Abstract
Background
Base excision repair (BER) pathway is a DNA repair pathway that is important in carcinogenesis and in response to DNA-damaging chemotherapy. XRCC1 is one of important molecular markers for BER. So far, the role of XRCC1 polymorphisms with clinical outcomes of advanced NSCLC treated with platinum-based chemotherapy is inconclusive. To explore the relationship between XRCC1 polymorphisms and platinum-based chemotherapy in advanced NSCLC patients, we performed this meta-analysis.
Methods
Crude odds ratios (ORs), Cox proportional hazard ratios (HRs) with the corresponding 95% confidence intervals (CIs) were adopted to assess the strength of association between XRCC1 polymorphisms and response rate, Overall survival (OS) and progression free survival (PFS) of advanced NSCLC treated with platinum-based chemotherapy. Q test and I 2 test were used for the assessment of heterogeneity. Subgroup analyses were conducted when heterogeneity exists. Begg’s funnel plots and Egger’s linear regression test were used to estimate publication bias. Sensitivity analysis was performed to evaluate the stability of the result.
Results
A total of 19 studies including 2815 individuals were eligible for the analysis, results showed XRCC1 194Arg allele was negatively associated with the objective response rate relative to 194Trp, and results of homozygous model, dominant model and heterozygous model suggested a gene dosage effect negative correlation between 194Arg allele and objective response rate(ArgArg vs TrpTrp: OR = 0.64(95%CI: 0.44-0.91); ArgArg + TrpArg vs TrpTrp: OR = 0.79(95%CI: 0.57-1.11); TrpArg vs TrpTrp: OR = 1.05(95%CI: 0.73-1.51)). XRCC1 399Gln may indicate favorable overall survival (GlnGln + GlnArg vs ArgArg: HR = 0.65(95%CI: 0.43–0.98)) and favorable PFS (GlnGln vs ArgArg: HR = 0.72(95%CI: 0.48–0.97)) in Asian patients; while in Caucasian patients, XRCC1 399Gln indicated poorer overall survival (GlnGln vs ArgArg: HR = 2.29(95%CI: 1.25–3.33)).
Conclusions
Our results indicated that in NSCLC patients treated with platinum-based regimen, XRCC1 194Arg allele suggest poor objective response rate, the GlnGln genotype of XRCC1 399 suggest poorer overall survival in Caucasian patients, and longer PFS in Asian patients.
Electronic supplementary material
The online version of this article (doi:10.1186/s12885-017-3487-y) contains supplementary material, which is available to authorized users.
Keywords: XRCC1, Polymorphism, Lung cancer, Platinum, Meta-analysis
Background
Lung cancer, with growing incidence, is becoming one of the most prevalent cancer types all over the world. And it’s the leading cause of cancer death in males and second leading cause of cancer death in females, approximate 17.6% of the cancer deaths was due to lung cancer. [1] It usually develop silently, with non-specific clinical symptoms in the early period, and is apt to be neglected, most patients developed to advanced stage when they had some symptoms, and lost the opportunity of radical surgery. [2] For decades, platinum-based combination chemotherapy has been established as the cornerstone of advanced non-small cell lung cancer (NSCLC) treatment [3, 4]. Although molecular-targeted therapy has been confirmed as first-line therapy option for those advanced NSCLC with driver gene mutations, including epidermal growth factor receptor (EGFR), anaplastic lymphoma receptor tyrosine kinase (ALK), and KRAS mutations in recent years, still majority of NSCLC patients are not indicated to adopt molecular-targeted therapy. For these patients, platinum-based combination remains the first choice. But some NSCLC patients benefit from the treatment, while others failed. That means, not all the advanced NSCLC patients can benefit from platinum-based chemotherapy. In the new era, it is very important to select suitable treatment program for individualized treatment.
Anti-tumor mechanism of cisplatin and carboplatin is generally acknowledged as follows: cisplatin and carboplatin enter cell nucleus, bind to DNA and form DNA adducts which lead to intrastrand or interstrand cross-links, result in DNA synthesis/replication dysfunction and DNA structure disruption, which ultimately brings about cell proliferation inhibition and cell apoptosis. [5, 6] Resistance to platinum agents is suggested to be the main reason for treatment failure. One proposed mechanism of platinum resistance is attributed to enhanced function of DNA repair system, which can repair and rescue the damaged DNA and help tumor cells survive. [7, 8] In other words, DNA repair pathway plays an important role in the treatment response to the platinum-based chemotherapy of NSCLC patients.
Base excision repair (BER) pathway is a DNA repair pathway that repairs damaged DNA throughout the cell cycle, and it is important in carcinogenesis and in response to DNA-damaging chemotherapy. X-ray repair cross-complementing protein 1 (XRCC1), which located on chromosome no. 19q13.2–13.3, undertook the DNA repair mission of single-strand breaks formed by ionizing radiation and alkylating agents. This protein interacts with DNA ligase III, polymerase beta and poly (ADP-ribose) polymerase, and forms a repair complex to participate in the BER pathway [9–12].
So far, a number of studies have investigated the role of XRCC1 polymorphisms with clinical outcomes of advanced NSCLC treated with platinum-based chemotherapy, but the results were quite controversial, some studies supported that there were some association between XRCC1 polymorphisms and clinical outcomes of advanced NSCLC treated with platinum-based chemotherapy (treatment response(TR), overall survival(OS) or progression-free survival(PFS)), [13–28] but others had different views. [29–31] To explore the association between XRCC1 polymorphisms with clinical outcomes of advanced NSCLC treated with platinum-based chemotherapy, we performed this meta-analysis under the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement guidelines [32].
Methods
We carried out this meta-analysis based on the PRISMA statement, all data was extracted from published papers, so that ethical approval was not required per our institutional ethics committee.
Search strategy and selection criteria
Eligible studies were identified by searching the PubMed, CNKI, EBSCO and Cochrane databases (prior to July 2015) using “XRCC1” or “X-ray repair cross-complementing protein 1”, “lung”, “polymorphism” and “platinum”. Additional articles were identified through the reference cited in the first series of articles selected. Only research articles with human subjects were included and the language was not limited. The included studies have to be designed to evaluate the XRCC1 polymorphisms and clinical outcomes of advanced NSCLC (no opportunity of surgery) treated with platinum-based chemotherapy. And a study was excluded if any of the following cases occurred: (i) the study did not report any clinical outcome; (ii) studies using XRCC1 polymorphisms either to predict lung cancer’s risk or to predict treatment toxicity; (iii) studies reported with the same data or overlapping data by the same authors; (iv) the response rate or overall survival reported in the study was either not specific to polymorphism or could not be attributed to a specific polymorphism; (v) the response rate or overall survival stratified by SNP was neither reported in nor derivable from the original article, and the principal investigator declined or was unable to provide this information on request.
Data extraction
Upon carefully reading through all articles of eligible studies, information was carefully extracted. For each study, characteristics such as name of first author, year of publication, original country and ethnicity of the patients, tumor stage, sample size (case no.), genotyping method, genotype distribution and clinical outcomes were collected. For studies including different subpopulations according to ethnicity or experiment design, we considered each subpopulation as a separate study in the meta-analysis. For example, the study carried out by Liao et al. [20] included training set and validation set two subpopulations, each set was used as a separate study in the meta-analysis. Ethnicity was categorized as “Caucasian” (European descendants) and “Asian”(mainland China, Taiwan and Korea).
