Abstract
Chromatin structure at the length scale encompassing local nucleosome–nucleosome interactions is thought to play a crucial role in regulating transcription and access to DNA1–3. However, this secondary structure of chromatin remains poorly understood compared with the primary structure of single nucleosomes or the tertiary structure of long-range looping interactions4. Here we report the first genome-wide map of chromatin conformation in human cells at the 1–3 nucleosome (50–500 bp) scale, obtained using ionizing radiation-induced spatially correlated cleavage of DNA with sequencing (RICC-seq) to identify DNA–DNA contacts that are spatially proximal. Unbiased analysis of RICC-seq signal reveals regional enrichment of DNA fragments characteristic of alternating rather than adjacent nucleosome interactions in tri-nucleosome units, particularly in H3K9me3-marked heterochromatin. We infer differences in the likelihood of nucleosome–nucleosome contacts among open chromatin, H3K27me3-marked, and H3K9me3-marked repressed chromatin regions. After calibrating RICC-seq signal to three-dimensional distances, we show that compact two-start helical fibre structures with stacked alternating nucleosomes are consistent with RICC-seq fragmentation patterns from H3K9me3-marked chromatin, while non-compact structures and solenoid structures are consistent with open chromatin. Our data support a model of chromatin architecture in intact interphase nuclei consistent with variable longitudinal compaction of two-start helical fibres.
The sequence-resolved three-dimensional (3D) folding of DNA at an intermediate length scale between single nucleosomes and approximately 10–100 kilobase (kb) looping interactions remains contentious owing to a dearth of methods capable of mapping the 3D folding of DNA with a spatial precision below ~10 nm and 10 base pairs (bp)4. Studies of purified or reconstituted chromatin have provided evidence supporting a longitudinally compacted 30-nm-diameter chromatin fibre model with a twisted zigzag (two-start helix)5–8 or solenoidal (one-start helix)9–11 arrangement of nucleosomes that depends on nucleosome spacing and linker histone binding1,2,12. In cells, fluorescence in situ hybridization cannot report directly on local DNA folding architecture3,13 and only one proximity ligation-based method, Micro-C, has begun to probe nucleosome organization at this local scale in yeast14. Thus, the extent to which chromatin in its native state adopts a structured 30 nm fibre2,15–17 or an alternative, unstructured ‘polymer melt’18,19 in different regions of the mammalian genome is largely unresolved2,14.
An approach to evaluating these models was pioneered in ref. 15, in which chromatin structure in living cells was probed with ionizing radiation. Compton scattering of X-ray and gamma photons in aqueous samples produces high-energy electrons that deposit their kinetic energy in discrete events, generating localized clusters of hydroxyl radicals20. These radicals cause spatially correlated breaks in DNA strands that pass through a cluster multiple times15,20 (Fig. 1a). The ends of the resulting single-stranded DNA (ssDNA) fragments represent pairwise distance constraints on the folded DNA because they arise from the same nanometre-scale cluster. Although uncorrelated breaks also occur, the average spacing between them is expected to be tens of kilobases for doses of ~100 Gy (ref. 21), and thus short fragments (<1 kb) are highly enriched for products of spatially correlated cleavage.
Figure 1. RICC-seq principle and assay.
a, Gamma rays scatter electrons, generating clusters of hydroxyl radicals dispersed throughout the cell sample (cyan)20. DNA passing through a cluster more than once is subject to spatially correlated strand breaks (red) that generate short ssDNA fragments. b, Workflow for irradiation and ssDNA capture for sequencing (see Methods). Cyan path: de-chromatinized genomic DNA control. c, Polyacrylamide gel electrophoresis (PAGE) analysis of end-labelled ssDNA, colours as in d. d, Line scans of c. e, FLD of sequenced library; ~78 and ~177 nt fragment peaks span single nucleosomes; the ~378 nt fragment spans second-nearest-neighbour nucleosomes. f, V-plot of RICC-seq fragments around occupied CTCF motifs (see Methods; data are pooled from three biological replicates). Grey ovals: cartoon of qualitative nucleosome positions.
To go beyond the genome-wide aggregate fragment length distribution (FLD) reported in ref. 15 and generate a sequence-resolved map of DNA–DNA contacts at the length scale of chromatin secondary structure, we combined irradiation of live cells with high-throughput sequencing (Fig. 1b, Extended Data Fig. 1, Supplementary Table 1 and Methods). We generated RICC-seq libraries (Fig. 1b) in human skin fibroblasts and sequenced 640 million read pairs (Supplementary Table 2). The size distribution of radiation-induced DNA fragments was consistent before and after library preparation (Fig. 1c–e), and between biological replicates (Extended Data Fig. 2a, b). We observed peaks, consistent with expectations about chromatin structure and those reported in ref. 15, at ~78, ~177, ~282, and ~373 nucleotides (nt) in the FLD (Fig. 1e)15. This fragmentation pattern was both irradiation- and chromatin-dependent (Fig. 1b–e). Crosslinked cells gave equivalent DNA fragmentation patterns (Extended Data Fig. 2e), demonstrating that RICC-seq signal is not substantially perturbed by DNA repair. We observed a slightly lower cleavage rate in heterochromatin, probably because of a higher density of radical-quenching associated proteins (Extended Data Fig. 2).
We asked whether structured RICC-seq signals arose from well-positioned nucleosomes flanking binding sites of the zinc finger protein CTCF and active transcription start sites. By plotting fragment length versus centre position (V-plot, see Methods), we observed a marked enrichment of both ~78 nt fragments (a single wrap around a nucleosome) and ~177 nt fragments (consistent with contacts between linker DNA exiting the nucleosome) at expected nucleosome positions (Fig. 1f and Extended Data Fig. 3a, b, d, e)22. We also observed CTCF footprints23,24 at motifs within CTCF peaks from chromatin immu-noprecipitation with sequencing (ChIP-seq) (Extended Data Fig. 3c and Methods), indicating that RICC-seq can detect protein binding on DNA in vivo.
To test whether the local RICC-seq FLD reflects differences in chromatin architecture, we calculated the enrichment of different RICC-seq fragment lengths (fragment length fold-enrichment (FLFE)) (Extended Data Fig. 4 and Methods) in 0.3–1 megabase (Mb) windows tiling the genome. We observed two major clusters with reciprocal patterns of enrichment, particularly in the 373 nt peak, expected to originate from second-nearest nucleosome interaction (Fig. 2a). The two clusters were respectively enriched in (1) open chromatin (associated with assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) and DNase-seq peaks) or euchromatin (associated with ChIP peaks for histone modifications such as histone 3 lysine 4 monomethylation (H3K4me1), H3K4me3, H3K27ac, and H3K36me3) or (2) transcriptionally repressed chromatin associated with H3K9me3 and H3K27me3 modifications1,2,25. Principal component analysis produced a single component (PC1) that captured the reproducible variation in enrichment across windows, and a linear baseline that captured overall enrichment towards longer fragments in repressed chromatin (Fig. 2b, c and Extended Data Figs 4 and 5a). The PC1 signal in each window was strongly anticorrelated with H3K9me3 ChIP-seq signal (Spearman’s ρ= −0.543, P < 2 × 10−16) and correlated with ATAC-seq signal (Spearman’s ρ = 0.479, P < 2 × 10−16) (Fig. 2c, d and Extended Data Fig. 6). Surprisingly, PC1 was very weakly correlated with H3K27me3-marked chromatin (Spearman’s ρ =0.0552, P ≈ 4 × 10−7), suggesting that it represents a different structural state or is heterogeneous within 300 kb windows.
