Abstract
Circular RNAs (CircRNAs) were first identified as a viroid and later found to also be an endogenous RNA splicing product in eukaryotes. In recent years, a series of RNA-sequencing analyses from a diverse range of eukaryotes have shed new light on these eukaryotic circRNAs, revealing dynamic expression patterns in various developmental stages and physiological conditions. In this review, we focus on circRNAs implicated in stress response pathways and explore potential mechanisms underlying their regulation. To date, circRNAs have been shown to act as scaffolds in the assembly of protein complexes, sequester proteins from native subcellular localization, activate transcription of parental genes, inhibit RNA–protein interactions, and function as regulators of microRNA activity. Although the mechanism modulating circRNA levels during stress remains unclear, circRNAs are shown to be regulated during biogenesis, degradation and exportation. As circRNAs do not have 5’ and 3’ ends, there are no entry points for exoribonucleases to initiate degradation. Such inherent stability makes this class of RNA a strong candidate to maintain homeostasis in the face of environmental challenges.
Keywords: Circular RNA, stress, circRNA biogenesis, circRNA degradation, environment, circularization, back-splicing, RNA stability
Introduction
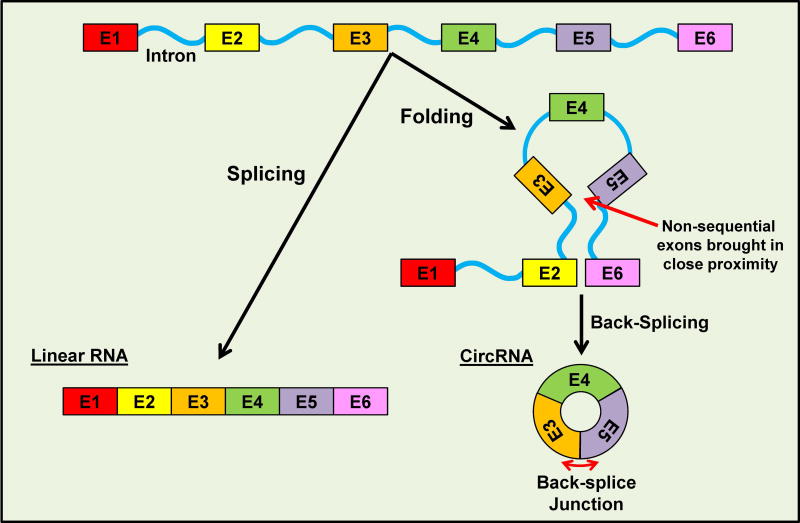
Circular RNAs (circRNAs) is a class of noncoding RNAs that form covalently closed circular loops. CircRNAs was first discovered as genomes for certain RNA viruses in 1970s and 1980s (Sanger et al. 1976; Hsu et al. 1979; Kos et al. 1986). In humans, the first endogenous circRNA was detected in 1991 while studying splicing pattern in a tumor suppressor candidate by the Vogelstein group (Nigro et al. 1991). They identified a small population of PCR products from reverse transcription in which the exons appeared scrambled (i.e., not in a sequential order as in primary transcripts). Nigro et al (1991) proposed that pre-mRNA formed a looped secondary structure, which facilitated the “scrambled” arrangement of exons, resulting in the formation of spliced RNA in circular form where the downstream 5’ splice site is joined to an upstream 3’ splice site, commonly referred as “back-splicing” (Figure 1). Further research revealed that these circRNAs were enriched in cytoplasmic poly(A) free fractions in both human and rodent cells (Nigro et al. 1991; Cocquerelle et al. 1992; Cocquerelle et al. 1993). The possibility for this class of noncoding RNAs to be produced in a regulatory manner was subsequently demonstrated in the testis-determining gene Sry, in which the circular form is expressed only in specific tissues and accounts for over 90% of Sry transcripts in mouse testes at a defined embryonic stage (Capel et al. 1993).
Figure 1. CircRNA biogenesis.
Simplified model illustrating the formation of circRNAs through the back-splicing of exons in a non-sequential order. Circularization of RNA can be generated by repetitive sequences within introns or RNA binding proteins to facilitate the back-splicing of non-sequential exons. The back-splice junction, an essential site for detecting circRNAs or for circRNA-specific knockdown experiments, is highlighted. A color version of the figure is available online.
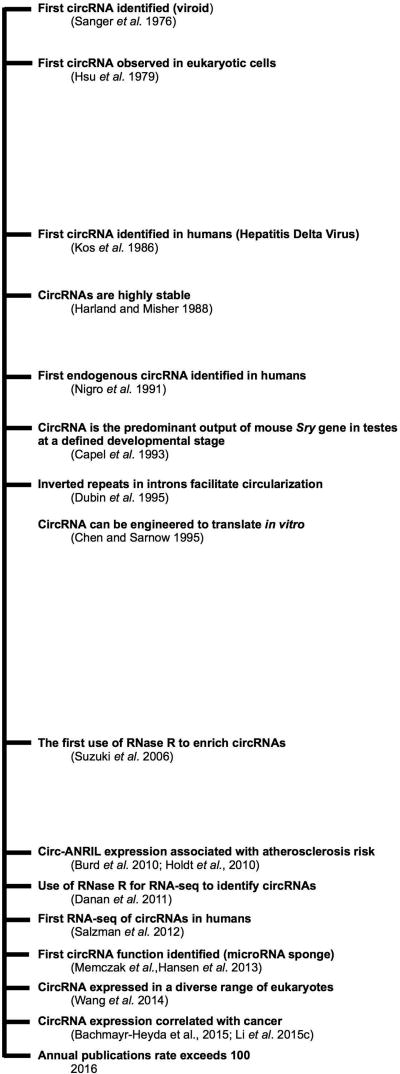
Although several circRNAs have been identified since the 1990s (Chao et al. 1998, Surono et al. 1999, Fluoriot et al. 2002, Houseley et al. 2006), the vast abundance and diversity of circRNAs were not fully appreciated until the development of RNA sequencing (RNA-seq) technologies in the twenty-first century, which profile circRNAs in a genome-wide manner (Danan et al. 2011, Salzman et al. 2012). In addition to RNA-seq, RNase R, an exoribonuclease with 3’ to 5’ activity, proved to be particularly useful in the characterization and identification of circRNAs (Suzuki et al. 2006). Given that circRNAs have no free 5’ or 3’ ends, the exoribonuclease RNase R specifically degrades linear RNAs, including any linear transcripts with scrambled exons due to trans-splicing or genomic rearrangements. The use of RNase R to enrich for circRNAs prior to RNA-seq was first used in archaea (Danan et al. 2011). In eukaryotes, RNA-seq was performed from total RNAs that was depleted of rRNAs and subjected to RNase R treatment, which resulted in the detection of over 25,000 distinct circRNAs in human cells (Jeck et al. 2013). RNA-seq has since been used to detect circRNA expression in various tissues, developmental stages, and organisms under a range of growth and stress conditions (e.g., Memczak et al. 2013, Salzman et al. 2013, Wang et al. 2014, Rybak-Wolf et al. 2015, Szabo et al. 2015, Veno et al. 2015, Dang et al. 2016; Table 1). Excellent reviews have extensively covered the mechanisms underlying their formation and function (reviewed in Lasda and Parker 2014, Chen 2016, Salzman 2016, Wilusz 2016). Here, we examine specific examples and discuss potential mechanisms underlying the regulatory roles of circRNA during stress. A timeline highlighting key discoveries on circRNAs relevant to our discussions is summarized in Figure 2.
Table 1.
CircRNAs regulated by stresses
| Stress | Specific CircRNAs | Reference |
|---|---|---|
| Over-growth | Circ-FOXO3 | Du et al. 2016b |
| Reactive Oxygen Species (ROS) and serum starvation | Circ-FOXO3 | Du et al. 2016a |
| Stress-ligand cell surface presentation | Circ-NFATC3, Circ-ANKRD17, and 32 other circRNAs | Schneider et al. 2016 |
| Antimicrobial response via lipopolysaccharide (LPS) | Circ-RasGEF1B | Ng et al. 2016 |
| Atherosclerosis and nucleolar stress | Circ-ANRIL | Holdt et al. 2016 |
| Chemotherapeutics | Fusion-CircRNA | Guarnerio et al. 2016 |
| Apoptosis induced by lead neurotoxicity | CircRar1 | Nan et al. 2016 |
| Hypoxia | Many CircRNAs | Boeckel et al. 2015 |
| Metabolic Stress and Inflammation | Circ-ITCH | Li et al. 2015a, Huang et al. 2015 |
| Oxygen and glucose deprivation | mmu-circRNA-015947 | Lin et al. 2016 |
| Nitrogen Starvation (Yeast) | Many CircRNAs | Wang et al. 2014 |
| Phosphate Starvation and Light exposure (Plants) | Many CircRNAs | Ye et al. 2015 |
| Chilling Injury (Plants) | Many CircRNAs | Zuo et al. 2016 |
Figure 2. A timeline of key events in circRNA research.
CircRNAs and Stress
To maintain cell homeostasis and allow organisms to flourish and reproduce, it is important to combat a variety of stresses that threaten their viability (reviewed in Kültz 2005). CircRNAs can, in theory, regulate cellular stress both positively and negatively: some may function to maintain homeostasis whereas dysregulation of others may enhance stress response to adapt chronic stress (e.g., during cancer development). In the following section, we provide specific examples of circRNAs involved in maintaining homeostasis, regulating cell growth and death, combating acute infection, and conferring resistance to chemotherapeutics (summarized in Table 1).
