Abstract
Background
Large-scale genome-wide association studies (GWAS) have so far identified 45 loci that are robustly associated with coronary heart disease (CHD) in data from adult men and women of European descent.
Objectives
To examine whether the CHD-associated loci are associated with measures of atherosclerosis in data from up to 9,582 individuals of European ancestry.
Methods
Forty-five SNPs representing the CHD-associated loci were genotyped in middle-aged to elderly individuals of European descent from four independent population-based studies (IMPROVE, MDC-CC, ULSAM and PIVUS). Intima-media thickness (IMT) was measured by external B-mode ultrasonography at the far wall of the bulb (sinus) and common carotid artery. Plaque presence was defined as a maximal IMT of the bulb >1.5 mm. We meta-analysed single-SNP associations across the four studies, and combined them in a genetic predisposition score. We subsequently examined the association of the genetic predisposition score with prevalent CHD and the three indices of atherosclerosis, adjusting for sex, age and Framingham risk factors.
Results
As anticipated, the genetic predisposition score was associated with prevalent CHD, with each additional risk allele increasing the odds of disease by 5.5% (p=4.1x10-6). Moreover, each additional CHD-risk allele across the 45 loci was associated with a 0.24% increase in IMT (p=4.0x10-3), and with a 2.8% increased odds of plaque presence (p=7.4x10-6) at the far wall of the bulb. The genetic predisposition score was not associated with IMT of the common carotid artery (p=0.47).
Conclusions
Our results suggest that the association between the 45 previously identified loci and CHD at least partly acts through atherosclerosis.
Keywords: genome-wide association, intima-media thickness, carotid artery, atherosclerosis, atherogenic plaque
Introduction
Coronary heart disease (CHD) is the result of life-long progression of atherosclerosis in the coronary vessels and is a major cause of death worldwide. Smoking, hypertension, hypercholesterolemia, diabetes, obesity, lack of physical activity, and family history have all been identified as major risk factors for CHD in the 1960s (1). More recently, twin and family studies have shown that genetic factors also contribute to CHD risk, explaining approximately 40-50% of its variance (2). Researchers have since aimed to identify genetic loci that are associated with CHD risk, first using the candidate gene and linkage approaches, and more recently using genome-wide arrays with improved coverage of genetic variation. Since the first genome-wide association study (GWAS) for CHD was published in 2006 (3), 46 loci have been identified as being robustly associated with CHD in data from individuals of European descent (4). Forty-five of these loci were identified in data from men and women combined, whilst one locus was only associated with CHD in men (4).
CHD is caused by atherosclerosis in the coronary arteries. Since an evaluation of coronary atherosclerosis with angiography is only performed in the clinical setting in the presence of strict indications, epidemiological studies on coronary atherosclerosis are hard to perform in the general population. However, since atherosclerosis usually affects more than one arterial bed, ultrasound of the carotid arteries has been widely used to assess the atherosclerotic burden in the epidemiological setting. The thickness of the intima-media complex (IMT), as well as the presence of overt plaques in the carotid arteries identified by ultrasound, have been shown to predict both myocardial infarction and stroke (5–8), and have been associated with plaque burden in the coronary circulation, as well as in other parts of the body (9–12). This suggests that atherosclerosis in the carotid arteries can be used as a proxy measure for coronary atherosclerosis. In line with this, a recent single-centre study showed that a genetic predisposition score (GPS), consisting of 13 CHD-associated loci, was associated with atherosclerosis in the carotid arteries (13).
In the present study, we examined whether the 45 CHD-associated loci that have so far been identified by GWAS in data from men and women combined are also associated with three indices of atherosclerosis in the carotid arteries as detected by ultrasound, i.e. 1) presence of atherosclerotic plaques; 2) IMT of the carotid bulb or sinus; and 3) IMT of the common carotid artery. We genotyped lead SNPs in the 45 loci in nearly 10,000 individuals from four population-based studies of middle-aged to elderly individuals of European descent, thereby increasing the power to find associations when compared with the previous effort (13). In the primary analysis, we meta-analysed the association of the 45 loci with CHD risk and the three indices of atherosclerosis across the four studies, and combined the associations of all 45 loci in a GPS. The hypothesis tested was that, in addition to CHD risk, the GPS would be associated with all three indices of atherosclerosis. In the secondary, exploratory analysis, we also examined associations with CHD risk and the indices of carotid atherosclerosis for the 45 CHD-associated loci individually.
Methods
Study populations
We examined associations of the 45 CHD-associated loci with CHD and carotid atherosclerosis in data from the IMPROVE (14), MDC-CC (15), ULSAM (16) and PIVUS (17) cohort studies. A detailed description of these studies, as well as on the way in which carotid atherosclerosis and CHD have been quantified is provided in Supplementary Table 1. Briefly, we performed B-mode ultrasound at the far wall of the common carotid artery in all studies, and calculated an average IMT of the left and right carotid artery (if available). IMT of the far wall of the bulb was calculated in a similar manner. We defined plaque presence as a maximal IMT of the far wall of the bulb >1.5 mm. IMT of the far wall of the bulb and plaque presence were not available in the ULSAM study.
