Abstract
Mechanical forces are key regulators of cell and tissue physiology. The basic molecular mechanism of fiber contraction by the sliding of actin filament upon myosin leading to conformational change has been known for decades. The regulation of force generation at the level of the cell, however, is still far from elucidated. Indeed, the magnitude of cell traction forces on the underlying extracellular matrix in culture is almost impossible to predict or experimentally control. The considerable variability in measurements of cell-traction forces indicates that they may not be the optimal readout to properly characterize cell contractile state and that a significant part of the contractile energy is not transferred to cell anchorage but instead is involved in actin network dynamics. Here we discuss the experimental, numerical, and biological parameters that may be responsible for the variability in traction force production. We argue that limiting these sources of variability and investigating the dissipation of mechanical work that occurs with structural rearrangements and the disengagement of force transmission is key for further understanding of cell mechanics.
INTRODUCTION
Mechanical forces are central to many physiological processes, including tissue morphogenesis (Levayer and Lecuit, 2012; Heisenberg and Bellaïche, 2013), patterning (Eaton and Jülicher, 2011), growth (Bosveld et al., 2016), renewal (Eisenhoffer et al., 2012), cell migration (Tambe et al., 2011), differentiation (Ruiz and Chen, 2008), and polarization (Pitaval et al., 2010). The mechanism of force generation has been studied for decades and attracted particular interest since it has been possible to measure the traction forces cells exert on their environment (Pelham and Wang, 1999). Despite these considerable efforts and the importance of mechanical forces in biology, we do not have a comprehensive understanding of the mechanisms that support and regulate force generation. Our ignorance is revealed by the variety of measures used to evaluate cell mechanical forces. This variability not only reflects the contribution of uncontrolled parameters but also limits our ability to detect differences between distinct conditions and thus our progress in the study of cell mechanics. To identify ways to solve this issue, we first describe the experimental and biological origins of intercellular variability in force generation. We then discuss the cellular mechanisms responsible for the latter and conclude that future efforts should focus on the investigation of the dissipation of contractile energy, which cannot be captured by current experimental methods.
VARIABILITY IN TRACTION FORCE MEASUREMENTS
The estimation of the traction force of a single cell is typically based on the measurement of the deformation of the cell-culture substrate in response to cell-generated force with either traction force microscopy (TFM; Dembo and Wang, 1999), which is based on a continuous substrate, or with the measurement of individual micropost deformation (Tan et al., 2003). Various readouts have been used to characterize cell mechanical state: the force per adhesion (Plotnikov et al., 2012), the force produced at cell apices (Rape et al., 2011b), the average traction force (Rape et al., 2011a), the integrated absolute forces (Reinhart-King et al., 2005), and the contractile energy (Butler et al., 2002), which we believe better characterize the mechanical effort generated by the cell. For all of these readouts, averaged values of several individual cells are typically used to represent cell traction forces. However, the wide range of forces, which has a non-Gaussian asymmetric distribution starting at very low values and ending with a long tail (e.g., Figure 1 in Milloud et al., 2017) tends to be overlooked but is manifest when all individual values are reported. A significant proportion of cells produce forces that are barely above detection threshold, whereas others produce three times more than the average value. Thus, regardless of how absolute or relative those measurements are, their broad range shows that we have not identified the major regulatory parameters that set the magnitude of these forces. It is critical to understand what lies behind this range in order to properly characterize cell contractile state. In addition, the range is not only troublesome per se, it is also a limitation when comparing two conditions in attempts to identify parameters influencing force production.
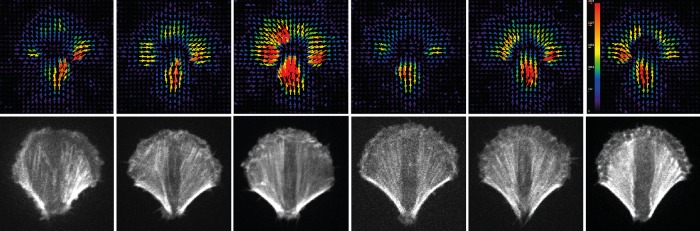
FIGURE 1:
Variability of traction forces in micropatterned cells. RPE1 cells were plated on a crossbow-shaped fibronectin micropattern on polyacrylamide gels. The expression of LifeAct–green fluorescent protein revealed the actin network architecture (bottom). TFM was used to measure the associated traction forces (top; color scale is the same for all images).
The most striking indicator of our ignorance of the key parameters regulating force generation is our inability to experimentally control traction force generation and reduce data dispersion. The magnitude of cell traction forces varies considerably as cells change shape during migration (Meili et al., 2010; Chang et al., 2013). The extent of the area over which the cell has spread clearly promotes traction forces (Tan et al., 2003; Tolić-Nørrelykke and Wang, 2005; Rape et al., 2011b), but the degree of elongation of the cell shape may have an opposing effect on traction forces (Oakes et al., 2014; Ye et al., 2014a). If it is assumed that such forces are being generated via the actin network, then it would be reasonable to argue that experimentally controlling the geometry of the actin network should also control force generation. However, controlling cell shape using extracelluar matrix (ECM) micropatterns and imposing a stringent subcellular adhesion pattern to direct stress fibers length, position, and orientation in an immortalized cell line was not sufficient to reduce significantly the intercellular variability in force generation (Tseng et al., 2011, 2012; Mandal et al., 2014; Oakes et al., 2014; Figure 1). Large variations remain in force measurements despite several attempts, including ours, to reduce it, notably with micropatterning and controlled integrin expression (Schiller et al., 2013). This suggests that parameters in addition to the actin network geometry regulate traction force generation. It should also be recognized that a static representation of the actin network geometry may mask variations in force generation that potentially arise from the underlying dynamics and the large diversity of molecular players involved in developing mechanical work in the cell. This will be discussed in detail later. An interesting possibility is that progression through the cell cycle also affects cell contractility. Inducing cells to exit the cell cycle and become quiescent controls force generation by reducing it to low levels (Tan et al., 2003; Rape et al., 2011a). However, it is difficult to know whether this effect on force generation is due to cell-cycle exit per se or from changes in growth factor signaling and requires further investigation.
We need to acknowledge this intrinsic variability and try to determine its origins in order to identify the key parameter regulating traction force magnitude and cell contractile state. In the following sections, we first discuss the technical sources of errors associated with cell manipulation and the processing of numerical data. We then discuss the biological variability and the intrinsic fluctuations due to the cellular mechanism of force generation.
EXPERIMENTAL VARIABILITY
When using TFM, the computation assumes that the gel is homogeneous, isotropic, and elastic. Hence any deviations from this assumed behavior will bias force estimation. Different types of soft materials are used with TFM and are mainly based on hydrogels (Pelham and Wang, 1997; Dembo and Wang, 1999; Tse and Engler, 2010) or elastomers (Harris et al., 1981; Balaban et al., 2001; Merkel et al., 2007). Gel formulation, the time taken and temperature for gelation, and the gel-storage parameters and duration all influence the reproducibility of the gel’s material properties (Denisin and Pruitt, 2016). In particular, hydrogels are susceptible to effects from drying, swelling, and aging (Dembo and Wang, 1999; Kara and Pekcan, 2001; Martiel et al., 2015). Polyacrylamide is usually chosen for its limited stress stiffening, reduced nonlinear properties (Storm et al., 2005), and instantaneous strain recovery (Dembo and Wang, 1999). However, care needs to be taken because stiffening has also been reported for polyacrylamide subjected to large deformations (Boudou et al., 2009). Gel deformation should also be of sufficient magnitude that the displacements of an embedded bead can be distinguished from movements related to thermal noise.
