Abstract
Bacteria use a process called quorum sensing to communicate and orchestrate collective behaviors including virulence factor secretion and biofilm formation. Quorum sensing relies on production, release, accumulation, and population-wide detection of signal molecules called autoinducers. Here, we develop concepts to coat surfaces with quorum-sensing-manipulation molecules as a method to control collective behaviors. We probe this strategy using Staphylococcus aureus. Pro- and anti-quorum-sensing molecules can be covalently attached to surfaces using click chemistry, where they retain their abilities to influence bacterial behaviors. We investigate key features of the compounds, linkers, and surfaces necessary to appropriately position molecules to interact with cognate receptors, and the ability of modified surfaces to resist long-term storage, repeated infections, host plasma components, and flow-generated stresses. Our studies highlight how this surface approach can be used to make colonization-resistant materials against S. aureus and other pathogens and how the approach can be adapted to promote beneficial behaviors of bacteria on surfaces.
Introduction
Bacteria frequently act as members of collectives to create surface-attached communities called biofilms and to activate production of virulence factors1–5. These and other bacterial group behaviors are regulated by a cell-cell chemical communication process called quorum sensing, which involves the production, release, and population-wide detection of extracellular signal molecules called autoinducers6–8. There is significant interest in developing synthetic strategies to manipulate quorum sensing. Potential applications include prevention of biofilm formation to reduce biofouling in industry and to halt recurrent disease in medicine, and suppression of virulence factor production to limit pathogen infectivity9–11. There is also interest in enhancing quorum-sensing-controlled behaviors in beneficial bacteria. For example, bacterial biofilm formation is essential in wastewater treatment and food processing, and, in the context of the microbiome, commensal bacteria play roles in fending off invading pathogens, presumably by relying on quorum-sensing-controlled traits12,13. Indeed, anti- and pro-quorum-sensing compounds have been developed and show promise in relevant model systems14–17.
Successful administration of anti- and pro-quorum-sensing compounds will require overcoming barriers such as lack of or inefficient compound transport to the site of action, for example, due to limited compound solubility or stability, lack of compound delivery to sites with complex topography (i.e., grooves/crevices), and resistance to chemical perturbations such as in pre-existing biofilms18–21. These issues complicate therapies, often necessitating delivery of large and frequent doses of the active compounds in the bulk phase, which in turn, can cause complications such as cytoxicity or off target effects. Moreover, chemotherapies are typically applied subsequent to bacterial colonization, often in life-threatening scenarios.
To circumvent the issues that commonly confound chemo- and other therapies, here we explore the biomedical promise of active quorum-sensing compounds tethered to surfaces, which would allow site-specific targeting appropriate for a wide range of applications. Previous investigations of this theme include surface-coatings designed to rapidly release quorum-sensing inhibitory compounds22 and attachment of dihydropyrolones to glass surfaces to suppress bacterial adhesion23. These earlier studies showed that such modified surfaces could reduce bacterial colonization, at least in the short-term. However, it is not known whether pro- and anti-quorum-sensing compounds covalently attached to surfaces function by influencing bacterial collective behaviors, and if so, what features of the compounds, linkers, and surfaces are key, if chemically-modified surfaces will remain resilient to long-term storage, to flow-generated stress, to the presence of host plasma components, and to repeated infection, and whether surfaces can be multi-functionalized with combinations of biomolecules with different activities. Here, we focus on surface coatings designed to manipulate quorum sensing in Staphylococcus aureus, the pathogen responsible for multiple fatal diseases including bacteremia, toxic shock syndrome, and medical device-related infections, many strains of which are multi-drug resistant (i.e., S. aureus MRSA)24,25. In this context, the Agr quorum-sensing system plays a central regulatory role in S. aureus pathogenicity and biofilm dynamics1,8.
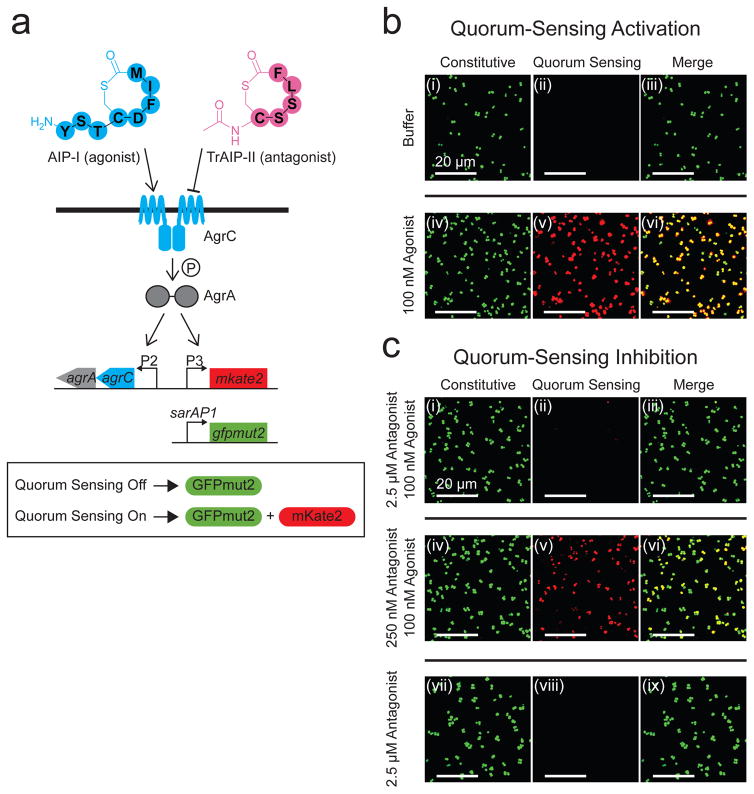
S. aureus Agr quorum sensing is driven by an autoinducer peptide (called AIP) harboring a thiolactone ring and an exocyclic tail at the N-terminus. AIP is processed from the precursor peptide AgrD by AgrB and other proteases, and the AIP is secreted26,27. Extracellular AIP is detected by a cognate transmembrane-bound receptor histidine kinase, AgrC, that upon AIP binding, autophosphorylates and subsequently funnels a phosphoryl group to the partner response regulator, AgrA28,29 (Fig. 1a). Phospho-AgrA activates the agrP3 promoter driving transcription of RNAIII that has multiple roles30. RNAIII functions as an mRNA that encodes δ-toxin (a membrane disrupting exo-protein that lyses eukaryotic host cells), and RNAIII also participates in regulation of other genes required for exo-toxin secretion and biofilm disassembly31. Detection of AIP launches the autoinduction positive feedback loop that increases AIP production, resulting in amplification of the quorum-sensing response8. There are four S. aureus Agr allelic variants (I to IV) that make four AIPs differing only in a few amino acid residues. AIPs activate quorum sensing in the S. aureus cells that produce them, and they generally inhibit quorum sensing in heterologous S. aureus cells possessing different AIP variants8. In the S. aureus agr-I strain that we study, AIP-I is the native autoinducer. TrAIP-II, a truncated AIP-II with the exocyclic tail replaced by an acetyl group, is a universal inhibitor for all four S. aureus Agr quorum-sensing systems32. TrAIP-II competes with the cognate AIPs for binding to the receptor32. Throughout this work, we focus on S. aureus agr-I because it possesses the most prevalent Agr type found world-wide in nosocomial infections. We use AIP-I as an autoinducer agonist and we use TrAIP-II as a competitive antagonist (Fig. 1a).
Figure 1. A strategy to quantify the Agr quorum-sensing responses of S. aureus to exogenously supplied agonists and antagonists.
a, The S. aureus reporter strain used in this study. b, Fluorescence images of the S. aureus reporter strain from panel a after 3 h incubation with buffer or buffer containing 100 nM AIP-I. Left panels (i) and (iv): constitutive sarAP1-gfpmut2 reporter. Middle panels (ii) and (v): quorum-sensing-controlled agrP3-mkate2 reporter. In S. aureus, sarAP1 is expressed constitutively and drives expression of the gene encoding the transcription factor SarA, and agrP3 is activated by AgrA~P in response to quorum sensing and AIP accumulation. Right panels (iii) and (vi): merged images from the left and middle panels. Top panels (i), (ii), and (iii): buffer. Bottom panels (iv), (v), and (vi): 100 nM AIP-I. c, Fluorescence images of the S. aureus reporter strain following 3 h incubation with TrAIP-II or combinations of AIP-I and TrAIP-II. Left panels (i), (iv), and (vii): constitutive sarAP1-gfpmut2 reporter. Middle panels (ii), (v), and (viii): quorum-sensing-controlled agrP3-mkate2 reporter. Right panels (iii), (vi), and (ix): merged images from the left and middle panels. Top panels (i), (ii), and (iii): 2.5 μM TrAIP-II + 100 nM AIP-I. Middle panels (iv), (v), and (vi): 250 nM TrAIP-II + 100 nM AIP-I. Bottom panels (vii), (viii), and (ix): 2.5 μM TrAIP-II. In panels b and c, images are based on n = 3 independent replicates. Scale bars: 20 μm.
Results
To follow the quorum-sensing status of S. aureus cells growing on unmodified and chemically-modified surfaces, we used confocal microscopy and an S. aureus reporter strain that produces the fluorescent protein mKate2 in response to exogenous addition of AIP-I. The reporter strain is a ΔagrBDCA ΔRNAIII S. aureus mutant harboring a multicopy plasmid carrying agrCA driven by the native agrP2 promoter. Thus, the strain does not produce endogenous AIP-I but plasmid restoration of AgrC-I and AgrA endows the strain with the capability to detect AIP-I if it is supplied exogenously. The plasmid also harbors the Agr-activated agrP3 promoter fused to mkate2. Therefore, in response to exogenously provided AIP-I, the reporter strain fluoresces red. This quorum-sensing response can be repressed by the administration of TrAIP-II (Fig. 1a). A constitutively expressed sarAP1-gfpmut2 gene was introduced onto the chromosome to enable normalization of quorum-sensing responses18. We validated the reliability of our reporter strain prior to examining the effects of surface-attached quorum-sensing modulators, noting some heterogeneity in expression level (Fig. S1 and accompanying text).
To demonstrate that the S. aureus reporter strain faithfully reports on Agr quorum sensing, we incubated it in the absence and presence of 100 nM AIP-I (i.e., the agonist). GFPmut2 was produced irrespective of the presence of AIP-I, however, mKate2 was produced only when AIP-I was supplied to the cells (Fig. 1b). When the reporter strain was simultaneously provided with 100 nM AIP-I and 2.5 μM TrAIP-II (i.e., the antagonist), little mKate2 production occurred, indicating inhibition of Agr quorum sensing (Fig. 1c (ii)). Reducing the TrAIP-II antagonist concentration by 10-fold caused a corresponding 9-fold decrease in quorum-sensing inhibition (Fig. 1c (v)). Addition of TrAIP-II alone elicited no mKate2 production (Fig. 1c (viii)).