Statistical analysis
The strength of association between XRCC1 polymorphisms and response rate of advanced NSCLC treated with platinum-based chemotherapy was assessed by Crude odds ratios (ORs) with the corresponding 95% confident intervals (CIs). [33] The odds of response rate were defined as the ratio of complete or partial response against stable or progressive disease [CR + PR vs. PD + SD, evaluated by the WHO criteria or the Response Evaluation Criteria in Solid Tumors criteria (RECIST)]. The pooled ORs were performed for an allele comparison (C vs. Y), a homozygous model (CC vs. YY), a heterozygous model (CY vs. YY), a recessive model (CC vs. CY + YY) and a dominant model (CC + CY vs. YY). [33] Overall survival (OS) and progression free survival (PFS) were evaluated by calculating pooled Cox proportional hazard ratios (HRs) and 95% confidence intervals (CIs) as relevant effect measures, HRs and 95% CIs were obtained directly from the raw data, or indirectly from the Kaplan-Meier curve of an article. [34] Q test and I 2 test were used for the assessment of heterogeneity. The fixed effects model was applied when the effects were assumed to be homogenous (Q test shown P value >0.1), otherwise a random effects model was applied for meta-analysis. When heterogeneity was present, and the number of the studies included was large enough to perform the multivariable regression analysis, a meta-regression analysis was employed to explore the sources of heterogeneity. Furthermore, subgroup analyses were conducted by ethnicity and sample size (n > 100). Hardy-Weinberg Equilibrium (HWE) were assessed by http://www.oege.org/software/hwe-mr-calc.shtml [35] on September 15, 2015. Begg’s funnel plots and Egger’s linear regression test were used to estimate publication bias. Sensitivity analysis was performed to evaluate the stability of the results in the procedure of re-analysis after interchange of fixed or random effects model and omitting each study one at a time, especially small sample studies.
All the statistical analyses were performed using STATA version 12.0 (STATA Corporation, College Station, TX).
Results
Overall 1569 potential studies were selected during the first step of systematic literature review, and a further review of the searched trials excluded 1448 studies, including 105 review articles, 258 studies on non-human beings, 1023 studies on other tumors, and 62 studies for other reasons. The remaining 121 studies were evaluated further, 96 studies were excluded, including 89 studies on lung cancer susceptibility, 3 studies on SCLC, 4 studies on treatment toxicity. Through detailed assessment, in the end, 19 follow-up studies were considered to meet all in inclusion criteria (Fig. 1). These were included in final analyses.
Fig. 1.

Literature search and selection of included studies
Study characteristics
A total of 19 studies including 2815 individuals were eligible for the final analysis, in which 18 studies (2815 individuals) were eligible for XRCC1 399 analysis, and 8 studies (1208 individuals) were eligible for XRCC1 194 analysis. Table 1 listed the main characteristics and genotype distribution of XRCC1 399 (rs 25487) and XRCC1 194 (rs 1799782) with respect to response rate and overall survival rate, including first author, published year, ethnicity, original country, source of controls and genotype distribution. Five of these studies were conducted on Caucasian patients, and 14 were conducted on Asian patients. Fifteen were published in English-language journals. Four were published in Chinese-language journals. The sample size of each report ranged from 45–382 individuals.
Table 1.
Main characteristics of studies included in this meta-analysis
| Study | Year | Country | Ethnicity | Case No. | Genotyping method | Stage | Treatment Response (Genotype distribution) | Outcome | SNPs of XRCC1 | HWE | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| XRCC1 Arg399Gln | XRCC1 Arg194Trp | ||||||||||||||||||||
| CR + PR | SD + PD | CR + PR | SD + PD | ||||||||||||||||||
| GG | GA | AA | GG | GA | AA | CC | CT | TT | CC | CT | TT | ||||||||||
| Peng [17] | 2014 | China | Asian | 235 | PCR-CTTP | IIIA-IV | 40 | 41 | 3 | 71 | 74 | 6 | − | − | − | − | − | − | TR/OS/PFS | Arg399Gln | <0.005 |
| Zhang [23] | 2014 | China | Asian | 375 | Sequenom MassARRAY | IIIA-IV | 49 | 54 | 24 | 125 | 94 | 29 | 60 | 44 | 23 | 118 | 90 | 41 | TR/OS/PFS | Arg194Trp Arg399Gln | <0.05 |
| Lee [24] | 2013 | S. Korea | Asian | 382 | Sequenome mass spectrometry-based genotyping assay | III-IV | 110 | 64 | 5 | 100 | 75 | 16 | − | − | − | − | − | − | TR/OS | Arg399Gln | >0.05 |
| Tiseo [30] | 2013 | Italy | Caucasian | 93 | TaqMan | IIIB-IV | 14 | 4 | 27 | 2 | − | − | − | − | − | − | TR/OS | Arg399Gln | − | ||
| Zhao [26] | 2013 | China | Asian | 147 | TaqMan allelic discrimination assay | IIIA-IV | 21 | 24 | 8 | 56 | 31 | 5 | 32 | 20 | 1 | 51 | 35 | 6 | TR/OS/PFS | Arg194Trp Arg399Gln | >0.05 |
| Li [18] | 2012 | China | Asian | 89 | PCR-RFLP | IIIA-IV | 20 | 6 | 24 | 39 | − | − | − | − | − | − | TR | Arg399Gln | − | ||
| Liao 1[20] | 2012 | Taiwan | Asian | 62 | SNPstream UHT | IIIB-IV | 9 | 9 | 1 | 17 | 22 | 4 | − | − | − | − | − | − | TR/OS | Arg399Gln | >0.05 |
| Liao 2[20] | 2012 | Taiwan | Asian | 45 | SNPstream UHT | IIIB-IV | − | − | − | − | − | − | − | − | − | − | − | − | OS | Arg399Gln | >0.05 |
| Joerger [22] | 2012 | Netherlands | Caucasian | 131 | BigDye Terminator Cycle Sequencing Ready Reaction Kit | IIIB-IV | 17 | 18 | 5 | 34 | 45 | 12 | − | − | − | − | − | − | TR/OS/PFS | Arg399Gln | >0.05 |
| Xu [14] | 2011 | China | Asian | 130 | PCR-RFLP | IIIB-IV | 30 | 14 | 0 | 36 | 40 | 10 | 12 | 14 | 18 | 42 | 26 | 18 | TR | Arg194Trp Arg399Gln | >0.05 |
| Zhou [21] | 2011 | China | Asian | 111 | PCR-RFLP | IV | 29 | 6 | 42 | 34 | − | − | − | − | − | − | TR/TTP | Arg399Gln | − | ||
| Yuan [31] | 2010 | China | Asian | 199 | PCR-RFLP | IIIA-IV | − | − | − | − | − | − | − | − | − | − | − | − | OS/PFS | Arg194Trp Arg399Gln | − |
| Hong [13] | 2009 | China | Asian | 164 | PCR-RFLP | III-IV | 26 | 28 | 3 | 44 | 53 | 10 | 19 | 31 | 7 | 54 | 42 | 11 | TR | Arg194Trp Arg399Gln | >0.05 |
| Kalikaki [19] | 2009 | Greece | Caucasian | 119 | PCR-RFLP | IIIA-IV | 11 | 26 | 21 | 60 | − | − | − | − | − | − | TR/OS | Arg399Gln | − | ||
| Sun [27] | 2009 | China | Asian | 87 | 3D DNA microarray | IV | 14 | 8 | 1 | 39 | 22 | 3 | 8 | 18 | 5 | 31 | 19 | 6 | TR | Arg194Trp Arg399Gln | >0.05 |
| Yao [29] | 2009 | China | Asian | 102 | PCR-RFLP | IIIB-IV | 13 | 9 | 32 | 48 | − | − | − | − | − | − | TR/OS | Arg399Gln | − | ||
| De las Penas R. [25] | 2006 | Spain | Caucasian | 135 | allelic discrimination assay | IIIB-IV | - | − | − | − | − | − | − | − | − | − | − | − | OS | Arg399Gln | − |
| Yuan [16] | 2006 | China | Asian | 200 | PCR-RFLP | Advanced | 24 | 38 | 10 | 69 | 46 | 13 | TR | Arg194Trp | >0.05 | ||||||
| Gurubhagavatula [28] | 2004 | USA | Caucasian | 103 | PCR-RFLP | IIIA-IV | - | − | − | − | − | − | − | − | − | − | − | − | OS | Arg399Gln | - |
| Wang [15] | 2004 | China | Asian | 105 | PCR-RFLP | IIIB-IV | 22 | 9 | 2 | 31 | 33 | 8 | 11 | 19 | 3 | 43 | 18 | 11 | TR | Arg194Trp Arg399Gln | >0.05 |
Footnote: TR treatment response, OS overall survival, PFS progression free survival, CR complete response, PR partial response, SD stable disease, PD progressive disease, SNPs single nucleotide polymorphisms, XRCC1 X-ray repair cross-complementing protein 1, HWE Hardy-Weinberg equilibrium, G = Arg, A = Gln, C = Arg, T = Trp
Meta-analysis results
XRCC1 194 polymorphism
Objective response
Seven studies including 1208 individuals referred the predictive value of XRCC1 Arg194Trp with respect to the sensitivity of lung cancer to platinum-based treatment. All the studies were carried out based on Asian population. In the homozygous model, the Arg genotype was inverse associated with objective response in all patients (ArgArg vs TrpTrp: OR = 0.64(95%CI: 0.44-0.91), p = 0.190, PBegg = 0.368, PEgger = 0.943, Fig. 2a). The ORs in homozygous model, dominant model and heterozygous model showed a gene dosage effect that Arg genotype is associated with worse response rates of platinum-based treatment (ArgArg vs TrpTrp: OR = 0.64(95%CI: 0.44-0.91), p = 0.190, PBegg = 0.368, PEgger = 0.943; ArgArg + TrpArg vs TrpTrp: OR = 0.79(95%CI: 0.56-1.11), p = 0.324, PBegg = 0.230, PEgger = 0.337; TrpArg vs TrpTrp: OR = 1.05(95%CI: 0.73-1.51), p = 0.347, PBegg = 0.035, PEgger = 0.088; Table 2). Recessive model also showed that the ArgArg genotype of XRCC1 194 was associated with worse objective response in all patients treated with platinum-basedregimen (ArgArg vs TrpArg + TrpTrp: OR = 0.55(95%CI: 0.36-0.84), p = 0.013, PBegg = 0.072, PEgger = 0.065; Fig. 2b). Analysis of allele comparison consistent with the results (Arg vs Trp: OR = 0.68(95%CI: 0.51-0.91), p = 0.028, PBegg = 0.368, PEgger = 0.317; Fig. 2c). This indicates that the 194Arg allele may be indicative of poorer response rates to platinum-based treatment than the 194Trp allele.
