Abstract
The α4β7 integrin present on host cells recognizes the V1V2 domain of the HIV-1 envelope protein. This interaction might be involved in virus transmission. Administration of α4β7-specific antibodies inhibit acquisition of SIV in a macaque challenge model. But the molecular details of V1V2:α4β7 interaction are unknown and its importance in HIV-1 infection remains controversial. Our biochemical and mutational analyses show that glycosylation is a key modulator of V1V2 conformation and binding to α4β7. Partially glycosylated, but not fully glycosylated, envelope proteins are preferred substrates for α4β7 binding. Surprisingly, monomers of the envelope protein bound strongly to α4β7 whereas trimers bound poorly. Our results suggest that a conformationally flexible V1V2 domain allows binding of the HIV-1 virion to the α4β7 integrin, which might impart selectivity for the poorly glycosylated HIV-1 envelope containing monomers to be more efficiently captured by α4β7 integrin present on mucosal cells at the time of HIV-1 transmission.
Keywords: HIV-1, virus capture, virus entry, α4β7 integrin, V1V2 domain, envelope glycoprotein, HIV vaccine
Graphical abstract
1. Introduction
The entry of viruses into a host cell is a complex and multistep process. While some viruses might “land and enter”, most viruses probably use a “land and seek” approach. The virus first attaches to a molecule that is either easily accessible or abundantly present on the cell surface, then seeks a primary receptor to which it binds specifically, and finally enters the cell (Boulant, Stanifer, & Lozach, 2015). This is perhaps best illustrated in the tailed bacteriophage T4, which contains six long fibers attached to the base of a tail. The tips of the fibers bind to lipopolysaccharide on the E. coli surface, allowing the virus to land on the host cell (adsorption). By reversible attachment and detachment of the tail fibers, the virus, still bound to the host cell, can move and scan for a specific and stable (irreversible) attachment to the primary receptor(s) on the cell surface. This strategy allows for high efficiency of infection which, in phage T4, reaches the theoretical maximum of one virus per host cell (Goldberg, 1983).
Although the components and mechanisms vary, the basic “land and seek” strategy appears well-conserved among viruses. Many mammalian viruses are known to move along the cell surface before binding to primary receptor and entering into the host cell. For instance murine leukemia virus (MLV) and vesicular stomatitis virus (VSV) have been described as “surfing” along cellular filopodia prior to entry (Lehmann, Sherer, Marks, Pypaert, & Mothes, 2005). This strategy is also essential for cell-to-cell transmission, an important feature of HIV-1 life cycle. HIV-1 has been reported to interact with a number of surface molecules that might aid in its attachment and entry into T-cells or cell-to-cell transmission (Mothes, Sherer, Jin, & Zhong, 2010). These include C-type lectin receptors such as dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin (DC-SIGN) (Geijtenbeek et al., 2000) and dendritic cell immunoreceptor (DCIR) (Lambert, Gilbert, Richard, Beaulieu, & Tremblay, 2008), heparin sulfate proteoglycan (HSPG) (Mondor, Ugolini, & Sattentau, 1998), sialic acid-binding immunoglobulin-type lectin-1 (Siglec-1) (Izquierdo-Useros et al., 2012; Jobe et al., 2016), and α4β7 integrin (Arthos et al., 2008) (Fig. 1A).
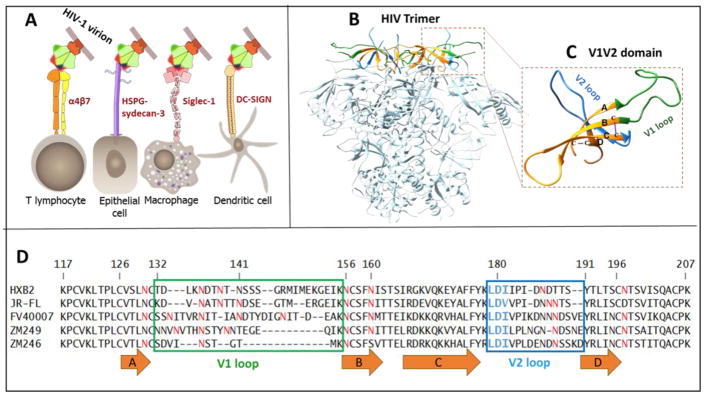
Fig. 1. Structure and function of the V1V2 domain of HIV-1 envelope glycoprotein.
A, Attachment of HIV-1 virion to host cells may be mediated by interaction of Env V1V2 domain with different surface molecules (attachment factors) present on different host cells. Siglec-1 binds sialic acid moieties on glycans in the V1V2 region. HSPG presented on sydecan-3 binds the V3 loop and has a binding site in the C-strand of the V1V2 region. DC-SIGN binds glycans on gp120 and enhancement of virus infection can be modulated by the V1V2 length, the overall V3 charge, and N-linked glycosylation patterns. One Env trimer of HIV-1 virion is shown; V1V2 domain is shown in red, CD4 binding site in green, and V3 domain in blue. B, Structure of the HIV-1 trimer (PDB: 4NCO (Ringe et al., 2013) showing the V1V2 domains at the apex. C, V1V2 domain is enlarged. V1 loop is shown in green, β-strands labeled A–D in orange, and V2 loop in blue. The residues missing in the structure are modeled using Swiss-Model web-server by homology with PDB:4NCO. D, Sequence alignment of the V1V2 domains used in this study. The β-strands are denoted as orange arrows. Potential N-linked glycosylation sites (NXT/S) are shown in red. The variable loops are boxed (colors correspond to C). The conserved LDI/V tripeptide is highlighted in blue. The numbers correspond to amino acids in HXB2 envelope glycoprotein.
The α4β7 integrin is a particularly intriguing molecule. Projecting out to ~22 nm from the cell surface, it has been implicated in enhancing the transmission competency of HIV-1 at the site of exposure during sexual transmission (C. Cicala, Arthos, & Fauci, 2011). The V1V2 domain, present at the apex of the HIV-1 envelope spike, is the site for α4β7 binding (Fig. 1B and 1C) (Jelicic et al., 2013). Although not essential for HIV-1 infection, the α4β7:V1V2 interaction has been reported to enhance the efficiency of infection in vitro and in vivo (Ansari et al., 2011; Li, 2015; Tjomsland et al., 2013). The RV144 trial, the only HIV-1 vaccine trial that showed a modest 31% efficacy, demonstrated correlation between elicitation of V2-specific antibodies and protection against HIV-1 infection (Haynes et al., 2012). In a macaque model of repeated low-dose vaginal challenge, animals treated with anti-α4β7 antibodies were >60% less likely to become infected at any one challenge than the untreated animals (Byrareddy et al., 2014). Furthermore, a direct correlation between the expression of α4β7 and susceptibility and progression of disease has been observed (Byrareddy et al., 2015). In the most recent study (Byrareddy et al., 2016), blocking of α4β7 in SIV infected monkeys maintained undetectable viral loads and normal CD4+ T cell counts even after all anti-retroviral therapy (ART) treatment was withdrawn.
The primary receptor and co-receptors of HIV-1 are CD4 and CCR5 (or CXCR4), respectively (Klatzmann et al., 1984; Rizzuto et al., 1998). Both receptors are reported to co-localize with α4β7 on the surface of CD4+ T-cells in the mucosa (Claudia Cicala et al., 2009). The trimeric envelope spike on the virion surface is the entry machine (Y. D. Kwon et al., 2015; Pancera et al., 2014). Each subunit (protomer) of the spike is a hetero-dimer of the glycoproteins gp120 and gp41 that are produced by cleavage of the envelope glycoprotein (Env) precursor, gp160. gp120 interacts with the CD4 receptor which causes a conformational change that exposes the CCR5 binding site in the V3 domain. Upon binding to CCR5, a series of conformational changes occur in the protomer, resulting in the insertion of the gp41 fusion peptide into the host cell membrane (Klasse, 2012; Weissenhorn, Dessen, Harrison, Skehel, & Wiley, 1997). The viral and host lipid bilayers fuse, resulting in entry of the nucleocapsid core into the target cell.
The interacting regions of Env, CD4, and CCR5 have been elucidated in atomic detail (Diskin, Marcovecchio, & Bjorkman, 2010; Y. D. Kwon et al., 2015; Kwong et al., 1998), leading to the design of vaccine candidates and antiviral therapeutics that can interfere with these interactions. However, it is unlikely that the HIV-1 virus lands on the CD4/CCR5 receptors. Nor is it likely that the virus can find the CD4 receptor by random interactions with the cell surface, which contains an intricate maze of molecules in which CD4 is probably buried. Therefore, attachment of HIV-1 virion to cell surface molecules such as α4β7 through the V1V2 domain probably provides a mechanism for virus capture, and to reach the CD4 receptor at the timescales of virus infection. However, very little is known about these putative pre-CD4 V1V2:α4β7 interactions. The highly conserved LDI/V tripeptide present at the junction of the C β-strand and the V2 loop (Fig. 1C and 1D) is thought to be important for binding to α4β7 (Arthos et al., 2008). But it is not clear if LDI/V is part of the binding site or if it is important for the conformation of the V1V2 domain (O’Rourke et al., 2010). Other residues of the V2 loop (Tassaneetrithep et al., 2014), and of the V3 loop (Peachman et al., 2015), were also linked to α4β7 binding. Complicating the issue is the glycosylation state of the V1V2 domain which contains a cluster of protein N-glycosylation sites. Furthermore, the number of glycosylation sites vary greatly in different strains and subtypes, and also depending on the length of the highly variable V1 and V2 loops. One or a very few transmitter/founder (T/F) viruses that establish the initial infection during HIV-1 transmission are reported to be poorly glycosylated (Derdeyn et al., 2004; Ping et al., 2013). How these T/F viruses are selected remained as an open question. If α4β7 were to be important for transmission of HIV-1 at the mucosa, as the evidence implicates (Byrareddy et al., 2014), a detailed understanding of the α4β7:V1V2 interactions is critical to elucidate the mechanisms and to design effective HIV-1 vaccines.
Here, we examined the α4β7 interaction of a series of HIV-1 envelope proteins (gp140) from different subtypes and strains, in different glycosylation and oligomeric states, and containing various mutations in the V1V2 domain. We found that glycosylation of the V1V2 domain affects the conformation, processing, and secretion of gp140. The fully glycosylated envelope proteins showed the least binding whereas partially deglycosylated V1V2 proteins showed the strongest binding. A strong correlation between the extent of deglycosylation and α4β7 binding was observed. Surprisingly, monomers of the envelope protein bound the strongest whereas trimers bound poorly to α4β7. Our results suggest that a flexible conformation of the V1V2 domain present at the apex of the trimer spike is essential for interaction with α4β7. We speculate that these findings may have biological significance in that the poorly glycosylated HIV-1 virions containing monomers of the envelope protein may present conformations that could be efficiently captured by α4β7 integrin present on the mucosal cells at the time of HIV-1 transmission.
2. Materials and methods
2.1. Cells and media
RPMI 8866 cells, a B lymphoid cell line (Sigma), were maintained in RPMI 1640 media (Gibco) supplemented with 2mM Glutamine and 10% fetal bovine serum (FBS; Quality Biologicals). HEK293F (Life Technologies) and HEK293S GnTI− (ATCC CRL-3022) were maintained in FreeStyle 293 expression medium (Life Technologies). GnTI− cells were supplemented with 1% FBS. All cells were grown in suspension on an orbital shaker (Infors HT) at 120 RPM, at 37°C, 80% humidity, and 8% CO2.