Figure 2. The RICC-seq FLD reveals variable chromatin compaction states.
a, Cluster averages of 1 Mb window FLDs hierarchically clustered and plotted with average z-scores of chromatin annotations. b, First principal component of variation (PC1) calculated from 1 Mb windows (black) or cluster averages (red). c, Spearman’s correlation of annotation signal within 1 Mb windows with baseline slope (positive slope reflects skew towards longer fragment lengths) and PC1 score. d, Track depicting the RICC-seq score (red; inner product of PC1 (b) with the FLFE (Extended Data Fig. 4a) in each 300 kb window), ATAC-seq insertion density (cyan), H3 lysine methylation ChIP-seq −log10(Poisson P value) (grey for H3K27me3 and black for H3K9me3; see Methods), and RICC-seq fragment density (blue). e, FLFE in region sets for three chromatin states. Data are pooled from two biological replicates.
We then analysed the FLD and FLFE across peaks of six histone modifications and peaks of chromatin accessibility (Fig. 2e and Extended Data Fig. 5). In agreement with results from principal component analysis, the FLFE profiles associated with open chromatin-associated modifications or ATAC-seq peaks resembled each other, with strong depletion in the 373 nt tri-nucleosome contact peak. In H3K9me3-marked regions, the tri-nucleosome contact peak was enriched. In open chromatin, we observed enrichment of 50–200 bp fragments, potentially caused by reduced radical quenching in open chromatin (Extended Data Fig. 2f, g). H3K27me3 regions showed little enrichment with a small peak at ~400 bp and may reflect heterogeneous structures. This structural difference between H3K9me3 and H3K27me3 peaks is consistent with recent observations of different scaling of chromatin compaction13 and different silencing kinetics25 associated with H3K27me3 or H3K9me3 modifications. Overall, differences among DNA–DNA contact profiles constitute a genome-wide map of variable chromatin architecture and support a model of chromatin regulation arising from local chromatin fibre compaction at the tri-nucleosome scale.
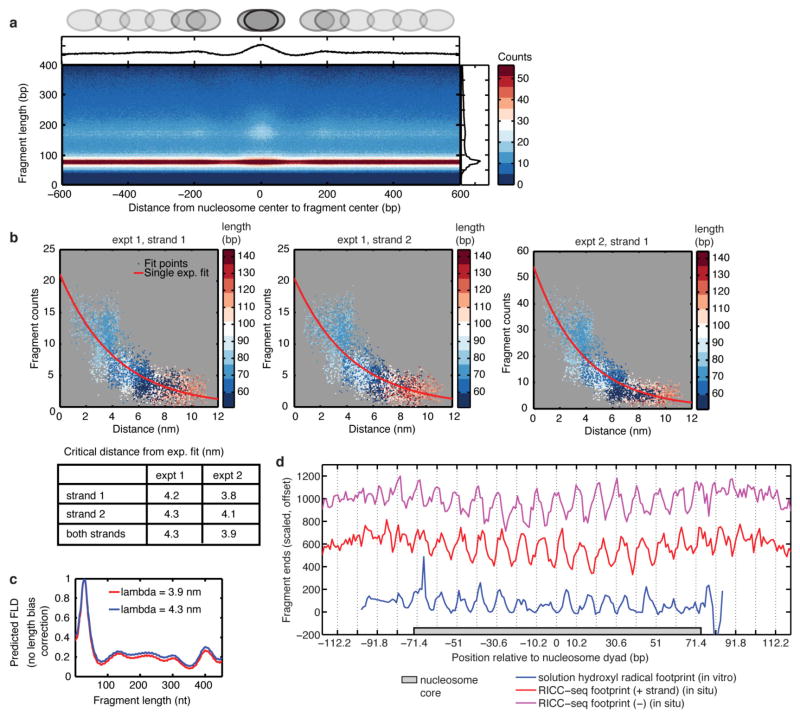
Existing models of chromatin fibre structure can be broadly classified into solenoids, zig-zags, and unstructured chromatin polymer melts1,2,18. We sought to compare RICC-seq results with quantitative predictions from these models. We took advantage of the mapping of DNA fragments to the genome to calibrate the RICC-seq cleavage frequency to 3D distance. We compared fragment counts around positioned nucleosomes with the distances between pairs of DNA backbone phosphates in mononucleosome crystal structure (Fig. 3a and Extended Data Figs 7 and 8; see Methods). Data were fitted with a single exponential with a length constant of 4.03 nm (95% CI: 3.97–4.10 nm), indicating that RICC-seq signal reports 3D distances in folded DNA on the nanometre-scale (Fig. 3b, c). The measured length constant was consistent with estimates of irradiation-generated radical diffusion (~3.5 nm)20 and comparable to the ~10 nm core nucleosome2,26. Deviations from the exponential probably resulted from variable protection from radical cleavage (Extended Data Fig. 8) or error in nucleosome position calls. The total RICC-seq cleavage signal was similar to solution hydroxyl radical footprinting data, indicating protection of DNA by histones (Extended Data Fig. 8).
Figure 3. Spatially correlated break frequency scales exponentially with 3D distance allowing calibration of signal.
a, Fragment 5′ and 3′ end locations plotted relative to nucleosome dyad (lower right triangle) and 3D distances between same pairs of phosphates on the basis of Protein Data Bank (PDB) 1KX5 (upper left triangle; see Methods). b, The 3′ end break frequency for fragments with a given 5′ end position (green arrow in a) plotted on one DNA strand of the nucleosome structure. c, Exponential fit of RICC-seq fragment counts to 3D distance between pairs of DNA backbone phosphates (both strands, pooled from two biological replicates).
To compare a diverse set of models with our in situ data, we generated 969 helical chromatin fibre models on the basis of minimal assumptions—the mechanical properties of DNA and the mononucleosome structure27 (Fig. 4a and Extended Data Fig. 9a–g). We predicted their fragment counts (Extended Data Fig. 9i) using the calibrated exponential model (Fig. 3) corrected for length bias (Extended Data Fig. 7).
Figure 4. RICC-seq identifies distinct classes of predicted fibre models correlated with epigenetic features.
a, Example model and simulation workflow. b, Fibre model phase space indicating six geometry classes based on clustering centre–centre nucleosome distance patterns (Extended Data Fig. 9h). Dots indicate coordinates of examples of each structure class (shown below) in the phase space. Literature values for characteristic nucleosome repeat lengths (NRLs)22 and compaction levels32 are indicated. c, f, Similarity scores (inner products) between experimental FLFE in ATAC-seq peaks (c) or H3K9me3 ChIP-seq peaks (f) and the predicted FLFE of each structure. Black outline: class 1. d, g, Mean similarity scores for each class. e, Three examples of FLFE comparisons shown after mean subtraction (subt.) of each FLFE. Experimental data are from two pooled biological replicates.
We clustered these structures on the basis of helical nucleosome contacts to obtain six classes that reflect established fibre geometries2 (Fig. 4b and Extended Data Fig. 9b). These structure classes possess ~10 bp periodicity in linker length because the DNA helix constrains the relative twist of consecutive nucleosomes27,28 (Fig. 4b and Extended Data Fig. 9b–j). Classes 1 and 2 encompass two-start (zig-zag) helices, class 3 comprises three-start helices, classes 4 and 5 comprise compact high-order-start helices and interdigitated solenoids, and class 6 consists of three compact structures that are true bent-linker solenoids.
