1:40 dilution of anti-Thy 1.1 antibody (Serotec, Raleigh, NC) and a 1:4 dilution of rabbit complement in medium. Cultures were passaged as needed and split 1:2 on pLL-coated plates when they exceeded 80% confluence. SC purity (>95%) was verified using immunohistochemistry to Thy1 and S100β. SCs were cultured using growth medium at 37°C in a water-jacketed incubator at 5% CO2, 20% O2.
Lentiviral vector construction
To generate transgenic SCs, lentivirus was produced and constructed by the Hope Center for Neurological Disorders Viral Vectors Core at Washington University in Saint Louis. The diagrams of two vectors (GDNF expression) are shown below (Figure 1). The plasmid packaging cell line, 293T was maintained in DMEM, supplemented with 10% fetal bovine serum, 100U/ml penicillin, 100μg/ml streptomycin in a 37°C incubator with 5% CO2. 293T cells were plated at 30-40% confluence 24hr before transfection (70-80% confluence when transfection). Ten μg of lentiviral vector with the appropriate insert, 5.8μg of pMD-Lg, 3.1μg of pCMV-G, and 2.5μg of RSV-REV were co-transfected into 293T cells using the calcium phosphate precipitation procedure. Six hours after transfection, the medium was replaced with the complete medium containing 6mM sodium butyrate. Culture supernatant was collected 42hr after transfection. The supernatant was passed through a 0.45 μm filter, concentrated by ultracentrifugation through a 20% sucrose cushion, and stored at -80°C until use. Vector titers were determined by transduction of HT1080 cells followed by qPCR assay.
Figure 1.
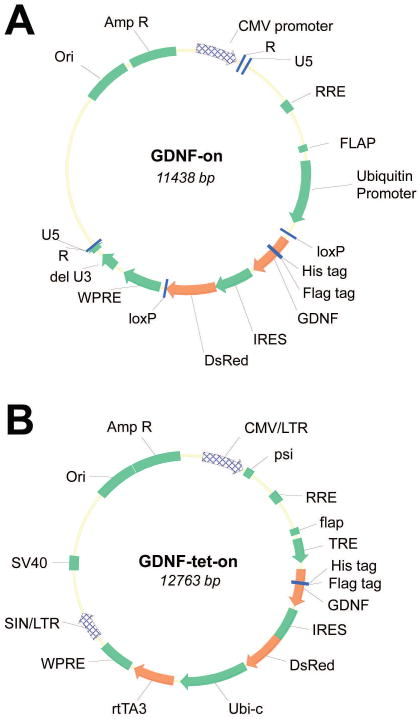
Diagrams of lentiviral constructs. Lentiviruses were designed and prepared to provide constitutively active GDNF expression (GDNF-on; A) or tetracycline derivative inducible GDNF expression (GDNF-tet-on; B).
Generation of transgenic SCs expressing GDNF
SC transduction was performed similar to previous methods (Wu-Fienberg et al. 2014). The day prior to transduction, normal SCs were seeded at 1×104 cells/well on pLL-coated 24-well plates and on the day of transduction, the number of cells in each well was determined by microscopy. Cells were incubated with 2μg/mL of polybrene (Santa Cruz Biotechnology, Dallas, TX) in SC culture medium for 1hr at 37°C, after which the vector was introduced. A multiplicity of infection (MOI) of ∼8-10 was used to transfect cells. To obtain a high purity of transduced cells, cells expressing intense DsRed fluorophore were collected by fluorescence-activated cell sorting (FACS) at the Site man Cancer Center of Washington University in Saint Louis on a FACScan Flow Cytometer (BD Biosciences, San Jose, CA). Data were analyzed with FlowJo software (FlowJo, Ashland, OR). SC transgene activation (GDNF-tet-on SCs) in culture was performed by adding doxycycline at 5μg/ml in growth medium.
Animal surgeries and design
Surgical procedures and peri-operative care measures were conducted in compliance with the Institutional Animal Care Use Committee (IACUC) and NIH guidelines. All animals were housed in a central animal care facility and provided with food (PicoLab rodent diet 20, Purina Mills Nutrition International, St. Louis, MO) and water ad libitum. All surgeries were performed under aseptic conditions and with the aid of an operating microscope under magnifications of 10-25X. Anesthesia was provided by intraperitoneal injection of ketamine (75 mg/kg, Fort Dodge Animal Health, Fort Dodge, IA) and dexmedetomidine (0.5 mg/kg, Pfizer Animal Health, Exton, PA).
A total of 48 adult male Lewis rats (250 g, Charles River Laboratories, Wilmington, MA) were randomized to one of six groups (Table 1). Animals were anesthetized as described above and the sciatic nerve was exposed using a gluteal muscle splitting technique. The sciatic nerve was transected 5 mm proximal to the trifurcation of the sciatic nerve and then immediately repaired with 14mm ANA nerve grafts by using four to five interrupted epineurial sutures (9–0 nylon) on both the proximal and distal nerve ends. Before securing the ANAs for groups indicated to contain SCs (Table 1), the ANA was injected with cells using a 26-gauge microsyringe (Hamilton Company, Reno, NV) oriented longitudinally toward distal nerve stump underneath the epineurium of the graft. Following these procedures, the muscle and skin were then closed in a layered fashion using vicryl and nylon suture (4-0), respectively. Animals were recovered with a subcutaneous injection of atipamezole HCl (1mg/kg, Antisedan®, Orion Corporation) and placed on a warming pad post-operatively. Following surgery, post-operative pain and hypersensitivity was managed using a single dose of Buprenorphine SR Lab (1.0 mg/kg, Zoo Pharm, Windsor, CO), and animals were returned to the central housing facility and closely monitored for infection, distress, and other morbidities.
Table I. Animal Experimental Design.
| Group name | Animal number (n) | Schwann cell (SC) description | Number of cells transplanted |
|---|---|---|---|
| Untreated | 8 | Untreated (normal culture conditions) | 1×106 SCs |
| GDNF-on | 8 | Transduced with constitutively active lentivirus containing transgene GDNF | 1×106 SCs |
| GDNF-tet-on (DOX) | 8 | Transduced with tetracycline inducible lentivirus containing transgene GDNF; Doxycycline for 6 weeks in vivo | 1×106 SCs |
| GDNF-tet-on (no DOX) | 8 | Transduced with tetracycline inducible lentivirus containing transgene GDNF; No Doxycycline | 1×106 SCs |
| GDNF-tet-on (1wk DOX) | 8 | Transduced with tetracycline inducible lentivirus containing transgene GDNF; Doxycycline for 1 week in vivo | 1×106 SCs |
| ANA | 8 | No cells added | None |
All animals were harvested for nerve tissue 6 weeks following procedures.
