Summary
Morphospaces are mathematical representations used for studying the evolution of morphological diversity and for the evaluation of evolved shapes among theoretically possible ones. Although widely used in zoology, they – with few exceptions – have been disregarded in plant science and in particular in the study of broad-scale patterns of floral structure and evolution. Here we provide basic information on the morphospace approach; we review earlier morphospace applications in plant science; and as a practical example, we construct and analyze a floral morphospace. Morphospaces are usually visualized with the help of ordination methods such as principal component analysis (PCA) or nonmetric multidimensional scaling (NMDS). The results of these analyses are then coupled with disparity indices that describe the spread of taxa in the space. We discuss these methods and apply modern statistical tools to the first and only angiosperm-wide floral morphospace published by Stebbins in 1951. Despite the incompleteness of Stebbins’ original dataset, our analyses highlight major, angiosperm-wide trends in the diversity of flower morphology and thereby demonstrate the power of this previously neglected approach in plant science.
Keywords: disparity, fitness landscape, floral structure, morphology, morphospace, multivariate statistics, pollination syndrome
The descriptive and synthetic value of morphospaces
Three main processes affect the evolution of species: variation, channeled by constraints (e.g. historical, developmental or mechanical); differential survival, channeled by selection (e.g. as exerted by biotic and abiotic conditions; Pearce, 2011); and chance (evolutionary drift). However, despite the vast diversity of plants that has arisen through these processes, not all of the possible phenotypic configurations are represented among extant plants. Some traits and trait combinations are vastly represented, while the majority of theoretically possible phenotypes were never accomplished in the course of evolution.
A widely used approach in zoology to study the evolution of realized phenotypes compared to those that are theoretically possible is to construct morphospaces: mathematical spaces describing and relating the phenotypes of organisms (Fig. 1). Because organisms differ in a large number of properties, these mathematical spaces typically are multidimensional (Raup & Michelson, 1965; Foote, 1997; McGhee, 1999; Mitteroecker & Huttegger, 2009). Visual descriptions of morphospaces hence are usually based on low-dimensional representations, so-called ordinations, of the space. In theoretical morphospaces, the axes of the reduced space are determined by a small set of parameters of morphogenetic or other biological models, derived from theoretical considerations rather than from the organisms themselves. For example, Raup & Michelson (1965) formulated a mathematical model comprising four variables (dimensions) that describe the theoretically possible gross geometries of coiled shell shapes. In empirical morphospaces, low-dimensional representations of a morphospace optimize the description of a measured sample of organisms. This approach, which is commonly used in morphometrics, is typically based on statistical ordination techniques such as principal component analysis or multidimensional scaling (MDS).
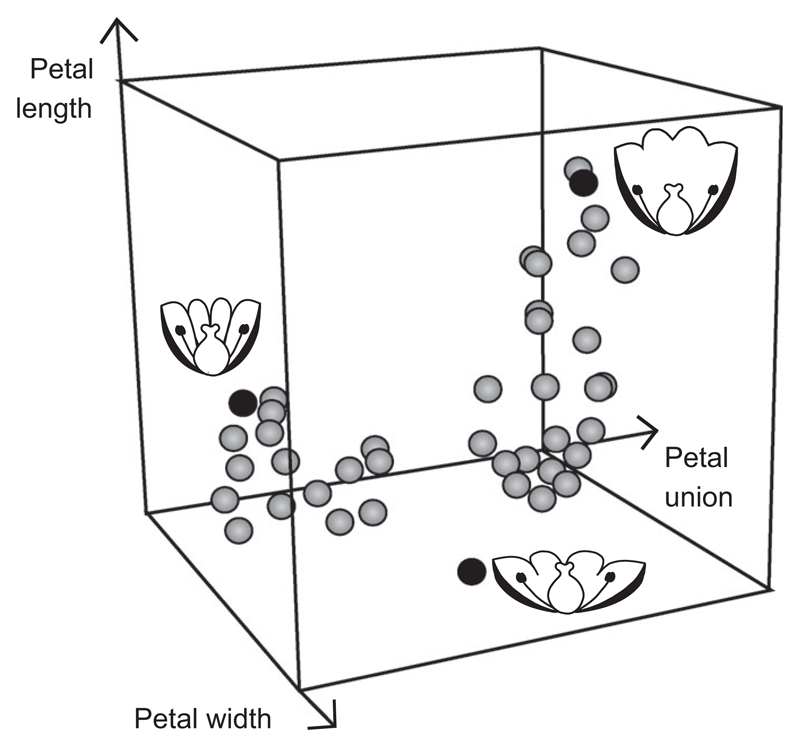
Fig. 1.
Illustration of a hypothetical 3D Euclidean flower morphospace. Each dimension is a variable describing a quantitative aspect of floral morphology (petal width, petal length, degree of petal union), and each point in this space is the position of a hypothetical flower in morphospace. Three of these flowers (black dots) are represented as sketches to illustrate that any displacement in this space that is not parallel to any of the axes can be directly interpreted as a simultaneous change in petal size and union.
The proportion of the morphospace occupied by extant species, the distribution of species and clades in the morphospace, and their variation through evolutionary time can be used to investigate various aspects of organismal evolution and diversity. For instance, Raup (1966, 1967) placed brachiopods, gastropods, bivalves and cephalopods in the morphospace of coiled shells (Raup & Michelson, 1965), showing that only a small part of the theoretical space was occupied. These clades clustered in different, relatively narrow parts of the space, indicating different functional, ecological and phylogenetic constraints.
The morphospace approach has been used to test a multitude of hypotheses in various systematic groups, such as the description of the temporal evolution of Crinoid fossils (Foote, 1995) or the phylogenetic and temporal distribution of morphological diversity in arthropods (Briggs et al., 1992). Other examples include the identification of modularity and integration in bat jaw structure (Monteiro & Nogueira, 2010), cichlid fish skulls (Cooper et al., 2010) and human crania (Mitteroecker et al., 2012). Ecological vs developmental constraints on the variation of butterflies wing eyespot color could be identified in a morphospace by showing that some artificially selected phenotypes could only diverge from the wild phenotype position to some specific areas in the space (Allen et al., 2008). Finally, morphospaces were also used to highlight trends of diversification in cichlid fish clades that spread in distinct parts and to different extents in the morphospace (Sidlauskas, 2008). So far, most such studies have focused on diverse features in groups of animals (Stone, 1997; McGhee, 1999; Erwin, 2007) such as mollusk and brachipod shells (Raup, 1966, 1967; McGhee, 1980; Wagner, 1995), hymenoptera wings (Perrard et al., 2012), fish skulls (Sidlauskas, 2008; Cooper et al., 2010), mammal skulls or skull parts (e.g. Carnivora, Drake & Klingenberg, 2010; mice, Klingenberg et al., 2004; bats, Monteiro & Nogueira, 2010; hominids, Mitteroecker et al., 2004; Gunz et al., 2008), and even women’s footprints (Domjanic et al., 2013). Few studies have focused on plants, investigating for instance the evolution and/or adaptive value of pollen type (Lupia, 1999; Ressayre et al., 2002) and seed plant conductive vessels (Wilson & Knoll, 2010), the fitness associated with different vegetative (Niklas & Kerchner, 1984; Niklas, 2004) or inflorescence (Prusinkiewicz et al., 2007) architectures, and various other functional traits (e.g. growth form, pollination and dispersal modes: Silva & Batalha, 2011). Morphospaces have also been used to investigate evolutionary trends in leaf shapes from fossil plant lineages (Boyce & Knoll, 2002), to compare the development of different shoot structural levels (Lacroix et al., 2003), to investigate the variation and the genetic determinism of leaf shape (Langlade et al., 2005; Chitwood et al., 2014), the plasticity of compound leaf shapes (Klingenberg et al., 2012) and the shape of organs in vegetative shoot systems (Jeune & Sattler, 1996; Jeune et al., 2006; Burns et al., 2008). With the exception of studies in pollination ecology (see section ‘The use of floral morphospaces in evolutionary ecology’), the morphospace of flowers has received little attention so far (but see Stebbins, 1951; and for flower color: Whibley et al., 2006 and Stournaras et al., 2013).
Here, we review and critically discuss the use of modern concepts of morphospace analysis to study floral structure and evolution. We first demonstrate how a morphospace approach can help to answer crucial questions on the evolution of flowers and suggest lines of work for flower morphologists and botanists. We then review the use of morphospaces in studies on pollinator-mediated selection and fitness variation in flowers, and finally apply up-to-date analytical methodology to reinterpret the seminal work of Stebbins (1951), who conducted the first and only broad-scale morphospace study focusing on angiosperm flowers. To conclude, we outline perspectives for future work integrating morphospace approaches with the study of flower diversity and evolution.
Morphospaces: a promising method to study flower morphological evolution
There is an extensive literature on the evolution of flowers, dealing with the physical and developmental constraints linked to floral organization, construction and mode (see Endress, 1994; for a conceptual framework of flower structure) and the selective pressures linked to the need to produce, protect and disperse plant gametes and seeds (Harder & Barrett, 2006). Traditional studies comprise morphological and developmental comparisons among clades of extant and extinct species (Endress, 2010, 2011; Schönenberger et al., 2010, 2012; Friis et al., 2011; von Balthazar & Schönenberger, 2013). During the past two decades, our knowledge of angiosperm evolution has grown dramatically due to great advances in technical and analytical methods in two main fields. Paleobotanical studies have greatly advanced our understanding of the early history of angiosperms. Particularly important in this respect was the recovery and detailed study of many well-preserved charcoalified fossil flowers from the Cretaceous, which yielded crucial information on early flowers and their diversification (Crane et al., 1995; Schönenberger, 2005; Friis et al., 2011). At the same time, molecular phylogenetics and cladistics have revolutionized our understanding of angiosperm systematics (Stevens, 2001; APG III, 2009; Soltis et al., 2011) and now constitute invaluable tools for further studies on the evolution of floral features by inferring species diversification rates and dating major events/innovations in the evolution of angiosperms (Magallón & Castillo, 2009; O’Meara, 2012). In addition, new insights from evolutionary developmental biology (Evo-Devo; Rosin & Kramer, 2009; Preston et al., 2011) and from modeling floral morphology (reviewed by Jabbour & Citerne, 2010) have critically advanced our understanding of the evolutionary history of floral structure and floral diversity.
These complementary fields provide us with a wealth of data and hypotheses to study the evolutionary history of flowers and vegetative structure of angiosperms. In this context, morphospaces will be particularly powerful to highlight the relationships among extant flower morphologies (horizontal sampling) as well as among extant and extinct ones (vertical sampling). By relating theoretically possible but nonexisting morphologies to rare or particularly successful floral morphologies, functional, developmental and phylogenetic constraints can be studied. For example, one can evaluate whether the evolution of perianth symmetry is less constrained in Ranunculales (basal eudicots) than in Asteridae (core eudicots) as has been suggested by Jabbour et al. (2009).
