SUMMARY
Most secreted growth factors and cytokines are functionally pleiotropic because their receptors are expressed on diverse cell types. While important for normal mammalian physiology, pleiotropy limits the efficacy of cytokines and growth factors as therapeutics. Stem cell factor (SCF) is a growth factor that acts through the c-Kit receptor tyrosine kinase to elicit hematopoietic progenitor expansion, but can be toxic when administered in vivo because it concurrently activates mast cells. We engineered a mechanism-based SCF partial agonist that impaired c-Kit dimerization, truncating downstream signaling amplitude. This SCF variant elicited biased activation of hematopoietic progenitors over mast cells in vitro and in vivo. Mouse models of SCF-mediated anaphylaxis, radioprotection, and hematopoietic expansion revealed that this SCF partial agonist retained therapeutic efficacy while exhibiting virtually no anaphylactic off-target effects. The approach of biasing cell activation by tuning signaling thresholds and outputs has applications to many dimeric receptor-ligand systems.
Keywords: Pleiotropy, stem cell factor, c-Kit, protein engineering, receptor signaling, receptor tyrosine kinase, mast cells, hematopoietic stem cells, anaphylaxis, cytokine
INTRODUCTION
Most secreted growth factors (GFs) and cytokines exert their effects through homo- or hetero-dimeric cell surface receptor complexes to initiate intracellular kinase-mediated signal transduction (Atanasova and Whitty, 2012; Klemm et al., 1998; Schlessinger and Ullrich, 1992; Stroud and Wells, 2004; Wang et al., 2009). Collectively, cytokines (e.g. Interleukins, Interferons, etc.) and GFs (e.g. EGF, NGF, TGF, etc.) that act through dimeric receptors hold great promise as therapeutics for a wide range of diseases. However, while antagonism of these molecules has been clinically successful, principally for immune diseases and cancer (Nepom et al., 2013), effective therapeutic exploitation of their agonistic properties has been limited to relatively few cases largely due to the difficulty of controlling agonism in vivo (Bendall and Bradstock, 2014; Floros and Tarhini, 2015).
Since GFs and cytokines act on multiple different cell types that express their receptors, they tend to exhibit pleiotropic activities, which has limited their clinical utility as agonists (Moraga et al., 2014; Nicola, 1994; Thomas et al., 2004; Yea et al., 2015). Pleiotropic actions can induce counterbalancing cellular activities, resulting in blunted therapeutic efficacy (Hart et al., 2014; Lin et al., 1995). Furthermore, pleiotropy exacerbates off-target toxicities through activation of undesired cell types (Baldo, 2014; Krieg et al., 2010). Previous studies have shown that natural and engineered ligands, which modulate receptor dimerization by altering receptor-ligand complex stability, affinity and/or geometry, have the capacity to modulate downstream signaling outputs and target cell selectivity (Levin et al., 2014; Macdonald-Obermann and Pike, 2014; Mitra et al., 2015; Moraga et al., 2015a; Moraga et al., 2015b; Riese, 2011; Yea et al., 2015). Engineered cytokine ligands that differentially engage their heterodimeric receptors can induce partial receptor activation that exhibit biased cell type-specific activities in vitro (Mitra et al., 2015), raising the possibility that such signal attenuation could serve as a therapeutic strategy for pleiotropic agonists (Levin et al., 2014; Riese, 2011).
Stem cell factor (SCF), also known as Kit ligand, mast cell growth factor or steel factor, is a four-helical bundle GF that exists as a non-covalent homodimer (Jiang et al., 2000; Zhang et al., 2000). SCF binds to and activates its cognate receptor, a proto-oncogene and type III receptor tyrosine kinase known as c-Kit or Kit, in a symmetric 2:2 stoichiometry (Figure 1A) (Liu et al., 2007; Yuzawa et al., 2007). In this complex architecture, the SCF dimer interface serves as the central attachment site between the c-Kit dimer (Figure 1A). A number of signaling pathways have been implicated downstream of SCF-induced c-Kit activation, including but not limited to the mitogen-activated protein kinase (MAP kinase)/ERK and phosphatidylinositol 3′-kinase (PI3-kinase)/AKT pathways (Ronnstrand, 2004). The SCF/c-Kit axis acts as a key survival and maintenance signal in the bone marrow niche of hematopoietic stem and progenitor cells (HSPCs) (Ding et al., 2012; Li and Johnson, 1994).