The predicted FLFEs of the zig-zag fibres of classes 1 and 2 are broadly consistent with genome-wide chromatin structure, as observed in ref. 15. They also agree with chemical crosslinking6,17, proximity ligation14, and in vitro reconstitution29 data (Extended Data Fig. 10). Importantly, RICC-seq data allow us to go beyond genome-wide averages to infer chromatin states correlated with regions of interest defined by other epigenomic features. Because RICC-seq data are averaged over large regions and millions of cells, resulting FLDs necessarily represent an average over an underlying ensemble of heterogeneous conformations1,2,10,12,14,17. FLFE profiles thus report on chromatin architectural features particularly enriched in the ensemble of structures at our loci of interest. Predicted and experimental FLFE comparisons show that fragmentation patterns expected from classes 3–6 (extended fibres and solenoids) are consistent with open chromatin FLFE (Fig. 4b–e) owing to their lack of a 380–400 bp peak. Fragment patterns generated from zig-zag helices in classes 1 and 2 are similar to the H3K9me3-marked chromatin FLFE (Fig. 4b, e–g) and have peak locations consistent with face-to-face stacked alternating nucleosomes, as seen in a reconstituted chromatin fibre29 (Extended Data Figs 9f and 10d–h). This nucleosome stacking may play a part in promoting binding of HP1-α, a chromatin architectural protein that preferentially binds compacted chromatin fibres30. The H3K27me3 FLFE is consistent with class 1 and 2 structures, but is also compatible with a broader range of structures including class 3 (Extended Data Fig. 10i–k), possibly reflecting a mix of architectures.
Detailed comparison of simulated fibres with a reconstituted fibre structure revealed an offset in the inferred linker length (Extended Data Fig. 10e), probably because of the absence of linker histone in our minimal model2,27–29,31. To explore the effects of linker histones on the FLFE, we adapted existing models of the nucleosome with linker histone (chromatosome) (Extended Data Fig. 10l; see Methods). The observed shift in the 177 bp peak between chromatin states was consistent with FLFE differences between zig-zag and bent-linker solenoid models rather than with differences in linker histone binding mode (Extended Data Fig. 10l–n). Although the low compaction of open chromatin2,3,23 makes a compact solenoid unlikely, a fluctuating or de-compacted bent-linker fibre may be plausible given factors that stabilize bent DNA. We attempted to enrich for long fragments, to potentially distinguish between these models (Extended Data Fig. 9k, l), but did not observe reproducible peaks beyond 500 bp.
Overall, our results support a role for intra-fibre zig-zag compaction in organizing condensed chromatin structure in the interphase nuclei of actively proliferating cells, in contrast to the polymer melt model2,18. RICC-seq is uniquely capable of reporting on base-pair-specific, short-range 3D contacts between nucleosomes, providing a sequence-resolved probe of DNA folding at the level of chromatin secondary structure in living cells. Using this method, we have extended the findings of ref. 15 to show that the stacking of alternating nucleosomes varies between H3K9me3-marked chromatin, H3K27me3-marked chromatin, and open chromatin. Our data support the long-held idea that DNA accessibility is regulated in part by longitudinal compaction of the chromatin fibre1,2, which, as suggested for yeast14, consists of locally ordered two-start zig-zag structure interrupted by disordered or heterogeneous domains. RICC-seq methodology is probably broadly extendable to the investigation of diverse condensed nucleic acid conformations in other biological systems, including viral capsids, bacterial spores, sperm chromatin, and other packaged DNA or RNA particles.
Online Content Methods, along with any additional Extended Data display items and Source Data, are available in the online version of the paper; references unique to these sections appear only in the online paper.
METHODS
No statistical methods were used to predetermine sample size. The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment.
Cell culture
BJ (passage 17–18) and BJ-5ta fibroblast cells were cultured in Eagle’s minimum essential medium (ATCC) with 10% heat-inactivated fetal bovine serum (Life Technologies), 100 U/mL penicillin–streptomycin (Life Technologies), in filter-cap flasks (Greiner BioOne Cellstar) at 37 °C with 5% CO2 and passaged at 80% confluence using 0.05% trypsin-EDTA (Life Technologies) or Accutase (Sigma-Aldrich). BJ cells were a gift from G. Wang and BJ-5ta cells were a gift from J. Cochran. Cell authenticity was authenticated using STR testing (Genetica Laboratories, Cincinnati, Ohio, USA) and compared with STR profiles from ATCC. Mycoplasma contamination was monitored by aligning sequenced reads to the Mycoplasma hominis complete genome sequence (NC_013511) using Bowtie 2 (ref. 33). Biological replicates were cultured and harvested on different days; technical replicates were RICC-seq libraries prepared in parallel from the same culture batch.
Cell irradiation and lysis
Cells were harvested at 80–100% confluence after washing with Ca2+ - and Mg2+ -free phosphate-buffered saline (PBS; Life Technologies) using Accutase (Sigma-Aldrich) and washed twice with room temperature PBS. Cell embedding into agarose plugs and irradiation were performed according to the procedure of Rydberg et al.15, with the following changes. Cell suspensions were brought to room temperature before mixing with low gelling temperature agarose (Sigma Type VII-A) at 37 °C. Agarose plugs were irradiated in 500 μL ice-cold PBS in a Mark I Cs-137 gamma irradiator (J. L. Shepherd) with a rotating turntable. Agarose plug washes were performed in 1 mL of TEE (10 mM Tris pH 8, 1 mM EDTA, 1 mM EGTA) at room temperature for 1 h, or 1 mL of TEE with 0.1 mg/mL RNase A (Qiagen) at 37 °C for 1 h, with gentle rocking. Genomic DNA controls (Fig. 1b, dotted cyan workflow) were prepared by lysing cells in agarose plugs as above, followed by soaking for 30 min in 0.5 M Tris pH 8, 1 mM EDTA, irradiation, and four washes in TEE as above. For fixed-cell experiments, cells were harvested using Accutase, then washed with PBS and crosslinked first for 20 min with 1.5 mM EGS in PBS at room temperature with gentle rotation, then with 1% formaldehyde at room temperature for 10 min, and quenched with 200 mM glycine for 5 min before washing in PBS and embedding in agarose. The standard irradiation protocol was followed, except that the 50 °C lysis incubation was extended to 4 days to reverse crosslinks.
Sequencing adaptor preparation
Adapters were annealed by mixing pairs of oligonucleotides (Supplementary Table 1) at a final concentration of 100 μM each in 10 mM Tris pH 8, 1 mM EDTA, and 50 mM NaCl, heating to 95 °C for 2 min, cooling to 60 °C at 1 °C/min, incubating 10 min, then cooling to 4 °C at 1 °C/min. Annealed adapters were gel-purified using a 10% Tris-Borate-EDTA non-denaturing polyacrylamide gel (Bio-Rad) and recovered via the crush and soak method34 without SDS or heating above room temperature. Custom oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, Iowa, USA) and the Stanford Protein and Nucleic Acid Facility (Stanford, California, USA).
