Abstract
Certain antibiotics detected in urine are associated with childhood obesity. In the current experimental study, we investigated two representative antibiotics detected in urine, florfenicol and azithromycin, for their early effects on adipogenesis, gut microbiota, short-chain fatty acids (SCFAs), and bile acids in mice. Thirty C57BL/6 mice aged four weeks were randomly divided into three groups (florfenicol, azithromycin and control). The two experimental groups were administered florfenicol or azithromycin at 5 mg/kg/day for four weeks. Body weight was measured weekly. The composition of the gut microbiota, body fat, SCFAs, and bile acids in colon contents were measured at the end of the experiment. The composition of the gut microbiota was determined by sequencing the bacterial 16S rRNA gene. The concentration of SCFAs and bile acids was determined using gas chromatography and liquid chromatography coupled to tandem mass spectrometry, respectively. The composition of the gut microbiota indicated that the two antibiotics altered the gut microbiota composition and decreased its richness and diversity. At the phylum level, the ratio of Firmicutes/Bacteroidetes increased significantly in the antibiotic groups. At the genus level, there were declines in Christensenella, Gordonibacter and Anaerotruncus in the florfenicol group, in Lactobacillus in the azithromycin group, and in Alistipes, Desulfovibrio, Parasutterella and Rikenella in both the antibiotic groups. The decrease in Rikenella in the azithromycin group was particularly noticeable. The concentration of SCFAs and secondary bile acids decreased in the colon, but the concentration of primary bile acids increased. These findings indicated that florfenicol and azithromycin increased adipogenesis and altered gut microbiota composition, SCFA production, and bile acid metabolism, suggesting that exposure to antibiotics might be one risk factor for childhood obesity. More studies are needed to investigate the specific mechanisms.
Introduction
According to data released by the World Health Organization (WHO) in 2008, more than one billion adults worldwide were overweight, and among those individuals, 300 million adults were obese[1,2]. Obesity is not only prevalent in adults but also in children[3,4]. Obesity is related to specific major chronic diseases, such as type II diabetes mellitus, coronary heart disease, atherosclerosis, and nonalcoholic fatty liver disease[5]. However, the etiology of obesity is not fully understood. Recently, the gut microbiota was found to be closely correlated with metabolic and immune function and an important contributor to adipogenesis. For example, Ley et al. found that obesity was related to intestinal microorganisms, with a decreased ratio of Bacteroidetes to Firmicutes observed in obese mice compared to lean mice from the same brood[6]. Severe obesity and insulin resistance occurred in mice one week after in vivo inoculation of Enterobacter cloacae B29 that could produce endotoxins[7]. The gut microbiota in human intestinal tracts was highly diverse in normal weight subjects compared with obese subjects, with Firmicutes more predominant in obese subjects [8]. Obese people with low microbial gene species richness tend to gain more weight with a high fat diet than obese people with high species richness[9].
The gut microbiota has a close relationship with the body's energy homeostasis and fat metabolism, but it also tends to be affected by factors such as breastfeeding, caesarean section, and dietary habits, as well as other factors [10,11]. Among them, exposure to antibiotics is a potential threat that cannot be ignored. Due to extensive use in humans and animals, antibiotics have frequently been found in aquatic environments and in food. Thus, they are listed as a class of emerging environmental pollutants [12]. Compared to other environmental pollutants, antibiotics can not only do direct harm to the human body but can also interfere with the human microbiome, possibly resulting in metabolic diseases, such as obesity[13]. For example, lab studies have found that certain antibiotics, such as vancomycin, penicillin and chlortetracycline, can alter the bacterial composition and endocrine activity of the gut microbiota, affect production of short-chain fatty acids (SCFAs) and the metabolism of bile acids, and increase body fat [14–18]. Several epidemiological studies have reported that antibiotic use in children was positively associated with obesity in children[19–22]. Recently, several studies have reported extensive exposure of school-aged children to antibiotics by measuring antibiotics in urine, and certain antibiotics, such as florfenicol and trimethoprim, were related to obesity [23,24].
Antibiotics have different effects on gut microbiota due to differences in their antimicrobial spectrum and thus differ in their capacity to interfere with fat metabolism. Florfenicol and azithromycin are two typical antibiotics frequently detected in urine, and they have a potential impact on fat metabolism, but there is still a lack of laboratory data regarding their effect on adipogenesis. Since florfenicol first came to the market in Japan in the 1990s, it has been used in a number of countries to treat bacterial disease in animals including cattle, pigs, chicken and fish. Florfenicol is an amphenicol antibiotic and has a broad spectrum of antimicrobial activity that includes a wide range of gram-positive and gram-negative bacteria and mycoplasma [25]. Because it is widely used in aquaculture, florfenicol is typically more abundant in water environments than other antibiotics. Azithromycin has relatively broad but shallow antibacterial activities. It inhibits some gram-positive bacteria, some gram-negative bacteria, and many atypical bacteria. Azithromycin is a second generation macrolide that is primarily used to treat respiratory tract and genital tract infections, and it has been recommended by a number of countries and regions as a first-line treatment for these types of infections [26]. To support the findings in human populations, the current study aimed to explore the effects of azithromycin and florfenicol on the gut microbiota and adipogenesis in mice.
Materials and methods
Mouse model, sample collection, and body measurement
The study was approved by the research ethics committee of Fudan University, and carried out in accordance with the relevant guidelines and regulations. To explore possible effects of early exposure to azithromycin and florfenicol on adipogenesis in mice, we chose to start the experiment when mice were four weeks old, which is in line with previous studies, because this time is considered to be a critical period for gut microbiota development in mice [14]. A total of 30 three-week-old C57BL/6 mice were obtained from the Animal Experimental Center of Fudan University. The mice were randomly divided into three groups (azithromycin, florfenicol, and control), and each group included ten mice (five males and five females). The mice were separately bred in polycarbonate cages based on sex and group. Before the experiment, the mice were acclimatized to standardized laboratory conditions at a temperature of 21±1°C, a relative humidity of 50±10%, and a 12 h light-dark cycle for one week. These conditions were maintained throughout the entire study. Antibiotics were orally administered in drinking water for four weeks starting the fourth week after birth. The drinking water was spiked with azithromycin and florfenicol at a concentration of 35 mg/L and 33 mg/L, respectively. We changed the solutions containing the antibiotics every day. According to the daily consumption by mice, the exposure dose was estimated to be 5 mg/kg/day for each of the two antibiotics[14]. Florfenicol and azithromycin dihydrate with a purity above 98% were purchased from Sigma-Aldrich Corporation (St Louis, MO, USA). During the experiment, no adverse events were observed. At the end of each week during antibiotic administration, the mice were weighed using an electronic scale. At the end of the fourth week during antibiotic administration, the body fat content of all mice was determined after excretion of feces and urine using a MesoQMR nuclear magnetic resonance analyzer for body composition analysis of conscious animals (Niumag Corporation). Mice were killed by CO2 inhalation and cervical dislocation. Then, mice were dissected, an incision was made in the colon with a sterile scalpel, and the contents of the colon were collected into sterilized centrifuge tubes using sterile forceps. All samples were immediately flash-frozen in liquid nitrogen and stored at -80°C until use.