2.2. Plasmids
The expression vector maps are shown in Fig 3. The furin-expressing plasmid, Furin:FLAG/pGEM7Zf(+), was obtained from Dr. Gary Thomas (Vollum Institute, Portland, OR). The furin fragment from this plasmid was subcloned into pcDNA3.1(−) (Life Technologies, Inc.) using EcoRI and HindIII restriction sites. gp140 clones were codon optimized and constructed as previously described (AlSalmi et al., 2015). gp140 mutations described in the “Results” were generated using NEB site-directed mutagenesis kit (NEB Q5) or by splicing by overlap extension (SOE)-PCR in the pcDNA3.1(-) vector. For gp16-scaffolded clones for mammalian expression, a modified gp16 pcDNA3.1 (−) vector was constructed with a Gluc (Gaussia Luciferase) signal peptide at the N-terminus followed by T4 gp16 small terminase, a small SASA linker and a Twin StrepTag. Restriction sites EcoRI and BamH1 were introduced between gp16 and the SASA linker. Codon optimized Zm249 and Zm246 sequences were constructed using gene assembly PCR (AlSalmi et al., 2015) with the appropriate restriction sites. The amplified DNAs were then digested with EcoR1 and BamH1 and ligated with the linearized gp16-pcDNA3.1(−). Additional mutations described in the “Results” were introduced by SOE-PCR and cloned into the pcDNA3.1(−) vector. The E. coli-expressed gp16-scaffold recombinants were constructed by SOE-PCR using primers containing NcoI and NotI restriction sites. Mutations were introduced into these clones using the NEB site-directed mutagenesis kit (NEB Q5) or by SOE-PCR.
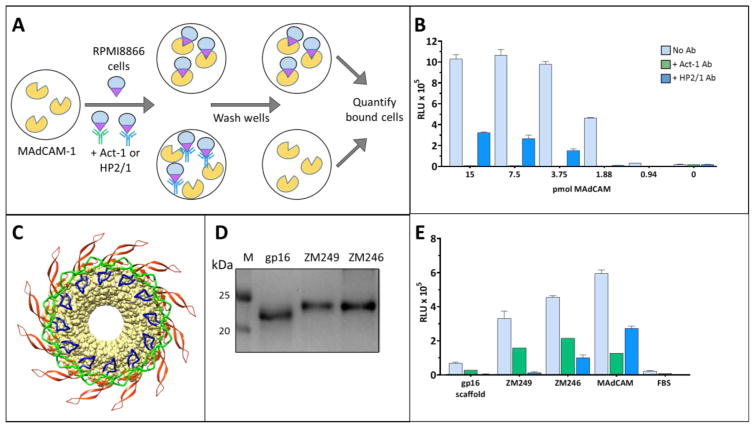
Fig. 3. Specificity of interaction between the V1V2 domain and the α4β7 integrin.
A, A schematic representation of the α4β7 binding assay. Wells of a 96-well plate are coated with the MAdCAM-1 and blocked with FBS to prevent nonspecific binding. The α4β7-expressing RPMI cells are added to the wells and the extent of binding is quantified by the luciferase-based CellTiterGlo kit (Promega). See Materials and Methods for more details. B, α4β7 binding of MAdCAM-1 over a range of concentrations. Binding of MAdCAM-1 (light blue bars) is specific as shown by the inhibition of binding by α4β7-specific mAbs, Act-1 (green bars) and HP2/1 (dark blue bars). C, A model depicting the multimeric display of the V1V2 domain (colored as in Fig. 1B) fused to the dodecameric phage T4 gp16-scaffold (yellow). The V1V2 domain is shown in ribbons and gp16 in surface view. D, SDS-PAGE (4–20% gradient) analysis of the E. coli-produced gp16 and gp16-V1V2 proteins. Lane M represents molecular weight markers. E, α4β7 binding activity of the proteins shown in D. Specificity of V1V2 binding to α4β7 was determined by the decrease in relative luminescence in the presence of the α4β7-specific mAbs, Act-1 (green bars) and HP2/1 (dark blue bars). Light blue bars show α4β7 binding activity in the absence of any mAb inhibitor. Data are shown as mean +/− SEM and are representative of at least three independent assays and three different preparations of proteins, done in triplicate.
2.3. α4β7 binding assay
Purified V1V2 and gp140 proteins were coated onto 96-well black, clear-bottom plate (Greiner), 15 pmoles per well, for 1 hour. The wells were subsequently washed 3 times with blocking buffer (1mM MnCl2, 0.1mM CaCl2, 10mM HEPES, 150mM NaCl, and 10% FBS) and then incubated with the blocking buffer for 1 hour. Wells were then washed 3 times with wash buffer (1mM MnCl2, 0.1mM CaCl2, 10mM HEPES, 150mM NaCl, and 1% FBS). RPMI cells (50 μl/well of 4×106 cells per ml) were added in cell dilution buffer (wash buffer containing 5% FBS) and allowed to bind for 1 hour. Wells were then washed 5 times with wash buffer and the remaining bound cells were detected with the CellTiterGlo kit (Promega) as per the manufacturer’s instructions.
For inhibition studies, the α4β7 blocking mAbs Act-1 and HP2/1 were used at 12μg/ml and 7μg/ml, respectively. Act-1 was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: α4-β7 monoclonal antibody (cat#11718) from Dr. A. A. Ansari (Lazarovits et al., 1984). HP2/1 was purchased from AbD Serotec.
2.4. Protein production and purification
2.4.1. Scaffolded V1V2 proteins (non-glycosylated)
The scaffolded V1V2 constructs were grown from a single E. coli colony in 5 ml LB media for overnight with antibiotics (kanamycin 50 μg L−1, chlorophenicol 34 μg L−1) at 37°C. Next day the culture was diluted (1:50) in Moor’s media with antibiotics and grown at 37°C until the O.D600 reached to 0.6. The recombinant protein expression was induced by adding 0.85 mM IPTG and the culture was grown at 20 °C for 5 hours. The cells were harvested at 7000 rpm for 15 minutes and were resuspended in Suspension buffer (50mM Tris-HCl pH 8, 300mM NaCl, and 20mM imidazole). DNase I and protease inhibiter were added and cells were lysed using French press (1000 psi). The lysate was centrifuged at 17,000 rpm for 30 minutes. The cell debris was discarded and the supernatant was passed through a .22 μM filter. Ni-NTA resin was added to the lysate and the mixture was incubated at 4°C in a rotating falcon tube for 1 hour. The Ni-NTA beads were collected in a column and were washed three times with Wash buffer (50mM Tris-HCl pH 8, 300mM NaCl, and 20mM imidazole). Bound proteins were subsequently eluted using the Elution buffer (50mM Tris-HCl pH 8, 250mM NaCl, and 250mM imidazole).
2.4.2. Glycosylated V1V2 and gp140 proteins expressed in mammalian cells
The V1V2 and gp140 proteins were produced in GnTI− cells as previously described (AlSalmi et al., 2015). For transfection, cells were grown overnight in 293 freestyle media to a density of 1 × 106 cells/ml in the presence of 1% FBS. Two hours prior to transfection, cultures were centrifuged at 100 × g for 5 min and resuspended in fresh medium to a density of 2 × 106 cells/ml in the absence of FBS. In Optimem (Gibco), gp16-expressing plasmid was added with polyethyleneimine (PEI25k, Polyscience Inc.) at a PEI/DNA (w/w) ratio of 3:1. After 5-minute incubation at room temperature, this mixture was added to cells. After 12 h, an equal volume of fresh medium and 1% FBS (v/v) was added. HyClone SFM4HEK293 medium was added to a final concentration of 10% and sodium butyrate was added to a final concentration of 1mM.
gp140 protomers were produced in 293F cells as previously described (AlSalmi et al., 2015). Briefly, for transfection, cells were grown overnight in 293 freestyle media to a density of 1 × 106 cells/ml. Two hours prior to transfection, cultures were centrifuged at 100 × g for 5 min and resuspended in fresh medium to a density of 2 × 106 cells/ml in the absence of FBS. The gp140-expression plasmid was mixed with the furin-expression plasmid at a 1:3 ratio and then it was mixed with OptiPro SFM (Invitrogen). In a separate tube, MAX transfection reagent (Invitrogen) was diluted in OptiPro SFM. These two mixtures were immediately combined and incubated at room temperature for 10 min and then added to the cells. After 4 hrs, 1mM sodium butyrate (Sigma-Aldrich) and 10% HyClone SFM4HEK293 (Hyclone Laboratories Inc) were added.
For all proteins, the supernatant was harvested and clarified using a 0.2 μm filter (Corning) on day 5. Protease inhibitor tablets (Roche Diagnostics) and Bio-Lock biotin blocking solution (IBA Life Sciences) were added to the supernatant containing the secreted proteins. After 30 minutes at 4°C, Strep-Tactin beads (Qiagen) were added and allowed to rotate overnight at 4 °C. The beads were pelleted at 200 rpm and then applied to a spin column (Pierce). They were briefly centrifuged to remove residual supernatant, and then washed twice with 50mm Tris-HCl, pH 8, and 300 mm NaCl. The bound gp140 or gp16-V1V2 proteins were eluted with Strep-Tactin elution buffer (2.5 mm d-desthiobiotin (Sigma), 25 mm Tris-HCl, pH 8, and 150 mm NaCl). The peak fractions were pooled and concentrated using 100 kDa MWCO Amicon Ultra-4 centrifugal filter units (Millipore).
The protomers were further purified by size exclusion column chromatography using the Hi-Load 16/600 Superdex-200 (prep-grade) column (GE Healthcare) equilibrated with the gel filtration buffer (25 mM Tris-HCl, pH 8, 150 mM NaCl). Chromatography was done using the ÄKTA FPLC system (GE Healthcare) and the separated trimer and monomer fractions were collected, concentrated, and stored at −80 °C.
2.5. Protein gels
Proteins were separated on either 4–20% gradient Tris-glycine gels (Life Technologies) or homemade 10% or 12% gels in the presence of DTT. The BLUEstain protein ladder 11–245 kDa (Gold Biotechnology) was used as a molecular weight marker. BN-PAGE was performed using the Novex NativePAGE BisTris gel system in 4–16% gradient gels according to the manufacturer’s instructions (Life Technologies). The NativeMark unstained protein standard (Life Technologies) was used as the molecular weight marker. All gels were stained with Coomassie Blue R-250 solution.
2.6. Deglycosylation
For preparation of proteins to be used in α4β7 binding assays and for analysis on native gels, the purified V1V2 and gp140 proteins were deglycosylated by treatment with 0.25U of PNGase (NEB) per 52.5 pmoles of protein under non-denaturing conditions (50 mM sodium phosphate, pH 7.5, and 5 mM DTT) and incubated overnight at room temperature. For deglycosylation under denaturing conditions, the proteins proteins were treated with PNGase (NEB) under denaturing conditions (0.5% SDS, 40 mM DTT, 50 mM sodium phosphate, and 1% NP-40) and incubated for about 1 hr at 37°C.
For gradual deglycosylation, the purified gp140 monomers or trimers were treated with 0.05U of PNGase per ~0.8 μg of protein in 50 mM sodium phosphate buffer (pH 7.5) and 5 mM DTT. PNGase was inactivated at different time intervals by changing the temperature of reaction mixture from 37°C to 3°C. The sample was split into two halves and one half (~4 μg) was used for SDS-PAGE to analyze the degree of deglycosylation and the other half for α4β7 binding.