Sequencing library preparation and analysis by end-labelling and PAGE
The procedure of Rydberg et al.15 was followed, except that phosphates were removed by incubating each 4.5 mm square plug piece with 80 μL of reaction mix containing 2.3 U of shrimp alkaline phosphatase (rSAP; New England Biolabs, Ipswitch, Massachusetts, USA) and 1 × CutSmart Buffer (New England Biolabs) for 1 h at 37 °C; and denaturation of agarose-embedded DNA was alternatively performed with 0.1 M NaOH15 or by heating at 95 °C for 8 min followed by 2 min of cooling on ice to re-solidify agarose with similar results.
For sequencing library preparation, DNA-containing agarose plugs were phosphatase-treated as above, and phosphatase was removed by digestion in 100 μL lysis buffer per 4.5 mm square plug piece (0.4M EDTA (Invitrogen UltraPure), 2% v/v Sarkosyl (Fisher BioReagents), 0.5 mg/mL Proteinase K (New England Biolabs), pH 8) for 18 h at 50 °C, then washed five times with 1 mL TE (10 mM Tris pH 8, 1 mM EDTA) at room temperature with gentle rocking, 1 h per wash; 5′ phosphates were added by soaking each 4.5 mm plug piece in 100 μL PNK buffer (70 mM Tris, 10 mM MgCl2, 5 mM dithiothreitol, 1 mM spermidine, pH 8) at 4 °C for 1 h, then soaking the plug piece in 70 μL reaction mix containing PNK buffer, 5 μM ATP (New England Biolabs), and 40 units of T4 polynucleotide kinase (Enzymatics; Beverly, Massachusetts, USA) at 4 °C for 2 h, then incubating it in the same reaction mix for 30 min at 37 °C. Plugs were washed four times in 1 mL TE at 4 °C. ssDNA fragments were recovered from the agarose plugs by thermal denaturation for 8 min at 95 °C followed by 2 min re-solidification on ice and passive diffusion into 140 μL TE per plug piece for ~16 h at room temperature with rotation. The supernatant was filtered using empty spin columns (Ambion AM10065) and the DNA was concentrated and further purified using an evaporator and a Qiagen Nucleotide Removal Kit (one column per two 4.5 mm agarose plugs), using 30 μL10 mM Tris pH 8 for elution.
Using a method adapted from Smith et al.35, purified ssDNA fragments were ligated to sequencing adapters with six-random-nucleotide overhangs (Supplementary Table 1) using 50 pmol of each adaptor, 1 × T4 Ligase Buffer (Enzymatics), and 450 U of T4 DNA Ligase (Enzymatics) at 15 °C for 16 h with mixing at 800 r.p.m. (Eppendorf Thermomixer) in a volume of 25 μL per 4.5 mm square plug equivalent. Excess adapters were removed with S-300-HR spin columns (Bio-Rad), MgCl2 was added to restore magnesium concentration to 10 mM, and the 3′ adaptor was extended using 0.02 U/μL Taq-B enzyme, 1 × PCR Buffer (both, Enzymatics) and 0.2 mM (each) dNTPs (New England Biolabs) at 65 °C for 30 min. The extension reactions were purified with a QIAQUICK PCR cleanup kit (Qiagen) and concentrated to 10–20 μL per sample. Libraries were size-selected between 100 and 1,000 bp to remove adaptor dimers using a 2.5% agarose gel and purified with a MinElute Gel Extraction Kit (Qiagen). Libraries were amplified for 13–16 cycles (up to 21 cycles for size-selected libraries) with primers that add the full Illumina sequencing adaptor sequences (an optimized variant of TruSeq adaptors, Supplementary Table 1) and NEBNext 2 × PCR Master Mix (New England Biolabs), using an aliquot of the sample to determine the optimal number of cycles as in ref. 23. Amplified libraries were size-selected two or three more times between 150 and 1,000 bp using 2.5% agarose gels, the MinElute Gel Extraction Kit (Qiagen), and additional amplification by five cycles between size selections, to remove remaining adaptor dimers. Sequencing was performed on an Illumina (San Diego, California, USA) MiSeq (75 bp paired end reads; shallow sequencing runs) or an Illumina HiSeq 2500 (Rapid Run Mode; 50 bp paired end reads; ELIM Biopharm, Hayward, California, USA).
ATAC-seq
BJ cells (passage 19) were harvested at 70% confluence using Accutase and washed twice with ice-cold PBS. Aliquots (8,000 cells) were pelleted for 5 min at 500g in 1.5 mL tubes at 4 °C, and resuspended in 50 μL of transposition mix containing 2.5 μL Nextera Tagment DNA Enzyme (Illumina), 1× Nextera Tagment DNA Buffer (Illumina), and 0.01% digitonin (Promega), using a modified ATAC-seq protocol36. Transposition reactions were performed at 37 °C with 300 r.p.m. mixing for 30 min. Libraries were purified with a QIAQUICK PCR cleanup kit, eluting with 20 μL, and amplified with NEBNext 2 × PCR Master Mix (New England Biolabs) using barcoded PCR primers (Supplementary Table 1) for variable numbers of cycles (~10) determined by using qPCR on an aliquot to avoid saturation. Amplified libraries were purified with a QIAQUICK PCR clean up kit and size-selected with a 2.5% agarose gel and MinElute Gel Extraction Kit before high-throughput sequencing on an Illumina HiSeq 2500 (50 bp paired end reads; ELIM Biopharm). Eight technical replicates were performed and data was pooled for peak calling and nucleosome calling (see below).
Sequencing data processing and alignment
Paired-end reads from both RICC-seq and ATAC-seq were trimmed of adaptor sequences using a custom Python script23 and aligned to hg19/GRCh37 using Bowtie 2 (version 2.1.0)33 with parameters -X2000 -p18. Alignments were filtered using SAMtools version 0.1.19 (ref. 37) to remove those aligning to all but chromosomes 1–22 and chromosome X and those with a mapping quality score below 30, and using BEDTools version 2.20.1 (ref. 38) to remove reads in the combined ENCODE project DAC Blacklisted Regions and Duke Excluded Regions39. Duplicate reads (PCR and optical) were removed using Picard tools (version 1.117) MarkDuplicates (https://broadinstitute.github.io/picard/). Genome-wide FLDs were calculated using Picard tools. RICC-seq data sets were evaluated for background nuclease contamination using the genome-wide FLD after pilot experiments. Biological replicate 3 was excluded from window-based and annotation-based FLFE analysis (see below) because the baseline signal exceeded one-third of the maximum 78 bp peak signal (Extended Data Fig. 2), indicating excessive non-specific DNA cleavage. It was included in break mapping analysis (see below) to improve sequencing depth. ATAC-seq data were further processed using custom Python code23 and bedGraphToBigWig (UCSC genome tools; http://hgdownload.cse.ucsc.edu/downloads.html#source_downloads) to obtain a track of transposase insertion density over the genome.
External data sets
CTCF ChIP-seq, mappability, and RNA-CAGE data for BJ cells were obtained from the ENCODE project39. CTCF motifs used were the ‘known 1’ motif determined from ChIP data and occupancy was evaluated using peaks from the same ENCODE ChIP data39,40. Transcription start sites were obtained from the RefSeq refGene table with a status of ‘reviewed’ or ‘validated.’ Histone post-translational modification ChIP-seq and DNase-seq data were obtained as tracks of −log10(Poisson P value) from consolidated epigenome E055 in the Roadmap Epigenomics project41.