Depending on its initial formulation, a polyacrylamide gel tends to present heterogeneities in its network structures that can induce variations in its stiffness over time or with its polymerization temperature (Denisin and Pruitt, 2016). Given that the magnitude of traction forces is highly sensitive to substrate stiffness (Plotnikov et al., 2012; Elosegui-Artola et al., 2016), the variations in gel stiffness may increase the range of traction force measurements. To reduce the intrabatch and interbatch stiffness variability, hydrogel polymerization by light (photopolymerization) has been used and is more rapid and homogeneous than polymerization by mixing free radical–producing chemicals (Nguyen and West, 2002). Micropillars have also been used instead of polyacrylamide gels because substrate stiffness is primarily related to the pillar aspect ratio (Tan et al., 2003; du Roure et al., 2005). However, the drawback with micropillars is that the variability in pillar deformation is also related to the way cells attach and spread on the pillars (Ghibaudo et al., 2011).
Coating of the gel surface with ECM proteins to promote cell adhesion and spreading is another source of variability. The composition and density of the ECM regulate cell adhesion signaling and the production of mechanical forces (Schwarz and Gardel, 2012; Lee et al., 2015). In addition, the anchorage of the adhesion ligand and the type of surface coating of the polyacrylamide surface also affect the amount of cell traction forces even without changing the overall cell shape (Pompe et al., 2011). Therefore inhomogeneities in ECM coating density can lead to intercellular variability. Such variations in ECM density can arise from the use of ultraviolet-activatable chemical cross-linkers to graft ECM proteins on polyacrylamide substrates. However, use of acrylamide polymerization to bind ECM proteins can improve the reproducibility of ECM grafting characteristics (Rape et al., 2011b; Vignaud et al., 2014).
COMPUTATIONAL VARIABILITY
Large errors can come from inaccurate estimation of the deformation field and image processing (Holenstein et al., 2017). First, it is generally assumed that traction forces are negligible perpendicular to the gel surface (and XY-plane) and hence that out-of-plane deformations need not be considered. Indeed, the Boussinesq theory predicts in-plane and out-of-plane deformations are independent at the surface of an incompressible substratum (Poisson ratio ∼0.5). The Poisson ratio of the gel substrate can be determined experimentally (Soiné et al., 2015; Gross and Kress, 2017), and recent work has highlighted that an error of 0.05 in the Poisson ratio can lead to a 38% variation in the force measurement (del Álamo et al., 2013). Hence out-of-plane effects are not always negligible (Maskarinec et al., 2009; Legant et al., 2013), and ignoring them can add error to the traction force estimation.
There is growing interest in improving the precision of the deformation field measurement to reveal subcellular regulation of traction forces. Current methods for measuring the gel-deformation field, that is, cross-correlation of bead displacement or the tracking of individual beads, do not contribute much to intercellular variability in force measurements. For both methods, bead density, homogeneity, and image quality are key parameters for the robustness and the resolution of the obtained data (Soiné et al., 2015; Holenstein et al., 2017). A combination of these methods can increase the resolution of the displacement field, reduce the error on the estimated force field, and improve the investigation of force regulation at the focal adhesion level (Sabass et al., 2008), but it is unlikely to decrease the intercellular variance of total traction force energies. Other parameters are more prone to influence the variability of the results and should be kept constant, such as 1) the correlation threshold used for selecting significant bead displacements over background noise, 2) the spatial displacement with sampling given by the range of bead numbers per size of the interrogation window, and 3) the erroneous displacements that can occur when beads disappear from the field of analysis (Martiel et al., 2015). Failures in bead tracking due to out-of-plane bead displacements can be limited by using a single-plane bead-embedding method or three-dimensional bead tracking (Maskarinec et al., 2009; Legant et al., 2013; Knoll et al., 2014).
The regularization procedure used to filter the image data or add additional constraints to the force estimation should be applied carefully because it affects the measurement of the force magnitude and the error in that measurement (Martiel et al., 2015; Soiné et al., 2015). Although it can be tempting to modulate the regularization factor in order to adapt the force-field dispersion to the exact cell shape, it is imperative to keep it constant from one analyzed cell to the next. Alternatively, it is possible to avoid using the regularization factor by applying finite-element modeling to the image of the cytoskeleton to restrict the space of possible solutions (Soiné et al., 2015).
From these considerations, we conclude that although solving some identified issues may further reduce the experimental and computational variability and improve the precision of force measurements, they are already quite accurate compared with the observed dispersion of traction force measurements. The main origin of intercellular force variability more likely stems from the force generation mechanism itself, which we will discuss further. In particular, we focus on the mechanical work performed by myosins, which is used to slide, displace, bend, and disassemble actin filaments rather than effectively pulling on focal adhesions, which we term dissipation, although it is not a true loss of energy by heat production but only a part of the energy consumption that is not turned into traction force generation.
MODULATION OF FORCE GENERATION IN ACTOMYOSIN BUNDLES
Numerous parameters have been shown to modulate force generation. In this section, we discuss how their multiplicity, interdependence, and nonlinear effect on traction force generation contribute to intercellular variations.
The main parameter underlying force generation is the ATPase activity of myosin, which results in a change in its conformation when it is bound to actin filaments (Adelstein and Eisenberg, 1980). Myosin ATPase activity and the stability of myosin filaments are also regulated by phosphorylation of its light chains (Watanabe et al., 2007), and chemical inhibition of these processes can dramatically reduce traction forces (Balaban et al., 2001; Labouesse et al., 2015, 2016). Of interest, some mechanical tension across a cell remains after treatment with such an inhibitor, potentially reflecting filament cross-linking in the actomyosin bundles (Labouesse et al., 2015). The partly overlapping roles of distinct myosin types add some complexity; myosin IIA tends to have a greater role during cell spreading, whereas myosin IIB tends to have a greater role in the motility of fully spread cells (Thomas et al., 2015). The complexity is further increased by the fact that myosins not only produce forces by pulling on actin filaments but also relax forces by disassembling actin filaments (Reymann et al., 2012; Vogel et al., 2013; Figure 2).
FIGURE 2:
Dissipation of mechanical work by stress fiber remodeling limits the traction forces applied on extracellular anchorages. Several sources of dissipation are schematized. The rolling tube represents the weak and fluctuating coupling of stress fiber (toilet paper) with the extracellular anchorages (the wall). It mimics the transient detachment of integrins (frictional slippage), as well as the disengagement of actin bundle from adhesions (clutch). Paper rolling out represents the nucleation of actin filaments by focal adhesions and filament translocation by myosins. Paper stretching represents the fiber elasticity and the energy that is lost in deforming it rather than pulling on the substrate. The character represents myosins at work, losing energy by pulling on a viscoelastic and ever-changing fiber and disassembling it in the meantime. (Drawing by “Benthos von Detritus,” http://benthos4.deviantart.com.)
Numerous actin-binding proteins modulate force generation. Perturbing filament nucleation by inhibiting formins affects the assembly of force-bearing structures and reduces traction forces (Oakes et al., 2012). By contrast, knocking down tropomyosins (Wolfenson et al., 2015) or cross-linkers (actinin or paladin) increases traction forces (Shao et al., 2010; Oakes et al., 2012; Azatov et al., 2016). This effect has been attributed to a specific reorganization of the actomyosin network (Oakes et al., 2012) or increased loading of myosins onto actin filaments, because actinin and myosins seem to compete for actin-binding sites (Peterson et al., 2004; Shao et al., 2010). However, the higher affinity of actinin for the actin filament is associated with higher traction forces of longer duration (Ehrlicher et al., 2015), suggesting that the relationship between actinin-mediated filament cross-linking and myosin-mediated force generation is not uniform but instead can be viewed as a bell-shaped curve (Ennomani et al., 2016). Other nonlinear effects stem from the tension-dependent loading of some actin-binding proteins such as cofilin, which bind less efficiently to actin bundles under tension (Hayakawa et al., 2011; Tojkander et al., 2015), or from the turnover of myosin, which is reduced as tension increases (Kobb et al., 2017). The multiplicity of players and nonlinear effects (Figure 2) is likely to make force generation highly sensitive to relatively small biochemical changes.