2.7. Western blotting
After separation by SDS-PAGE, proteins were transferred to a PVDF membrane using the Trans-Blot® Turbo™ Transfer System and Trans-Blot® Turbo™ RTA Mini PVDF Transfer Kit (Bio-rad Laboratories Inc) for 10 min at 25 volts. After protein transfer, using vacuum-driven technologyUsing SNAP i.d 2.0 (EMD Milipore), blots were blocked for 30 s with 30 ml of 1% Bovine Albumin (Amresco LLC) followed by incubation with 5 ml of StrepMAB-Classic HRP conjugated Ab (iba Life Sciences, dilution 1:10,000 in PBS) for 15 min. The membrane was washed four times with 30 ml of wash buffer for 30 s (0.1% Tween-20 in 1x PBS). Signal was detected using Clarity™ Western ECL Blotting substrate (Bio-rad Laboratories Inc).
3. Results
3.1. Preparation of V1V2 recombinant proteins
To characterize the V1V2:α4β7 interactions, we have produced a large number of V1V2 recombinant proteins in two forms (Supplementary file 1: Supplementary Fig. S1A and S1B): i) scaffolded V1V2 in which the V1V2 domain was fused to a self-assembling bacteriophage T4 protein, gene product 16 (gp16), and ii) V1V2 domain as part of the entire ectodomain of HIV-1 envelope glycoprotein, gp140. The latter contained gp120 and gp41 truncated at amino acid (aa) 664 (HXB2 numbering), which is at the junction of the membrane proximal external region (MPER) and the transmembrane domain. Importantly, V1V2 proteins from several HIV-1 subtypes including subtypes A (BG505), AE (FV40007, FV40100), B (JR-FL, SF162), and C (ZM249, ZM246) were produced to determine their ability to bind to α4β7, which also allowed inclusion of several internal controls to validate the findings.
The scaffolded V1V2 proteins were constructed by fusing the V1V2 domain (aa 118–207; Fig. 1C and 1D) to the carboxy terminus of gp16 from phage T4. Also known as small terminase (TerS), the 18 kDa protein is a DNA packaging protein that self-assembles into stable 12-mers (and 11-mers) (Sun et al., 2012). Consequently, the oligomeric gp16-V1V2 would display a symmetric and high density array of V1V2 motifs for interaction with α4β7(see below in Fig. 3C). The V1V2 domains from two subtype C founder viruses, ZM249 and Zm246 (Salazar-Gonzalez et al., 2009), were produced either in the non-glycosylated form by expression from a prokaryotic expression vector or in the glycosylated form a mammalian expression vector (Supplementary file 1: Supplementary Fig. S1A and S1B). Either a hexa-histidine tag or a twin Strep-tag was added to the termini for rapid purification of the recombinant proteins (see Materials and Methods for details).
The gp140 clones were constructed by inserting the gp140 sequences from BG505, SF162, JR-FL, FV40007, and FV40100 into our mammalian expression vector (AlSalmi et al., 2015) (Supplementary file 1: Supplementary Fig. S1C). The FV4007 and FV40100 gp140 sequences were from the CRF_01 subtype AE founder virus (FV) sequences isolated from the early-capture RV217 trial patients 40007 and 40100, respectively (AlSalmi et al., 2015; Robb et al., 2016). In-frame insertion of the gp140 DNA resulted in the fusion of human CD5 signal peptide at the N-terminus for secretion of gp140 into the culture medium (AlSalmi et al., 2015; Ringe et al., 2013) and a twin Strep-tag at the C-terminus of gp140 for rapid purification by Strep-Tactin chromatography (AlSalmi et al., 2015). In addition, the R6 mutation was introduced to enhance furin cleavage (Sanders et al., 2002) and the SOSIP mutations to stabilize the gp140 structure through inter-subunit disulfide crosslinking between gp120 and gp41 (Binley et al., 2000). The gp140 proteins were produced in different glycosylated forms by transfecting the recombinant DNA into different mammalian cell lines, 293F, GnTI−, or CHO. Secreted gp140 proteins were purified by passing the culture medium through a Strep-Tactin affinity column followed by size-exclusion chromatography where necessary (AlSalmi et al., 2015) (see Materials and Methods for details).
Reactivity with well-characterized V1V2 monoclonal antibodies (mAbs) demonstrated that the V1V2 domain of gp16-V1V2 and gp140 proteins displayed a “native-like” conformation. First, CH58, a mAb that recognizes a linear epitope in the V2 domain (Liao et al., 2013; Nicely et al., 2015) showed as strong reactivity with the gp16-V1V2 and the FV40007 gp140, consistent with the fact that both the proteins retained nine of the ten binding residues of the epitope (Fig. 2A and 2C). Both the non-glycosylated and glycosylated gp16-V1V2 proteins bound well, in agreement with the previous report that binding of CH58 mAb is not dependent on glycosylation (Liao et al., 2013). On the other hand, JR-FL gp140 that contained only six of the ten binding residues bound poorly. Second, CH59, a conformational V1V2 mAb (Liao et al., 2013) bound only with the FV40007 gp140 as it retained all four key aa residues that are essential for binding (Fig. 2B and 2D). X-ray structure of the CH59-V2 complex showed that the residues K169 and H173 are critical for binding (Liao et al., 2013). Both the gp16-V1V2 and FV40007 gp140 in which H173 was replaced by Y lost reactivity with CH59. Third, the quaternary broadly neutralizing antibody (BnAb) PG9, which recognizes the mannose glycans at positions N160 and N156 and the overall cationic nature of the C β-strand (McLellan et al., 2011) reacted with gp16-V1V2 and FV40007 gp140 as both the proteins retained these features (Fig. 2A and 2E). But the reactivity was lower than that reported with the gp140 trimers (Walker et al., 2009), consistent with the fact that PG9 requires the quaternary (trimeric) structure for stable binding. On the other hand, JRFL gp140 having an E mutation at K168 in the C β-strand lost its reactivity, in agreement with the previous reports (Walker et al., 2009) (Fig. 2A and 2E). None of the control proteins lacking V1V2 showed any reactivity with any of these antibodies. Collectively, therefore, these sets of data indicated that our purified proteins retained a “native-like” conformation of the V1V2 domain.
Fig. 2. gp16-V1V2 scaffold and gp140 protomer proteins binding to CH58, CH59 and PG9.
A, CH58 binding residues in V2 sequence (156–187) of JR-FL, FV40007, Zm249 are denoted with blue letters. B, CH59 binding residues in V2 sequence (164–187) of JR-FL, FV40007, Zm249 is denoted with orange letters. C–E gp16-V1V2 scaffold and gp140 protomer proteins binding to CH58 C, CH59 D, and PG9 E. The proteins used in binding are gp16 expressed in E. coli (pink line), Zm249 gp16-V1V2 expressed in E. coli (red line), gp16 expressed in GnTI− (khaki line), Zm249 gp16-V1V2 expressed in GnTI− (black line), JR-FL gp140 expressed in 293F (blue line), and FV40007 gp140 expressed in 293F (green line). Binding data are representative of at least three independent assays, done in triplicate.
3.2. Specificity of interaction between the V1V2 domain and the α4β7 integrin
We have developed a high-throughput assay to analyze the V1V2:α4β7 interactions (Fig. 3A). MAdCAM-1 (mucosal addressin cell adhesion molecule-1), the natural ligand for α4β7 (Berlin et al., 1993), was coated on a 96-well ELISA plate and incubated with RPMI-8866 B-cells (Matsuoka, Moore, Yagi, & Pressman, 1967) that constitutively express α4β7 on the cell surface but not the HIV-1 primary receptor CD4 and co-receptor CCR5 (Peachman et al., 2015). After washing off the unbound cells, the MAdCAM-1-bound cells were quantified using the luciferase-based CellTiterGlo detection system (Fig. 3B; see Materials and Methods for details). The specificity of binding was ascertained by the inhibition of binding by α4β7-specific mAbs (Fig. 3B; see below). This direct binding assay is similar to the one recently reported (Peachman et al., 2015) except that the luciferase system provided a robust ~100-fold dynamic range to evaluate differences in α4β7 binding. In a typical experiment, ~7.5 pmol (0.4μg) of MAdCAM-1 captured ~2 × 105 cells, generating a signal of ~1 × 106 relative light units (RLU) (Fig. 3B). Negative controls lacking MAdCAM-1 or wells blocked with FBS prior to adding MAdCAM-1 gave a background of ~104 RLUs. In the subsequent experiments for testing V1V2:α4β7 binding, ~1μg of purified V1V2 proteins were coated on a 96-well ELISA plate and compared with the MAdCAM-1 binding used as a positive control.
We first tested the non-glycosylated scaffolded V1V2 domains from subtype C ZM249 and ZM246 viruses for binding to α4β7 (Fig. 3C; see below for binding of the glycosylated forms). Both the purified proteins (Fig. 3D) bound to α4β7 at slightly less but comparable levels as its natural ligand MAdCAM-1 (Fig. 3E). The interaction was specific, as established by several criteria. First, the negative controls, gp16 alone or FBS wells did not show significant binding. Second, on a molar basis, binding of the gp16-V1V2 proteins is comparable to that of MAdCAM-1. Third, pre-incubation of cells with α4β7-specific mAbs ACT-1 and HP2/1 inhibited the binding of the gp16-V1V2 proteins to the α4β7-expressing RPMI 8866 cells.
Significantly, we found that the antibody inhibition pattern of V1V2:α4β7 binding was distinct from that of MAdCAM-1:α4β7 binding. HP2/1 inhibited V1V2:α4β7 binding more strongly than Act-1, whereas the reverse was true for MAdCAM-1:α4β7 binding (Fig. 3E). This was consistently observed for all the scaffolded V1V2 and gp140 constructs tested in this study (see below). X-ray structures of α4β7-Ab complexes have established that Act-1 recognizes the β7 subunit and sterically inhibits the binding of MAdCAM-1 whereas HP2/1 recognizes the α4 subunit with its epitope located on the opposite side of the α4β7 binding cleft (Yu, Schurpf, & Springer, 2013). This suggests that while both MAdCAM-1 and V1V2 domain interacted with α4β7, they did so in a different manner.
3.3. Deglycosylated, but not glycosylated, V1V2 domain binds α4β7
Structure-function studies have established that the V1V2 domain is heavily glycosylated and glycosylation is essential for neutralization by several BnAbs, including the quaternary BnAbs PG9 and PG16 (Doores & Burton, 2010). The ZM249 V1V2 domain contains seven N-linked glycosylation motifs, NXT/S. (Fig. 1D). To determine the importance of glycosylation in the V1V2:α4β7 interaction, gp16-V1V2 proteins were expressed in GnTI− cells, which add Man5–9GlcNAc2 to Asn of the N-glycosylation motif (Reeves, Callewaert, Contreras, & Khorana, 2002). The glycosylated gp16-V1V2 proteins secreted into the medium were purified by Strep-Tactin affinity chromatography and tested for binding to α4β7. Surprisingly, the glycosylated gp16-V1V2 proteins did not show significant binding to α4β7 (Fig. 4A, see ZM249 untreated). Similar results were obtained when the scaffold proteins were produced in 293F cells that introduce complex glycosylations (Chang et al., 2007) (data not shown; production of gp16-V1V2 proteins was low in 293F cells, hence GnTI− cells were used for further analyses).
