Abstract
Nocardioides dokdonensis, belonging to the class Actinobacteria, was first isolated from sand sediment of a beach in Dokdo, Korea, in 2005. In this study, we determined the genome sequence of FR1436, the type strain of N. dokdonensis, and analyzed its gene contents. The genome sequence is the second complete one in the genus Nocardioides after that of Nocardioides sp. JS614. It is composed of a 4,376,707-bp chromosome with a G + C content of 72.26%. From the genome sequence, 4,104 CDSs, three rRNA operons, 51 tRNAs, and one tmRNA were predicted, and 71.38% of the genes were assigned putative functions. Through the sequence analysis, dozens of genes involved in steroid metabolism, especially its degradation, were detected. Most of the identified genes were located in large gene clusters, which showed high similarities with the gene clusters in Pimelobacter simplex VKM Ac-2033D. Genomic features of N. dokdonensis associated with steroid catabolism indicate that it could be used for research and application of steroids in science and industry.
Electronic supplementary material
The online version of this article (doi:10.1186/s40793-017-0257-z) contains supplementary material, which is available to authorized users.
Keywords: Nocardioidaceae, Propionibacteria, Corynebacteria, Cholesterol, Steroid medicine
Introduction
Bacteria in the genus Nocardioides were first isolated from soil in 1976 [1] and currently more than 90 validly published Nocardioides species are available from diverse terrestrial and aquatic environments such as soil, wastewater, plant roots, groundwater, beach sand, and marine sediment [2–10]. Originally, the genus was classified as a member of the order Actinomycetales in the phylum Actinobacteria, but recently was reclassified to the order Propionibacteriales [11]. Actinobacteria, also called Gram-positive high G + C bacteria, contain diverse bacterial groups that are capable of a variety of secondary metabolism including biosynthesis of antibiotics and degradation of harmful compounds [12, 13]. The genus Nocardioides is also known to utilize several kinds of non-degradable materials such as alkane compounds [14], atrazine [15], phenanthrene [16], trinitrophenol [17], and vinyl chloride [18]. Despite almost 100 species with validly published names and their useful features associated with secondary metabolism, only draft genome sequences are publically available for the genus besides that of Nocardioides sp. JS614.
N. dokdonensis was isolated from beach sand in Dokdo, a volcanic island located in the East Sea of Korea, in 2005 [19]. The East Sea is called a “mini-ocean” due to its oceanological properties [20] and is known to have a high microbial diversity [21]. To reveal distinguishing genomic features of Nocardioides species, we determined and analyzed the genome sequence of N. dokdonensis FR1436T.
Organism information
Classification and features
Nocardioides dokdonensis FR1436T, a Gram-positive, non-motile, and strictly aerobic bacterium, was isolated from sand sediment of the Dokdo island in Korea [19]. The strain grows at the temperature range of 4 to 30 °C (optimum, 25 °C), pH range of 5.0 to 10.0 (optimum, 7.0), and NaCl concentration of 0 to 7% (w/v) (optimum, 0 to 3) [19]. Its colony size is about 1.0–2.0 mm on TSA medium after incubation for 3 days at 25 °C. Cells are 1.2–1.8 μm long and 0.6–0.9 μm wide in size [19] (Fig. 1). FR1436 can utilize adonitol, glycerol, melezitose, melibiose, ribose, sodium acetate, sodium citrate, sodium propionate, and sodium pyruvate as a sole carbon source [19]. Minimum information about the genome sequence (MIGS) for FR1436 is described in Table 1.
Fig. 1.
Transmission electron microscopic image of N. dokdonensis FR1436
Table 1.