Fig. 2.
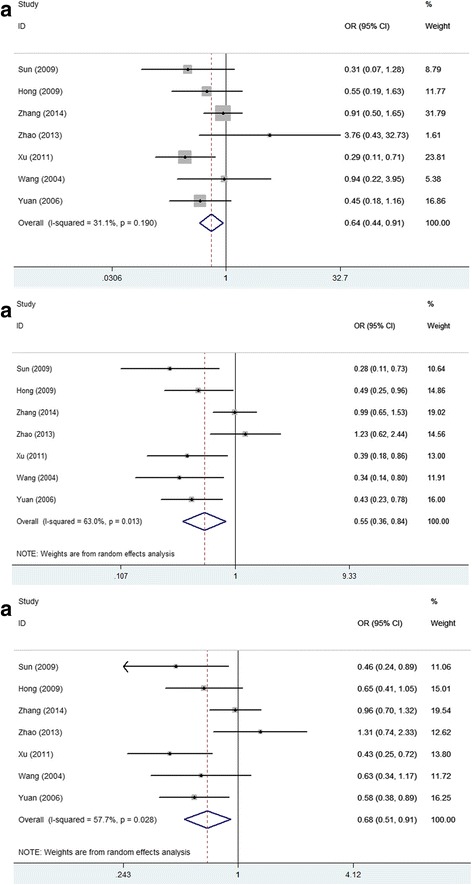
Forest plots of XRCC1 Arg194Trp polymorphisms and objective response in platinum-based chemotherapy by different allele contrast models. a homozygous model, b recessive model, c allele comparison. An OR >1 (or <1) indicates that the 194Arg is more (or less) likely to show response than 194Trp
Table 2.
The association between XRCC1 Arg194Trp and Arg399Gln polymorphisms and objective response, OS and PFS
| XRCC1 | Objective response | Overall survival | Progression-free survival | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Studies(refs.) | Pooled OR | Ph /I 2(%) | Studies(refs.) | Pooled HR | Ph /I 2(%) | Studies(refs.) | Pooled HR | Ph /I 2(%) | |
| Arg194Trp | |||||||||
| ArgArg vs. TrpTrp | |||||||||
| All | 7 [13–16, 23, 26, 27] | 0.64(0.44-0.91) | 0.19/31.1 | 3 [23, 26, 31] | 1.19(0.73–1.65) | 0.92/0.0 | 3 [23, 26, 31] | 1.28(0.74–1.82) | 0.92/0.0 |
| Sample size>100 | 6 [13–16, 23, 26] | 0.67(0.46-0.97) | 0.17/35.4 | 3 [23, 26, 31] | 1.19(0.73–1.65) | 0.92/0.0 | 3 [23, 26, 31] | 1.28(0.74–1.82) | 0.92/0.0 |
| Year ≥ 2009 | 5 [13, 14, 23, 26, 27] | 0.65(0.44-0.99) | 0.09/49.8 | 3 [23, 26, 31] | 1.19(0.73–1.65) | 0.92/0.0 | 3 [23, 26, 31] | 1.28(0.74–1.82) | 0.92/0.0 |
| TrpArg vs. TrpTrp | |||||||||
| All | 7 [13–16, 23, 26, 27] | 1.05(0.73–1.51) | 0.35/10.8 | - | - | - | - | - | - |
| Sample size>100 | 6 [13–16, 23, 26] | 1.04(0.71–1.52) | 0.24/25.4 | - | - | - | - | - | - |
| Year ≥ 2009 | 5 [13, 14, 23, 26, 27] | 0.91(0.60–1.39) | 0.56/0.0 | - | - | - | - | - | - |
| ArgArg + TrpArg vs. TrpTrp | |||||||||
| All | 7 [13–16, 23, 26, 27] | 0.79(0.56–1.11) | 0.32/13.9 | 1 [26] | - | - | 1 [26] | - | - |
| Sample size>100 | 6 [13–16, 23, 26] | 0.80(0.57–1.14) | 0.23/27.1 | 1 ([26]) | - | - | 1 [26] | - | - |
| Year ≥ 2009 | 5 [13, 14, 23, 26, 27] | 0.75(0.51–1.10) | 0.26/24.6 | 1 ([26]) | - | - | 1 [26] | - | - |
| ArgArg vs. TrpArg + TrpTrp | |||||||||
| All | 7 [13–16, 23, 26, 27] | 0.55(0.36-0.84) | 0.01/63.0 | 3 [23, 26, 31] | 1.07(0.79–1.35) | 0.79/0.0 | 3 [23, 26, 31] | 0.94(0.72–1.17) | 0.648/0.0 |
| Sample size>100 | 6 [13–16, 23, 26] | 0.60(0.39-0.92) | 0.02/62.7 | 3 [23, 26, 31] | 1.07(0.79–1.35) | 0.79/0.0 | 3 [23, 26, 31] | 0.94(0.72–1.17) | 0.648/0.0 |
| Year ≥ 2009 | 5 [13, 14, 23, 26, 27] | 0.63(0.37–1.05) | 0.02/65.9 | 3 [23, 26, 31] | 1.07(0.79–1.35) | 0.79/0.0 | 3 [23, 26, 31] | 0.94(0.72–1.17) | 0.648/0.0 |
| Arg399Gln | |||||||||
| GlnGln vs. ArgArg | |||||||||
| All | 10 [13–15, 17, 20, 22–24, 26, 27] | 0.77(0.38–1.57) | 0.005/62.0 | 7 [17, 19, 22–24, 28, 31] | 1.15(0.61–1.69) | 0.006/66.6 | 5 [17, 22, 23, 26, 31] | 0.72(0.48-0.97) | 0.136/42.9 |
| Sample size>100 | 8 [13–15, 17, 22–24, 26] | 0.77(0.35–1.73) | 0.002/69.9 | 7 [17, 19, 22–24, 28, 31] | 1.15(0.61–1.69) | 0.006/66.6 | 5 [17, 22, 23, 26, 31] | 0.72(0.48-0.97) | 0.136/42.9 |
| Asian | 9 [13–15, 17, 20, 23, 24, 26, 27] | 0.75(0.33–1.68) | 0.003/66.1 | 4 [17, 23, 24, 31] | 0.86(0.41–1.30) | 0.052/61.2 | 4 [17, 23, 26, 31] | 0.67(0.40-0.94) | 0.106/51.0 |
| Caucasian | 1 [22] | - | - | 3 [19, 22, 28] | 2.29(1.25-3.33) | 0.423/0.0 | 1 [22] | - | - |
| Year ≥ 2009 | 9 [13, 14, 17, 22–24, 26, 27] | 0.83(0.39–1.77) | 0.005/63.5 | 6 [17, 19, 22–24, 31] | 1.06(0.54–1.57) | 0.011/66.2 | 5 [17, 22, 23, 26, 31] | 0.72(0.48-0.97) | 0.136/42.9 |
| GlnArg vs. ArgArg | |||||||||
| All | 10 [13–15, 17, 20, 22–24, 26, 27] | 0.90(0.67–1.21) | 0.053/46.2 | 7 [17, 19, 23–25, 28, 31] | 0.93(0.68–1.18) | 0.003/69.3 | 4 [17, 23, 26, 31] | 0.96(0.79–1.14) | 0.416/0.0 |
| Sample size>100 | 8 [13–15, 17, 22–24, 26] | 0.89(0.63–1.26) | 0.02/57.8 | 7 [17, 19, 23–25, 28, 31] | 0.93(0.68–1.18) | 0.003/69.3 | 4 [17, 23, 26, 31] | 0.96(0.79–1.14) | 0.416/0.0 |