Fiber structure also contributes to force generation. Muscle cells have a sarcomeric organization, with repeating alternating units of relatively wide bands of aligned myosin filaments and relatively thin bands of α-actinin molecules positioned orthogonally to the actin filaments they cross-link (Bray et al., 2008). The sarcomeric organization produces and transmits force independently of the length of the myosin bands (Rassier, 1999; Rassier and Pavlov, 2010). Contractile fibers in nonmuscle cells display alternating units of myosin and α-actinin, but the banding is not systematically as regular as in muscle cells. The relationship between myosin band length and contraction is not clear. In some nonmuscle cells, myosin band length decreases as tension increases (Aratyn-Schaus et al., 2011), but in others, the shortening of some bands is compensated by the elongation of others (Peterson et al., 2004; Chapin et al., 2012), making the net effect on traction forces difficult to predict. The effect of sarcomeric versus random filament organization in contractile bundles is not clear. Sarcomeric organization seems to optimize force transmission by segregating regions where myosin and actin filaments overlap, and force is generated from regions where actin filaments are connected to each other and force is transmitted. However, the force generated by a sarcomere is dependent on its size, and the optimum force generated reflects a theoretical point at which a given myosin filament is fully engaged with actin filaments from opposing ends of the sarcomere and these opposing actin filaments do not overlap (Rassier et al., 1999). In nonmuscle cells, disorganized structures may allow higher amount of myosin filaments in series to engage with actin filaments and thus generate larger forces. This high level of work is produced at the expense of efficiency, however, because part of the work is dissipated in noneffective filament translocation (Figure 2). Moreover, the net traction force would also become less predictable.
Dynamic reorganization of the bundle structure of contractile fibers (engaged actin and myosin filaments) also contributes to modulate the magnitude of force generation. Actin filament nucleation at contractile fiber ends (Russell et al., 2011; Skau et al., 2015; Tee et al., 2015) opposes the transmission of tensional forces in the fiber to the ECM (Tojkander et al., 2015). Newly nucleated actin filaments at focal adhesions move inward and are subjected to local variations in internal tension. Stretched sarcomeric-like structures can elongate and generate new sarcomeres (Chapin et al., 2012). Sarcomere length fluctuations have been proposed to buffer the variations in local tension and maintain a constant tension on anchorage points (Russell et al., 2011; Chapin et al., 2012; Figure 2). Overall and to maintain tension homeostasis, the reorganization of contractile bundle structures may follow a form of elastic deformation response to small fluctuation in mechanical load but a form of a plastic deformation in response to large fluctuations in mechanical load (Bonakdar et al., 2016). These plastic deformations are likely to be associated with ruptures in actin-filament cross-links and with the disengagement of myosin and actin filaments. They protect cells against mechanical damage by allowing the contractile fibers to elongate while maintaining the net traction force of the contractile bundle (Bonakdar et al., 2016).
The conservation of mass implies that the inward flow is coupled to an equivalent process of contractile fiber disassembly. Analyses of turnover rates for actin and myosin filaments reveal relatively high turnover rates along the length of the contractile fiber (characteristic lifetime, 1 min) compared with the contractile bundle lifetime (1 h; Hu et al., 2017). Turnover is uneven and faster at the center of the fiber, where sarcomeric-like structures are stretched (Peterson et al., 2004). The association of viscoelastic and plastic reorganizations in irregular, motile, and permanently renewing structures makes force generation and transmission to the ECM particularly difficult to discern and characterize (Figure 2).
Finally, the idea that contractile bundles operate independently in the generation of traction forces is probably an oversimplification. First, various structures—meshworks and fibers—can exert forces with distinct force/dynamics relationships (Aratyn-Schaus et al., 2011). Second, these structures move under tension, bind to, and sometimes partially fuse with adjacent fibers, leading to complex force-producing networks (Tojkander et al., 2015). Moreover, it seems that all fibers are interconnected rather than being independent entities, and the local ablation of a given fiber can trigger relaxation throughout the cell (Hu et al., 2003; Chang and Kumar, 2013; Kassianidou et al., 2017). The interconnections of aligned fibers are highlighted by the orthogonal alignment of sarcomeric units in register among diverging contractile fibers (Fenix et al., 2016; Beach et al., 2017; Hu et al., 2017). Conspicuous bundles even seem connected to the surrounding cortical meshwork, which contributes to force generation in the bundle (Labouesse et al., 2015, 2016). Such a continuous meshwork with embedded contractile fibers may account for force propagation and global force regulation at the level of the cell (Oakes et al., 2014, 2017).
MODULATION OF FORCE TRANSMISSION TO SITES OF CELL ANCHORAGE
Rigid anchors fully transmit the traction forces to which they are submitted, but focal adhesions are subtler structures. Their dynamics modulate the proportion of forces that is conveyed from the intracellular space to the extracellular matrix. Focal adhesions are active sensors and regulators of traction forces (Geiger et al., 2009). They convey bidirectional signaling between extracellular cues and the architecture of the actin cytoskeleton (Parsons et al., 2010).
The attractive concept that focal adhesion size is directly correlated to the force applied to the focal adhesion (Balaban et al., 2001) appears to be applicable to focal adhesion assembly in the early growing phase (first few minutes; Stricker et al., 2011) but not to mature focal adhesions. However, focal adhesion composition is a key regulator of traction force transmission and force generation. A strong reduction of traction forces can occur on removal of any of the key elements of the focal adhesion, such as paxillin (Plotnikov et al., 2012; Lee et al., 2015), vinculin (Plotnikov et al., 2012), talin (Austen et al., 2015), and kindlin (Bharadwaj et al., 2017), or by preventing the recruitment and clustering of integrins (Liu et al., 2014). The exact integrin composition of the focal adhesions also finely modulates specific downstream signaling pathways and the magnitude of the traction force (Schiller et al., 2013; Bharadwaj et al., 2017; Milloud et al., 2017).
Focal adhesion maturation is controlled by positive feedback loops related to the traction force applied and involves the promotion of focal adhesion growth and the recruitment of new molecules (Kuo et al., 2011; Schiller et al., 2013; Ye et al., 2014b). Integrin β1 engagement activates myosin II, and integrin αV activates contractile bundle enlargement. Together the two integrin types synergize so that the traction force adapts to the substrate stiffness (Schiller et al., 2013). The positive feedback loop also relies on the removal of inactivators of force generation such as betaPix and the Rac pathway (Kuo et al., 2011). Superimposed negative feedback loops also exist. One counteractive tendency is that actomyosin contractility also stimulates proteolysis and endocytosis of integrins (Kuo et al., 2011). In addition, tension on actin filaments stimulates their nucleation by formins associated to focal adhesions (Courtemanche et al., 2013; Jégou et al., 2013), which reduces the transmission of tension from the actin filaments to the ECM. Interference with this nucleation strongly increases traction forces (Elkhatib et al., 2014). The interplay between positive and negative feedback loops generates oscillating forces that are instrumental in substrate stiffness sensing (Plotnikov et al., 2012; Wu et al., 2017). However, these variations and instabilities are also likely to contribute to intercellular variability.