Classification and general features of N. dokdonensis FR1436 according to the MIGS recommendations [39]
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Classification | Domain Bacteria | TAS [40] | |
| Phylum Actinobacteria | TAS [41] | ||
| Class Actinobacteria | TAS [42] | ||
| Order Propionibacteriales | TAS [11] | ||
| Family Nocardioidaceae | TAS [11] | ||
| Genus Nocardioides | TAS [43] | ||
| Species Nocardioides dokdonensis | TAS [19] | ||
| Strain FR1436 | TAS [19] | ||
| Gram stain | Gram-positive | TAS [19] | |
| Cell shape | Rod | TAS [19] | |
| Motility | Non-motile | TAS [19] | |
| Sporulation | Nonsporulating | TAS [19] | |
| Temperature range | 4 to 30 °C | TAS [19] | |
| Optimum temperature | 25 °C | TAS [19] | |
| pH range; Optimum | 5.0 to 10.0, 7.0 | TAS [19] | |
| Carbon source | Adonitol, glycerol, melezitose, melibiose, ribose, sodium acetate, sodium citrate, sodium propionate, sodium pyruvate | TAS [19] | |
| MIGS-6 | Habitat | Sand sediment | TAS [19] |
| MIGS-6.3 | Salinity | 0 to 7% (w/v) | TAS [19] |
| MIGS-22 | Oxygen requirement | Strictly aerobic | TAS [19] |
| MIGS-15 | Biotic relationship | Free-living | TAS [19] |
| MIGS-14 | Pathogenicity | Unknown | NAS |
| MIGS-4 | Geographic location | Republic of Korea | TAS [19] |
| MIGS-5 | Sample collection | 2008 | TAS [19] |
| MIGS-4.1 | Latitude | 37° 05′ N | TAS [19] |
| MIGS-4.2 | Longitude | 131° 13′ E | TAS [19] |
| MIGS-4.4 | Altitude | Not reported | NAS |
aEvidence codes - IDA Inferred from Direct Assay, TAS Traceable Author Statement (i.e., a direct report exists in the literature), NAS Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [44]
Phylogenetically, N. dokdonensis belongs to the family Nocardioidaceae of the order Propionibacteriales, and a phylogenetic tree based on the 16S rRNA genes of the type strains in the genus Nocardioides shows that N. dokdonensis FR1436 forms a sister clade with N. lianchengensis (Fig. 2), which was isolated from soil, and shares common ancestor with N. marinisabuli, N. basaltis, and N. salaries.
Fig. 2.
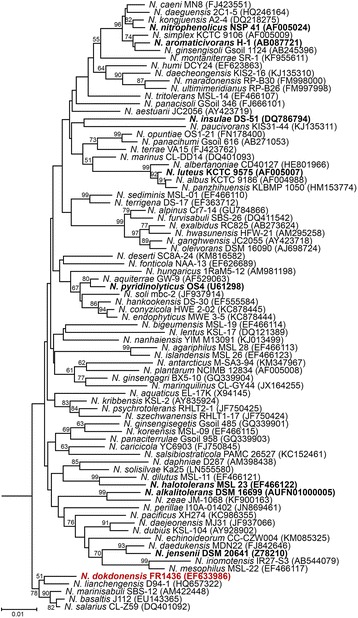
Phylogenetic relationship of the species in Nocardioides. A neighbor-joining tree based on the 16S rRNA gene was generated using MEGA 5 and Jukes-Cantor model was used for calculation of evolutionary distance based on the comparison of 1275 nucleotides. Bootstrap values (percentages of 1000 replications) greater than 50% are shown at each node; Nocardia asteroids NBRC 15531 (BAFO01000006) was used as an out-group. Scale bar represents 0.01 nucleotide substitutions per site. Accession numbers of the 16S rRNA gene are presented in the parentheses. Species for which genome sequences are available are indicated in bold
Genome sequencing information
Genome project history
As part of the project that investigates the genomic and metabolic features of bacterial isolates in and around Dokdo, the genome sequencing and analysis of N. dokdonensis FR1436 were performed at the Laboratory of Microbial Genomics and Systems/Synthetic Biology at Yonsei University. The complete genome sequence of N. dokdonensis FR1436T (= KCTC 19309 T = JCM 14815 T) has been deposited in GenBank under the accession number CP015079. The Bioproject accession number is PRJNA191956. A summary of the genome project is provided in Table 2.
Table 2.