Nucleosome calling
ATAC-seq narrow peaks were called using MACS2 (version 2.1.0.20140616)42 using parameters ‘–nomodel–nolambda–keep-dup all –broad–call-summits–slocal 10000’ and extending each peak by 120 bp on both sides, filtered for peaks of fewer than 500 bp, then merged peaks within 120 bp using BEDTools (version 2.20.1)38. Nucleosome positions within these peaks were called using NucleoATAC with default parameters using a sequence bias track calculated on the basis of Tn5 transposition in human genomic DNA43.
V-plots
V-plots44 were calculated by using custom Python code to plot alignments by fragment length versus fragment centre position with respect to the centre of the CTCF motif or the TSS. CTCF motifs were filtered for occupancy using ChIP data39, and TSSs were classified as active if they were within a RNA-CAGE peak in BJ fibroblasts39 or inactive if they did not overlap an ATAC-seq peak in BJ fibroblasts. V-plot matrices for 0 Gy control and 300 Gy irradiated cell samples were scaled for equal sequencing depth relative to library complexity, and the former was subtracted from the latter to account for background DNA fragmentation. The same background subtraction was performed for dechromatinized DNA sample V-plots.
Window-based fragment size distribution analysis
The FLD between 1 and 700 nt was calculated in 100 kb to 1 Mb windows throughout the genome, excluding gaps and blacklisted regions (as in read filtering), using a custom Python script. Background FLDs were calculated similarly from non-irradiated cell data, then subtracted from irradiated sample FLDs. Both background and signal FLDs were scaled by the number of cells in each sample and by the number of unique molecules in the library (Picard MarkDuplicates version 1.117). We controlled for bias due to local nucleotide composition and alignability (Fig. 2 and Extended Data Figs 2 and 4) by comparing regions of interest to GC-content-matched control regions and normalizing by the signal from dechromatinized DNA (Extended Data Figs 4 and 5). The FLD was calculated in tenfold more shuffled windows of each size and background-subtracted. FLDs were then sorted by GC nucleotide content, divided into 300 bins and averaged within each bin to create a lookup table. To calculate FLFE for a given window, its background-subtracted FLD was divided by the interpolated histogram for the GC-content-matched window from the lookup table. The region of interest FLFE from irradiated cells was then divided by the FLFE calculated from irradiated genomic DNA smoothed with a 30-point moving average. Two-step clustering of normalized FLFEs was performed using k-means clustering with k =20 and 5 replicates followed by hierarchical clustering of the 20 cluster averages (MATLAB version 2013a) using a Euclidean distance metric; k-means cluster averages were median filtered with a 15 bp window and one cluster was dropped for only representing a single window before hierarchical clustering. Principal component analysis (MATLAB version 2013a) was used on raw FLFEs or on averages of the k-means clusters, after smoothing with a 30-point moving average and subtraction of a linear baseline fit by least squares. Annotation track values39,41 were averaged over 1 Mb windows and the averages were standardized as z-scores.
For RefSeq refGene, the number of annotated genes within each window was used as the value before calculating the z-score. These z-scores were averaged within clusters. Spearman’s correlation over windows was calculated between principal component score and baseline slope and the window average z-score of each annotation. The RICC-seq genome track was calculated as the inner product between the linear-baseline-subtracted FLFE in each 100 or 300 kb window and the first principal component of the 1 Mb window cluster averages.
Annotation-based fragment size distribution analysis
Control regions were randomly selected throughout the genome to match the size of broad peaks of histone modifications40 and narrow ATAC-seq peaks. FLD histograms were calculated in peaks and control regions for non-irradiated cells, irradiated cells, and irradiated genomic DNA. The non-irradiated background was scaled and subtracted from the other two signals. FLFE was calculated as the ratio between the peak and control region background-subtracted FLDs. To account for sequence bias, the irradiated cell FLFE was normalized by the irradiated genomic DNA FLFE.
Fragment length bias estimation
Length bias in sequenced library composition (which can arise from ssDNA fragment capture, adaptor ligation, PCR, and sequencing biases) was estimated by comparing the genome-wide FLD detected by 5′ 32P labelling and denaturing PAGE with the genome-wide FLD of sequenced fragments. Gel profiles were scanned with a storage phosphor screen (Molecular Devices) and imaged with a Typhoon 9400 Imager (GE Healthcare) and fragment lengths were interpolated from a denatured low molecular mass DNA ladder (New England Biolabs). The ratio between the sum-normalized sequencing and gel fragment length histograms was calculated and smoothed with a 100 bp moving average filter, then fitted to a single exponential. Fitted exponentials were calculated for three sequencing experiments and averaged to obtain the length bias profile to multiply by predicted FLDs for comparison with experimental length distributions.
RICC-seq signal to physical distance calibration model
Fragment 5′ and 3′ end positions were plotted relative to nucleosome dyads called with NucleoATAC41 with a minimum occupancy score of 0.5. Fragment counts were scaled for cell number and unique fragment counts, background-subtracted and length bias normalized. Three-dimensional distances were measured between 5′ phosphate phosphorus atom positions in the PDB 1KX5 structure26. Aggregated distance versus joint cleavage frequency data were fitted with a single exponential using nonlinear least squares with a trust region reflective algorithm (‘fit’, model ‘exp1’, using a trust-region algorithm, MATLAB version 2013a). Fits were also computed for individual strands and individual replicates.
Nucleosome footprinting
RICC-seq nucleosome footprinting signal was calculated as the total number of fragment ends at any base. For comparison, we reconstituted mononucleosomes from bacterially expressed Xenopus laevis histones on short dsDNA containing a single copy of the Widom 601 sequence45. Double-stranded DNA or reconstituted nucleosomes (2 nM) were incubated in 6 mM Tris-HCl, pH 8.0 with 0.5 mM Fe(III)Cl and 6 mM ascorbic acid. Reactions were incubated for 60 min at room temperature, then quenched with 1/10 volume of 50% glycerol. Reactions were ethanol precipitated and resuspended in a solution of in HiDi Formamide containing 500 ROX size standard (Fisher Scientific), then loaded onto a capillary sequencer (Applied Biosystems). Resulting traces were aligned using HiTrace46 and analysed by fitting a Gaussian peak and plotting the area under this curve as the intensity at each base.
Chromatin fibre structure simulation
Chromatin fibre structures spanning 0.8–4.4 nm of height per nucleosome and linker lengths of 20–70 bp were simulated using fibreModel27 in a basin-hopping simulation with 1,000 Monte Carlo steps, 30 nucleosomes, 10 segments per linker, 3 tail segments, a tolerance for the mean square force on each local minimization of 10−6, and a nucleosome flipping probability of 50%. Height per nucleosome was fixed. Resulting structures were energy-minimized using the fibreModel databaseparse operation, and the lowest-energy structure was retained for each combination of height per nucleosome and linker length. Nucleosome steric parameters were based on the PDB 1KX5 structure26. PDB files were generated from the database of structures using the beadbranch2pdb.py script in the fibreModel package27 and visualized with PyMOL(https://www.pymol.org/). Because this package can only place integral number of base pairs per linker segment, but simulated linker segments for some structures corresponded to non-integral multiples of true base pairs, the number of pseudo-base-pairs for the PDB per segment was rounded to the next highest integer.