The dynamic regulation of focal adhesions is intimately linked to the key role of focal adhesion molecule turnover. Traction forces are controlled by the rates of binding/unbinding of integrins to the ECM (Elosegui-Artola et al., 2014). One consequence is that the contacts focal adhesions have with the ECM can transiently rupture and impair the transmission of traction force, leading to a process called frictional slippage occurring at low forces and low ECM stiffness (Aratyn-Schaus and Gardel, 2010; Figure 2). This rupture process can also occur between the actin network and the focal adhesion in a process similar to “clutch” engagement or disengagement (Figure 2). It would not affect the mechanical force generated by actomyosin contraction but would affect whether it is transmitted to the ECM (and converted into measurable traction forces) or dissipated and lost (Parsons et al., 2010; Swaminathan and Waterman, 2016). The correlation between traction force magnitude at focal adhesions and the internal actin retrograde flow is a signature of the clutch engagement between the two structures (Gardel et al., 2008). Similarly, traction force magnitude has been associated with the inward translocation of bundled actin filaments at contractile fiber ends (Russell et al., 2011; Elkhatib et al., 2014). Because of the weakness of the connections between actin network and focal adhesions and between focal adhesions and the ECM, strong internal actin retrograde flow or high ECM substrate stiffness place too high a load on the clutch, causing disengagement of the connections and interruption of the transmission of force. Hence there is an optimum for traction force generation, but it is highly sensitive to the rate of actin retrograde flow, the number of components involved in the clutch, and the stiffness of the ECM substrate (Bangasser et al., 2013). This sensitivity can generate high intercellular variability. Furthermore, the clutch effect may be overwhelmed by additional mechanotransduction effects; for example, in a high-force regime, clutch disengagement may be overridden by the unfolding of talin, which in turn mediates the recruitment of additional linkers that reinforce the adhesion and allows the transmission of traction forces to increase (Elosegui-Artola et al., 2016). Furthermore, clutch disengagement can also be promoted by the action of Kank, which can detach talin from actin and hence reduce force transmission (Sun et al., 2016).
CONCLUSION AND PERSPECTIVES
Although not absolute, the relative traction forces that cells transmit to the ECM can be precisely measured experimentally. However, all studies so far have been limited by the large intercellular variability of the magnitude of these force measurements, and this variability has considerably hindered identifying the mechanisms regulating force generation (Figure 1). Several sources of experimental variability can be envisaged, and most seem to result from the intercellular variability in the force-producing mechanism. The expression levels of the numerous proteins involved in force generation contribute to intercellular variability, but other factors specific to cell mechanics are also likely to be involved. Of note, it appears that a significant part of the mechanical work produced by the actomyosin contraction is dissipated (Balland et al., 2005; Mitrossilis et al., 2009). This dissipated work is not transmitted to the ECM and therefore cannot be estimated by measuring substrate deformation. Numerous examples described here showed that filament translocation, sliding, disassembly, fraying, reorganization, and turnover are likely to represent the major sources of mechanical work dissipation (Figure 2). All potential outlets of dissipation should be taken into account in future models of cell mechanics (Hoffman and Crocker, 2009). The intercellular variability in the magnitude of the traction force is likely to result from modulation of these dissipation processes, which seem to be the main missing piece in our understanding of the regulation of the traction forces that cells apply to their environment.
New biophysical methods are required to measure the entire mechanical work produced by the cell rather than simply the part that is transmitted to sites of extracellular anchorage. These methods would shine some light on the mechanisms by which mechanical work is dissipated and on cell mechanical efficiency. The fluctuation-dissipation theorem has been used to study the fluctuation spectrum of beads in or at the surface of cells to estimate the entire mechanical energy and compare it to traction energy (Mizuno et al., 2009; Robert et al., 2010; Bohec et al., 2013; Schlosser et al., 2015). Another possibility could be to use force gauges all along actomyosin bundles. Use of α-actinin fluorescence resonance energy transfer (FRET) sensors is a technique of choice to measure stress throughout the entire cell (Meng and Sachs, 2011; Rahimzadeh et al., 2011; Gayrard and Borghi, 2016). The loss/gain of FRET signal anywhere along the actin bundle is a direct readout of local tension increase/release. Subcellular changes in force generation can be detected in various contexts, such as cell spreading (Ye et al., 2014b; Suffoletto et al., 2015) and differentiation (Guo et al., 2014). These FRET measurements require careful calibration in order to be converted into exact force measurements (Meng and Sachs, 2011; Gayrard and Borghi, 2016). How the evaluation of all of these intracellular forces can be used to estimate the global contractile energy of the cell and compare it to the traction energy remains a challenge, but it seems a promising avenue for future research.
Future progress in our understanding of the mechanisms regulating force generation should focus on examining individual parameters while keeping all of the others parameters constant. Two methods seem well suited for this. The first is to work with individual cells rather than cell populations and vary one single parameter while measuring the traction forces. New optogenetic approaches provide a way to modulate in real time a specific parameter (connectivity, filament disassembly, crowding, stiffness, etc.) in the same cellular background. Recently, optogenetic tools have been established to study the main pathways regulating cell contractility (Rao et al., 2013; Guglielmi et al., 2015; Oakes et al., 2017; Valon et al., 2017), and these should be of great help in this direction. The second involves in vitro approaches aimed at reconstituting the force generation mechanism in precisely controlled biochemical conditions (as opposed to cells in which they are not known and possibly variable). Coassembly of actin filaments and myosin revealed variations of contraction rates with bundle length reminiscent of sarcomeric organization (Thoresen et al., 2011). Controlled hydrodynamic forces on growing actin filaments have been shown to affect filament growth rate (Jégou et al., 2013). Finally, the geometric manipulation of actomyosin networks showed that several key features of cellular actomyosin networks can be recapitulated in vitro: the specific action of myosin on defined architectures and its capacity to regulate the disassembly of the architectures (Reymann et al., 2012), the nonlinear effect of cross-linkers (Ennomani et al., 2016), the amplification of network deformation with the size of the contracting region within the contractile fiber (Linsmeier et al., 2016), and the feedback loops between tension and protein organization at sites of contractile fiber anchorage (Ciobanasu et al., 2014). These experiments point toward the feasibility of investigating actomyosin network dynamics and identifying mechanisms responsible for force dissipation in living cells.
Acknowledgments
We have greatly benefited from longstanding discussions and collaborations with Alex Mogilner (New York University). We thank Jacky Goetz and Antoine Jegou for discussions pointing at the use of the actinin-FRET sensor for global cellular force measurements. We also thank Daisuke Inoue for the drawing in Figure 2. Work in the CytoMorpho Lab dedicated to actin network contraction is supported by the Agence Nationale pour la Recherche (ANR Max Force, ANR-14-CE11-0003-01).