| Asian | 9 [13–15, 17, 20, 23, 24, 26, 27] | 0.90(0.65–1.25) | 0.035/51.8 | 4 [17, 23, 24, 31] | 0.91(0.56–1.27) | 0.003/78.1 | 4 [17, 23, 26, 31] | 0.96(0.79–1.14) | 0.416/0.0 |
| Caucasian | 1 [22] | - | - | 3 [19, 25, 28] | 1.01(0.51–1.51) | 0.057/65.2 | - | - | - |
| Year ≥ 2009 | 9 [13, 14, 17, 22–24, 26, 27] | 0.97(0.79–1.20) | 0.112/38.4 | 5 [17, 19, 23, 24, 31] | 0.97(0.64–1.31) | 0.003/74.7 | 4 [17, 23, 26, 31] | 0.96(0.79–1.14) | 0.416/0.0 |
| GlnGln + GlnArg vs. ArgArg | |||||||||
| All | 13 [13–15, 17–24, 26, 27] | 0.72(0.50–1.04) | 0.000/70.9 | 6 [17, 19, 20, 23, 26, 31] | 0.73(0.50–1.05) | 0.001/72.5 | 4 [17, 23, 26, 31] | 0.94(0.77–1.10) | 0.274/22.8 |
| Sample size>100 | 10 [13–15, 17, 19, 21–24, 26] | 0.78(0.53–1.16) | 0.000/72.5 | 5 [17, 19, 23, 26, 31] | 0.85(0.55–1.15) | 0.02/65.9 | 4 [17, 23, 26, 31] | 0.94(0.77–1.10) | 0.274/22.8 |
| Asian | 11 [13–15, 17, 18, 20, 21, 23, 24, 26, 27] | 0.70(0.46–1.07) | 0.000/75.7 | 5 [17, 20, 23, 26, 31] | 0.65(0.43-0.98) | 0.003/71.7 | 4 [17, 23, 26, 31] | 0.94(0.77–1.10) | 0.274/22.8 |
| Caucasian | 2 [19, 22] | 0.82(0.46–1.44) | 0.966/0.0 | 1 [19] | - | - | - | - | - |
| Year ≥ 2009 | 12 [13, 14, 17–24, 26, 27] | 0.93(0.65–1.34) | 0.004/64.3 | 6 [17, 19, 20, 23, 26, 31] | 0.73(0.50–1.05) | 0.001/72.5 | 4 [17, 23, 26, 31] | 0.94(0.77–1.10) | 0.274/22.8 |
| GlnGln vs. GlnArg + ArgArg | |||||||||
| All | 12 [13–15, 17, 20, 22–24, 26, 27, 29, 30] | 0.85(0.50–1.45) | 0.028/49.0 | 5 [19, 24, 26, 29, 30] | 1.19(0.53–1.84) | 0.01/68.9 | 2 [26, 30] | 0.94(0.37–1.52) | 0.435/0.0 |
| Sample size>100 | 9 [13–15, 17, 22–24, 26, 29] | 0.78(0.43–1.41) | 0.015/58.0 | 4 [19, 24, 26, 29] | 1.44(1.01-1.88) | 0.686/0.0 | 1 [26] | - | - |
| Asian | 10 [13–15, 17, 20, 23, 24, 26, 27, 29] | 0.75(0.41–1.37) | 0.023/53.4 | 3 [24, 26, 29] | 1.40(0.96–1.84) | 0.999/0.0 | 1 [26] | - | - |
| Caucasian | 2 [22, 30] | 1.58(0.42–5.98) | 0.194/40.6 | 2 [19, 30] | 1.43(−1.05–3.91) | 0.058/72.1 | 1 [30] | - | - |
| Year ≥ 2009 | 11 [13, 14, 17, 20, 22–24, 26, 27, 29, 30] | 0.87(0.47–1.62) | 0.038/51.0 | 5 [19, 24, 26, 29, 30] | 1.19(0.53–1.84) | 0.01/68.9 | 2 [26, 30] | 0.94(0.37–1.52) | 0.435/0.0 |
Footnote: OR odds ratio, HR hazard ratio, refs references, OR/HR with the corresponding 95% CIs >/<1 means significance
Overall survival and progression-free survival
Data from 3 included studies (including 721 patients) were applicable for the analysis. As shown in Table 2, there is no association between the 194Arg genotype and a significant increase of hazard for death in all patients (ArgArg vs TrpTrp: HR = 1.19(95%CI: 0.82–1.73), p = 0.90). The pooled results showed no significant association between XRCC1 Arg194Trp polymorphism and PFS under either kinds of genetic model (Table 2). No significant heterogeneity was detected among the predictive values.
XRCC1 399 polymorphism
Objective response
Eighteen studies including 2814 individuals were qualified for analyzing the association between XRCC1 Arg399Gln polymorphism and treatment response to platinum-based treatment of NSCLC. No significant association was found between the 399Gln allele and response rate relative to the 399Arg allele in either kinds of genetic model (Table 2). However, the results show a suspected borderline association between the 399Gln allele and poorer treatment response in dominant model (GlnGln + GlnArg vs ArgArg: OR = 0.72(95%CI: 0.50–1.04), p = 0.000, PBegg = 0.077, PEgger = 0.093; Fig. 3a). And stratified analysis by ethnicity showed that the borderline association mainly derived from Asian population. Sensitive analysis further confirms the robustness of our results, so we considered that there was no significant association between XRCC1 399Gln allele and the objective response rate relative to 399Arg allele.
Fig. 3.