CircRNAs that regulate activity of microRNAs involved in homeostasis
Sequence analyses revealed that some circRNAs had multiple predicted microRNA (miRNA) binding sites, leading to the hypothesis that this subset acts as a sponge to reduce the levels of available miRNAs, which would otherwise inhibit the translation and decay of target mRNAs. As miRNAs played an important role in the cellular response to stress (reviewed in Leung and Sharp 2010), it is possible that specific circRNAs regulate various stress response pathways by inhibiting miRNA activity. Two seminal studies in 2013 demonstrated that a circRNA termed CDR1as/ciRS-7, with over 70 predicted miR-7 binding sites, inhibited the function of miR-7 in the brain by acting as a miRNA sponge (Memczak et al. 2013, Hansen et al. 2013a). These back-to-back studies of CDR1as pointed to the first biological function ascribed to circRNAs (i.e., a miRNA sponge). Subsequently, studies proposed a similar function for some other circRNAs (Lukiw 2013, Wang et al. 2015, Liu et al. 2016, Geng et al. 2016, Zheng et al. 2016, Werfel et al. 2016, Wang et al. 2016b). However, it should be noted that only one additional circRNAs, circ-SRY, contain substantial number of miRNA binding sites (Capel et al. 1993, Dubin et al. 1995, Hansen et al. 2013a), raising the question whether miRNA inhibition is a general feature of circRNAs (see Cautionary notes below for further discussions). Here, specific examples of circRNAs regulating the activity of miRNAs involved in maintaining homeostasis and stress pathways are discussed below.
miR-7 sponge CDR1as regulates insulin secretion
Insulin secretion is an important mechanism for maintaining homeostasis of blood glucose levels. Diabetic patients are regularly exposed to both high and low glucose levels, which induce a variety of stress responses, such as the production of reactive oxygen species (ROS) and inflammation (reviewed in Evans et al. 2002). In a study of islet cells (responsible for insulin secretion) treated with various agents that stimulated the transcription and secretion of insulin, the levels of the miR-7 sponge CDR1as significantly increased when the cells were treated with forskolin or phorbol myristate acetate (PMA) but not glucose (Xu et al. 2015). Additionally, overexpression of CDR1as led to an increase in insulin secretion, and two predicted miR-7 targets involved in insulin secretion, Myrip and Pax6m, were both inhibited by miR-7 and up-regulated by CDR1as overexpression. Although the results suggested that CDR1as played a role in insulin secretion, Xu et al. (2015) did not perform any knockdown experiments. Therefore, it is not clear whether endogenous CDR1as is expressed at sufficient levels to regulate insulin secretion. However, the results provide promising evidence that ectopic expression of circRNAs at sufficient levels may modulate miRNA activity and help regulate miRNAs involved in glucose homeostasis.
Possible roles of circRNAs as miRNA sponges in tumorigenesis
Tumor cells are constantly exposed to multiple stress conditions, providing an opportunity to identify factors that combat stress and restore homeostasis (Dong et al. 2016). Using RNA-seq, circRNAs were shown to be differentially regulated in response to stresses, including hypoxia, which is common in the tumor microenvironment (e.g., Boeckel et al. 2015). A study of the expression profile of circRNAs in normal and tumor cells found several circRNAs to be differentially expressed (Bachmayr-Heyda et al. 2015). For example, the miR-7 sponge CDR1as was significantly up-regulated in tumor samples. Notably, miR-7 was also shown to regulate various oncogenes, including p21-activated kinase 1 (PAK1) (Bachmayr-Heyda et al. 2015). PAK1 is a kinase that is commonly activated by DNA-damaging agents (radiation or etoposide), and trigger DNA repair and inhibit apoptosis (Advani et al. 2015). It is possible that CDR1as is up-regulated to help cancer cells cope during chronic exposure to stress. However, it is unclear whether such overexpression of CDR1as in tumor cells results in radiation resistance and cell survival.
Another characterized miRNA sponge is circ-SRY (Capel et al. 1993 and Dubin et al. 1995), which has 16 miR-138 binding sites (Hansen et al. 2013a). Although Sry is normally considered a testis-specific gene, linear Sry mRNA is expressed in other tissues (Clepet et al. 1993), and its expression is correlated with a poor prognosis in hepatocellular carcinoma (Xue et al. 2015). Previous work identified miR-138 as an inhibitor of tumor cell invasion by inhibiting HIF-1α, a transcription factor involved in the hypoxia stress response (Yeh et al. 2013). It will be of interest to determine whether circ-SRY is also expressed in cancers, and if so, whether it is expressed at sufficient levels to act as a miR-138 sponge to inhibit the hypoxia stress response.
Cautionary notes about miRNA sponge studies during stress
While many studies have reported the role of circRNAs as miRNA sponges, bioinformatics analyses of predicted circRNA sequences found no enrichment of miRNA binding sites compared to linear mRNA counterparts (Guo et al. 2014, Conn et al. 2015, You et al. 2015). Additionally, the expression of most circRNAs was very low compared to that of linear mRNAs (Enuka et al. 2016). Such low expression levels of circRNAs are unlikely to sufficiently reduce miRNA levels and have significant biological implications. Thus, when considering the possible role of a specific circRNA as a miRNA sponge, quantitation of the amounts of circRNA and miRNA is necessary. One characteristic of miRNA sponges is their association with Argonaute, which is a core protein that binds and executes miRNA functions (Leung and Sharp, 2010). To determine the role of circRNAs as miRNA sponges, genome-wide screening is needed to determine how many circRNAs are associated with Argonaute. Notably, a recent study used RNA-seq to identify circRNAs in mice exposed to lead poisoning and found that circRar1 was up-regulated in the hippocampus and cerebral cortex to control apoptosis in neurons (Nan et al. 2016). The study revealed that circRar1 contained one miR-671 binding site: a miRNA that inhibits the expression of pro-apoptotic proteins caspase-8 and p38, both are predicted miR-671 targets. Inhibition of circ-Rar1 using small interfering RNAs (siRNA) against the back-splice junction (cf. Figure 3), intriguingly, increased the levels of miR-671 and decreased the expression of caspase-8 and p38 protein. Conversely, overexpression of circRar1 resulted in an increased expression of caspase-8 and p38 proteins, and induced apoptosis (Nan et al. 2016). Given that there is only one miR-671 binding site on circRar1, it is unlikely that this circRNA acts as a sponge for miR-671, but the mechanism warrants further investigation.
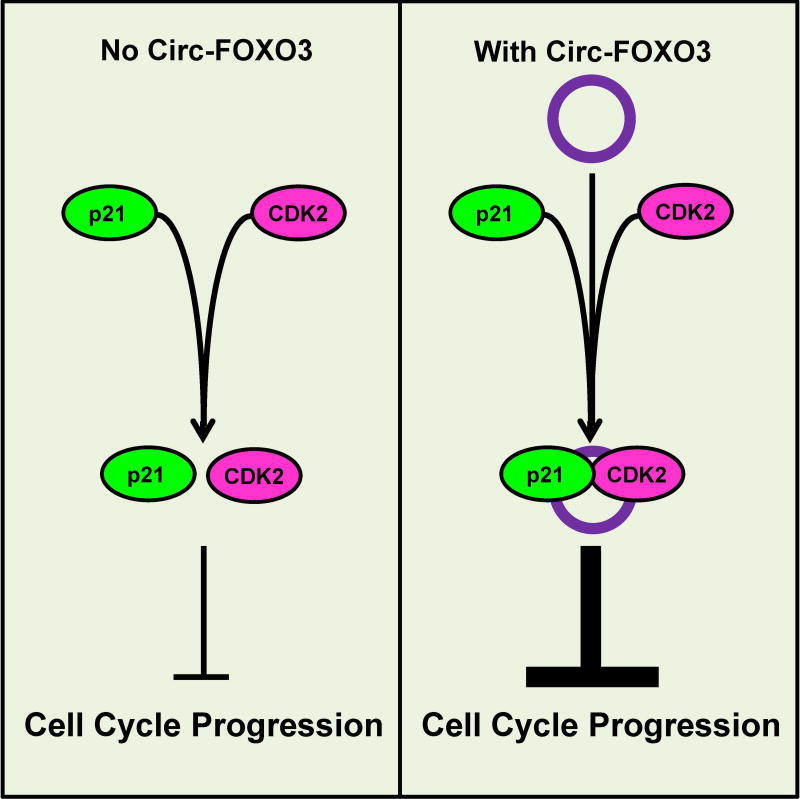
Figure 3. CircRNA acts as a scaffold for the formation of a protein complex that disrupts cell cycle progression upon cell overgrowth.
Left: When circ-FOXO3 is not present, the interaction between p21 and CDK2 is weak and unable to fully inhibit cell cycle progression. Right: when circ-FOXO3 is present, it acts as a scaffold to increase the interaction between p21 and CDK2 and inhibit cell cycle progression. A color version of the figure is available online.
Lastly, many experiments that demonstrated the roles of specific circRNAs as miRNA sponges involved the overexpression of cDNA constructs that generate circRNAs (Hansen et al. 2013a, Liang and Wilusz 2014). Using this approach, linear mRNA is, however, also generated from the same cDNA construct at measurable levels (Liang and Wilusz 2014). To demonstrate that the observed phenotype is specific to circRNAs but not the linear counterparts, constructs with a single splice site mutation that disrupts the formation of circRNA should be used as a negative control. To ascertain whether these circRNAs act as miRNA sponges, it is critical to test whether siRNA knockdown targeting the back-splice junction results in the opposite phenotype of circRNA sponge overexpression. As illustrated below, many circRNAs can act in a manner independent of miRNA sponges; therefore, other possible functions of circRNAs should be considered.