Abbreviations used:
- ECM
extracellular matrix
- TFM
traction force microscopy
Footnotes
REFERENCES
- Adelstein RS, Eisenberg E. Regulation and kinetics of the actin-myosin-ATP interaction. Annu Rev Biochem. 1980;49:921–956. doi: 10.1146/annurev.bi.49.070180.004421. [DOI] [PubMed] [Google Scholar]
- Aratyn-Schaus Y, Gardel ML. Transient frictional slip between integrin and the ECM in focal adhesions under myosin II tension. Curr Biol. 2010;20:1145–1153. doi: 10.1016/j.cub.2010.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aratyn-Schaus Y, Oakes P W, Gardel ML. Dynamic and structural signatures of lamellar actomyosin force generation. Mol Biol Cell. 2011;22:1330–1339. doi: 10.1091/mbc.E10-11-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austen K, Ringer P, Mehlich A, Chrostek-Grashoff A, Kluger C, Klingner C, Sabass B, Zent R, Rief M, Grashoff C. Extracellular rigidity sensing by talin isoform-specific mechanical linkages. Nat Cell Biol. 2015;17:1597–1609. doi: 10.1038/ncb3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azatov M, Goicoechea SM, Otey CA, Upadhyaya A. The actin crosslinking protein palladin modulates force generation and mechanosensitivity of tumor associated fibroblasts. Sci Rep. 2016;6:28805. doi: 10.1038/srep28805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban NQ, Schwarz US, Riveline D, Goichberg P, Tzur G, Sabanay I, Mahalu D, Safran S, Bershadsky A, Addadi L, Geiger B. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat Cell Biol. 2001;3:466–472. doi: 10.1038/35074532. [DOI] [PubMed] [Google Scholar]
- Balland M, Richert A, Gallet F. The dissipative contribution of myosin II in the cytoskeleton dynamics of myoblasts. Eur Biophys J. 2005;34:255–261. doi: 10.1007/s00249-004-0447-7. [DOI] [PubMed] [Google Scholar]
- Bangasser BL, Rosenfeld SS, Odde DJ. Determinants of maximal force transmission in a motor-clutch model of cell traction in a compliant microenvironment. Biophys J. 2013;105:581–592. doi: 10.1016/j.bpj.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach JR, Bruun KS, Shao L, Li D, Swider Z, Remmert K, Zhang Y, Conti MA, Adelstein RS, Rusan NR, et al. Actin dynamics and competition for myosin monomer govern the sequential amplification of myosin filaments. Nat Cell Biol. 2017;19:85–93. doi: 10.1038/ncb3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharadwaj M, Strohmeyer N, Colo GP, Helenius J, Beerenwinkel N, Schiller HB, Fässler R, Müller DJ. αV-class integrins exert dual roles on α5β1 integrins to strengthen adhesion to fibronectin. Nat Commun. 2017;8:14348. doi: 10.1038/ncomms14348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohec P, Gallet F, Maes C, Safaverdi S, Visco P, van Wijland F. Probing active forces via a fluctuation-dissipation relation: application to living cells. Europhys Lett. 2013;102:50005. [Google Scholar]
- Bonakdar N, Gerum R, Kuhn M, Spörrer M, Lippert A, Schneider W, Aifantis KE, Fabry B. Mechanical plasticity of cells. Nat Mater. 2016;15:1090–1094. doi: 10.1038/nmat4689. [DOI] [PubMed] [Google Scholar]
- Bosveld F, Markova O, Guirao B, Martin C, Wang Z, Pierre A, Balakireva M, Gaugue I, Ainslie A, Christophorou N, et al. Epithelial tricellular junctions act as interphase cell shape sensors to orient mitosis. Nature. 2016;530:495–498. doi: 10.1038/nature16970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudou T, Ohayon J, Picart C, Pettigrew RI, Tracqui P. Nonlinear elastic properties of polyacrylamide gels: Implications for quantification of cellular forces. Biorheology. 2009;46:191–205. doi: 10.3233/BIR-2009-0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray M, Sheehy SP, Parker KK. Sarcomere alignment is regulated by myocyte shape. Cell Motil Cytoskeleton. 2008;651:641–651. doi: 10.1002/cm.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JP, Tolic‘-Nørrelykke IM, Fabry B, Fredberg JJ. Traction fields, moments, and strain energy that cells exert on their surroundings. Am J Physiol Cell Physiol. 2002;282:C595–C605. doi: 10.1152/ajpcell.00270.2001. [DOI] [PubMed] [Google Scholar]
- Chang C, Kumar S. Vinculin tension distributions of individual stress fibers within cell-matrix adhesions. J Cell Sci. 2013;126:3021–3030. doi: 10.1242/jcs.119032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SS, Guo W, Kim Y, Wang Y. Guidance of cell migration by substrate dimension. Biophys J. 2013;104:313–321. doi: 10.1016/j.bpj.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapin LM, Blankman E, Smith MA, Shiu Y, Beckerle MC. Lateral communication between stress fiber sarcomeres facilitates a local remodeling response. Biophys J. 2012;103:2082–2092. doi: 10.1016/j.bpj.2012.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciobanasu C, Faivre B, Le Clainche C. Actomyosin-dependent formation of the mechanosensitive talin-vinculin complex reinforces actin anchoring. Nat Commun. 2014;5:3095. doi: 10.1038/ncomms4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtemanche N, Lee JY, Pollard TD, Greene EC. Tension modulates actin filament polymerization mediated by formin and profilin. Proc Natl Acad Sci USA. 2013;110:9752–9757. doi: 10.1073/pnas.1308257110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Álamo JC, Meili R, Álvarez-González B, Alonso-Latorre B, Bastounis E, Firtel R, Lasheras JC. Three-dimensional quantification of cellular traction forces and mechanosensing of thin substrata by fourier traction force microscopy. PLoS One. 2013;8:e69850. doi: 10.1371/journal.pone.0069850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembo M, Wang Y. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys J. 1999;76:2307–2316. doi: 10.1016/S0006-3495(99)77386-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisin AK, Pruitt BL. Tuning the range of polyacrylamide gel stiffness for mechanobiology applications. ACS Appl Mater Interfaces. 2016;8:21893–21902. doi: 10.1021/acsami.5b09344. [DOI] [PubMed] [Google Scholar]
- du Roure O, Saez A, Buguin A, Austin RH, Chavrier P, Siberzan P, Ladoux B. Force mapping in epithelial cell migration. Proc Natl Acad Sci USA. 2005;102:2390–2395. doi: 10.1073/pnas.0408482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton S, Jülicher F. Cell flow and tissue polarity patterns. Curr Opin Genet Dev. 2011;21:747–752. doi: 10.1016/j.gde.2011.08.010. [DOI] [PubMed] [Google Scholar]
- Ehrlicher AJ, Krishnan R, Guo M, Bidan CM, Weitz DA, Pollak MR. Alpha-actinin binding kinetics modulate cellular dynamics and force generation. Proc Natl Acad Sci USA. 2015;112:6619–6624. doi: 10.1073/pnas.1505652112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhoffer GT, Loftus PD, Yoshigi M, Otsuna H, Chien C-B, Morcos PA, Rosenblatt J. Crowding induces live cell extrusion to maintain homeostatic cell numbers in epithelia. Nature. 2012;484:546–549. doi: 10.1038/nature10999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkhatib N, Neu MB, Zensen C, Schmoller KM, Louvard D, Bausch AR, Betz T, Vignjevic DM. Fascin plays a role in stress fiber organization and focal adhesion disassembly. Curr Biol. 2014;24:1492–1499. doi: 10.1016/j.cub.2014.05.023. [DOI] [PubMed] [Google Scholar]