16S rRNA gene sequencing and data processing
Total genomic DNA was extracted from thawed colon content samples using a Powersoil DNA Extraction Kit (MoBio, Carlsbad, CA, USA) in 96-well format, and the 16S rRNA gene was amplified with barcoded fusion primers targeting the V3, V4, and V5 regions. Amplicon pools were sequenced on a 2×150 bp Illumina MiSeq platform. Two reads were paired, and paired-end reads were assembled with an overlapping region of at least 20 bp guaranteed; reads containing N were removed. Primers and adaptor sequences, bases with a quality of less than 20 at both ends, and sequences with a length of less than 400 bp were excluded. After pairing the abovementioned sequences, high-quality sequences were classified into multiple operational taxonomic units (OTUs) according to sequence similarity (> 97%). To make the data more interpretable, we edited the OTUs according to their representation among the samples. We ranked the abundance of OTUs from high to low, and OTUs after the top 30 were classified into “Others”. This classification reduced the noise of amplicon datasets and avoided spurious associations when there was a preponderance of zero counts. Taxonomic assignment and diversity calculations were also conducted. The distance between samples was calculated using the evolution and abundance information in the sequence of each sample to reflect whether there was a significant difference in the microbial community between samples. Linear discriminant analysis effect size (LEfSe) was employed to detect significant differences in relative abundance of microbial taxa between groups at the species level.
Analysis of SCFAs and bile acids in colonic contents
The SCFA standards included acetic acid (AA), propionic acid (PPA), isobutyric acid (IBA), n-butyric acid (NBA), isovaleric acid (IVA), n-valeric acid (NVA), and 2-ethylbutyric acid (EBA). The bile acid standards included cholic acid (CA), deoxycholic acid (DCA), chenodeoxycholic acid (CDCA), lithocholic acid (LCA), ursodeoxycholic acid (UDCA), hyodeoxycholic acid (HDCA), and two isotope-labeled bile acids (LCA-d4 and CA-d5). All these standards were obtained from Sigma-Aldrich Corporation, Dr. Ehrenstorfer (Augsburg, Germany), and Anpel Laboratory Technologies Inc. (Shanghai, China). The SCFAs and bile acids in the colon contents were determined in our lab using modified methods published previously [27].
For SCFAs, after 50.0 mg of the colonic contents was weighed and EBA was added as an internal standard, the sample was completely homogenized with 1.0 ml of 0.5 M oxalic acid for 5 min and centrifuged at 8000 r/min for 10 min. The supernatant was filtered with a 0.22 μm nylon membrane, and 1 μl of the filtrate was analyzed using a capillary gas chromatography instrument equipped with a flame ionization detector (FID) (GC-2010 Plus, Shimadzu). The SCFAs were separated on a fused-silica capillary column with a free fatty acid phase (DB-FFAP) and dimensions of 30 m × 0.53 mm i.d. coated with 1.0 μm film thickness. Nitrogen was used as the carrier gas at an initial flow rate of 6.44 ml/min in the constant pressure mode. The initial oven temperature was 50°C. This temperature was maintained for 1.0 min, raised to 180°C at 20°C/min, held for 1.0 min, and then increased to 200°C at 20°C/min and held for 2 min. Glass wool was inserted into the glass liner of the injection port, and the split injection mode was used with a split ratio of 30 to 1. The FID temperature and injection port was 240 and 200°C, respectively. The flow rates of hydrogen, air and nitrogen that composed the gas for the FID were 40, 400 and 30 ml/min, respectively. The SCFAs were quantified using the internal standard curve method.
For bile acids, after 50.0 mg of colonic content was weighed and isotope-labeled internal standards (LCA-d4 and CA-d5) were added, the bile acids were completely homogenized with 1 ml of 0.2 M NaOH for 20 min and centrifuged at 8000 r/min for 10 min. The extraction step was repeated three times, and the supernatants were combined. After the Oasis Prime HLB cartridge (60 mg/3 cc, Waters) was conditioned with 2 ml of methanol and 2 ml of water, the combined supernatants were loaded. The cartridge was washed with 2 ml of 20% methanol in water solution, and bile acids were eluted with 3 ml of acetonitrile:methanol (90:10) containing 1% formic acid. The eluent was evaporated with a weak nitrogen flow, and the bile acids were reconstituted with 0.5 ml of 50% methanol water solution. After the reconstituted solution was filtered through a 0.22 μm nylon membrane, 10 μl of the filtrate was analyzed using ultra-performance liquid chromatography coupled with high-resolution quadrupole time-of-flight mass spectrometry (SYNAPT G2, Waters Micromass, Manchester, UK). Bile acids were separated on an analytical column (Acquity UPLC HSS T3 column, 100 mm × 3.0 mm × 1.8 μm) at a column temperature of 50°C with a mobile phase of methanol and water containing 0.1% formic acid at a flow rate of 0.55 ml/min. The linear elution program was as follows: from 0 to 3.00 min, increased to 35% from 5% methanol; from 3.00 to 6.00 min, increased to 65% methanol; from 6.00 to 8.00 min, increased to 80% methanol; from 8.00 to 9.00 min, increased to 90% methanol; from 9.00 to 10.00 min, increased to 95% methanol; from 10.00 to 10.50 min, increased to 98% methanol; from 10.50 to 11.00 min, decreased to 5% methanol and maintained until 12.00 min. For mass spectrometry, nitrogen and argon were used as the desolvation and collision gas, respectively. The flow rate of the desolvation gas was set to 800 L/h with a temperature of 400°C. The capillary voltage was set to 2.8 kV, the cone gas was set to 30 L/h, the source temperature was set to 110°C, and the sampling cone voltage was set to 30 v. The bile acids were quantified using the isotope-dilution method, and the calibration curve was based on the peak area of the extracted precursor ion chromatogram for each bile acid from the total ion chromatogram at a mass window of 0.02 Da.
Statistical analysis
Quantitative data are reported as the mean ± standard error. The Kolmogorov—Smirnov D test was used to ascertain the normality of the data. Statistical differences in body composition and the colonic SCFA and bile acid content between the groups were tested with one-way analysis of variance (one-way ANOVA) and multiple t-tests with Bonferroni correction for continuous variables. All analysis was conducted for males and females separately. A p-value less than 0.05 was considered statistically significant. All statistical analyses were performed using R 3.1.1 software[28]. Reads were assembled using PANDAseq (v. 2.7) [29]. Trimmomatic (v. 0.30) was used to filter primers and adapter sequences[30]. USEARCH (v. 8.0) was employed to pair assembled and filtered reads[31]. The QIIME pipeline with RDP classifier Bayesian algorithm was used for taxonomic assignment with the SILVA_119 16S rRNA database [32,33]. OTU classification, UniFrac analysis, and calculation of diversity metrics were also conducted with QIIME pipeline. Linear discriminant analysis effect size (LEfSe) was conducted using Galaxy Online software [34,35].