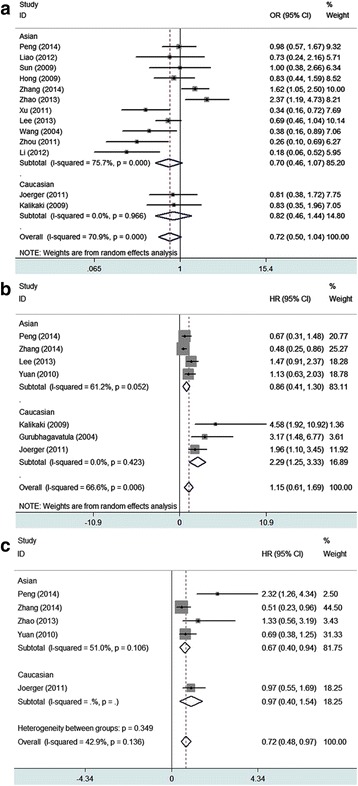
Forest plots of XRCC1 Arg399Gln polymorphisms and clinical outcomes in platinum-based chemotherapy. a Dominant model of association between 399Gln and objective response relative to 399Arg; An OR >1 (or <1) indicates that the 399Gln is more (or less) likely to show response than 399Arg. b Homozygous model of association between 399Gln and overall survival relative to 399Arg; An HR > 1 (or <1) indicates that the 399Gln is more (or less) likely to show worse overall survival than 399Arg. c Homozygous model of association between 399Gln and progression free survival relative to 399Arg. An HR > 1 (or <1) indicates that the 399Gln is more (or less) likely to show worse progression free survival than 399Arg
Overall survival
Thirteen studies covering a total of 2128 patients were eligible for the analysis. The results show neither in dominant model nor in homozygous model, no association was detected between the 399Gln allele and overall survival. In dominant model (GlnGln + GlnArg vs ArgArg: HR = 0.73(95%CI: 0.50–1.05), p = 0.001, PBegg = 0.133, PEgger = 0.169; see Figure, Additional file 1); in homozygous model (GlnGln vs ArgArg: HR = 1.15(95%CI: 0.61–1.69), p = 0.006, PBegg = 0.764, PEgger = 0.594; Fig. 3b). Results of stratified analysis by ethnicity showed as follows: in dominant model, the GlnGln + GlnArg genotypes of XRCC1 399 indicated better overall survival than the ArgArg genotype in Asian patients treated with platinum-based regimen (GlnGln + GlnArg vs ArgArg: HR = 0.65(95%CI: 0.43–0.98), p = 0.003, PBegg = 0.260, PEgger = 0.178; see Figure, Additional file 2); and in homozygous model, the GlnGln genotype of XRCC1 399 showed no association with overall survival in Asian patients (GlnGln vs ArgArg: HR = 0.86(95%CI: 0.41–1.30), p = 0.052, PBegg = 0.308, PEgger = 0.287; Fig. 3b); while in Caucasian patients, the result showed the GlnGln genotype of XRCC1 399 was associated with poorer overall survival (GlnGln vs ArgArg: HR = 2.29(95%CI: 1.25–3.33), p = 0.423, PBegg = 0.296, PEgger = 0.045; Fig. 3b). No publication bias was detected according to the results of funnel plot and the Egger’s test (Fig. 4).
Fig. 4.
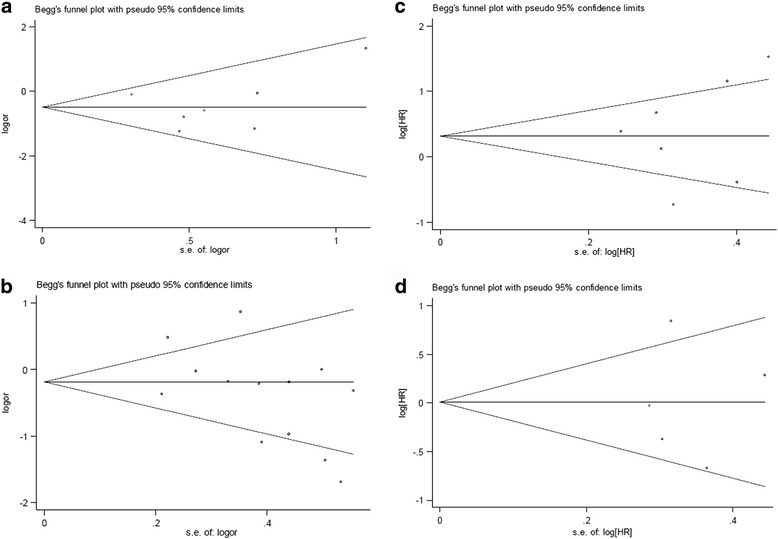
Begg’s funnel plot for publication bias test.Homozygous model of association between XRCC1 Arg194Trp and objective response (ArgArg vs TrpTrp); a Homozygous model of association between XRCC1 Arg399Gln and objective response (GlnGln vs ArgArg); b Homozygous model of association between XRCC1 Arg399Gln and overall survival (GlnGln vs ArgArg); c Homozygous model of association between XRCC1 Arg399Gln and PFS (GlnGln vs ArgArg)
Progression-free survival
Only six studies (including 1180 individuals) were applicable for the analysis. The results show GlnGln genotype of XRCC1 399 was associated with longer PFS than ArgArg genotype in patients treated with platinum-based regimen (GlnGln vs ArgArg: HR = 0.72(95%CI: 0.48–0.97), p = 0.136; Fig. 3c). No publication bias was detected according to the results of funnel plot and the Egger’s test (GlnGln vs ArgArg: PBegg = 1, PEgger = 0.989; Fig. 4).
Sensitivity analysis
Examining genotype frequencies in the controls, significant deviation from HWE was detected in the two articles. [17, 23] When these studies were excluded, the conclusion remain unchanged in the meta-analysis. When the study of small sample (sample size ≤ 100) was excluded, the conclusion remain unchanged in the meta-analysis (Table 2). Additionally, we performed a sensitivity analysis on time of published years. We excluded the papers published before 2009, and performed meta-analysis, the conclusion remain unchanged. Detailed results were showed in Table 2.
Discussion
Platinum-based combination chemotherapy remains the first-line treatment regimen for advanced NSCLC. However, platinum (cisplatin or carboplatin) may cause severe toxic side effect, such as gastrointestinal reaction, neutropenia, anemia, renal toxicity and hepatic toxicity ect. Studies were carried out to explore whether non-platinum-based chemotherapy could achieve comparable efficacy as platinum-based chemotherapy. [36, 37] Meta-analysis’ results show gemcitabine plus docetaxel (GD) acquired similar survival with platinum-based regimens in first-line treatment of advanced NSCLC, platinum-based regimens had an advantage in time to progression (TTP) and overall response rate (ORR) with more grade 3–4 nausea/vomiting, anemia, neutropenia and febrile neutropenia compared with GD. [38] Besides, patients with platinum resistance may not benefit from platinum-based chemotherapy.
Finding predictive markers to guide personalized treatment is essential. The XRCC1 polymorphisms have been widely investigated in lung cancer, and it was reported that different genotype of XRCC1 could predict different lung cancer risk, also it was reported that different genotype of XRCC1 could predict different clinical outcomes (different response rate to platinum-based regimen, different overall survival and different progression-free survival). Meanwhile, others might have different opinions. To explore the relationship between XRCC1 polymorphism and clinical outcomes to platinum-based regimen, and further guide our clinical strategic decision, we conduct this analysis.
Our results showed that XRCC1 194Arg allele was negatively associated with the objective response rate relative to 194Trp, and interestingly, results of homozygous model, dominant model and heterozygous model suggested a gene dosage effect negative correlation between 194Arg allele and objective response rate. This further confirms the robustness of our results. But no association was found between 194Arg allele and either overall survival or PFS. That may due to too few eligible studies. Hence, the conclusions drawn in this meta-analysis about the association between 194Arg allele and both overall survival and PFS should be cautiously considered. More studies need to be carried out and applied for further analysis.
Analysis showed that XRCC1 399Gln allele played different roles in different ethnicity. Although under fixed models, association was found between XRCC1 399Gln allele and not only overall survival, but also objective response rate and progress free survival. As heterogeneity was detected in the analysis of overall survival and objective response rate, after random model was adopted, no significant association was found between XRCC1 399Gln allele and the objective response rate relative to 399Arg allele. XRCC1 399Gln allele indicated better overall survival in Asian patients in dominant model; while in Caucasian patients, the GlnGln genotype of XRCC1 399 was associated with poorer overall survival in homozygous model. Furthermore, in homozygous model, 399GlnGln genotype was associated with longer PFS than 399ArgArg genotype in Asian patients treated with platinum-based regimen. According to the analysis results, it seemed that XRCC1 339Gln allele had contradictory results on different ethnic groups. Maybe there were other reasons that may influence the results of OS and PFS. Because we know that response rate more directly reflects pharmacogenomics roles, while OS and PFS may be influenced by many other factors, such as supportive care, dietary habits, living habits, constitutional factors and so on. In addition to those factors, more studies with much larger sample size are required to be able to draw more definitive conclusions.