To generate a cellular nerve allografts (ANAs), a total of 24 Sprague-Dawley rats (250°g, Charles River Laboratories, Wilmington, MA) were used as donors to obtain nerve for processing. Sciatic nerve was exposed as described above and harvested bilaterally from donor rats. These nerves were processed and decellularized using a series of detergents as previously described (Poppler et al. 2016). Following tissue harvest, animals were euthanized with an intraperitoneal injection of Euthasol® (150 mg/kg, Delmarva Laboratories, Des Moines, IA).
Tissue harvesting and histology
At 6 weeks post-operatively, animals were re-anesthetized and prepped to expose the right sciatic nerve and graft. Following tissue harvest, animals were euthanized. En bloc specimens of the graft and distal sciatic nerve underwent histomorphometric analysis as previously described (Hunter et al. 2007). Briefly, nerve was harvested and stored in 3% glutaraldehyde (Polysciences Inc., Warrington, PA). The nerves were post-fixed in 1% osmium tetroxide and serially dehydrated in ethanol and toluene. The nerves were then embedded in epoxy (Polysciences), and sectioned on an ultramicrotome into 1μm cross sections. Slides were counter-stained with 1% toluidine blue dye. The slides were then analyzed at 1000× on a Leitz Laborlux S microscope. The Leco IA32 Image Analysis System (Leco, St. Joseph, MI) was utilized to quantify nerve fiber counts and percent neural tissue. All analysis was done by an observer blinded to the experimental groups.
RNA Isolation from SCs for gene expression analysis
For microarray and qRT-PCR analysis, cultured cell RNA was extracted using Trizol (Life Technologies, Carlsbad, CA), chloroform and RNeasy Mini Kit (Qiagen, Valencia, CA) according to manufacturer's instructions, where 4 samples each were collected SC cultures at the end of 7 days where groups included: untreated SCs; GDNF-on SCs; and GDNF-tet-on SCs (DOX; induced with doxycycline). RNA concentration was determined on a Nano Drop 1000 Spectrophotometer (Thermo Scientific, Wilmington, DE) and purity and integrity were assessed on an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) according to manufacturer's recommendations.
Microarray: amplification and labeling of RNA transcripts, hybridization, and analysis
For microarray analysis, RNA transcripts were first amplified by T7 linear amplification using the Message Amp II aRNA Amplification Kit (cat# AM 1751, Life Technologies, Carlsbad, CA). Four hundred (400) ng of RNA (11μl) was primed with an oligo-dT T7 primer (1μl) by heating to 70°C for 10 minutes, then cooled on ice for 3 minutes. For 1st strand cDNA synthesis, each RNA received 10× reaction buffer (2μl), dNTP mix (4μl), RNase Inhibitor (1μl), and reverse transcriptase (1μl) (Superscript III; Life Technologies). Reverse transcription was carried out for 2 hours at 50°C. After a three minute incubation on ice, the cDNA underwent 2nd strand synthesis by adding water (63μl), 10× 2nd strand buffer (10μl), dNTP mix (4μl), DNA polymerase (2μl) and RNase H (1μl). This cocktail was incubated at 16°C for 2 hours. Following a short column cleanup according to manufacturer's protocol (DNA Clean and Concentrator 5; Zymo Research), in vitro-transcription was carried out by adding non-modified bases (ATP, CTP, GTP, and UTP) (4μl of each), 10× T7 reaction buffer (4μl), and T7 RNA polymerase enzyme mix (4μl). The IVT reaction was carried out for 14 hours. Following reaction termination with water (60μl), the RNAs were cleaned with RNeasy columns (Qiagen) according to manufacturer's protocol.
Balanced RNAs were suspended in Agilent 2X Gene Expression buffer (55μl), Agilent 10X Blocking agent (11μl), and KREAblockTM (6μl). The hybridization solutions were applied to Agilent Human v1 4x44K microarrays (G2519F-014850). Hybridization was carried out at 65°C for 20 hours. Washing procedures were carried out according to Agilent gene expression protocols. Slides were scanned on an Agilent C-class Microarray scanner to detect Cy5 fluorescence, according to manufacturer's specifications. Gridding and analysis of images was performed using Feature Extraction (v11.5.1.1, Agilent Technologies, Santa Clara, CA).
Gene and protein expression analysis of SC cultures
To verify select gene and protein expression from cultured cells, qRT-PCR and ELISA were performed. RNA was isolated as described and reverse transcribed into complementary DNA per manufacturer's instructions for the High Capacity RNA to DNA by Superscript II Reverse Transcriptase (Life Technologies, Carlsbad, CA). Taqman Probes and Taqman Fast Advanced Master Mix (Life Technologies) were used for qPCR analysis according to manufacturer's recommendations. Gene expression levels were normalized to a housekeeping gene (β-actin (ACTB)). Analyses were performed on a Step One Plus thermocycler (Applied Biosystems, Foster City, CA) and the data analyzed on Step One Software v2.2.2 (Applied Biosystems). Quantification of GDNF production was determined by performing enzyme-linked immune-sorbent assay (ELISA) on cell culture medium containing doxycycline at 5μg/ml in growth medium incubated with confluent cells. Cells were seeded and grown with the same medium 72 hours prior to this incubation on 24 well plates at a density of 1×104 cells/well. Confluent cell density was measured (∼100,000 cells/well) and 1 mL of culture medium was incubated with the cells and collected after 1 hour for ELISA measurement. ELISA was performed on this medium according to the manufacturer's instructions (R&D Systems, Minneapolis, MN). Plates were read using a BioTek EL800 (BioTek, Winooski, VT) plate reader and BioTek software.
Fibroblast and SC co-culture
Co-culture model systems of SC and fibroblasts (FBs) were generated using SCs, derived as previously described, with a rat FB cell line (CRL-1764, ATCC, Manassas, VA). First, SC culture was generated under the stated conditions. GDNF-tet-on (DOX) SCs were treated with 5μg/ml doxycycline in growth media for 72 hours to induce robust GDNF expression. This media was then removed, and the cells were trypsinized and seeded with the FBs at a ratio of SC:FB 2:1 using 10% FBS-DMEM culture medium for 48 hours. SCs transduced with a Td-tomato reporter served as a vehicle control group as active fluorescent reporter was necessary for SC sorting. Following this co-culture for 48 hours, the cells were trypsinized to select for FBs using a negative selection process (SCs expressing reporter) via fluorescence-activated cell sorting (FACS). The sorted fibroblasts were then washed twice and seeded to non-PLL coated 6 wells plate for two hours to select for viable cells which attach. Finally, the attached cells were washed twice with cold PBS and RNA was isolated using a NucleoSpin RNA kit (Macherey-Nagel GmbH & Co. KG, Germany) according to the manufacturer's instruction.