Results
Body weight and composition
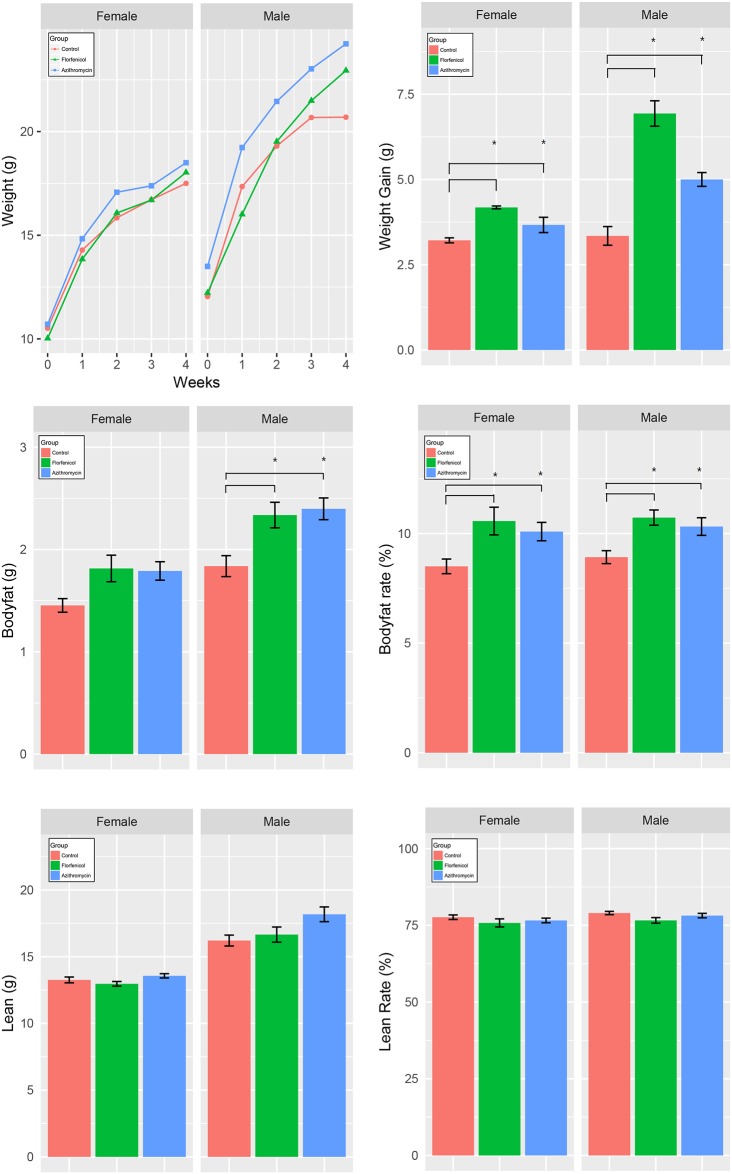
As shown in Fig 1, there was no significant difference in weight among the 3 groups at the beginning of the experiment. The antibiotic groups gained more weight than the control group during the 4-week period, especially the males. The weight gains in the florfenicol group between week 4 and week 1 were larger than in the azithromycin group. The average percent body fat in the florfenicol group (10.62% ± 0.34%) and azithromycin group (10.20% ± 0.28%) was significantly higher than that in the control group (8.73% ± 0.22%). The differences in body fat between the antibiotic groups and the control group were larger in males than in females. The difference in weight gain between the florfenicol group and the control group was also larger in males than in females. The absolute value or rate of lean mass was similar for antibiotic-treated and control mice.
Fig 1. The weight change within four weeks and body composition after four weeks.
One-way analysis of variance (one-way ANOVA) and multiple t-tests with Bonferroni correction were employed to measure differences in weight gain, body fat (g), body fat rate (%), lean weight (g), and lean rate (%) between the three groups. * indicates that the difference between the two line-linked groups was statistically significant (p < 0.05).
General OTU information
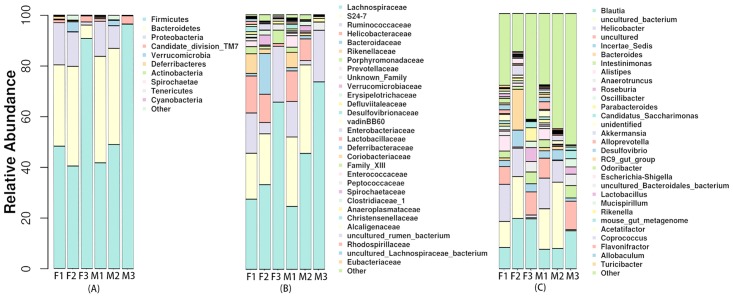
A total of 14,226,262 16S rDNA sequence reads from V3, V4, and V5 regions were obtained from the 30 samples. The number of sequence reads for each sample ranged from 368,026 to 620,382 with an average of 474,208. The average length of the sequence reads was 445 bp. The taxon abundance of each sample was generated into 12 phyla, 23 classes, 33 orders, 61 families, 125 genera and 255 species. Overall, twelve phyla were identified in the study groups. Among them, five phyla were commonly found in each group, and three phyla were dominant at > 1% concentration but varied in relative abundance with sex and antibiotic treatment. The majority of sequencing reads at the phyla level were Firmicutes, Bacteroidetes, and Proteobacteria, which dominated the bacterial community in each of the studied groups. A total of 61 families were detected. Among them, nine families were commonly present, and Lachnospiraceae, Ruminococcaceae, and Helicobacteraceae were the dominant families detected. A total of 125 OTUs were identified at the genus level. The highest number of genera (125) was detected in the male control group, whereas the lowest (76) was found in the female mice treated with azithromycin. Antibiotics significantly decreased the number of genera in each of the antibiotic treatment groups. A total of 255 different species were found, and the highest number of species presented in the control group.
Diversity of gut microbiota
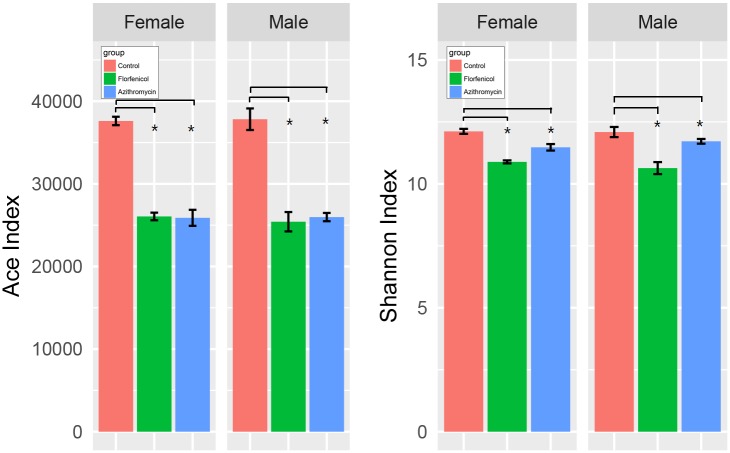
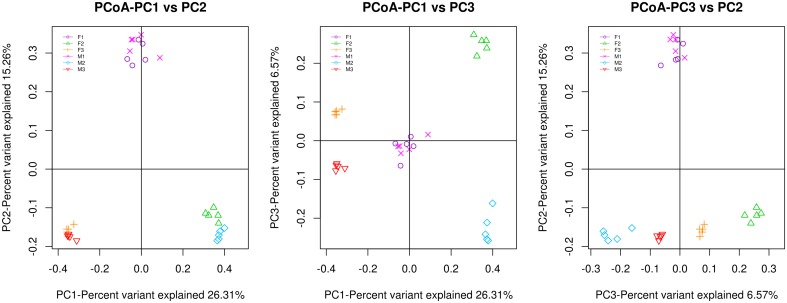
There are two types of alpha diversity indexes, community richness indexes (Chao, ACE) and community diversity indexes (Shannon). Fig 2 shows that the control group had higher richness and diversity index levels than the antibiotic groups (both males and females), and there were no significant sex-related differences. Good’s coverage, a measure of sampling completeness, ranged between 91.86% and 96.89% at a 97% similarity level. Beta diversity reflects diversity differences among samples. UniFrac principal coordinates analysis (PCoA) of 15,034 OTUs (grouped at 97% sequence similarity) showed a clear separation between the control and antibiotic samples using an unweighted analysis (Fig 3). The percentages of variation represented by PC1 and PC2 were 26.31% and 15.26%, respectively. In the two antibiotic groups, the male and female groups were also well separated.