Project information
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | Complete |
| MIGS-28 | Libraries used | A 20-kb library |
| MIGS-29 | Sequencing platforms | PacBio RS II system |
| MIGS-31.2 | Fold coverage | 355.4× |
| MIGS-30 | Assemblers | SMRTpipe HGAP 3.0 |
| MIGS-32 | Gene calling method | Prokka |
| Locus Tag | I601 | |
| Genbank ID | CP015079 | |
| Genbank Date of Release | March 31, 2016 | |
| GOLD ID | Gp0037383 | |
| BIOPROJECT | PRJNA191956 | |
| MIGS-13 | Source Material Identifier | FR1436 |
| Project relevance | Environmental, soil bacterium |
Growth conditions and genomic DNA preparation
N. dokdonensis FR1436 was streaked on trypticase soy agar medium (Difco, 236,950) and incubated at 25 °C for 3 days. A single colony was inoculated in trypticase soy broth and incubated at 25 °C for 2 days. Cells in the exponential phase were harvested and genomic DNA was extracted using Wizard Genomic DNA Purification Kit (Promega, USA) according to the manufacturer’s protocol.
Genome sequencing and assembly
Genome sequencing of N. dokdonensis FR1436 was performed using the PacBio RS II System (Macrogen, Inc., Republic of Korea). A 20-kb library and C4-P6 chemistry were used for the genome sequencing. A total of 200,435 continuous long reads and 1,551,246,448 base pairs were generated after genome sequencing and quality trimming of the sequencing reads. De novo assembly was conducted with SMRTpipe HGAP and scaffolding and gap filling were performed with SMRTpipe AHA. Finally, consensus sequences were generated with SMRTpipe Quiver.
Genome annotation
Structural gene prediction and functional annotation were conducted using the Prokka program [22]. Additionally, we performed a functional assignment of the predicted protein-coding sequences using blastp against Pfam, Uniref90, KEGG, COG, and GenBank NR databases for more accurate annotation. tRNAscan-SE [23] and RNAmmer [24] were used for prediction of transfer RNAs and ribosomal RNAs, respectively. Assignment of the Clusters of Orthologous Groups was conducted with RPS-BLAST against COG database with an e-value cutoff of less than 1e-02. Clustered regularly interspaced short palindromic repeats were predicted with CRISPR Finder [25]. Proteins containing signal peptide and transmembrane helices were predicted using SignalP [26] and TMHMM [27], respectively. Secondary metabolite biosynthetic genes were predicted using AntiSMASH program [28].
Genome properties
N. dokdonensis FR1436 has a single chromosome of 4,376,707 bp in length, and consists of 72.26% of G + C content (Fig. 3 and Table 3). The genome has 4165 genes that are comprised of 4104 CDSs, three rRNA operons, 51 tRNAs, and one tmRNA. Results from the analysis of KEGG pathways indicated that, in the genome of FR1436, all of the genes involved in glycolysis, gluconeogenesis, and citrate cycle are present and well conserved. Among the predicted genes, 71.38% of the genes were assigned putative functions and 2832 CDSs was functionally assigned to the COG categories (Table 4). Also in the genome, ten putative CRISPR repeats were predicted using the CRISPRFinder program, but there were no CRISPR-associated proteins next to the predicted repeat sequences. Two gene clusters, possibly associated with secondary metabolism, were predicted using the AntiSMASH program. One cluster (accession numbers ANH38050 to ANH38087) has genes associated with the phenylacetate catabolic pathway [29] and another cluster (accession numbers ANH40163 to ANH40204) has genes of type 3 polyketide synthases.
Fig. 3.

Circular representation of the genome of N. dokdonensis FR1436. The first and second circles from inside indicate COG-assigned genes in color codes. Black circle represents the G + C content and red-yellow circle is for the G + C skew. Innermost, blue-scattered spots are tRNA genes and red-scattered spots indicate rRNA genes
Table 3.
Genome statistics
| Attribute | Value | % of total |
|---|---|---|
| Genome size (bp) | 4,376,707 | 100 |
| DNA coding (bp) | 4,059,326 | 92.75 |
| DNA G + C (bp) | 3,162,427 | 72.26 |
| DNA scaffolds | 1 | |
| Total genes | 4165 | 100 |
| Protein coding genes | 4104 | 98.54 |
| RNA genes | 61 | 1.46 |
| Pseudogenes | 0 | 0 |
| Genes in internal clusters | ND* | ND* |
| Genes with function prediction | 2973 | 71.38 |
| Genes assigned to COGs | 2832 | 69.01 |
| Genes with Pfam domains | 2584 | 62.04 |
| Genes with signal peptides | 343 | 8.24 |
| Genes with transmembrane helices | 1011 | 24.27 |
| CRISPR repeats | 10 | 10 |
*ND not determined
Table 4.