Data and statistical analysis
All data are represented as mean ± standard deviation. For animal studies, each animal was considered an ‘n,’ while for cell culture studies each culture well was treated as an ‘n’ and experiments were performed in triplicate. Statistical analyses were performed using GraphPad Prism version 5 (GraphPad Software, Inc., La Jolla, CA). Student's t test or ANOVA (for multiple comparisons) followed by Tukey post hoc tests were used to determine statistical significance, where each group was compared to all other groups and p<0.05 considered statistically significant. For data presented in figures, lowercase letters were used to indicate which groups were not statistically different from one another (p>0.05; each group has the same letter) or statistically different from one another (p<0.05; differing group(s) will have a letter that is different from the other group(s)).
Results and Discussion
Transplanted SCs expressing elevated levels of GDNF decrease axon regeneration
Two sets of transgenic SCs capable of expressing GDNF at high levels were developed. These SCs were transduced with lentiviruses for constitutive expression of GDNF transgene (GDNF-on) or doxycycline-inducible GDNF transgene expression (GDNF-tet-on). Using qRT-PCR analysis and ELISA, we verified that elevated quantities of GDNF were expressed by either of these GDNF-overexpressing transgenic SC groups (Figure 2).
Figure 2.
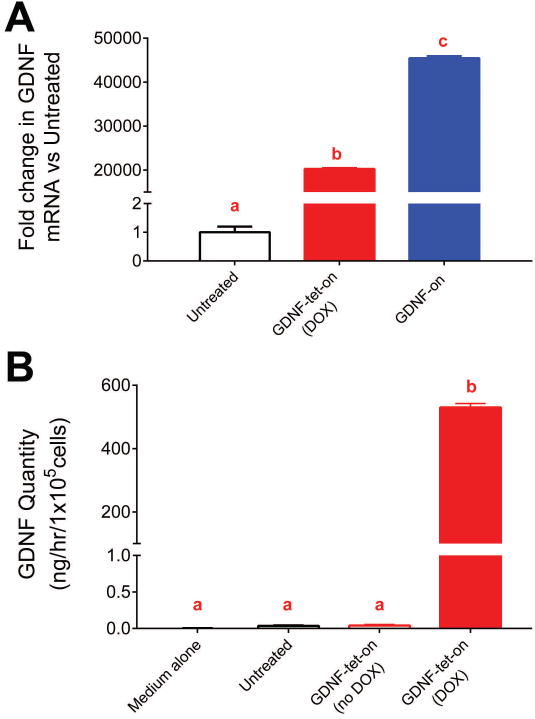
GDNF expression from GDNF transgenic SCs. SCs transduced with lentivirus to overexpress GDNF had higher Gdnf mRNA expression (A). Additionally, GDNF transgenic SCs were capable of expressing >500 ng/mL/hr of secreted GDNF protein (B). Data represented as means with standard deviation error bars (n≥3) and letters indicate which groups were not statistically different from one another (p>0.05; each group has the same letter) or statistically different from one another (p<0.05; differing groups will have a letter that is different from the other groups).
The regenerative impact these GDNF-overexpressing transgenic SCs have on nerve regeneration was assessed by transplanting them into short (14mm) ANAs used to repair a sciatic nerve defect. Aspects critical for nerve regeneration were measured using histomorphometric analysis of distal nerve and ANAs at a 6 week endpoint. Myelinated axon counts in the distal nerve were decreased for either GDNF-overexpressing transgenic SC group (GDNF-on and GDNF-tet-on (DOX)) compared to untreated SCs (Untreated) (Figure 3A). This decrease in axon regeneration across the ANA was not due to the lentiviral transduction of the SCs or related genotoxicity to the SCs, as animals not administered doxycycline (i.e. no transgene activation; GDNF-tet-on (no DOX)) had myelinated axon counts in the distal nerve not different than the Untreated group (p>0.05). Alternatively, conditional activation of GDNF-overexpression for only one week as opposed to 6 weeks (GDNF-tet-on (1wk DOX)) still resulted in decreased myelinated axon counts in the distal nerve. This overall outcome was also reflected in percent neural tissue, a measure of the quality of axonal regeneration, where again any GDNF-overexpressing transgenic SC group had decreased percent neural tissue compared to transgenic SCs with no GDNF transgene activation (GDNF-tet-on (no DOX))) (Figure 3B). No group demonstrated differences to any other group in myelinated axon counts (between 5,000-10,000 myelinated axons) within the mid-graft of ANAs, which suggests that axon regeneration into the ANA was not generally effected (Figure 3C).
Figure 3.
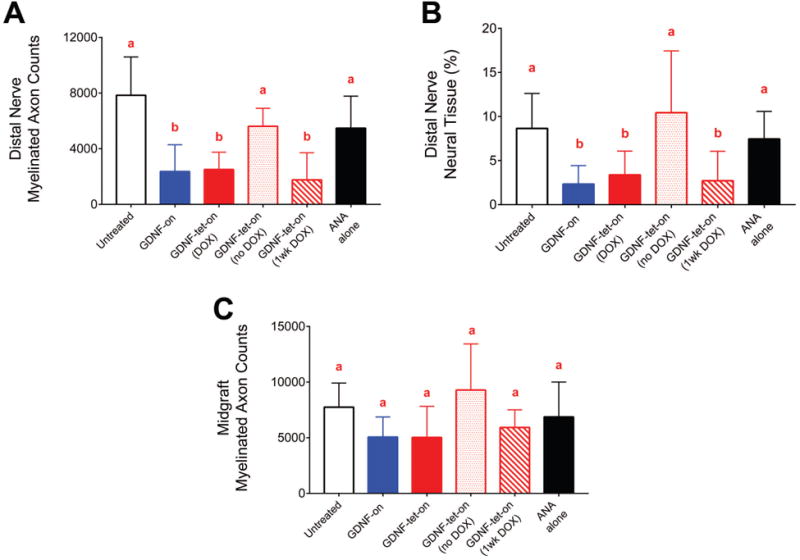
Morphometric analysis of axonal regeneration across ANAs containing GDNF-overexpressing transgenic SCs. Six weeks following implantation of SCs into ANAs, nerve distal to ANAs was harvested for analysis of axonal regeneration. Groups containing elevated GDNF levels contained significantly fewer myelinated axons (A) and percent neural tissue (B) compared to controls. No groups demonstrated differences in myelinated axon counts (C) within the ANAs. Data represented as means with standard deviation error bars (n=8) and letters indicate which groups were not statistically different from one another (p>0.05; each group has the same letter) or statistically different from one another (p<0.05; differing groups will have a letter that is different from the other groups).