Fig 2. Ace index and Shannon index in the samples from the normal and antibiotic-treated groups.
One-way analysis of variance (one-way ANOVA) and multiple t-tests with Bonferroni correction were employed to measure differences between the three groups. The ACE index represents the community richness of the gut microbiota, and the Shannon index represents the community diversity of the gut microbiota. * indicates that the difference between the two line-linked groups was statistically significant (p < 0.05).
Fig 3. The multivariate principal coordinate data analysis of the groups.
F1, female control group; F2, female mice treated with florfenicol; F3, female mice treated with azithromycin; M1, male control group; M2, male mice treated with florfenicol; M3, male mice treated with azithromycin. Each group has ten mice (5 male and 5 female subjects). PC1 and PC2 are the two principal coordinate components. PC1 indicates the maximum possible interpretation of the main components of the data variations. PC2 represents the largest proportion of interpretation of remaining variations, etc.
Composition of gut microbiota
There were substantial variations in the composition of gut microbiota associated with antibiotics and sex. Scoring and ranking were carried out for these differences. In females, Fig 4a shows that at the level of phylum, the relative abundance of Firmicutes increased and the relative abundance of Bacteroidetes and Proteobacteria decreased significantly in the azithromycin group compared with the control group. The relative abundance of Verrucomicrobia increased and that of Deferribacteres decreased significantly in the florfenicol group compared with the control group. In males, the relative abundance of Firmicutes increased while that of Bacteroidetes decreased to nearly zero in the azithromycin group compared with the control group. For florfenicol-treated mice, the relative abundance of Firmicutes was slightly higher in males than in females. For azithromycin-treated mice, females had higher relative abundance levels of Bacteroidetes and Proteobacteria and a lower relative abundance level of Firmicutes than males.
Fig 4. Microbial distribution at three different levels in samples from the normal and antibiotic-treated groups.
(A) Taxonomic distribution of the top 10 phyla; (B) Taxonomic distribution of the top 30 families; (C) Taxonomic distribution of the top 30 genera. Abbreviations are the same as those described for Fig 3. Each group has ten mice (5 male and 5 female subjects).
At the family level, Fig 4b shows that in females the relative abundance of Lachnospiraceae, Ruminococcaceae, and Porphyromonadaceae increased, while that of Helicobacteraceae, Rikenellaceae, and Bacteroidaceae decreased significantly in the azithromycin group compared with the control group. In the florfenicol group, the relative abundance of Bacteroidaceae and Verrucomicrobiaceae increased, whereas the relative abundance of Ruminococcaceae, Helicobacteraceae, Rikenellaceae and Porphyromonadaceae decreased compared with the control group. After florfenicol treatment, males had a lower relative abundance of Bacteroidaceae than females. After azithromycin treatment, the relative abundance of Porphyromonadaceae decreased in males but increased in females compared with the control group.
At the genus level, Fig 4c shows that in females the relative abundance of Blautia and Parabacteroides increased, but that of Helicobacter and Alistipes decreased significantly in the azithromycin group compared with the control group. The relative abundances of Blautia and Bacteroides increased, while that of Alistipes decreased in the florfenicol group. In males, the relative abundance of Parabacteroides did not change significantly compared with the control group. Moreover, the relative abundance of Blautia and of Bacteroides was not notably different between the florfenicol and control groups.
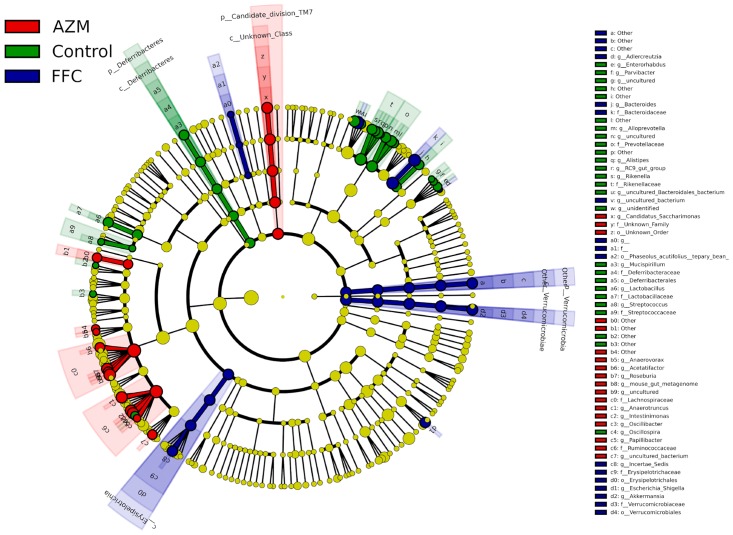
The LEfSe determines the features most likely to explain differences between groups by coupling standard tests for statistical significance with additional tests encoding biological consistency and effect relevance. At the genus level, Christensenella, Gordonibacter, and Anaerotruncus decreased in the florfenicol group; Lactobacillus decreased in the azithromycin group; Alistipes, Desulfovibrio, Parasutterella and Rikenella declined in both the antibiotic groups; and the decrease of Rikenella in the azithromycin group was particularly noticeable. Furthermore, LEfSe analysis identified other representative species of each group as biomarkers to distinguish different groups (Fig 5).
Fig 5. LEfSe results in the three groups, representing the relevant features on taxonomic trees.
AZM, mice treated with azithromycin; FFC, mice treated with florfenicol. Due to the length of bacterial names, abbreviations are employed under the family level. Each group has 10 mice (5 male and 5 female subjects).
SCFAs and bile acids
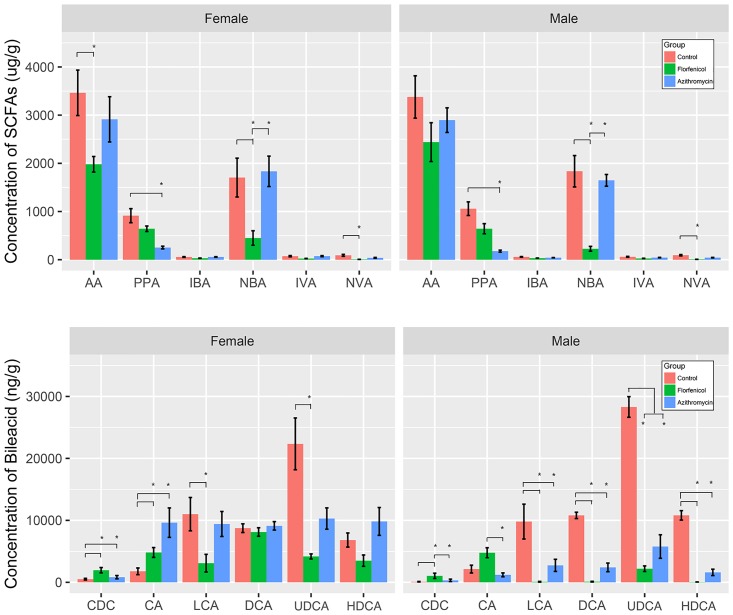
As shown in Fig 6, the two antibiotics decreased the concentration of some SCFAs in the colonic contents, and florfenicol showed a stronger effect compared with azithromycin. Florfenicol decreased the concentration of some SCFAs in the colonic contents, especially AA, NBA, and NVA, with a 10-fold lower concentration than that in the control group. However, for azithromycin, only PPA and NVA showed decreased concentrations. There were no significant differences between the male and female groups.