Circ-FOXO3 acts as a scaffold in the assembly of protein complexes to disrupt cell cycle progression in response to cell overgrowth
Du et al. (2016b) recently found that a circRNA derived from the transcription factor FOXO3, the dysregulation of which contributes to tumor development, was up-regulated in overconfluent cancer cell culture. Reducing the circ-FOXO3 level by using an siRNA against the back-splice junction led to increased cell growth, whereas ectopic expression of circ-Foxo3 repressed cell cycle progression at the G1 stage (Du et al. 2016b) (Figure 3). By performing immunoprecipitation against a panel of cell cycle proteins, the same study showed that circ-FOXO3 was physically associated with cyclin dependent kinase 2 (CDK2) and p21, a cell cycle inhibitor of CDK2 activity. Native gradient gel electrophoresis further demonstrated that circ-FOXO3 overexpression resulted in an increased interaction between CDK2 and p21 and that siRNA-mediated knockdown decreased the protein–protein interaction (Du et al. 2016b). The circ-FOXO3-induced association of p21 with CDK2 resulted in the inhibition of CDK2 activity and blocking of cell cycle progression. This study illustrates for the first time how a circRNA can act as a scaffold to regulate protein–protein interactions.
Circ-FOXO3 alters the localization of proteins by sequestration during cardiac senescence
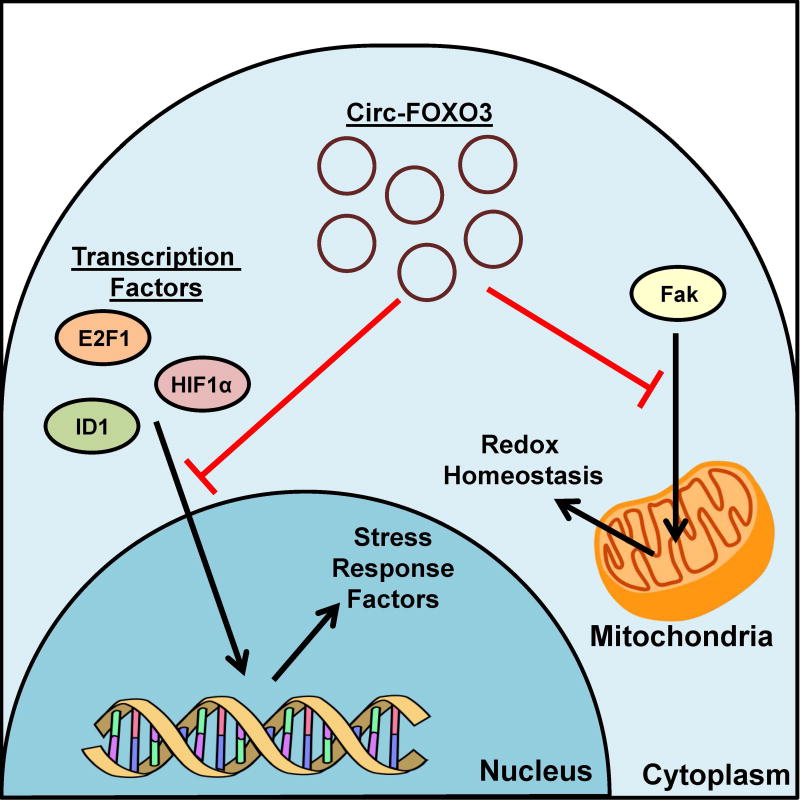
Interestingly, the same circ-FOXO3 also sequestered proteins involved in cellular stress pathways and induced cardiac senescence (Du et al. 2016a) (Figure 4). In the study by Du et al. (2016a), the level of circ-FOXO3 was up-regulated when mouse embryonic fibroblasts were exposed to ROS or serum starvation and when mice were treated with doxorubicin, which induced cardiac senescence through the induction of ROS. The up-regulation of circ-FOXO3 promoted senescence by interacting with several proteins, including ID-1, E2F1, FAK, and HIF-1α, involved in cell survival and stress response pathways. In the same study, the association of circ-FOXO3 with these four proteins was demonstrated by enriching circ-FOXO3 using antibodies against these proteins, and, conversely, by detecting these proteins via pulldown using a biotin-labeled probe that bound to circ-FOXO3. The induction of circ-FOXO3, either via transient overexpression in cell culture or doxorubicin treatment in mice, reduced the nuclear translocation of ID-1, E2F1, and HIF1α, as well as mitochondria localization of FAK. Instead, these proteins were sequestered in the cytoplasm co-localized with circ-FOXO3. Consistently, knockdown of circ-FOXO3 caused an increase in the respective nuclear and mitochondrial localizations of these four proteins (Du et al. 2016a). Taken together, these data suggest that one potential function of circRNAs is to reduce the subcellular localization of proteins via sequestration.
Figure 4. CircRNA sequesters proteins during cardiac senescence.
Circ-FOXO3 expression is able to bind to the transcription factors E2F1, ID1, and HIF1α and inhibit their translocation into the nucleus. The lack of nuclear translocation decreases the transcription of stress response factors. Cic-FOXO3 is also able to bind to FAK and inhibit its localization into the mitochondria. Reduced FAK levels in the mitochondria decrease the ability for cells to re-establish redox homeostasis. A color version of the figure is available online.
Many circRNAs are predicted to interact with RNA binding proteins (RBPs) (Hentze and Preiss 2013), although bioinformatic analyses of circRNA sequences found very little enrichment in binding sites of RBPs compared with those of its linear counterpart (You et al. 2015). Yet, unlike mRNAs, circRNAs are not normally translated, and therefore RBPs bound to circRNAs are not displaced by ribosomes (Jeck et al. 2013, Guo et al. 2014). The more stable association of circRNAs with RBPs potentially allows the former to act as sponges and reduce the levels of available RBPs. In addition, circRNAs could act as a reservoir of RBPs that are released in response to certain signals, such as cellular stresses. Recently, a set of 34 circRNAs was found to be associated with IMP3, an RBP involved in post-transcriptional regulation and a potential driver of tumor development (Schneider et al. 2016). Therefore, it will be of interest to test whether these circRNAs can act as sponges and modulate the activity of RBPs, such as IMP3.
Circ-RasGEF1B increases the stability of mRNAs involved in the antimicrobial response
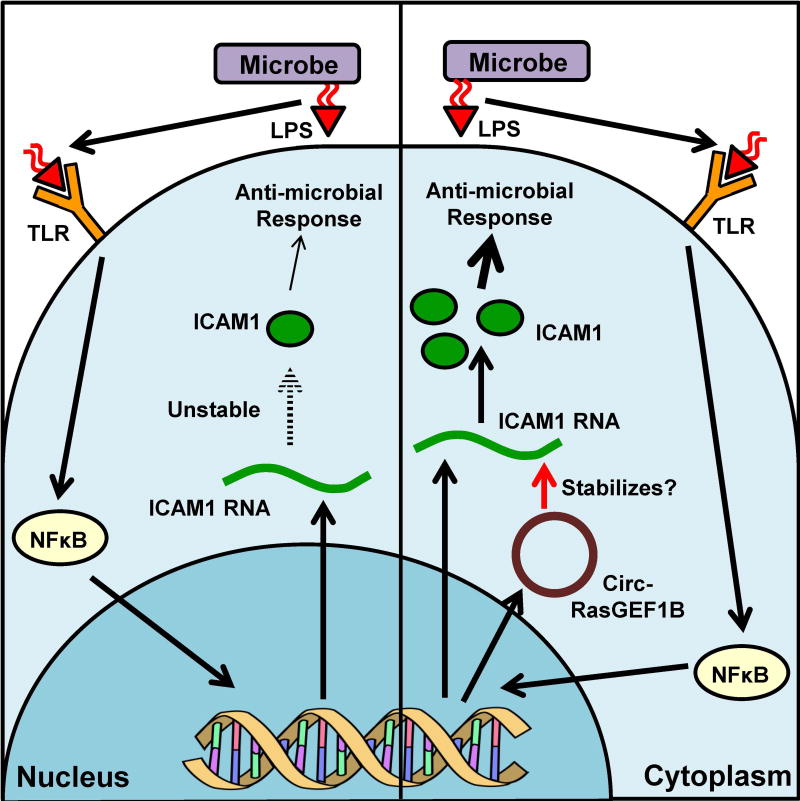
While many stresses are a constant battle to maintain homeostasis, many other stresses, such as microbe infection, are acute in nature and require a rapid and efficient response. For example, lipopolysaccharides (LPS) on the surface of bacteria trigger the NF-κB signaling cascade, which promotes transcription of factors involved in the antimicrobial response. Upon LPS treatment, circ-RasGEF1B, a circRNA conserved in humans and mice, was up-regulated via the NF-κB pathway (Ng et al. 2016) (Figure 5). Of the LPS-induced genes examined, the expression of the intracellular adhesion molecule (ICAM) ICAM-1, which is involved in immune response signaling, was reduced at the RNA and protein level upon knockdown of circ-RasGEF1B. In addition, circ-RasGEF1B was localized in the cytoplasm and not co-purified with ribosomal fractions, indicating its noncoding nature. Interestingly, knockdown of this circRNA resulted in a small but significant decrease in the stability of mature mRNAs but not that of pre-mRNA. Informatics analyses revealed no shared miRNA binding site between circ-RasGEF1B and the ICAM-1 transcript, ruling out the possibility that circ-RasGEF1B acting directly via miRNA-based regulation. It remains unclear how circ-RasGEF1B regulates ICAM-1 mRNA stability.