Mononucleosome models
Approximate models of mononucleosomes with 30 bp linkers were generated by manually docking two straight 10 bp B-DNA segments (PDB 1DCV structure)47 onto the DNA ends of the 146 bp core nucleosome structure (PDB accession number 1KX3)26 or three 10 bp B-DNA segments onto the DNA ends of the linker histone H5 chromatosome structure (PDB accession number 4QLC)31 using PyMOL. The linkers were either docked to closely match the base pairing, which was slipped, leaving a 1 bp overhang (fine dock) or to approximately match the linker DNA tangent vector (rough dock). The H1 chro-matosome was obtained by selecting nucleosome 6 with 30 bp linkers from the docked fibre model fitted to cryo-electron microscopy density29. Molecular model visualization was performed with PyMOL and PDB files were edited with PyMOL and PDBEditor (http://bioinformatics.org/pdbeditor/PDBEditor.jar).
Fibre structure model comparisons
The locations of true base-pair phosphates were linearly interpolated from pseudo-base-pair coordinates using custom MATLAB (version 2013a) code and used to calculate pairwise 3D Euclidean distances for each structure. The exponential model for cleavage frequency versus distance calculated from DNA break mapping analysis was used to produce a probability distribution for each fragment spanning a pair of phosphates, and these fragments were then binned by length to generate a predicted FLD that was then multiplied by the approximated fragment length bias. Predicted FLDs were then scaled by the maximum value between 51 and 500 bp. Inner products were calculated between the predicted FLD for each structure and the maximum-scaled genome-wide experimental FLD over the range 148–500 bp. Predicted FLFEs were calculated as the ratio of the scaled FLD to the genome-wide experimental FLD. Their means were subtracted before calculating an inner product with the experimental FLFEs for ATAC-seq or chromatin modification peak sets. Structures were hierarchically clustered (MATLAB version 2013a, ‘clustergram’) on the basis of Euclidean distances between pairwise centres of nucleosomes within the fibre, on the basis of the relative strength of each contact type (N + 1 to N + 7) for each structure.
Accession numbers and data availability
Raw RICC-seq and BJ fibroblast ATAC-seq data, as well as an ATAC-seq insertion density track, peaks, nucleosome positions, RICC-seq V-plots, principal component score similarity tracks, and particular fragment size distribution histograms used to evaluate the crosslinked control and the size-selected libraries, have been deposited in the NCBI Gene Expression Omnibus48 under accession number GSE81807. All other data are available from the corresponding author upon reasonable request.
Code availability
Custom MATLAB and Python code is available upon request via GitHub repository (https://github.com/GreenleafLab/RICCseqTools).
Extended Data
Extended Data Figure 1. RICC-seq library preparation workflow.
Cells lysed in agarose plugs after irradiation are processed within the plugs to avoid mechanical shear of DNA. The 3′ phosphate ends (and all phosphates) are removed by alkaline phosphatase treatment, then 5′ phosphates are added back by polynucleotide kinase (T4 PNK) in preparation for ligation. Short 5′ phosphorylated ssDNA fragments are then passively eluted from agarose plugs after thermal denaturation and incorporated into paired-end sequencing libraries by ligation of random hexamer overhang adapters. Adaptor dimers are removed by size selection before and after PCR amplification of libraries.
Extended Data Figure 2. Reproducibility, sequence bias, robustness to crosslinking, and genome coverage of RICC-seq data sets.
a, Irradiated cell samples (300 Gy and 0 Gy) compared between two technical replicates and 300 Gy fragment size distributions compared after subtraction of scaled 0 Gy histograms. b, Fragment size distributions compared between three biological replicates. Owing to their low random DNA cleavage baselines, experiments 1 and 2 were used for genome-wide fragment length profile analysis and calibration of the exponential cleavage model (Figs 2–4). Experiment 3 has a higher background of non-specific breaks, probably because of mild nuclease contamination during the plug processing steps of the protocol. Although the low signal-to-background ratio of this experiment led us to exclude it from genome-wide distribution analysis, we did include it in V-plot and footprint generation to obtain higher sequencing coverage of ~78 bp fragments (Fig. 1 and Extended Data Fig. 3). Background subtraction of 0 Gy signal is as in a. c, Fragment end sequence bias for all irradiation conditions in an experiment with machine-mixed random hexamer adaptor oligonucleotides. d, Fragment end sequence bias in biological replicate processed with hand-mixed random hexamer adaptor oligonucleotides. e, Raw fragment size distributions obtained from live and formaldehyde/EGS-crosslinked cells (subsampled to same sequencing depth). f, Normalized expected RICC-seq fragment density from cellular chromatin (red) or dechromatinized DNA (black) estimated by dividing the number of fragments within annotated peaks by the number within size-matched control regions elsewhere in the genome (error bars represent s.e.m. with n =3 independent experiments; centre value is mean). g, Ratio of cellular fragment density ratio to genomic DNA fragment density ratio from f. h, P values for comparisons in g (Mann–Whitney U test; n =3 biological replicates).
Extended Data Figure 3. RICC-seq produces characteristic 78 nt fragments around positioned nucleosomes and protection footprints at CTCF-bound sites.
a, V-plot of dechromatinized DNA control for Fig. 1f. b, V-plot of RICC-seq fragments from cells around unoccupied CTCF motifs without a CTCF ChIP-seq peak (see Methods). c, Difference in counts of RICC-seq fragment ends piled up over aggregated CTCF motifs within (bound) or outside (not bound) ChIP peaks. d, V-plot of cell and dechromatinized DNA around active (associated with RNA-CAGE peaks) transcription start sites (see Methods). e, V-plot of cell fragments around inactive TSSs (lacking an ATAC-seq peak in BJ fibroblasts). Grey ovals are cartoon representations of nucleosome positions and phasing. Data for all panels were pooled from three biological replicates.
Extended Data Figure 4. Calculation of FLFE normalizes sequence bias in the RICC-seq fragment size distribution throughout the genome.
a, Workflow for calculating and normalizing FLFE. b, Fragment length distributions in 1 Mb windows sorted by GC content. c, FLFE with respect to lookup table of GC content-binned genome-wide average FLD in equal-sized windows. d, Z-scores of chromatin annotations in same 1 Mb windows. See Methods for annotation details. Data are pooled from two biological replicates.
Extended Data Figure 5. FLFE PC1 and regional profiles are reproducible across biological replicates.
a, PC1 calculated independently from each of two biological replicates. b, Region FLFE profiles calculated for additional annotations. ATAC-seq, H3K9me3-only, and H3K27me3 (excluding H3K9me3 or H3K4me1) are as in Fig. 2. Euchromatin-associated annotations are on a blue background. c, For two annotation regions, reproducibility of regional FLFE profiles between two biological replicates is shown in terms of the raw FLFE profiles (top), the FLFE in de-chromatinized DNA samples that quantifies sequence bias (middle), and the GC baseline normalized FLFE (bottom). Grey shaded areas represent peaks as in Fig. 1.
Extended Data Figure 6. PC1 signal is correlated with ATAC-seq signal and anti-correlated with H3K9me3 ChIP signal.
a, b, Example tracks of RICC-seq PC1 signal for combined biological replicates and each replicate separately in the same region of chromosome 5 as in Fig. 2 (at higher zoom) and in a region of chromosome 3. c, Correlation between replicates for PC1 score. d, Correlation of PC1 score from combined replicates with euchromatic (ATAC-seq) and heterochromatic (H3K9me3 or H3K27me3) annotations in 300 kb windows. ATAC-seq data are shown as insertion density, while H3K9me3 and H3K27me3 signals are shown as −log10(Poisson P value over local background) (see Methods).