Yet another use of morphospace methods is to identify factors that might have led to the repeated evolution of particular trait combinations. Such factors may be functional, biogeographic, ecological (e.g. breeding system, growth form, pollination mode; see section ‘The use of floral morphospaces in evolutionary ecology’) or historical. Adding phylogenetic relationships to a morphospace enables the investigation of the relationship between taxonomic diversity (the number of species) and disparity (morphological diversity). For example, in the case of an adaptive radiation, clades with high species numbers may occupy a large portion of morphospace. By contrast, species-rich clades may cluster in a narrow part of morphospace if they share key innovations of high adaptive value (Harmon et al., 2003; Erwin, 2007). In angiosperms, such questions have never been addressed using quantitative tools. The application of a morphospace approach thus promises novel insights into patterns and processes of flower evolution.
The alternative horizontal approach comprises the construction and comparison of morphospaces for different sets of characters (e.g. perianth, androecium or gynoecium), which are involved in different functions (e.g. pollinator attraction, pollen dispersal or ovule protection) and therefore are under different selective pressures (Wagner, 1996). Hypotheses that can be tested and quantified in this way are, for example, the limited variation in stamen size and shape compared to sepals and petals (Endress, 1994), or the great variation of pollinator-attractive traits in deceptive species (see Ackerman et al., 2011).
Finally, morphospaces allow for studying the unfolding of clade disparity during ontogeny or over geologic timescales (vertical sampling strategies) through comparison of flowers from different developmental stages (constructing developmental morphospaces; Gerber et al., 2007) or fossils from different geological periods (Wagner, 1996). The latter type of analysis can be complemented by inferences from ancestral state reconstructions of characters (Sidlauskas, 2008; Roelants et al., 2011). In the presence of a useful underlying model of trait evolution, these reconstructions can be helpful to study variation of morphological diversity in relation to variation of taxonomic diversity, for example in the case of extinction events (Fig. 2; Wagner, 1996; Lupia, 1999).
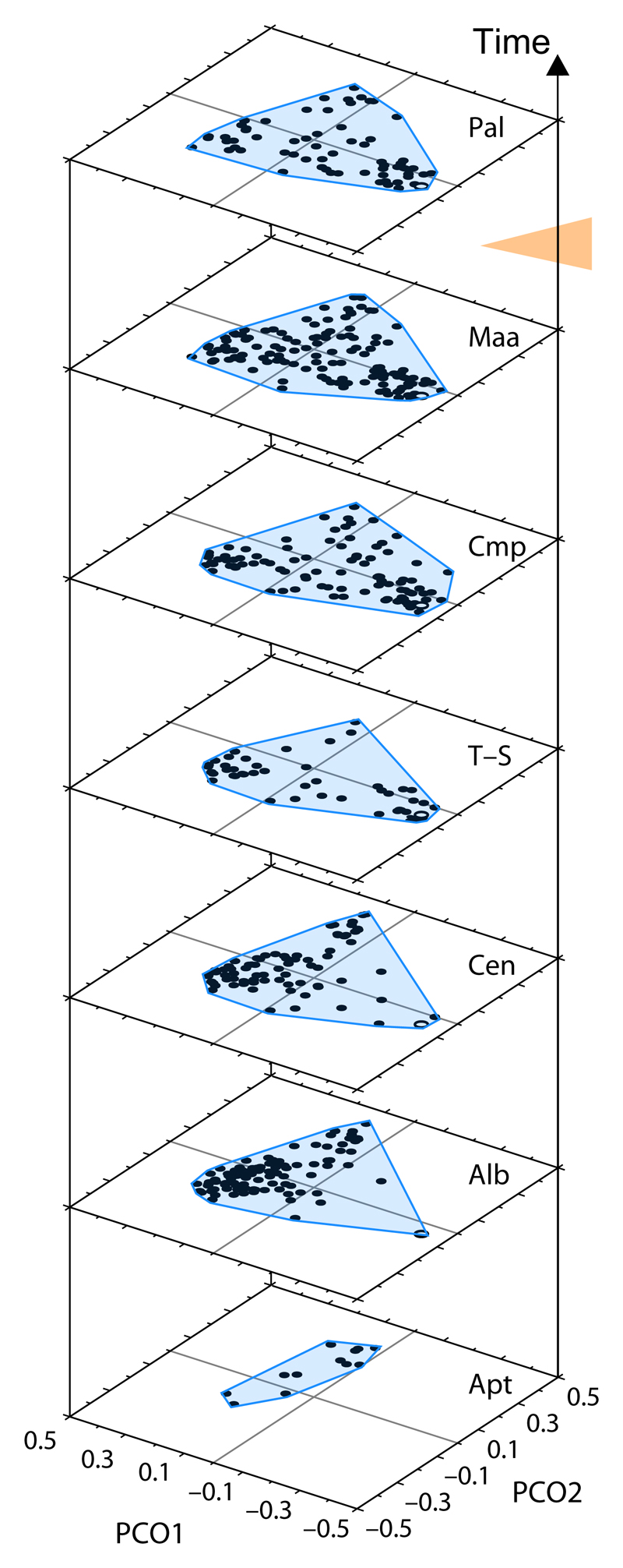
Fig. 2.
Angiosperm pollen morphospace (built from 29 characters such as aperture type and position) through the Cretaceous and the Paleocene, based on North American fossils. The morphospace was constructed by plotting the scores of each species on the first two principal coordinate axes for each time interval. The evolution of morphospace occupation of angiosperm pollen over time is represented here by the size of the blue-shaded areas (convex hulls). Angiosperm taxonomic diversity decreased during the Cretaceous/Tertiary boundary extinction (orange arrowhead). Lupia (1999) showed that this event was not accompanied by a decrease in pollen disparity in the fossil record. Pal, Paleocene; Maa, Maastrichtian; Cmp,Campanian; T–S, Turonian to Santonian; Cen, Cenomanian; Alb, Albian; Apt, Aptian. Figure modified, with permission, from Lupia (1999) © 1999 The Paleontological Society.
From theory to practice
What morphospaces can and cannot do
The structure of a morphospace is determined by the choice of the characters to include and by the way these characters are coded. This selection of morphological properties, which is based on the research question, the systematic group and the material available for study, can affect the assessment of the occupied vs empty parts of the morphospace (Maclaurin, 2003; Huttegger & Mitteroecker, 2011). The actual coding of characters further influences the way morphological (dis)similarity is quantified (the metric imposed on the space) and what the possible transitions between character states are (Mitteroecker & Huttegger, 2009). The choice of a particular type of variable and metric that properly reflect the studied developmental, functional or evolutionary properties of the organisms is often not trivial. For example, in flowers, the evolution toward an increase in petal (or other organ) number may be achieved by the addition of one petal (e.g. from 4 to 5 petals) in a clade or by its multiplication (e.g. from 3 to 6) in another clade. The numbers originate from counts, but these types of characters might rather be treated as nominal (categories) or ordinal data (ranked categories), depending on the evolutionary model employed.
For characters that can be represented by a real number with an interval or ratio scale, a morphospace can be constructed by taking the measurements or parameters as Cartesian coordinates of an Euclidean space (Fig. 1). This is the common approach in morphometrics and for many theoretical morphospaces (Herrera, 1993; Medel et al., 2003; Shipunov & Bateman, 2005; Yoshioka et al., 2006; Gómez et al., 2009; van der Niet et al., 2010). In such a morphospace, the distance between points can be interpreted as morphological dissimilarity. Several points on a straight line in morphospace (e.g. a developmental or evolutionary trajectory) differ in the same morphological pattern, and the angle between two such linear directions can be interpreted as the magnitude of divergence between the corresponding morphological patterns or trajectories (Mitteroecker & Huttegger, 2009). However, Euclidean morphospaces with their wealth of interpretable geometric relationships are only possible for relatively small degrees of morphological diversity and for geometrically independent variables with comparable units. For example, in a space constituted by variables of different units, distances and angles cannot be interpreted – only linearity, intermediacy and patterns such as clusters of organisms (so-called affine geometry; Huttegger & Mitteroecker, 2011). For organisms differing in qualitative properties, such as the presence or absence of organs, or organ arrangement (phyllotaxis), no unique quantitative notion of similarity is possible because the variables possess a nominal scale only. Spaces with weak geometric relations (topological or pretopological spaces) may still be possible to construct (Stadler et al., 2001). In general, the wider the studied morphological diversity, the weaker the mathematical and geometric properties of the morphospace (Mitteroecker & Huttegger, 2009).
Finally, assessing organ homology can be challenging when dealing with characters of distantly related clades. As a typical example in flowers, petaloid structures may be derived either from bracts, sepals or stamens, and the distinction between sepals and petals is often unclear (Kramer & Jaramillo, 2005; Ronse de Craene & Brockington, 2013).
How to visualize morphospaces
Depending on the type of data (nominal, ordinal or scale), morphospace representations are built using different methods. When dealing with two or three continuous scale characters (e.g. petal length and petal width), a morphospace can be simply represented as a two- (2D) or three-dimensional (3D) diagram in which each axis represents one of the variables (Allen et al., 2008; Fig. 1). When dealing with more than three characters (e.g. the length and width of three different kinds of floral organs), ordination methods are used to reduce the number of variables in such a way that relationships between individuals can be adequately represented in a 2D or 3D diagram. Widely used ordination methods are Principal Component Analysis (PCA), which creates a low-dimensional ordination that maximizes the variation between the measured specimens, and between-group PCA, which maximizes the variation between group means (Dommergues et al., 1996; Drake & Klingenberg, 2010; Córdoba & Cocucci, 2011; Mitteroecker & Bookstein, 2011; Mitteroecker et al., 2013).
While PCA can only be used for variables at an interval scale, metric and nonmetric multidimensional scaling (MDS and NMDS) are more general techniques that are also suited for ordinal and nominal data as long as a notion of (dis)similarity can be expressed numerically (Ollerton et al., 2009; Xiao & Laflamme, 2009; Bröderbauer et al., 2013). MDS, which is also referred to as principal coordinate analysis (PCoA), yields an ordination in which the Euclidean distances between the specimens in the low-dimensional representation approximate the original measures of (dis)similarity in a least-squares sense. If these original measures are Euclidean distances, then MDS leads to the same ordination as PCA. By contrast, NMDS ordinates the original distances or dissimilarity by maximizing the correlation between the ranked distances (Rabinovitz, 1986).
Ordination methods are powerful tools for the exploration of high-dimensional data, but they can also lead to misinterpretation, especially when single components are interpreted separately or in the presence of polynomial relationships between variables. For more details see Swan (1970), Mardia et al. (1979), Mitteroecker et al. (2005) and Bookstein (2013, 2014).
The statistical significance of group differences in mean or variance in the morphospace are usually tested by multivariate nonparametric or randomization tests, such as nonparametric analysis of variance (Anderson, 2001), analysis of similarity (Clarke, 1993), or permutation tests (Good, 2000).