Fig 6. Concentration of SCFAs and bile acids in samples from the normal and antibiotic-treated groups.
One-way analysis of variance (one-way ANOVA) and multiple t-tests with Bonferroni correction were employed to measure the concentration differences in bile acids and SCFAs between the three groups. * indicates that the difference between the two line-linked groups was statistically significant (p < 0.05).
In males, the mice treated with florfenicol had a higher concentration of CDCA and lower concentrations of LCA, DCA, UDCA and HDCA compared with the control group. The concentrations of all bile acids except CDCA and CA in the mice treated with azithromycin decreased compared with the control group, and the mice treated with azithromycin showed no significant difference compared with those treated with florfenicol. There were some differences between the male and female groups. Compared with females, males had lower concentrations of all bile acids in the azithromycin group and lower concentrations of secondary bile acids (LCA, DCA, UDCA, HDCA) in the florfenicol group.
Discussion
In this study, azithromycin and florfenicol were found to increase adipogenesis in mice and alter the overall abundance, diversity and composition of the gut microbiota, the production of SCFAs, and the metabolism of bile acids in colonic contents. The changes in gut microbiota showed some antibiotic- or sex-specific differences. To the best of our knowledge, this is the first time the effects of azithromycin and florfenicol on gut microbiota composition and adipogenesis in mice have been explored.
We observed that antibiotics disturbed the gut microbiota composition. Most previous studies support these finding. For example, oral administration of amoxicillin, cefotaxime, and vancomycin decreased the abundance and altered the gut microbiota composition in rats[17]. The gut microbiota diversity indicated by the Shannon index was lower in both the antibiotic groups than in the control group, suggesting that the antibiotics reduced not only the richness but also the diversity of the gut microbiota, which is in line with results from previous studies[17,36]. Our study also found that azithromycin decreased the relative abundance of Bacteroidetes and Proteobacteria and increased the relative abundance of Firmicutes, which corresponds with the results of most other studies [6,37]. However, in Khan’s study, the Bacteroidetes/Firmicutes ratio increased[36]. Differences in dose, type of mice and the duration of experiments may be the reason for this inconsistency. Azithromycin increased the relative abundance of Parabacteroides and decreased the relative abundance of Desulfovibrio in the female group, but the opposite results were observed in the male group. This might be related to the differences in endocrine activity and the genetic background between male and female mice and needs to be further explored.
Both florfenicol and azithromycin decreased the concentrations of SCFAs in the colons of mice, but some differences were seen between the two antibiotics. This may be related to the changes in relative abundance and number of SCFA-producing bacteria in the gut microbiota induced by these antibiotics. For example, the decrease in the relative abundance of dominant propionate-producing Bacteroidetes in the group treated with azithromycin could have caused a reduced concentration of propionate in the colonic sample[38]. However, the relative abundance of acetate-producing Blautia increased in all the antibiotic groups except for the group of male mice group treated with florfenicol, but the concentration of acetate in these groups did not increase[39]. This may be explained by the possibility that the decrease in other acetate-producing bacteria, such as Ruminococcaceae, or the effects of the decreased absolute amount of Blautia on the production of acetate exceeded the effect of the increased relative abundance of acetate-producing Blautia [38]. Moreover, the relative abundance of butyrate-producing Roseburia and Anaerotruncus increased in mice treated with azithromycin but not in mice treated with florfenicol, which might explain the discrepancy in butyrate concentration between the two experimental groups[40].
Some human studies reported a decreased concentration of SCFAs induced by antibiotics, which supports our findings. Høverstad et al. reported that oral intake of particular antibiotics reduced the concentration of SCFAs in fecal samples under a therapeutic dose[41]. Young also found that amoxicillin under a therapeutic dose decreased the concentration of butyrate in fecal samples from men[42]. However, Cho’s result was inconsistent with our findings; in his study, penicillin and vancomycin at an exposure dose of 1 μg per g body weight increased most of the SCFAs in mice[14]. The possible reason for this could be the different exposure dose and type of antibiotic. In our study, a therapeutic dose was used, which reduced the total number of SCFA-producing bacteria and then decreased the concentration of SCFAs in the gut.
We found that florfenicol and azithromycin increased the concentrations of primary bile acids in the colon but decreased the concentrations of secondary bile acids, suggesting down-regulation of the 7-dehydroxylation process. Some studies have also reported similar effects for other antibiotics, such as vancomycin. In a single-blinded randomized controlled trial with 20 male subjects, 7 days of vancomycin t.i.d. was found to decrease fecal secondary bile acids and increase primary bile acids in plasma and fecal samples under the therapeutic dose [43,44]. The 7-dehydroxylation activity of Clostridium and Eubacterium in the gut microbiota is the key process that turns primary bile acid into secondary bile acid [45]. However, these two bacterial genera had extremely low relative abundances in the gut microbiota of our experimental mice, suggesting there might be other undiscovered bacteria responsible for 7-dehydroxylation. For example, other Clostridium family microbiota, such as Ruminococcaceae and Lachnospiraceae, might share similar functions with Clostridium scindens, and the relative abundances of these bacteria were also decreased by florfenicol and azithromycin [46]. Another possible explanation could be that the florfenicol and azithromycin decreased the total level of gut microbiota in our study as indicated by the Chao and ACE index, and they reduced the number of all bacteria involved in secondary bile acid synthesis. Sayin et al. found that germ-free mice had a higher gene expression level of primary bile acid biosynthesis compared with normal C57BL/6 mice, which supports the above explanation[27]. Furthermore, there are other reactions related to bile acid metabolism, such as deconjugation, oxidation and epimerization, and Bacteroidaceae and Lachnospiraceae are the main taxa that participate in these reactions [47]. In a human study, after subjects were treated with rifaximin b.i.d. for 8 weeks, the relative abundance of Bacteroidaceae and Lachnospiraceae in fecal samples was negatively correlated with primary bile acids and secondary/primary bile acid ratios in serum samples[46]. This finding is consistent with our results, indicating that in addition to the 7-dehydroxylation process other reactions, possibly involving Bacteroidaceae and Lachnospiraceae, also affect the production of secondary bile acid[48].
In our study, male mice in the florfenicol group had lower concentrations of secondary bile acids and male mice in the azithromycin group had lower concentrations of all bile acids. Gut microbiota and sex hormones may contribute to this difference. Males in the florfenicol and azithromycin group exhibited lower concentrations of Bacteroides and Lactobacillus, respectively. These taxa have been associated with bile acid homeostasis, and decreased relative abundance may result in dysfunction of bile acid synthesis[49]. In addition, the sex hormone in males is correlated with bile acid concentration, which might result in a decrease in the excretion of bile acid[50]. The reason for the sex-associated differences in the alteration of bile acids caused by antibiotics is not clear and should be investigated in future studies.