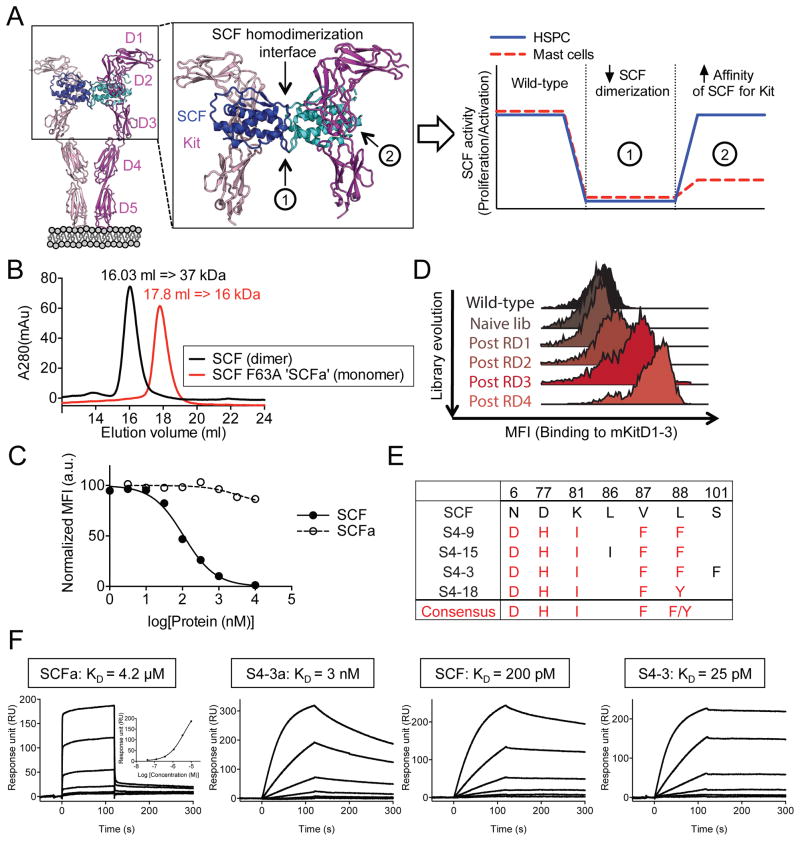
Figure 1. Engineering of mouse SCF partial agonist.
(A) Schematic strategy to decouple SCF efficacy and toxicity. Crystal structure of SCF/c-Kit complex was obtained from PDB: 2E9W. (B) Overlay of size exclusion chromatograms. SCF and SCF F63A (SCFa) elution volumes were used to approximate their respective molecular weights based on a standard curve. (C) On-yeast competitive blocking of mouse SCF/c-Kit interaction by soluble wild-type SCF or SCFa. Yeast expressing wild-type SCF were stained with 20 nM fluorescently-labeled mKitD1-3 tetramers in the presence of unlabeled soluble SCF variants. Data represent the mean ± SEM in duplicates and are representative of two independent experiments. (D) Histogram overlays assessing mKit staining of the “second-generation” SCF library. The post-selection libraries were stained with 100 nM labeled mKitD1-3 then competed with 1 μM unlabeled mKitD1-3 for 60 min. (B and C) MFI = mean fluorescence intensity. (E) Summary of amino acid sequences of engineered high-affinity SCF variants. The position of mutated residues and their corresponding sequences in wild-type SCF are denoted at the top of the table. Red text color indicates the consensus mutations. (F) Representative SPR sensorgrams of the indicated SCF variants binding to immobilized mKitD1-3. See also Figure S1 and S2.
Several lines of evidence suggest that the biological activity of SCF on HSPCs could be exploited for therapeutic uses. Treatment with SCF induces HSPC expansion, radioprotection and subsequent repopulation of hematopoietic lineages, leading to improved recovery and survival after irradiation (Gardner et al., 1998; Tong et al., 1993; Zsebo et al., 1992). However, pleiotropic SCF actions also play critical roles in mast cell development, maturation and activation (Galli et al., 1993; Taylor et al., 1995). The therapeutic potential of SCF is thus limited by the concurrent activation of mast cells, which results in allergic and anaphylactic responses in mice and humans (Costa et al., 1996).