Phylomorphospaces, in which (time-calibrated) phylogenetic frameworks are plotted within morphospaces, provide temporal landmarks for transitions identified in a morphospace. In addition, they allow us to highlight trends in the morphological diversification of taxa and to compare the magnitude and the direction of their morphological evolution (Macholán, 2006; Clabaut et al., 2007; Sidlauskas, 2008). Phylogenetic relationships are usually added to the morphospace by reconstructing possible ancestral states for all nodes of the phylogeny and including them in the morphospace (Sidlauskas, 2008; Klingenberg & Gidaszewski, 2010), or by linking extant taxa and their related fossils in the morphospace (Macholán, 2006; Benson & Choiniere, 2013). This method remains limited by phylogenetic and ancestral state reconstruction uncertainties.
How to measure morphological diversity (disparity)?
Disparity indices quantify the morphological diversity of a clade, that is the distribution of its constitutive taxa into vast or limited regions of the morphospace (Wills et al., 1994; Foote, 1997; Erwin, 2007). Examples of such indices are the sum of the variances of all characters (usually referred to as total variance), the mean pairwise distance between individuals (Foote, 1992, 1995; Wills et al., 1994), or the determinant of the sample variance-covariance matrix (generalized variance; Huttegger & Mitteroecker, 2011). All these indices are scalar summary statistics of multivariate distributions. They crucially depend on assumptions about common scales and the geometric independence of the variables (Huttegger & Mitteroecker, 2011). Reliable scientific inference (i.e. unaffected by factors such as sample size and the number and type of variables) may require the comparison of different disparity indices and ordinations (Foote, 1997; Ciampaglio et al., 2001).
Temporal changes in disparity values can be related to variation in taxonomic diversity. The evolution of taxonomic diversity in time (‘clade shape’) has, for example, been quantified by Gould et al. (1987) for marine invertebrates. They investigated whether the extant diversity of a clade evolved from a larger ancestral diversity (bottom heavy distribution) or, instead, increased steadily over time during evolution (top heavy distribution). Similar approaches have later been applied to quantify clade shape in the context of morphologic diversity (Wagner, 1995; Foote, 1997; Lupia, 1999; Hughes et al., 2013). The evolution of disparity through time and patterns of diversification can also be inferred on phylogenies by calculating the disparity through time (DTT) index, a measure of subclades’ disparity relative to their age (Harmon et al., 2003).
The evolution of disparity is central to understanding the evolutionary history of taxa. For example, it has been suggested that a high disparity might allow taxa to overcome extinction events more easily, as they have a broader spectrum of possibilities for adaptation to changing environments (Foote, 1997; Chevin et al., 2010).
The use of floral morphospaces in evolutionary ecology
Pollination syndromes
Pollination syndromes, or floral syndromes, are sets of floral traits, like shape, color, scent or rewards, which flowers have evolved as adaptations to different pollen vectors and which, theoretically, may appear repeatedly in unrelated lineages as a result of convergent evolution. A classification of plants according to pollination syndromes was first proposed by Delpino (Vogel, 1954, 2012), and later elaborated by Vogel (1954, 2012), van der Pijl (1961), Faegri & Van Der Pijl (1966), Endress (1994) and Proctor et al. (1996).
Morphospaces have been used in studies on pollination syndromes in three main ways. (1) As tools for describing pollination systems, that is, identifying the floral traits associated with various pollen vectors. Plant species under the same pollination regime are expected to cluster together in a space constructed with these traits (Wilson et al., 2004; Córdoba & Cocucci, 2011; Ortega-Olivencia et al., 2012; Bröderbauer et al., 2013; Maia et al., 2013). For example, Ortega-Olivencia et al. (2012) used a morphospace made of 35 floral traits to show that the bird pollination syndrome in Scrophularia was related to the showy colors of the corolla and staminodes, the lateral narrowing of the corolla tube and the absence of nectar guides. (2) Morphospaces were used to test the relevance of previously described pollination syndromes by investigating whether a given plant species’ distribution in the morphospace corresponds to the definition of the pollination syndrome. For example, Ollerton et al. (2009) ordinated 537 floral trait combinations corresponding to 11 theoretical pollination syndromes into a 3D space (a theoretical morphospace). They added plant species from six plant communities to this space, in which they had made observations of the most effective pollinators. Few of these species fell into the area corresponding to the predicted theoretical pollination syndrome, because these syndromes were not relevant, or because some of the plotted species were not specialized enough to their pollinators to exhibit the corresponding floral syndromes (see also Hingston & McQuillan, 2000; Ollerton & Watts, 2000). (3) Morphospaces were used to infer the pollen vectors of nondocumented pollination systems by including such species in the morphospace and evaluating whether they fall in an area of the space already occupied by closely related species belonging to documented pollination systems (Gibernau et al., 2010; Bröderbauer et al., 2013).
Instead of focusing on trait convergences, morphospace approaches and the calculation of disparity indices have also been used to demonstrate floral morphological divergence. It has been shown that in certain plant communities floral divergence was positively selected for, so that pollinators discriminate between different plant species and do not transfer heterospecific pollen by indiscriminate foraging (Eaton et al., 2012).
Finally, pollination syndromes have also been investigated on the basis of nonmorphological phenotypic characters, for instance scent, nectar composition and time of anthesis, by constructing phenotype spaces (Ollerton & Watts, 2000; Ollerton & Raguso, 2006; Cortis et al., 2009; Jürgens et al., 2013).
The sensory color space
Sensory color spaces are theoretical spaces used to evaluate the pollinators’ ability to discriminate between flower colors, an important prerequisite for studying the evolution of visual communication within small (e.g. co-occurring Oxalis species; de Jager et al., 2011) or large taxonomic groups (e.g. 876 plant species from all over the world; Stournaras et al., 2013).
Sensory color spaces were first proposed by Burkhardt (1989) and Goldsmith (1990) for bird vision, and have been used in studies investigating the ability of birds to discriminate between congeners’ plumages (Endler et al., 2005; Stoddard & Prum, 2008) or egg colors (Stoddard & Stevens, 2011). Each color is described as a point in a polyhedron or a polygon, whose position is determined by the relative stimulation of the different receiver photoreceptors. For example, in birds the sensory color space is a tetrahedron in which positions from each tip correspond to the relative stimulation of one of the four retinal cone types (UV-, small-, medium- and long-wavelength sensitive; Fig. 3).
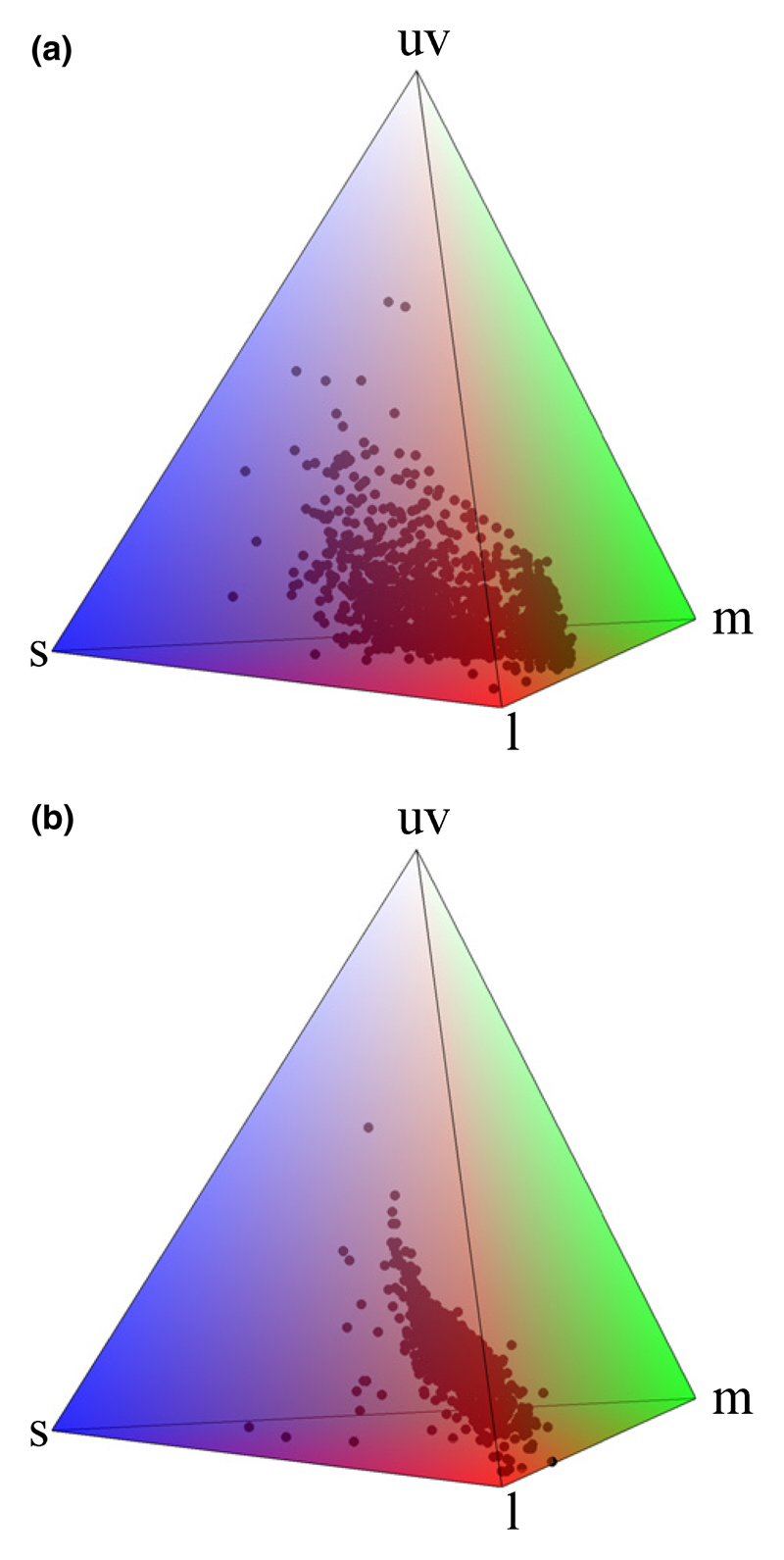
Fig. 3.
Avian sensory color space filled with (a) 1300 reflectance data from the flowers of 876 plant species (as some flowers present several colors, several points belong to the same species); (b) reflectance data of fruits from 948 plant species. uv, l, m and s represent the maximum stimulation of uv-, long-, medium- and short-wavelength sensitivity cones of birds’ eyes. The authors showed that color diversity of fruits, measured as the volume of convex hulls in the space and corrected for the difference in sample size, was almost half the color diversity of flowers. Figure reproduced, with permission, from Stournaras et al. (2013) © 2013 John Wiley & Sons Inc.