In this study, both azithromycin and florfenicol were observed to increase adipogenesis in mice, and growth was accelerated in male mice, which is supported by most previous studies. For example, both of the antibiotics decreased the relative abundance of Lactobacillus, and this was consistent with the finding by M. Cox et al. In their study, when the same amount of food was ingested, the penicillin group with a lower relative abundance of Lactobacillus exhibited more weight gain than the control group[51]. We found that the increased relative abundance of Rumen caused by azithromycin was associated with adipogenesis in mice. A previous study demonstrated that individuals with a higher abundance of Rumen bacteria had a higher incidence of insulin resistance, fatty liver and low-grade inflammation[52]. Azithromycin decreased the relative abundance of Akkermansia muciniphila, which could lead to an increase in intestinal inflammation and obesity [53]. The Shannon and Chao index and the body fat measurements of mice in our study also suggested that individuals with low bacterial richness had higher overall body fat than those with high bacterial richness [54,55]. Moreover, some human studies have also found a strong association between antibiotic exposure and obesity in boys[20,23,56,57].
SCFAs and bile acids are important metabolites in fat metabolism. They not only adjust the body's energy intake but also affect fat metabolism [58] [59]. Our study found that body fat rate was significantly higher and the SCFA content was significantly lower in both of the antibiotic groups compared with the control group. The decreased concentration of SCFAs may contribute to obesity as follows. First, because SCFAs can serve as not only a source of energy but also the main nutrients for intestinal cells to maintain intestinal homeostasis, a decrease in SCFAs may lead to nutrition deficiency in intestinal cells and decreased defense, which leads to increased susceptibility to low-level inflammation, eventually leading to the occurrence of obesity [60]. Second, some SCFAs are negatively correlated with the occurrence of obesity. For example, butyric acid and propionic acid can promote gluconeogenesis through the cyclic AMP pathway and brain-gut neural circuits and inhibit the accumulation of fat in adipose tissue[61,62]. Therefore, a decrease in the production of these SCFAs may lead to down-regulation of related metabolic pathways, resulting in the occurrence of obesity.
An increased ratio of primary/secondary bile acids was found in our study, and this increase may lead to obesity via several pathways. First, primary bile acids have been reported to have strong antimicrobial activity and could cause a major shift in gut microbiota composition toward Firmicutes and against Bacteroidetes [63]. The ratio of Firmicutes to Bacteroidetes increased significantly in our study, which is a sign of obesity [64,65]. Second, the increase in the primary/secondary bile acid ratio could also lead to a decrease in basal metabolic rate [58,66]. In the case of the same energy intake and daily activities, a lower basal metabolic rate leads to more fat accumulation in the body. In addition to the well-established roles of bile acids in dietary lipid absorption and cholesterol homeostasis, they act as signaling molecules with systemic endocrine functions [67,68]. Alterations in bile acid metabolism can lead to insulin resistance and obesity [43]. For example, bile acids activate mitogen-activated protein kinase pathways and are ligands for the G-protein-coupled receptor TGR5, and secondary bile acids have a much stronger activating capacity for TGR5 than primary bile acids[69]. Down-regulation of TGR5 could disrupt energy metabolism and lead to obesity [70].
Conclusions
We found that azithromycin and florfenicol altered gut microbiota composition, the production of SCFAs, and the metabolism of bile acids in colonic contents and increased adipogenesis in mice. Some effects showed a sex difference. These findings support the hypothesis that exposure to antibiotics is one important risk factor for childhood obesity, and the effects may be mediated by gut microbiota. More lab studies are needed to investigate specific mechanisms and to confirm the role of gut microbiota in adipogenesis induced by antibiotics.
Supporting information
(PDF)
(CSV)
(TXT)
(TRE)
Acknowledgments
This work was supported by the Natural Science Foundation of Shanghai (No. 16ZR1403400), the Shanghai Key Discipline Construction Project of Shanghai Municipal Public Health (Grant No. 15GWZK0801), The Fourth Round of Three-year Public Health Action Plan of Shanghai (No. 15GWZK0202) and the Starting Foundation for New Teachers of Fudan University (No. JIF201111).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Natural Science Foundation of Shanghai (No. 16ZR1403400), the Shanghai Key Discipline Construction Project of Shanghai Municipal Public Health (Grant No. 15GWZK0801), The Fourth Round of Three-year Public Health Action Plan of Shanghai (No. 15GWZK0202) and the Starting Foundation for New Teachers of Fudan University (No. JIF201111). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.James WPT. WHO recognition of the global obesity epidemic. International Journal of Obesity. Nature Publishing Group; 2008;32: S120–S126. doi: 10.1038/ijo.2008.247 [DOI] [PubMed] [Google Scholar]
- 2.James PT, Leach R, Kalamara E. The worldwide obesity epidemic. Obesity research. 2001. [DOI] [PubMed] [Google Scholar]
- 3.Ogden CL, Carroll MD, Kit BK, Flegal KM. Prevalence of Obesity and Trends in Body Mass Index Among US Children and Adolescents, 1999–2010. JAMA. American Medical Association; 2012;307: 483–490. doi: 10.1001/jama.2012.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krebs NF, Himes JH, Jacobson D, Nicklas TA. Assessment of child and adolescent overweight and obesity. Pediatrics. 2007. [DOI] [PubMed] [Google Scholar]
- 5.Waxman A. Why a Global Strategy on Diet, Physical Activity and Health? Nutrition and Fitness: Mental Health, Aging, and the Implementation of a Healthy Diet and Physical Activity Lifestyle. Basel: Karger Publishers; 2005;95: 162–166. doi: 10.1159/000088302 [DOI] [PubMed] [Google Scholar]
- 6.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006. [DOI] [PubMed] [Google Scholar]
- 7.Fei N, Zhao L. An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. The ISME Journal. Nature Publishing Group; 2013;7: 880–884. doi: 10.1038/ismej.2012.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang H, DiBaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu Y, et al. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci USA. National Acad Sciences; 2009;106: 2365–2370. doi: 10.1073/pnas.0812600106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E, et al. Dietary intervention impact on gut microbial gene richness. Nature. Nature Research; 2013;500: 585–588. doi: 10.1038/nature12480 [DOI] [PubMed] [Google Scholar]
- 10.Pihl AF, Fonvig CE, Stjernholm T, Hansen T, Pedersen O, Holm J-C. The Role of the Gut Microbiota in Childhood Obesity. Child Obes. 2016. doi: 10.1089/chi.2015.0220 [DOI] [PubMed] [Google Scholar]