The transplantation of GDNF transgenic SCs to ANAs used to repair a nerve defect confirmed elevated levels of GDNF from GDNF transgenic SCs cause axon entrapment, as previously determined (Santosa et al. 2013). However, we have expanded on this result to determine that even temporal regulation can still lead to entrapment within an ANA. By using SCs with conditional activation of GDNF, it was revealed that elevated GDNF from transgenic SCs can cause axon entrapment with as little as 1 week of elevated GDNF levels within an ANA.
Transplanted SCs expressing elevated levels of GDNF lead to extracellular matrix remodeling and unmyelinated axon sprouting
Based on the impact that transplanted GDNF-overexpressing transgenic SCs had on axonal regeneration across the ANA, the graft was assessed for possible causes for this outcome. Both GDNF-overexpressing transgenic SC groups (GDNF-on and GDNF-tet-on (DOX)) and the GDNF transgenic SC control (GDNF-tet-on (no DOX)) were compared for qualitative histological assessment of the nerve graft including nerve architecture and extracellular matrix (ECM). Either GDNF-overexpressing transgenic SC group contained areas of disorganized axonal regeneration within the graft compared to GDNF-tet-on (no DOX) (Figure 4A-C). These GDNF-overexpressing transgenic SC groups (GDNF-on and GDNF-tet-on (DOX)) had large areas devoid of myelinated axons and densely stained for ECM structures. In these GDNF-overexpressing transgenic SC groups, the areas containing myelinated axons were densely packed with decreased spacing between axonal regenerating units compared to GDNF-tet-on (No DOX). Indeed, electron microscopy revealed that both GDNF-overexpressing transgenic SC groups had extensive ECM remodeling with tightly packed collagen fiber bundles in areas devoid of myelinated axons (Figure 4D-I). Additionally for both GDNF-overexpressing transgenic SC groups, the areas with myelinated axons contained large numbers of clustered, neighboring unmyelinated axons suggesting extensive axonal sprouting. Overall, both GDNF-overexpressing transgenic SC groups demonstrated a similar impact to nerve regeneration, which included decreased axon regeneration across the GDNF-overexpressing transgenic SC region and extensive ECM remodeling within the ANA suggesting a structural “blockade” to axon regeneration across these environments.
Figure 4.
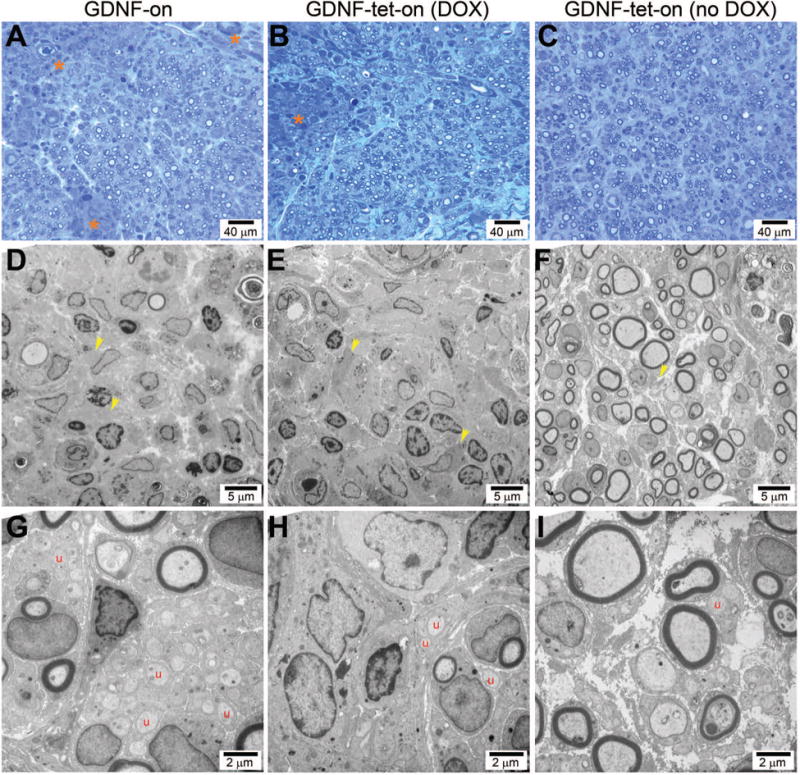
Transplanted GDNF-overexpressing transgenic SCs contained disorganized nerve regeneration including ECM remodeling and unmyelinated axonal sprouts. Histological micrographs taken within ANAs revealed the GDNF-on (A,D,G) and GDNF-tet-on (DOX) (B,E,H) groups had disorganized nerve regeneration compared to the control (GDNF-tet-on (no DOX); C,F,I). Asterisks indicate large regions devoid of myelinated axons with poor ECM structure (A,B). Yellow arrows indicate collagen fibrils, which are tightly clustered in elevated GDNF groups (D,E). Letter ‘u’ indicates unmyelinated axons, which are increased in elevated GDNF groups (G,H).
GDNF-overexpressing transgenic SCs demonstrate changes to their gene expression beyond the intended GDNF transgene expression
To understand whether SCs transduced to express high levels of GDNF have additional modifications to their function and/or gene expression, unbiased, differential microarray analysis was performed. We compared SCs cultured for 1 week while expressing high levels of GDNF (GDNF-on and GDNF-tet-on (DOX)) to SCs cultured for 1 week with no exposure to GDNF (Untreated). A total of 469 probes were found to be a common set differentially expressed in GDNF-overexpressing transgenic SC groups compared to untreated SCs (Figure 5). These 469 genes were refined based on literature indicating their potential relevance in nerve injury and regeneration (i.e. studies demonstrating their role in the nervous system, cell adhesion, cell proliferation, extracellular matrix (ECM) remodeling, or growth factor signaling) leading to a set of 20 shared genes for GDNF-overexpressing transgenic SCs (Table 2). From this set, Neuropeptide Y (Npy) was chosen as a candidate for microarray validation based on high expression (>10 fold absolute change) in either GDNF-overexpressing transgenic SC group. Using qRT-PCR analysis of cell culture and immunohistochemistry of in vivo tissues containing GDNF-overexpressing transgenic SCs, elevated NPY expression from either GDNF-overexpressing transgenic SC group was verified (data not shown).