Sensory spaces have been used to investigate plant color variation patterns in (bumble) bee- (Chittka, 1992, 1997; Tastard et al., 2008; Arnold et al., 2009), fly- (Troje, 1993) and bird-pollinated flowers, and bird-dispersed fruits (Burns et al., 2009; Stournaras et al., 2013). In most of the studies on bee-pollinated species, no pollinator-driven divergence (or convergence) in color characters was found (see also Gumbert et al., 1999; Arnold et al., 2009; de Jager et al., 2011), suggesting that there is no bee-driven selection of flower color. Alternatively, color variation may be constrained and thus hidden by other effects, such as pleiotropy, selection by herbivores or responses to environmental stress (Rausher, 2008; Schaefer & Ruxton, 2011). However, Stournaras et al. (2013) have shown that the volume of the avian color space occupied by fruits is much smaller than the one occupied by flowers, suggesting that bird pollinators and dispersers might exert different selective pressures on flower and fruit colors, or that the constraints on color pigment production are higher on fruits than on flowers (Fig. 3).
The latter study (see also Chittka et al. (1994) for a comparison of leaves and flowers) illustrates that sensory spaces are ‘standardized, generalizable and transportable’, meaning that different biological models can be ordinated in the same space during successive independent analyses (Stoddard & Prum, 2008).
Fitness landscapes
Fitness landscapes, first introduced by Sewall Wright in 1932 are spaces in which allele combinations (of individuals) or gene frequencies (of populations) are associated with fitness values. Simpson’s (1944) adaptive landscapes are an extension of Wright’s concept to phenotypic traits; adaptive landscapes hence are phenotype spaces or morphospaces, augmented with a fitness dimension (cited in Mitteroecker & Huttegger, 2009). Linking fitness values to floral phenotypes has been used to understand the mechanisms underlying speciation and the maintenance of hybrid zones (Whibley et al., 2006). It has also been used to quantify and represent correlative selection on floral traits (Schluter & Nychka, 1994; Campbell, 2009). For a 2D phenotype space, the adaptive landscape is represented as a surface with ‘peaks’ and ‘valleys’ that correspond to phenotypes of high and low fitness, respectively. Fitness landscapes and adaptive landscapes are central concepts in evolutionary ecology, both as metaphors and as actual mathematical models (Arnold et al., 2001). In Prusinkiewicz et al. (2007), for instance, an axis corresponding to a life history or climatic parameter was added to a 2D theoretical morphospace of inflorescence architecture. Fitness variation was modeled according to this parameter and highlighted by colors so that trajectories through paths of high adaptive value could explain evolution from a type of inflorescence to another in a changing environment. Such high fitness paths are called evolutionary wormholes by the authors (Prusinkiewicz et al., 2007).
Note that inferring fitness only from the distribution of groups in morphospace, without any independent fitness estimate, can be highly misleading. For example, even in the absence of any selection (i.e. equal fitness for all morphologies), the groups are unlikely to be equally distributed across morphospace. Clustering in morphospace is possible even in the absence of fitness peaks and valleys (Pie & Weitz, 2005).
Fitness is the sum of survival and reproductive success (Schluter, 1988). For plants, a good approximation of fitness is the sum of female (seed set) and male (seeds sired) reproductive success (Strauss, 1997; Dafni et al., 2005). These measures can be rather difficult to obtain and the fitness associated with a phenotype is thus often estimated by one or several of its components, such as pollen/pollinium removal (male success), pollinium reception and fruit/seed set (female success; O’Connell & Johnston, 1998; Maad, 2000; Conner et al., 2009; Benitez-Vieyra et al., 2010), pollinator visitation rates (Campbell, 2009), or simulations of population growth from one year to the next (Prusinkiewicz et al., 2007).
Morphospaces and the evolution of flower morphology: a new approach to Stebbins’ data
Stebbins’ topological space
The first floral morphospace, and to our knowledge the only one assessing the proportion of floral shapes among possible ones that actually evolved in nature, was created by Stebbins in 1951. Stebbins’ aim was to find evidence of the role of natural selection on floral trait evolution by demonstrating that some forms repeatedly appeared among angiosperms. He assumed that the adaptive values of floral traits were interdependent and that selection should therefore be studied using character combinations rather than single characters. Stebbins constructed a floral morphospace with eight binary characters related to the perianth, the androecium and the gynoecium (Table 1), and included data for 288 angiosperm families (as circumscribed in 1951). To visualize this morphospace, he organized the 256 possible combinations (28) into a table of 16 rows and 16 columns (Fig. 4a,b). The number of angiosperm families in which these combinations occurred was then recorded in the corresponding element of the table to highlight the contrast among character state combinations with high and low frequency of occurrence in angiosperms.
Table 1.
List of the eight floral characters used by Stebbins (1951) to create his topological floral morphospace
| Character | State ‘0’ | State ‘1’ |
|---|---|---|
| 1 | Calyx and corolla both present (dichlamydous) | Corolla or entire perianth absent (mono- or achlamydous) |
| 2 | Petals separate (polypetalous) | Petals (sepals in monochlamydous forms) united (sympetalous) |
| 3 | Calyx and corolla regular (actinomorphic) | Perianth irregular (zygomorphic) |
| 4 | Stamens more numerous than perianth members (polystemonous) | Stamens the same number as or fewer than perianth members (oligostemonous) |
| 5 | Carpels separate (apocarpous) | Carpels united (syncarpous) |
| 6 | Seeds more than one per carpel (many) | Seeds not more than one per carpel (few) |
| 7 | Placentation axial | Placentation parietal, basal or free central |
| 8 | Carpels not united with the receptacle or perianth (hypogynous or perigynous) | Carpels united with receptacle, perianth or both (epigynous) |
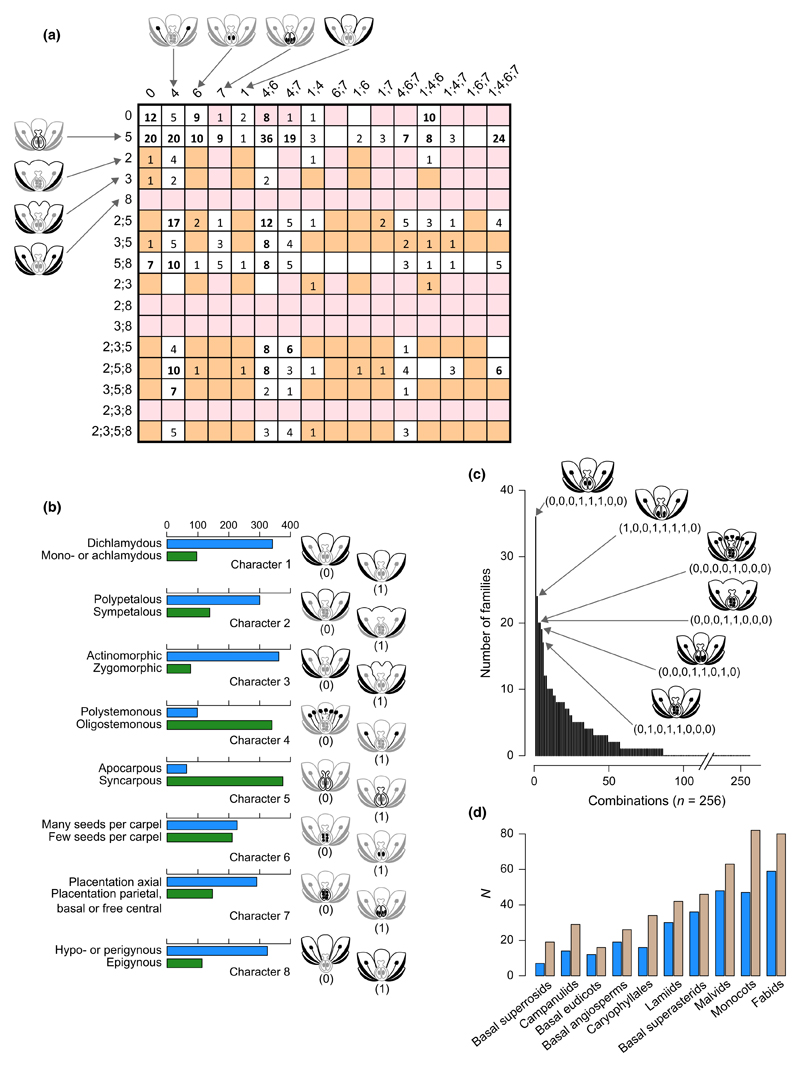
Fig. 4.
Stebbins’ (1951) floral morphospace. (a) An adaptation of Stebbins’ original chart. Numbers in front of rows and on top of columns indicate characters under state ‘1’ (called ‘advanced’ in Stebbins’ article). For example, the top left square corresponds to the combination in which all characters are under state ‘0’ (called ‘primitive’ in Stebbins’ article); the square formed by the second row and the sixth column corresponds to the combination were characters 5, 4 and 6 are in state ‘1’ and all other characters are in state ‘0’; the bottom right square corresponds to the combination where all eight characters are in state ‘1’. Numbers in each square represent the number of angiosperm families for which the corresponding combination has been recorded. Combinations that were interpreted as ‘nearly impossible’ by Stebbins are highlighted in pink; the ones associated with ‘low survival’ are highlighted in orange (see Stebbins (1951) for further details and full interpretation). Figure redrawn, with permission, from Stebbins (1951) © John Wiley & Sons Inc. (b) Frequency of the 16 character states (2 per character) in angiosperm families according to Stebbins’ dataset. Each of the character states is schematized. Horizontal bars give the number of angiosperm families for which each character state was recorded by Stebbins (1951; see Supporting Information Table S1). (c) Frequency diagram of the 256 morphological character state combinations from Stebbins (1951). Binary vectors and schematizations of the six most successful combinations are given on the right part of the plot. (d) Number (N) of angiosperm families per angiosperm subgroup: as recorded in the sampling of Stebbins (1951) (blue bars) and as present in the APG III (2009); Stevens, 2001) classification (brown bars).
Among the 256 possible character state combinations, only 86 were realized in angiosperm families (Fig. 4a,b). The six most common combinations were recorded in 17 to 36 different families, while 37 combinations were found in only one or two families (Fig. 4c). Stebbins interpreted most of the empty squares of the morphospace as ‘structurally impossible’ combinations (n = 96; for instance, the association of an inferior ovary and free carpels, and/or a parietal placentation and free carpels) or ‘apparently unadaptive combinations’ (n = 128; for instance, the union of petals and high stamen number, the presence of zygomorphy and high stamen number, and/or the absence of petals in zygomorphic flowers; Fig. 4a). His explanation for the success of highly represented combinations was that they might evolve easily and confer high ecological advantages. Among the realized combinations, five were represented by only a few but species-rich groups. These groups are Caesalpiniaceae (paraphyletic, now part of Fabaceae, see APG III (2009) for an up-to-date classification of angiosperms), Papilionaceae (= Fabaceae), Orchidaceae, Gramineae (= Poaceae) and Compositae (= Asteraceae). Stebbins hypothesized that direct competition might have prevented other groups from acquiring these trait combinations.