Number of protein coding genes of N. dokdonensis FR1436 associated with the general COG functional categories
| Code | Value | Percentage* | Description |
|---|---|---|---|
| J | 151 | 3.68 | Translation, ribosomal structure and biogenesis |
| A | 1 | 0.02 | RNA processing and modification |
| K | 212 | 5.17 | Transcription |
| L | 164 | 4.00 | Replication, recombination, and repair |
| B | 1 | 0.02 | Chromatin structure and dynamics |
| D | 25 | 0.61 | Cell cycle control, cell division and chromosome partitioning |
| V | 41 | 1.00 | Defense mechanisms |
| T | 116 | 2.83 | Signal transduction mechanisms |
| M | 124 | 3.02 | Cell wall/membrane/envelope biogenesis |
| N | 3 | 0.07 | Cell motility |
| U | 29 | 0.71 | Intracellular trafficking, and secretion |
| O | 111 | 2.70 | Posttranslational modification, protein turnover, chaperones |
| C | 240 | 5.85 | Energy production and conversion |
| G | 145 | 3.53 | Carbohydrate transport and metabolism |
| E | 317 | 7.72 | Amino acid transport and metabolism |
| F | 75 | 1.83 | Nucleotide transport and metabolism |
| H | 106 | 2.58 | Coenzyme metabolism |
| I | 232 | 5.65 | Lipid metabolism |
| P | 133 | 3.24 | Inorganic ion transport and metabolism |
| Q | 86 | 2.10 | Secondary metabolites biosynthesis, transport, and catabolism |
| R | 321 | 7.82 | General function prediction only |
| S | 199 | 4.85 | Function unknown |
| - | 1272 | 30.99 | Not in COGs |
*The percentages are based on the total number of protein-coding genes in the genome
Insights from the genome sequence
In the genome of N. dokdonensis FR1436, dozens of steroid-degrading genes were detected (Additional file 1). Major functions of steroids, essential biomolecules in living organisms, include maintaining membrane fluidity as a component of the cell membrane and controlling cell metabolism as signaling molecules [30]. Moreover, steroid medicines are used for treatment of a number of diseases from inflammation to cancer [31]. The molecular backbone of steroids is composed of three cyclohexanes and one cyclopentane. To the backbone, diverse side chains are attached to endow them with diverse functions [32]. Catabolic pathways of steroid degradation or modification have been analyzed in depth for some genera in the order Corynebacteriales [33–35]. In Nocardioidaceae, several large gene clusters, which have potential binding sites of the transcriptional regulator associated with steroid catabolism in their promoters, were predicted in the genome of Pimelobacter simplex VKM Ac-2033D [36]. In the genome of FR1436, gene cluster A, which is known to be involved in degrading steroid rings A/B, and gene cluster B, which is involved in degrading side chains, were detected (Fig. 4). However, in FR1436, cluster A is separated into two large gene clusters and an additional mce gene cluster, which is involved in steroid uptake [37], was detected (Additional file 1). In VKM Ac-2033D, cluster A is located approximately 350-kb downstream of cluster B, whereas in FR1436, cluster A is located 6 kb downstream. Moreover, two kstR and 11 kstR2 genes, which encode the TetR family of transcriptional regulators and are reported to regulate cholesterol metabolism in mycobacteria [38], were detected (Additional file 1). Besides the genes in clusters A and B, genes encoding 3-beta-hydroxysteroid dehydrogenase (ANH36717 and ANH37882), 3-alpha-hydroxysteroid dehydrogenase (ANH37023 and ANH37488), and steroid delta-isomerase (ANH36955) were also detected in the genome of FR1436. Additionally, all genes involved in degradation of cholesterol to HIP-CoA were identified (Fig. 5). These results indicate that the genus Nocardioides can be useful for research and utilization of steroid metabolism.