Surface-attached AIP-I activates quorum sensing
Our overarching goal is to investigate whether we can positively and negatively manipulate S. aureus Agr quorum sensing using surface-attached compounds. To achieve this goal, we employed copper-catalyzed azide-alkyne cyclo-addition click chemistry to attach active compounds to surfaces33. We chose polyethylene glycol (PEG) polymers decorated with azide groups as the surface linkers because PEG is flexible, hydrophilic, and non-bulky, and we used glass as a model surface material.
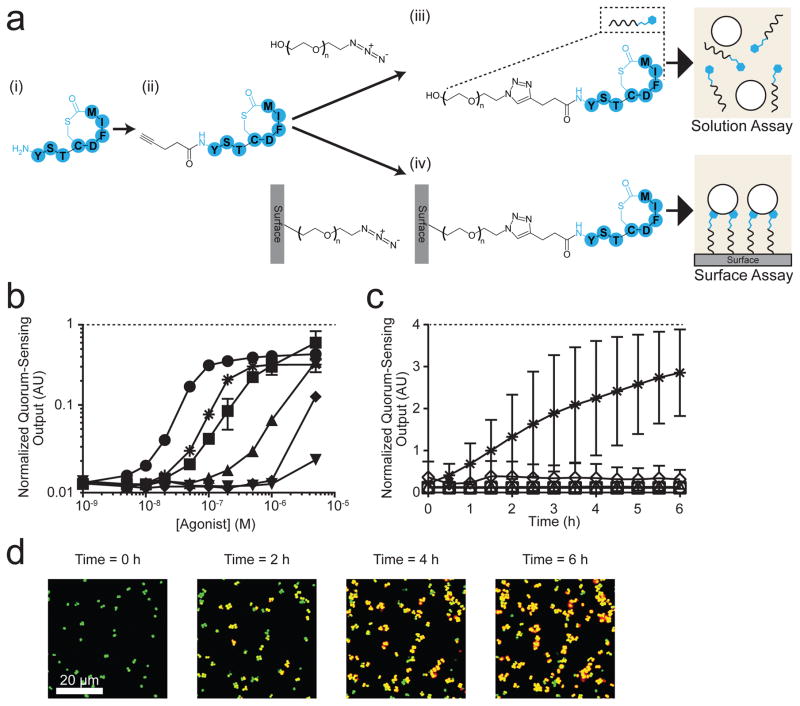
To make the AIP-I amenable to surface attachment, we synthesized AIP-I containing an alkyne at the N-terminus (Fig. S2a and Fig. S3a, and Methods). We call this compound “Alkyne-AIP-I” (Fig. 2a). Prior to attaching the Alkyne-AIP-I to the surface linkers, we carried out the click reaction between the Alkyne-AIP-I and a surface-free version of the PEG330-azide linker to test if the AIP-I derivatives retained their activation capability following the click reaction. The reaction between the alkyne and azide produces a triazole ring linking AIP-I to the PEG330 polymer. We call this compound “PEG330-triazole-AIP-I” (Fig. 2a (iii), Fig. S2a and Fig. S3b). In solution, AIP-I, Alkyne-AIP-I, and PEG330-triazole-AIP-I activated Agr quorum sensing in the S. aureus reporter strain with similar efficacy albeit with different potencies (see “Solution Assay”, Fig. 2a (iii) and Fig. 2b). The calculated EC50 values were 28 nM (±3) for AIP-I, 190 nM (±40) for Alkyne-AIP-I, and 1.1 μM (±0.20) for PEG330-triazole-AIP-I (Table 1). Addition of 2.5 μM TrAIP-II repressed the Agr quorum-sensing response to all three compounds by a comparable magnitude (Fig. 2b). We interpret these results to mean that, analogous to AIP-I, both Alkyne-AIP-I and the PEG330-triazole-AIP-I activate quorum sensing by targeting the AgrC-I receptor.
Figure 2. S. aureus Agr quorum sensing is activated by surface-attached AIP-I.
a, Strategy for modification of AIP-I and attachment to the surface; (i) AIP-I, (ii) Alkyne-AIP-I, (iii) PEG330-triazole-AIP-I, and (iv) Surface-PEG10000-triazole-AIP-I. The PEG subscript denotes the molecular weight. b, The normalized S. aureus Agr quorum-sensing output, which is defined as the quorum-sensing-controlled reporter output divided by the output from the constitutive reporter, in response to AIP-I (circles), Alkyne-AIP-I (squares), PEG330-triazole-AIP-I (triangles), AIP-I + 2.5 μM TrAIP-II (asterisks), Alkyne-AIP-I + 2.5 μM TrAIP-II (diamonds), and PEG330-triazole-AIP-I + 2.5 μM TrAIP-II (upside down triangles). Data points indicate means and error bars denote standard deviations from triplicate experiments. c, The normalized Agr quorum-sensing output from the S. aureus reporter strain measured as a function of time in the presence of Surface-PEG10000-triazole-AIP-I (asterisks), Surface-PEG10000-triazole-AIP-I + 2.5 μM TrAIP-II in solution (diamonds), Surface-PEG10000 (squares), Surface-PEG10000-azide (circles), and Surface-PEG10000-triazole-Linear-AIP-I (triangles). See Fig. S5 for details of the chemical procedures in each panel. Normalized quorum-sensing outputs were measured as means from 1000 – 4000 individual cells in each experiment. Data points indicate means and error bars denote standard deviations from triplicate experiments. An ANOVA test with Tukey-Kramer post hoc analysis was used to assess the statistical significance between the means for the AIP-I coated surface and the control surfaces using the T = 6 h data (P < 0.0001 for all pairwise comparisons). d, Representative merged fluorescence images of the S. aureus reporter strain on the Surface-PEG10000-triazole-AIP-I at T = 0, 2, 4, and 6 h. Images are based on n = 3 independent experiments; one representative image for each condition was chosen from ~ 50 images acquired from different regions of each surface. Scale bars: 20 μm. AU: arbitrary units.
Table 1.
EC50 values (from n = 3 experiments) for AIP-I and its derivatives and IC50 values (from n = 4 experiments) for TrAIP-II and its derivatives when AIP-I is present at 100 nM (i.e., the EC95).
| EC50 (nM) | AIP-I | 28 (± 3) |
| Alkyne-AIP-I | 190 (± 40) | |
| PEG330-triazole-AIP-I | 1100 (± 200) | |
| IC50 (μM) | TrAIP-II | 1.5 (± 0.5) |
| Alkyne-TrAIP-II | 0.21 (± 0.05) | |
| PEG330-triazole-TrAIP-II | 3.1 (± 1.5) |
For the surface modification work, we used silanization and maleimide-thiol chemistry to decorate the surface with PEG10000 polymers carrying azide moieties at one end (Fig. S4 and Methods). We name this unit the “Surface-PEG10000-azide”. Calculations of the height of the PEG brush34 suggest that a PEG10000 polymer would have sufficient length to span the peptidoglycan layer and position the attached AIP-I to interact with the AgrC-I receptor located on the cell membrane. We could not, however, carry out our companion solution assay with the PEG10000 polymer attached to AIP-I due to limitations in purification and characterization of soluble PEG10000 entities. In solution, PEG10000 polymers adopt a random-coiled conformation that does not mimic the extended conformation they possess when they are attached to surfaces with high grafting density34. We carried out the click reaction to covalently attach the Alkyne-AIP-I to the Surface-PEG10000-azide (Fig. 2a (iv)), generating “Surface-PEG10000-triazole-AIP-I”. To examine whether the PEG-triazole-AIP-I moiety remained functional when attached to the surface, we provided the S. aureus reporter strain to the Surface-PEG10000-triazole-AIP-I in microfluidic chambers. The Agr quorum-sensing response was induced (see “Surface Assay”, Fig 2a (iv) and Fig. 2c). Compared to T = 0 h when cells were in the quorum-sensing-off mode, the response was activated over time and reached an average of 25-fold activation at T = 6 h (Fig. 2c,d and S5a, and Supplementary Movie 1). These results suggest that the surface-attached AIP-I is indeed recognized as an autoinducer by the cognate membrane-bound AgrC-I receptor. To confirm our interpretation of surface-tethered AIP-I eliciting the Agr quorum-sensing response in S. aureus, 2.5 μM TrAIP-II antagonist was provided in solution to the S. aureus cells residing on the AIP-I coated surface. The Agr quorum-sensing response was repressed (Fig. 2c and Fig. S5b (i)).
To reinforce the above results, we show that S. aureus did not activate a quorum-sensing response when introduced onto the identical surface lacking the triazole-AIP-I decoration (Surface-PEG10000) or that had not undergone the click reaction (Surface-PEG10000-azide) (Fig. 2c, Fig. S5b (ii) and (iii)). Furthermore, the reporter strain that was presented a surface coated with the identical PEG10000 polymer attached, via a triazol ring, to a ring-opened version of AIP-I did not elicit a response (see compound 6 in Fig. S2a, Fig. 2c and Fig. S5b (iv); called Surface-PEG10000-triazole-Linear-AIP-I). Identical results were obtained using the Surface-PEG10000-triazole-AIP-I in which the thioester ring was opened via treatment with 10 mM cysteine (Fig. S5b (v)). Consistent with these results, an S. aureus reporter strain lacking AgrC-I did not respond to the surface-attached AIP-I (Fig. S5b (vi)). Thus, surface-attached AIP-I specifically and reversibly binds to AgrC-I, indicating that it functions as an autoinducer.
Crucial features of surface-attached quorum-sensing molecules
We investigated the requirements for the surface-attached AIP-I to bind AgrC-I and activate Agr quorum sensing in S. aureus by changing particular features of the tethered molecules. Presumably, AgrC-I diffuses freely in the membrane35. In S. aureus, the plasma membrane is covered by a 15–30 nm peptidoglycan layer with 4–5 nm diameter pores36. Peptidoglycan is considered to be an elastic mesh-network that can expand, contract, and tolerate transport of globular molecules of up to 100 kDa36. First, we examined how altering the length of the polymer attaching the AIP-I to the surface influenced the S. aureus Agr quorum-sensing response. We attached AIP-I with two different surface linkers: Surface-PEG10000-azide and Surface-PEG400-azide. Using atomic force microscopy, the heights of the linkers were measured to be 26.5 nm (±2.5) and 1.9 nm (±0.2), respectively. The calculated Flory radius of PEG10000 in solution is ~10 nm34, indicating that the surface-attached PEG10000 exists in an extended configuration due to local crowding. This configuration suggests that the terminal functional groups are exposed and thus could properly position attached AIP-I molecules to interact with AgrC-I receptors. Indeed, we confirmed that the azide moieties on both polymers were amenable to the click reaction using alkyne-functionalized dyes (Surface-PEG10000-azide, Fig. S4c and Surface-PEG400-azide, Fig. S6a). Finally, we attached the Alkyne-AIP-I to both surfaces, and found that the Surface-PEG10000-triazole-AIP-I activated the S. aureus Agr quorum-sensing response whereas the Surface-PEG400-triazole-AIP-I did not (Fig. S6b,c). We presume that the latter result is due to a geometrical restriction related to the PEG400 length, and this shorter linker does not appropriately position the AIP-I to access AgrC-I receptors on the plasma membrane. As a control, we show that the S. aureus reporter strain could activate Agr quorum sensing on the Surface-PEG400-triazole-AIP-I if it was also provided with 50 nM AIP-I in solution (Fig. S6b and Fig. S6c (iii)).