Figure 5. CircRNA increases stability of mRNA involved in antimicrobial response.
Left: Lipopolysaccharide (LPS) acts as a ligand and binding to Toll-Like Receptors (TLR). TLR activates the NFκB pathway leading to transcription of proteins involved in the antimicrobial response, including ICAM1. Right: When circ-RasGEF1B is induced by NFκB, this circRNA stabilizes the mature ICAM1 mRNA via unknown mechanism, leading to its translation and antimicrobial response. A color version of the figure is available online.
Circ-ANRIL regulates ribosomal maturation and induces apoptosis to confer atheroprotection
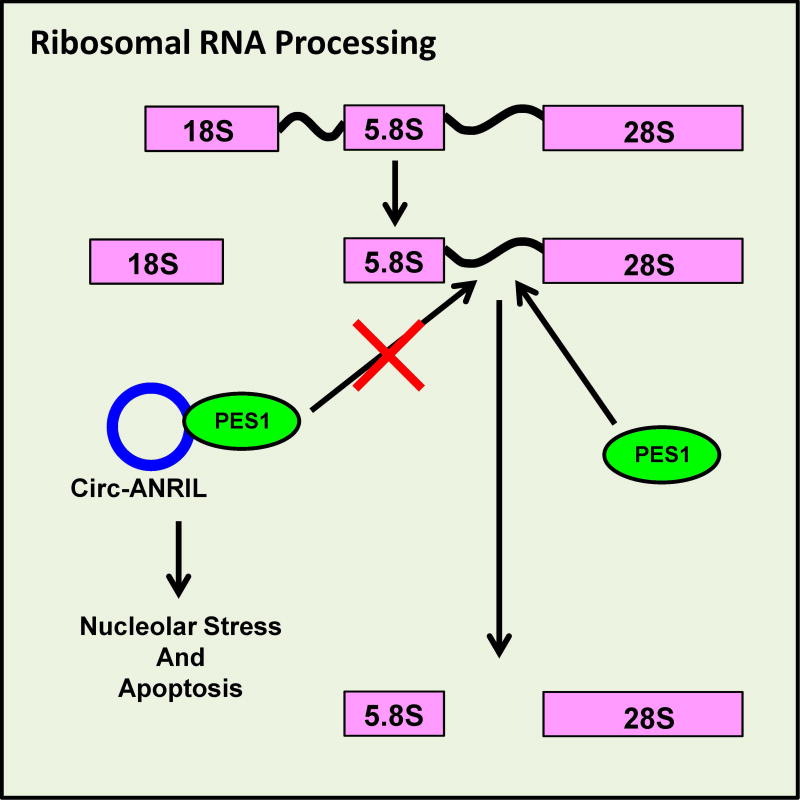
Besides mRNA stability, a circRNA derived from the exon 5, 6, and 7 of the non-coding RNA ANRIL (circ-ANRIL) has been shown to regulate rRNA maturation and induce nucleolar stress and this circRNA is associated with atherosclerosis risk (Holdt et al. 2016). In atherosclerosis, the arterial walls thicken from white blood cell accumulation and smooth muscle proliferation, eventually resulting in the clogging of arteries. Holdt et al. (2016) reported that circ-ANRIL expression was associated with a reduced risk of atherosclerosis (atheroprotection). Increasing circ-ANRIL expression in mammalian cells to the level of atheroprotection observed in humans (∼3-fold) inhibited rRNA maturation and induced nucleolar stress. The prolonged nucleolar stress eventually induced apoptosis, which is believed to reduce the build-up of white blood cells in arterial plaque. Upon circ-ANRIL knockdown, nucleolar stress and rRNA processing were restored. In the same study, proteomics analyses revealed that circ-ANRIL associated strongly with proteins involved in ribosome biogenesis and assembly. Further characterization revealed that circ-ANRIL physically interacted with the rRNA processing protein PES1 in the same binding domain as pre-rRNA. Circ-ANRIL thus competes with rRNA for binding to PES1 and inhibits rRNA processing, thereby inducing nucleolar stress and apoptosis (Figure 6). It is unclear how circ-ANRIL acts as a competitive inhibitor whereas the linear isoform is unable to perform the same function. Interestingly, linear ANRIL and additional circRNAs from ANRIL were previously shown to have the opposite effect and promote atherosclerosis (Burd et al. 2010, Holdt et al. 2010). Future studies will be required to understand how the balance of linear and circular forms of ANRIL influences atherosclerosis.
Figure 6. CircRNA inhibits ribosomal RNA maturation and induces apoptosis for atheroprotection.
PES1 is an enzyme involved in the final steps of ribosomal RNA (rRNA) maturation. When circ-ANRIL is expressed at high levels, the circRNA is able to act as a competitive inhibitor and prevent the PES1–rRNA interaction, leading to nucleolar stress and eventually apoptosis. A color version of the figure is available online.
Many experiments that demonstrated the roles of specific circRNAs as miRNA sponges involved the circRNAs (f-circRNAs) confer resistance to chemotherapeutics
A study of fusion proteins involved in leukemia found that several fusion genes (e.g., PML/RARα and MLL/AF9) that promoted tumorigenesis produced circRNAs (f-circPR and f-circM9, respectively) (Guarnerio et al. 2016). Knockdown of individual f-circRNAs triggered apoptosis in leukemic cells. Intriguingly, ectopic expression of the f-circRNA conferred protection against standard chemotherapeutics (arsenic trioxide and cytarabine) that are commonly used to treat leukemia by inducing oxidative damage and apoptosis in rapidly dividing cells (Guarnerio et al. 2016). Both f-circPR and f-circM9 led to increased proliferation and transformation upon ectopic expression, but only f-circM9 triggered a specific cell growth signaling pathway mediated via Akt. These data indicated that the proliferative function of these f-circRNAs was likely mediated by distinct mechanisms within cancer cells. Although the mechanism of f-circRNAs and their role in tumorigenesis warrant further investigation (e.g., the amount of endogenous level of f-circRNAs and the identity of targets) (Croce 2016), their ability to promote tumorigenesis and confer resistance to existing therapeutics could potentially make them a target for treatment. On the other hand, because circRNAs are readily detectable in blood and differentially expressed in cancer, circRNAs also have the potential to be used as predictive or diagnostic biomarkers (reviewed in Wang et al. 2016a).
Is circRNA induction a by-product of increased gene expression during stress?
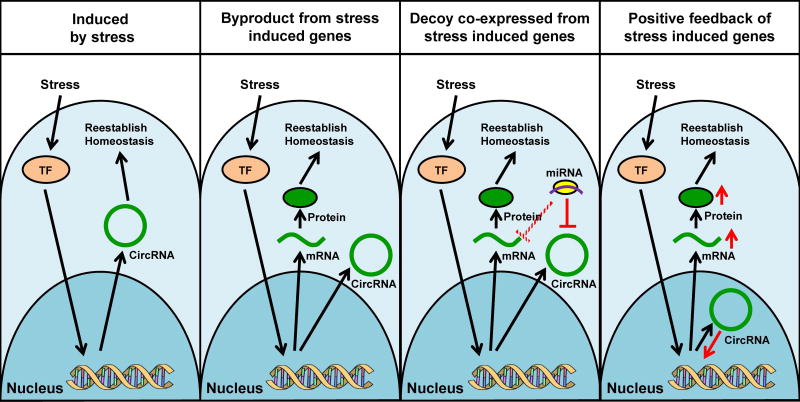
Plants provide a good model system to study stress responses due to their continual adaptation to a changing environment. Previous studies demonstrated that circRNAs were differentially expressed under phosphate starvation, reduced light exposure, and chilling (Ye et al. 2015, Zuo et al. 2016). Although the biological implications of these findings were not identified, a bioinformatics analysis of chilling-induced circRNAs found that the circRNAs originated from genes involved in various stress-response pathways, such as redox homeostasis, metabolism, and heat shock cascades (Zuo et al. 2016). As noted by Ye et al. (2015), many circRNAs showed a positive correlation with their linear counterparts, and none had a negative correlation, suggesting that these circRNAs may be up-regulated as a consequence of increased transcription of the parental genes. Although many circRNAs may simply be up-regulated as a by-product of increased transcription, many other circRNAs may have defined roles in stress response, such as having a direct stress response function, protecting linear RNA by acting as a decoy, and enhancing transcription of stress response genes (Figure 7 and 8). For example, by sharing the binding sites for eight miRNAs, the same previously mentioned circ-FOXO3 acted as a decoy for linear FOXO3 mRNA, where ectopic expression of circ-FOXO3 resulted in increased translation of FOXO3 (Yang et al. 2016). As a result, the up-regulation of both linear and circular RNAs from the same gene may help tune the final expression of linear mRNAs.
Figure 7. Potential models to explain circRNA induction in response to stress.
In response to stress, activated signaling cascades will induce the nuclear translocation of a transcription factor (TF) resulting in the transcription of stress-induced genes. Far left: Stress-induced circRNAs perform specific functions to help re-establish homeostasis. Second to left: CircRNAs are by-products of stress-induced gene expression. Second to right: Stress induces the production of linear and circular RNAs from the same stress-induced genes, where circRNA acts as a decoy against miRNAs or RBPs. As a result, the circRNA decoy modulates the expression of stress-induced gene by titrating factors that bind onto the same RNA sequences. Far right: CircRNAs from stress-induced genes enhance transcription to increase expression of stress response genes (cf. Figure 8). A color version of the figure is available online.