Figure 5.
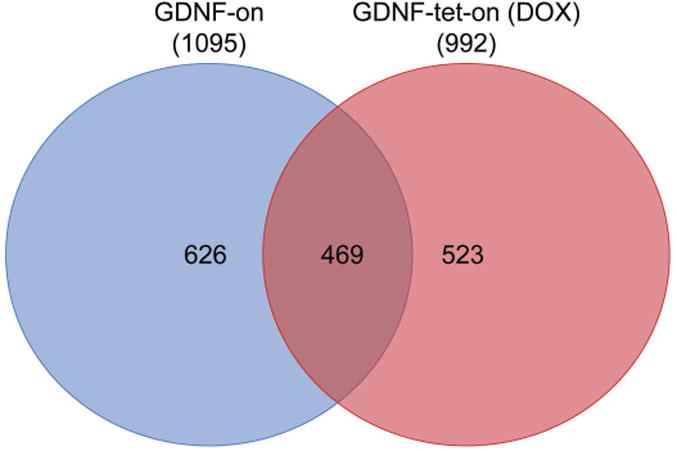
SCs expressing GDNF have differential gene expression patterns. Venn diagram representing the number of differentially regulated genes in SCs expressing GDNF in culture for 1 week compared to untreated, cultured SCs (total number of differentially regulated gene in parenthesis next to group name). SCs expressing GDNF contained a distinct and overlapping set of genes dysregulated. Microarray data obtained from n=4 culture experiments per group, where genes differentially expressed were considered when p<0.05.
Table II. mRNA expression for select shared genes between GDNF-overexpressing transgenic SC groups from microarray datasets.
| Gene symbol | GDNF-on vs Untreated | GDNF-tet-on (DOX) vs Untreated |
|---|---|---|
| Cited1 | 16.8305 | 3.04396 |
| Dclre1b | 2.26544 | 2.13106 |
| Ubash3b | 2.20503 | 11.7411 |
| Dmkn | 4.88226 | 6.59956 |
| Dpys | 7.05057 | 7.68614 |
| Grk6 | 2.12918 | 2.79704 |
| Il18 | 2.90913 | 6.06152 |
| Npy | 45.0458 | 36.8527 |
| Pcna | 2.41008 | 2.28184 |
| Plaur | 3.70414 | 2.15399 |
| Racgap1 | 3.79474 | 2.51704 |
| Timeless | 3.96162 | 5.43282 |
| Col14a1 | -7.01508 | -4.13905 |
| Figf | -4.17877 | -4.33677 |
| Gfra1 | -4.19517 | -3.93374 |
| Il3ra | -2.44535 | -2.53645 |
| Il6st | -2.81296 | -4.34935 |
| Mst1 | -9.85052 | -5.26302 |
| Rgcc | -4.42464 | -6.21813 |
| Wif1 | -2.43653 | -2.39099 |
Twenty genes were selected from the microarray analysis (n=4 per group); the values shown indicate the fold difference in mRNA levels (p<0.05).
Elevated levels of GDNF impact FB gene expression in SC-FB co-culture
While the impact of short-term elevated GDNF delivery initially seems to conflict with other studies demonstrating that short-term elevated GDNF delivery from transgenic SCs improves nerve regeneration (Marquardt et al. 2015; Shakhbazau et al. 2013), these previous studies provided elevated GDNF to nerve by transplanting transgenic SCs to nerve distal to the injury and repair site. Our current studies transplanted transgenic SCs to a cellular scaffolds (ANAs), which are repopulated with a cellular distribution similar to nerve but contain an increased number of stromal cells, such as FBs (Poppler et al. 2016).
To consider whether GDNF-overexpressing transgenic SCs impact stromal cells (primarily fibroblasts (FBs)), we assessed changes to a panel of ECM associated genes in a SC-FB co-culture model, where the SC to FB ratio was 2:1, closely mimicking the cell ratio for injured nerve (Poppler et al. 2016; Salonen et al. 1988). The co-culture of FBs with transgenic SCs conditionally expressing a fluorescent reporter (SCs with FBs) did not have an impact on FB gene expression for a small panel of ECM related genes (absolute fold change <2), which included collagens 1a1, 1a2, 4a1, and 18a1. However, the co-culture of GDNF-overexpressing transgenic SCs with FBs resulted in increased expression of Col1a2 and Col4a1 from FBs compared to FBs alone or SCs with FBs (Figure 6). These results demonstrate that the GDNF-overexpressing transgenic SCs impact FB gene expression and also offers further potential insight regarding the failure of temporal GDNF regulation to avoid axon entrapment in our in vivo regeneration model.
Figure 6.
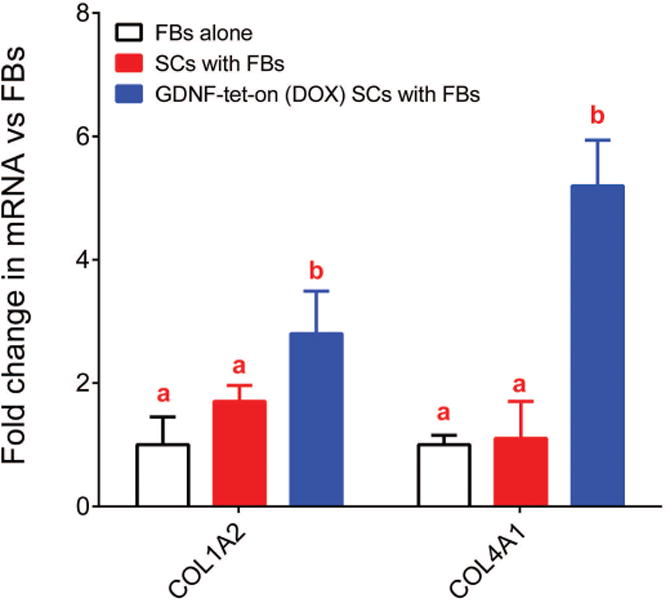
Fibroblast gene expression is impacted due to co-culture with GDNF-overexpressing transgenic SCs. Fibroblasts (FBs) were cultured with transgenic SCs, with or without GDNF expression. ECM FB gene expression was increased when FBs were cultured with GDNF-overexpressing transgenic SCs expressing GDNF. Data represented as means with standard deviation error bars (n=4) and letters indicate which groups were not statistically different from one another (p>0.05; each group has the same letter) or statistically different from one another (p<0.05; differing groups will have a letter that is different from the other groups).