To examine how surface coverage density of PEG10000-triazole-AIP-I affects activation of AgrC-I-directed quorum sensing, we measured the number of reacted azides in a unit area on the Surface-PEG10000-azide following the click reaction. To do this, we clicked a fluorescent dye harboring an alkyne to the Surface-PEG10000-azide (Fig. S4c (ii)). The intensity of fluorescence from the surface is directly proportional to the number of reacted azide moieties. We obtained an average intensity for each dye molecule (Fig. S6d), which enabled us to calculate the coverage density of reacted azides by dividing the total integrated intensity in a unit surface area by the average single-molecule intensity (see Methods). We calculate the coverage density of the clicked azide to be 2.1 × 104 μm−2 (±0.11). We assume that the alkyne dye and the Alkyne-AIP-I have identical reactivity in the click reaction33, thus rendering the same surface coverage density, which is sufficient to stimulate the S. aureus Agr quorum-sensing response (Fig. S6e). We reduced the surface coverage density of active AIP-I by mixing the Alkyne-AIP-I and the Alkyne-Linear-AIP-I (i.e., the inactive counterpart) at different ratios prior to attachment to the surface. We found that Agr quorum-sensing activation depends on the surface-attached AIP-I coverage density (Fig. S6e). At a coverage density below 2.1 × 102 μm−2, the surface-attached AIP-I did not elicit the quorum-sensing response in S. aureus, and above a coverage density of 1.6 × 104 μm−2 the response was saturated (Fig. S6e). Together, our results show that the length of the PEG polymer coupled with the coverage density of the surface-bound AIP-I molecules are key factors for AgrC-I-directed quorum-sensing activation.
Quorum sensing is inhibited by surface-attached TrAIP-II
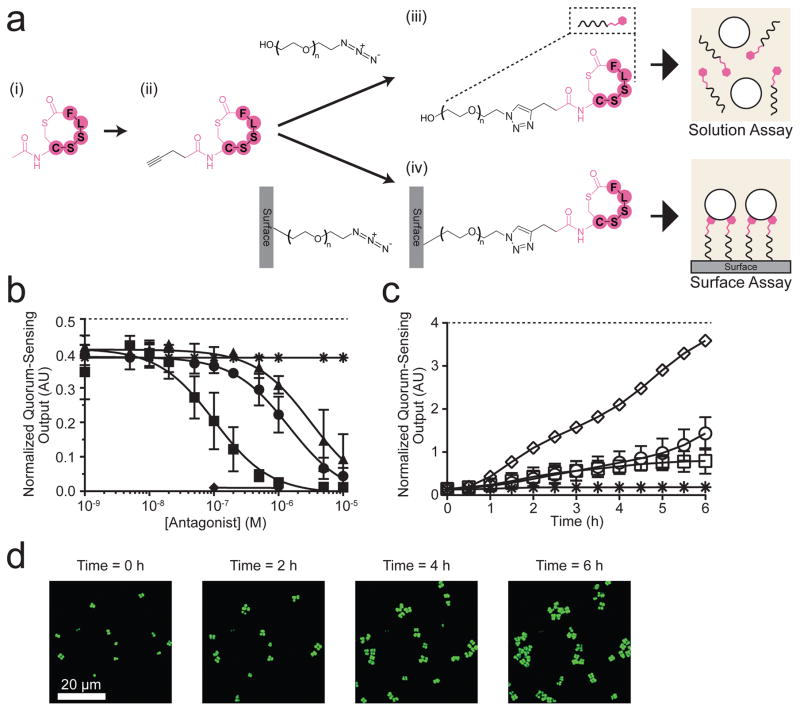
We investigated whether a quorum-sensing antagonist, when immobilized on a surface, could interfere with Agr quorum-sensing signal transduction. Using the above strategy, we synthesized a clickable quorum-sensing antagonist, the Alkyne-TrAIP-II (Fig. S2b and Fig. S3c), and attached it to the PEG330-azide in solution and to the Surface-PEG10000-azide (Fig. 3a), rendering PEG330-triazole-TrAIP-II and Surface-PEG10000-triazole-TrAIP-II, respectively. We characterized the products as outlined above for AIP-I (Fig. S3d). In solution, in the absence of AIP-I, the PEG330-triazole-TrAIP-II elicited no Agr quorum-sensing activation. When AIP-I was supplied at 0.1 μM (i.e., at its EC95), all of the TrAIP-II derivatives (i.e., Alkyne-TrAIP-II and PEG330-triazole-TrAIP-II) showed dose-dependent inhibition of Agr quorum sensing (Fig. 3b). The Alkyne-TrAIP-II was the most potent of the compounds (Table 1). We presume that modification of the inhibitor with the alkyne at the N-terminus endows the inhibitor with enhanced accessibility to the receptor and/or tighter binding.
Figure 3. A surface-attached Agr quorum-sensing antagonist, TrAIP-II, inhibits the S. aureus quorum-sensing response to AIP-I.
a, Strategy for modification of TrAIP-II and attachment to the surface; (i) TrAIP-II, (ii) Alkyne-TrAIP-II, (iii) PEG330-triazole-TrAIP-II, and (iv) Surface-PEG10000-triazole-TrAIP-II. b, The S. aureus Agr quorum-sensing reporter assay in solution with 100 nM AIP-I (asterisks), TrAIP-II + 100 nM AIP-I (circles), Alkyne-TrAIP-II + 100 nM AIP-I (squares), PEG330-triazole-TrAIP-II + 100 nM AIP-I (triangles), PEG330-triazole-TrAIP-II (diamonds). Normalized Agr quorum-sensing outputs were measured as a function of the concentrations of TrAIP-II and its derivatives. PEG330-triazole-TrAIP-II (diamonds) was measured at only the two highest concentrations due to limitations in available quantities of the reagent. Data points indicate means and error bars denote standard deviations from n = 4 experiments. c, The Agr quorum-sensing surface inhibition assay was performed with 30 nM AIP-I in solution and the following surfaces: Surface-PEG10000-triazole-TrAIP-II (asterisks), Surface-PEG10000-azide (circles), and Surface-PEG10000-triazole-Linear-TrAIP-II (squares). We also added 1 μM AIP-I in solution with the Surface-PEG10000-triazole-TrAIP-II (diamonds). The normalized Agr quorum-sensing outputs were measured for 1000 – 4000 individual cells in each experiment. Data points indicate means and error bars denote standard deviations from triplicate experiments. An ANOVA test with Tukey-Kramer post hoc analysis was used to assess the statistical significance between the means for the TrAIP-II coated surface and the control surfaces using the T = 6 h data (P < 0.0001 for all pairwise comparisons). d, Merged fluorescence images of the S. aureus reporter strain on the Surface-PEG10000-triazole-TrAIP-II + 30 nM AIP-I in bulk at T = 0, 2, 4, and 6 h. Images are based on n = 3 independent experiments; one representative image for each condition was chosen from ~ 50 images acquired from different regions of each surface. Scale bars: 20 μm.
We next examined quorum-sensing inhibition by surface-bound TrAIP-II. Introduction of the S. aureus reporter strain to the Surface-PEG10000-azide in the presence of AIP-I in solution at its EC50 (30 nM) resulted in activation of Agr quorum sensing (Fig. 3c and Fig. S7). However, if in the presence of 30 nM AIP-I, the strain was presented to the Surface-PEG10000-triazole-TrAIP-II, quorum-sensing inhibition occurred (Fig. 3c,d and Supplementary Movie 2), albeit with modest activation occurring at T = 6 h (Fig. S7 (i)). As controls, we increased the AIP-I concentration to 1 μM in solution and found that this was sufficient to outcompete the surface-bound TrAIP-II, causing strong activation of Agr quorum sensing over the 6 h period of the experiment (Fig. 3c and Fig. S7 (ii)). This result indicates that inhibition by surface-bound TrAIP-II is competitive, as it is in solution. Furthermore, Agr quorum-sensing inhibition did not significantly occur when S. aureus cells were added to the Surface-PEG10000-triazole-Linear-TrAIP-II (Fig. 3c and Fig. S7(iv)), demonstrating the requirement for specific structural elements in the antagonist.