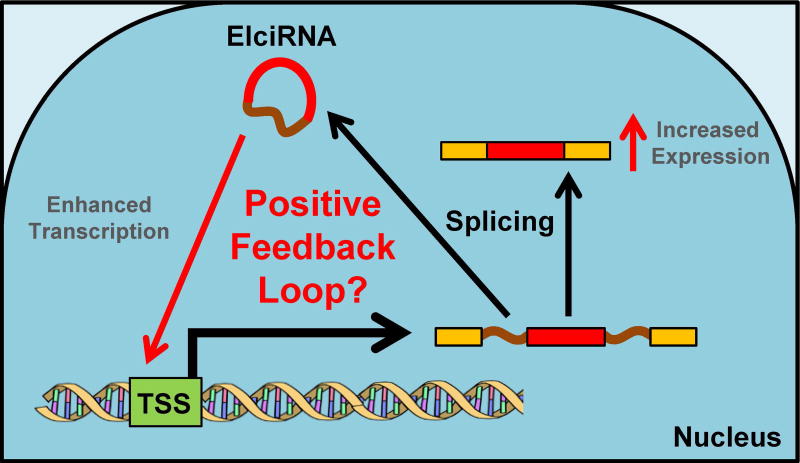
Figure 8. Exon-intron circRNAs (EIciRNA) enhance transcription of parental gene.
Transcription of pre-mRNA from select genes generates both linear mRNA and EIciRNAs. EIciRNAs localize to the transcriptional start site (TSS) and enhance the transcription of the parental gene creating a positive feedback loop. A color version of the figure is available online.
Intron containing circRNAs from stress response genes activate mRNA transcription
Notably, one subclass of circRNAs, exon-intron circRNAs (EIciRNAs), are nuclear circRNAs that retain intronic sequences (Li et al. 2015b) and enhance the expression of the transcription of parental genes. These EIciRNAs are localized at the transcriptional start site of its parental gene and interacts with RNA Polymerase II and other transcription initiation machinery. Li et al. (2015b) found that knockdown of the EIciRNAs, led to a decrease in the levels of the linear mRNA species derived from the same parental genes. These EIciRNAs may have potential roles in the cellular regulation of stress. For instances, circPAIP2 is derived from the gene Poly(A)-binding Protein-interacting Protein 2 (PAIP2), which has been implicated in various stress response pathways, including the regulation of Vascular Endothelial Growth Factor (VEGF) mRNA levels under hypoxic conditions (Onesto et al. 2004) and acting as an antiviral restriction factor (McKinney et al. 2013). Potentially, circPAIP2 expression could enhance the transcriptional levels of PAIP2, thereby acting in a positive feedback manner to increase PAIP2 levels, to regulate viral replication or VEGF expression in different stress conditions (Figure 8).
Regulation of CircRNAs Involved in the Stress Response
The aforementioned studies have shown that circRNAs have various functions and are differentially regulated. However, the exact mechanisms underlying their regulation upon stress remain poorly understood. As illustrated in the following section, these circRNAs could be regulated at the level of biogenesis, degradation, or exportation.
Modulating circRNA levels through biogenesis
One potential way to regulate the expression of circRNAs is by modulating their biogenesis (reviewed in Lasda and Parker 2014, Chen 2016, Salzman 2016, Wilusz 2016). As early as 1995, research had shown that inverted repeats in the introns of the mouse Sry gene were required for circularization (Dubin et al. 1995). Subsequently, computational methods revealed that introns surrounding circularized exons were enriched with repetitive sequences, such as Alu elements, with, on average, three Alu elements upstream and another three in the inverse orientation downstream (Zhang et al. 2014; Liang and Wilusz 2014; Li et al., 2015b). These inverted repetitive sequences, which required as few as 30 nucleotides for complementation, facilitated back-splicing (Liang and Wilusz 2014). Therefore, factors that bind and modulate these intronic sequences could potentially alter the circRNA level upon stress.
ADAR modulates circRNA biogenesis by editing intronic sequences
Introns that complement to form double-stranded RNAs are prime targets for the RNA-editing enzymes ADAR1 and ADAR2, which convert adenosines into inosines through de-amination (Ivanov et al. 2015). Upon activation of ADAR1/2, RNA editing of intronic sequences decreased complementation and hence circRNA production. When both ADAR1 and ADAR2 were depleted via siRNA, the levels of circRNA were significantly up-regulated, as shown by both RNA-seq data and qRT-PCR (Ivanov et al. 2015, Rybak-Wolf et al., 2015). Notably, ADAR1 was previously shown to be essential for regulating endoplasmic reticulum stress (Qiu et al. 2013). It will be of interest to test whether the activation of ADAR1 regulates circRNA biogenesis during endoplasmic reticulum stress.
RBPs induced in response to stress may regulate circularization
The first RBP implicated in the formation of circRNAs was a splicing factor called Muscleblind (Mbl) (Houseley et al. 2006). Mbl binding sites were shown to be enriched in select introns of the Mbl gene and regulate the generation of circRNAs from the Mbl gene (Ashwal-Fluss et al. 2014). However, the Mbl protein only regulates Mbl circRNAs (Kramer et al. 2015). During the epithelial-mesenchymal transition, another RBP known as Quaking/QKI was shown to be differentially expressed and responsible for generating many circRNAs (Conn et al. 2015). The same study showed that QKI binding sites found within introns close to circularizing exons facilitated RNA looping and back-splicing via protein–protein dimerization (Conn et al. 2015). Notably, QKI down-regulated proapoptotic proteins in response to irradiation treatment (Guo et al. 2011). It remains unclear whether the regulation of these proapoptotic proteins could be due, at least in part, to the induction of QKI-regulated circRNAs.
RBM20 is an RBP that has been associated with alterative splicing of the Titin gene (Guo et al. 2012), as well as the circularization of select Titin exons (Khan et al. 2016), although the global impact on RBM20 regulated circRNAs has not been characterized. A crosslinking-immunoprecipitation sequencing (CLIP-seq) study of RBM20 detected a significant number of RBM20 RNA binding sites throughout the genome (Maatz et al. 2014). These binding sites may help pinpoint genomic locations of circRNAs generated by RBP20. Similar CLIP-seq studies of RBPs, particularly those that are induced upon stress, may provide further avenues for predicting production of stress-associated circRNAs in a genome-wide manner.
Modulation of the circRNA level via degradation and exportation
A key property of circRNAs that has generated significant interest is their stability due to their lack of free 5’ or 3’ ends, which are the common entry points of exoribonucleases. Harland and Misher (1988) were the first to demonstrate that circRNA was more stable than linear RNA using exogenously produced circRNAs injected into Xenopus embryos. They used circRNA as a control when studying RNA sequences that reduced mRNA stability of TFIIIA by recruiting 3’ to 5’ exoribonucleases. When the TFIIIA sequence was inserted into a linear reporter RNA, the stability of the reporter was drastically reduced. However, when inserted into a circRNA, the stability of the circRNA remained unchanged. Their study was the first to show that circRNAs are extremely stable in vivo, with half-lives of over 40 h. As discussed earlier, such resistance to exoribonucleases was later exploited in subsequent circRNA sequencing papers, wherein RNase R exoribonuclease was used to remove linear RNAs, resulting in enrichment of circRNAs (e.g., Danan et al., 2011. Jeck et al. 2013, Rybak-Wolf et al. 2015).
The stability of endogenous mammalian circRNAs remained relatively unexplored until 2013 (Jeck et al. 2013). Using a small molecule inhibitor of transcription called actinomycin D, Jeck et al. (2013) showed that circRNAs were extremely stable, persisting for several days. Many later studies using actinomycin D replicated their findings (Ashwal-Fluss et al. 2014, Zheng et al. 2016, Holdt et al. 2016), all of which found that most circRNAs were resistant to degradation. However, one caveat of using actinomycin D is that its prolonged inhibition of transcription, which alters normal physiology and ultimately induces cell death, may confound the measurement of stability. Recently, the stability of circRNA was confirmed by another type of measurement using 4-thiouridine, which labels transcribing RNAs without drastically modulating cellular physiology (Enuka et al. 2016). After a short exposure to 4-thiouridine, RNA can be harvested at defined time points. The labeled RNAs can then be enriched by the chemical attachment of biotin and pull-down with streptavidin beads. Enuka et al. (2016) reported that epidermal growth factor (EGF), a cell proliferation signal, had little to no effect on the stability of circRNAs. Intriguingly, they also found that although circRNAs were much more stable than their linear counterparts, the half-lives of circRNAs were highly variable, ranging from 8 to 50 h (Enuka et al. 2016). This variability in stability suggests that additional properties of circRNA regulate their half-lives. Further research is required to identify potential mechanisms of circRNA removal and degradation.
Cellular export of circRNAs can reduce endogenous levels
One way cells can decrease the levels of circRNAs is through exportation (Li et al. 2015c, Lasda and Parker 2016). As circRNAs are readily detectable in blood, their abundance in exosomes in the circulatory system has been examined. These studies showed that specific circRNAs, but not their linear RNA counterparts, were enriched in exosomes (Li et al. 2015c, Lasda and Parker 2016). In addition, no global exportation of circRNAs was observed by RNA-seq, pointing to the existence of a mechanism for allowing specific circRNAs to be retained intracellularly and/or selectively exported. Li et al. (2015c) found that the miR-7 sponge CDR1as was only present in exosomes under certain conditions and that its exportation was dependent on miR-7 levels. When miR-7 was present at low levels within the cell, CDR1as was detectable in exosomes. However, when miR-7 was overexpressed, CDR1as was not detected within exosomes (Li et al. 2015c). The correlation between CDR1as and miR-7 expression points to a specific mechanism for circRNA enrichment, possibly mediated by miR-7 binding. Further studies are required to detect the signal that leads to their accumulation in exosomes.