We ascertained CHD status in the IMPROVE, ULSAM and PIVUS studies using information from the most recently updated version of the medical registry that was available to us. We included participants as CHD cases in the analysis if they had been hospitalized for myocardial infarction, angina, percutaneous transluminal coronary angioplasty, and/or coronary artery bypass grafting. CHD status was not available in the MDC-CC study.
All studies were approved by the local scientific committees and were performed in accordance with the declaration of Helsinki. Written informed consents were obtained from all participants. Descriptive information for participants of the four studies is shown in Table 1.
Table 1. Descriptive characteristics of adults per study stratified by sex.
| MDC-CC | IMPROVE | ULSAM | PIVUS | ||||
|---|---|---|---|---|---|---|---|
| Men | Women | Men | Women | Men | Men | Women | |
| nmax | 1915 | 2821 | 1666 | 1780 | 1120 | 474 | 475 |
| Age (years) | 57.7 ± 6.0 | 57.5 ± 5.9 | 64.0 ± 5.4 | 64.4 ± 5.5 | 71.0 ±0.6 | 70.1 ± 0.2 | 70.3 ± 0.1 |
| BMI (kg/m2) | 26.1 ± 3.5 | 25.3 ± 4.2 | 27.4 ± 3.6 | 27.1 ± 4.8 | 26.3 ± 3.4 | 27.0 ± 3.7 | 27.1 ± 4.9 |
| Diastolic blood pressure (mmHg) | 89 ± 9 | 86 ± 9 | 83 ± 10 | 81 ± 10 | 84 ± 9 | 79 ± 10 | 78 ± 10 |
| Systolic blood pressure (mmHg) | 143 ± 19 | 140 ± 19 | 142 ± 18 | 142 ± 19 | 147 ± 19 | 146 ± 22 | 153 ± 23 |
| LDL-cholesterol (mmol/l) | 4.1 ± 0.9 | 4.2 ± 1.0 | 3.4 ± 0.9 | 3.7 ± 1.1 | 3.9 ± 0.9 | 3.2 ± 0.9 | 3.5 ± 0.9 |
| HDL-choleserol (mmol/l) | 1.1 (1.0, 1.4) | 1.5 (1.2, 1.7) | 1.1 (0.9, 1.3) | 1.3 (1.1, 1.6) | 1.2 (1.0, 1.5) | 1.3 (1.1, 1.5) | 1.6 (1.3, 1.9) |
| Current smoking (nsmokers / nnon-smokers) | 510 / 1405 | 716 / 2105 | 279 / 1387 | 238 / 1368 | 222 / 898 | 47 / 426 | 57 / 418 |
| Type 2 diabetes status (cases / controls) | 210 / 1705 | 160 / 2661 | 522 / 1115 | 387 / 1368 | 121 / 998 | 65 / 409 | 46 / 429 |
| IMT CCA (mm) | 0.74 (0.64, 0.85) | 0.70 (0.63, 0.79) | 0.74 (0.66, 0.83) | 0.70 (0.64, 0.77) | 0.71 (0.63, 0.81) | 0.88 (0.79, 1.00) | 0.85 (0.76, 0.97) |
| IMT bulb (mm) | 1.33 (1.02, 1.82) | 1.19 (0.97, 1.56) | 1.13 (0.91, 1.45) | 1.00 (0.80, 1.25) | na | 0.98 (0.89, 1.12) | 0.94 (0.84, 1.07) |
| Plaque presence (cases / controls) | 529 / 827 | 537 / 1372 | 1112 / 547 | 967 / 799 | na | 272 / 202 | 233 / 242 |
| CHD status (cases / controls) | na | na | 76 / 1590 | 45 / 1735 | 316 / 804 | 112 / 362 | 54 / 421 |
IMT, intima-media thickness; CCA, common carotid artery; bulb, also known as sinus; CHD, coronary heart disease; na, not available
Genotyping
Participants of the MDC-CC study were genotyped using Illumina’s OmniExpress chip; participants of the ULSAM and PIVUS studies with Illumina’s OmniExpress and Cardio-Metabochip; and those participating in the IMPROVE study using the Cardio-Metabochip. We prioritized lead SNPs of the 45 CHD-associated loci as reported in the most recent paper of the CARDIoGRAMplusC4D consortium (4), provided those SNPs had been directly genotyped. We replaced lead SNPs by genotyped proxies in high linkage disequilibrium (LD) with lead SNPs if the latter had not been directly genotyped. We identified proxies using the SNP Annotation and Proxy search (SNAP) tool of the BROAD Institute (http://www.broadinstitute.org/mpg/snap/ldsearch.php), which uses genotype data from the first pilot study of the 1000 Genomes Project (18). We included proxy SNPs for 3 of the 45 loci in IMPROVE, 24 of the 45 loci in MDC-CC, and 1 of the 45 loci in ULSAM and PIVUS (Supplementary Table 2).