In this study, we report that partial agonism of c-Kit by an engineered SCF, selectively activates HSPCs versus mast cells in vivo, resulting in narrowed pleiotropy and reduced toxicity (Figure 1A). That we have achieved this result through a ligand engineering strategy that is highly applicable to many dimeric GF systems suggests that engineering cytokine and GF partial agonists could have broad applications in other systems.
RESULTS
Engineering high-affinity SCF variant with a crippled homodimerization interface
We took a two-step approach to modulate c-Kit signaling: 1- decrease the propensity of SCF to self-dimerize by targeting its dimer interface, and 2- increase the affinity of SCF monomers for c-Kit (Figure 1A). We focused our engineering efforts on mouse SCF because of its cross-reactivity with mouse c-Kit (mKit) and human c-Kit (hKit). To ‘cripple’ SCF homodimerization, we introduced an alanine mutation within the SCF dimer interface at Phe63 (F63A), an amino acid residue previously shown to be critical to SCF dimerization (Hsu et al., 1997). While wild-type SCF behaves as a dimer in solution (MW~ 37 kDa), the SCF F63A (SCFa) mutant behaves as a monomer (MW~ 16 kDa) (Figure 1B). Due to the loss of avidity afforded by the dimer, the monomeric SCFa binds to mKit with approximately 20,000-fold lower affinity compared to wild-type homodimeric SCF and competitively antagonizes neither mouse nor human SCF/c-Kit interactions (Figure 1C, 1F and S2B). Yeast surface display was used to affinity-mature the monomeric SCF interaction with mKit (Boder and Wittrup, 1997). We used a structure-guided approach to create mutant libraries of SCF that were then selected with a soluble version of mKit consisting of the 3 N-terminal extracellular domains (mKitD1-3) (Figure 1D, 1E and S1). Two generations of in vitro evolution progressively enriched for SCF variants with a consensus set of mutations that confer higher-affinity binding to mKit (Figure 1D, 1E and S1).
The affinity of the SCF variants for c-Kit was measured by surface plasmon resonance (SPR) (Figure 1F, S1E, S1F and S2A). S4-3a, which is the highest affinity monomeric variant tested, showed approximately a 1000-fold increase in affinity compared to the “wild-type” but monomeric (F63A) SCFa (Figure 1F, S2A and data not shown). Compared to the dimeric wild-type SCF, the high-affinity monomeric S4-3a bound with a 10-fold reduction in affinity while the dimeric high-affinity S4-3 variant showed a 10-fold higher apparent affinity for mKit (Figure 1F). In addition, a yeast-based competition assay revealed that the affinity-enhanced monomeric S4-3a mutant effectively inhibits SCF/c-Kit interaction with similar potency as wild-type SCF (Figure S2B). Thus, this high-affinity monomeric version of SCF would be expected to bind to each subunit of c-Kit with high affinity, yet attenuate the ability of c-Kit to form a signaling-competent dimer, and antagonize endogenous SCF.
High-affinity monomeric variant S4-3a behaves as a partial agonist of c-Kit
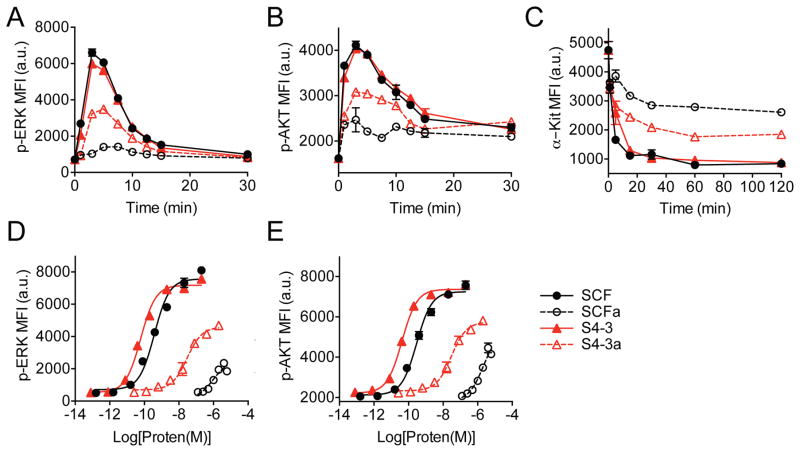
We analyzed downstream activation of the ERK and AKT signaling pathways in c-Kit-expressing mouse MC/9 mast cells. All variants of SCF in high versus low affinity and dimer versus monomer states induced time-dependent phospho-ERK and phospho-AKT responses at 1 μM (Figure 2A and 2B). The monomeric variants (SCFa and S4-3a) exhibited slightly delayed signal initiation and induced significantly lower peak amplitudes of ~15% and ~50%, respectively (Figure 2A and 2B). Correspondingly, stimulation with SCF dimers resulted in the most extensive c-Kit internalization followed by the high-affinity monomer S4-3a and, finally, the “wild-type” monomer SCFa (Figure 2C). Signaling dose-response curves confirmed that the dimers of wild-type SCF and the high-affinity variant S4-3 exhibited similar agonistic behavior, reaching a similar maximal response (Emax) and potency (EC50) (Figure 2D and 2E). However, SCFa and S4-3a monomers acted as partial agonists that induce 25–40% and 50–70% of the Emax achieved by the dimers, respectively (Figure 2D and 2E). Although the potency of both SCFa and S4-3a have been reduced compared to the dimers, S4-3a was found to be two-orders of magnitude more potent than SCFa (EC50, SCFa = ~ 1–2 μM; EC50, S4-3a = ~20–30 nM) (Figure 2D and 2E).