As a final note, a current limitation within our studies is whether the co-expression of DsRed with GDNF contributed to the described circumstances leading to axon entrapment. DsRed has been shown to be cytotoxic to mammalian cells (Strack et al. 2008; Tao et al. 2007), which through cell death could potentially impact nerve regeneration. However, previous experiments from our group have utilized similar GDNF-overexpressing transgenic SCs that co-express DsRed with GDNF. When these cells were transplanted to nerve distal to the injury and repair site, there was no negative impact on nerve regeneration (Marquardt et al. 2015). Therefore, it is unlikely that DsRed toxicity could cause the observed effects and more likely that GDNF-overexpression is the main contributing factor to the observed changes within an ANA associated with the transplanted GDNF-overexpressing transgenic SCs.
Conclusions
The current studies demonstrate that axon entrapment can occur despite temporal regulation of transgenic SCs expressing GDNF. ANAs transplanted with transgenic SCs expressing GDNF-IRES-DsRed lead to significant ECM remodeling of ANA basal lamina associated with axon entrapment, while the GDNF-overexpressing transgenic SCs also demonstrate changes to their gene expression beyond the intended GDNF transgene expression, which may contribute to the outcome. Overall, our studies suggest that a mechanism partially responsible for axon entrapment due to elevated GDNF associated with transgenic SCs is independent of the impact of GDNF on axons.
Acknowledgments
This work was supported in part by National Institutes of Neurological Disorders and Stroke of the National Institutes of Health under award number 2 R01 NS051706. Lentivirus preparation and construction in this study were supported by the Alafi Neuroimaging Laboratory, the Hope Center for Neurological Disorders, and the National Institutes of Neurological Disorders and Stroke of the National Institutes of Health under award number P30 NS057105 to Washington University. We would like to thank Dr. Mingjie Li and his lab (Washington University) for producing the lentiviruses and technical assistance. We thank the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine for help with genomic analysis. We thank the Alvin J. Siteman Cancer Center at Washington University School of Medicine and Barnes-Jewish Hospital in St. Louis, Mo., for the use of the Siteman Flow Cytometry Core, which provided cell sorting service. The Siteman Cancer Center is supported in part by an NCI Cancer Center Support Grant P30 CA91842, while the Genome Technology Access Center is partially supported by this NCI Cancer Center Support Grant and by ICTS/CTSA Grant UL1 TR000448 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and NIH Roadmap for Medical Research. The authors declare no conflict of interest regarding this work. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or Washington University.
Footnotes
Conflict of Interest: The authors declare no competing financial interests. No benefit of any kind will be received either directly or indirectly by the author(s).
References
- Bennett DL, Michael GJ, Ramachandran N, Munson JB, Averill S, Yan Q, McMahon SB, Priestley JV. A distinct subgroup of small DRG cells express GDNF receptor components and GDNF is protective for these neurons after nerve injury. J Neurosci. 1998;18(8):3059–72. doi: 10.1523/JNEUROSCI.18-08-03059.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd JG, Gordon T. Glial cell line-derived neurotrophic factor and brain-derived neurotrophic factor sustain the axonal regeneration of chronically axotomized motoneurons in vivo. Exp Neurol. 2003a;183(2):610–9. doi: 10.1016/s0014-4886(03)00183-3. [DOI] [PubMed] [Google Scholar]
- Boyd JG, Gordon T. Neurotrophic factors and their receptors in axonal regeneration and functional recovery after peripheral nerve injury. Mol Neurobiol. 2003b;27(3):277–324. doi: 10.1385/MN:27:3:277. [DOI] [PubMed] [Google Scholar]
- de Winter F, Hoyng S, Tannemaat M, Eggers R, Mason M, Malessy M, Verhaagen J. Gene therapy approaches to enhance regeneration of the injured peripheral nerve. Eur J Pharmacol. 2013;719(1–3):145–52. doi: 10.1016/j.ejphar.2013.04.057. [DOI] [PubMed] [Google Scholar]
- Eggers R, de Winter F, Hoyng SA, Roet KC, Ehlert EM, Malessy MJ, Verhaagen J, Tannemaat MR. Lentiviral vector-mediated gradients of GDNF in the injured peripheral nerve: effects on nerve coil formation, Schwann cell maturation and myelination. PLoS One. 2013;8(8):e71076. doi: 10.1371/journal.pone.0071076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers R, Hendriks WT, Tannemaat MR, van Heerikhuize JJ, Pool CW, Carlstedt TP, Zaldumbide A, Hoeben RC, Boer GJ, Verhaagen J. Neuroregenerative effects of lentiviral vector-mediated GDNF expression in reimplanted ventral roots. Mol Cell Neurosci. 2008;39(1):105–17. doi: 10.1016/j.mcn.2008.05.018. [DOI] [PubMed] [Google Scholar]
- Ellerbroek SM, Wennerberg K, Burridge K. Serine phosphorylation negatively regulates RhoA in vivo. J Biol Chem. 2003;278(21):19023–31. doi: 10.1074/jbc.M213066200. [DOI] [PubMed] [Google Scholar]
- Fu SY, Gordon T. Contributing factors to poor functional recovery after delayed nerve repair: prolonged axotomy. J Neurosci. 1995a;15(5 Pt 2):3876–85. doi: 10.1523/JNEUROSCI.15-05-03876.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu SY, Gordon T. Contributing factors to poor functional recovery after delayed nerve repair: prolonged denervation. J Neurosci. 1995b;15(5 Pt 2):3886–95. doi: 10.1523/JNEUROSCI.15-05-03886.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu SY, Gordon T. The cellular and molecular basis of peripheral nerve regeneration. Mol Neurobiol. 1997;14(1–2):67–116. doi: 10.1007/BF02740621. [DOI] [PubMed] [Google Scholar]