All SNPs passed quality control criteria with a call rate > 95% and a blind duplicate concordance rate of 100%. The distributions of all variants were in Hardy-Weinberg equilibrium, as determined by a Chi-squared test with one degree of freedom (p>1.1x10-3).
Statistical analyses
To put the results of the genetic association study into context, we first examined the association of plaque presence and IMT of the bulb and common carotid artery with CHD risk. IMT of the bulb and common carotid artery were natural log transformed in all analyses to achieve normal distributions. We performed study-specific analyses in the IMPROVE, ULSAM (IMT of the common carotid artery only) and PIVUS studies using logistic regression, adjusting for sex, age and Framingham risk factors, i.e. low-density lipoprotein (LDL)-cholesterol, natural log transformed high-density lipoprotein (HDL)-cholesterol, systolic blood pressure, diabetes status, current smoking status. We subsequently meta-analysed results across studies using a fixed-effects model.
The association of the 45 loci with CHD status and plaque presence was assessed using logistic regression in each study separately, assuming an additive genetic model. We ran all association models twice, once with adjustment for sex, age and multi-dimensional scaling components, to adjust for population structure (in IMPROVE), and once with additional adjustment for Framingham risk factors. The association of the 45 loci with IMT of the bulb and common carotid artery was examined using linear regression as described for CHD status and plaque presence above.
After completing the single SNP association analysis within each of the studies, we performed a fixed effects meta-analysis for each SNP - trait association across studies using the inverse variance method. We subsequently calculated the GPS to examine the association of all 45 CHD-associated loci combined for each of the four traits, using single SNP summary statistics from the meta-analyses as described previously (http://cran.r-project.org/web/packages/gtx/vignettes/ashg2012.pdf) (19). Each SNP had an equal weight in the GPS. Weighting the contribution of each SNP by its previously reported effect size for CHD risk (4) yielded very similar results (Table 2). In order to assess heterogeneity in effect size of the GPS across studies, we also calculated unweighted study specific predisposition scores, followed by a fixed effect meta-analysis across studies using the inverse variance method.
Table 2. Associations of a genetic predisposition score of 45 coronary heart disease-associated loci with coronary heart disease, plaque presence and intima-media thickness of the bulb and common carotid artery after meta-analysis of results from the IMPROVE, MDC-CC, ULSAM and PIVUS studies.
| Model 1 | Model 2 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR/beta | 95% CI | p_uw | p_w | N | I2 | OR/beta | 95% CI | p_uw | p_w | N | I2 | |||
| Coronary heart disease | 1.053 | 1.030 | 1.077 | 5.43x10-6 | - | 5,512 | 60 | 1.055 | 1.031 | 1.079 | 4.13x10-6 | - | 5,370 | 56 |
| Plaque presence | 1.030 | 1.018 | 1.043 | 6.63x10-7 | 4.51x10-8 | 7,636 | 63 | 1.028 | 1.016 | 1.041 | 7.35x10-6 | 1.35x10-6 | 7,503 | 56 |
| IMT bulb | 2.93x10-3 | 1.25x10-3 | 4.61x10-3 | 6.26x10-4 | 8.55x10-5 | 7,480 | 68 | 2.44x10-3 | 7.77x10-4 | 4.11x10-3 | 4.03x10-3 | 1.39x10-3 | 7,350 | 66 |
| IMT CCA | 7.66x10-4 | -8.28x10-5 | 1.61x10-3 | 0.08 | 1.68x10-2 | 9,582 | 59 | 3.11x10-4 | -5.27x10-4 | 1.15x10-3 | 0.47 | 0.21 | 9,445 | 52 |
Model 1 shows associations adjusted for sex and age; model 2 shows associations adjusted for sex, age and Framingham risk factors, i.e. LDL-cholesterol, natural log transformed HDL-cholesterol, systolic blood pressure, diabetes and current smoking; odds ratios (OR) are shown for the association of a genetic predisposition score of the 45 coronary heart disease-associated loci with coronary heart disease and plaque presence at the far wall of the bulb, also known as sinus; betas are shown for the association of a genetic predisposition score with intima-media thickness (IMT) at the far wall of the bulb and common carotid artery (CCA); ORs, betas and their 95% confidence intervals show the effect size per risk allele across the 45 loci - the effect size for association with CHD as described earlier was not taken into account (i.e. an unweighted score); p_uw shows the p-value for the unweighted genetic predisposition score; p_w shows the p-value for a genetic predisposition score in which the effect size of each individual SNP for CHD risk is taken into account (Deloukas et al., 2013 [PMID 23202125]); I2 indicates the proportion of variation in effect size across studies that is due to true heterogeneity. Associations were considered significant if p<0.05 for coronary heart disease, and p<0.017 for carotid atherosclerosis traits, i.e. an α of 0.05 adjusted for three traits.