Extended Data Figure 7. Fragment length bias correction. Bias was corrected by comparing fragment size distributions between sequencing data.
(a) and intensity profiles from 5′ end-labelled denaturing gel electrophoresis of RICC-seq ssDNA fragments before sequencing library preparation (b). c, End-labelled fragments have a length distribution weighted more towards long fragments. d, The ratio of the two distributions (sequencing/gel) was calculated and smoothed, then (e) fitted with a single exponential to obtain an estimate of the length bias. Bias estimate is shown for three biological replicates.
Extended Data Figure 8. RICC-seq signal from positioned nucleosomes reflects known nucleosome structure.
a, V-plot of RICC-seq fragments around ATAC-seq called nucleosome positions. b, Exponential fits of background-subtracted and length bias corrected (Extended Data Fig. 7) fragment counts to the 3D distance between the end positions of each fragment shown for individual replicates and individual strands. c, Example showing the magnitude of predicted FLD changes for a simulated fibre structure (see Methods) between the calibration values obtained from biological replicate experiments. d, Cleavage locations (fragment ends) were mapped around nucleosome dyad centres called by ATAC-seq (see Methods). Background-subtracted (0 Gy), sequencing depth, and cell-number-scaled fragment end counts are shown for the positive and negative reference genome strand. Blue trace shows cleavage frequency from in vitro hydroxyl-radical footprinted reconstituted nucleosome (scaled to comparable amplitude).
Extended Data Figure 9. Clustering fibre structures into five classes on the basis of helical symmetry. a, Example fibre structure illustrating nucleosome and fibre coordinates.
b, Energy (log10(kBT)) per nucleosome for each minimum-energy structure. c, Distribution of linker DNA crossing distance for each minimum-energy structure at every linker length and compaction level combination. d, e, Maps of nucleosome tilt angles (nucleosome axis relative to fibre axis) (d) and rotation angles around fibre for adjacent nucleosomes (e). f, g, Distance between centres of second-nearest-neighbour (f, N to N + 2) and third-nearest-neighbour (g, N to N + 3) nucleosomes in each fibre model. h, Hierarchical clustering of the 969 candidate structures into five classes on the basis of the distribution of nucleosome centre distances for intervals between N to N + 1 (adjacent nucleosomes) and N to N + 7. The matrix was row-standardized before clustering to measure the relative intensity of contact distances for each structure. Classes are coloured as in Fig. 4b. i, Mean with bands (one s.d.) of predicted FLFE plotted for each class of structure. j, Mean energy of the structures in each class. k, Comparison of predicted FLD between a solenoid structure and an extended zig-zag structure without length bias correction for 51–2,000 bp fragments. l, Measured RICC-seq FLD beyond 500 nt before (left) and after (right) subtraction of an exponential baseline. k, Comparison of predicted FLD between a solenoid structure and an extended zig-zag structure without length bias correction for 51–2,000 bp fragments. m, Example FLFEs of structure models with varying linker length. n, Example FLFEs of structure models with varying compaction level.
Extended Data Figure 10. Two-start helix structures (class 1) are consistent with genome-wide average chromatin structure, facultative heterochromatin, and a reconstituted fibre containing linker histone, while bent linkers are consistent with open chromatin.
a, Map of similarity scores between the predicted FLFEs of simulated fibre structures and the genome-wide experimental FLFE, shown in c, measured as the ratio of the genome-wide FLD to the genome-wide dechromatinized DNA FLD. b, Mean similarity scores as calculated in a for each structure class. c, Genome-wide experimental FLFE calculated with respect to the dechromatinized DNA FLD after a one-parameter fit to obtain equivalent scaling of read counts. d, Top view and side view of reconstituted chromatin fibre with NRL =187 bp (40 bp linker length)29. e, Similarity score between predicted FLDs for each simulated fibre structure and the FLFE of the reconstituted fibre structure. Linker length of the reconstituted fibre is indicated. f, Mean similarity scores from e, for each structure class. g, Predicted FLD for the reconstituted fibre and genome-wide experimental FLD. h, Fragments with characteristic lengths (red) labelled on the tetranucleosome unit of the reconstituted fibre. i, Similarity score between predicted FLFE for the fibre structure candidates and the experimental FLFE from H3K27me3 peaks with no H3K9me3 or H3K4 me1 ChIP peak overlap, with (j) averages of similarity score within clusters as in Fig. 4b. Class 1 regions (Fig. 4b) are outlined in black in a, e, and i. k, Comparison of FLFEs from H3K27me3 ChIP peaks with H3K9me3 FLFE and predicted FLFE from a high-similarity-score structure. l, Mononucleosome models built by adding 10 bp straight DNA linker pieces to achieve 30 bp linker lengths. Structures are based on the core nucleosome crystal structure, chromatosome crystal structure with linker histone H5, and a chromatin fibre with linker histone H1 (see Methods). The predicted FLD and FLFE with respect to the genome-wide aggregate FLD is shown for each structure. m, The FLFE and FLD in three chromatin states shown for comparison. n, Predicted chromatosome FLDs for extended zig-zag and solenoid structures. Curves in l–n are scaled by the maximum of the 78 bp peak.
Supplementary Material
Acknowledgments
We acknowledge R. Das for conversations and help with capillary sequencing, E. Koslover, A. Spakowitz, and A. Schep for sharing code and sample data, discussions, and reading the manuscript, A. Kathiria for technical assistance, J. Buenrostro and C. Araya for advice and sharing code, I. Whitehouse for sharing a protocol, C. Fuller for nucleosome reconstitution help, P. Zhu and G. Li for sharing their chromatin fibre model, T. Phillips, G. J. Gu, O. Rando and W. Johnson for discussions, and G. Wang for BJ cells. BJ-5ta cells were a gift from J. Cochran. V.I.R. acknowledges the support of the Walter V. and Idun Berry Postdoctoral Fellowship. S.K.D. acknowledges support from a National Institutes of Health (NIH) Predoctoral Molecular Biophysics Training Program grant to Stanford University and from a National Science Foundation Graduate Fellowship. A.F.S. acknowledges support from National Institutes of Health (NIH) grant R01GM106005. W.J.G. acknowledges NIH grants R21HG007726, and P50HG00773501. This work was supported by the Rita Allen Foundation, the Baxter Foundation, and the Human Frontier Science Program.
Footnotes
Supplementary Information is available in the online version of the paper.
Author Contributions V.I.R., S.K.D., A.F.S., and W.J.G. designed experiments. V.I.R. and S.K.D. performed experiments. V.I.R. and S.K.D. analysed data. V.I.R., A.F.S., and W.J.G. interpreted the results and wrote the paper.
The authors declare competing financial interests: details are available in the online version of the paper.
Readers are welcome to comment on the online version of the paper.
Reviewer Information Nature thanks S. Grigoryev and the other anonymous reviewer(s) for their contribution to the peer review of this work.