Figure 2. Partial c-Kit activation induced by SCF variants with crippled dimerization interface.
(A–C) Induction kinetics of (A) ERK1/2 phosphorylation, (B) AKT phosphorylation and (C) c-Kit internalization after stimulation with 1 μM of indicated SCF variants. (D and E) Dose response of (D) ERK1/2 phosphorylation and (E) AKT phosphorylation at 5 min of stimulation with indicated SCF variants. (A–E) Experiments were performed using mouse MC/9 mast cells. Data represent the mean ± SEM in triplicates and are representative of at least two independent replicates. MFI = mean fluorescence intensity; a.u. = arbitrary units.
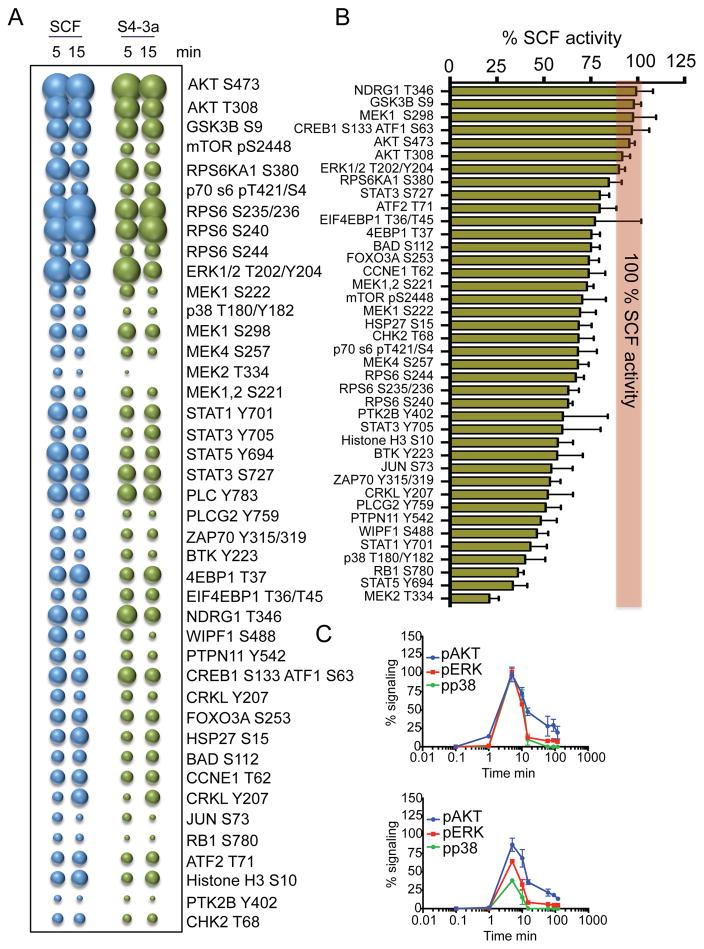
SCF activates additional pathways to AKT and ERK (Ronnstrand, 2004), so we studied the activation of 118 unique signaling molecules by phospho-flow cytometry in MC/9 cells (Table S1). At a saturating concentration of 1 μM, SCF and S4-3a activated 42 signaling proteins, including members of the STAT family (STAT1, 3 and 5), MAP kinase family (ERK, MEK and p38) and PI3K family (AKT, RSK1, RPS6). S4-3a exhibited a lower overall maximal activation (Emax) than that elicited by SCF (Figure 3A). Interestingly, the partial signal activation by S4-3a as a proportion of that by SCF was different for many of the 42 signaling proteins. S4-3a activated some molecules to comparable levels as SCF (e.g. MEK1, NDRG1, AKT); other molecules were activated to 60–70% of the levels induced by SCF (e.g. MEK4, STAT3, RPS6KA1); and yet other signaling proteins were activated to levels lower than 50% of the levels induced by SCF (e.g. p38, MK2, RB1) (Figure 3B). Kinetics studies in MC/9 cells confirmed these observations and showed that S4-3a activated ~90% AKT, ~70% ERK and ~50% p38 compared to their respective activation levels induced by SCF (Figure 3C).