We performed power calculations in order to appreciate the single-SNP associations in light of the power we had to detect them (Supplementary Table 3). For associations with CHD and plaque presence, we examined the statistical power to detect the odds ratio for association with CHD risk that was previously reported in stage 2 of the CARDIoGRAMplusC4D effort (4). Power calculations were otherwise based on the effect allele frequency of the SNPs after meta-analysis of data from the four studies included here, the number of available cases, i.e. 603 for CHD and 3,650 for plaque presence; the number of controls per case, i.e. 8.1 for CHD and 1.9 for plaque presence; and the baseline disease risk, i.e. 0.109 for CHD and 0.478 for plaque presence in our data.
A meta-analysis of GWAS previously identified two loci for plaque presence and three for IMT of the common carotid artery (20). Two of these loci, rs1878406 near EDNRA on chromosome 4 (associated with plaque presence) and rs445925 near the apolipoprotein cluster on chromosome 19 (associated with IMT of the common carotid artery), were also identified as being associated with CHD by GWAS (4), and were amongst the 45 loci included in the present analysis. We calculated the power we had to detect the association of these two loci with carotid atherosclerosis using the effect size of the replication stage reported previously.
A two-sided p-value <0.05 was considered statistically significant for the association of the GPS and individual loci with CHD risk. For the three indices of carotid atherosclerosis, a p-value <0.017 was considered significant for associations with the GPS (0.05/3 traits). Alpha was set to 1.1x10-3 (0.05/45 loci) in the more exploratory secondary analysis, in which we examined associations of the individual SNPs with the three indices of atherosclerosis. Effect sizes are reported as OR (95% CI) for dichotomous outcomes and beta ± SE for continuous outcomes.
We used SAS version 9.3 for windows (SAS Institute, Cary, NC, USA) for data cleaning and quality control analyses. Association analyses were performed using PLINK version 1.07 (21) in IMPROVE and MDC-CC, and using SNPTEST version 2.4.1 (22) in ULSAM and PIVUS. We performed the meta-analyses using METAL (23), and subsequently used a publically available R-package (http://cran.r-project.org/web/packages/gtx/index.html) applied in STATA (STATA version 12, StataCorp, College Station, TX) to combine single SNP associations in a GPS. We used Quanto version 1.2.4 (24) for the power calculations.
Results
Meta-analysis of the available data showed that plaque presence increased the odds of CHD by 1.49 (95% CI 1.13-1.97, p=5x10-3). Each unit increase in natural log-transformed IMT of the bulb and common carotid artery increased the odds of CHD by 1.92 (95% CI 1.14-3.23, p=0.015) and 2.37 (95% CI 1.43-3.92, p=1x10-3), respectively.
Primary aim: Associations of the GPS with CHD, plaque presence and IMT of the bulb and common carotid artery
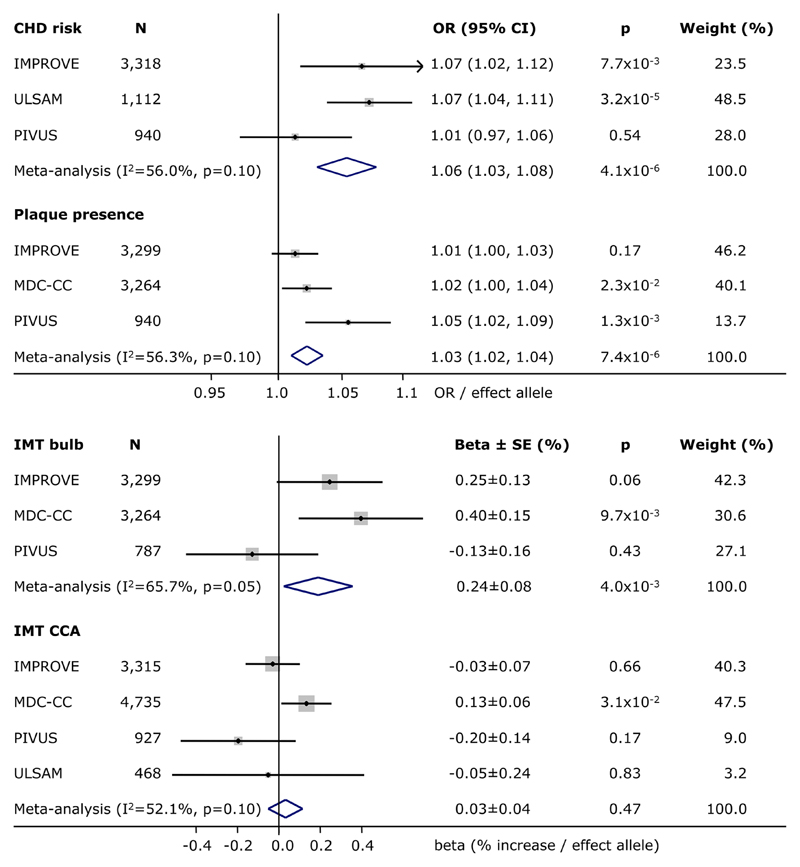
In line with results of the most recent meta-analysis of GWAS, the GPS of 45 established CHD-associated loci was significantly associated with CHD risk (p=4.1x10-6). On average, each additional risk allele across the 45 loci was associated with a 5.5% increased odds of CHD (Table 2, Figure 1). Besides being related to CHD, the GPS was also associated with plaque presence (p=7.4x10-6) and with IMT of the bulb (p=4.0x10-3). Each additional CHD risk increasing allele across the 45 loci was associated with a 2.8% increased odds of plaque, and with a 0.24% increased IMT of the bulb. The GPS was not associated with IMT of the common carotid artery (p=0.47) (Table 2, Figure 1). Heterogeneity in effect size across studies was moderate for all four traits (I2=52-66%).