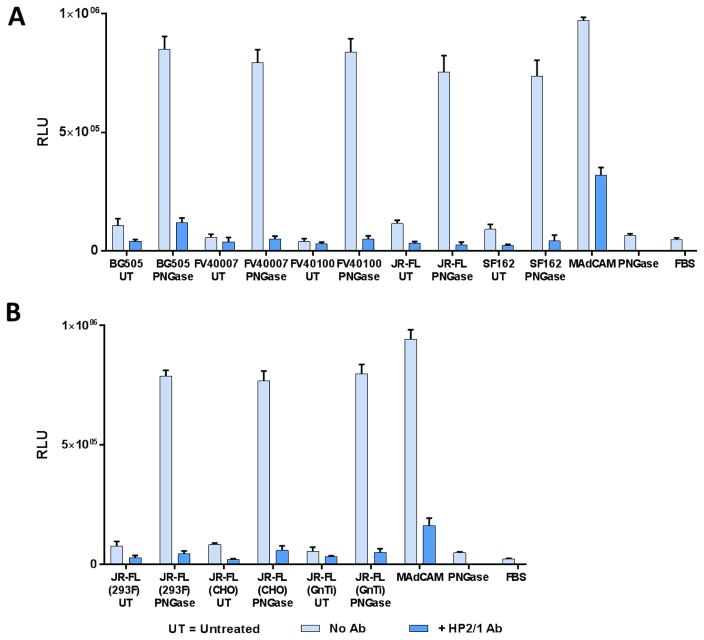
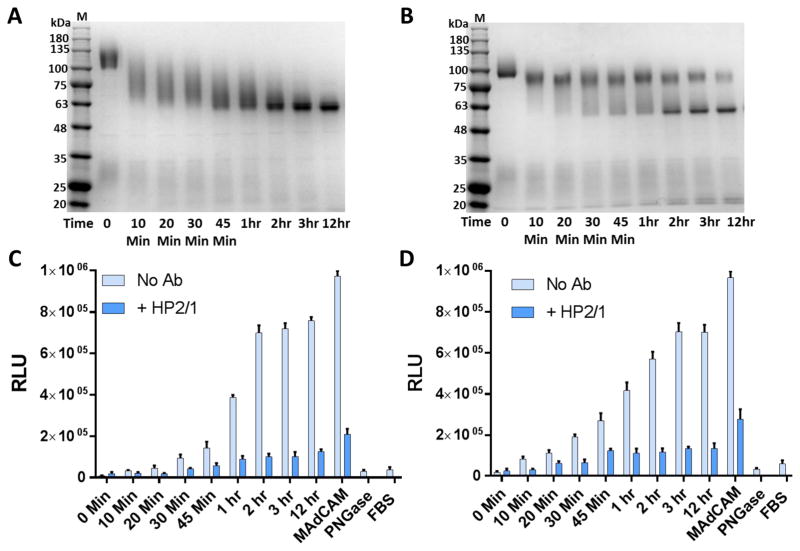
Fig. 4. Deglycosylated, but not glycosylated, V1V2 domain binds α4β7.
A, α4β7 binding of ZM249 gp16-V1V2 with (PNGase) and without (untreated) PNGase treatment. B, SDS-PAGE (4–20% gradient) analysis of ZM249 gp16-V1V2 treated with PNGase in the presence (+) or absence (−) of SDS. C, Native-PAGE (4–16% gradient) analysis of JR-FL gp140 and ZM249 gp16-V1V2 with and without PNGase treatment. D, SDS-PAGE (4–20% gradient) analysis of JR-FL and FV40007 gp140s treated with PNGase, with and without SDS. E and F α4β7 binding activity of FV40007 E and JR-FL F, gp140s with and without PNGase treatment. Lanes M represents molecular weight markers. α4β7 binding assays in the absence of mAb inhibitor (light blue bars) or in the presence of Act-1 mAb (green bars) or HP2/1 mAb (dark blue bars) were performed as described in the Fig. 3 legend. Binding was expressed as percent of PNGase-treated gp140 from FV40007 E or JR-FL F. Data are shown as mean +/− SEM and are representative of at least three independent assays and three different preparations of proteins, done in triplicate.
We hypothesized that failure of the glycosylated V1V2 proteins to bind to α4β7 may be because the glycans rigidified the V1V2 conformation and/or hindered α4β7’s access to the V1V2 binding region. This was tested by deglycosylation of gp16-V1V2 recombinant proteins (ZM249) with PNGase in the presence of 5mM DTT (non-denaturing deglycosylation, no SDS was added) and evaluation of the deglycosylated proteins for α4β7-binding. PNGase cleaves asparagine-linked complex glycans including the hybrid or high mannose oligosaccharides, unless the core contains an α(1→3)-fucose linkage (Tretter, Altmann, & Marz, 1991). That the glycans were removed by deglycosylation was confirmed by gel analysis (Maley, Trimble, Tarentino, & Plummer, 1989). As shown in Fig. 4B, the deglycosylated gp16-V1V2 band under non-denaturing conditions (−SDS; lane 2) migrated much faster than the glycosylated band (lane 3) but it migrated slightly above the fully deglycosylated gp16-V1V2 band under denaturing conditions (+SDS; lane 1). Moreover, deglycosylation did not cause de-oligomerization or alter the oligomeric state of the gp16-dodecamer (Fig. 4C Blue-native gel, compare ZM249 lanes 3 and 4, + and − PNGase). Similar pattern was observed with the other V1V2 and gp140 proteins (Fig. 4C and D; see below, and data not shown). Thus, deglycosylation under non-denaturing conditions removed most, but not all, of the glycans and retained the quaternary structure of the envelope proteins. Importantly, with respect to binding to α4β7, in contrast to the glycosylated V1V2 that showed no significant binding, the deglycosylated V1V2 bound strongly and this binding was inhibited by the anti-α4β7 mAbs, Act-1 and HP2/1 (Fig. 4A, ZM249-PNGase bars), in a similar pattern as the E. coli produced nonglycosylated V1V2 proteins (Fig. 3E). These data confirmed the specificity of V1V2: α4β7 interaction.
To determine if the deglycosylation-dependent α4β7 binding was also true for the V1V2 that is naturally scaffolded as part of gp140 structure, the JR-FL and FV40007 gp140 proteins were expressed in 293F cells as these produced greater amounts of the recombinant proteins. Purification by Strep-Tactin chromatography yielded gp140 protomers consisting of a mixture of monomers, dimers, and trimers (AlSalmi et al., 2015) (Fig. 4C Blue-native gel, lane 1). The protomers were tested for α4β7-binding before and after non-denaturing PNGase treatment, which resulted in removal of most of the glycans, as evident from the mobility on SDS-gel (Fig. 4D, compare lanes with and without SDS), and retention of the oligomeric state of the protomers, as evident from the mobility on native gel (Fig. 4C, compare lanes 1 and 2). Similar to the scaffolded V1V2 domains, the glycosylated gp140 protomers did not bind to α4β7 (Fig. 4E and 4F, untreated bars), whereas the deglycosylated ones bound strongly (PNGase bars). Furthermore, binding was inhibited by Act-1 and HP2/1 mAbs in a manner similar to that of gp16-V1V2 (greater inhibition by HP2/1 than by Act-1) confirming the specificity.
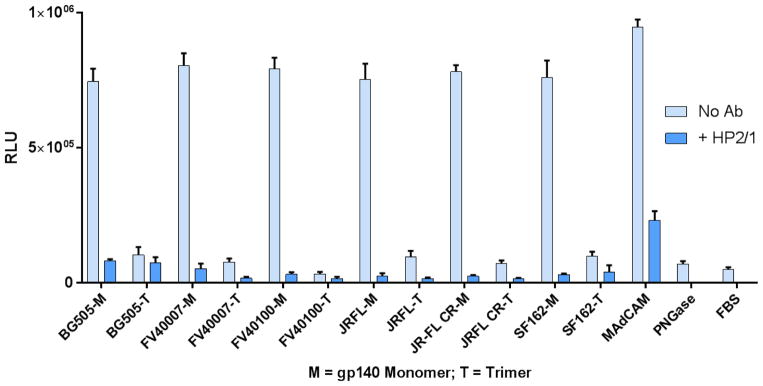
To establish this point further, we generated purified gp140 protomers from several subtype strains including BG505, SF162, and FV40100 (Fig. 5A). In addition, we have also produced the JR-FL gp140 protein in three different glycosylations states by expressing the protein in 293F, GnTI−, or CHO cells (Fig. 5B). All the gp140 proteins regardless the subtype or the type of glycosylation showed poor binding to α4β7 in the glycosylated form and strong binding following deglycosylation, and binding was inhibited by the α4β7 mAbs, Act-1 and HP2/1. Overall, the scaffolded V1V2 domain and gp140 envelope proteins exhibited comparable levels of α4β7 binding activity.
Fig. 5. The α4β7 integrin binds to deglycosylated (poorly glycosylated) gp140 HIV-1 envelope protein, regardless of the type of glycosylation or the HIV-1 subtype.
A, α4β7 binding with gp140 proteins of five strains of HIV-1 (BG505, FV40007, FV40100, JR-FL, and SF162). B, α4β7 binding activity of the JR-FL gp140 proteins produced in three different cell lines (293F, CHO, and GnTI−). α4β7 binding activity in the absence of any mAb inhibitor was shown in light blue bars. Specificity of V1V2 binding to α4β7 was determined by the decrease in relative luminescence in the presence of the α4β7-specific HP2/1 mAb (dark blue bars). Data are shown as mean +/− SEM and are representative of at least three independent assays and three different preparations of proteins, done in triplicate.
3.4. Glycosylation modulates V1V2 conformation and interaction with α4β7
A series of gp16-V1V2 and gp140 glycosylation mutants were constructed and the recombinant proteins were purified to determine how glycosylation at different sites of the V1V2 domain might affect α4β7 binding (Fig. 6A–C, Supplementary file 1: Supplementary Fig. S2A–C). These studies led to several findings:
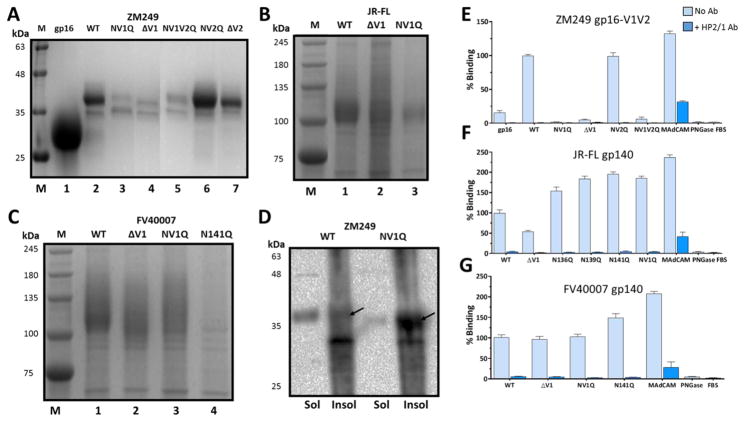
Fig. 6. Glycosylation of V1V2 domain modulates protein conformation, processing, and α4β7 binding.