CircRNAs can be diluted by rapid proliferation
Bachmayr-Heyda et al. (2015) noted that circRNA expression levels were low in rapidly dividing cells and that the ratio of circRNA to linear RNA was higher in normal colon cells compared to that of cancer cells. They attributed this difference to rapid cell division preventing the accumulation of circRNAs in cancer cells (Bachmayr-Heyda et al. 2015). Studies also demonstrated that circRNAs were expressed at higher than normal levels in neurons, which are nondividing (Rybak-Wolf et al., 2015, You et al. 2015). Typically, in response to stress, cells halt proliferation. Therefore, in the absence of division, cells could, in theory, also accumulate circRNAs, which may in turn regulate stress responses.
Similar to neurons, circRNAs are enriched in platelet cells (Alhasan et al. 2016). Platelets are small circulating cells that lack nuclei and are therefore unable to transcribe additional RNA. The lack of transcription reduces the overall complexity of the cell’s response to stress stimuli. Given that platelet cells are regularly exposed to various stresses (e.g., ROS production in response to inflammation (Chakrabarti et al. 2005), these cells may be an excellent model system for investigating the functions and stability of circRNA during stress.
Degradation of circRNA through miRNA-mediated endoribonuclease activity
A study of the miR-7 sponge CDR1as, which contains a nearly perfect complementary binding site for miR-671, identified one specific mechanism of circRNA degradation (Hansen et al. 2011). When miRNAs bind with target RNAs with perfect or nearly perfect complementation, the RNA silencing complex activated the endoribonculease (slicer) activity of a core miRNA binding protein, AGO2, to cleave RNA (Hammond 2005). Hansen et al. (2011) confirmed the role of slicer-mediated degradation of CDR1as by mutating the miR-671 binding site to a miR-769 site. Northern blot analysis indicated that the mutated CDR1as was more stable than the wild-type CDR1as. However, when miR-769 was overexpressed, the mutant CDR1as was degraded. To date, miRNA-mediated degradation of circRNAs has not been observed in any other circRNAs. Thus, a more general regulator of circRNA stability will need to be identified.
Future Perspectives
Although circRNAs were first identified as early as 1976, the bulk of the research has been performed in the last few years (cf. Figure 2). Current research has shown that they are very stable as compared to linear RNA, found within all eukaryotes, and dynamically expressed in a tissue-specific manner. Beyond characterizing the general properties of circRNAs, a major emphasis for the recent research has been on identifying their biological functions. In this review, we used various examples to illustrate the emergent roles of circRNAs in apoptosis, oxidative stress, protection against microbe infections, and other conditions that disrupt cellular homeostasis (Table 1). Some circRNAs act as sponges or decoys to sequester RNA/proteins while others serve as scaffolds for the assembly of protein complexes. Given the low abundance of most circRNAs (Enuka et al. 2015, Zhang et al. 2016a), it is unclear how many molecules are required for executing these specific functions. For example, circANRIL is estimated to have 800–1000 copies per cell (Holdt et al. 2016), raising the question of whether this would be sufficient to inhibit the rRNA processing factor PES1, which is present at 10-fold higher levels than circANRIL. Similarly, it is questionable whether one molecule of circ-RasGEF1B could affect the stability of ICAM-1, the level of which is 2,580-fold higher than that of circ-RasGEF1B (Ng et al. 2016). A quantitative study of circRNAs and their potential targets (RNA/protein) is needed to shed light on the potential roles of circRNAs during stress. Importantly, the recent introduction of the genome-editing techniques, such as CRISPR/Cas9 (e.g., Zhang et al., 2016a), raises the possibility of introducing single-point mutations at splice sites or introns that abrogate circularization to probe the physiological significance of these circRNAs during stress.
To date, circRNAs are considered noncoding, given that no endogenous circRNAs are associated with ribosomes (Jeck et al. 2013, Guo et al., 2014). Nevertheless, engineered circRNAs containing viral internal ribosome entry sites (IRES) and appropriate reading frames can result in translation in vitro and in cells (Chen and Sarnow 1995, Wang and Wang 2015, Kramer et al. 2015, Abe et al. 2015). Given that IRES-mediated initiation of translation is favored under stress conditions (Hellen and Sarnow 2001, Holcik and Soneberg 2005, Komar and Hatzoglou 2011), it is theoretically possible that certain circRNAs containing cellular IRES and open reading frames may be translated under such conditions when cap-dependent translation is suppressed. Upon stress, these IRES-containing circRNAs may potentially be better competitors for the available pool of ribosomes and initiation factors for translation.
As the splicing that results in circRNAs is in competition with the linear RNA counterpart (Ashwal-Fluss et al. 2014), one function of circRNAs during stress could simply be altering splicing patterns from sequential order to back-splicing. In this case, the resulting circRNAs may not necessarily be functional, and the production of circRNAs may merely be a by-product of alternative mode of splicing during stress. Yet, it is clear that over hundreds of circRNAs are expressed at 10-fold higher than their linear counterparts (e.g., Jeck et al., 2013; reviewed in Wilusz 2016) and some, such as circ-SRY, are the predominant output of the parental gene at a specific developmental stage (Capel et al. 1993). In addition, emerging evidence suggests that circRNAs can themselves undergo further alternative splicing that is unique compared to the linear counterparts (Gao et al. 2016, Zhang et al. 2016b). Therefore, it will be critical to better understand how alternative splicing regulates circRNA biogenesis during stress. Notably, a few circRNAs have been found to retain intronic sequences (Gardner et al. 2012, Li et al. 2015b, Gao et al. 2016). These intron-containing circRNAs were localized in the nucleus, as opposed to most circRNAs, which are found in the cytoplasm (Gardner et al. 2012, Li et al. 2015b). They co-localized at the transcriptional start sites of the genes from which they originated and up-regulated the gene expression of the linear mRNAs that these genes transcribed (Gardner et al. 2012, Li et al. 2015b). Besides back-splicing, other methods of biogenesis involving lariat precursors, exon skipping, and tRNA processing have been discovered in lower eukaryotic organisms (Barrett et al. 2015, Lu et al. 2015). The variability in circRNAs produced through alternative splicing, intron retention and other modes of biogenesis could thus provide unique sequences that may have specific biological roles.
Regarding their potential as regulators of homeostasis, the most important property of circRNAs is likely their stability. As circRNAs can persist for days, such stability could allow them to act as a buffer against stresses. This idea was proposed by Hansen et al. (2013b) in relation to the possible roles of circRNAs as miRNA sponges. As miRNA levels fluctuate in response to stress, abundant circRNAs potentially “sponge” up excess miRNAs. In a similar fashion, circRNAs could potentially sponge up RBPs during stress. Alternatively, the long half-lives of circRNAs could allow them to “remember” (stable memory) the cellular state or “forget” the cellular state when certain circRNAs are degraded. Notably, the expression pattern of one-cell and two-cell embryos of the nematode Caenorhabditis elegans varies considerably (Memczak et al. 2013) and the one-cell to two-cell transition is completed in less than an hour (Greenstein and Lee 2006), suggesting that circRNAs may be regulated in short time periods. If specific circRNAs functioned as a stable memory of a cellular state, stress-induced changes in the expression or stability of circRNAs could provide a mechanism to adjust homeostasis to a new cellular state.
So far, only a handful of studies, which are likely to be the first examples of many to come, reporting the roles of circRNAs in stress responses and how stress modulates circRNA level through biogenesis, degradation and exportation. Given that stress responses are commonly mediated via signaling molecules (e.g., cytokines) and transcription factors, it is expected these two classes would be likely targets, as in the case for microRNAs during stress (Tsang et al., 2007, Inui et al., 2010, Leung and Sharp, 2010). Depending on how they are derived, circRNAs can incorporate defined RNA elements, such as introns and binding sites for miRNAs or RBPs, and potentially IRES elements and RNA localization signals. Such flexibility in generating circRNAs makes this class of RNA regulator a strong candidate to regulate the full spectrum of gene expression— from transcription to translation—during stress.
Acknowledgments
The authors thank Dr. Yoshinari Ando and other members of the lab for discussion, Dr. Jeremy Wilusz for advice on the timeline of circRNA discovery.
Funding
Authors acknowledge support an NIGMS training grant T32GM007814 and an NCI training grant T32CA009110 to J.W.F and the Johns Hopkins Bloomberg School of Public Health Start up fund to Anthony. RNA work in the Leung lab is supported by American Cancer Society Research Scholar Award [129539-RSG-16-062-01-RMC] and National Institute of Health [R01GM104135].
Biographies
Joseph W. Fischer is a Ph.D student in the McKusick-Nathans Institute of Genetic Medicine at Johns Hopkins University School of Medicine. His research is focused on the regulation of circular RNA biogenesis and degradation.
Anthony K.L. Leung is an Assistant Professor from Johns Hopkins University, with primary appointment at the Department of Biochemistry and Molecular Biology at Bloomberg School of Public Health and secondary appointment in the Department of Oncology at the School of Medicine. Dr. Leung's research program focuses on understanding the relationship between noncoding RNAs and stress, using cellular imaging, genomics and proteomics approaches.
Footnotes
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of this article.