Figure 1. Association of the 45 CHD-associated loci with carotid atherosclerosis.
Summary statistics for the association of a genetic predisposition score of 45 coronary heart disease (CHD)-associated loci with CHD risk, plaque presence and intima-media thickness (IMT) of the bulb – also known as sinus - and common carotid artery (CCA) in the IMPROVE, MDC-CC, ULSAM and PIVUS studies, as well as after fixed effects, inverse variance weighted meta-analysis. OR and 95% confidence intervals per additional risk allele were provided for associations with dichotomous outcomes, beta and SE expressed in % increase per additional risk allele were provided for continuous outcomes. Weights in % were based on the inverse of the variance. P-values were provided for associations in individual studies, as well as after meta-analysis. I2 and p-values for heterogeneity in effect size across studies were provided for results of the meta-analysis. Studies were ordered by sample size.
Secondary aim: Associations of the individual loci with CHD, plaque and IMT
Associations with CHD were directionally consistent with previously reported results for 34 of the 45 loci (pbinomial=4.0x10-4), and reached significance for four loci in MRAS (rs9818870), in KCNK5 (rs10947789), near LPAL2 and LPA (rs2048327) and near ADAMTS7 (rs7173743) (p<0.05). There was no heterogeneity in effect size across studies for these loci (I2=0%), with the exception of the locus near LPAL2 and LPA (I2=69%) (Supplementary Table 4). The low number of loci for which associations reached significance reflects the low statistical power we had to detect such associations, with a median power based on effect sizes reported previously for CHD risk of 13% (Supplementary Table 3). The statistical power to detect associations with CHD risk only exceeded 80% for the locus near CDKN2B/A on chromosome 9p21.3, for which we did not detect an association with CHD (Supplementary Table 4).
Associations with plaque presence were directionally consistent with previously reported associations with CHD for 34 of the 45 loci (pbinomial=4.0x10-4). After stringent Bonferroni correction, the association with plaque presence only reached statistical significance for the locus near CDKN2B/A on chromosome 9p21.3 (p=2.7x10-4), without evidence of heterogeneity in effect size across studies (I2=0%) (Supplementary Table 5). Each additional risk allele at the 9p21.3 locus was associated with a 13.9% increased odds of plaque presence (p=2.7x10-4). The 9p21.3 locus was one of the two loci for which we had >80% power to detect an association with plaque presence with the effect size reported previously for CHD risk; the median power across the 45 loci was 31% (Supplementary Table 3). We could not confirm the association of rs1878406 near EDNRA with plaque presence (OR=1.08, 95% CI=0.97, 1.20, p=0.15), for which we had 99% power to detect the OR of 1.31 reported earlier (20).
We did not observe an enrichment of directionally consistent associations with IMT of the bulb and common carotid artery when compared with previously reported associations with CHD; 27 of the 45 associations with IMT of the bulb were in the same direction (pbinomial=0.12), as compared with 19 of the 45 associations with IMT of the common carotid artery (pbinomial=0.19). None of the individual loci were significantly associated with IMT of the bulb or common carotid artery after correcting for multiple testing (Supplementary Tables 6 and 7). However, rs445925 near the apolipoprotein gene cluster on chromosome 19 has been identified as associated with IMT of the common carotid artery by GWAS (20), and the association of rs445925 with IMT of the common carotid artery does meet the alpha of 0.05 that is more appropriate for confirmation of established loci in the present study, with a beta that is comparable with the previously reported 1.2% per effect allele (beta=1.3±0.4%, p=1.9x10-3).
Discussion
The present meta-analysis of results from four individual studies clearly showed that a genetic predisposition score, consisting of 45 previously identified CHD-associated loci, was associated with carotid atherosclerosis. The genetic predisposition score was associated with overt plaque presence and IMT of the bulb, independently of traditional cardiovascular risk factors, but not with IMT of the common carotid artery. The fact that effect sizes were not attenuated by adjusting for lipids, systolic blood pressure, smoking and diabetes suggests that these loci influence IMT and plaque formation at the bulb at least in large part via mechanisms other than these traditional risk factors. The genetic predisposition score was also associated with prevalent CHD in the present meta-analysis.
IMT of the bulb and the common carotid artery represent two physiologically separate entities; the former has been associated with CHD, whereas the latter has mainly been associated with stroke and blood pressure control (17,25). We therefore chose to treat IMT at the two locations as separate outcomes, in spite of comparable effect sizes for association with CHD in our study. No uniform definition of plaque occurrence or standardized way of reporting IMT results exists. We defined prevalent plaque on the basis of a cut-off value for IMT of the bulb, since atherosclerosis usually starts in the bulb area. Therefore, plaque presence and IMT of the bulb are two closely related outcomes in our study.