A–C, SDS-PAGE (12%) analysis of the glycosylation mutants of Zm249 gp16-V1V2 A, JR-FL gp140 B, and FV40007 gp140 C. D, Western blot analysis using gp16-antibodies of the soluble secreted protein (sol) and the insoluble cell pellet (insol) following transfection with WT or various glycosylation mutants of ZM249 gp16-V1V2. E–G, α4β7 binding activity of the glycosylation mutants of Zm249 gp16-V1V2 E, FV40007 gp140 F, and JR-FL gp140 G after treatment with PNGase under non-denaturing conditions (no SDS). Lanes M represents molecular weight markers. NV1Q and NV2Q refer to mutants in which all asparagine residues were mutated to glutamine residues in the V1 or V2 loop, respectively, and NV1V2Q refers to the mutant in which all asparagine residues were mutated to glutamine residues in both the V1 and the V2 loops. ΔV1 refers to the deletion of the entire V1 loop. α4β7 binding assays in the absence (light blue bars) or presence (dark blue bars) of HP2/1 mAb were performed as described in the Fig. 3 legend. Binding was expressed as percent of PNGase-treated: ZM249 gp16-V1V2 E, WT JR-FL gp140 F, or WT FV40007 gp140 G. Data are shown as mean +/− SEM and are representative of at least three independent assays and three different preparations of proteins, done in triplicate.
First, glycosylation of the V1 loop is essential for secretion of the HIV-1 envelope protein. Mutation of the three N-linked glycosylation sites in the V1 loop (NV1Q) or deletion of the entire V1 loop (ΔV1) resulted in poor production of gp16-V1V2 (Fig. 6A; compare lanes 2 with 3 and 4). Removal of all glycosylation in the V1 loop (ΔV1) (lane 4) and the V2 loop (NV1V2Q) (lane 5) similarly reduced expression (Fig. 6A). This defect was specific to V1 loop because analogous mutations in the V2 loop, removal of all glycosylation sites (NV2Q) or deletion of the entire V2 loop (ΔV2), did not impair protein production (lanes 6 and 7). Likewise, removal of glycosylation sites in the B β-strand (NBQ) did not alter protein production (Supplementary file 1: Supplementary Fig. S2A). Similar trends were also observed when the glycosylation sites were mutated in the V1 loop of JR-FL (Fig. 6B) or FV40007 gp140s (Fig. 6C). However, the defect was less severe in the context of these “gp140-scaffolded” V1V2 than the gp16-scaffolded V1V2. The defect appears to be due to differences in post-translational processing because, unlike the WT protein, most of the mutant protein remained associated with the cell pellet and failed to secrete into the culture medium (Fig. 6D).
Second, mutations in the middle of the V1 loop appear to be more essential for secretion of gp140. In particular, lack of glycosylation at residue 141 (N141Q) of FV40007 gp140 resulted in near complete loss of gp140 production (Fig. 6C, lane 4).
Third, glycosylation of V1 loop is essential for proper folding and conformation of the V1V2 domain. In gp16-V1V2 mutant proteins lacking glycosylation sites in the V1 loop (NV1Q), or the entire V1 loop (ΔV1), α4β7-binding activity was completely abolished (Fig. 6E; all the proteins were deglycosylated in order to detect α4β7 binding). Analogous mutations in the V2 loop (NV2Q), however, did not affect α4β7 binding (Fig. 6E). These results suggest that lack of V1 loop glycosylation in the scaffolded-V1V2 severely impairs the conformation of the V1V2 domain and renders it incompetent to bind to α4β7.
Fourth, mutation of some of the glycosylation sites in the background of gp140 differentially modulates the V1V2 conformation (Fig. 6F and G, Supplementary file 1: Supplementary Fig. S2B and S2C, and Supplementary file 1: Supplementary Fig. S3A and S3B). For instance, in contrast to gp16-V1V2, removal of V1 loop glycosylation sites in JR-FL gp140 (Fig. 6F; NV1Q) resulted in 1.5 to 2-fold increase in its binding to α4β7 whereas a 50% decrease was observed when the V1 loop was deleted (ΔV1). On the other hand, no significant change in α4β7 binding was observed when similar mutations were introduced into FV40007 gp140, with the exception of a modest increase in the N141Q mutant (Fig. 6G). A series of glycosylation mutants in the V2 loop and the B β-strand of gp140 proteins either showed no change or a modest increase in α4β7 binding (Fig. 6F and 6G, Supplementary file 1: Supplementary file 1: Supplementary Fig. S3A and S3B).
Collectively, the above data showed that the V1V2 domain conformation is modulated by glycosylation, which in turn affected post-translational processing, secretion, and α4β7 binding.
3.5. Degree of deglycosylation correlates with α4β7 binding
The above data showing that the deglycosylated gp140 proteins bound strongly to α4β7 integrin is consistent with the observation that the T/F viruses are poorly glycosylated and contain shorter V1V2 loops when compared to chronic viruses (Chohan et al., 2005; Derdeyn et al., 2004; Ping et al., 2013; Sagar, Wu, Lee, & Overbaugh, 2006). Thus, an argument can be made that a HIV-1 virion containing a poorly glycosylated envelope would be more efficiently captured by α4β7, leading to efficient transmission into the new host. One way to address this question is to create an ensemble of glycosylation states by gradual deglycosylation of gp140 and ask if α4β7 binding correlates with the degree of deglycosylation.
Deglycosylation of gp140 was optimized by treatment of the purified gp140 proteins with different concentrations of PNGase for different times. A PNGase concentration and a set of time points that showed gradual removal of glycans were selected for each gp140 construct by analyzing the samples on SDS and Blue native PAGE. The extent of deglycosylation was characterized by the appearance of partially deglycosylated species which migrated as a diffuse band. These species were converted to a single sharp fully deglycosylated band that accumulated with time (Fig. 7A and 7B, Supplementary file 1: Supplementary Fig. S4D–F). Testing of these samples for α4β7 binding showed that the binding activity increased gradually with increasing deglycosylation, reaching a maximum when gp140 is nearly completely deglycosylated (equivalent to poorly glycosylated). The same pattern was observed with five different gp140 proteins belonging to three different HIV-1 subtypes (FV40007, Fig. 7C; JR-FL, Fig. 7D; FV40100, Supplementary file 1: Supplementary Fig. S4A; BG505, Supplementary Fig. S4B; SF162, Supplementary Fig. S4C). These data demonstrate that the α4β7 integrin preferentially binds to partially or poorly glycosylated HIV-1 envelope proteins when compared to the fully glycosylated proteins.
Fig. 7. Degree of deglycosylation correlates with α4β7 binding.
A and B, SDS-PAGE (4–20% gradient) analysis of FV40007 gp140 A, and JR-FL gp140 B. gp140 Env monomers were treated with PNGase for different time intervals (0 Min to 12 hr). C and D, Binding of α4β7 to FV40007 gp140 C, and JR-FL gp140 D after deglycosylation for the time intervals indicated in A and B. Lanes M represent molecular weight markers. α4β7 binding assays in the absence of mAb inhibitor (light blue bars) or in the presence of HP2/1 mAb (dark blue bars) were performed as described in the Fig. 3 legend. Data are shown as mean +/− SEM and are representative of at least three independent assays done in triplicate.
3.6. Monomers, but not trimers, of gp140 bind to α4β7
Monomers of gp140 would expose the V1V2 domain better than the trimers. Trimers, due to constraints imposed by the quaternary structure would be more rigid. In addition, trimers would impose steric constraints in most angles for the approach of the 280 kDa α4β7 towards the binding site located in the V1V2 domain. To determine if α4β7 discriminates between monomers and trimers, gp140 monomers and trimers were prepared from five different HIV-1 viruses (BG505, SF162, FV40007, FV40100, JR-FL), using our recently described purification protocol (Supplementary file 1: Supplementary Fig. S5A–D, and data not shown) (AlSalmi et al., 2015). These gp140 proteins are cleaved and stabilized by SOSIP mutations and are in a “native-like” conformation, as determined by a series of biochemical and antigenicity analyses(AlSalmi et al., 2015; Ringe et al., 2013). In addition, we have also produced JR-FL gp140 trimers in both uncleaved and cleaved states to determine if there is a difference in α4β7 binding between the uncleaved and cleaved conformations. Surprisingly, the gp140 trimers showed poor, if any, binding to α4β7 whereas the monomers showed strong binding. All five monomer-trimer pairs exhibited the same behavior, and no significant difference was observed between the uncleaved and cleaved trimers (Fig. 8). The specificity of α4β7 binding was ascertained by inhibition with the HP2/1 mAb.
Fig. 8. Monomers, but not trimers, of gp140 bind to α4β7.
α4β7 binding activity of the monomers and trimers of BG505, FV40007, FV40100, JR-FL, JR-FL CR and SF162 before and after PNGase treatment under non-denaturing conditions (see Materials and Methods section for additional details). “M” represents monomers. “T” represents trimers. “CR” represents cleavage resistant (uncleaved). All except JR-FL were cleaved. See Supplementary file 1: Supplementary Fig. S5A–D for the purification profiles of monomers and trimers. α4β7 binding in the absence of mAb inhibitor (light blue bars) or in the presence of HP2/1 mAb (dark blue bars) were performed as described in the Fig. 3 legend. Data are shown as mean +/− SEM and are representative of at least three independent assays done in triplicate.
4. Discussion
Efficient entry of a virus into a host cell requires that the virus be first captured on the cell surface. Capture is often mediated by an attachment factor(s) that is abundantly present on the surface, but one that might be different from the primary receptor (Geijtenbeek et al., 2000; Izquierdo-Useros et al., 2014; D. S. Kwon, Gregorio, Bitton, Hendrickson, & Littman, 2002; Petrovic et al., 2004). This is especially critical for cell-to-cell transmission where a virus binds to a host cell but does not enter that cell, a significant aspect of HIV-1 life cycle (Stewart & Nemerow, 2007). The 22 nm-long α4β7 integrin present on CD4+ T-cells is thought to be one such “capture receptor” for HIV-1 (C. Cicala et al., 2011; Claudia Cicala et al., 2009). Studies indicate that it might be important for acquisition of HIV-1 during sexual transmission (Ansari et al., 2011; Byrareddy et al., 2016; Byrareddy et al., 2014; Byrareddy et al., 2015; Tjomsland et al., 2013). Furthermore, it has been demonstrated that the α4β7 integrin recognizes the V1V2 domain of the HIV-1 envelope protein (Arthos et al., 2008) but the molecular details of this interaction are unknown. Knowledge on the nature and dynamics of this interaction could help design effective vaccines that can interfere with this initial encounter between HIV-1 virus and host cell.
We have developed a high-throughput α4β7 binding assay and a series of V1V2 constructs to analyze the V1V2:α4β7 interactions. These include a number of gp140 proteins in which the V1V2 domain is part of the ectodomain of the HIV-1 envelope glycoprotein, and a scaffolded V1V2 domain which is free of influence by the envelope protein structure. The sequences were from seven different HIV-1 strains and four subtypes; subtypes A (BG505), B (JR-FL, SF162), C (ZM249, ZM246) and CRF_01 AE (FV4007, FV40100). The gp140 proteins were prepared in different oligomeric states; monomers, protomers, and trimers, and in different glycosylation states by expressing them in 293F, GnTI−, or CHO cell lines. Being cleavage-proficient and stabilized by SOSIP mutations, these proteins exhibit a native-like conformation (Ringe et al., 2013). Furthermore, their reactivity with various mAbs that recognize linear (CH58), conformational (CH59), or conformational plus glycan-specific (PG9) V1V2 epitopes suggest that the V1V2 domain in these proteins are in a native-like conformation. Additionally, several positive, negative, and internal controls including the α4β7’s natural ligand MAdCAM-1 and two α4β7-specific mAbs Act-1 and/or HP2/1 that inhibit binding were included in each experiment to validate the specificity of binding. Finally, a number of mutations were introduced into the V1V2 domain and where necessary, the same mutation(s) was introduced into three different versions to verify the results.