Wild-type S. aureus responds to surface-attached quorum-sensing molecules
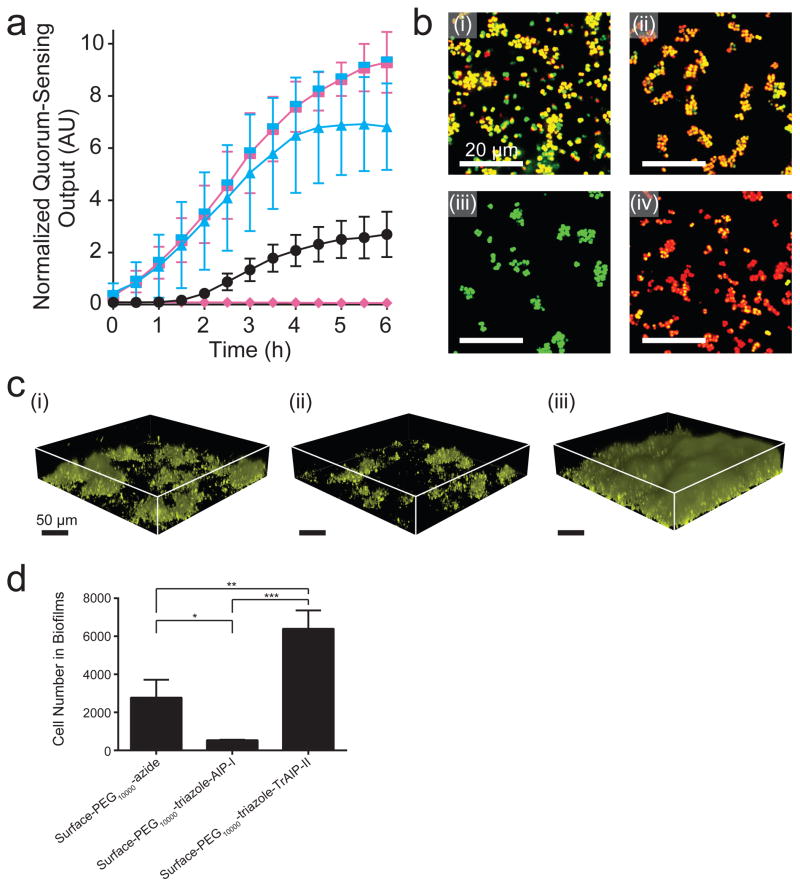
We investigated whether surface-attached pro- and anti-quorum-sensing molecules could control S. aureus behaviors in the more natural and clinically-relevant context of S. aureus strains that are capable of producing AIP-I37, unlike the reporter strain used above. For this analysis, we introduced the fluorescent quorum-sensing reporter genes into wild-type S. aureus agr-I (RN6390b) and S. aureus MRSA agr-I. In the presence of the Surface-PEG10000-azide, S. aureus agr-I activated Agr quorum sensing in response to the accumulation of endogenously produced AIP-I (Fig. 4a,b (i) and Supplementary Movie 3). The Surface-PEG10000-triazole-AIP-I caused an earlier and higher magnitude induction than did the Surface-PEG10000-azide, showing that S. aureus agr-I responded to both the surface-tethered AIP-I and to its endogenously produced AIP-I (Fig.4a,b (ii) and Supplementary Movie 3). The attached wild-type cells clearly responded to the surface attached AIP-I (Fig. 4b (ii)). This event elicited the positive autoinduction feedback loop causing the surface-adhered S. aureus cells to induce production and release of endogenous, soluble, AIP-I autoinducer. That autoinducer, in turn, activated the Agr quorum-sensing response in unattached S. aureus cells residing in solution above the surface as shown by their red fluorescence emission (Fig. S8a (ii)). Thus, attached cells, if they have the capacity to make autoinducer, can rapidly propagate the signal to neighboring, non-surface-adhered cells. The Surface-PEG10000-triazole-TrAIP-II repressed the Agr quorum-sensing response of the surface-adhered S. aureus cells over the 6 h period of the experiment (Fig.4a,b (iii) and Supplementary Movie 3). Furthermore, the surface-attached TrAIP-II also repressed the quorum-sensing response of the neighboring, non-surface-adhered S. aureus cells (Fig. S8a (iii)). Presumbly, surface-attached TrAIP-II, by repressing quorum sensing in the surface-adhered S. aureus cells, decreased their endogenous production of AIP-I, retarding the autoinduction feedback loop which, in turn, delayed signal propagation beyond the surface. Consistent with this interpretation, exogenous provision of 1 μM AIP-I in solution relieved inhibition from the Surface-PEG10000-triazole-TrAIP-II (Fig.4a,b (iv) and Supplementary Movie 3), confirming, as above, that the Surface-PEG10000-triazole-TrAIP-II functions competitively. Analogous results were obtained using the clinical pathogen S. aureus MRSA (Fig. S8b), which highlights the generality of our results and approach for regulating quorum sensing by surface modification.
Figure 4. Wild-type S. aureus responds to surface-attached AIP-I and TrAIP-II.
a, Normalized Agr quorum-sensing outputs were measured for Surface-PEG10000-azide (black circles), Surface-PEG10000-triazole-AIP-I (blue triangles), and Surface-PEG10000-triazole-TrAIP-II (pink diamonds). The split pink-blue square symbols show the result when 1 μM AIP-I was supplied in solution in the Surface-PEG10000-triazole-TrAIP-II experiments. In each experiment, normalized quorum-sensing outputs were measured as means from ~2000 individual cells. Data points indicate means and error bars denote standard deviations from triplicate experiments. An ANOVA test with Tukey-Kramer post hoc analysis was used to assess the statistical significance between the means from the Surface-PEG10000-azide and the quorum-sensing molecule coated surfaces using the T = 6 h data (P < 0.0001 for all pairwise comparisons). b, Merged fluorescent images of wild-type S. aureus agr-I (RN6390b), on modified surfaces at T = 6 h. (see Fig. S1 for details; green: constitutive fluorescence, red: quorum-sensing-controlled fluorescence). (i) Surface-PEG10000-azide, (ii) Surface-PEG10000-triazole-AIP-I, (iii) Surface-PEG10000-triazole-TrAIP-II, and (iv) Surface-PEG10000-triazole-TrAIP-II + 1 μM AIP-I in bulk. Images are based on n = 3 independent experiments; one representative image for each condition was chosen from ~ 50 images taken from different regions of each surface. Scale bars: 20 μm. c, Three-dimensional renderings of biofilms (225 × 225 × 44 μm3) of the S. aureus agr-I strain (RN6390b) constitutively expressing mKO. Biofilms were grown for 18 h on the following modified surfaces: (i) Surface-PEG10000-azide, (ii) Surface-PEG10000-triazole-AIP-I, and (iii) Surface-PEG10000-triazole-TrAIP-II. Images are based on n = 3 independent experiments with 6 regions imaged in each replicate; one representative image for each condition was chosen from the 18 total images for each surface type. Scale bars: 50 μm. d, Number of S. aureus agr-I (strain RN6390b) cells in biofilms on the designated modified surfaces (225 × 225 × 44 μm3) at T = 18 h (8 h under flow and 10 h with no flow; see Materials and Methods for protocol details). Data points indicate means and error bars denote standard deviations from triplicate experiments. * = P < 0.05, ** = P < 0.005, and *** = P < 0.0005 for an ANOVA test with Tukey-Kramer post hoc analysis for pairwise comparisons.
Next, we explored how the modified surfaces influence S. aureus biofilm colonization dynamics. We introduced a constitutively expressed mKO fluorescent reporter into the wild-type S. aureus agr-I strain and grew the strain on the Surface-PEG10000-azide, the Surface-PEG10000-triazole-AIP-I, and the Surface-PEG10000-triazole-TrAIP-II (Fig. 4c and 4d); all surfaces had comparable initial cell attachment (Fig. S8c (i)). Consistent with our above quorum-sensing transcriptional reporter results and with the known role for Agr quorum sensing in triggering biofilm dispersal in S. aureus, when grown on the Surface-PEG10000-triazole-AIP-I, the S. aureus agr-I strain induced biofilm dispersal, which resulted in an ~80% reduction in biofilm coverage compared to when the strain was grown on the Surface-PEG10000-azide. When the S. aureus agr-I strain was grown on the Surface-PEG10000-triazole-TrAIP-II, repression of Agr quorum sensing increased biofilm coverage ~2-fold compared to when the strain was grown on the Surface-PEG10000-azide. Analogous results were obtained using the wild-type S. aureus agr-II (i.e, subgroup II: RN6607) strain grown on the Surface-PEG10000-azide, the Surface-PEG10000-triazole-AIP-II, and the Surface-PEG10000-triazole-TrAIP-II (Fig. S8c (ii)). Specifically, in the S. aureus agr-II strain, Agr quorum sensing was activated by the surface-attached AIP-II, which results in biofilm dispesal, and quorum sensing was inhibited by the Surface-PEG10000-triazole-TrAIP-II, which resulted in increased biofilm coverage of the surface.
Applications of surface-attached quorum-sensing molecules
The analyses presented here focused on glass surfaces. However, other surfaces are relevant in industry and medicine such as metals and plastics. To expand our results, we chemically modified gold and polydimethylsiloxane (PDMS) surfaces with the dye-triazole-PEG moieties (Fig. S8d). Given the effectiveness and specificity of the click reaction, our preliminary results with the dye molecules suggest that AIP-I or TrAIP-II can be attached to these two surfaces. These results suggest that the logic used here is general and could be broadened to other materials as well as other molecules of interest.
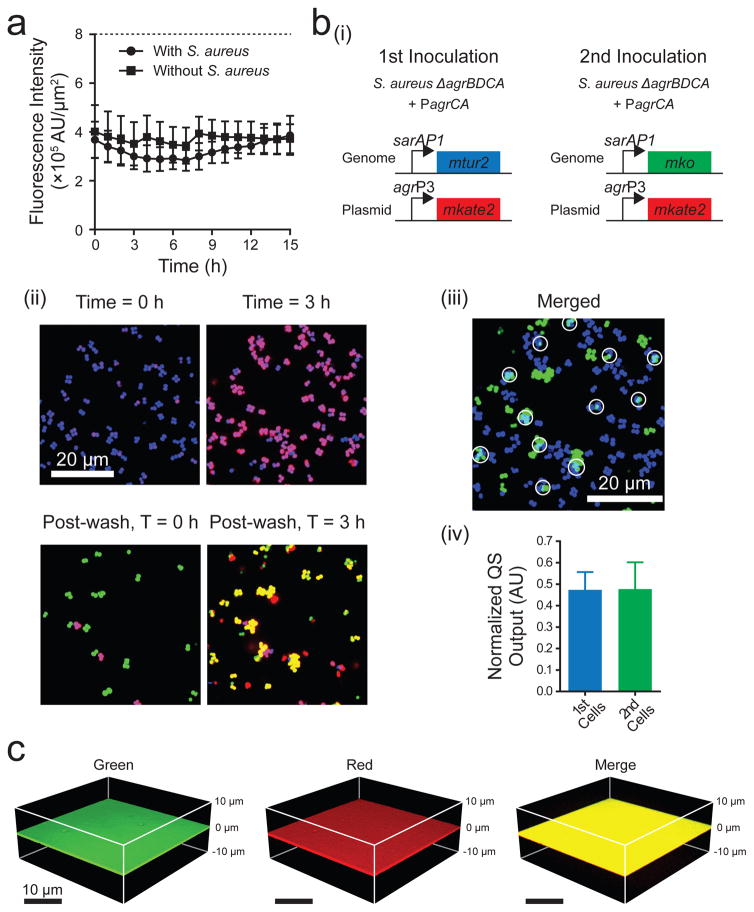
Having a reasonable shelf lifetime will also be crucial for chemically coated surfaces in medical and industrial applications. We examined the long-term stability of the Surface-PEG10000-triazole-dye and the Surface-PEG10000-triazole-AIP-I as a proxy for modified surface longevity. Irrespective of the presence or absence of S. aureus cells, the intensity of the surface-attached dye did not change for 15 h (Fig. 5a). In storage, in the absence of cells, the intensity did not change for 40 days (Fig. S8e (i)), and furthermore, the response of S. aureus cells to AIP-I-attached surfaces that had been stored for 40 days was identical to that of freshly generated surfaces (Fig. S8e (ii)). We interpret these findings to mean that the surface-tethered entities do not detach over time and the components remain stable for long time periods. These results are especially important in the context of materials such as submerged prosthetics, as our results suggest that concerns about cytotoxicity due to leaching or instability may not be highly relevant to the strategy we have developed.