Fig. 4.
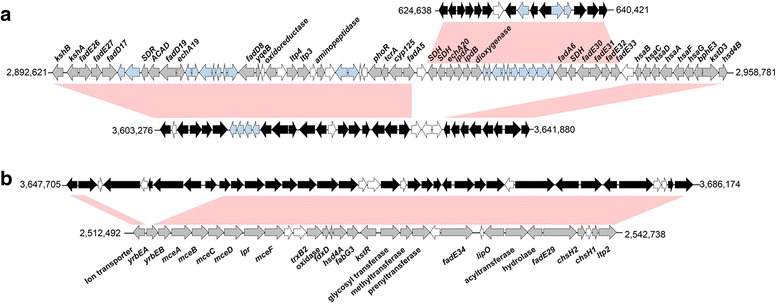
Steroid degrading gene clusters. Gene clusters were referred from the ones of P. simplex VKM Ac-2033D [35], for which genes associated with steroid degradation are indicated in grey arrows. Genes associated with steroid degradation in N. dokdonensis FR1436 are represented by black arrows. Sky blue indicates genes located in the cluster, but little information associated with steroid degradation. White arrows indicate genes encoding hypothetical protein. a. Gene cluster A involved in degradation of steroid ring A and B [35]. Accession numbers of the genes in P. simplex VKM Ac-2033D are AIY19941 to AIY17666. Accession numbers of the genes in N. dokdonensis FR1436 are ANH39848 to ANH39880 and ANH37060 to ANH37075. b. Gene cluster B involved in degradation of side chains of steroids [35]. Accession numbers of the genes are AIY19891 to AIY17347 for P. simplex VKM Ac-2033D and ANH39925 to ANH39888 for N. dokdonensis FR1436
Fig. 5.
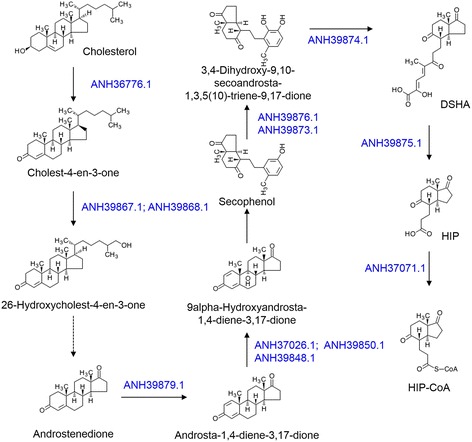
Cholesterol degradation pathway. Metabolic pathway was referred from the KEGG pathway map 00984. Blue indicates gene accession numbers involved in the cholesterol degradation in N. dokdonensis FR1436. DSHA, 3-hydroxy-5,9,17-trioxo-4,5:9,10-disecoandrosta-1(10),2-dien-4-oate; HIP, 9,17-dioxo-1,2,3,4,10,19-hexanorandrostan-5-oic acid
Conclusions
Steroids are important biomolecules in living organisms and carry out diverse roles as components of the cell membrane to signaling molecules [30]. Moreover, steroids are being used to treat various diseases from inflammation to cancer [31]. These indicate that research on modification of steroid compounds has infinite possibilities to improve human health. To date, studies on bacterial steroid metabolism have been mainly focused on the order Corynebacteriales [33–35]. Recently, genome analysis of the genus Nocardioides in the order Propionibacteriales revealed several kinds of gene clusters associated with steroid degradation [36]. In this study, we determined the complete genome sequence of N. dokdonensis FR1436 and analyzed the genome sequence to detect the presence of genes related to steroid metabolism. In the genome of FR1436, dozens of genes associated with steroid catabolism were detected in large gene clusters. These results demonstrate that bacteria in the genus Nocardioides can be used as promising candidates for steroid research and related fields of industry.
Acknowledgements
We recognize Ju Yeon Song for her involvement during the early stages of the project.
Funding
This work was financially supported by the National Research Foundation (NRF-2014M3C9A3068822 and NRF-2011-0017670) of the Ministry of Science, ICT and Future Planning, Republic of Korea.