- Gavazzi I, Kumar RD, McMahon SB, Cohen J. Growth responses of different subpopulations of adult sensory neurons to neurotrophic factors in vitro. Eur J Neurosci. 1999;11(10):3405–14. doi: 10.1046/j.1460-9568.1999.00756.x. [DOI] [PubMed] [Google Scholar]
- Gordon T, Tyreman N, Raji MA. The basis for diminished functional recovery after delayed peripheral nerve repair. J Neurosci. 2011;31(14):5325–34. doi: 10.1523/JNEUROSCI.6156-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm L, Holinski-Feder E, Teodoridis J, Scheffer B, Schindelhauer D, Meitinger T, Ueffing M. Analysis of the human GDNF gene reveals an inducible promoter, three exons, a triplet repeat within the 3′-UTR and alternative splice products. Hum Mol Genet. 1998;7(12):1873–86. doi: 10.1093/hmg/7.12.1873. [DOI] [PubMed] [Google Scholar]
- Hoke A. A (heat) shock to the system promotes peripheral nerve regeneration. J Clin Invest. 2011;121(11):4231–4. doi: 10.1172/JCI59320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoke A, Cheng C, Zochodne DW. Expression of glial cell line-derived neurotrophic factor family of growth factors in peripheral nerve injury in rats. Neuroreport. 2000;11(8):1651–4. doi: 10.1097/00001756-200006050-00011. [DOI] [PubMed] [Google Scholar]
- Hoke A, Gordon T, Zochodne DW, Sulaiman OA. A decline in glial cell-line-derived neurotrophic factor expression is associated with impaired regeneration after long-term Schwann cell denervation. Exp Neurol. 2002;173(1):77–85. doi: 10.1006/exnr.2001.7826. [DOI] [PubMed] [Google Scholar]
- Hoke A, Ho T, Crawford TO, LeBel C, Hilt D, Griffin JW. Glial cell line-derived neurotrophic factor alters axon schwann cell units and promotes myelination in unmyelinated nerve fibers. J Neurosci. 2003;23(2):561–7. doi: 10.1523/JNEUROSCI.23-02-00561.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyng SA, de Winter F, Tannemaat MR, Blits B, Malessy MJ, Verhaagen J. Gene therapy and peripheral nerve repair: a perspective. Front Mol Neurosci. 2015;8:32. doi: 10.3389/fnmol.2015.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyng SA, Tannemaat MR, De Winter F, Verhaagen J, Malessy MJ. Nerve surgery and gene therapy: a neurobiological and clinical perspective. J Hand Surg Eur Vol. 2011;36(9):735–46. doi: 10.1177/1753193411420348. [DOI] [PubMed] [Google Scholar]
- Hunter DA, Moradzadeh A, Whitlock EL, Brenner MJ, Myckatyn TM, Wei CH, Tung TH, Mackinnon SE. Binary imaging analysis for comprehensive quantitative histomorphometry of peripheral nerve. J Neurosci Methods. 2007;166(1):116–24. doi: 10.1016/j.jneumeth.2007.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase T, Jung CG, Bae H, Zhang M, Soliven B. Glial cell line-derived neurotrophic factor-induced signaling in Schwann cells. J Neurochem. 2005;94(6):1488–99. doi: 10.1111/j.1471-4159.2005.03290.x. [DOI] [PubMed] [Google Scholar]
- Jesuraj NJ, Marquardt LM, Kwasa JA, Sakiyama-Elbert SE. Glial cell line-derived neurotrophic factor promotes increased phenotypic marker expression in femoral sensory and motor-derived Schwann cell cultures. Exp Neurol. 2014;257:10–8. doi: 10.1016/j.expneurol.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PJ, Wood MD, Moore AM, Mackinnon SE. Tissue engineered constructs for peripheral nerve surgery. Eur Surg. 2013;45(3) doi: 10.1007/s10353-013-0205-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaewkhaw R, Scutt AM, Haycock JW. Integrated culture and purification of rat Schwann cells from freshly isolated adult tissue. Nat Protoc. 2012;7(11):1996–2004. doi: 10.1038/nprot.2012.118. [DOI] [PubMed] [Google Scholar]
- Kim HA, DeClue JE, Ratner N. cAMP-dependent protein kinase A is required for Schwann cell growth: interactions between the cAMP and neuregulin/tyrosine kinase pathways. J Neurosci Res. 1997;49(2):236–47. [PubMed] [Google Scholar]
- Kinameri E, Matsuoka I. Autocrine action of BMP2 regulates expression of GDNF-mRNA in sciatic Schwann cells. Brain Res Mol Brain Res. 2003;117(2):221–7. doi: 10.1016/s0169-328x(03)00326-7. [DOI] [PubMed] [Google Scholar]
- Klemke RL, Cai S, Giannini AL, Gallagher PJ, de Lanerolle P, Cheresh DA. Regulation of cell motility by mitogen-activated protein kinase. J Cell Biol. 1997;137(2):481–92. doi: 10.1083/jcb.137.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi J, Mackinnon SE, Watanabe O, Ball DJ, Gu XM, Hunter DA, Kuzon WM., Jr The effect of duration of muscle denervation on functional recovery in the rat model. Muscle Nerve. 1997;20(7):858–66. doi: 10.1002/(sici)1097-4598(199707)20:7<858::aid-mus10>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Lang P, Gesbert F, Delespine-Carmagnat M, Stancou R, Pouchelet M, Bertoglio J. Protein kinase A phosphorylation of RhoA mediates the morphological and functional effects of cyclic AMP in cytotoxic lymphocytes. EMBO J. 1996;15(3):510–9. [PMC free article] [PubMed] [Google Scholar]
- Leclere PG, Norman E, Groutsi F, Coffin R, Mayer U, Pizzey J, Tonge D. Impaired axonal regeneration by isolectin B4-binding dorsal root ganglion neurons in vitro. J Neurosci. 2007;27(5):1190–9. doi: 10.1523/JNEUROSCI.5089-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackinnon SE. Nerve surgery 2015 [Google Scholar]