References
- 1.Li G, Reinberg D. Chromatin higher-order structures and gene regulation. Curr Opin Genet Dev. 2011;21:175–186. doi: 10.1016/j.gde.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luger K, Dechassa ML, Tremethick DJ. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nature Rev Mol Cell Biol. 2012;13:436–447. doi: 10.1038/nrm3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilbert N, et al. Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell. 2004;118:555–566. doi: 10.1016/j.cell.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 4.Risca VI, Greenleaf WJ. Unraveling the 3D genome: genomics tools for multiscale exploration. Trends Genet. 2015;31:357–372. doi: 10.1016/j.tig.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bednar J, et al. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc Natl Acad Sci USA. 1998;95:14173–14178. doi: 10.1073/pnas.95.24.14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dorigo B, et al. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306:1571–1573. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- 7.Horowitz RA, Agard DA, Sedat JW, Woodcock CL. The three-dimensional architecture of chromatin in situ: electron tomography reveals fibers composed of a continuously variable zig-zag nucleosomal ribbon. J Cell Biol. 1994;125:1–10. doi: 10.1083/jcb.125.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schalch T, Duda S, Sargent DF, Richmond TJ. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 9.Finch JT, Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci USA. 1976;73:1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robinson PJ, Fairall L, Huynh VA, Rhodes D. EM measurements define the dimensions of the “30-nm” chromatin fiber: evidence for a compact, interdigitated structure. Proc Natl Acad Sci USA. 2006;103:6506–6511. doi: 10.1073/pnas.0601212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Widom J, Klug A. Structure of the 300A chromatin filament: X-ray diffraction from oriented samples. Cell. 1985;43:207–213. doi: 10.1016/0092-8674(85)90025-x. [DOI] [PubMed] [Google Scholar]
- 12.Routh A, Sandin S, Rhodes D. Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc Natl Acad Sci USA. 2008;105:8872–8877. doi: 10.1073/pnas.0802336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boettiger AN, et al. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature. 2016;529:418–422. doi: 10.1038/nature16496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsieh THS, et al. Mapping nucleosome resolution chromosome folding in yeast by micro-C. Cell. 2015;162:108–119. doi: 10.1016/j.cell.2015.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rydberg B, Holley WR, Mian IS, Chatterjee A. Chromatin conformation in living cells: support for a zig-zag model of the 30 nm chromatin fiber. J Mol Biol. 1998;284:71–84. doi: 10.1006/jmbi.1998.2150. [DOI] [PubMed] [Google Scholar]
- 16.Scheffer MP, Eltsov M, Frangakis AS. Evidence for short-range helical order in the 30-nm chromatin fibers of erythrocyte nuclei. Proc Natl Acad Sci USA. 2011;108:16992–16997. doi: 10.1073/pnas.1108268108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grigoryev SA, et al. Hierarchical looping of zigzag nucleosome chains in metaphase chromosomes. Proc Natl Acad Sci USA. 2016;113:1238–1243. doi: 10.1073/pnas.1518280113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eltsov M, Maclellan KM, Maeshima K, Frangakis AS, Dubochet J. Analysis of cryo-electron microscopy images does not support the existence of 30-nm chromatin fibers in mitotic chromosomes in situ. Proc Natl Acad Sci USA. 2008;105:19732–19737. doi: 10.1073/pnas.0810057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishino Y, et al. Human mitotic chromosomes consist predominantly of irregularly folded nucleosome fibres without a 30-nm chromatin structure. EMBO J. 2012;31:1644–1653. doi: 10.1038/emboj.2012.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alpen EL. Radiation Biophysics. 2. Academic; 1998. pp. 104–131. [Google Scholar]
- 21.Roots R, Kraft G, Gosschalk E. The formation of radiation-induced DNA breaks: the ratio of double-strand breaks to single-strand breaks. Int J Radiat Oncol Biol Phys. 1985;11:259–265. doi: 10.1016/0360-3016(85)90147-6. [DOI] [PubMed] [Google Scholar]
- 22.Valouev A, et al. Determinants of nucleosome organization in primary human cells. Nature. 2011;474:516–520. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tullius TD, Dombroski BA. Hydroxyl radical “footprinting”: high-resolution information about DNA-protein contacts and application to lambda repressor and Cro protein. Proc Natl Acad Sci USA. 1986;83:5469–5473. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bintu L, et al. Dynamics of epigenetic regulation at the single-cell level. Science. 2016;351:720–724. doi: 10.1126/science.aab2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J Mol Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 27.Koslover EF, Fuller CJ, Straight AF, Spakowitz AJ. Local geometry and elasticity in compact chromatin structure. Biophys J. 2010;99:3941–3950. doi: 10.1016/j.bpj.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Widom J. A relationship between the helical twist of DNA and the ordered positioning of nucleosomes in all eukaryotic cells. Proc Natl Acad Sci USA. 1992;89:1095–1099. doi: 10.1073/pnas.89.3.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song F, et al. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science. 2014;344:376–380. doi: 10.1126/science.1251413. [DOI] [PubMed] [Google Scholar]
- 30.Fan JY, Rangasamy D, Luger K, Tremethick DJ. H2A.Z alters the nucleosome surface to promote HP1 α-mediated chromatin fiber folding. Mol Cell. 2004;16:655–661. doi: 10.1016/j.molcel.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 31.Zhou BR, et al. Structural mechanisms of nucleosome recognition by linker histones. Mol Cell. 2015;59:628–638. doi: 10.1016/j.molcel.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grigoryev SA, Arya G, Correll S, Woodcock CL, Schlick T. Evidence for heteromorphic chromatin fibers from analysis of nucleosome interactions. Proc Natl Acad Sci USA. 2009;106:13317–13322. doi: 10.1073/pnas.0903280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J, Russell DW. Isolation of DNA fragments from polyacrylamide gels by the crush and soak method. CSH Protoc. 2006 doi: 10.1101/pdb.prot2936. http://dx.doi.org/10.1101/pdb.prot2936. [DOI] [PubMed]
- 35.Smith DJ, Whitehouse I. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature. 2012;483:434–438. doi: 10.1038/nature10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corces MR, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nature Genet. 2016;48:1193–1203. doi: 10.1038/ng.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kheradpour P, Kellis M. Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic Acids Res. 2014;42:2976–2987. doi: 10.1093/nar/gkt1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kundaje A, et al. Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schep AN, et al. Structured nucleosome fingerprints enable high-resolution mapping of chromatin architecture within regulatory regions. Genome Res. 2015;25:1757–1770. doi: 10.1101/gr.192294.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henikoff JG, Belsky JA, Krassovsky K, MacAlpine DM, Henikoff S. Epigenome characterization at single base-pair resolution. Proc Natl Acad Sci USA. 2011;108:18318–18323. doi: 10.1073/pnas.1110731108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guse A, Fuller CJ, Straight AF. A cell-free system for functional centromere and kinetochore assembly. Nature Protocols. 2012;7:1847–1869. doi: 10.1038/nprot.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoon S, et al. HiTRACE: high-throughput robust analysis for capillary electrophoresis. Bioinformatics. 2011;27:1798–1805. doi: 10.1093/bioinformatics/btr277. [DOI] [PubMed] [Google Scholar]
- 47.Eichman BF, Vargason JM, Mooers BH, Ho PS. The Holliday junction in an inverted repeat DNA sequence: sequence effects on the structure of four-way junctions. Proc Natl Acad Sci USA. 2000;97:3971–3976. doi: 10.1073/pnas.97.8.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.