Extensive analysis of the V1V2:α4β7 interactions using the above system showed that glycosylation is a key and complex modulator of V1V2 conformation and α4β7 binding. First, the mutant data show that glycosylation of V1V2 domain is important for processing, secretion, and α4β7 binding properties of the HIV-1 envelope glycoprotein. This was most strikingly observed with the scaffolded V1V2. While the α4β7 binding conformation was preserved in the “wild-type” (WT) V1V2 domain, the NV1Q mutant that lacked glycosylation in the V1 loop was severely impaired in processing, secretion, and α4β7 binding. On the other hand, mutating the V2 loop glycosylation sites including a deletion of the loop did not lead to significant defects. In the context of the gp140 structure, analogous glycosylation mutations in V1 loop showed variable effects. Some showed inefficient processing, secretion and/or decreased α4β7 binding whereas others showed increased α4β7 binding. Analyses of a large collection of mutants suggest that the glycosylation pattern of the V1V2 region, particularly the V1 loop, affects its conformation and efficiency of binding to α4β7.
Our data show that the poorly glycosylated envelope protein is the most preferred substrate for α4β7 binding whereas the fully glycosylated protein is the least preferred. Indeed, a striking correlation was observed between the degree of deglycosylation and α4β7 binding. Gradual deglycosylation of an ensemble of gp140 protomers with time resulted in progressive increase in α4β7 binding, regardless of the type of glycosylation, subtype, or strain used. These data are consistent with the previous reports that synthetic (non-glycosylated) peptides covering the V1V2 regions bound to α4β7 (Nawaz et al., 2011; Peachman et al., 2015; Tassaneetrithep et al., 2014), and the 293F-glycosylated gp120 AN1(subtype B) did not bind to α4β7 whereas the gp120 produced in CHO lec1 cells lacking complex glycosylation showed binding (Nawaz et al., 2011). Furthermore, the gp120 deglycosylated with endoglycosidase H showed the strongest α4β7 binding (Nawaz et al., 2011). Our biochemical analyses showed that the deglycosylation done under non-denaturing conditions did not affect the oligomeric state of gp140. This means that while glycosylation of V1V2 domain is essential for achieving the right conformation necessary for processing and secretion, removing the glycans after the envelope protein is matured did not destabilize the oligomeric structure. On the other hand, it apparently made the V1V2 domain conformationally more flexible to bind to the α4β7 integrin. Studies have shown that glycosylation rigidifies protein conformation and deglycosylation increases flexibility (Lee, Qi, & Im, 2015). Collectively, therefore, our data suggest that the partially or poorly glycosylated envelope protein is efficiently captured by the α4β7 integrin.
To delineate if α4β7 binding is sensitive to the oligomeric state of gp140, the gp140 protomers were separated by size-exclusion chromatography and tested for α4β7 binding. The results demonstrated, surprisingly, that the monomers, but not the trimers, bound to α4β7. This was observed with five different monomer-trimer pairs produced from A, B, or CRF_01 AE subtypes. Although it was generally assumed that the trimer spike present on the HIV-1 envelope is the active structure involved in all aspects of virus entry, from the structural and conformational standpoint, the monomer would be better disposed to interact with the α4β7 integrin. The V1V2 domain of monomeric gp140 would be more accessible than the trimeric spike constrained by the quaternary structure. The SOSIP trimers might be even more constrained because of the covalent disulfide bonds created between gp120 and gp41 subunits (Beddows et al., 2005; Klasse et al., 2013; Sanders et al., 2004). Moreover, our in silico docking attempts indicate that approach of trimer towards α4β7 would likely encounter clashes in almost every angle. These arguments are consistent with the discussion above in that a conformationally flexible deglycosylated gp140 binds to α4β7.
It has been reported that the HIV-1 envelope contains a relatively small number of trimer spikes, on average about fourteen. In addition, it also contains other forms of the envelope protein, including monomers (Moore et al., 2006; Stieh et al., 2015; Yuan, Bazick, & Sodroski, 2006). Indeed, the gp140 prepared from the CRF_01 AE T/F viruses FV40007 and FV40100 produced a large portion of the protein as monomers (in preparation). That this apparently nonfunctional, i.e. fusion incompetent, form of the envelope protein was efficiently captured by the cell-bound α4β7 integrin raises the possibility if the monomers present in the HIV-1 envelope might play a role in HIV-1 capture by α4β7 integrin during transmission. Coincidentally, it has been observed that the T/F viruses contain envelopes with low levels of glycosylation and shorter V1V2 loops, which would also mean fewer glycans (Curlin et al., 2010; Sagar et al., 2006). This type of capture by α4β7 might thus be more relevant at the site of exposure where transmission competence of a variant at a fast timescale would be critical to establish the initial infection. During chronic infection or in in vitro experiments, however, this may not be important, as was observed in previous studies (Parrish et al., 2012; Perez, Chen, Liao, & Montefiori, 2014) because the virus concentration is high and the time available for infection is not as stringent. Our data, thus, suggest a plausible linkage between poorly glycosylated envelope containing monomeric envelope glycoprotein, capture of V1V2 domain by α4β7 integrin, and HIV-1 transmission. It is also consistent with the observed correlation between certain V2 domain binding antibodies and reduced risk of HIV-1 infection in humans in the RV144 vaccine efficacy trial (Haynes et al., 2012) and in macaque SIV challenge trials (Vaccari et al., 2016) where monomeric gp120 envelope protein was used as an immunogen. These V2 antibodies might potentially interfere with capture by α4β7 integrin. Experiments are underway to test this hypothesis.
An alternative hypothesis is that poor, or near lack of, binding of trimers or glycosylated envelope proteins to α4β7 integrin might mean that α4β7 integrin plays no significant role in HIV-1 infection, as was reported in studies where infectious molecular clones (IMCs) and pseudoviruses were used for infection in in vitro (Parrish et al., 2012; Perez et al., 2014). However, two points should be kept in mind. First, our results would also predict this because the virions produced by transfection of 293 cells in vitro will contain glycosylated envelopes, hence would not be able to interact with the α4β7 integrin. This does not necessarily mean that the primary HIV-1 virions are not captured by α4β7 because the pattern and degree of glycosylation of the envelope protein produced during a human infection in vivo might be different from that produced by 293 cells in vitro. Therefore, it is critical to examine the role of α4β7 in HIV-1 infection using primary viruses produced by PBMCs or isolated from patient sera. Such studies have not yet been performed. Second, our assay can report only the strong interaction between α4β7 and envelope protein but not the weak interactions. The weakly associated envelope protein would have been removed during the washes. Therefore, lack of binding of trimers and glycosylated proteins under our assay conditions does not necessarily mean that the envelope trimers do not dynamically interact with α4β7 integrin through weak interactions, which might be very significant for virus capture and entry at the time of transmission (see model below).
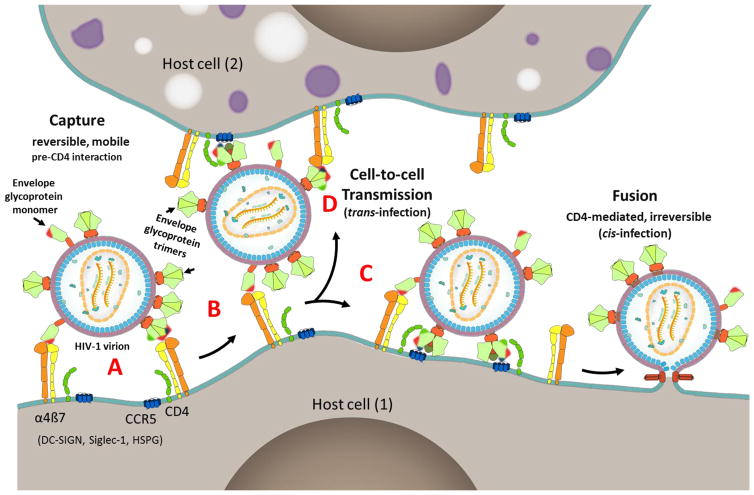
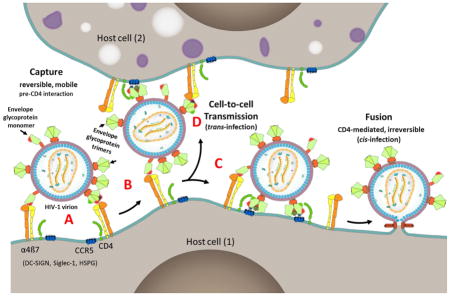
Finally, our findings and the above analyses on the envelope protein-α4β7 integrin interactions suggest a speculative model for the capture of HIV-1 virions by host cells. The V1V2 domain, located at the apex of the trimeric envelope spike, may have evolved as a “capture domain” for attachment of HIV-1 to host cells through molecules such as α4β7 (Pena-Cruz et al., 2013), Siglec-1 (Izquierdo-Useros et al., 2014; Izquierdo-Useros et al., 2012), and DC-SIGN (Geijtenbeek et al., 2000; D. S. Kwon et al., 2002). As demonstrated in this study and others (Arthos et al., 2008; Hong, Nguyen, Young, Su, & Lee, 2007; Jobe et al., 2016) (Fig. 1 and Fig. 9), these molecules recognize the V1V2 domain of the HIV-1 envelope protein. Hovering at about 20 to 30 nm from the host cell surface, these attachment factors, unlike the much shorter 7 nm-long primary receptor CD4 are better disposed to capture the HIV-1 virion. Capture is dependent on the conformational flexibility and accessibility of the V1V2 domain, which, in the case of the α4β7 integrin, might be efficicent if the HIV-1 envelope is poorly glycosylated and contains monomers. The virus inoculum an individual is exposed to during transmission would contain a highly heterogeneous mixture of chronic HIV-1 viruses with respect to length of the V1 and V2 loops, extent of glycosylation of the V1V2 domain, and ratio of monomers to trimers in the envelope. Of these, according to this model, only a small fraction of the viruses are expected to be efficiently captured by the α4β7 integrin.
Fig. 9. A model for HIV-1 virion capture.
A, HIV-1 virion attaches to host cell (1) through reversible multipoint attachment between the V1V2 domain and α4β7 integrin, leading to virus capture. Other cell surface molecules such as DC-SIGN, Siglec-1, and HSPG (Fig. 1A) might also serve as attachment factors leading to virus capture by a variety of host cells. Poorly glycosylated envelope and presence of monomers of the envelope protein enhance virion capture by α4β7 integrin. Patches of red, green, and blue shown on the interacting envelope proteins represent V1V2 domain, CD4 binding site, and V3 domain, which serve as binding sites for α4β7 integrin, CD4 receptor, and CCR5 receptor, respectively. B and C, Capture allows the virus to move on the cell surface and find its primary receptor CD4 and co-receptor CCR5 while remained bound to the host cell. The curvature of the virion might also be a factor in reaching the receptors that may otherwise be buried in the surface maze (C). A series of conformational changes follow resulting in membrane fusion and entry (cis-infection). D, Alternatively, the virus engages with the primary and co-receptors present on another host cell (2) resulting in cell-to-cell transmission (trans-infection).
Once captured (Fig. 9A), the local concentration of the virus in the microenvironment of the primary CD4 receptor raises. Due to the multipoint attachments that are relatively weak and reversible, the virus, still attached to the cell, could scan the cell surface (Fig. 9B) and find the primary CD4 receptor (Fig. 9C). The curvature of the “spherical” HIV-1 virion might also be a factor for movement of the virus and successful engagement with the receptor (cis-infection) (Fig. 9C). Alternatively, the virus could engage with a CD4 receptor on another host cell resulting in cell-to-cell transmission (trans-infection) (Fig. 9D).