Figure 3. “Biased” signaling activation induced by S4-3a.
(A) Bubble plot representation of the signaling pathways activated by SCF and S4-3a at the indicated times in MC/9 mast cells. The size of the bubble represents the intensity of the signaling activated. (B) The levels of signal activation induced by S4-3a at 5 min of stimulation were normalized to those induced by SCF and order based on signaling potency. The red line represents the SCF signaling potency normalized to 100%. (C) pAKT, pERK and pp38 kinetics experiments performed in MC/9 cells stimulated with SCF (top) or S4-3a (bottom). (A–C) Stimulation was performed with 1 μM SCF or S4-3a. Data represent mean ± SD and are from three independent experiments. See also Table S1.
The engineered S4-3a variant impairs c-Kit dimerization on the cell surface
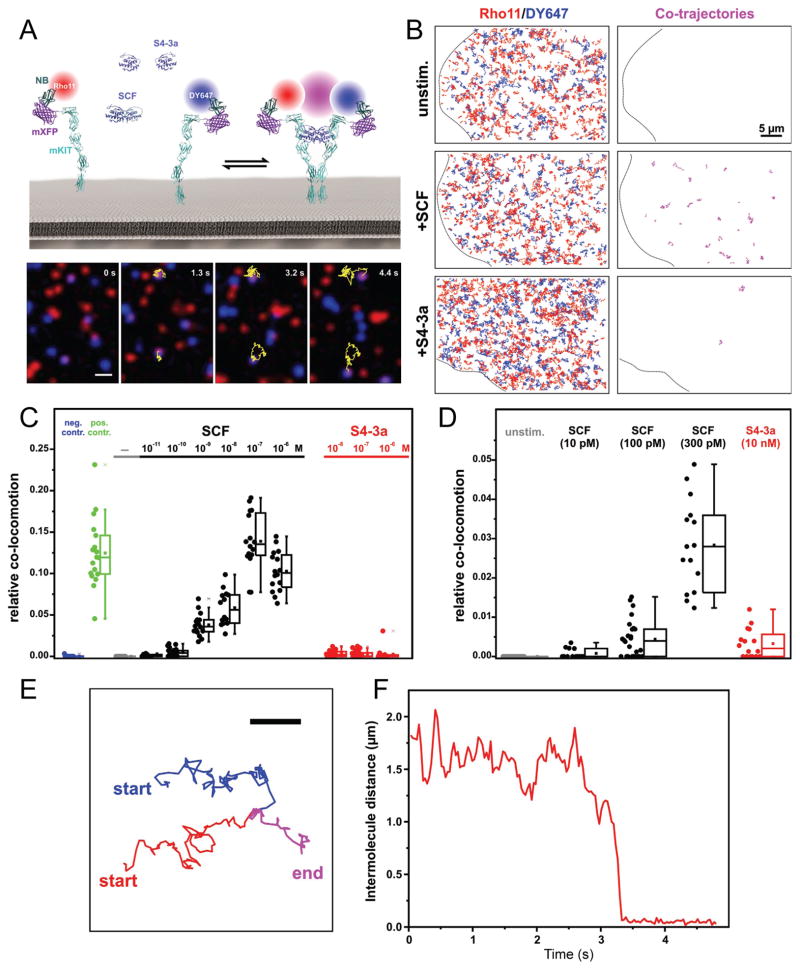
The spatiotemporal dynamics and dimerization of c-Kit by SCF and S4-3a at the plasma membrane were investigated via dual-color single molecule total internal reflection fluorescence (TIRF) microscopy. mKit fused to an N-terminal monomeric non-fluorescent green fluorescent protein (mXFP-mKit) was expressed in HeLa cells and labeled by addition of Rho11 or DY647 conjugated anti-GFP nanobodies (NB) (Figure 4A). Well-balanced cell surface labeling of mXFP-mKit was achieved to allow simultaneous dual color imaging at the single molecule level with minor photobleaching (Movie S1; Figure S3A and S3B). Random diffusion of mXFP-mKit at the plasma membrane with an average diffusion constant of (0.140 ± 0.002) μm2/s was observed (Figure S3C and S3D). While quantitative single molecule co-localization and co-tracking analysis did not reveal any dimerization of mXFP-mKit in absence of ligand, addition of SCF induced receptor dimer formation in a dose-dependent manner, reaching saturation at approximately 10 – 100 nM SCF (Movie S1 and S2; Figure 4B–4D). A corresponding decrease in mobility to an average diffusion constant of (0.12 ± 0.002) μm2/s was observed (Figure S3C and S3D). By comparison, statistically significant yet much weaker mXFP-mKit dimerization occurred in the presence of S4-3a (Movie S3; Figure 4B–4D). In line with the formation of only few mKit dimers, the overall average diffusion constant of (0.142 ± 0.002) μm2/s in the presence of S4-3a was not significantly different than that of unliganded receptor (Figure S3C and S3D). Furthermore, we found that 100 pM of SCF induced a similar degree of receptor dimerization as S4-3a can maximally achieve (Figure 4C and 4D).