References
- Abe N, Matsumoto K, Nishihara M, Nakano Y, Shibata A, Maruyama H, Shuto S, Matsuda A, Yoshida M, Ito Y, Abe H. Rolling Circle Translation of Circular RNA in Living Human Cells. Sci Rep. 2015;5:16435. doi: 10.1038/srep16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Advani SJ, Camargo MF, Seguin L, Mielgo A, Anand S, Hicks AM, Aguilera J, Franovic A, Weis SM, Cheresh DA. Kinase-independent role for CRAF-driving tumour radioresistance via CHK2. Nat Commun. 2015;6:8154. doi: 10.1038/ncomms9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhasan AA, Izuogu OG, Al-Balool HH, Steyn JS, Evans A, Colzani M, Ghevaert C, Mountford JC, Marenah L, Elliott DJ, Santibanez-Koref M, Jackson MS. Circular RNA enrichment in platelets is a signature of transcriptome degradation. Blood. 2016;127:e1–e11. doi: 10.1182/blood-2015-06-649434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW, Zeillinger R, Pils D. Correlation of circular RNA abundance with proliferation--exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057. doi: 10.1038/srep08057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett SP, Wang PL, Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife. 2015;4:e07540. doi: 10.7554/eLife.07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckel JN, Jae N, Heumuller AW, Chen W, Boon RA, Stellos K, Zeiher AM, John D, Uchida S, Dimmeler S. Identification and Characterization of Hypoxia-Regulated Endothelial Circular RNA. Circ Res. 2015;117:884–90. doi: 10.1161/CIRCRESAHA.115.306319. [DOI] [PubMed] [Google Scholar]
- Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6:e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P, Lovell-Badge R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–30. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- Chakrabarti S, Varghese S, Vitseva O, Tanriverdi K, Freedman JE. CD40 ligand influences platelet release of reactive oxygen intermediates. Arterioscler Thromb Vasc Biol. 2005;25:2428–2434. doi: 10.1161/01.ATV.0000184765.59207.f3. [DOI] [PubMed] [Google Scholar]
- Chao CW, Chan DC, Kuo A, Leder P. The mouse formin (Fmn) gene: abundant circular RNA transcripts and gene-targeted deletion analysis. Mol Med. 1998;4:614–28. [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268:415–7. doi: 10.1126/science.7536344. [DOI] [PubMed] [Google Scholar]
- Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205–11. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- Clepet C, Schafer AJ, Sinclair AH, Palmer MS, Lovell-Badge R, Goodfellow PN. The human SRY transcript. Hum Mol Genet. 1993;2:2007–12. doi: 10.1093/hmg/2.12.2007. [DOI] [PubMed] [Google Scholar]
- Cocquerelle C, Daubersies P, Majerus MA, Kerckaert JP, Bailleul B. Splicing with inverted order of exons occurs proximal to large introns. EMBO J. 1992;11:1095–8. doi: 10.1002/j.1460-2075.1992.tb05148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular RNA molecules. Faseb j. 1993;7:155–60. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–34. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- Croce CM. Genetics: Are circRNAs involved in cancer pathogenesis? Nat Rev Clin Oncol. 2016;13:658. doi: 10.1038/nrclinonc.2016.113. [DOI] [PubMed] [Google Scholar]
- Danan M, Schwartz S, Edelheit S, Sorek R. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 2011;40:3131–42. doi: 10.1093/nar/gkr1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Yan L, Hu B, Fan X, Ren Y, Li R, Lian Y, Yan J, Li Q, Zhang Y, Li M, Ren X, Huang J, Wu Y, Liu P, Wen L, Zhang C, Huang Y, Tang F, Qiao J. Tracing the expression of circular RNAs in human pre-implantation embryos. Genome Biol. 2016;17:130. doi: 10.1186/s13059-016-0991-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Xu J, Wang X, Jin B. Influence of the interaction between long noncoding RNAs and hypoxia on tumorigenesis. Tumour Biol. 2016;37:1379–85. doi: 10.1007/s13277-015-4457-0. [DOI] [PubMed] [Google Scholar]
- Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z, Li X, Yang BB. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. 2016a doi: 10.1093/eurheartj/ehw001. [DOI] [PubMed] [Google Scholar]
- Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016b;44:2846–58. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin RA, Kazmi MA, Ostrer H. Inverted repeats are necessary for circularization of the mouse testis Sry transcript. Gene. 1995;167:245–8. doi: 10.1016/0378-1119(95)00639-7. [DOI] [PubMed] [Google Scholar]
- Evans JL, Goldfine ID, Maddux BA, Grodsky GM. Oxidative stress and stress-activated signaling pathways: a unifying hypothesis of type 2 diabetes. Endocr Rev. 2002;23:599–622. doi: 10.1210/er.2001-0039. [DOI] [PubMed] [Google Scholar]
- Enuka Y, Lauriola M, Feldman ME, Sas-Chen A, Ulitsky I, Yarden Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016;44:1370–83. doi: 10.1093/nar/gkv1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flouriot G, Brand H, Seraphin B, Gannon F. Natural trans-spliced mRNAs are generated from the human estrogen receptor-alpha (hER alpha) gene. J Biol Chem. 2002;277:26244–51. doi: 10.1074/jbc.M203513200. [DOI] [PubMed] [Google Scholar]
- Gao Y, Wang J, Zheng Y, Zhang J, Chen S, Zhao F. Comprehensive identification of internal structure and alternative splicing events in circular RNAs. Nat Commun. 2016;7:12060. doi: 10.1038/ncomms12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner EJ, Nizami ZF, Talbot CC, Jr, Gall JG. Stable intronic sequence RNA (sisRNA), a new class of noncoding RNA from the oocyte nucleus of Xenopus tropicalis. Genes Dev. 2012;26:2550–9. doi: 10.1101/gad.202184.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng HH, Li R, Su YM, Xiao J, Pan M, Cai XX, Ji XP. The Circular RNA CDR1as Promotes Myocardial Infarction by Mediating the Regulation of miR-7a on Its Target Genes Expression. PLoS One. 2016;11:e0151753. doi: 10.1371/journal.pone.0151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenstein D, Lee LA. Oocyte-to-embryo transition: kinase cabal plots regime change. Curr Biol. 2006;16:R93–5. doi: 10.1016/j.cub.2006.01.028. [DOI] [PubMed] [Google Scholar]
- Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH, Pandolfi PP. Oncogenic Role of Fusion-circRNAs Derived from Cancer-Associated Chromosomal Translocations. Cell. 2016;165:289–302. doi: 10.1016/j.cell.2016.03.020. [DOI] [PubMed] [Google Scholar]
- Guo W, Shi X, Liu A, Yang G, Yu F, Zheng Q, Wang Z, Allen DG, Lu Z. RNA binding protein QKI inhibits the ischemia/reperfusion-induced apoptosis in neonatal cardiomyocytes. Cell Physiol Biochem. 2011;28:593–602. doi: 10.1159/000335755. [DOI] [PubMed] [Google Scholar]
- Guo W, Schafer S, Greaser ML, Radke MH, Liss M, Govindarajan T, Maatz H, Schulz H, Li S, Parrish AM, Dauksaite V, Vakeel P, Klaassen S, Gerull B, Thierfelder L, Regitz-Zagrosek V, Hacker TA, Saupe KW, Dec GW, Ellinor PT, Macrae CA, Spallek B, Fischer R, Perrot A, Ozcelik C, Saar K, Hubner N, Gotthardt M. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat Med. 2012;18:766–73. doi: 10.1038/nm.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM. Dicing and slicing: the core machinery of the RNA interference pathway. FEBS Lett. 2005;579:5822–9. doi: 10.1016/j.febslet.2005.08.079. [DOI] [PubMed] [Google Scholar]
- Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. Embo j. 2011;30:4414–22. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013a;495:384–8. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013b;73:5609–12. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- Harland R, Misher L. Stability of RNA in developing Xenopus embryos and identification of a destabilizing sequence in TFIIIA messenger RNA. Development. 1988;102:837–52. doi: 10.1242/dev.102.4.837. [DOI] [PubMed] [Google Scholar]
- Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- Hentze MW, Preiss T. Circular RNAs: splicing's enigma variations. Embo j. 2013;32:923–5. doi: 10.1038/emboj.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318–27. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- Holdt LM, Beutner F, Scholz M, Gielen S, Gabel G, Bergert H, Schuler G, Thiery J, Teupser D. ANRIL expression is associated with atherosclersois risk at chromosome 9p21. Arterioscler Thromb Vasc Biol. 2010;30:620–7. doi: 10.1161/ATVBAHA.109.196832. [DOI] [PubMed] [Google Scholar]
- Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou A, Gabel G, Beutner F, Scholz M, Thiery J, Musunuru K, Krohn K, Mann M, Teupser D. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016;7:12429. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley JM, Garcia-Casado Z, Pascual M, Paricio N, O'dell KM, Monckton DG, Artero RD. Noncanonical RNAs from transcripts of the Drosophila muscleblind gene. J Hered. 2006;97:253–60. doi: 10.1093/jhered/esj037. [DOI] [PubMed] [Google Scholar]
- Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339–40. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol. 2010;11:252–63. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M, Dieterich C, Rajewsky N. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170–7. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna. 2013;19:141–57. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MA, Reckman YJ, Aufiero S, Van Den Hoogenhof MM, Van Der Made I, Beqqali A, Koolbergen DR, Rasmussen TB, Van Der Velden J, Creemers EE, Pinto YM. RBM20 Regulates Circular RNA Production From the Titin Gene. Circ Res. 2016 doi: 10.1161/CIRCRESAHA.116.309568. [DOI] [PubMed] [Google Scholar]