This capture mechanism may also provide survival advantages to the virus, following the initial infection, to adapt to host defenses and to different cellular environments. For instance, if the host mounts an immune response to prevent capture, escape variants can be selected, in which either the glycosylation pattern or the epitope signature is altered rendering the virus resistant to the antibody but not necessarily perturbing the envelope protein structure. Alternatively, the variant V1V2 conformation may be recognized by another surface molecule, such as DC-SIGN, Siglec-1, or HSPG, and/or co-opt another entry pathway such as endocytosis (Melikyan, 2014). Furthermore, efficient capture by these capture molecules may no longer be essential to propagate the infection in the chronic state. Given this promiscuity of HIV-1 virus, a combinatorial approach would be necessary to design vaccines to effectively prevent HIV transmission. In addition to the BnAbs antibodies that block CD4-mediated fusion and entry events, it may be necessary that the vaccine also induce capture-blocking antibodies that can interfere with the pre-CD4 events that lead to HIV-1 attachment and capture. The future vaccine approaches, therefore, should focus on designing an envelope protein immunogen, or a cocktail of immunogens including the monomeric envelope glycoprotein that can elicit both capture-blocking as well as entry-blocking antibody responses.
Highlights.
The V1V2 domain of HIV-1 envelope protein binds α4β7 integrin on host cells
Glycosylated or trimeric envelope proteins bind poorly to α4β7 integrin
Deglycosylated envelope protein monomers efficiently bind to α4β7 integrin
HIV-1 virions having monomers may be efficiently captured during transmission
Acknowledgments
This work was funded by the NIAID, NIH grant AI102725 (to VBR) and in part by F32AI115912 (to ELM) and HJF subaward W81XWH-11-2-0174 (to VBR).
The authors thank Dr. Ayça Akal-Strader for her critical feedback and generous help in manuscript preparation, and Dr. James Arthos from NIAID, NIH, for useful discussions. FV40007 gp140 sequence was kindly provided by Drs. Sodsai Tovanabutra and Jerome Kim of the U.S. Military HIV Research Program, Silver Spring, MD.
Abbreviations
- DC-SIGN
dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin
- DCIR
dendritic cell immunoreceptor
- HSPG
heparin sulfate proteoglycan
- mAbs
monoclonal antibodies
- MAdCAM-1
mucosal addressin cell adhesion molecule-1
- Siglec-1
sialic acid-binding immunoglobulin-type lectin-1
- T/F
transmitter/founder
- gp16
gene product 16
- MPER
membrane proximal external region
- FV
founder virus
- BnAb
broadly neutralizing antibody
- RLU
relative light units
- IMC
infectious molecular clones
Footnotes
Conflicts of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Author contributions
SC, ELM, WA, GG, NA, GU, QY and MM designed the recombinant constructs. SC, WA, GG, and QY purified proteins and did biochemical analyses. SC, ELM and KKP did α4β7 binding assays. VP did structural modeling. MLR and MR provided materials and analyzed the results. SC, ELM and VBR wrote the manuscript. VBR directed the project.
Disclaimer
The views expressed in this article are those of the authors and do not necessarily reflect the policy or the position of the Department of the Army, Department of Defense, nor the U.S. Government.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- AlSalmi W, Mahalingam M, Ananthaswamy N, Hamlin C, Flores D, Gao G, Rao VB. A New Approach to Produce HIV-1 Envelope Trimers: BOTH CLEAVAGE AND PROPER GLYCOSYLATION ARE ESSENTIAL TO GENERATE AUTHENTIC TRIMERS. J Biol Chem. 2015;290(32):19780–19795. doi: 10.1074/jbc.M115.656611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari AA, Reimann KA, Mayne AE, Takahashi Y, Stephenson ST, Wang R, … Villinger F. Blocking of alpha4beta7 gut-homing integrin during acute infection leads to decreased plasma and gastrointestinal tissue viral loads in simian immunodeficiency virus-infected rhesus macaques. J Immunol. 2011;186(2):1044–1059. doi: 10.4049/jimmunol.1003052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthos J, Cicala C, Martinelli E, Macleod K, Van Ryk D, Wei D, … Fauci AS. HIV-1 envelope protein binds to and signals through integrin alpha4beta7, the gut mucosal homing receptor for peripheral T cells. Nat Immunol. 2008;9(3):301–309. doi: 10.1038/ni1566. [DOI] [PubMed] [Google Scholar]
- Beddows S, Schulke N, Kirschner M, Barnes K, Franti M, Michael E, … Moore JP. Evaluating the immunogenicity of a disulfide-stabilized, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol. 2005;79(14):8812–8827. doi: 10.1128/JVI.79.14.8812-8827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin C, Berg EL, Briskin MJ, Andrew DP, Kilshaw PJ, Holzmann B, … Butcher EC. Alpha 4 beta 7 integrin mediates lymphocyte binding to the mucosal vascular addressin MAdCAM-1. Cell. 1993;74(1):185–195. doi: 10.1016/0092-8674(93)90305-a. [DOI] [PubMed] [Google Scholar]
- Binley JM, Sanders RW, Clas B, Schuelke N, Master A, Guo Y, … Moore JP. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol. 2000;74(2):627–643. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulant S, Stanifer M, Lozach PY. Dynamics of virus-receptor interactions in virus binding, signaling, and endocytosis. Viruses. 2015;7(6):2794–2815. doi: 10.3390/v7062747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrareddy SN, Arthos J, Cicala C, Villinger F, Ortiz KT, Little D, … Ansari AA. Sustained virologic control in SIV+ macaques after antiretroviral and alpha4beta7 antibody therapy. Science. 2016;354(6309):197–202. doi: 10.1126/science.aag1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrareddy SN, Kallam B, Arthos J, Cicala C, Nawaz F, Hiatt J, … Ansari AA. Targeting alpha(4)beta(7) integrin reduces mucosal transmission of simian immunodeficiency virus and protects gut-associated lymphoid tissue from infection. Nature Medicine. 2014;20(12):1397–1400. doi: 10.1038/nm.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrareddy SN, Sidell N, Arthos J, Cicala C, Zhao C, Little DM, … Ansari AA. Species-specific differences in the expression and regulation of alpha4beta7 integrin in various nonhuman primates. J Immunol. 2015;194(12):5968–5979. doi: 10.4049/jimmunol.1402866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang VT, Crispin M, Aricescu AR, Harvey DJ, Nettleship JE, Fennelly JA, … Davis SJ. Glycoprotein structural genomics: solving the glycosylation problem. Structure. 2007;15(3):267–273. doi: 10.1016/j.str.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chohan B, Lang D, Sagar M, Korber B, Lavreys L, Richardson B, Overbaugh J. Selection for human immunodeficiency virus type 1 envelope glycosylation variants with shorter V1–V2 loop sequences occurs during transmission of certain genetic subtypes and may impact viral RNA levels. Journal of Virology. 2005;79(10):6528–6531. doi: 10.1128/JVI.79.10.6528-6531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicala C, Arthos J, Fauci AS. HIV-1 envelope, integrins and co-receptor use in mucosal transmission of HIV. Journal of Translational Medicine. 2011:9. doi: 10.1186/1479-5876-9-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicala C, Martinelli E, McNally JP, Goode DJ, Gopaul R, Hiatt J, … Arthos J. The integrin alpha4beta7 forms a complex with cell-surface CD4 and defines a T-cell subset that is highly susceptible to infection by HIV-1. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(49):20877–20882. doi: 10.1073/pnas.0911796106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curlin ME, Zioni R, Hawes SE, Liu Y, Deng W, Gottlieb GS, … Mullins JI. HIV-1 envelope subregion length variation during disease progression. PLoS pathogens. 2010;6(12):e1001228. doi: 10.1371/journal.ppat.1001228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derdeyn CA, Decker JM, Bibollet-Ruche F, Mokili JL, Muldoon M, Denham SA, … Hunter E. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science (New York, N Y) 2004;303(5666):2019–2022. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- Diskin R, Marcovecchio PM, Bjorkman PJ. Structure of a clade C HIV-1 gp120 bound to CD4 and CD4-induced antibody reveals anti-CD4 polyreactivity. Nature structural & molecular biology. 2010;17(5):608–613. doi: 10.1038/nsmb.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doores KJ, Burton DR. Variable loop glycan dependency of the broad and potent HIV-1-neutralizing antibodies PG9 and PG16. J Virol. 2010;84(20):10510–10521. doi: 10.1128/JVI.00552-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geijtenbeek TBH, Kwon DS, Torensma R, van Vliet SJ, van Duijnhoven GCF, Middel J, … van Kooyk Y. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell. 2000;100(5):587–597. doi: 10.1016/S0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- Goldberg E. Recognition, Attachment and Injection. In: Mathews KECK, Mosig G, Berget PB, editors. Bacteriophage. T4. Washington DC: American Society for Microbiology; 1983. pp. 32–39. [Google Scholar]
- Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, … Kim JH. Immune-Correlates Analysis of an HIV-1 Vaccine Efficacy Trial. New England Journal of Medicine. 2012;366(14):1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong PW, Nguyen S, Young S, Su SV, Lee B. Identification of the optimal DC-SIGN binding site on human immunodeficiency virus type 1 gp120. J Virol. 2007;81(15):8325–8336. doi: 10.1128/JVI.01765-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo-Useros N, Lorizate M, McLaren PJ, Telenti A, Krausslich HG, Martinez-Picado J. HIV-1 capture and transmission by dendritic cells: the role of viral glycolipids and the cellular receptor Siglec-1. PLoS Pathog. 2014;10(7):e1004146. doi: 10.1371/journal.ppat.1004146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo-Useros N, Lorizate M, Puertas MC, Rodriguez-Plata MT, Zangger N, Erikson E, … Martinez-Picado J. Siglec-1 Is a Novel Dendritic Cell Receptor That Mediates HIV-1 Trans-Infection Through Recognition of Viral Membrane Gangliosides. Plos Biology. 2012;10(12) doi: 10.1371/journal.pbio.1001448. ARTN e1001448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelicic K, Cimbro R, Nawaz F, Huang da W, Zheng X, Yang J, … Fauci AS. The HIV-1 envelope protein gp120 impairs B cell proliferation by inducing TGF-beta1 production and FcRL4 expression. Nat Immunol. 2013;14(12):1256–1265. doi: 10.1038/ni.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobe O, Trinh HV, Kim J, Alsalmi W, Tovanabutra S, Ehrenberg PK, … Rao M. Effect of cytokines on Siglec-1 and HIV-1 entry in monocyte-derived macrophages: the importance of HIV-1 envelope V1V2 region. Journal of Leukocyte Biology. 2016;99(6):1089–1106. doi: 10.1189/jlb.2A0815-361R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasse PJ. The molecular basis of HIV entry. Cell Microbiol. 2012;14(8):1183–1192. doi: 10.1111/j.1462-5822.2012.01812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasse PJ, Depetris RS, Pejchal R, Julien JP, Khayat R, Lee JH, … Moore JP. Influences on trimerization and aggregation of soluble, cleaved HIV-1 SOSIP envelope glycoprotein. J Virol. 2013;87(17):9873–9885. doi: 10.1128/JVI.01226-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, … Montagnier L. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature. 1984;312(5996):767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- Kwon DS, Gregorio G, Bitton N, Hendrickson WA, Littman DR. DC-SIGN-mediated internalization of HIV is required for trans-enhancement of T cell infection. Immunity. 2002;16(1):135–144. doi: 10.1016/s1074-7613(02)00259-5. [DOI] [PubMed] [Google Scholar]