Abbreviations
- BLAST
Basic Local Alignment Search Tool
- CDS
Coding sequence
- COG
Clusters of Orthologous Groups
- CRISPR
Clustered regularly interspaced short palindromic repeat
- DSHA
3-Hydroxy-5,9,17-trioxo-4,5:9,10-disecoandrosta-1(10),2-dien-4-oate
- HIP
9,17-Dioxo-1,2,3,4,10,19-hexanorandrostan-5-oic acid
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MIGS
Minimum information about a genome sequence
- NR
Non-redundant
- Pfam
Protein families
- RPS-BLAST
Reversed position specific-BLAST
- Uniref
UniProt reference clusters
Additional file
Genes associated with steroid metabolism. (XLSX 14 kb)
Authors’ contributions
JFK conceived, organized and supervised the project, interpreted the results, and edited the manuscript. SKK prepared the high-quality genomic DNA and performed the sequence assembly, gene prediction, gene annotation. MJK analyzed the genome information and drafted the manuscript. All of the authors read and approved the final version of the manuscript before submission.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s40793-017-0257-z) contains supplementary material, which is available to authorized users.
References
- 1.Prauser H. Nocardioides, a new genus of the order Actinomycetales. Int J Syst Evol Microbiol. 1976;26:58–65. [Google Scholar]
- 2.Lim JM, Kim SJ, Hamada M, Ahn JH, Weon HY, Suzuki K, Ahn TY, Kwon SW. Nocardioides daecheongensis sp. nov., isolated from soil. Int J Syst Evol Microbiol. 2014;64(Pt 12):4109–4114. doi: 10.1099/ijs.0.063610-0. [DOI] [PubMed] [Google Scholar]
- 3.Deng S, Chang X, Zhang Y, Ren L, Jiang F, Qu Z, Peng F. Nocardioides antarcticus sp. nov., isolated from marine sediment. Int J Syst Evol Microbiol. 2015;65(8):2615–2621. doi: 10.1099/ijs.0.000309. [DOI] [PubMed] [Google Scholar]
- 4.Singh H, Yin CS. Nocardioides flava sp. nov., isolated from rhizosphere of poppy plant, Republic of Korea. Arch Microbiol. 2016;198(3):279–285. doi: 10.1007/s00203-015-1178-0. [DOI] [PubMed] [Google Scholar]
- 5.Han JH, Kim TS, Joung Y, Kim MN, Shin KS, Bae T, Kim SB. Nocardioides endophyticus sp. nov. and Nocardioides conyzicola sp. nov., isolated from herbaceous plant roots. Int J Syst Evol Microbiol. 2013;63(Pt 12):4730–4734. doi: 10.1099/ijs.0.054619-0. [DOI] [PubMed] [Google Scholar]
- 6.Cui Y, Woo SG, Lee J, Sinha S, Kang MS, Jin L, Kim KK, Park J, Lee M, Lee ST. Nocardioides daeguensis sp. nov., a nitrate-reducing bacterium isolated from activated sludge of an industrial wastewater treatment plant. Int J Syst Evol Microbiol. 2013;63(Pt 10):3727–3732. doi: 10.1099/ijs.0.047043-0. [DOI] [PubMed] [Google Scholar]
- 7.Yoon JH, Kang SJ, Park S, Kim W, Oh TK. Nocardioides caeni sp. nov., isolated from wastewater. Int J Syst Evol Microbiol. 2009;59(Pt 11):2794–2797. doi: 10.1099/ijs.0.010124-0. [DOI] [PubMed] [Google Scholar]
- 8.Kim KH, Roh SW, Chang HW, Nam YD, Yoon JH, Jeon CO, Oh HM, Bae JW. Nocardioides basaltis sp. nov., isolated from black beach sand. Int J Syst Evol Microbiol. 2009;59(Pt 1):42–47. doi: 10.1099/ijs.0.65785-0. [DOI] [PubMed] [Google Scholar]
- 9.Kubota M, Kawahara K, Sekiya K, Uchida T, Hattori Y, Futamata H, Hiraishi A. Nocardioides aromaticivorans sp. nov., a dibenzofuran-degrading bacterium isolated from dioxin-polluted environments. Syst Appl Microbiol. 2005;28(2):165–174. doi: 10.1016/j.syapm.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Yoon JH, Kim IG, Kang KH, Oh TK, Park YH. Nocardioides aquiterrae sp. nov., isolated from groundwater in Korea. Int J Syst Evol Microbiol. 2004;54(Pt 1):71–75. doi: 10.1099/ijs.0.02585-0. [DOI] [PubMed] [Google Scholar]