HWE states that allele and genotype frequencies in a population will remain constant from generation to generation in the absence of other evolutionary influences. It is not uncommon that quality of studies may vary in meta-analysis of genetic association studies in genetic epidemiology. As reports showed that XRCC1 was associated with lung cancer risk, we considered that violation of HWE was not necessarily be excluded in this analysis, and furthermore, no genotyping error was detected in those studies.
Heterogeneity was detected in parts of the analysis, through random effects model, some of them generated a significant OR/HR results, and all the results were confirmed by sensitive analysis. The existence of heterogeneity indicated variability, which may have been caused by different characteristics, such as ethnicity, region, sample size, gender, method of genotyping used among patient populations. Hence, stratified analyses of subpopulations are needed to reduce such variability, and much larger studies should be undertaken to ensure sufficient statistical power.
Many proteins involved in DNA damage repair system have a role in repairing the cross links by platinum. Nucleotide excision repair (NER) and base excision repair (BER) pathway are major DNA repair systems. Besides XRCC1, other DNA base excision repair genes including OGG1, [17] APE1, [17, 39–41] XRCC3, [42] PARP1, [43] were reported to be associated with clinical outcomes in NSCLC treated with platinum-based regimen. Genes involved in NER system such as ERCC1 [41, 43–45], ERCC2(XPD), [44, 46] BAG1, [46] XPA, [47] were also reported to be associated with clinical outcomes in NSCLC treated with platinum-based regimen. In addition, association between BRCA1, [41, 45] MDR1, [48] eIF3a, [49, 50] PKM2, [51] and clinical outcomes in NSCLC treated with platinum-based regimen were also investigated. Li P’s research demonstrated the combined effects of BAG1 and XPD polymorphisms on chemotherapy sensitivity and survival in patients with advanced NSCLC. [46] Huang ZL’s study showed the expression of ERCC1 and BRCA1 was significantly associated with the disease free survival (DFS) time in patients with NSCLC treated with adjuvant cisplatin-based chemotherapy, respectively. The combination of the ERCC1 and BRCA1 expression levels may be a promising prognostic prediction for adjuvant cisplatin-based chemotherapy. [45].
DNA repair genetic polymorphisms may be better used in the future to predict clinical outcomes from treatments in cancer care [52] and help to improve therapeutic regimen plan setting and patient care. More studies with much larger sample size are required to be able to draw definitive conclusions about the role of DNA repair variants and treatment outcome.
Although interesting results have been achieved in this meta-analysis, there are several limitations. First, a systematic review should ideally be conducted using individual patient data. However, it’s not practical because individual patient data from studies are not always easily obtainable, lacking of the original data of the included studies limited our further evaluation of potential interactions. Second, we only included the studies published in English and Chinese, studies published in other languages were difficult to get. Last, our results were based on unadjusted published estimates. Without data limitations, we could adjust them such as age, smoking condition, pathological type, gender et al., and get more definitive and detailed conclusions. Overall, our results indicated that in NSCLC patients treated with platinum-based regimen, XRCC1 194Arg allele suggest poor objective response rate, the GlnGln genotype of XRCC1 399 suggest poorer overall survival in Caucasian patients, and longer PFS in Asian patients.
Additional files
Dominant model of association between 399Gln and overall survival relative to 399Arg. (DOCX 73 kb)
Dominant model of association between 399Gln and overall survival relative to 399Arg in Asian population. (DOCX 62 kb)
Acknowledgements
Not applicable.
Funding
This study funded by the National Natural Science Foundation of China (NSFC, GrantNo.81101535) and the Special Foundation for National Clinical Specialties of China (to Department of Oncology, Nanfang Hospital). The former plays role in collection, analysis of data and in writing the manuscript, the latter plays role in the design of the study.
Availability of data and materials
The datasets analysed during the current study are available in the PubMed, CNKI, EBSCO and Cochrane databases repository. Weblinks of each study included in the datasets have been provided in the “Reference” part.
Abbreviations
- BER
Base excision repair
- CI
Confidence interval
- CR
Complete response
- HR
Hazard ratio
- HWE
Hardy-Weinberg Equilibrium
- NSCLC
Non-small cell lung cancer
- OR
Odds ratio
- OS
Overall survival
- PD
Progressive disease
- PFS
Progression free survival
- PR
Partial response,
- PRISMA
Preferred Reporting Items for Systematic Reviews and Meta-Analyses
- RCT
Randomized controlled trial
- RECIST
Response Evaluation Criteria in Solid Tumors criteria,
- RR
Risk ratio
- SCLC
Small cell lung cancer
- SD
Stable disease
- SNP
Single nucleotide polymorphisms
- TR
Treatment response
- XRCC1
X-ray repair cross-complementing protein 1
Authors’ contributions
DJL plays role in study design, collection, analysis of data and writing the manuscript. DX plays role in collection, analysis of data. All authors have read and approved the final version of this manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s12885-017-3487-y) contains supplementary material, which is available to authorized users.
Contributor Information
Dan-Juan Li, Email: miamili@foxmail.com.
Dong Xiao, Email: xiao_d@hotmail.com.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Li DJ, Deng G, Xiao ZQ, Yao HX, Li C, Peng F, Li MY, Zhang PF, Chen YH, Chen ZC. Identificating 14-3-3 sigma as a lymph node metastasis-related protein in human lung squamous carcinoma. Cancer Lett. 2009;279(1):65–73. doi: 10.1016/j.canlet.2009.01.028. [DOI] [PubMed] [Google Scholar]
- 3.Pfister DG, Johnson DH, Azzoli CG, Sause W, Smith TJ, Baker SJ, Olak J, Stover D, Strawn JR, Turrisi AT, et al. American Society of Clinical Oncology treatment of unresectable non-small-cell lung cancer guideline: update 2003. J Clin Oncol. 2004;22(2):330–353. doi: 10.1200/JCO.2004.09.053. [DOI] [PubMed] [Google Scholar]
- 4.NCCN . The NCCN non-small cell lung cancer clinical practice guidelines in oncology (version 4.2017) [EB/OL] NCCN: Fort Washington; 2017. [Google Scholar]
- 5.Boulikas T, Vougiouka M. Cisplatin and platinum drugs at the molecular level. (review) Oncol Rep. 2003;10(6):1663–1682. [PubMed] [Google Scholar]
- 6.Martin LP, Hamilton TC, Schilder RJ. Platinum resistance: the role of DNA repair pathways. Clin Cancer Res. 2008;14(5):1291–1295. doi: 10.1158/1078-0432.CCR-07-2238. [DOI] [PubMed] [Google Scholar]