In the present study, we showed an association of the GPS of 45 CHD-associated loci with IMT of the bulb and with prevalent plaques, but not with IMT of the common carotid artery. Based on the before mentioned differences between IMT of the bulb and common carotid artery, a stronger association with IMT of the bulb is perhaps not surprising. A GPS consisting of 13 CHD-associated loci was previously significantly associated with IMT of the bulb, and showed a trend towards an association with common carotid artery in the MDC-CC study, the largest population-based study included in the present meta-analysis (13). In line with these results, we observed a borderline significant association of the GPS of 45 loci with IMT of the common carotid artery in the MDC-CC study after adjusting for traditional risk factors. Assuming the association in MDC-CC is genuine, then the lack of association in the remaining studies may reflect a difference in sampling strategy. For example, participants of the MDC-CC study were younger on average than participants of the other three studies.
We used a GPS that was based on the number of CHD risk increasing alleles in the 45 GWAS-identified loci as our primary exposure variable. Weighting the number of CHD risk-increasing alleles by their effect size for CHD risk as previously reported (4) yielded similar results. Using a GPS results in a higher statistical power when compared with the power to detect associations with individual loci (26). However, using a GPS also has its limitations, since the chain of events leading to CHD and overt myocardial infarction is complex. It is therefore unlikely that all 45 CHD-associated loci influence CHD risk by influencing the risk of atherosclerosis.
Only rs1333049 on chromosome 9p21.3 was significantly associated with plaque presence after stringent Bonferroni correction in our data. Importantly, 9p21.3 was also the only locus for which we had >80% power to detect an association with plaque presence with the same effect size as reported previously for CHD risk. More CHD-associated loci that at least partly influence CHD risk via atherosclerosis remain to be identified, since the GPS was significantly associated with plaque presence even after exclusion of the 9p21.3 locus (p=9.2x10-5), and since we observed an enrichment of SNP - plaque presence associations that were directionally consistent with the SNP - CHD risk associations reported earlier (4). Fourteen of the loci that showed directionally consistent associations across traits were also characterized by a larger effect size for plaque presence in the current study than the effect size reported previously for CHD risk (4).
Based on the results of an earlier GWAS for carotid atherosclerosis, one would conclude that the loci represented by rs1878406 near EDNRA, and rs445925 near APOE, are amongst the loci that at least partly influence CHD risk via atherosclerosis (20). However, we were able to replicate the association of rs445925 with IMT of the common carotid artery, but not the association of rs1878406 with plaque presence, in spite of having adequate statistical power. Still, since the association of rs1878406 with plaque presence was directionally consistent with that reported earlier for CHD risk, and additionally showed a stronger effect size for association with plaque presence than CHD risk, in our study as well as in the literature, we feel that future efforts with larger samples are required to formally confirm or refute the relevance of this locus for plaque presence. Such efforts are anticipated to also identify other CHD-associated loci that at least partly exert their effect on disease risk by influencing atherosclerosis.
The locus on chromosome 9p21.3 that was significantly associated with plaque presence in our study has repeatedly been reported as the locus that is most strongly associated with CHD risk in GWAS (27–29). It has also been identified as being associated with atherosclerosis and other types of cardiovascular diseases (30). This locus has recently been associated with a reduced ANRIL expression, which in turn was associated with common carotid artery stenosis. ANRIL expression was suggested to affect cell growth, possibly via CDKN2A/B regulation (31), but the exact mechanism by which the locus influences atherosclerosis remains to be established.
In spite of being adequately powered (89%), we were unable to detect the established association between the 9p21.3 locus and CHD risk after meta-analysis of data from the IMPROVE, PIVUS and ULSAM studies. This lack of association may reflect survival bias, since participants of the included studies are middle-aged to elderly, and the 9p21.3 locus was previously shown to be more strongly associated with CHD risk in younger individuals (32).
A major strength of the present study is the large number of individuals for whom detailed phenotypic data were available from four studies that represent a rather homogenous population. Only the IMPROVE study also included data from non-Swedish participants, i.e. from individuals of Finnish, Dutch, French and Italian backgrounds. The homogenous study population implies that our results require confirmation in individuals of other ethnic backgrounds. The use of a GPS of 45 CHD-associated loci gave us adequate statistical power to detect associations with CHD risk and three different atherosclerotic outcomes. The power of the secondary association analysis, in which we examined associations of the individual loci with the three atherosclerotic outcomes, was considerably lower, since it required correction for performing 45 independent tests. This most likely resulted in false negative results.
In conclusion, a genetic predisposition score of 45 CHD-associated loci was associated with prevalent CHD and plaques, as well as with IMT of the bulb, independently of traditional risk factors for cardiovascular disease. In contrast, we found no association of the genetic predisposition score with IMT of the common carotid artery.