- Elosegui-Artola A, Bazellières E, Allen MD, Andreu I, Oria R, Sunyer R, Gomm JJ, Marshall JF, Jones JL, Trepat X, Roca-Cusachs P. Rigidity sensing and adaptation through regulation of integrin types. Nat Mater. 2014;13:631–637. doi: 10.1038/nmat3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elosegui-Artola A, Oria R, Chen Y, Kosmalska A, Pérez-González C, Castro N, Zhu C, Trepat X, Roca-Cusachs P. Mechanical regulation of a molecular clutch defines force transmission and transduction in response to matrix rigidity. Nat Cell Biol. 2016;18:540–548. doi: 10.1038/ncb3336. [DOI] [PubMed] [Google Scholar]
- Ennomani H, Letort G, Guérin C, Martiel J, Cao W, Nédélec F, De La Cruz EM, Théry M, Blanchoin L. Architecture and connectivity govern actin network contractility. Curr Biol. 2016;26:616–626. doi: 10.1016/j.cub.2015.12.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenix AM, Taneja N, Buttler CA, Lewis J, Van Engelenburg SB, Ohi R, Burnette DT. Expansion and concatenation of nonmuscle myosin IIA filaments drive cellular contractile system formation during interphase and mitosis. Mol Biol Cell. 2016;27:1465–1478. doi: 10.1091/mbc.E15-10-0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardel ML, Sabass B, Ji L, Danuser G, Schwarz US, Waterman CM. Traction stress in focal adhesions correlates biphasically with actin retrograde flow speed. J Cell Biol. 2008;183:999–1005. doi: 10.1083/jcb.200810060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayrard C, Borghi N. FRET-based molecular tension microscopy. Methods. 2016;94:33–42. doi: 10.1016/j.ymeth.2015.07.010. [DOI] [PubMed] [Google Scholar]
- Geiger B, Spatz JP, Bershadsky AD. Environmental sensing through focal adhesions. Nat Rev Mol Cell Biol. 2009;10:21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- Ghibaudo M, Di Meglio J-M, Hersen P, Ladoux B. Mechanics of cell spreading within 3 D-micropatterned environments. Lab Chip. 2011;11:805–812. doi: 10.1039/c0lc00221f. [DOI] [PubMed] [Google Scholar]
- Gross W, Kress H. Simultaneous measurement of the Young’s modulus and the Poisson ratio of thin elastic layers. Soft Matter. 2017;13:1048–1055. doi: 10.1039/c6sm02470j. [DOI] [PubMed] [Google Scholar]
- Guglielmi G, Barry JD, Huber W, De Renzis S. An optogenetic method to modulate cell contractility during tissue morphogenesis. Dev Cell. 2015;35:646–660. doi: 10.1016/j.devcel.2015.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Wang Y, Sachs F, Meng F. Actin stress in cell reprogramming. Proc Natl Acad Sci USA. 2014;111:E5252–E2561. doi: 10.1073/pnas.1411683111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AK, Stopak D, Wild P. Fibroblast traction as a mechanism for collagen morphogenesis. Nature. 1981;290:249–251. doi: 10.1038/290249a0. [DOI] [PubMed] [Google Scholar]
- Hayakawa K, Tatsumi H, Sokabe M. Actin filaments function as a tension sensor by tension-dependent binding of cofilin to the filament. J Cell Biol. 2011;195:721–727. doi: 10.1083/jcb.201102039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisenberg C-P, Bellaïche Y. Forces in tissue morphogenesis and patterning. Cell. 2013;153:948–962. doi: 10.1016/j.cell.2013.05.008. [DOI] [PubMed] [Google Scholar]
- Hoffman BD, Crocker JC. Cell mechanics: dissecting the physical responses of cells to force. Annu Rev Biomed Eng. 2009;11:259–288. doi: 10.1146/annurev.bioeng.10.061807.160511. [DOI] [PubMed] [Google Scholar]
- Holenstein CN, Silvan U, Snedeker JG. High-resolution traction force microscopy on small focal adhesions - improved accuracy through optimal marker distribution and optical flow tracking. Sci Rep. 2017;7:41633. doi: 10.1038/srep41633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Chen J, Fabry B, Numaguchi Y, Gouldstone A, Ingber DE, Fredberg JJ, Butler JP, Wang N. Intracellular stress tomography reveals stress focusing and structural anisotropy in cytoskeleton of living cells. Am J Physiol Cell Physiol. 2003;285:C1082–C1090. doi: 10.1152/ajpcell.00159.2003. [DOI] [PubMed] [Google Scholar]
- Hu S, Dasbiswas K, Guo Z, Tee Y, Thiagarajan V, Hersen P, Chew T, Safran SA, Zaidel-Bar R, Bershadsky AD. Long-range self-organization of cytoskeletal myosin II filament stacks. Nat Cell Biol. 2017;19:133–141. doi: 10.1038/ncb3466. [DOI] [PubMed] [Google Scholar]
- Jégou A, Carlier M-F, Romet-Lemonne G. Formin mDia1 senses and generates mechanical forces on actin filaments. Nat Commun. 2013;4:1883. doi: 10.1038/ncomms2888. [DOI] [PubMed] [Google Scholar]
- Kara S, Pekcan Ö. Drying of heterogeneous hydrogels formed with various water contents: a photon transmission study. J Appl Polym Sci. 2001;82:1944–1951. [Google Scholar]
- Kassianidou E, Brand CA, Schwarz US, Kumar S. Geometry and network connectivity govern the mechanics of stress fibers. Proc Natl Acad Sci USA. 2017;114:2622–2627. doi: 10.1073/pnas.1606649114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll SG, Ali MY, Saif MTA. A novel method for localizing reporter fluorescent beads near the cell culture surface for traction force microscopy. J Vis Exp. 2014;2014(91):51873. doi: 10.3791/51873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobb AB, Zulueta-Coarasa T, Fernandez-Gonzalez R. Tension regulates myosin dynamics during Drosophila embryonic wound repair. J Cell Sci. 2017;130:689–696. doi: 10.1242/jcs.196139. [DOI] [PubMed] [Google Scholar]
- Kuo J-C, Han X, Hsiao C-T, Yates JR, Waterman CM. Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for β-Pix in negative regulation of focal adhesion maturation. Nat Cell Biol. 2011;13:383–393. doi: 10.1038/ncb2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labouesse C, Gabella C, Meister J-J, Vianay B, Verkhovsky AB. Microsurgery-aided in-situ force probing reveals extensibility and viscoelastic properties of individual stress fibers. Sci Rep. 2016;6:23722. doi: 10.1038/srep23722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labouesse C, Verkhovsky AB, Meister J-J, Gabella C, Vianay B. Cell shape dynamics reveal balance of elasticity and contractility in peripheral arcs. Biophys J. 2015;108:2437–2447. doi: 10.1016/j.bpj.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Abdeen AA, Tang X, Saif TA, Kilian KA. Geometric guidance of integrin mediated traction stress during stem cell differentiation. Biomaterials. 2015;69:174–183. doi: 10.1016/j.biomaterials.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legant WR, Choi CK, Miller JS, Shao L, Gao L, Betzig E, Chen CS. Multidimensional traction force microscopy reveals out-of-plane rotational moments about focal adhesions. Proc Natl Acad Sci USA. 2013;110:881–886. doi: 10.1073/pnas.1207997110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levayer R, Lecuit T. Biomechanical regulation of contractility: spatial control and dynamics. Trends Cell Biol. 2012;22:61–81. doi: 10.1016/j.tcb.2011.10.001. [DOI] [PubMed] [Google Scholar]
- Linsmeier I, Banerjee S, Oakes PW, Jung W, Kim T, Murrell MP. Disordered actomyosin networks are sufficient to produce cooperative and telescopic contractility. Nat Commun. 2016;7:12615. doi: 10.1038/ncomms12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Medda R, Liu Z, Galior K, Yehl K, Spatz JP, Cavalcanti-Adam EA, Salaita K. Nanoparticle tension probes patterned at the nanoscale: Impact of integrin clustering on force transmission. Nano Lett. 2014;14:5539–5546. doi: 10.1021/nl501912g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal K, Wang I, Vitiello E, Orellana LAC, Balland M. Cell dipole behaviour revealed by ECM sub-cellular geometry. Nat Commun. 2014;5:5749. doi: 10.1038/ncomms6749. [DOI] [PubMed] [Google Scholar]