- 11.Zhi XY, Li WJ, Stackebrandt E. An update of the structure and 16S rRNA gene sequence-based definition of higher ranks of the class Actinobacteria, with the proposal of two new suborders and four new families and emended descriptions of the existing higher taxa. Int J Syst Evol Micr. 2009;59:589–608. doi: 10.1099/ijs.0.65780-0. [DOI] [PubMed] [Google Scholar]
- 12.Barka EA, Vatsa P, Sanchez L, Gaveau-Vaillant N, Jacquard C, Klenk HP, Clement C, Ouhdouch Y, van Wezel GP. Taxonomy, physiology, and natural products of Actinobacteria. Microbiol Mol Biol Rev. 2016;80(1):1–43. doi: 10.1128/MMBR.00019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park HJ, Kim ES. An inducible Streptomyces gene cluster involved in aromatic compound metabolism. FEMS Microbiol Lett. 2003;226(1):151–157. doi: 10.1016/S0378-1097(03)00585-8. [DOI] [PubMed] [Google Scholar]
- 14.Hamamura N, Yeager CM, Arp DJ. Two distinct monooxygenases for alkane oxidation in Nocardioides sp. strain CF8. Appl Environ Microbiol. 2001;67(11):4992–4998. doi: 10.1128/AEM.67.11.4992-4998.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Topp E, Mulbry WM, Zhu H, Nour SM, Cuppels D. Characterization of S-triazine herbicide metabolism by a Nocardioides sp isolated from agricultural soils. Appl Environ Microb. 2000;66(8):3134–3141. doi: 10.1128/AEM.66.8.3134-3141.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwabuchi T, Inomata-Yamauchi Y, Katsuta A, Harayama S. Isolation and characterization of marine Nocardioides capable of growing and degrading phenanthrene at 42 degrees C. J Mar Biotechnol. 1998;6(2):86–90. [Google Scholar]
- 17.Rajan J, Valli K, Perkins RE, Sariaslani FS, Barns SM, Reysenbach AL, Rehm S, Ehringer M, Pace NR. Mineralization of 2,4,6-trinitrophenol (picric acid): characterization and phylogenetic identification of microbial strains. J Ind Microbiol. 1996;16(5):319–324. doi: 10.1007/BF01570041. [DOI] [PubMed] [Google Scholar]
- 18.Coleman NV, Mattes TE, Gossett JM, Spain JC. Phylogenetic and kinetic diversity of aerobic vinyl chloride-assimilating bacteria from contaminated sites. Appl Environ Microb. 2002;68(12):6162–6171. doi: 10.1128/AEM.68.12.6162-6171.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park SC, Baik KS, Kim MS, Chun J, Seong CN. Nocardioides dokdonensis sp nov., an actinomycete isolated from sand sediment. Int J Syst Evol Micr. 2008;58:2619–2623. doi: 10.1099/ijs.0.65835-0. [DOI] [PubMed] [Google Scholar]
- 20.Choi Y. Open-ocean convection in the Japan (east) sea. La mer. 1996;34:259–272. [Google Scholar]
- 21.Kim YE, Yoon H, Kim M, Nam YJ, Kim H, Seo Y, Lee GM, Ja Kim Y, Kong WS, Kim JG, Seu YB. Metagenomic analysis of bacterial communities on Dokdo Island. J Gen Appl Microbiol. 2014;60(2):65–74. doi: 10.2323/jgam.60.65. [DOI] [PubMed] [Google Scholar]
- 22.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 23.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25(5):955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35(9):3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35(Web Server issue):W52–W57. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8(10):785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 27.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 28.Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, Lee SY, Fischbach MA, Muller R, Wohlleben W, et al. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015;43(W1):W237–W243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teufel R, Mascaraque V, Ismail W, Voss M, Perera J, Eisenreich W, Haehnel W, Fuchs G. Bacterial phenylalanine and phenylacetate catabolic pathway revealed. Proc Natl Acad Sci U S A. 2010;107(32):14390–14395. doi: 10.1073/pnas.1005399107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bloch K. Sterol molecule - structure, biosynthesis, and function. Steroids. 1992;57(8):378–383. doi: 10.1016/0039-128X(92)90081-J. [DOI] [PubMed] [Google Scholar]