Stebbins’ representation can be easily interpreted in terms of state combination success. However, as Stebbins himself noted, this type of representation has a few caveats. The most important one may be the lack of meaning of the displacements in the chart, as similar morphologies are not grouped together in the representation. In fact, Stebbins’ representation is not even a space in the sense introduced above, as it does not offer any relations or measures of similarity between the different morphologies (see also Mitteroecker & Huttegger, 2009). In addition, this chart does not display any systematic information.
A new analysis of Stebbins’ dataset
Since Stebbins’ work, a range of new methods have been developed to construct morphospaces (see section ‘From theory to practice’). We created an NMDS ordination from Stebbins’ original dataset (see Supporting Information Methods S1), which allowed us to apply a simple distance metric for binary data (the number of variables in which two families differ) to Stebbins’ variables. We then measured disparity among angiosperm subgroups with three different indexes. Our goal was to construct an empirical morphospace in which we could investigate the spatial distribution of the 288 angiosperm families as sampled by Stebbins and to demonstrate the potential of the morphospace approach to study the evolution of angiosperms as a whole.
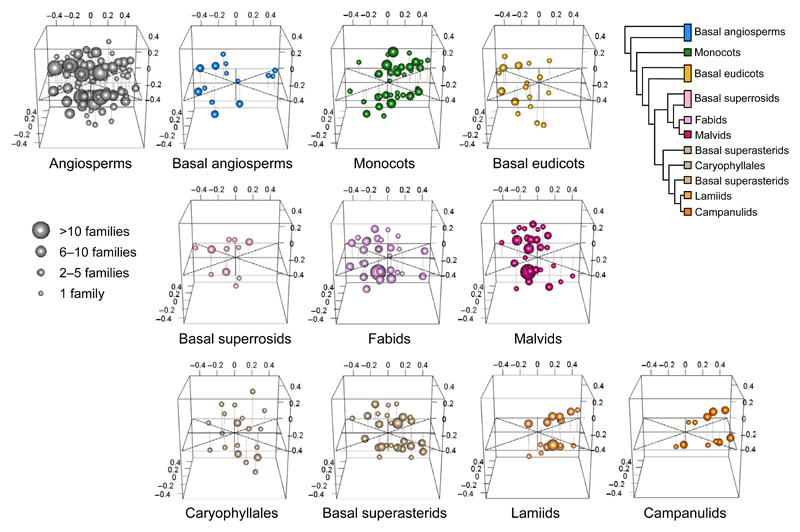
Our analyses of Stebbins’ dataset suggest that, when grouped into subgroups (basal angiosperms, monocots, basal eudicots, basal superrosids, fabids, malvids, Caryophyllales, basal superasterids, lamiids and campanulids; adapted from Soltis et al., 2011), angiosperms show significant differences in floral morphology (positions of the occupied areas in morphospace, npMANOVA overall test: df = 9, F = 11.3, r2 = 0.19, P = 0.001, Fig. 5) and in disparity (spread of the occupied areas in morphospace, Fig. 6a). The NMDS ordination produces a good representation of the data when three dimensions were retained (stress value = 0.092; see Clarke (1993) for stress value significance; Fig. 5). Lamiids were significantly different from all other groups with a tendency to have dichlamydous flowers (with both petals and sepals), united petals, few stamens and united carpels. Monocots, basal superasterids and campanulids tended to group together, sharing a majority of dichlamydous flowers with united carpels. No apparent morphological grouping could be found among the remaining groups (basal angiosperms, basal eudicots, basal superrosids, fabids, malvids and Caryophyllales; see Fig. S1 and Methods S1 for character state frequencies and post hoc test results).
Fig. 5.
Three-dimensional nonmetric multidimensional scaling (NMDS) representation of 10 angiosperm subgroups adapted from APG III (2009; Stevens, 2001). Tree modified, with permission, from Soltis et al. (2011) © 2011 Botanical Society of America. Each plot shows in the same morphospace the particular realizations of character state combinations in one of the subgroups. Symbol size is proportional to the number of families exhibiting the corresponding combination (see legend in the figure). Raw data are from Stebbins (1951) and presented in Table S1. NMDS stress value = 0.0915.
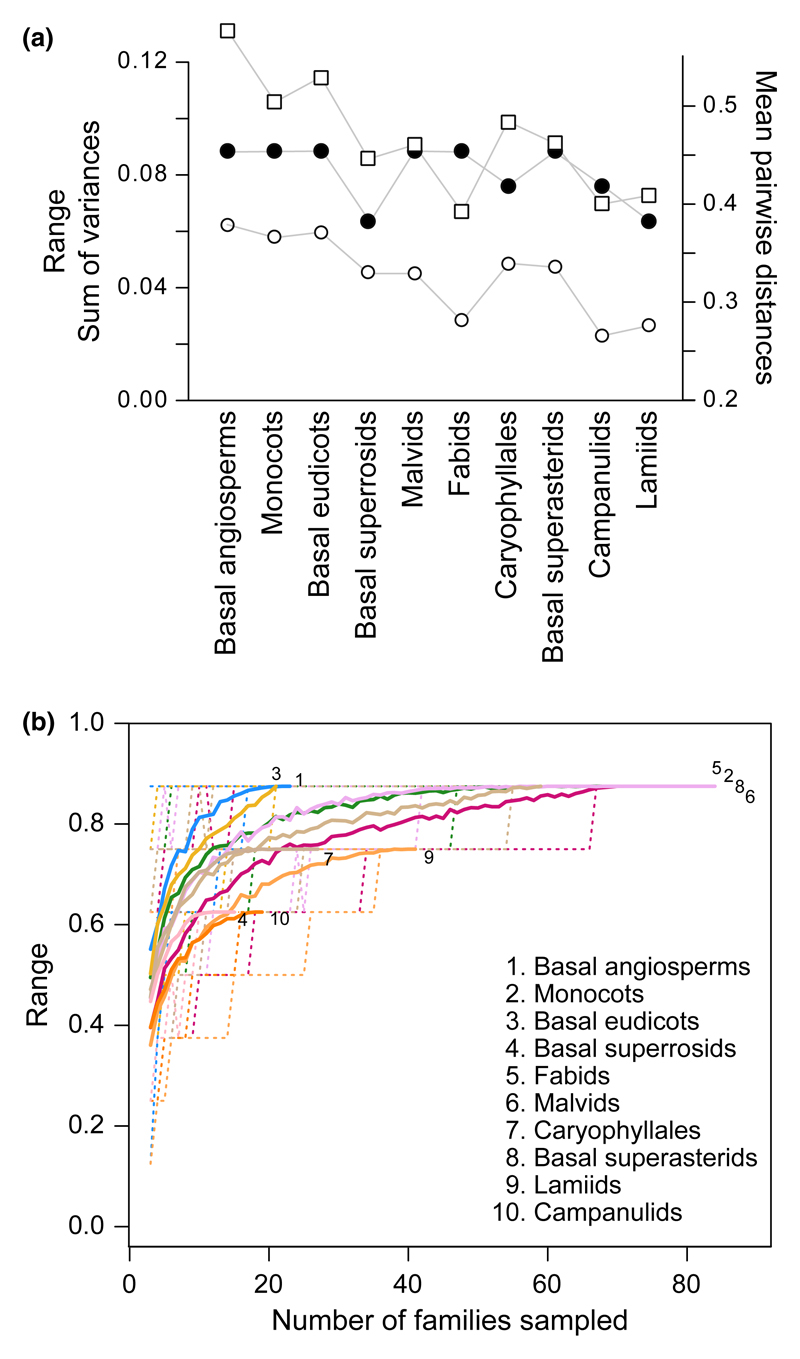
Fig. 6.
(a) Disparity indices calculated for 10 angiosperm subgroups out of Stebbins’ (1951) dataset. Mean pairwise distances (open circles), range (closed circles), and sum of variances (open squares). (b) Rarefaction curves for the range. Curves represent the mean (solid lines) and 90% confidence intervals (dashed lines) of the indices calculated from 200random samplings without replacement for each sampling size.
According to the mean pairwise distances and the sum of variances, malvids, lamiids and campanulids showed the lowest disparity, while basal superrosids, basal superasterids and fabids showed an intermediate level of disparity, and monocots, basal eudicots and basal angiosperms showed the highest disparity (Fig. 6a). According to the range (Fig. 6a,b), the highest disparity was reached by basal eudicots, then basal angiosperms, monocots, fabids, malvids and basal superasterids, whereas Caryophyllales and lamiids, then basal superrosids and campanulids showed less disparity.
Stebbins’ representation of his data is a theoretical morphospace, in which a number of extant angiosperm families were added to each character combination. It allows for the identification of character combinations that currently exist as compared to the ones that do not. Remember, however, that Stebbins’ approach is primarily a tabulation of data, not a space in the mathematical sense, because it lacks any relations or measures of (dis)similarity between morphologies. By constrast, our ordination analysis of the same data (using a simple metric) in order to construct an empirical morphospace, allowed us to visualize the relative positions of 10 angiosperm subgroups and their differences in disparity. These two approaches together provide a comprehensive picture of the proportion of realized trait combinations and of the distribution of floral disparity within angiosperms. The families sampled by Stebbins represent angiosperm diversity to an acceptable degree (compared to the number of families per category in APG III, 2009; Stevens, 2001; Fig. 4d); thus our analysis gives an idea of what will be possible with a more detailed and more comprehensive sampling strategy, more adequate character definitions and an incorporation of fossil data.
The floral morphospace: challenges and perspectives
Very few studies have used morphospaces to investigate the evolution of broad-scale patterns of angiosperm floral morphology and diversity. Morphospaces are mathematical abstractions, based on the researcher’s choice of the number and type of characters, and on the (metric) quantification of morphological similarity. Careful interpretations of such broad-scale analyses may lead to a more comprehensive understanding of the driving forces and constraints that have acted on floral evolution throughout different parts of the angiosperms’ phylogeny. They allow one to investigate relative morphospace distributions among different angiosperm clades and to identify particularly common and successful character combinations. Furthermore, it is possible to investigate how the occupied areas of morphospace evolved over geologic time, and how selection has acted on the different types of floral organs. Many mathematical and statistical tools are available, routinely used for the study of animal morphospaces, and the time has come to apply them to plants and in particular to flowers. Recent progress in paleobotany (reviewed by Friis et al., 2011) has yielded a number of excellently preserved fossils from angiosperm clades, such as the Laurales, Fagales and Ericales. An important area for the future will be to explore how the incorporation of fossils may modify current perceptions of the morphospace occupied by angiosperm flowers based solely on extant taxa. The rapid developments in bioinformatics and morphometrics during the last decades has led to numerous databases of various types of phenotypic data: FReD for colour (Arnold et al., 2010), Pherobase for scent (El-Sayed, 2012) and FLOWer for flow cytometry (Loureiro et al., 2008). One of them, PROTEUS (Sauquet, 2013), deals with flower morphology and will be a particularly valuable source for novel large-scale morphospace studies of flower evolution.
Supporting Information
Additional supporting information may be found in the online version of this article.