Perspectives
Genome-wide association studies (GWAS) have so far identified 45 loci as being robustly associated with CHD. With few exceptions (i.e. LDLR, PCSK9, APOB), the causal genes and mechanisms by which these loci influence disease risk are yet unknown. Such information is essential before we can use results from GWAS in the clinic, e.g. for drug development and biomarker identification.
In the present study, we identified two CHD-associated loci for which associations with (subclinical) atherosclerosis reach statistical significance, independently of traditional risk factors. Knowing the mechanism by which these loci influence CHD risk, we recommend prioritizing these loci for functional follow-up studies, aiming to identify the causal genes. Our results indicate that more CHD-associated loci that influence disease risk via atherogenic pathways remain to be identified. Hence, further targeted genetic association studies in larger samples are worthwhile.
Supplementary Material
Acknowledgments
IMPROVE is funded by European Commission (LSHM-CT- 2007- 037273), the Swedish Heart-Lung Foundation, the Swedish Research Council (8691), the Knut and Alice Wallenberg Foundation, the Foundation for Strategic Research, the Torsten and Ragnar Söderberg Foundation, the Strategic Cardiovascular Programme of Karolinska Institutet, the Stockholm County Council and the Stockholm County Council (560183).
PIVUS and ULSAM genotyping was performed by the SNP&SEQ Technology Platform in Uppsala (www.genotyping.se), which is supported by Uppsala University, Uppsala University Hospital, the Swedish Research Council for Infrastructures and the Wellcome Trust (WT098017, WT090532, and WT064890).
Marcel den Hoed is supported by the Swedish Heart-Lung Foundation (20140543).
Rona J Strawbridge is supported by the Swedish Heart-Lung Foundation (20120600), the Tore Nilsson, Gamla Tjänarinnor and Fredrik and Ingrid Thurings foundations.
Steve Humphries is a British Heart Foundation Chair holder (CH/92025) and is funded by RG/08/008 and RG/10/12/28456.
Cecilia Lindgren is a Wellcome Trust Research Career Development Fellow (086596/Z/08/Z)
Erik Ingelsson is supported by grants from the Knut and Alice Wallenberg Foundation, the European Research Council (ERC Starting Grant, 335395), the Swedish Heart-Lung Foundation (20120197), and the Swedish Research Council (2012-1397).
Olle Melander was funded by European Research Council (StG-282255), Swedish Medical Research Council, the Swedish Heart and Lung Foundation, the Novo Nordisk Foundation, the Medical Faculty of Lund University, Malmö University Hospital, the Albert Påhlsson Research Foundation, the Crafoord Foundation, the Ernhold Lundströms Research Foundation, the Region Skane, Hulda and Conrad Mossfelt Foundation and King Gustaf V and Queen Victoria Foundation, the Lennart Hanssons Memorial Fund and the Marianne and Marcus Wallenberg Foundation.
None of the authors declare a relationship with industry.
List of Abbreviations
- CHD
Coronary heart disease
- GWAS
Genome-wide association study
- IMT
Intima-media thickness
- GPS
Genetic predisposition score
- LD
Linkage disequilibrium
- SNAP
SNP Annotation and Proxy Search
- LDL
low-density lipoprotein
- HDL
High-density lipoprotein
- OR
Odds ratio
- RNA
Ribonucleic acid
References
- 1.Kannel WB. Some lessons in cardiovascular epidemiology from Framingham. Am J Cardiol. 1976;37:269–82. doi: 10.1016/0002-9149(76)90323-4. [DOI] [PubMed] [Google Scholar]
- 2.Peden JF, et al. Thirty-five common variants for coronary artery disease: the fruits of much collaborative labour. Human molecular genetics. 2011;20:R198–205. doi: 10.1093/hmg/ddr384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ozaki K, et al. Genome-wide association study to identify single-nucleotide polymorphisms conferring risk of myocardial infarction. Methods in molecular medicine. 2006;128:173–80. doi: 10.1007/978-1-59745-159-8_12. [DOI] [PubMed] [Google Scholar]
- 4.The CARDIoGRAMplusC4D consortium et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nature genetics. 2012;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lorenz MW, et al. Prediction of clinical cardiovascular events with carotid intima-media thickness: a systematic review and meta-analysis. Circulation. 2007;115:459–67. doi: 10.1161/CIRCULATIONAHA.106.628875. [DOI] [PubMed] [Google Scholar]
- 6.O'Leary DH, et al. Thickening of the carotid wall. A marker for atherosclerosis in the elderly? Cardiovascular Health Study Collaborative Research Group. Stroke. 1996;27:224–31. doi: 10.1161/01.str.27.2.224. [DOI] [PubMed] [Google Scholar]
- 7.Rosvall M, et al. Incident coronary events and case fatality in relation to common carotid intima-media thickness. Journal of internal medicine. 2005;257:430–7. doi: 10.1111/j.1365-2796.2005.01485.x. [DOI] [PubMed] [Google Scholar]