- Kwon YD, Pancera M, Acharya P, Georgiev IS, Crooks ET, Gorman J, … Kwong PD. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nature structural & molecular biology. 2015;22(7):522–531. doi: 10.1038/nsmb.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393(6686):648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert AA, Gilbert C, Richard M, Beaulieu AD, Tremblay MJ. The C-type lectin surface receptor DCIR acts as a new attachment factor for HIV-1 in dendritic cells and contributes to trans- and cis-infection pathways. Blood. 2008;112(4):1299–1307. doi: 10.1182/blood-2008-01-136473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarovits AI, Moscicki RA, Kurnick JT, Camerini D, Bhan AK, Baird LG, … Colvin RB. Lymphocyte activation antigens. I. A monoclonal antibody, anti-Act I, defines a new late lymphocyte activation antigen. Journal of immunology (Baltimore, Md: 1950) 1984;133(4):1857–1862. [PubMed] [Google Scholar]
- Lee HS, Qi YF, Im W. Effects of N-glycosylation on protein conformation and dynamics: Protein Data Bank analysis and molecular dynamics simulation study. Scientific Reports. 2015;5 doi: 10.1038/srep08926. ARTN 8926 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann MJ, Sherer NM, Marks CB, Pypaert M, Mothes W. Actin- and myosin-driven movement of viruses along filopodia precedes their entry into cells. Journal of Cell Biology. 2005;170(2):317–325. doi: 10.1083/jcb.200503059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. Binding of HIV-1 virions to alpha(4)beta(7) expressing cells and impact of antagonizing alpha(4)beta(7) on HIV-1 infection of primary CD4(+) T cells (vol 29, pg 381, 2014) Virologica Sinica. 2015;30(2):162–162. doi: 10.1007/s12250-014-3525-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Bonsignori M, Alam SM, McLellan JS, Tomaras GD, Moody MA, … Haynes BF. Vaccine induction of antibodies against a structurally heterogeneous site of immune pressure within HIV-1 envelope protein variable regions 1 and 2. Immunity. 2013;38(1):176–186. doi: 10.1016/j.immuni.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maley F, Trimble RB, Tarentino AL, Plummer TH., Jr Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal Biochem. 1989;180(2):195–204. doi: 10.1016/0003-2697(89)90115-2. [DOI] [PubMed] [Google Scholar]
- Matsuoka Y, Moore GE, Yagi Y, Pressman D. Production of free light chains of immunoglobulin by a hematopoietic cell line derived from a patient with multiple myeloma. Proceedings of the Society for Experimental Biology and Medicine Society for Experimental Biology and Medicine (New York, N Y) 1967;125(4):1246–1250. doi: 10.3181/00379727-125-32327. [DOI] [PubMed] [Google Scholar]
- McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, … Kwong PD. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480(7377):336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melikyan GB. HIV entry: a game of hide-and-fuse? Current opinion in virology. 2014;4:1–7. doi: 10.1016/j.coviro.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondor I, Ugolini S, Sattentau QJ. Human immunodeficiency virus type 1 attachment to HeLa CD4 cells is CD4 independent and gp120 dependent and requires cell surface heparans. J Virol. 1998;72(5):3623–3634. doi: 10.1128/jvi.72.5.3623-3634.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PL, Crooks ET, Porter L, Zhu P, Cayanan CS, Grise H, … Binley JM. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. Journal of Virology. 2006;80(5):2515–2528. doi: 10.1128/Jvi.80.5.2515-2528.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mothes W, Sherer NM, Jin J, Zhong P. Virus Cell-to-Cell Transmission. Journal of Virology. 2010;84(17):8360–8368. doi: 10.1128/Jvi.00443-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz F, Cicala C, Van Ryk D, Block KE, Jelicic K, McNally JP, … Arthos J. The genotype of early-transmitting HIV gp120s promotes alpha (4) beta(7)-reactivity, revealing alpha (4) beta(7) +/CD4+ T cells as key targets in mucosal transmission. PLoS pathogens. 2011;7(2):e1001301. doi: 10.1371/journal.ppat.1001301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicely NI, Wiehe K, Kepler TB, Jaeger FH, Dennison SM, Rerks-Ngarm S, … Haynes BF. Structural analysis of the unmutated ancestor of the HIV-1 envelope V2 region antibody CH58 isolated from an RV144 vaccine efficacy trial vaccinee. EBioMedicine. 2015;2(7):713–722. doi: 10.1016/j.ebiom.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke SM, Schweighardt B, Phung P, Fonseca DPAJ, Terry K, Wrin T, … Berman PW. Mutation at a single position in the V2 domain of the HIV-1 envelope protein confers neutralization sensitivity to a highly neutralization-resistant virus. Journal of Virology. 2010;84(21):11200–11209. doi: 10.1128/JVI.00790-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, … Kwong PD. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514(7523):455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish NF, Wilen CB, Banks LB, Iyer SS, Pfaff JM, Salazar-Gonzalez JF, … Doms RW. Transmitted/founder and chronic subtype C HIV-1 use CD4 and CCR5 receptors with equal efficiency and are not inhibited by blocking the integrin alpha4beta7. PLoS Pathog. 2012;8(5):e1002686. doi: 10.1371/journal.ppat.1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peachman KK, Karasavvas N, Chenine AL, McLinden R, Rerks-Ngarm S, Jaranit K, … Rao M. Identification of New Regions in HIV-1 gp120 Variable 2 and 3 Loops that Bind to alpha4beta7 Integrin Receptor. PloS one. 2015;10(12):e0143895. doi: 10.1371/journal.pone.0143895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena-Cruz V, Etemad B, Chatziandreou N, Nyein PH, Stock S, Reynolds SJ, … Sagar M. HIV-1 envelope replication and alpha 4 beta 7 utilization among newly infected subjects and their corresponding heterosexual partners. Retrovirology. 2013;10 doi: 10.1186/1742-4690-10-162. Artn 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez LG, Chen H, Liao HX, Montefiori DC. Envelope glycoprotein binding to the integrin alpha4beta7 is not a general property of most HIV-1 strains. J Virol. 2014;88(18):10767–10777. doi: 10.1128/JVI.03296-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic A, Alpdogan O, Willis LM, Eng JM, Greenberg AS, Kappel BJ, … van den Brink MR. LPAM (alpha 4 beta 7 integrin) is an important homing integrin on alloreactive T cells in the development of intestinal graft-versus-host disease. Blood. 2004;103(4):1542–1547. doi: 10.1182/blood-2003-03-0957. [DOI] [PubMed] [Google Scholar]
- Ping LH, Joseph SB, Anderson JA, Abrahams MR, Salazar-Gonzalez JF, Kincer LP … the Center for, H. I. V. A. V. I. C. Comparison of viral Env proteins from acute and chronic infections with subtype C human immunodeficiency virus type 1 identifies differences in glycosylation and CCR5 utilization and suggests a new strategy for immunogen design. J Virol. 2013;87(13):7218–7233. doi: 10.1128/JVI.03577-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci U S A. 2002;99(21):13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringe RP, Sanders RW, Yasmeen A, Kim HJ, Lee JH, Cupo A, … Moore JP. Cleavage strongly influences whether soluble HIV-1 envelope glycoprotein trimers adopt a native-like conformation. Proc Natl Acad Sci U S A. 2013;110(45):18256–18261. doi: 10.1073/pnas.1314351110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuto CD, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong PD, Hendrickson WA, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science (New York, N Y) 1998;280(5371):1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- Robb ML, Eller LA, Kibuuka H, Rono K, Maganga L, Nitayaphan S … Team, R. V. S. Prospective Study of Acute HIV-1 Infection in Adults in East Africa and Thailand. N Engl J Med. 2016;374(22):2120–2130. doi: 10.1056/NEJMoa1508952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar M, Wu X, Lee S, Overbaugh J. Human immunodeficiency virus type 1 V1–V2 envelope loop sequences expand and add glycosylation sites over the course of infection, and these modifications affect antibody neutralization sensitivity. Journal of Virology. 2006;80(19):9586–9598. doi: 10.1128/JVI.00141-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar-Gonzalez JF, Salazar MG, Keele BF, Learn GH, Giorgi EE, Li H, … Shaw GM. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. The Journal of experimental medicine. 2009;206(6):1273–1289. doi: 10.1084/jem.20090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Dankers MM, Busser E, Caffrey M, Moore JP, Berkhout B. Evolution of the HIV-1 envelope glycoproteins with a disulfide bond between gp120 and gp41. Retrovirology. 2004;1:3. doi: 10.1186/1742-4690-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Vesanen M, Schuelke N, Master A, Schiffner L, Kalyanaraman R, … Moore JP. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol. 2002;76(17):8875–8889. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PL, Nemerow GR. Cell integrins: commonly used receptors for diverse viral pathogens. Trends in microbiology. 2007;15(11):500–507. doi: 10.1016/j.tim.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Stieh DJ, King DF, Klein K, Aldon Y, McKay PF, Shattock RJ. Discrete partitioning of HIV-1 Env forms revealed by viral capture. Retrovirology. 2015;12:81. doi: 10.1186/s12977-015-0207-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Gao S, Kondabagil K, Xiang Y, Rossmann MG, Rao VB. Structure and function of the small terminase component of the DNA packaging machine in T4-like bacteriophages. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(3):817–822. doi: 10.1073/pnas.1110224109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassaneetrithep B, Tivon D, Swetnam J, Karasavvas N, Michael NL, Kim JH, … Cardozo T. Cryptic determinant of alpha4beta7 binding in the V2 loop of HIV-1 gp120. PloS one. 2014;9(9):e108446. doi: 10.1371/journal.pone.0108446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjomsland V, Ellegard R, Kjolhede P, Wodlin NB, Hinkula J, Lifson JD, Larsson M. Blocking of integrins inhibits HIV-1 infection of human cervical mucosa immune cells with free and complement-opsonized virions. European Journal of Immunology. 2013;43(9):2361–2372. doi: 10.1002/eji.201243257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretter V, Altmann F, Marz L. Peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase F cannot release glycans with fucose attached alpha 1----3 to the asparagine-linked N-acetylglucosamine residue. Eur J Biochem. 1991;199(3):647–652. doi: 10.1111/j.1432-1033.1991.tb16166.x. [DOI] [PubMed] [Google Scholar]
- Vaccari M, Gordon SN, Fourati S, Schifanella L, Liyanage NP, Cameron M, … Franchini G. Adjuvant-dependent innate and adaptive immune signatures of risk of SIVmac251 acquisition. Nature Medicine. 2016;22(7):762–770. doi: 10.1038/nm.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, … Burton DR. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326(5950):285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387(6631):426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- Yu Y, Schurpf T, Springer TA. How natalizumab binds and antagonizes alpha4 integrins. J Biol Chem. 2013;288(45):32314–32325. doi: 10.1074/jbc.M113.501668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Bazick J, Sodroski J. Characterization of the multiple conformational States of free monomeric and trimeric human immunodeficiency virus envelope glycoproteins after fixation by cross-linker. J Virol. 2006;80(14):6725–6737. doi: 10.1128/JVI.00118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]