Figure 5. Features of autoinducer-attached surfaces.
a, Stability of surface-attached molecules. Mean surface fluorescence intensities were measured from the Surface-PEG10000-triazole-dye over time in the presence and absence of S. aureus cells. Alexa Fluor 555 alkyne dye (Thermo Fisher, MA) was attached to the surface using the click reaction. Data points indicate means and error bars denote standard deviations from n = 4 experiments. b, Stability of surface-attached AIP-I and its activity following repeated introduction of S. aureus cells. (i) The relevant genotypes and fluorescent reporters used for the multiple inoculations; mtur2 denotes mturquoise2. (ii) Representative merged fluorescence images of the S. aureus reporter strains on the Surface-PEG10000-triazole-AIP-I. At T = 0 h, the S. aureus reporter strain (quorum-sensing-off: blue, quorum-sensing-on: purple) was introduced. At T = 3 h, S. aureus cells were washed off the surface and the second reporter strain (quorum-sensing-off: green, quorum-sensing-on: yellow) was introduced to the same surface. Scale bar: 20 μm. (iii) Merged fluorescence images of the area colonized by both strains. The constitutive fluorescent colors from the two S. aureus reporter strains (blue, T = 3 h and green, post-wash, T = 3 h) were artificially aligned. The light blue area with the white circles shows that cells from both the first and second inoculations colonized the same region. In panel b (ii) and b (iii), images are based on n = 3 independent experiments; one representative image for each condition was chosen from ~10 images taken from different regions of each surface. (iv) Normalized Agr quorum-sensing outputs were measured at 3 h after each strain was introduced onto the same surface. Data points indicate means and error bars denote standard deviations from triplicate experiments. c, Mixtures of dye molecules containing alkyne moieties can be simultaneously attached to the Surface-PEG10000-azide. Dyes: Alexa Fluor 488 alkyne (green) and Alexa Fluor 594 alkyne (red). Three-dimensional renderings of the surface that underwent the click reaction with a mixture of the two dyes are shown. Images are based on n = 4 independent experiments.
To further explore issues pertaining to medical applications of modified surfaces, we performed the quorum-sensing analysis in the presence of human blood plasma because S. aureus frequently causes bacteremia. Having blood plasma present throughout the experiment did not alter the ability of the AIP-I-coated surface to stimulate S. aureus quorum sensing (Fig. S8f). We also investigated the long-term effectiveness of surface-attached quorum-sensing molecules to repeated “infection”. We introduced the S. aureus reporter strain to the Surface-PEG10000-triazole-AIP-I (Fig. 5b (i)) and after 3 h, we removed the S. aureus cells by repeatedly introducing flow (σshear = 0.03–0.3 Pa) and air-bubbles38. We subsequently added a second dose of S. aureus reporter cells to the surface (Fig. 5b (i,ii)). The two S. aureus reporter strains were labeled with different constitutive fluorescent colors so we could monitor each of them. Cells from both the first and second inoculations colonized the same region (Fig. 5b (iii)), and both responded to the modified surface, activating Agr quorum sensing to a comparable magnitude (Fig. 5b (iv)). Thus, the surface-attached molecules remained effective following mechanical shear. This feature is especially important in the biomedical arena in which long-term activity of surface-attached molecules will be essential to combat repeated infection. Finally, we demonstrate the potential to generate multi-functional surfaces by simultaneously attaching an equal mixture of red and green dyes to the same surface (Fig. 5c). As noted, we estimate that there are 2.1 × 104 μm−2 sites on the surface available for attachment to biomolecules (Fig. S6e). However, an AIP-I surface coverage density of 1.6 × 104 μm−2 is sufficient to fully induce S. aureus Agr quorum sensing (Fig. S6e). Thus, approximately 25% of the sites (or 0.5 × 104 μm−2) are not required to achieve the maximal quorum-sensing response. These sites could conceivably be used to attach a different biomolecule, for example, one with an orthogonal activity. Thus, our preliminary results suggest surprising versatility in this approach: quorum-sensing molecules could be simultaneously or sequentially attached in combination with other biomolecules, such as antimicrobial agents, enzymes, or perhaps quorum-sensing compounds that target other bacteria39–41.
Discussion
We have discovered and documented that surface-attached molecules can successfully be used to control bacterial quorum sensing, which has potential implications for the development of anti-biofouling or anti-colonization materials in industry and medicine, respectively. As our test case, we have considered a crucial medical example: S. aureus Agr quorum sensing. In S. aureus, Agr quorum-sensing activation leads to the production of a battery of virulence factors that are reponsible for invasion and dissemination in host tissues10. Agr quorum sensing in S. aureus also activates the biofilm disassembly process42. Thus, precisely manipulating Agr quorum sensing using synthetic strategies that rely on soluble or surface-bound compounds will require tuning to, for example, terminate virulence while not enabling biofilm formation. Although deploying such strategies is not perfectly straightforward, we nonetheless use S. aureus as our proof of principle because of the urgent medical need for its control.
Here, we explored variations in surface coatings to target S. aureus Agr quorum sensing for different applications. First, antagonist-coated surfaces could be used in scenarios such as acute infections, e.g. staphylococcal scalded skin syndrome and toxic shock syndrome where it is essential to halt production of exo-toxins. TrAIP-II is a global antagonist that represses quorum sensing in all four S. aureus Agr subgroups. In the current work, we demonstrated that surface-attached TrAIP-II represses Agr quorum sensing in strains of Agr subgroups I and II. Presumably, subgroup III and IV strains would be likewise repressed, although we did not test them in this study. In the opposite vein, autoinducer-coated surfaces could have merit in scenarios including chronic infection, e.g. pneumonia or medical device-related infections, where S. aureus biofilms are the major issue. Indeed, S. aureus cells residing in surface-bound biofilms are more resistant to antibiotics and host immune defenses than are their planktonic counterparts43. AIP-I-coated surfaces, by triggering biofilm dispersal and transitioning the S. aureus cells to the planktonic lifestyle, could render surfaces resistant to biofilm colonization and, furthermore, render the dispersing cells more susceptible to antibiotics and to host immune defenses. In these contexts, it could be ideal to attach a global quorum-sensing agonist to the surface, if one existed. Such a strategy could disperse biofilms composed of any Agr subgroup. With these ideas in mind, our research raises the exciting, but now plausible possibility that surfaces decorated with quorum-sensing-modulating molecules can be assessed for anti-infective properties in animal models harboring indwelling devices. We note that instances of Agr-deficient virulent S. aureus strains have been reported44. Infections by Agr-defective strains primarily occur in patients that have been subjected to extreme courses of antibiotics and/or that have impaired immune functions44. Such strains would not be vulnerable to the strategies proposed here.
Our strategy for coating surfaces with pro- or anti-quorum-sensing molecules and using them to influence bacterial behaviors is not limited to the model bacterium S. aureus. Indeed, other bacteria could provide simpler cases for deployment. For example, the Gram-positive bacterium Enterococcus faecalis causes life-threatening urinary tract infections, bacteremia, endocarditis, and meningitis in humans45. Pathogenicity of E. faecalis relies on the Fsr quorum-sensing system, which is homologus to the S. aureus Agr quorum-sensing system46. However, importantly, in the case of E. faecalis, activation of Fsr quorum sensing promotes both biofilm formation and virulence factor production46. Thus, surfaces harboring Fsr quorum-sensing antagonists (i.e., ZBzl-YAA591147) could have a dual benefit in preventing biofilms and reducing exo-toxin production. Such dual benefits can also be imagined for other bacteria such as Listeria monocytogenes and Streptococcus pyogenes, as both pathogens possess Agr-type quorum-sensing systems that activate biofilm formation and virulence factor expression at high cell density48. Moreover, the beneficial bacterium Lactobacillus plantarum, which is important in the dairy and fermented food industries, also has an Agr-type quorum-sensing system called Lam that could be manipulated using surfaces in applications in food production49.
In summary, we have shown that surface-attached molecules can be used to manipulate a particular bacterial signaling system. Our strategy has the potential to be expanded to other systems with known ligands and with accessible cognate receptors. The strategy we have developed is primarily useful for Gram-positive bacteria because they do not possess an outer membrane. Strategies for Gram-negative bacteria can be explored perhaps by exploiting surfaces coated with quorum-sensing-manipulation compounds together with molecules that form pores in the outer membrane such as holins, endolysins, or bacteriocins50. Likewise, surface-attached quorum-sensing molecules could be examined with other orthogonal approaches that exploit surface release of active compounds22.
Materials and Methods
Bacterial strains and plasmids
The strains and plasmids used are listed in Supplementary Table 1. Staphylococcus aureus strains RN4220, RN9011, RN6390b, RN6911, and RN6607 and the plasmids pJL1111 and pRN7062 were gifts from Dr. Richard Novick’s group (New York University). S. aureus strains MK12118 (RN6390b carrying pMK021; agrP3-gfpmut2, sarAP1-mkate2) and S. aureus MRSA strain MK13118 (BAA1680 carrying pMK021) were used. DNA polymerase, dNTPs, and restriction enzymes were purchased from New England Biolabs (NEB, Ipswich, MA). DNA extraction and purification kits were acquired from Qiagen (Valencia, CA). DNA oligonucleotides were purchased from Integrated DNA Technologies (Coralville, Iowa). Sequences of plasmids were verified by Genewiz (South Plainfield, NJ).
Plasmids carrying constitutively expressed fluorescent fusions were constructed by replacing the mkate2 gene from pMK014 (sarAP1-mkate2) with genes encoding different fluorescent proteins (gfpmut2, mturquoise2, and mko). To make these plasmids, the gfpmut2 gene was amplified by PCR from pMK02118 using primers MKF013/MKR013, the mturquoise2 gene51 was amplified by PCR from pDP428, a generous gift from Daniel B. Kearns and Ethan Garner using primers MKF011/MKR011 and the mko gene was amplified by PCR from pCN00552 using primers MKF014/MKR014. The amplified genes were used to replace mkate2 by overlap extension PCR cloning. These plasmids are called pMK012 (sarAP1-gfpmut2), pMK011 (sarAP1-mturquoise2), and pMK013 (sarAP1-mko).