- 8.Rosvall M, et al. Incidence of stroke is related to carotid IMT even in the absence of plaque. Atherosclerosis. 2005;179:325–31. doi: 10.1016/j.atherosclerosis.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 9.Amato M, et al. Carotid intima-media thickness by B-mode ultrasound as surrogate of coronary atherosclerosis: correlation with quantitative coronary angiography and coronary intravascular ultrasound findings. Eur Heart J. 2007;28:2094–101. doi: 10.1093/eurheartj/ehm244. [DOI] [PubMed] [Google Scholar]
- 10.Graner M, et al. Association of carotid intima-media thickness with angiographic severity and extent of coronary artery disease. Am J Cardiol. 2006;97:624–9. doi: 10.1016/j.amjcard.2005.09.098. [DOI] [PubMed] [Google Scholar]
- 11.Hansen T, et al. A total atherosclerotic score for whole-body MRA and its relation to traditional cardiovascular risk factors. European radiology. 2008;18:1174–80. doi: 10.1007/s00330-008-0864-6. [DOI] [PubMed] [Google Scholar]
- 12.Kablak-Ziembicka A, et al. Diagnostic value of carotid intima-media thickness in indicating multi-level atherosclerosis. Atherosclerosis. 2007;193:395–400. doi: 10.1016/j.atherosclerosis.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 13.Hamrefors V, et al. A myocardial infarction genetic risk score is associated with markers of carotid atherosclerosis. Journal of internal medicine. 2012;271:271–81. doi: 10.1111/j.1365-2796.2011.02472.x. [DOI] [PubMed] [Google Scholar]
- 14.Baldassarre D, et al. Measurements of carotid intima-media thickness and of interadventitia common carotid diameter improve prediction of cardiovascular events: results of the IMPROVE (Carotid Intima Media Thickness [IMT] and IMT-Progression as Predictors of Vascular Events in a High Risk European Population) study. J Am Coll Cardiol. 2012;60:1489–99. doi: 10.1016/j.jacc.2012.06.034. [DOI] [PubMed] [Google Scholar]
- 15.Magnusson M, et al. A diabetes-predictive amino acid score and future cardiovascular disease. Eur Heart J. 2013;34:1982–9. doi: 10.1093/eurheartj/ehs424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wohlin M, et al. Apolipoprotein E epsilon4 genotype is independently associated with increased intima-media thickness in a recessive pattern. Lipids. 2007;42:451–6. doi: 10.1007/s11745-007-3045-5. [DOI] [PubMed] [Google Scholar]
- 17.Andersson J, et al. Echogenecity of the carotid intima-media complex is related to cardiovascular risk factors, dyslipidemia, oxidative stress and inflammation: the Prospective Investigation of the Vasculature in Uppsala Seniors (PIVUS) study. Atherosclerosis. 2009;204:612–8. doi: 10.1016/j.atherosclerosis.2008.10.038. [DOI] [PubMed] [Google Scholar]
- 18.The 1000 Genomes Project consortium et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burgess S, et al. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37:658–65. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bis JC, et al. Meta-analysis of genome-wide association studies from the CHARGE consortium identifies common variants associated with carotid intima media thickness and plaque. Nature genetics. 2011;43:940–7. doi: 10.1038/ng.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marchini J, et al. A new multipoint method for genome-wide association studies by imputation of genotypes. Nature genetics. 2007;39:906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 23.Willer CJ, et al. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gauderman WJ, Morrison JM. QUANTO 1.1: A computed program for power and sample size calculations for genetic-epidemiology studies. 2006 http://hydrauscedu/gxe. [Google Scholar]
- 25.Ebrahim S, et al. Carotid plaque, intima media thickness, cardiovascular risk factors, and prevalent cardiovascular disease in men and women: the British Regional Heart Study. Stroke. 1999;30:841–50. doi: 10.1161/01.str.30.4.841. [DOI] [PubMed] [Google Scholar]
- 26.Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLoS genetics. 2013;9:e1003348. doi: 10.1371/journal.pgen.1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helgadottir A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science (New York, NY) 2007;316:1491–3. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 28.McPherson R, et al. A common allele on chromosome 9 associated with coronary heart disease. Science (New York, NY) 2007;316:1488–91. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samani NJ, et al. Genomewide association analysis of coronary artery disease. The New England journal of medicine. 2007;357:443–53. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holdt LM, et al. Recent studies of the human chromosome 9p21 locus, which is associated with atherosclerosis in human populations. Arterioscler Thromb Vasc Biol. 2012;32:196–206. doi: 10.1161/ATVBAHA.111.232678. [DOI] [PubMed] [Google Scholar]
- 31.Congrains A, et al. Genetic variants at the 9p21 locus contribute to atherosclerosis through modulation of ANRIL and CDKN2A/B. Atherosclerosis. 2012;220:449–55. doi: 10.1016/j.atherosclerosis.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 32.Palomaki GE, et al. Association between 9p21 genomic markers and heart disease: a meta-analysis. JAMA. 2010;303:648–56. doi: 10.1001/jama.2010.118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.