- 7.Galluzzi L, Senovilla L, Vitale I, Michels J, Martins I, Kepp O, Castedo M, Kroemer G. Molecular mechanisms of cisplatin resistance. Oncogene. 2012;31(15):1869–1883. doi: 10.1038/onc.2011.384. [DOI] [PubMed] [Google Scholar]
- 8.Amable L. Cisplatin resistance and opportunities for precision medicine. Pharmacol Res. 2016;106:27–36. [DOI] [PubMed]
- 9.Caldecott KW. XRCC1 and DNA strand break repair. DNA Repair (Amst) 2003;2(9):955–969. doi: 10.1016/S1568-7864(03)00118-6. [DOI] [PubMed] [Google Scholar]
- 10.Vidal AE, Boiteux S, Hickson ID, Radicella JP. XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein-protein interactions. EMBO J. 2001;20(22):6530–6539. doi: 10.1093/emboj/20.22.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitehouse CJ, Taylor RM, Thistlethwaite A, Zhang H, Karimi-Busheri F, Lasko DD, Weinfeld M, Caldecott KW. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell. 2001;104(1):107–117. doi: 10.1016/S0092-8674(01)00195-7. [DOI] [PubMed] [Google Scholar]
- 12.Rice PA. Holding damaged DNA together. Nat Struct Biol. 1999;6(9):805–806. doi: 10.1038/12257. [DOI] [PubMed] [Google Scholar]
- 13.Hong CY, Xu Q, Yue Z, Zhang Y, Yuan Y. Correlation of the sensitivity of NP chemotherapy in non-small lung cancer with DNA repair gene XRCC1 polymorphism. Ai Zheng. 2009;28(12):1291–1297. doi: 10.5732/cjc.009.10139. [DOI] [PubMed] [Google Scholar]
- 14.Xu C, Wang X, Zhang Y, Li L. Effect of the XRCC1 and XRCC3 genetic polymorphisms on the efficacy of platinum-based chemotherapy in patients with advanced non-small cell lung cancer. Zhongguo Fei Ai Za Zhi. 2011;14(12):912–917. doi: 10.3779/j.issn.1009-3419.2011.12.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang ZH, Miao XP, Tan W, Zhang XR, Xu BH, Lin DX. Single nucleotide polymorphisms in XRCC1 and clinical response to platin-based chemotherapy in advanced non-small cell lung cancer. Ai Zheng. 2004;23(8):865–868. [PubMed] [Google Scholar]
- 16.Yuan P, Miao XP, Zhang XM, Wang ZH, Tan W, Sun Y, Zhang XR, Xu BH, Lin DX. XRCC1 and XPD genetic polymorphisms predict clinical responses to platinum-based chemotherapy in advanced non-small cell lung cancer. Zhonghua Zhong Liu Za Zhi. 2006;28(3):196–199. [PubMed] [Google Scholar]
- 17.Peng Y, Li Z, Zhang S, Xiong Y, Cun Y, Qian C, Li M, Ren T, Xia L, Cheng Y, et al. Association of DNA base excision repair genes (OGG1, APE1 and XRCC1) polymorphisms with outcome to platinum-based chemotherapy in advanced nonsmall-cell lung cancer patients. Int J Cancer. 2014;135(11):2687–2696. doi: 10.1002/ijc.28892. [DOI] [PubMed] [Google Scholar]
- 18.Li D, Zhou Q, Liu Y, Yang Y, Li Q. DNA repair gene polymorphism associated with sensitivity of lung cancer to therapy. Med Oncol. 2012;29(3):1622–1628. doi: 10.1007/s12032-011-0033-7. [DOI] [PubMed] [Google Scholar]
- 19.Kalikaki A, Kanaki M, Vassalou H, Souglakos J, Voutsina A, Georgoulias V, Mavroudis D. DNA repair gene polymorphisms predict favorable clinical outcome in advanced non-small-cell lung cancer. Clin Lung Cancer. 2009;10(2):118–123. doi: 10.3816/CLC.2009.n.015. [DOI] [PubMed] [Google Scholar]
- 20.Liao WY, Shih JY, Chang GC, Cheng YK, Yang JC, Chen YM, Yu CJ. Genetic polymorphism of XRCC1 Arg399Gln is associated with survival in non-small-cell lung cancer patients treated with gemcitabine/platinum. J Thorac Oncol. 2012;7(6):973–981. doi: 10.1097/JTO.0b013e31824fe98c. [DOI] [PubMed] [Google Scholar]
- 21.Zhou F, Yu Z, Jiang T, Lv H, Yao R, Liang J. Genetic polymorphisms of GSTP1 and XRCC1: prediction of clinical outcome of platinum-based chemotherapy in advanced non-small cell lung cancer (NSCLC) patients. Swiss Med Wkly. 2011;141:w13275. doi: 10.4414/smw.2011.13275. [DOI] [PubMed] [Google Scholar]
- 22.Joerger M, Burgers SA, Baas P, Smit EF, Haitjema TJ, Bard MP, Doodeman VD, Smits PH, Vincent A, Huitema AD, et al. Germline polymorphisms in patients with advanced nonsmall cell lung cancer receiving first-line platinum-gemcitabine chemotherapy: a prospective clinical study. Cancer. 2012;118(9):2466–2475. doi: 10.1002/cncr.26562. [DOI] [PubMed] [Google Scholar]
- 23.Zhang L, Ma W, Li Y, Wu J, Shi GY. Pharmacogenetics of DNA repair gene polymorphisms in non-small-cell lung carcinoma patients on platinum-based chemotherapy. Genet Mol Res. 2014;13(1):228–236. doi: 10.4238/2014.January.14.2. [DOI] [PubMed] [Google Scholar]
- 24.Lee SY, Kang HG, Yoo SS, Kang YR, Choi YY, Lee WK, Choi JE, Jeon HS, Shin KM, Oh IJ, et al. Polymorphisms in DNA repair and apoptosis-related genes and clinical outcomes of patients with non-small cell lung cancer treated with first-line paclitaxel-cisplatin chemotherapy. Lung Cancer-J Iaslc. 2013;82(2):330–339. doi: 10.1016/j.lungcan.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 25.de Las PR, Sanchez-Ronco M, Alberola V, Taron M, Camps C, Garcia-Carbonero R, Massuti B, Queralt C, Botia M, Garcia-Gomez R, et al. Polymorphisms in DNA repair genes modulate survival in cisplatin/gemcitabine-treated non-small-cell lung cancer patients. Ann Oncol. 2006;17(4):668–675. doi: 10.1093/annonc/mdj135. [DOI] [PubMed] [Google Scholar]
- 26.Zhao W, Hu L, Xu J, Shen H, Hu Z, Ma H, Shu Y, Shao Y, Yin Y. Polymorphisms in the base excision repair pathway modulate prognosis of platinum-based chemotherapy in advanced non-small cell lung cancer. Cancer Chemother Pharmacol. 2013;71(5):1287–1295. doi: 10.1007/s00280-013-2127-8. [DOI] [PubMed] [Google Scholar]
- 27.Sun X, Li F, Sun N, Shukui Q, Baoan C, Jifeng F, Lu C, Zuhong L, Hongyan C, YuanDong C, et al. Polymorphisms in XRCC1 and XPG and response to platinum-based chemotherapy in advanced non-small cell lung cancer patients. Lung Cancer-J Iaslc. 2009;65(2):230–236. doi: 10.1016/j.lungcan.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 28.Gurubhagavatula S, Liu G, Park S, Zhou W, Su L, Wain JC, Lynch TJ, Neuberg DS, Christiani DC. XPD and XRCC1 genetic polymorphisms are prognostic factors in advanced non-small-cell lung cancer patients treated with platinum chemotherapy. J Clin Oncol. 2004;22(13):2594–2601. doi: 10.1200/JCO.2004.08.067. [DOI] [PubMed] [Google Scholar]
- 29.Yao CY, Huang XE, Li C, Shen HB, Shi MQ, Feng JF, Pan LX, Tang JH. Lack of influence of XRCC1 and XPD gene polymorphisms on outcome of platinum-based chemotherapy for advanced non small cell lung cancers. Asian Pac J Cancer Prev. 2009;10(5):859–864. [PubMed] [Google Scholar]
- 30.Tiseo M, Bordi P, Bortesi B, Boni L, Boni C, Baldini E, Grossi F, Recchia F, Zanelli F, Fontanini G, et al. ERCC1/BRCA1 expression and gene polymorphisms as prognostic and predictive factors in advanced NSCLC treated with or without cisplatin. Br J Cancer. 2013;108(8):1695–1703. doi: 10.1038/bjc.2013.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan P, Liu L, Wu C, Zhong R, Yu D, Wu J, Xu Y, Nie S, Miao X, Sun Y, et al. No association between XRCC1 polymorphisms and survival in non-small-cell lung cancer patients treated with platinum-based chemotherapy. Cancer Biol Ther. 2010;10(9):854–859. doi: 10.4161/cbt.10.9.13238. [DOI] [PubMed] [Google Scholar]