- Martiel J-L, Leal A, Kurzawa L, Balland M, Wang I, Vignaud T, Tseng Q, Théry M. Measurement of cell traction forces with Image. J Methods Cell Biol. 2015;125:269–287. doi: 10.1016/bs.mcb.2014.10.008. [DOI] [PubMed] [Google Scholar]
- Maskarinec SA, Franck C, Tirrell DA, Ravichandran G. Quantifying cellular traction forces in three dimensions. Proc Natl Acad Sci USA. 2009;106:22108–22113. doi: 10.1073/pnas.0904565106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meili R, Alonso-Latorre B, del Alamo JC, Firtel RA, Lasheras JC. Myosin II is essential for the spatiotemporal organization of traction forces during cell motility. Mol Biol Cell. 2010;21:405–417. doi: 10.1091/mbc.E09-08-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F, Sachs F. Visualizing dynamic cytoplasmic forces with a compliance-matched FRET sensor. J Cell Sci. 2011;124:261–269. doi: 10.1242/jcs.071928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkel R, Kirchgessner N, Cesa CM, Hoffmann B. Cell force microscopy on elastic layers of finite thickness. Biophys J. 2007;93:3314–3323. doi: 10.1529/biophysj.107.111328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milloud R, Destaing O, de Mets R, Bourrin-Reynard I, Oddou C, Delon A, Wang I, Albigès-Rizo C, Balland M. αvβ3 Integrins negatively regulate cellular forces by phosphorylation of its distal NPXY site. Biol Cell. 2017;109:127–137. doi: 10.1111/boc.201600041. [DOI] [PubMed] [Google Scholar]
- Mitrossilis D, Fouchard J, Guiroy A, Desprat N, Rodriguez N, Fabry B, Asnacios A. Single-cell response to stiffness exhibits muscle-like behavior. Proc Natl Acad Sci USA. 2009;106:18243–18248. doi: 10.1073/pnas.0903994106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno D, Bacabac R, Tardin C, Head D, Schmidt CF. High-resolution probing of cellular force transmission. Phys Rev Lett. 2009;102:1–4. doi: 10.1103/PhysRevLett.102.168102. [DOI] [PubMed] [Google Scholar]
- Nguyen KT, West JL. Photopolymerizable hydrogels for tissue engineering applications. Biomaterials. 2002;23:4307–4314. doi: 10.1016/s0142-9612(02)00175-8. [DOI] [PubMed] [Google Scholar]
- Oakes PW, Banerjee S, Marchetti MC, Gardel ML. Geometry regulates traction stresses in adherent cells. Biophys J. 2014;107:825–833. doi: 10.1016/j.bpj.2014.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes PW, Beckham Y, Stricker J, Gardel M L. Tension is required but not sufficient for focal adhesion maturation without a stress fiber template. J Cell Biol. 2012;196:363–374. doi: 10.1083/jcb.201107042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes PW, Wagner E, Brand CA, Probst D, Linke M, Schwarz US, Glotzer M, Gardel ML. Harnessing optogenetics to probe sub-cellular mechanics. bioRxiv doi: 10.1101/109595. 2017 [Google Scholar]
- Parsons JT, Horwitz AR, Schwartz MA. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat Rev Mol Cell Biol. 2010;11:633–643. doi: 10.1038/nrm2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham RJ, Wang Y-L. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci USA. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham RJ, Wang Y-L. High resolution detection of mechanical forces exerted by locomoting fibroblasts on the substrate. Mol Biol Cell. 1999;10:935–945. doi: 10.1091/mbc.10.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson LJ, Rajfur Z, Maddox AS, Freel CD, Chen Y, Edlund M, Otey C, Burridge K. Simultaneous stretching and contraction of stress fibers in vivo. Mol Biol Cell. 2004;15:3497–3508. doi: 10.1091/mbc.E03-09-0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitaval A, Tseng Q, Bornens M, Théry M. Cell shape and contractility regulate ciliogenesis in cell cycle-arrested cells. J Cell Biol. 2010;191:303–312. doi: 10.1083/jcb.201004003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikov SV, Pasapera AM, Sabass B, Waterman CM. Force fluctuations within focal adhesions mediate ecm-rigidity sensing to guide directed cell migration. Cell. 2012;151:1513–1527. doi: 10.1016/j.cell.2012.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompe T, Kaufmann M, Kasimir M, Johne S, Glorius S, Renner L, Bobeth M, Pompe W, Werner C. Friction-controlled traction force in cell adhesion. Biophys J. 2011;101:1863–1870. doi: 10.1016/j.bpj.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimzadeh J, Meng F, Sachs F, Wang J, Verma D, Hua SZ. Real-time observation of flow-induced cytoskeletal stress in living cells. Am J Physiol Cell Physiol. 2011;301:C646–C652. doi: 10.1152/ajpcell.00099.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao MV, Chu P-H, Hahn KM, Zaidel-Bar R. An optogenetic tool for the activation of endogenous diaphanous-related formins induces thickening of stress fibers without an increase in contractility. Cytoskeleton. 2013;70:394–407. doi: 10.1002/cm.21115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rape A, Guo W-H, Wang Y-L. Microtubule depolymerization induces traction force increase through two distinct pathways. J Cell Sci. 2011a;124:4233–4240. doi: 10.1242/jcs.090563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rape AD, Guo W-H, Wang Y-L. The regulation of traction force in relation to cell shape and focal adhesions. Biomaterials. 2011b;32:2043–2051. doi: 10.1016/j.biomaterials.2010.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassier DE, MacIntosh BR, Herzog W. Length dependence of active force production in skeletal muscle. J Appl Physiol. 1999;86:1445–1457. doi: 10.1152/jappl.1999.86.5.1445. [DOI] [PubMed] [Google Scholar]
- Rassier DE, Pavlov I. Contractile characteristics of sarcomeres arranged in series or mechanically isolated from myofibrils. Adv Exp Med Biol. 2010;682:123–140. doi: 10.1007/978-1-4419-6366-6_7. [DOI] [PubMed] [Google Scholar]
- Reinhart-King CA, Dembo M, Hammer DA. The dynamics and mechanics of endothelial cell spreading. Biophys J. 2005;89:676–689. doi: 10.1529/biophysj.104.054320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymann A-C, Boujemaa-Paterski R, Martiel J-L, Guérin C, Cao W, Chin HF, De La Cruz EM, Théry M, Blanchoin L. Actin network architecture can determine myosin motor activity. Science. 2012;336:1310–1314. doi: 10.1126/science.1221708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert D, Nguyen T-H, Gallet F, Wilhelm C. In vivo determination of fluctuating forces during endosome trafficking using a combination of active and passive microrheology. PLoS One. 2010;5:e10046. doi: 10.1371/journal.pone.0010046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz SA, Chen CS. Emergence of patterned stem cell differentiation within multicellular structures. Stem Cells. 2008;26:2921–2927. doi: 10.1634/stemcells.2008-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RJ, Grubbs AY, Mangroo SP, Nakasone SE, Dickinson RB, Lele TP. Sarcomere length fluctuations and flow in capillary endothelial cells. Cytoskeleton (Hoboken) 2011;68:150–156. doi: 10.1002/cm.20501. [DOI] [PubMed] [Google Scholar]