We integrated the constitutively expressed reporter fusions onto the S. aureus chromosome. To do this, we used a site-specific integration suicide vector, pJC1111, carrying a cadmium resistance cassette and the SaPI-1 attS sequence which integrates into the S. aureus chromosomal attachment site (attC) of pathogenicity island 1 (SaPI-1)53. This plasmid is integrated in single copy and maintained stably53. We digested pJC1111 using restriction enzymes NarI/SphI. The sarAP1-gfpmut2 gene was amplified by PCR from pMK012 using primers MKF031/MKR031 followed by digestion with NarI/SphI and ligation into digested pJC1111. This plasmid is called pMK032 (sarAP1-gfpmut2 in the suicide vector). We used the same procedure for other fluorescent genes; pMK031 (sarAP1-mturquoise2 in the suicide vector), and pMK033 (sarAP1-mko in the suicide vector). The plasmids were introduced into Escherichia coli DH5α using chemical transformation (New England Biolabs, Ipswich, MA) followed by selection with ampicillin. The plasmids were purified from E. coli, introduced by electroporation into S. aureus strain RN9011, which expresses the SaPI-1 integrase, and colonies containing the fusions integrated onto the chromosome were selected with cadmium. Subsequently, the chromosomal integrants were transduced into S. aureus strain RN6911 using standard phage transduction techniques with phage 80α. These strains are called MK232 (sarAP1-gfpmut2 in the genome), MK231 (sarAP1-mturquoise2 in the genome), and MK233 (sarAP1-mko in the genome).
A plasmid carrying a transcriptional fusion to monitor S. aureus Agr quorum-sensing activity was constructed by replacing the lacZ gene from vector pRN706254 (agrP3-lacZ) with the mkate2 gene. pRN7062 also harbors the genes encoding the Agr quorum-sensing detection components agrCA under their native agrP2 promoter but driven in the opposite direction. To make this plasmid, the lacZ gene was removed from pRN7062 by digestion with EcoRI/NarI. The mkate2 gene was obtained from pMK014 by EcoRI/NarI digestion. The digested mkate2 gene was ligated into digested pRN7062. This plasmid is called pMK051 (agrP2-agrCA, agrP3-mkate2). This construct was first introduced into E. coli, purified, and subsequently introduced into S. aureus strain RN4220 using selection with erythromycin. Subsequently, using phage transduction, the plasmid was introduced into S. aureus strains MK232, MK231, and MK233. The resultant strains are called MK242 (sarAP1-gfpmut2 in the genome and pMK051), MK241 (sarAP1-mtor2 in the genome and pMK051), MK243 (sarAP1-mko in the genome and pMK051). To construct the S. aureus ΔagrBDCA strain harboring agrP3-mkate2 on a plasmid, the vector pMK00418 (agrP3-mkate2) was introduced into S. aureus strain MK232 (sarAP1-gfpmut2 in the genome of S. aureus ΔagrBDCA), leading to strain MK245. Introduction of these plasmids into S. aureus strains does not alter growth or quorum-sensing phenotypes55.
We constructed control strains to study heterogeneity of the Agr quorum-sensing response. The first control strain has the agrP2-agrCA and agrP3-mkate2 genes inserted onto the genome of RN6911, and harbors sarAP1-gfpmut2 on a plasmid. To make this strain, we amplified the agrP2-agrCA, and agrP3-mkate2 genes from pMK051 using primers MKF032/MKR032, and inserted this fragment into the suicide vector pJC1111 by overlap extension PCR cloning. This plasmid is called pMK064 (agrP2-agrCA, agrP3-mkate2 in the suicide vector). The gene was integrated into the S. aureus strain RN6911 chromosome as described above. We call this strain MK264 (agrP2-agrCA, agrP3-mkate2 in the genome). The vector pMK012 (sarAP1-gfpmut2) was introduced into MK264, leading to strain MK265 (agrP2-agrCA, agrP3-mkate2 in the genome and pMK012). The second control strain was constructed by introducing pMK014 (sarAP1-mkate2) into strain MK232, leading to MK244 (sarAP1-gfpmut2 on the genome and pMK014). The third control strain has the agrP2-agrCA gene inserted onto the genome of RN6911, and harbors agrP3-mkate2 on a plasmid. To construct this strain, we amplified the agrP2-agrCA gene from pMK051 using primers MKF033/MKR033, and inserted this fragment into the suicide vector pJC1111 by overlap extension PCR cloning. This plasmid is called pMK060 (agrP2-agrCA in the suicide vector). The gene was integrated into the S. aureus strain RN6911 chromosome as described above. We call this strain MK260 (agrP2-agrCA in the genome). The vector pMK00418 (agrP3-mkate2) was introduced into MK260, leading to strain MK261 (agrP2-agrCA in the genome and pMK004). Finally, a constitutively expressed mKO fluorescent reporter (sarAP1-mko in pMK013) into wild-type S. aureus agr-I (strain RN6390b) and wild-type S. aureus agr-II (strain RN6607) to measure the number of cells in biofilms on surfaces.
Growth conditions
S. aureus RN6911 derivatives were grown overnight at 37 °C with shaking in Tryptic Soy Broth (TSB; Difco, Franklin Lakes, NJ) with 10 μg/ml tetracycline and 10 μg/ml erythromycin to maintain plasmids, back-diluted 1:200, and re-grown for 3 h (to OD600 ~ 0.05–0.1). S. aureus MK121, MK131, MK125, and MK126 were grown overnight at 37 °C with shaking in TSB with 10 μg/ml erythromycin, back-diluted 1:2000, and re-grown for 3 h (to OD600 ~ 0.05–0.1).
Synthesis of AIP-I, AIP-II, and TrAIP-II derivatives
AIP-I, AIP-II, and TrAIP-II derivatives were synthesized using a combined solid-phase/solution-phase approach. Linear peptide α-thioester precursors were generated using Fmoc-solid phase peptide synthesis employing a hydrazine linker system. The peptides were then cyclized in solution to install the thiolactone macrocyclic. Synthetic details are provided in Supplementary Methods.
Fluorescence reporter assay
Transcription from fluorescence reporter genes was measured in S. aureus strain MK242. Overnight cultures were diluted 1:200 into fresh TSB with 10 μg/ml tetracycline and 10 μg/ml erythromycin, re-grown, and 90 μl of these cultures were distributed into wells of 96 well plates (MatTek, Ashland, MA), followed by addition of 10 μl of AIP-I and/or TrAIP-II and/or derivatives. Subsequently, 50 μl of mineral oil was added (Sigma, St. Louis, MO) to prevent evaporation. Using a Synergy 2 plate reader (Biotek, Winooski, VT), GFPmut2 and mKate2 levels were measured at 484 nm/528 nm and 588 nm/633 nm, respectively. Measurements were conducted with 15 min intervals at 37 °C with shaking. This assay is called “Solution Assay” in the main text.
Surface fabrication
Surface-PEG10000-azide, Surface-PEG400-azide, Surface-PEG10000, and Surface-PEG400 were fabricated as follows (see Fig. S4a). Glass slides (36 × 60 mm2, Ted Pella, Redding, CA) were boiled for 2 h in 5% sodium peroxodisulfate (Sigma) at 50 °C and washed twice with Millipore water. The washed slides were submerged in piranha etch solution (3:1 H2SO4:H2O2, Fisher Scientific, Hampton, NH) for 6 h. Next, the slides were washed twice with Millipore water followed by rinsing with acetone (Sigma) three times to remove any trace of water. The hydroxylated glass substrate was incubated in 25% (3-mercaptopropyl) trimethoxysilane (Sigma) in acetone for 60 min to decorate the surface with thiol (-SH) functional groups. The thiol-decorated surface was rinsed with acetone and then Millipore water. After drying the substrates, the surfaces were incubated with a polymer-containing solution such as maleimide-PEG10000-azide, maleimide-PEG400-azide, maleimide-PEG10000, or maleimide-PEG400 (100 g/L, Nanocs, New York, NY) in 0.9 M sodium sulfate (Sigma) and 10 mM pH = 7.2 Tris-HCl buffer for 12 h to undergo the thiol-maleimide addition reaction34. The substrates were rinsed with Millipore water to yield Surface-PEG10000-azide, Surface-PEG400-azide, Surface-PEG10000, and Surface-PEG400, respectively. To fabricate the PDMS-based Surface-PEG10000-azide, glass slides were spin-coated with a PDMS mixture from the Sylgard 184 elastomer kit (Dow Corning, Midland, MI) at 2000 rpm for 30 sec. PDMS-coated surfaces were allowed to solidify, and then submerged in H2O:H2O2:HCl (5:1:1) for 1 h. Next, the slides were washed twice with Millipore water followed by rinsing with acetone (Sigma) three times to remove any trace of water, rendering the hydroxylated PDMS-based substrate. The remainder of the procedures are identical to those described above for glass surfaces. To fabricate the gold-based Surface-PEG5000-azide, coverslips coated with 10 nm thickness of gold (Amsbio, Cambridge, MA) were washed three times with acetone, and subsequently washed with isopropanol for 15 min. After drying the gold-coated substrates, thiol-PEG5000-azide (100 g/L, Nanocs) in 0.9 M sodium sulfate was added to the surface for 12 h to undergo the thiol-gold reaction. Following the reaction, the surfaces were washed thoroughly with Millipore water.
The azides decorating the surface-attached PEG400 and PEG10000 were subjected to the click reaction to link molecules carrying alkyne groups, such as Alkyne-AIP-I, Alkyne-AIP-II, Alkyne-TrAIP-II, and Alkyne-dye. The solution for the click reaction contained: 1 mM CuSO4, 50 mM ascorbic acid, 5 mM Tris (3-hydroxypropyltriazolylmethyl)amine, and 100 μM alkyne molecule in 100 mM phosphate buffer (pH = 7). Surface-PEG10000-azide and Surface-PEG400-azide were incubated with the click solution containing the alkyne molecules for 3 h. After rinsing the substrates three times with dimethyl sulfoxide (50% in water, Sigma) to remove any unreacted chemicals, the surfaces had become linked by the triazole rings to the AIP-I, AIP-II, TrAIP-II, or the dyes.
To investigate long-term surface stability, the Surface-PEG10000-triazole-AIP-I and the Surface-PEG10000-triazole-dye were stored at 4 °C for 40 days in a wet container covered with aluminum foil to prevent desiccation and exposure to light.
Surface characterization
The modified surfaces were characterized using a Fourier transform infrared (FTIR) spectrometer, fluorophore labeling, or atomic force microscopy (AFM) as follows: An FTIR spectra for the Surface-PEG10000-azide was obtained as an averaged signal by scanning the sample 128 times at 4 cm−1 resolution using a Thermo Nicolet Nexus 670 FTIR spectrometer (Thermo Electron Corp., Waltham, MA) equipped with a liquid nitrogen-cooled MCT/A detector (Fig. S4b). Prior to taking the spectra of the Surface-PEG10000-azide, the unmodified glass surface was scanned to provide the background spectrum. Background and sample measurements were taken only after the sample chamber was purged sufficiently with dry nitrogen to reduce the levels of carbon dioxide and water vapor.