- Marquardt LM, Ee X, Iyer N, Hunter D, Mackinnon SE, Wood MD, Sakiyama-Elbert SE. Finely Tuned Temporal and Spatial Delivery of GDNF Promotes Enhanced Nerve Regeneration in a Long Nerve Defect Model. Tissue Eng Part A. 2015;21(23–24):2852–64. doi: 10.1089/ten.tea.2015.0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt LM, Sakiyama-Elbert SE. Engineering peripheral nerve repair. Curr Opin Biotechnol. 2013;24(5):887–92. doi: 10.1016/j.copbio.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt LM, Sakiyama-Elbert SE. GDNF preconditioning can overcome Schwann cell phenotypic memory. Exp Neurol. 2015;265:1–7. doi: 10.1016/j.expneurol.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheson CR, Carnahan J, Urich JL, Bocangel D, Zhang TJ, Yan Q. Glial cell line-derived neurotrophic factor (GDNF) is a neurotrophic factor for sensory neurons: comparison with the effects of the neurotrophins. J Neurobiol. 1997;32(1):22–32. [PubMed] [Google Scholar]
- Meintanis S, Thomaidou D, Jessen KR, Mirsky R, Matsas R. The neuron-glia signal beta-neuregulin promotes Schwann cell motility via the MAPK pathway. Glia. 2001;34(1):39–51. [PubMed] [Google Scholar]
- Morgan L, Jessen KR, Mirsky R. The effects of cAMP on differentiation of cultured Schwann cells: progression from an early phenotype (04+) to a myelin phenotype (P0+, GFAP-, N-CAM-, NGF-receptor-) depends on growth inhibition. J Cell Biol. 1991;112(3):457–67. doi: 10.1083/jcb.112.3.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey TK, Kleitman N, Bunge RP. Isolation and functional characterization of Schwann cells derived from adult peripheral nerve. J Neurosci. 1991;11(8):2433–42. doi: 10.1523/JNEUROSCI.11-08-02433.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveilhan P, ElShamy WM, Ernfors P. Differential regulation of mRNAs for GDNF and its receptors Ret and GDNFR alpha after sciatic nerve lesion in the mouse. Eur J Neurosci. 1997;9(7):1450–60. doi: 10.1111/j.1460-9568.1997.tb01499.x. [DOI] [PubMed] [Google Scholar]
- Poppler L, Ee X, Schellhardt L, Hoben G, Pan D, Hunter D, Yan Y, Moore A, Snyder-Warwick A, Stewart S. Axonal growth arrests after an increased accumulation of Schwann cells expressing senescence markers and stromal cells in a cellular nerve allografts. Tissue Eng Part A. 2016 doi: 10.1089/ten.tea.2016.0003. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonen V, Aho H, Roytta M, Peltonen J. Quantitation of Schwann cells and endoneurial fibroblast-like cells after experimental nerve trauma. Acta Neuropathol. 1988;75(4):331–6. doi: 10.1007/BF00687785. [DOI] [PubMed] [Google Scholar]
- Santosa KB, Jesuraj NJ, Viader A, MacEwan M, Newton P, Hunter DA, Mackinnon SE, Johnson PJ. Nerve allografts supplemented with schwann cells overexpressing glial-cell-line-derived neurotrophic factor. Muscle Nerve. 2013;47(2):213–23. doi: 10.1002/mus.23490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakhbazau A, Mohanty C, Shcharbin D, Bryszewska M, Caminade AM, Majoral JP, Alant J, Midha R. Doxycycline-regulated GDNF expression promotes axonal regeneration and functional recovery in transected peripheral nerve. J Control Release. 2013;172(3):841–51. doi: 10.1016/j.jconrel.2013.10.004. [DOI] [PubMed] [Google Scholar]
- Strack RL, Strongin DE, Bhattacharyya D, Tao W, Berman A, Broxmeyer HE, Keenan RJ, Glick BS. A noncytotoxic DsRed variant for whole-cell labeling. Nat Methods. 2008;5(11):955–7. doi: 10.1038/nmeth.1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannemaat MR, Eggers R, Hendriks WT, de Ruiter GC, van Heerikhuize JJ, Pool CW, Malessy MJ, Boer GJ, Verhaagen J. Differential effects of lentiviral vector-mediated overexpression of nerve growth factor and glial cell line-derived neurotrophic factor on regenerating sensory and motor axons in the transected peripheral nerve. Eur J Neurosci. 2008;28(8):1467–79. doi: 10.1111/j.1460-9568.2008.06452.x. [DOI] [PubMed] [Google Scholar]
- Tao W, Evans BG, Yao J, Cooper S, Cornetta K, Ballas CB, Hangoc G, Broxmeyer HE. Enhanced green fluorescent protein is a nearly ideal long-term expression tracer for hematopoietic stem cells, whereas DsRed-express fluorescent protein is not. Stem Cells. 2007;25(3):670–8. doi: 10.1634/stemcells.2006-0553. [DOI] [PubMed] [Google Scholar]
- Trupp M, Ryden M, Jornvall H, Funakoshi H, Timmusk T, Arenas E, Ibanez CF. Peripheral expression and biological activities of GDNF, a new neurotrophic factor for avian and mammalian peripheral neurons. J Cell Biol. 1995;130(1):137–48. doi: 10.1083/jcb.130.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker BA, Rahimtula M, Mearow KM. Laminin and growth factor receptor activation stimulates differential growth responses in subpopulations of adult DRG neurons. Eur J Neurosci. 2006;24(3):676–90. doi: 10.1111/j.1460-9568.2006.04963.x. [DOI] [PubMed] [Google Scholar]
- Verity AN, Wyatt TL, Hajos B, Eglen RM, Baecker PA, Johnson RM. Regulation of glial cell line-derived neurotrophic factor release from rat C6 glioblastoma cells. J Neurochem. 1998;70(2):531–9. doi: 10.1046/j.1471-4159.1998.70020531.x. [DOI] [PubMed] [Google Scholar]
- Wood MD, Gordon T, Kemp SW, Liu EH, Kim H, Shoichet MS, Borschel GH. Functional motor recovery is improved due to local placement of GDNF microspheres after delayed nerve repair. Biotechnol Bioeng. 2013a;110(5):1272–81. doi: 10.1002/bit.24800. [DOI] [PubMed] [Google Scholar]
- Wood MD, Gordon T, Kim H, Szynkaruk M, Phua P, Lafontaine C, Kemp SW, Shoichet MS, Borschel GH. Fibrin gels containing GDNF microspheres increase axonal regeneration after delayed peripheral nerve repair. Regen Med. 2013b;8(1):27–37. doi: 10.2217/rme.12.105. [DOI] [PubMed] [Google Scholar]
- Wu-Fienberg Y, Moore AM, Marquardt LM, Newton P, Johnson PJ, Mackinnon SE, Sakiyama-Elbert SE, Wood MD. Viral transduction of primary Schwann cells using a Cre-lox system to regulate GDNF expression. Biotechnol Bioeng. 2014;111(9):1886–94. doi: 10.1002/bit.25247. [DOI] [PMC free article] [PubMed] [Google Scholar]