- Sabass B, Gardel ML, Waterman CM, Schwarz US. High resolution traction force microscopy based on experimental and computational advances. Biophys J. 2008;94:207–220. doi: 10.1529/biophysj.107.113670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller HB, Hermann MR, Polleux J, Vignaud T, Zanivan S, Friedel CC, Sun Z, Raducanu A, Gottschalk KE, Théry M, et al. β1- and αv-class integrins cooperate to regulate myosin II during rigidity sensing of fibronectin-based microenvironments. Nat Cell Biol. 2013;15:625–636. doi: 10.1038/ncb2747. [DOI] [PubMed] [Google Scholar]
- Schlosser F, Rehfeldt F, Schmidt CF. Force fluctuations in three-dimensional suspended fibroblasts. Philos Trans R Soc Lond B Biol Sci. 2015;370:20140028. doi: 10.1098/rstb.2014.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz US, Gardel ML. United we stand—integrating the actin cytoskeleton and cell-matrix adhesions in cellular mechanotransduction. J Cell Sci. 2012;125:3051–3060. doi: 10.1242/jcs.093716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, Wang JH-C, Pollak MR, Wells A. α-Actinin-4 is essential for maintaining the spreading, motility and contractility of fibroblasts. PLoS One. 2010;5:e13921. doi: 10.1371/journal.pone.0013921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skau CT, Plotnikov SV, Doyle AD, Waterman CM. Inverted formin 2 in focal adhesions promotes dorsal stress fiber and fibrillar adhesion formation to drive extracellular matrix assembly. Proc Natl Acad Sci USA. 2015;112:E2447–E2456. doi: 10.1073/pnas.1505035112. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Soiné JRD, Brand CA, Stricker J, Oakes PW, Gardel ML, Schwarz US. Model-based traction force microscopy reveals differential tension in cellular actin bundles. PLoS Comput Biol. 2015;11:e1004076. doi: 10.1371/journal.pcbi.1004076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm C, Pastore JJ, MacKintosh FC, Lubensky TC, Janmey PA. Nonlinear elasticity in biological gels. Nature. 2005;435:191–194. doi: 10.1038/nature03521. [DOI] [PubMed] [Google Scholar]
- Stricker J, Aratyn-Schaus Y, Oakes PW, Gardel ML. Spatiotemporal constraints on the force-dependent growth of focal adhesions. Biophys J. 2011;100:2883–2893. doi: 10.1016/j.bpj.2011.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suffoletto K, Ye N, Meng F, Verma D, Hua SZ. Intracellular forces during guided cell growth on micropatterns using FRET measurement. J Biomech. 2015;48:627–635. doi: 10.1016/j.jbiomech.2014.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Tseng HY, Tan S, Senger F, Kurzawa L, Dedden D, Mizuno N, Wasik AA, Thery M, Dunn AR, et al. Kank2 activates talin, reduces force transduction across integrins and induces central adhesion formation. Nat Cell Biol. 2016;18:941–953. doi: 10.1038/ncb3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan V, Waterman CM. The molecular clutch model for mechanotransduction evolves. Nat Cell Biol. 2016;18:459–461. doi: 10.1038/ncb3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tambe DT, Hardin CC, Angelini TE, Rajendran K, Park CY, Serra-Picamal X, Zhou EH, Zaman MH, Butler JP, Weitz DA, et al. Collective cell guidance by cooperative intercellular forces. Nat Mater. 2011;10:469–475. doi: 10.1038/nmat3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS. Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proc Natl Acad Sci USA. 2003;100:1484–1489. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee YH, Shemesh T, Thiagarajan V, Hariadi RF, Anderson KL, Page C, Volkmann N, Hanein D, Sivaramakrishnan S, Kozlov MM, Bershadsky AD. Cellular chirality arising from the self-organization of the actin cytoskeleton. Nat Cell Biol. 2015;17:445–457. doi: 10.1038/ncb3137. [DOI] [PubMed] [Google Scholar]
- Thomas DG, Yenepalli A, Denais CM, Rape A, Beach JR, Wang Y, Schiemann WP, Baskaran H, Lammerding J, Egelhoff TT. Non-muscle myosin IIB is critical for nuclear translocation during 3D invasion. J Cell Biol. 2015;210:583–594. doi: 10.1083/jcb.201502039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoresen T, Lenz M, Gardel ML. Reconstitution of contractile actomyosin bundles. Biophys J. 2011;100:2698–2705. doi: 10.1016/j.bpj.2011.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tojkander S, Gateva G, Husain A, Krishnan R, Lappalainen P. Generation of contractile actomyosin bundles depends on mechanosensitive actin filament assembly and disassembly. Elife. 2015;4:e06126. doi: 10.7554/eLife.06126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolic’-Nørrelykke IM, Wang N. Traction in smooth muscle cells varies with cell spreading. J Biomech. 2005;38:1405–1412. doi: 10.1016/j.jbiomech.2004.06.027. [DOI] [PubMed] [Google Scholar]
- Tse JR, Engler AJ. Preparation of hydrogel substrates with tunable mechanical properties. Curr Protoc Cell Biol. 2010 doi: 10.1002/0471143030.cb1016s47. Chapter 10, Unit 10.16. [DOI] [PubMed] [Google Scholar]
- Tseng Q, Duchemin-Pelletier E, Deshiere A, Balland M, Guillou H, Filhol O, Théry M. Spatial organization of the extracellular matrix regulates cell-cell junction positioning. Proc Natl Acad Sci USA. 2012;109:1506–1511. doi: 10.1073/pnas.1106377109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng Q, Wang I, Duchemin-Pelletier E, Azioune A, Carpi N, Gao J, Filhol O, Piel M, Théry M, Balland M. A new micropatterning method of soft substrates reveals that different tumorigenic signals can promote or reduce cell contraction levels. Lab Chip. 2011;11:2231–2240. doi: 10.1039/c0lc00641f. [DOI] [PubMed] [Google Scholar]
- Valon L, Marín-Llauradó A, Wyatt T, Charras G, Trepat X. Optogenetic control of cellular forces and mechanotransduction. Nat Commun. 2017;8:14396. doi: 10.1038/ncomms14396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignaud T, Ennomani H, Théry M. Polyacrylamide hydrogel micropatterning. Methods Cell Biol. 2014;120:93–116. doi: 10.1016/B978-0-12-417136-7.00006-9. [DOI] [PubMed] [Google Scholar]
- Vogel SK, Petrasek Z, Heinemann F, Schwille P. Myosin motors fragment and compact membrane-bound actin filaments. Elife. 2013;2:1–18. doi: 10.7554/eLife.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Hosoya H, Yonemura S. Regulation of myosin II dynamics by phosphorylation and dephosphorylation of its light chain in epithelial cells. Mol Biol Cell. 2007;18:605–616. doi: 10.1091/mbc.E06-07-0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfenson H, Meacci G, Liu S, Stachowiak MR, Iskratsch T, Ghassemi S, Roca-Cusachs P, O’Shaughnessy B, Hone J, Sheetz MP. Tropomyosin controls sarcomere-like contractions for rigidity sensing and suppressing growth on soft matrices. Nat Cell Biol. 2015;18:33–42. doi: 10.1038/ncb3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Plotnikov SV, Moalim AY, Waterman CM, Liu J. Two distinct actin networks mediate traction oscillations to confer focal adhesion mechanosensing. Biophys J. 2017;112:780–794. doi: 10.1016/j.bpj.2016.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye GJC, Aratyn-Schaus Y, Nesmith AP, Pasqualini FS, Alford PW, Parker KK. The contractile strength of vascular smooth muscle myocytes is shape dependent. Integr Biol (Camb) 2014a;6:152–163. doi: 10.1039/c3ib40230d. [DOI] [PubMed] [Google Scholar]
- Ye N, Verma D, Meng F, Davidson MW, Suffoletto K, Hua SZ. Direct observation of α-actinin tension and recruitment at focal adhesions during contact growth. Exp Cell Res. 2014b;327:57–67. doi: 10.1016/j.yexcr.2014.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]