Please note: Wiley Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
We thank Richard Lupia, Douglas E. Soltis and Kalliope E. Stournaras for permission to reproduce figures, FWF for financial support, two anonymous reviewers for their helpful comments on the manuscript, and Ursula Schachner for spell-checking.
References
- Ackerman JD, Cuevas AA, Hof D. Are deception-pollinated species more variable than those offering a reward? Plant Systematics and Evolution. 2011;293:91–99. [Google Scholar]
- Allen CE, Beldade P, Zwaan BJ, Brakefield PM. Difference in the selection response of serially repeated color pattern characters: standing variation, development, and evolution. BMC Evolutionary Biology. 2008;8:94. doi: 10.1186/1471-2148-8-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecology. 2001;26:32–46. [Google Scholar]
- APG III. An update of the Angiosperm Phylogeny Group plants: APG III. Botanical Journal of the Linnean Society. 2009;161:105–121. [Google Scholar]
- Arnold SEJ, Faruq S, Savolainen V, McOwan PW, Chittka L. FReD: The Floral Reflectance Database — a web portal for analyses of flower colour. PLoS ONE. 2010;5:e14287. doi: 10.1371/journal.pone.0014287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold SEJ, Le Comber SC, Chittka L. Flower color phenology in European grassland and woodland habitats, through the eyes of pollinators. Israel Journal of Plant Sciences. 2009;57:211–230. [Google Scholar]
- Arnold SEJ, Pfrender ME, Jones A. The adaptive landscape as a conceptual bridge between micro- and macroevolution. Genetica. 2001;112–113:9–32. [PubMed] [Google Scholar]
- von Balthazar M, Schönenberger J. Comparative floral structure and systematics in the balsaminoid clade including Balsaminaceae, Marcgraviaceae and Tetrameristaceae. Botanical Journal of the Linnean Society. 2013;173:325–386. [Google Scholar]
- Benitez-Vieyra S, Ordano M, Fornoni J, Boege K, Domínguez CA. Selection on signal−reward correlation: limits and opportunities to the evolution of deceit in Turnera ulmifolia L. Journal of Evolutionary Biology. 2010;23:2760–2767. doi: 10.1111/j.1420-9101.2010.02132.x. [DOI] [PubMed] [Google Scholar]
- Benson RBJ, Choiniere JN. Rates of dinosaur limb evolution provide evidence for exceptional radiation in Mesozoic birds. Proceedings of the Royal Society B. 2013;280:20131780. doi: 10.1098/rspb.2013.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bookstein FL. Random walk as a null model for high-dimensional morphometrics of fossil series: geometrical considerations. Paleobiology. 2013;39:52–74. [Google Scholar]
- Bookstein FL. Measuring and reasoning: numerical Inference in the Sciences. NewYork, NY, USA: Cambridge University Press; 2014. [Google Scholar]
- Boyce K, Knoll AH. Evolution of developmental potential and the multiple independent origins of leaves in Paleozoic vascular plants. Paleobiology. 2002;28:70–100. [Google Scholar]
- Briggs DEG, Fortey RA, Wills MA. Morphological disparity in the Cambrian. Science. 1992;256:1670–1673. doi: 10.1126/science.256.5064.1670. [DOI] [PubMed] [Google Scholar]
- Bröderbauer D, Weber A, Anita D. The design of trapping devices in pollination traps of the genus Arum (Araceae) is related to insect type. Botanical Journal of the Linnean Society. 2013;172:385–397. doi: 10.1111/boj.12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhardt D. UV vision: a bird’s eye view of feathers. Journal of Comparative Physiology A. 1989;164:787–796. [Google Scholar]
- Burns JH, Munguia P, Nomann BE, Braun SJ, Terhorst CP, Miller TE. Vegetative morphology and trait correlations in 54 species of Commelinaceae. Botanical Journal of the Linnean Society. 2008;158:257–268. [Google Scholar]
- Burns KC, Cazetta E, Galetti M, Valido A, Schaefer HM. Geographic patterns in fruit colour diversity: do leaves constrain the colour of fleshy fruits? Oecologia. 2009;159:337–343. doi: 10.1007/s00442-008-1227-3. [DOI] [PubMed] [Google Scholar]
- Campbell DR. Using phenotypic manipulations to study multivariate selection of floral trait associations. Annals of Botany. 2009;103:1557–1566. doi: 10.1093/aob/mcp032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevin L-M, Lande R, Mace GM. Adaptation, plasticity, and extinction in a changing environment: towards a predictive theory. PLoS Biology. 2010;8:e1000357. doi: 10.1371/journal.pbio.1000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittka L. The colour hexagon: a chromaticity diagram based on photoreceptor excitations as a generalized representation of colour opponency. Journal of Comparative Physiology A. 1992;170:533–543. [Google Scholar]
- Chittka L. Bee color vision is optimal for coding flower color, but flower colors are not optimal for being coded-Why? Israel Journal of Plant Sciences. 1997;45:115–127. [Google Scholar]
- Chittka L, Shmida A, Troje N, Menzel R. Ultraviolet as a component of flower reflections, and the colour perception of Hymenoptera. Vision Research. 1994;34:1489–1508. doi: 10.1016/0042-6989(94)90151-1. [DOI] [PubMed] [Google Scholar]
- Chitwood DH, Ranjan A, Martinez CC, Headland LR, Thiem T, Kumar R, Covington MF, Hatcher T, Naylor DT, Zimmerman S, et al. A modern ampelography: a genetic basis for leaf shape and venation patterning in grape. Plant Physiology. 2014;164:259–272. doi: 10.1104/pp.113.229708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciampaglio CN, Kemp M, McShea DW. Detecting changes in morphospace occupation patterns in the fossil record: characterization and analysis of measures of disparity. Paleobiology. 2001;27:695–715. [Google Scholar]
- Clabaut C, Bunje PME, Salzburger W, Meyer A. Geometric morphometric analyses provide evidence for the adaptive character of the Tanganyikan cichlid fish radiations. Evolution. 2007;61:560–578. doi: 10.1111/j.1558-5646.2007.00045.x. [DOI] [PubMed] [Google Scholar]
- Clarke KR. Non-parametric multivariate analyses of changes in community structure. Australian Journal of Ecology. 1993;18:117–143. [Google Scholar]
- Conner JK, Heather FS, Karoly K. Tests of adaptation: functional studies of pollen removal and estimates of natural selection on anther position in wild radish. Annals of Botany. 2009;103:1547–1556. doi: 10.1093/aob/mcp071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper WJ, Parsons K, McIntyre A, Kern B, McGee-Moore A, Albertson RC. Bentho-pelagic divergence of cichlid feeding architecture was prodigious and consistent during multiple adaptive radiations within African Rift-lakes. PLoS ONE. 2010;5:e9551. doi: 10.1371/journal.pone.0009551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Córdoba AS, Cocucci AA. Flower power: its association with bee power and floral functional morphology in papilionate legumes. Annals of Botany. 2011;108:919–931. doi: 10.1093/aob/mcr196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortis P, Vereecken NJ, Schiestl FP, Lumaga MRB, Scrugli A, Cozzolino S. Pollinator convergence and the nature of species’ boundaries in sympatric Sardinian Ophrys (Orchidaceae) Annals of Botany. 2009;104:497–506. doi: 10.1093/aob/mcn219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane PR, Friis EM, Pederson KR. The origin and early diversification of angiosperms. Nature. 1995;374:27–33. [Google Scholar]
- Dafni A, Kevan PG, Husband BC. Practical pollination biology. Cambridge, ON, Canada: Enviroquest Ltd; 2005. [Google Scholar]
- Domjanic J, Fieder M, Seidler H, Mitteroecker P. Geometric morphometric footprint analysis of young women. Journal of Foot and Ankle Research. 2013;6:27. doi: 10.1186/1757-1146-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dommergues J-L, Laurin B, Meister C. Evolution of ammonoid morphospace during the Early Jurassic radiation. Paleobiology. 1996;22:219–240. [Google Scholar]
- Drake AG, Klingenberg CP. Large-scale diversification of skull shape in domestic dogs: disparity and modularity. The American Naturalist. 2010;175:289–301. doi: 10.1086/650372. [DOI] [PubMed] [Google Scholar]
- Eaton DAR, Fenster CB, Hereford J, Huang SQ, Ree RH. Floral diversity and community structure in Pedicularis (Orobanchaceae) Ecology. 2012;93:S182–S194. [Google Scholar]
- El-Sayed AM. [accessed 1 January 2014];The Pherobase: Database of Pheromones and Semiochemicals. 2012 [WWW document] URL http://www.pherobase.com/ [Google Scholar]
- Endler JA, Westcott DA, Madden JR, Robson T. Animal visual systems and the evolution of color patterns: sensory processing illuminates signal evolution. Evolution. 2005;59:1795–1818. doi: 10.1554/04-669.1. [DOI] [PubMed] [Google Scholar]
- Endress PK. Diversity and evolutionary biology of tropical flowers. Cambridge, UK: Cambridge University Press; 1994. [Google Scholar]
- Endress PK. Flower structure and trends of evolution in eudicots and their major subclades. Annals of the Missouri Botanical Garden. 2010;97:541–583. [Google Scholar]
- Endress PK. Evolutionary diversification of the flowers in angiosperms. American Journal of Botany. 2011;98:370–396. doi: 10.3732/ajb.1000299. [DOI] [PubMed] [Google Scholar]
- Erwin DH. Disparity: morphological pattern and developmental context. Palaeontology. 2007;50:57–73. [Google Scholar]
- Faegri K, Van Der Pijl L. The principles of pollination ecology. New York, NY, USA: Pergamon Press; 1966. [Google Scholar]
- Foote M. Rarefaction analysis of morphological and taxonomic diversity. Paleobiology. 1992;18:1–16. [Google Scholar]
- Foote M. Morphological diversification of Paleozoic crinoids. Paleobiology. 1995;21:273–299. [Google Scholar]
- Foote M. The evolution of morphological diversity. Annual Review of Ecology and Systematics. 1997;28:129–152. [Google Scholar]
- Friis EM, Crane P, Pedersen KR. Early flowers and angiosperm evolution. Cambridge, UK: Cambridge University Press; 2011. [Google Scholar]
- Gerber S, Neige P, Eble GJ. Combining ontogenetic and evolutionary scales of morphological disparity: a study of early Jurassic ammonites. Evolution & Development. 2007;9:472–482. doi: 10.1111/j.1525-142X.2007.00185.x. [DOI] [PubMed] [Google Scholar]