To characterize the Surface-PEG10000-azide, Alexa Fluor 555 dye functionalized with an alkyne moiety (Thermo Fisher, MA) was used. The Surface-PEG10000 was used as the negative control. The surfaces were treated with the click solution containing the Alexa Fluor 555 alkyne dye. After washing away the unreacted dyes, the surfaces were imaged by confocal microscopy (Fig. S4c). Surface-PEG400-azide (Fig. S6a), gold-based Surface-PEG5000-azide, and PDMS-based Surface-PEG10000-azide (Fig. S8c) were characterized using the identical procedure.
AFM was used to measure the heights of the polymer brushes on the Surface-PEG10000-azide and the Surface-PEG400-azide. The surfaces were allowed to dry via evaporation in air. AFM topographical images of the boundaries between the PEG-attached regions and the unmodified regions were obtained in air using a Bruker Dimension Icon AFM (Bruker Biosciences, Billerica, MA) equipped with AFM tips (Bruker Biosciences) in Scanasyst-air peak force tapping mode. The scan regions were 2 × 2 μm2. The heights of surface-attached polymers were measured as the difference between the unmodified region and the PEG-attached region. We scanned several regions from two independently fabricated surfaces.
Quorum-sensing gene expression analyses on surfaces
The chemically modified surfaces were bonded to microfluidic chambers (400 μm × 100 μm × 2 cm) using an epoxy glue (Fisher Scientific). The assembled chambers were inoculated with the S. aureus agr-I reporter strain cultures, and cells were allowed to settle onto surfaces for 10 min, after which sterile M9 medium containing 10 μg/ml erythromycin, 0.5 % glucose, 0.5 % casamino acids, 5 mM NaCl, 50 mM MgSO4, and 0.1 mM CaCl2 was flowed steadily into the devices for 30 min to remove planktonic cells. After this step, chambers were placed on the microscope and the fluorescent reporter output measured. In experiments containing autoinducer or antagonist in solution, molecules were added in the wash medium. In experiments containing human blood plasma (Biological Specialty Corp, Colmar, PA), 100% blood plasma solution was initially flowed into the chambers containing the chemically modified surfaces for 10 min. Subsequently, S. aureus agr-I reporter cells were seeded for 10 min, and then 20% or 50% blood plasma diluted in the above medium was flowed into the chambers for 30 min to remove the planktonic cells. After this step, chambers were placed on the microscope and the fluorescent reporter output measured.
Biofilm analyses on surfaces
Chambers containing modified surfaces were seeded with wild-type S. aureus strains harboring constitutively expressed mko on a plasmid, and the cells were allowed to settle onto the surfaces for 10 min, after which sterile TSB containing 3% NaCl and 10 μg/ml erythromycin was flowed steadily into the devices for 8 h. This procedure was sufficient to produce three-dimensional S. aureus biofilms of 10–30 μm thickness. Flow was terminated for 10 h, and subsequently, planktonic cells were removed by steady flow for 10 min. The remaining biofilms were measured by confocal microscopy. Cells in biofilms covering a surface area of 225 × 225 μm2 were imaged at six different regions for each surface type in n = 3 independent experiments.
Microscopy and imaging
Imaging was performed using a Nikon Eclipse Ti inverted microscope (Melville, NY) fitted with a Yokogawa CSU X-1 confocal spinning disk scanning unit (Biovision Technologies, Exton, PA) and DU-897 X-9351 camera (Andor, Concord, MA). Laser lines at 445, 488, 543, and 592 nm were used to excite the mTurquoise2, GFPmut2, mKO, and mKate2 fluorescent proteins, respectively. Laser lines at 488, 543, and 592 nm were used to excite Alexa Fluor 488 fluorophore, Alexa Fluor 555 fluorophore, and Alexa Fluor 594 fluorophore, respectively. In order to obtain single-cell resolution, both a 100× oil objective with N.A. 1.4 (Nikon, Melville, NY) and a 1.5× lens placed between the CSU X-1 and the Nikon microscope side port were used. Consequently, the magnification of 0.1 μm per pixel in the XY plane was obtained. For single-cell analysis, custom code was written in Matlab. Briefly, the area of an individual cell was recognized and segmented using a watershed-based algorithm. In this process, we removed cells if they were on the edge of the image or if they were smaller than 30% of the average cell size, suggesting they were out of focus. In the area of an individual cell, both the constitutive GFPmut2 fluorescence and the quorum-sensing controlled mKate2 fluorescence were measured, subtracted from background signals and summed. The normalized quorum-sensing output was calculated as the quorum-sensing controlled mKate2 intensity divided by the constitutively expressed GFPmut2 intensity in individual cells. In each experiment, images of many regions on the surfaces were taken to include 1000 – 4000 individual cells. Each replicate was performed using independent bacterial cultures and independent surfaces at room temperature. Identical procedures were performed for the strains harboring different constitutive fluorescent proteins such as mTurquoise2 and mKO (Fig. 5b). Custom code was used to count the cells in the biofilms. Each image was segmented in the z-plane and assessed independently (Fig. 4d and S8c).
Quantitation of surface coverage density and single-molecule microscopy
The coverage density of “clicked” azides on the Surface-PEG10000-azide was quantified based on fluorophore labeling. We assume that the alkyne-dye, the Alkyne-AIP-I, and the Alkyne-TrAIP-II have identical reactivity in the click reaction, rendering the same surface coverage density. Thus, surface coverage density can be quantified using fluorophore labeling as a proxy for surface-attached quorum-sensing compounds. In the fluorophore labeling assay, the surface-attached azides underwent the click reaction with 100 μM Alexa Fluor 555 alkyne fluorophore. After conjugation, the total intensity of fluorescence from the surface was measured (at 10% laser power), subtracted from the background signal, and divided by the average single-molecule intensity. The average single-molecule intensity was obtained by measuring the intensity of single Alexa Fluor 555 molecules as follows: We treated the Surface-PEG10000-azide with 0.1 – 10 nM Alexa Fluor 555 alkyne fluorophore, and this low concentration of alkyne-dye produced individually discernable fluorescent spots (20–300 in a 51 × 51 μm2) (Fig. S6d (i)). The individual fluorescent spots were subjected to photobleaching. They displayed stepwise decreases in intensity. Background subtracted intensities of a few thousand spots across 5 independent dye-linked surfaces were measured and analyzed using our custom Matlab code. Briefly, the obtained images were smoothed and filtered to produce a zero-based image in which bright fluorescent spots were located with pixel level accuracy by a peak-finding algorithm. With the high spatial resolution of the microscope (100 nm/pixel), the intensity of each spot was measured from an 8 pixel × 8 pixel region centered at each of these peaks. The integrated fluorescence emission within each region was measured over consecutive frames while exposing the sample to high intensity illumination (at 100% laser power with 500 msec exposure time) as the photobleaching process. The photobleaching intensity trace revealed the number of fluorophores present in that spot, and the size of these intensity steps yielded the intensity of emission from the fluorophores in the spot. Most fluorescent spots decreased in one step following bleaching (Fig. S6d (ii)). A small number of spots bleached in two steps (Fig. S6d (ii)). By fitting the bleaching step sizes for over 3,600 spots with a normal distribution (Fig. S6d (iii)), we were able to arrive at the average single-molecule intensity (7,000 (AU)), which was further verified by counting fluorescent spots from the surface. Unlike the reaction of the surface with the low concentration of alkyne-dye, the surface intensity from the typical surface that had reacted with 100 μM alkyne-dye was above the maximum detection limit of the camera when exposed to 100% laser power. Thus, we used a two-step calibration process in which we measured the total intensity of uniform surfaces that had reacted with 1 nM, 5 nM, and 10 nM alkyne-dye at 100% laser power and compared them to that obtained with 10% laser power, yielding an average ratio of 49. For the final surface measured at 10% laser power with 500 msec exposure time, we multiplied the integrated intensity by 49, and divided that by the average single-molecule intensity (7,000 (AU)), finally giving a surface coverage density.
Supplementary Material
Acknowledgments
We thank the Novick laboratory for generously providing S. aureus strains and plasmids. We thank N. Wingreen for discussions about heterogeniety, J. Yan for image analysis, B. Wang for helpful discussion and mentoring in S. aureus genetic techniques, B. Bratton for discussions about single-molecule microscopy, I. Pelczer and K. Conover for the NMR measurements, and D. Dabbs for help with the FTIR measurement. We are grateful to members of the B.L.B., H.A.S., and T.W.M. laboratories for suggestions. This work was supported by NSF grant MCB-1344191 (B.L.B. and H.A.S.), the Howard Hughes Medical Institute, NIH grant 2R37GM065859 and NSF grant MCB-0948112 (B.L.B.), NIH grant R01 AI042783 (TWM) and a STX fellowship (M.K.K.).
Footnotes
Supplementary Information is available.
Author Contributions M.K.K., H.A.S., and B.L.B. conceived the idea. M.K.K., A.Z., T.W.M., H.A.S., and B.L.B. designed the experiments. M.K.K. and A.Z. performed the majority of the experiments. A.W. helped with the solution assay. M.K.K., A.Z., A.W., and Z.Z.B. contributed new reagents/analytic tools. M.K.K., A.Z., T.W.M., H.A.S., and B.L.B. analyzed the data. M.K.K., A.Z., H.A.S., and B.L.B. wrote the manuscript.
The authors declare no competing financial interests.
Data availablity All data sets that support the findings of this study are available in the present manuscript in either the main text or the supplementary information.