- Komar AA, Hatzoglou M. Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle. 2011;10:229–40. doi: 10.4161/cc.10.2.14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos A, Dijkema R, Arnberg AC, Van Der Meide PH, Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323:558–60. doi: 10.1038/323558a0. [DOI] [PubMed] [Google Scholar]
- Kramer MC, Liang D, Tatomer DC, Gold B, March ZM, Cherry S, Wilusz JE. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015;29:2168–82. doi: 10.1101/gad.270421.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kultz D. Molecular and evolutionary basis of the cellular stress response. Annu Rev Physiol. 2005;67:225–57. doi: 10.1146/annurev.physiol.67.040403.103635. [DOI] [PubMed] [Google Scholar]
- Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20:1829–42. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasda E, Parker R. Circular RNAs Co-Precipitate with Extracellular Vesicles: A Possible Mechanism for circRNA Clearance. PLoS One. 2016;11:e0148407. doi: 10.1371/journal.pone.0148407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Sharp PA. MicroRNA functions in stress responses. Mol Cell. 2010;40:205–15. doi: 10.1016/j.molcel.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Zhang L, Li W, Deng J, Zheng J, An M, Lu J, Zhou Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015a;6:6001–13. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, Zhu P, Chang Z, Wu Q, Zhao Y, Jia Y, Xu P, Liu H, Shan G. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015b;22:256–64. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015c;25:981–4. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–47. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SP, Ye S, Long Y, Fan Y, Mao HF, Chen MT, Ma QJ. Circular RNA expression alterations are involved in OGD/R-induced neuron injury. Biochem Biophys Res Commun. 2016;471:52–6. doi: 10.1016/j.bbrc.2016.01.183. [DOI] [PubMed] [Google Scholar]
- Liu Q, Zhang X, Hu X, Dai L, Fu X, Zhang J, Ao Y. Circular RNA Related to the Chondrocyte ECM Regulates MMP13 Expression by Functioning as a MiR-136 'Sponge' in Human Cartilage Degradation. Sci Rep. 2016;6:22572. doi: 10.1038/srep22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Filonov GS, Noto JJ, Schmidt CA, Hatkevich TL, Wen Y, Jaffrey SR, Matera AG. Metazoan tRNA introns generate stable circular RNAs in vivo. Rna. 2015;21:1554–65. doi: 10.1261/rna.052944.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukiw WJ. Circular RNA (circRNA) in Alzheimer's disease (AD) Front Genet. 2013;4:307. doi: 10.3389/fgene.2013.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maatz H, Jens M, Liss M, Schafer S, Heinig M, Kirchner M, Adami E, Rintisch C, Dauksaite V, Radke MH, Selbach M, Barton PJ, Cook SA, Rajewsky N, Gotthardt M, Landthaler M, Hubner N. RNA-binding protein RBM20 represses splicing to orchestrate cardiac pre-mRNA processing. J Clin Invest. 2014;124:3419–30. doi: 10.1172/JCI74523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mckinney C, Yu D, Mohr I. A new role for the cellular PABP repressor Paip2 as an innate restriction factor capable of limiting productive cytomegalovirus replication. Genes Dev. 2013;27:1809–20. doi: 10.1101/gad.221341.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, Le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–8. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Nan A, Chen L, Zhang N, Liu Z, Yang T, Wang Z, Yang C, Jiang Y. A novel regulatory network among LncRpa, CircRar1, MiR-671 and apoptotic genes promotes lead-induced neuronal cell apoptosis. Arch Toxicol. 2016 doi: 10.1007/s00204-016-1837-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WL, Marinov GK, Liau ES, Lam YL, Lim YY, Ea CK. Inducible RasGEF1B circular RNA is a positive regulator of ICAM-1 in the TLR4/LPS pathway. RNA Biol. 2016;13:861–71. doi: 10.1080/15476286.2016.1207036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B. Scrambled exons. Cell. 1991;64:607–13. doi: 10.1016/0092-8674(91)90244-s. [DOI] [PubMed] [Google Scholar]
- Onesto C, Berra E, Grepin R, Pages G. Poly(A)-binding protein-interacting protein 2, a strong regulator of vascular endothelial growth factor mRNA. J Biol Chem. 2004;279:34217–26. doi: 10.1074/jbc.M400219200. [DOI] [PubMed] [Google Scholar]
- Qiu W, Wang X, Buchanan M, He K, Sharma R, Zhang L, Wang Q, Yu J. ADAR1 is essential for intestinal homeostasis and stem cell maintenance. Cell Death Dis. 2013;4:e599. doi: 10.1038/cddis.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, Herzog M, Schreyer L, Papavasileiou P, Ivanov A, Ohman M, Refojo D, Kadener S, Rajewsky N. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell. 2015;58:870–85. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J. Circular RNA Expression: Its Potential Regulation and Function. Trends Genet. 2016;32:309–16. doi: 10.1016/j.tig.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73:3852–6. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider T, Hung LH, Schreiner S, Starke S, Eckhof H, Rossbach O, Reich S, Medenbach J, Bindereif A. CircRNA-protein complexes: IMP3 protein component defines subfamily of circRNPs. Sci Rep. 2016;6:31313. doi: 10.1038/srep31313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surono A, Takeshima Y, Wibawa T, Ikezawa M, Nonaka I, Matsuo M. Circular dystrophin RNAs consisting of exons that were skipped by alternative splicing. Hum Mol Genet. 1999;8:493–500. doi: 10.1093/hmg/8.3.493. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006;34:e63. doi: 10.1093/nar/gkl151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, Jiang C, Parast MM, Murry CE, Laurent LC, Salzman J. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015;16:126. doi: 10.1186/s13059-015-0690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang J, Zhu J, Van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell. 2007;26:753–67. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veno MT, Hansen TB, Veno ST, Clausen BH, Grebing M, Finsen B, Holm IE, Kjems J. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 2015;16:245. doi: 10.1186/s13059-015-0801-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, Dinneny JR, Brown PO, Salzman J. Circular RNA is expressed across the eukaryotic tree of life. PLoS One. 2014;9:e90859. doi: 10.1371/journal.pone.0090859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21:172–9. doi: 10.1261/rna.048272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YH, Yu XH, Luo SS, Han H. Comprehensive circular RNA profiling reveals that circular RNA100783 is involved in chronic CD28-associated CD8(+)T cell ageing. Immun Ageing. 2015;12:17. doi: 10.1186/s12979-015-0042-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Nazarali AJ, Ji S. Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am J Cancer Res. 2016a;6:1167–76. [PMC free article] [PubMed] [Google Scholar]
- Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, Zhou LY, Sun T, Wang M, Yu T, Gong Y, Liu J, Dong YH, Li N, Li PF. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016b;37:2602–11. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- Werfel S, Nothjunge S, Schwarzmayr T, Strom TM, Meitinger T, Engelhardt S. Characterization of circular RNAs in human, mouse and rat hearts. J Mol Cell Cardiol. 2016;98:103–107. doi: 10.1016/j.yjmcc.2016.07.007. [DOI] [PubMed] [Google Scholar]
- Wilusz JE. Circular RNAs: Unexpected outputs of many protein-coding genes. RNA Biol. 2016:1–11. doi: 10.1080/15476286.2016.1227905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Guo S, Li W, Yu P. The circular RNA CDR1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep. 2015;5:12453. doi: 10.1038/srep12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue TC, Zhang L, Ren ZG, Chen RX, Cui JF, Ge NL, Ye SL. Sex-determination gene SRY potentially associates with poor prognosis but not sex bias in hepatocellular carcinoma. Dig Dis Sci. 2015;60:427–35. doi: 10.1007/s10620-014-3377-y. [DOI] [PubMed] [Google Scholar]
- Yang W, Du WW, Li X, Yee AJ, Yang BB. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2016;35:3919–31. doi: 10.1038/onc.2015.460. [DOI] [PubMed] [Google Scholar]
- Ye CY, Chen L, Liu C, Zhu QH, Fan L. Widespread noncoding circular RNAs in plants. New Phytol. 2015;208:88–95. doi: 10.1111/nph.13585. [DOI] [PubMed] [Google Scholar]
- Yeh YM, Chuang CM, Chao KC, Wang LH. MicroRNA-138 suppresses ovarian cancer cell invasion and metastasis by targeting SOX4 and HIF-1alpha. Int J Cancer. 2013;133:867–78. doi: 10.1002/ijc.28086. [DOI] [PubMed] [Google Scholar]
- You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, Wang X, Hou J, Liu H, Sun W, Sambandan S, Chen T, Schuman EM, Chen W. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18:603–10. doi: 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–47. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xue W, Li X, Zhang J, Chen S, Zhang JL, Yang L, Chen LL. The Biogenesis of Nascent Circular RNAs. Cell Rep. 2016a;15:611–24. doi: 10.1016/j.celrep.2016.03.058. [DOI] [PubMed] [Google Scholar]
- Zhang XO, Dong R, Zhang Y, Zhang JL, Luo Z, Zhang J, Chen LL, Yang L. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016b;26:1277–87. doi: 10.1101/gr.202895.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, Liang L, Gu J, He X, Huang S. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Wang Q, Zhu B, Luo Y, Gao L. Deciphering the roles of circRNAs on chilling injury in tomato. Biochem Biophys Res Commun. 2016 doi: 10.1016/j.bbrc.2016.07.032. [DOI] [PubMed] [Google Scholar]