- Gibernau M, Chartier M, Barab'e D. Recent advances towards an evolutionary comprehension of Araceae pollination. In: Seberg O, Peterson G, Barfod AS, Davis JI, editors. Diversity, phylogeny and evolution in the monocotyledons. Fourth International Conference on the Comparative Biology of the Monocotyledons Proceedings; Aarhus, Denmark: Aarhus University Press; 2010. pp. 101–104. [Google Scholar]
- Goldsmith TH. Optimization, constraint, and history in the evolution of eyes. Quarterly Review of Biology. 1990;65:281–322. doi: 10.1086/416840. [DOI] [PubMed] [Google Scholar]
- Gómez JM, Abdelaziz M, Camacho JPM, Munõz-Pajares AJ, Perfectti F. Local adaptation and maladaptation to pollinators in a generalist geographic mosaic. Ecology Letters. 2009;12:672–682. doi: 10.1111/j.1461-0248.2009.01324.x. [DOI] [PubMed] [Google Scholar]
- Good P. Permutation tests: a practical guide to resampling methods for testing hypotheses. New York, NY, USA: Springer; 2000. [Google Scholar]
- Gould SJ, Gilinksy NL, German RZ. Asymmetry of lineages and the direction of evolutionary time. Science. 1987;236:1437–1441. doi: 10.1126/science.236.4807.1437. [DOI] [PubMed] [Google Scholar]
- Gumbert A, Kunze J, Chittka L. Floral colour diversity in plant communities, bee colour space and a null model. Proceedings of the Royal Society B: Biological Sciences. 1999;266:1711–1716. [Google Scholar]
- Gunz P, Bookstein FL, Mitteroecker P, Stadlmayr A, Seidler H, Weber GW. Early modern human diversity suggests subdivided population structure and a complex out-of-Africa scenario. Proceedings of the National Academy of Sciences, USA. 2008;106:6094–6098. doi: 10.1073/pnas.0808160106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder LD, Barrett SCH. Ecology and evolution of flowers. Oxford, UK: Oxford University Press; 2006. [Google Scholar]
- Harmon LJ, Schulte JA, Larson A, Losos JB. Tempo and mode of evolutionary radiation in iguanian lizards. Science. 2003;301:961–964. doi: 10.1126/science.1084786. [DOI] [PubMed] [Google Scholar]
- Herrera CM. Selection on floral morphology and environmental determinants of fecundity in a hawk moth-pollinated violet. Ecological Monographs. 1993;63:251–275. [Google Scholar]
- Hingston AB, McQuillan PB. Are pollination syndromes useful predictors of floral visitors in Tasmania? Austral Ecology. 2000;25:600–609. [Google Scholar]
- Hughes M, Gerber S, Wills MA. Clades reach highest morphological disparity early in their evolution. Proceedings of the National Academy of Sciences, USA. 2013;110:13875–13879. doi: 10.1073/pnas.1302642110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttegger SM, Mitteroecker P. Invariance and meaningfulness in phenotype spaces. Evolutionary Biology. 2011;38:335–351. [Google Scholar]
- Jabbour F, Citerne HL. Modeling flowers and inflorescences. International Journal of Plant Developmental Biology. 2010;4:38–46. [Google Scholar]
- Jabbour F, Nadot S, Damerval C. Evolution of floral symmetry: a state of the art. Comptes Rendus Biologies. 2009;332:219–231. doi: 10.1016/j.crvi.2008.07.011. [DOI] [PubMed] [Google Scholar]
- de Jager ML, Dreyer LL, Ellis AG. Do pollinators influence the assembly of flower colours within plant communities? Oecologia. 2011;166:543–553. doi: 10.1007/s00442-010-1879-7. [DOI] [PubMed] [Google Scholar]
- Jeune B, Barabé D, Lacroix C. Classical and dynamic morphology: toward a synthesis through the space of forms. Acta Biotheoretica. 2006;54:277–293. doi: 10.1007/s10441-007-9007-8. [DOI] [PubMed] [Google Scholar]
- Jeune B, Sattler R. Quelques aspects d’une morphologie continuiste et dynamique. Canadian Journal of Botany. 1996;74:1023–1039. [Google Scholar]
- Jürgens A, Wee S-L, Shuttleworth A, Johnson SD. Chemical mimicry of insect oviposition sites: a global analysis of convergence in angiosperms. Ecology Letters. 2013;16:1157–1167. doi: 10.1111/ele.12152. [DOI] [PubMed] [Google Scholar]
- Klingenberg CP, Duttke S, Whelan S, Kim M. Developmental plasticity, morphological variation and evolvability: a multilevel analysis of morphometric integration in the shape of compound leaves. Journal of Evolutionary Biology. 2012;25:115–129. doi: 10.1111/j.1420-9101.2011.02410.x. [DOI] [PubMed] [Google Scholar]
- Klingenberg CP, Gidaszewski N. Testing and quantifying phylogenetic signals and homoplasy in morphometric data. Systematic Biology. 2010;59:245–261. doi: 10.1093/sysbio/syp106. [DOI] [PubMed] [Google Scholar]
- Klingenberg CP, Leamy LJ, Cheverud JM. Integration and modularity of quantitative trait locus effects on geometric shape in the mouse mandible. Genetics. 2004;166:1909–1921. doi: 10.1534/genetics.166.4.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EM, Jaramillo AM. Genetic basis for innovations in floral organ identity. Journal of Experimental Zoology. 2005;304B:526–535. doi: 10.1002/jez.b.21046. [DOI] [PubMed] [Google Scholar]
- Lacroix C, Jeune B, Purcell-MacDonald S. Shoot and compound leaf comparisons in eudicots: dynamic morphology as an alternative approach. Botanical Journal of the Linnean Society. 2003;143:219–230. [Google Scholar]
- Langlade NB, Feng X, Dransfield T, Copsey L, Hanna AI, Thébaud C, Bangham A, Hudson A, Coen E. Evolution through genetically controlled allometry space. Proceedings of the National Academy of Sciences, USA. 2005;102:10 221–10 226. doi: 10.1073/pnas.0504210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loureiro J, Rodriguez E, Santos C, Doležel J, Suda J. [accessed 1 January 2014];FLOWer: A Plant DNA Flow Cytometry Database. 2008 [WWW document] URL http://flower.web.ua.pt/ [Google Scholar]
- Lupia R. Discordant morphological disparity and taxonomic diversity during the Cretaceous angiosperm radiation: North American pollen record. Paleobiology. 1999;25:1–28. [Google Scholar]
- Maad J. Phenotypic selection in hawkmoth-pollinated Platanthera bifolia: targets and fitness surfaces. Evolution. 2000;54:112–123. doi: 10.1111/j.0014-3820.2000.tb00012.x. [DOI] [PubMed] [Google Scholar]
- Macholán M. A geometric morphometric analysis of the shape of the first upper molar in mice of the genus Mus (Muridae, Rodentia) Journal of Zoology. 2006;270:672–681. [Google Scholar]
- Maclaurin J. The good, the bad and the impossible: a critical notice of theoretical morphology: the concept and its applications by George McGhee. Biology and Philosophy. 2003;18:463–476. [Google Scholar]
- Magallón S, Castillo A. Angiosperm diversification through time. American Journal of Botany. 2009;96:349–365. doi: 10.3732/ajb.0800060. [DOI] [PubMed] [Google Scholar]
- Maia ACD, Gibernau M, Carvalho AT, Gonçalves AG, Schlindwein C. The cowl does not make the monk: scarab beetle pollination of the Neotropical aroid Taccarum ulei (Araceae: Spathicarpeae) Biological Journal of the Linnean Society. 2013;108:22–34. [Google Scholar]
- Mardia KV, Kent JT, Bibby JM. Multivariate Analysis. London, UK: Academic Press; 1979. [Google Scholar]
- McGhee GR. Shell form in the biconvex articulate Brachipoda: a geometric analysis. Paleobiology. 1980;6:57–76. [Google Scholar]
- McGhee GR. Theoretical morphology. The concept and its applications. New York, NY, USA: Columbia University Press; 1999. [Google Scholar]
- Medel R, Botto-Mahan C, Kalin-Arroyo M. Pollinator-mediated selection on the nectar guide phenotype in the Andean monkey flower, Mimulus luteus. Ecology. 2003;84:1721–1732. [Google Scholar]
- Mitteroecker P, Bookstein FL. Linear discrimination, ordination, and the visualization of selection gradients in modern morphometrics. Evolutionary Biology. 2011;38:100–114. [Google Scholar]
- Mitteroecker P, Gunz P, Bernhard M, Schaefer K, Bookstein FL. Comparison of cranial ontogenetic trajectories among great apes and humans. Journal of Human Evolution. 2004;46:679–698. doi: 10.1016/j.jhevol.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Mitteroecker P, Gunz P, Bookstein FL. Heterochrony and geometric morphometrics: a comparison of cranial growth in Pan paniscus versus Pan troglodytes. Evolution & Development. 2005;7:244–258. doi: 10.1111/j.1525-142X.2005.05027.x. [DOI] [PubMed] [Google Scholar]
- Mitteroecker P, Gunz P, Neubauer S, Müller GB. How to explore morphological integration in human evolution and development? Evolutionary Biology. 2012;39:536–553. [Google Scholar]
- Mitteroecker P, Gunz P, Windhager S, Schaefer K. A brief review of shape, form, and allometry in geometric morphometrics, with applications to human facial morphology. Hystrix. 2013;24:59–66. [Google Scholar]
- Mitteroecker P, Huttegger SM. The concept of morphospaces in evolutionary and developmental biology: mathematics and metaphors. Biological Theory. 2009;4:54–67. [Google Scholar]
- Monteiro LR, Nogueira MR. Adaptive radiations, ecological specialization, and the evolutionary integration of complex morphological structures. Evolution. 2010;64:724–744. doi: 10.1111/j.1558-5646.2009.00857.x. [DOI] [PubMed] [Google Scholar]
- van der Niet T, Zollikofer CPE, Ponce de León MS, Johnson SD, Linder HP. Three-dimensional geometric morphometrics for studying floral shape variation. Trends in Plant Science. 2010;15:424–426. doi: 10.1016/j.tplants.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Niklas KJ. Computer models of early land plant evolution. Annual Review of Earth and Planetary Sciences. 2004;32:47–66. [Google Scholar]
- Niklas KJ, Kerchner V. Mechanical and photosynthetic constraints on the evolution of plant shape. Paleobiology. 1984;10:79–101. [Google Scholar]
- O’Connell LM, Johnston MO. Male and female pollination success in a deceptive orchid, a selection study. Ecology. 1998;79:1246–1260. [Google Scholar]
- Ollerton J, Alarcón R, Waser NM, Price MV, Watts S, Cranmer L, Hingston A, Peter CI, Rotenberry J. A global test of the pollination syndrome hypothesis. Annals of Botany. 2009;103:1471–1480. doi: 10.1093/aob/mcp031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollerton J, Raguso RA. The sweet stench of decay. New Phytologist. 2006;172:379–381. doi: 10.1111/j.1469-8137.2006.01903.x. [DOI] [PubMed] [Google Scholar]