References
- 1.Wang B, Muir TW. Regulation of virulence in Staphylococcus aureus: molecular mechanisms and remaining puzzles. Cell Chem Biol. 2016;23:214–224. doi: 10.1016/j.chembiol.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kong KF, Vuong C, Otto M. Staphylococcus quorum sensing in biofilm formation and infection. Int J Med Microbiol. 2006;296:133–139. doi: 10.1016/j.ijmm.2006.01.042. [DOI] [PubMed] [Google Scholar]
- 3.Drescher K, et al. Architectural transitions in Vibrio cholerae biofilms at single-cell resolution. Proc Natl Acad Sci U S A. 2016;113:2066–2072. doi: 10.1073/pnas.1601702113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan J, Sharo AG, Stone HA, Wingreen NS, Bassler BL. Vibrio cholerae biofilm growth program and architecture revealed by single-cell live imaging. Proc Natl Acad Sci U S A. 2016;113:5337–5343. doi: 10.1073/pnas.1611494113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nadell CD, Drescher K, Foster KR. Spatial structure, cooperation and competition in biofilms. Nat Rev Microbiol. 2016;14:589–600. doi: 10.1038/nrmicro.2016.84. [DOI] [PubMed] [Google Scholar]
- 6.Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 7.Papenfort K, Bassler BL. Quorum sensing signal-response systems in Gram-negative bacteria. Nat Rev Microbiol. 2016;14:576–588. doi: 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Novick RP, Geisinger E. Quorum sensing in Staphylococci. Annu Rev Genet. 2008;42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 9.Dinges MM, Orwin PM, Schlievert PM. Exotoxins of Staphylococcus aureus. Clin Microbiol Rev. 2000;13:16–34. doi: 10.1128/cmr.13.1.16-34.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arvidson S, Tegmark K. Regulation of virulence determinants in Staphylococcus aureus. Int J Med Microbiol. 2001;291:159–170. doi: 10.1078/1438-4221-00112. [DOI] [PubMed] [Google Scholar]
- 11.Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol. 2003;50:101–104. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- 12.Hsiao A, et al. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature. 2014;515:423–426. doi: 10.1038/nature13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ismail AS, Valastyan JS, Bassler BL. A host-produced autoinducer-2 mimic activates bacterial quorum sensing. Cell Host Microbe. 2016;19:470–480. doi: 10.1016/j.chom.2016.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Loughlin CT, et al. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc Natl Acad Sci USA. 2013;110:17981–17986. doi: 10.1073/pnas.1316981110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sully EK, et al. Selective chemical inhibition of agr quorum sensing in Staphylococcus aureus promotes host defense with minimal impact on resistance. Plos Pathog. 2014;10 doi: 10.1371/journal.ppat.1004174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Otto M. Quorum-sensing control in Staphylococci - a target for antimicrobial drug therapy? Fems Microbiol Lett. 2004;241:135–141. doi: 10.1016/j.femsle.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 17.Cegelski L, Marshall GR, Eldridge GR, Hultgren SJ. The biology and future prospects of antivirulence therapies. Nature Reviews Microbiology. 2008;6:17–27. doi: 10.1038/nrmicro1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim MK, Ingremeau FB, Zhao A, Bassler BL, Stone HA. Local and global consequence of flow on bacterial quorum sensing. Nature Microbiology. 2016;1:15005. doi: 10.1038/nmicrobiol.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu HD, et al. Modulating Vibrio cholerae quorum-sensing-controlled communication using autoinducer-loaded nanoparticles. Nano Lett. 2015;15:2235–2241. doi: 10.1021/acs.nanolett.5b00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Persat A, et al. The mechanical world of bacteria. Cell. 2015;161:988–997. doi: 10.1016/j.cell.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim MK, Drescher K, Pak OS, Bassler BL, Stone HA. Filaments in curved streamlines: rapid formation of Staphylococcus aureus biofilm streamers. New J Phys. 2014;16 doi: 10.1088/1367-2630/16/6/065024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Broderick AH, et al. Surface coatings that promote rapid release of peptide-based AgrC inhibitors for attenuation of quorum sensing in Staphylococcus aureus. Adv Healthc Mater. 2014;3:97–105. doi: 10.1002/adhm.201300119. [DOI] [PubMed] [Google Scholar]
- 23.Ho KKK, et al. Quorum sensing inhibitory activities of surface immobilized antibacterial dihydropyrrolones via click chemistry. Biomaterials. 2014;35:2336–2345. doi: 10.1016/j.biomaterials.2013.11.072. [DOI] [PubMed] [Google Scholar]
- 24.Lowy FD. Staphylococcus aureus infections. New Engl J Med. 1998;339:2026–2027. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 25.Gordon RJ, Lowy FD. Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis. 2008;46:S350–S359. doi: 10.1086/533591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang LS, Gray L, Novick RP, Ji GY. Transmembrane topology of AgrB, the protein involved in the post-translational modification of AgrD in Staphylococcus aureus. J Biol Chem. 2002;277:34736–34742. doi: 10.1074/jbc.M205367200. [DOI] [PubMed] [Google Scholar]
- 27.Wang BY, Zhao AS, Novick RP, Muir TW. Key driving forces in the biosynthesis of autoinducing peptides required for staphylococcal virulence. P Natl Acad Sci USA. 2015;112:10679–10684. doi: 10.1073/pnas.1506030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lina G, et al. Transmembrane topology and histidine protein kinase activity of AgrC, the agr signal receptor in Staphylococcus aureus. Mol Microbiol. 1998;28:655–662. doi: 10.1046/j.1365-2958.1998.00830.x. [DOI] [PubMed] [Google Scholar]
- 29.Wang BY, Zhao AS, Novick RP, Muir TW. Activation and inhibition of the receptor histidine kinase AgrC occurs through opposite helical transduction motions. Mol Cell. 2014;53:929–940. doi: 10.1016/j.molcel.2014.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koenig RL, Ray JL, Maleki SJ, Smeltzer MS, Hurlburt BK. Staphylococcus aureus AgrA binding to the RNAIII-agr regulatory region. J Bacteriol. 2004;186:7549–7555. doi: 10.1128/JB.186.22.7549-7555.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fechter P, Caldelari I, Lioliou E, Romby P. Novel aspects of RNA regulation in Staphylococcus aureus. Febs Lett. 2014;588:2523–2529. doi: 10.1016/j.febslet.2014.05.037. [DOI] [PubMed] [Google Scholar]
- 32.Lyon GJ, Wright JS, Muir TW, Novick RP. Key determinants of receptor activation in the agr autoinducing peptides of Staphylococcus aureus. Biochemistry-Us. 2002;41:10095–10104. doi: 10.1021/bi026049u. [DOI] [PubMed] [Google Scholar]
- 33.Hong V, Presolski SI, Ma C, Finn MG. Analysis and optimization of copper-catalyzed azide-alkyne cycloaddition for bioconjugation. Angew Chem Int Edit. 2009;48:9879–9883. doi: 10.1002/anie.200905087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emilsson G, et al. Strongly stretched protein resistant poly(ethylene glycol) brushes prepared by grafting-to. Acs Appl Mater Inter. 2015;7:7505–7515. doi: 10.1021/acsami.5b01590. [DOI] [PubMed] [Google Scholar]
- 35.Cisar EA, Geisinger E, Muir TW, Novick RP. Symmetric signalling within asymmetric dimers of the Staphylococcus aureus receptor histidine kinase AgrC. Mol Microbiol. 2009;74:44–57. doi: 10.1111/j.1365-2958.2009.06849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. Fems Microbiol Rev. 2008;32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 37.Traber KE, et al. agr function in clinical Staphylococcus aureus isolates. Microbiol-Sgm. 2008;154:2265–2274. doi: 10.1099/mic.0.2007/011874-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khodaparast S, Kim MK, Silpe J, Stone HA. Bubble-driven detachment of bacteria from confined microgeometries. Environ Sci Technol. 2017;51:1340–1347. doi: 10.1021/acs.est.6b04369. [DOI] [PubMed] [Google Scholar]
- 39.Vasilev K, Cook J, Griesser HJ. Antibacterial surfaces for biomedical devices. Expert Rev Med Devic. 2009;6:553–567. doi: 10.1586/erd.09.36. [DOI] [PubMed] [Google Scholar]
- 40.Gallo J, Holinka M, Moucha CS. Antibacterial surface treatment for orthopaedic implants. Int J Mol Sci. 2014;15:13849–13880. doi: 10.3390/ijms150813849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee JJ, et al. Synthetic ligand-coated magnetic nanoparticles for microfluidic bacterial separation from blood. Nano Lett. 2014;14:1–5. doi: 10.1021/nl3047305. [DOI] [PubMed] [Google Scholar]
- 42.Boles BR, Horswill AR. agr-mediated dispersal of Staphylococcus aureus biofilms. Plos Pathog. 2008;4 doi: 10.1371/journal.ppat.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davies D. Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov. 2003;2:114–122. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- 44.Painter KL, et al. What role does the quorum-sensing accessory gene regulator system play during Staphylococcus aureus bacteremia? Trends in Microbiology. 2014;22:676–685. doi: 10.1016/j.tim.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 45.Huycke MM, Spiegel CA, Gilmore MS. Bacteremia caused by hemolytic, high-level gentamicin-resistant Enterococcus faecalis. Antimicrob Agents Ch. 1991;35:1626–1634. doi: 10.1128/aac.35.8.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cook LC, Federle MJ. Peptide pheromone signaling in Streptococcus and Enterococcus. Fems Microbiol Rev. 2014;38:473–492. doi: 10.1111/1574-6976.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakayama J, et al. Development of a peptide antagonist against fsr quorum sensing of Enterococcus faecalis. ACS Chem Biol. 2013;8:804–811. doi: 10.1021/cb300717f. [DOI] [PubMed] [Google Scholar]
- 48.Gray B, Hall P, Gresham H. Targeting agr- and agr-like quorum sensing systems for development of common therapeutics to treat multiple Gram-positive bacterial infections. Sensors-Basel. 2013;13:5130–5166. doi: 10.3390/s130405130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fujii T, et al. Two homologous agr-like quorum-sensing systems cooperatively control adherence, cell morphology, and cell viability properties in Lactobacillus plantarum WCFS1. J Bacteriol. 2008;190:7655–7665. doi: 10.1128/JB.01489-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang IN, Smith DL, Young R. Holins: The protein clocks of bacteriophage infections. Annu Rev Microbiol. 2000;54:799–825. doi: 10.1146/annurev.micro.54.1.799. [DOI] [PubMed] [Google Scholar]
- 51.Goedhart J, et al. Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93% Nature Communications. 2012;3 doi: 10.1038/ncomms1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nadell CD, Drescher K, Wingreen NS, Bassler BL. Extracellular matrix structure governs invasion resistance in bacterial biofilms. Isme Journal. 2015;9:1700–1709. doi: 10.1038/ismej.2014.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen J, Yoong P, Ram G, Torres VJ, Novick RP. Single-copy vectors for integration at the SaPI1 attachment site for Staphylococcus aureus. Plasmid. 2014;76:1–7. doi: 10.1016/j.plasmid.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lyon GJ, Mayville P, Muir TW, Novick RP. Rational design of a global inhibitor of the virulence response in Staphylococcus aureus, based in part on localization of the site of inhibition to the receptor-histidine kinase, AgrC. P Natl Acad Sci USA. 2000;97:13330–13335. doi: 10.1073/pnas.97.24.13330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Charpentier E, et al. Novel cassette-based shuttle vector system for Gram-positive bacteria. Appl Environ Microbiol. 2004;70:6076–6085. doi: 10.1128/AEM.70.10.6076-6085.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.