- 32.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He XF, Su J, Zhang Y, Huang X, Liu Y, Ding DP, Wang W, Arparkorn K. Association between the p53 polymorphisms and breast cancer risk: meta-analysis based on case-control study. Breast Cancer Res Treat. 2011;130(2):517–529. doi: 10.1007/s10549-011-1583-2. [DOI] [PubMed] [Google Scholar]
- 34.Cao Z, Song J, Wang J, Guo X, Yu S, Dong W. Association between polymorphisms in XRCC1 gene and treatment outcomes of patients with advanced gastric cancer: a systematic review and meta-analysis. PLoS One. 2014;9(1) doi: 10.1371/journal.pone.0085357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez S, Gaunt TR, Day IN. Hardy-Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studies. Am J Epidemiol. 2009;169(4):505–514. doi: 10.1093/aje/kwn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kubota K, Kawahara M, Ogawara M, Nishiwaki Y, Komuta K, Minato K, Fujita Y, Teramukai S, Fukushima M, Furuse K. Vinorelbine plus gemcitabine followed by docetaxel versus carboplatin plus paclitaxel in patients with advanced non-small-cell lung cancer: a randomised, open-label, phase III study. Lancet Oncol. 2008;9(12):1135–1142. doi: 10.1016/S1470-2045(08)70261-4. [DOI] [PubMed] [Google Scholar]
- 37.Rubio JC, Vazquez S, Vazquez F, Amenedo M, Firvida JL, Mel JR, Huidobro G, Alvarez E, Lazaro M, Alonso G, et al. A phase II randomized trial of gemcitabine-docetaxel versus gemcitabine-cisplatin in patients with advanced non-small cell lung carcinoma. Cancer Chemother Pharmacol. 2009;64(2):379–384. doi: 10.1007/s00280-008-0884-6. [DOI] [PubMed] [Google Scholar]
- 38.Yu Y, Xu X, Du Z, Shi M. Non-platinum regimens of gemcitabine plus docetaxel versus platinum-based regimens in first-line treatment of advanced non-small cell lung cancer: a meta-analysis on 9 randomized controlled trials. Cancer Chemother Pharmacol. 2012;69(5):1265–1275. doi: 10.1007/s00280-012-1833-y. [DOI] [PubMed] [Google Scholar]
- 39.Dai N, Cao XJ, Li MX, Qing Y, Liao L, Lu XF, Zhang SH, Li Z, Yang YX, Wang D. Serum APE1 autoantibodies: a novel potential tumor marker and predictor of chemotherapeutic efficacy in non-small cell lung cancer. PLoS One. 2013;8(3):e58001. doi: 10.1371/journal.pone.0058001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang D, Xiang DB, Yang XQ, Chen LS, Li MX, Zhong ZY, Zhang YS. APE1 overexpression is associated with cisplatin resistance in non-small cell lung cancer and targeted inhibition of APE1 enhances the activity of cisplatin in A549 cells. Lung Cancer-J Iaslc. 2009;66(3):298–304. doi: 10.1016/j.lungcan.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 41.Li Z, Qing Y, Guan W, Li M, Peng Y, Zhang S, Xiong Y, Wang D. Predictive value of APE1, BRCA1, ERCC1 and TUBB3 expression in patients with advanced non-small cell lung cancer (NSCLC) receiving first-line platinum-paclitaxel chemotherapy. Cancer Chemother Pharmacol. 2014;74(4):777–786. doi: 10.1007/s00280-014-2562-1. [DOI] [PubMed] [Google Scholar]
- 42.Shen XY, Lu FZ, Wu Y, Zhao LT, Lin ZF. XRCC3 Thr241Met polymorphism and clinical outcomes of NSCLC patients receiving platinum-based chemotherapy: a systematic review and meta-analysis. PLoS One. 2013;8(8):e69553. doi: 10.1371/journal.pone.0069553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie KJ, He HE, Sun AJ, Liu XB, Sun LP, Dong XJ. Expression of ERCC1, MSH2 and PARP1 in non-small cell lung cancer and prognostic value in patients treated with platinum-based chemotherapy. Asian Pac J Cancer Prev. 2014;15(6):2591–2596. doi: 10.7314/APJCP.2014.15.6.2591. [DOI] [PubMed] [Google Scholar]
- 44.Yang Y, Xian L. The association between the ERCC1/2 polymorphisms and the clinical outcomes of the platinum-based chemotherapy in non-small cell lung cancer (NSCLC): a systematic review and meta-analysis. Tumour Biol. 2014;35(4):2905–2921. doi: 10.1007/s13277-013-1493-5. [DOI] [PubMed] [Google Scholar]
- 45.Huang ZL, Cao X, Luo RZ, Chen YF, Zhu LC, Wen Z. Analysis of ERCC1, BRCA1, RRM1 and TUBB3 as predictors of prognosis in patients with non-small cell lung cancer who received cisplatin-based adjuvant chemotherapy: a prospective study. Oncol Lett. 2016;11(1):299–305. doi: 10.3892/ol.2015.3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li P, Wang YD, Cheng J, Chen JC, Ha MW. Association between polymorphisms of BAG-1 and XPD and chemotherapy sensitivity in advanced non-small-cell lung cancer patients treated with vinorelbine combined cisplatin regimen. Tumour Biol. 2015;36(12):9465–9473. doi: 10.1007/s13277-015-3672-z. [DOI] [PubMed] [Google Scholar]
- 47.Cheng H, Qin Q, Sun X, Li F, Sun N, Cheng L, Lu Z, Chen B. Predictive effect of XPA and XPD polymorphisms on survival of advanced NSCLC patients treated with platinum-based chemotherapy: a three-dimensional (3-D), polyacrylamide gel-based DNA microarray method. Technol Cancer Res Treat. 2013;12(5):473–482. doi: 10.7785/tcrt.2012.500337. [DOI] [PubMed] [Google Scholar]
- 48.Yan PW, Huang XE, Yan F, Xu L, Jiang Y. Influence of MDR1 gene codon 3435 polymorphisms on outcome of platinum-based chemotherapy for advanced non small cell lung cancer. Asian Pac J Cancer Prev. 2011;12(9):2291–2294. [PubMed] [Google Scholar]
- 49.Xu X, Han L, Yang H, Duan L, Zhou B, Zhao Y, Qu J, Ma R, Zhou H, Liu Z. The a/G allele of eIF3a rs3740556 predicts platinum-based chemotherapy resistance in lung cancer patients. Lung Cancer-J Iaslc. 2013;79(1):65–72. doi: 10.1016/j.lungcan.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 50.Yin JY, Shen J, Dong ZZ, Huang Q, Zhong MZ, Feng DY, Zhou HH, Zhang JT, Liu ZQ. Effect of eIF3a on response of lung cancer patients to platinum-based chemotherapy by regulating DNA repair. Clin Cancer Res. 2011;17(13):4600–4609. doi: 10.1158/1078-0432.CCR-10-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Papadaki C, Sfakianaki M, Lagoudaki E, Giagkas G, Ioannidis G, Trypaki M, Tsakalaki E, Voutsina A, Koutsopoulos A, Mavroudis D, et al. PKM2 as a biomarker for chemosensitivity to front-line platinum-based chemotherapy in patients with metastatic non-small-cell lung cancer. Br J Cancer. 2014;111(9):1757–1764. doi: 10.1038/bjc.2014.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bartsch H, Arab K, Nair J. Biomarkers for hazard identification in humans. Environ Health. 2011;10(Suppl 1):S11. doi: 10.1186/1476-069X-10-S1-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dominant model of association between 399Gln and overall survival relative to 399Arg. (DOCX 73 kb)
Dominant model of association between 399Gln and overall survival relative to 399Arg in Asian population. (DOCX 62 kb)
Data Availability Statement
The datasets analysed during the current study are available in the PubMed, CNKI, EBSCO and Cochrane databases repository. Weblinks of each study included in the datasets have been provided in the “Reference” part.