- Ollerton J, Watts S. Phenotype space and floral typology: towards an objective assessment of pollination syndromes. Scandinavian Association for Pollination Ecology honours Knut Faegri. 2000;39:149–159. [Google Scholar]
- O’Meara BC. Evolutionary inferences from phylogenies: a review of methods. Annual Review of Ecology, Evolution, and Systematics. 2012;43:267–285. [Google Scholar]
- Ortega-Olivencia A, Rodríguez-Riaño T, Pérez-Bote JL, López J, Mayo C, Valtueña FJ, Navarro-Pérez M. Insects, birds and lizards as pollinators of the largest-flowered Scrophularia of Europe and Macaronesia. Annals of Botany. 2012;109:153–167. doi: 10.1093/aob/mcr255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce T. Evolution and constraints on variation: variant specification and range of assessment. Philosophy of Science. 2011;8:739–751. [Google Scholar]
- Perrard A, Villemant C, Carpenter JM, Baylac M. Differences in caste dimorphism among three hornet species (Hymenoptera: Vespidae): forewing size, shape and allometry. Journal of Evolutionary Biology. 2012;25:1389–1398. doi: 10.1111/j.1420-9101.2012.02527.x. [DOI] [PubMed] [Google Scholar]
- Pie MR, Weitz JS. A null model of morphospace occupation. American Naturalist. 2005;166:E1–E13. doi: 10.1086/430727. [DOI] [PubMed] [Google Scholar]
- van der Pijl L. Ecological aspects of flower evolution. II. Zoophilous flower classes. Evolution. 1961;15:44–59. [Google Scholar]
- Preston JC, Hileman LC, Cubas P. Reduce, reuse, and recycle: developmental evolution of trait diversification. American Journal of Botany. 2011;98:397–403. doi: 10.3732/ajb.1000279. [DOI] [PubMed] [Google Scholar]
- Proctor M, Leo P, Lack A. Natural history of pollination. London, UK: Harper Collins; 1996. [Google Scholar]
- Prusinkiewicz P, Erasmus Y, Lane B, Harder LD, Coen E. Evolution and development of inflorescence architectures. Science. 2007;316:1452–1456. doi: 10.1126/science.1140429. [DOI] [PubMed] [Google Scholar]
- Rabinovitz GB. An introduction to nonmetric multidimensional scaling. American Journal of Political Science. 1986;19:343–390. [Google Scholar]
- Raup DM. Geometric analysis of shell coiling: general problems. Journal of Paleontology. 1966;40:1178–1190. [Google Scholar]
- Raup DM. Analysis of shell coiling: coiling in ammonoids. Journal of Paleontology. 1967;41:43–65. [Google Scholar]
- Raup DM, Michelson A. Theoretical morphology of the coiled shell. Science. 1965;147:1294–1295. doi: 10.1126/science.147.3663.1294. [DOI] [PubMed] [Google Scholar]
- Rausher MD. Evolutionary transitions in floral color. International Journal of Plant Sciences. 2008;169:7–21. [Google Scholar]
- Ressayre A, Godelle B, Raquin C, Gouyon PH. Aperture pattern ontogeny in angiosperms. Journal of Experimental Zoology. 2002;294:122–135. doi: 10.1002/jez.10150. [DOI] [PubMed] [Google Scholar]
- Roelants K, Haas A, Bossuyt F. Anuran radiations and the evolution of tadpole morphospace. Proceedings of the National Academy of Sciences, USA. 2011;108:8731–8736. doi: 10.1073/pnas.1100633108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronse de Craene LP, Brockington SF. Origin and evolution of petals in angiosperms. Plant Ecology and Evolution. 2013;146:5–25. [Google Scholar]
- Rosin FM, Kramer EM. Old dogs, new tricks: regulatory evolution in conserved genetic modules leads to novel morphologies in plants. Developmental Biology. 2009;332:25–35. doi: 10.1016/j.ydbio.2009.05.542. [DOI] [PubMed] [Google Scholar]
- Sauquet H. [accessed 1 January 2014];PROTEUS: A database for recording morphological data and creating NEXUS matrices Version 1.20. 2013 [WWW document] URL http://eflower.myspecies.info/proteus. [Google Scholar]
- Schaefer HM, Ruxton GD. Plant–animal communication. New York, NY, USA: Oxford University Press; 2011. [Google Scholar]
- Schluter D. Estimating the form of natural selection on a quantitative trait. Evolution. 1988;42:849–861. doi: 10.1111/j.1558-5646.1988.tb02507.x. [DOI] [PubMed] [Google Scholar]
- Schluter D, Nychka D. Exploring fitness surfaces. The American Naturalist. 1994;143:597–616. [Google Scholar]
- Schönenberger J. Rise from the ashes – reconstruction of charcoal fossil flowers. Trends in Plant Science. 2005;10:436–443. doi: 10.1016/j.tplants.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Schönenberger J, von Balthazar M, Sytsma KJ. Diversity and evolution of floral structure among early diverging lineages in the Ericales. Philosophical Transactions of the Royal Society B. 2010;365:437–448. doi: 10.1098/rstb.2009.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönenberger J, von Balthazar M, Takahashi M, Xiao X, Crane PR, Herendeen PS. Glandulocalyx upatoiensis, a fossil flower of Ericales (Actinidiaceae/Clethraceae) from the Late Cretaceous (Santonian) of Georgia, USA. Annals of Botany. 2012;109:921–936. doi: 10.1093/aob/mcs009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipunov AB, Bateman RM. Geometric morphometrics as a tool for understanding Dactylorhiza (Orchidaceae) diversity in European Russia. Biological Journal of the Linnean Society. 2005;85:1–12. [Google Scholar]
- Sidlauskas B. Continuous and arrested morphological diversification in sister clades of characiform fishes: a phylomorphospace apprach. Evolution. 2008;62:3135–3156. doi: 10.1111/j.1558-5646.2008.00519.x. [DOI] [PubMed] [Google Scholar]
- Silva IA, Batalha MA. Plant functional types in Brazilian savannas: the niche partitioning between herbaceous and woody species. Perspectives in Plant Ecology, Evolution and Systematics. 2011;13:201–206. [Google Scholar]
- Soltis DE, Smith SA, Cellinese N, Wurdack KJ, Tank DC, Brockington SF, Refulio-Rodriguez NF, Walker JB, Moore MJ, Carlsward BS, et al. Angiosperm phylogeny: 17 genes, 640 taxa. American Journal of Botany. 2011;98:704–730. doi: 10.3732/ajb.1000404. [DOI] [PubMed] [Google Scholar]
- Stadler BM, Stadler PF, Wagner GP, Fontana W. The topology of the possible: formal spaces underlying patterns of evolutionary change. Journal of Theoretical Biology. 2001;213:241–274. doi: 10.1006/jtbi.2001.2423. [DOI] [PubMed] [Google Scholar]
- Stebbins GL. Natural selection and the differentiation of angiosperm families. Evolution. 1951;5:299–324. [Google Scholar]
- Stevens PF. [accessed 1 January 2014];Angiosperm Phylogeny Website Version 12. 2001 July 2012. [WWW document] URL http://www.mobot.org/MOBOT/research/APweb/welcome.html. [Google Scholar]
- Stoddard MC, Prum RO. Evolution of avian plumage color in a tetrahedral color space: a phylogenetic analysis of New World buntings. The American Naturalist. 2008;171:755–776. doi: 10.1086/587526. [DOI] [PubMed] [Google Scholar]
- Stoddard MC, Stevens M. Avian vision and the evolution of egg color mimicry in the common cuckoo. Evolution. 2011;65:2004–2013. doi: 10.1111/j.1558-5646.2011.01262.x. [DOI] [PubMed] [Google Scholar]
- Stone JR. The spirit of D’Arcy Thomson dwells in empirical morphospace. Mathematical Biosciences. 1997;142:13–30. doi: 10.1016/s0025-5564(96)00186-1. [DOI] [PubMed] [Google Scholar]
- Stournaras KE, Lo E, Böhning-Gaese K, Cazetta E, Dehling DM, Schleuning M, Stoddard MC, Donoghue MJ, Prum RO, Schaefer HM. How colorful are fruits? Limited color diversity in fleshy fruits on local and global scales. New Phytologist. 2013;198:617–629. doi: 10.1111/nph.12157. [DOI] [PubMed] [Google Scholar]
- Strauss SY. Floral characters link herbivores, pollinators, and plant fitness. Ecology. 1997;78:1640–1645. [Google Scholar]
- Swan JMA. An examination of some ordination problems by use of simulated vegetational data. Ecology. 1970;51:89–102. [Google Scholar]
- Tastard E, Andalo C, Giurfa M, Burrus M, Thébaud C. Flower colour variation across a hybrid zone in Antirrhinum as perceived by bumblebee pollinators. Arthropod-Plant Interactions. 2008;2:237–246. [Google Scholar]
- Troje N. Spectral categories in the learning behaviour of blowflies. Zeitschrift für Naturforschung. 1993;48c:96–104. [Google Scholar]
- Vogel S. Blütenbiologische Typen als Elemente der Sippengliederung, dargestellt anhand der Flora Südafrikas. Botanische Studien. 1954;1:1–338. [Google Scholar]
- Vogel S. Floral-biological syndromes as elements of diversity within tribes in the flora of South Africa. Aachen, Germany: Shaker Verlag GmbH; 2012. [Google Scholar]
- Wagner PJ. Testing evolutionary constraint hypotheses with early Paleozoic gastropods. Paleobiology. 1995;21:248–272. [Google Scholar]
- Wagner PJ. Contrasting the underlying patterns of active trends in morphologic evolution. Evolution. 1996;50:990–1007. doi: 10.1111/j.1558-5646.1996.tb02341.x. [DOI] [PubMed] [Google Scholar]
- Whibley AC, Langlade NB, Andalo C, Hanna AI, Bangham A, Thébaud C, Coen E. Evolutionary paths underlying flower color variation in Antirrhinum. Science. 2006;313:963–966. doi: 10.1126/science.1129161. [DOI] [PubMed] [Google Scholar]
- Wills MA, Briggs DEG, Fortey RA. Disparity as an evolutionary index: a comparison of Cambrian and recent arthropods. Paleobiology. 1994;20:93–130. [Google Scholar]
- Wilson JP, Castellanos MC, Hogue JN, Thomson JD, Armbruster WS. A multivariate search for pollination syndromes among penstemons. Oikos. 2004;104:345–361. [Google Scholar]
- Wilson JP, Knoll AH. A physiologically explicit morphospace for tracheid-based water transport in modern and extinct seed plants. Paleobiology. 2010;36:335–355. [Google Scholar]
- Wright S. The roles of mutation, inbreeding, crossbreeding, and selection in evolution. Proceedings of the Sixth International Congress of Genetics. 1932;1:356–366. [Google Scholar]
- Xiao S, Laflamme M. On the eve of animal radiation: phylogeny, ecology and evolution of the Ediacara biota. Trends in Ecology and Evolution. 2009;24:31–40. doi: 10.1016/j.tree.2008.07.015. [DOI] [PubMed] [Google Scholar]
- Yoshioka Y, Ohsawa R, Iwata H, Nimomiya S, Fukuta N. Quantitative evaluation of petal shape and picotee color pattern in Lisianthus by image analysis. Journal of the American Society for Horticultural Science. 2006;131:261–266. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.