Figure 4. Dimerization of mKit quantified by dual-color single molecule TIRF imaging.
(A) Single molecule dimerization of mXFP-mKit expressed in HeLa cells and labelled by anti-GPF nanobodies conjugated with Rho11 (red) and DY647 (blue), respectively. Magenta represents co-localization. Top: cartoon of the assay; bottom: raw image series of mXFP-mKit in presence of SCF and trajectory of a dimer. Yellow lines indicate the co-trajectories of mKit dimers. Scale bar: 1 μm. (B) Trajectories (from 100 frames, 3.2 s) of individual Rho11-labeled (red) and Dy647-labeled mKit (blue) and co-trajectories (magenta) for an unstimulated cell, as well as cells after stimulation with SCF (100 nM) and S4-3a (100 nM), respectively. Scale bar: 5 μm. (C) Relative numbers of co-trajectories for mXFP-mKit in absence of ligand (unstim.) and after stimulation with indicated concentrations of SCF or S4-3a, compared to negative and positive control experiments. (D) A close-up comparison of mXFP-mKit dimerization shown in (C). (E) Formation of an individual mKit dimer in the presence of SCF probed by co-tracking Rho11 (red), Dy647 (blue), co-trajectory (magenta). Scale bar: 1 μm. (F) Distance plot of (E) identifying the formation of an individual mKit dimer. See also Figure S3.
High-affinity monomeric partial agonist S4-3a exhibits HSPC-selective functional activity compared to wild-type SCF in vitro
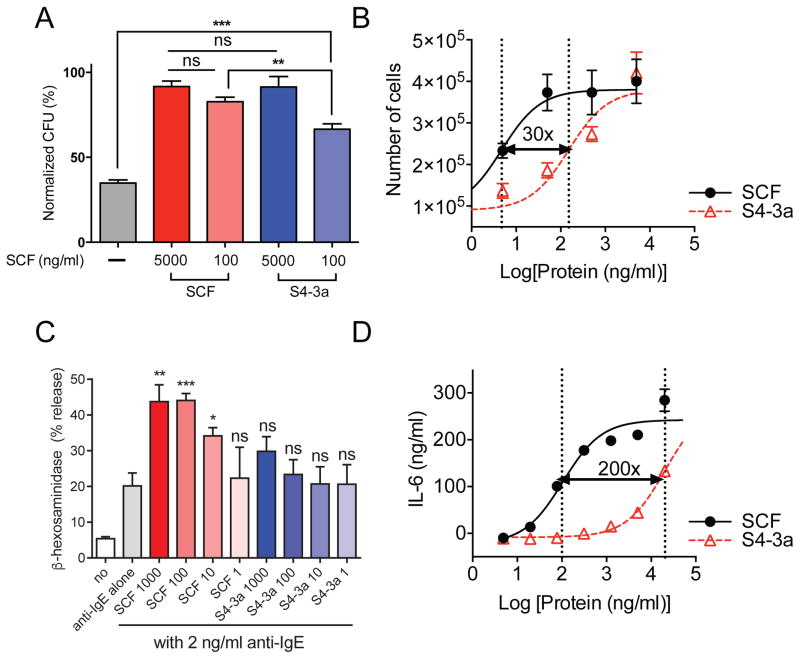
The in vitro biological functions of wild-type SCF versus high-affinity partial agonist were compared in the two cell populations of interest, HSPCs and mast cells. For HSPCs, we found that S4-3a and SCF induced comparable CFU-GM (colony formation unit-granulocyte/macrophage) colonies (Williams et al., 1992) from sorted mouse bone marrow Lin−c-Kit+Sca-1+ (LSK) HSPCs cultured in semi-solid medium at 5000 ng/ml (Figure 5A). At 100 ng/ml, S4-3a showed slightly reduced colony-forming capacity compared to 100 ng/ml SCF though still significantly higher than the control (Figure 5A). To further examine the ex vivo proliferative effects on HSPCs, sorted LSK HSPCs were cultured with SCF variants on stromal cells. By quantifying the total number of hematopoietic cells generated in a 6-day period, we found that S4-3a, again, stimulated the same maximal response as SCF (i.e. cell proliferation) in a dose-dependent manner albeit with approximately 30-fold lower potency (Figure 5B).