- 11.Mayorga Reyes L, González Vázquez R, Cruz Arroyo SM, Melendez Avalos A, Reyes Castillo PA, Chavaro Pérez DA, et al. Correlation between diet and gut bacteria in a population of young adults. Int J Food Sci Nutr. 2016;67: 470–478. doi: 10.3109/09637486.2016.1162770 [DOI] [PubMed] [Google Scholar]
- 12.Jin Y, Wu S, Zeng Z, Fu Z. Effects of environmental pollutants on gut microbiota. Environ Pollut. 2017;222: 1–9. doi: 10.1016/j.envpol.2016.11.045 [DOI] [PubMed] [Google Scholar]
- 13.Mikkelsen KH, Knop FK, Vilsbøll T, Frost M, Hallas J, Pottegård A. Use of antibiotics in childhood and risk of Type 1 diabetes: a population-based case-control study. Diabet Med. 2017;34: 272–277. doi: 10.1111/dme.13262 [DOI] [PubMed] [Google Scholar]
- 14.Cho I, Yamanishi S, Cox L, Methé BA, Zavadil J, Li K, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. Nature Publishing Group; 2012;488: 621–626. doi: 10.1038/nature11400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Looft T, Johnson TA, Allen HK, Bayles DO, Alt DP, Stedtfeld RD, et al. In-feed antibiotic effects on the swine intestinal microbiome. Proc Natl Acad Sci USA. National Acad Sciences; 2012;109: 1691–1696. doi: 10.1073/pnas.1120238109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson CJ, Young VB. Antibiotic administration alters the community structure of the gastrointestinal micobiota. Gut Microbes. Taylor & Francis; 2010;1: 279–284. doi: 10.4161/gmic.1.4.12614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tulstrup MV-L, Christensen EG, Carvalho V, Linninge C, Ahrné S, Højberg O, et al. Antibiotic Treatment Affects Intestinal Permeability and Gut Microbial Composition in Wistar Rats Dependent on Antibiotic Class. Loh G, editor. PLoS ONE. Public Library of Science; 2015;10: e0144854 doi: 10.1371/journal.pone.0144854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin Y, Wu Y, Zeng Z, Jin C, Wu S, Wang Y, et al. From the Cover: Exposure to Oral Antibiotics Induces Gut Microbiota Dysbiosis Associated with Lipid Metabolism Dysfunction and Low-Grade Inflammation in Mice. Toxicol Sci. Oxford University Press; 2016;154: 140–152. doi: 10.1093/toxsci/kfw150 [DOI] [PubMed] [Google Scholar]
- 19.Trasande L, Blustein J, Liu M, Corwin E, Cox LM, Blaser MJ. Infant antibiotic exposures and early-life body mass. International Journal of Obesity. Nature Publishing Group; 2013;37: 16–23. doi: 10.1038/ijo.2012.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saari A, Virta LJ, Sankilampi U, Dunkel L, Saxen H. Antibiotic exposure in infancy and risk of being overweight in the first 24 months of life. Pediatrics. 2015;135: 617–626. doi: 10.1542/peds.2014-3407 [DOI] [PubMed] [Google Scholar]
- 21.Yallapragada SG, Nash CB, Robinson DT. Early-Life Exposure to Antibiotics, Alterations in the Intestinal Microbiome, and Risk of Metabolic Disease in Children and Adults. Pediatr Ann. 2015;44: e265–9. doi: 10.3928/00904481-20151112-09 [DOI] [PubMed] [Google Scholar]
- 22.Mansi Y, Abdelaziz N, Ezzeldin Z, Ibrahim R. Randomized controlled trial of a high dose of oral erythromycin for the treatment of feeding intolerance in preterm infants. Neonatology. 2011;100: 290–294. doi: 10.1159/000327536 [DOI] [PubMed] [Google Scholar]
- 23.Wang H, Wang N, Bin Wang, Fang H, Fu C, Tang C, et al. Antibiotics detected in urines and adipogenesis in school children. Environment International. Elsevier B.V; 2016;89–90: 204–211. doi: 10.1016/j.envint.2016.02.005 [DOI] [PubMed] [Google Scholar]
- 24.Wang H, Wang N, Wang B, Zhao Q, Fang H, Fu C, et al. Antibiotics in Drinking Water in Shanghai and Their Contribution to Antibiotic Exposure of School Children. Environ Sci Technol. 2016;50: 2692–2699. doi: 10.1021/acs.est.5b05749 [DOI] [PubMed] [Google Scholar]
- 25.Varma KJ, Sams RA, Lobell RD, Lockwood PW. Pharmacokinetics and efficacy of a new broad spectrum antibiotic, florfenicol in cattle. Acta Veterinaria Scandinavica Supplementum (Denmark). Acta Veterinaria …; 1991. [Google Scholar]
- 26.Retsema J, Girard A, Schelkly W, Manousos M, Anderson M, Bright G, et al. Spectrum and mode of action of azithromycin (CP-62,993), a new 15-membered-ring macrolide with improved potency against gram-negative organisms. Antimicrob Agents Chemother. American Society for Microbiology; 1987;31: 1939–1947. doi: 10.1128/AAC.31.12.1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall H-U, Bamberg K, et al. Gut Microbiota Regulates Bile Acid Metabolism by Reducing the Levels of Tauro-beta-muricholic Acid, a Naturally Occurring FXR Antagonist. Cell Metabolism. 2013;17: 225–235. doi: 10.1016/j.cmet.2013.01.003 [DOI] [PubMed] [Google Scholar]
- 28.Team RC. R: A language and environment for statistical computing. 2013. [Google Scholar]
- 29.Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. PANDAseq: paired-end assembler for illumina sequences. BMC Bioinformatics. BioMed Central; 2012;13: 1 doi: 10.1186/1471-2105-13-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. Oxford University Press; 2014;30: btu170–2120. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edgar R. Usearch. Lawrence Berkeley National Laboratory (LBNL), Berkeley, CA (United States); 2010.
- 32.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K. QIIME allows analysis of high-throughput community sequencing data. Nature. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucl Acids Res. Oxford University Press; 2013;41: D590–6. doi: 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paulson JN, Stine OC, Bravo HC, Pop M. Differential abundance analysis for microbial marker-gene surveys. Nature methods. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Čech M, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucl Acids Res. Oxford University Press; 2016;44: gkw343–W10. doi: 10.1093/nar/gkw343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khan I, Azhar EI, Abbas AT, Kumosani T, Barbour EK, Raoult D, et al. Metagenomic Analysis of Antibiotic-Induced Changes in Gut Microbiota in a Pregnant Rat Model. Front Pharmacol. 2016;7: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murphy EF, Cotter PD, Healy S, Marques TM, O'Sullivan O, Fouhy F, et al. Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut …. BMJ Publishing Group Ltd and British Society of Gastroenterology; 2010;59: 1635–1642. doi: 10.1136/gut.2010.215665 [DOI] [PubMed] [Google Scholar]
- 38.Flint HJ, Scott KP, Louis P, Duncan SH. The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol. 2012;9: 577–589. doi: 10.1038/nrgastro.2012.156 [DOI] [PubMed] [Google Scholar]
- 39.Murugesan S, Ulloa-Martínez M, Martínez-Rojano H, Galván-Rodríguez FM, Miranda-Brito C, Romano MC, et al. Study of the diversity and short-chain fatty acids production by the bacterial community in overweight and obese Mexican children. Eur J Clin Microbiol Infect Dis. Springer Berlin Heidelberg; 2015;34: 1337–1346. doi: 10.1007/s10096-015-2355-4 [DOI] [PubMed] [Google Scholar]
- 40.Polansky O, Sekelova Z, Faldynova M, Sebkova A, Sisak F, Rychlik I. Important Metabolic Pathways and Biological Processes Expressed by Chicken Cecal Microbiota. Dozois CM, editor. Applied and …. American Society for Microbiology; 2016;82: 1569–1576. doi: 10.1128/AEM.03473-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Høverstad T, Carlstedt-Duke B, Lingaas E, Norin E, Saxerholt H, Steinbakk M, et al. Influence of Oral Intake of Seven Different Antibiotics on Faecal Short-Chain Fatty Acid Excretion in Healthy Subjects. Scandinavian Journal of Gastroenterology. Taylor & Francis; 2009;21: 997–1003. doi: 10.3109/00365528608996411 [DOI] [PubMed] [Google Scholar]