- 31.Bai C, Schmidt A, Freedman LP. Steroid hormone receptors and drug discovery: therapeutic opportunities and assay designs. Assay Drug Dev Technol. 2003;1(6):843–852. doi: 10.1089/154065803772613471. [DOI] [PubMed] [Google Scholar]
- 32.Hanukoglu I. Steroidogenic enzymes: structure, function, and role in regulation of steroid hormone biosynthesis. J Steroid Biochem Mol Biol. 1992;43(8):779–804. doi: 10.1016/0960-0760(92)90307-5. [DOI] [PubMed] [Google Scholar]
- 33.Van der Geize R, Yam K, Heuser T, Wilbrink MH, Hara H, Anderton MC, Sim E, Dijkhuizen L, Davies JE, Mohn WW, Eltis LD. A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc Natl Acad Sci U S A. 2007;104(6):1947–1952. doi: 10.1073/pnas.0605728104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uhia I, Galan B, Kendall SL, Stoker NG, Garcia JL. Cholesterol metabolism in Mycobacterium smegmatis. Environ Microbiol Rep. 2012;4(2):168–182. doi: 10.1111/j.1758-2229.2011.00314.x. [DOI] [PubMed] [Google Scholar]
- 35.Wilbrink MH, Petrusma M, Dijkhuizen L, van der Geize R. FadD19 of Rhodococcus rhodochrous DSM43269, a steroid-coenzyme a ligase essential for degradation of C-24 branched sterol side chains. Appl Environ Microb. 2011;77(13):4455–4464. doi: 10.1128/AEM.00380-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shtratnikova VY, Schelkunov MI, Fokina VV, Pekov YA, Ivashina T, Donova MV. Genome-wide bioinformatics analysis of steroid metabolism-associated genes in Nocardioides simplex VKM ac-2033D. Curr Genet. 2016;62(3):643–656. doi: 10.1007/s00294-016-0568-4. [DOI] [PubMed] [Google Scholar]
- 37.Mohn WW, Wilbrink MH, Casabon I, Stewart GR, Liu J, van der Geize R, Eltis LD. Gene cluster encoding cholate catabolism in Rhodococcus spp. J Bacteriol. 2012;194(24):6712–6719. doi: 10.1128/JB.01169-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kendall SL, Burgess P, Balhana R, Withers M, ten Bokum A, Lott JS, Gao C, Uhia-Castro I, Stoker NG. Cholesterol utilization in mycobacteria is controlled by two TetR-type transcriptional regulators: kstR and kstR2. Microbiol-Sgm. 2010;156:1362–1371. doi: 10.1099/mic.0.034538-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol. 2008;26(5):541–547. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms - proposal for the domains Archaea, bacteria, and Eucarya. P Natl Acad Sci USA. 1990;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garrity G, Holt J. The road map to the manual. In: Garrity GM, Boone DR, Castenholz RW, editors. Bergey's manual of systematic bacteriology. Volume 1. Second. New York: Springer; 2001. pp. 119–169. [Google Scholar]
- 42.Stackebrandt E, Rainey FA, WardRainey NL. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int J Syst Bacteriol. 1997;47(2):479–491. doi: 10.1099/00207713-47-2-479. [DOI] [Google Scholar]
- 43.Yoon JH, Lee ST, Park YH. Genetic analyses of the genus Nocardioides and related taxa based on 16S-23S rDNA internally transcribed spacer sequences. Int J Syst Bacteriol. 1998;48:641–650. doi: 10.1099/00207713-48-3-641. [DOI] [PubMed] [Google Scholar]
- 44.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]