- 42.Young VB, Schmidt TM. Antibiotic-associated diarrhea accompanied by large-scale alterations in the composition of the fecal microbiota. J Clin Microbiol. American Society for Microbiology; 2004;42: 1203–1206. doi: 10.1128/JCM.42.3.1203-1206.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vrieze A, Vrieze A, Out C, Out C, Fuentes S, Fuentes S, et al. Impact of oral vancomycin on gut microbiota, bile acid metabolism, and insulin sensitivity. Journal of Hepatology. 2014;60: 824–831. doi: 10.1016/j.jhep.2013.11.034 [DOI] [PubMed] [Google Scholar]
- 44.Weingarden AR, Weingarden AR, Chen C, Chen C, Bobr A, Bobr A, et al. Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. American Journal of Physiology—Gastrointestinal and Liver Physiology. American Physiological Society; 2014;306: G310–G319. doi: 10.1152/ajpgi.00282.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gérard P. Metabolism of Cholesterol and Bile Acids by the Gut Microbiota. Pathogens. 2014;3: 14–24. doi: 10.3390/pathogens3010014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kakiyama G, Kakiyama G, Pandak WM, Pandak WM, Gillevet PM, Gillevet PM, et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. Journal of Hepatology. 2013;58: 949–955. doi: 10.1016/j.jhep.2013.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hirano S, Nakama R, Tamaki M, Masuda N, Oda H. Isolation and characterization of thirteen intestinal microorganisms capable of 7 alpha-dehydroxylating bile acids. Applied and …. American Society for Microbiology; 1981;41: 737–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wahlström A, Sayin SI, Marschall H-U, Bäckhed F. Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metabolism. 2016;24: 41–50. doi: 10.1016/j.cmet.2016.05.005 [DOI] [PubMed] [Google Scholar]
- 49.Org E, Mehrabian M, Parks BW, Shipkova P, Liu X, Drake TA, et al. Sex differences and hormonal effects on gut microbiota composition in mice. Gut Microbes. Taylor & Francis; 2016;7: 313–322. doi: 10.1080/19490976.2016.1203502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frommherz L, Rist MJ, Roth A, Bub A, Hummel E, Watzl B, et al. Bile acid plasma concentrations are associated with age, sex and lipid metabolism in healthy humans.: 149/355. Annals of Nutrition and Metabolism. Annals of Nutrition and Metabolism; 2015;67: 269–270. [Google Scholar]
- 51.Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158: 705–721. doi: 10.1016/j.cell.2014.05.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. Nature Publishing Group; 2010;464: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dao MC, Everard A, Aron-Wisnewsky J, Sokolovska N, Prifti E, Verger EO, et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut …. BMJ Publishing Group Ltd and British Society of Gastroenterology; 2016;65: 426–436. [DOI] [PubMed] [Google Scholar]
- 54.Lin S-W, Freedman ND, Shi J, Gail MH, Vogtmann E, Yu G, et al. Beta-diversity metrics of the upper digestive tract microbiome are associated with body mass index. Obesity (Silver Spring). 2015;23: 862–869. doi: 10.1002/oby.21020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. Nature Research; 2013;500: 541–546. doi: 10.1038/nature12506 [DOI] [PubMed] [Google Scholar]
- 56.Azad MB, Bridgman SL, Becker AB, Kozyrskyj AL. Infant antibiotic exposure and the development of childhood overweight and central adiposity. Int J Obes (Lond). Nature Publishing Group; 2014;38: 1290–1298. doi: 10.1038/ijo.2014.119 [DOI] [PubMed] [Google Scholar]
- 57.Ajslev TA, Andersen CS, Gamborg M, Sørensen TIA, Jess T. Childhood overweight after establishment of the gut microbiota: the role of delivery mode, pre-pregnancy weight and early administration of antibiotics. Int J Obes (Lond). Nature Publishing Group; 2011;35: 522–529. doi: 10.1038/ijo.2011.27 [DOI] [PubMed] [Google Scholar]
- 58.Lefebvre P, Lefebvre P, Cariou B, Cariou B, Lien F, Lien F, et al. Role of Bile Acids and Bile Acid Receptors in Metabolic Regulation. Physiological Reviews. American Physiological Society; 2009;89: 147–191. doi: 10.1152/physrev.00010.2008 [DOI] [PubMed] [Google Scholar]
- 59.Kumari M, Kozyrskyj AL. Gut microbial metabolism defines host metabolism: an emerging perspective in obesity and allergic inflammation. Obesity Reviews. 2017;18: 18–31. doi: 10.1111/obr.12484 [DOI] [PubMed] [Google Scholar]
- 60.Ha CW. Mechanistic links between gut microbial community dynamics, microbial functions and metabolic health. World J Gastroenterol. 2014;20: 16498 doi: 10.3748/wjg.v20.i44.16498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tolhurst G, Heffron H, Lam YS, Parker HE, Habib AM, Diakogiannaki E, et al. Short-Chain Fatty Acids Stimulate Glucagon-Like Peptide-1 Secretion via the G-Protein–Coupled Receptor FFAR2. Diabetes. American Diabetes Association; 2012;61: 364–371. doi: 10.2337/db11-1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kimura I, Inoue D, Hirano K, Tsujimoto G. The SCFA Receptor GPR43 and Energy Metabolism. Front Endocrinol. Frontiers; 2014;5: 840 doi: 10.3389/fendo.2014.00085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Islam KBMS, Fukiya S, Hagio M, Fujii N, Ishizuka S, Ooka T, et al. Bile Acid Is a Host Factor That Regulates the Composition of the Cecal Microbiota in Rats. Gastroenterology. Elsevier; 2011;141: 1773–1781.: doi: 10.1053/j.gastro.2011.07.046 [DOI] [PubMed] [Google Scholar]
- 64.Flint HJ. Obesity and the Gut Microbiota. Journal of Clinical Gastroenterology. 2011;45: S128–S132. doi: 10.1097/MCG.0b013e31821f44c4 [DOI] [PubMed] [Google Scholar]
- 65.Chang CJ, Lin CS, Lu CC, Martel J, Ko YF. Ganoderma lucidum reduces obesity in mice by modulating the composition of the gut microbiota. Nature. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Swann JR, Swann JR, Want EJ, Want EJ, Geier FM, Geier FM, et al. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc Natl Acad Sci USA. National Acad Sciences; 2011;108 Suppl 1: 4523–4530. doi: 10.1073/pnas.1006734107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Houten SM, Houten SM, Watanabe M, Watanabe M, Auwerx J, Auwerx J. Endocrine functions of bile acids. The EMBO Journal. EMBO Press; 2006;25: 1419–1425. doi: 10.1038/sj.emboj.7601049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Prawitt J, Prawitt J, Caron S, Caron S, Staels B, Staels B. Bile Acid Metabolism and the Pathogenesis of Type 2 Diabetes. Curr Diab Rep. Current Science Inc; 2011;11: 160–166. doi: 10.1007/s11892-011-0187-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thomas C, Thomas C, Gioiello A, Gioiello A, Noriega L, Noriega L, et al. TGR5-Mediated Bile Acid Sensing Controls Glucose Homeostasis. Cell Metabolism. 2009;10: 167–177. doi: 10.1016/j.cmet.2009.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen X, Lou G, Meng Z, Huang W. TGR5: a novel target for weight maintenance and glucose metabolism. Exp Diabetes Res. Hindawi Publishing Corporation; 2011;2011: 853501–5. doi: 10.1155/2011/853501 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(CSV)
(TXT)
(TRE)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.