Figure 5. In vitro HSPC-biased activity by high-affinity monomer S4-3a compared to wild-type SCF.
(A) Ex vivo CFU-GM colony formation assay. Sorted mouse LSK HSPCs were cultured in semi-solid methylcellulose medium with 100 ng/ml recombinant mouse IL-3 and the indicated concentration of SCF variants. The number of CFU-GM colonies were scored after a 7-day period and normalized to the top data point in the 5000 ng/ml SCF group. (B) Ex vivo proliferation of purified LSK HSPCs from mouse bone marrow. Cells were cultured on irradiated OP9 stromal cells for 6 days in the presence of titrating concentrations of SCF variants, and the total number of hematopoietic cells generated was quantified. (C) β-hexosaminidase release by human PBCMCs treated with anti-IgE alone or with anti-IgE together with SCF or S4-3a at indicated concentrations (ng/ml) for 30 min in vitro. See also Figure S4. (D) Dose-response curves of IL-6 release by mouse BMCMCs stimulated for 6 h with SCF variants in vitro. (A and C) Data represent mean ± SEM and are pooled from at least two independent experiments with a total of n 6. *p<0.05, **p<0.01, ***p<0.001, and ns = not significant (i.e., p>0.05) compared to the PBS-treated control group by unpaired, two-tailed Student’s t test. (B and D) Data represent mean ± SEM and are representative of at least two independent experiments with n = 3 per experiment. The vertical dotted lines indicate the EC50 values of the dose-response curves. The numbers on top of the black arrows indicate the fold difference between the EC50s.
For mast cells, we first determined whether SCF variants could activate human mast cells and/or enhance their degranulation in the context of FcεRI-induced activation (Gilfillan et al., 2009). Activation of human peripheral blood-derived cultured mast cells (PBCMCs) (Gaudenzio et al., 2016) was measured by quantifying the release of granule-associated enzyme β-hexosaminidase (Akahoshi et al., 2011). While IgE/anti-IgE stimulation efficiently induced β-hexosaminidase release in PBCMCs, neither of the SCF variants tested alone induced a detectable response (Figure S4). Minimal β-hexosaminidase release by mast cells stimulated in vitro with SCF as a single agent has been reported previously (Hill et al., 1996). For IgE-dependent activation, we found that SCF considerably enhanced the degranulation of IgE-sensitized/anti-IgE-stimulated human PBCMCs whereas S4-3a treatment did not induce a statistically significant effect compared to PBS at the concentrations examined (Figure 5C). We also characterized the effect of SCF variants on IL-6 release by mouse bone marrow-derived cultured mast cells (BMCMCs) because in vitro stimulation of mouse mast cells with recombinant SCF has been shown to result in more extensive IL-6 production compared to other mediators such as histamine (Gagari et al., 1997). In this experiment, we covered a greater concentration range (up to 20 μg/ml) and found that S4-3a induced a similar maximal cellular response (i.e., IL-6 production) as SCF but with significantly lower potency (~200-fold) in BMCMCs (Figure 5D).
S4-3a has similar proliferative and radioprotective effects on HSPCs as SCF in vivo
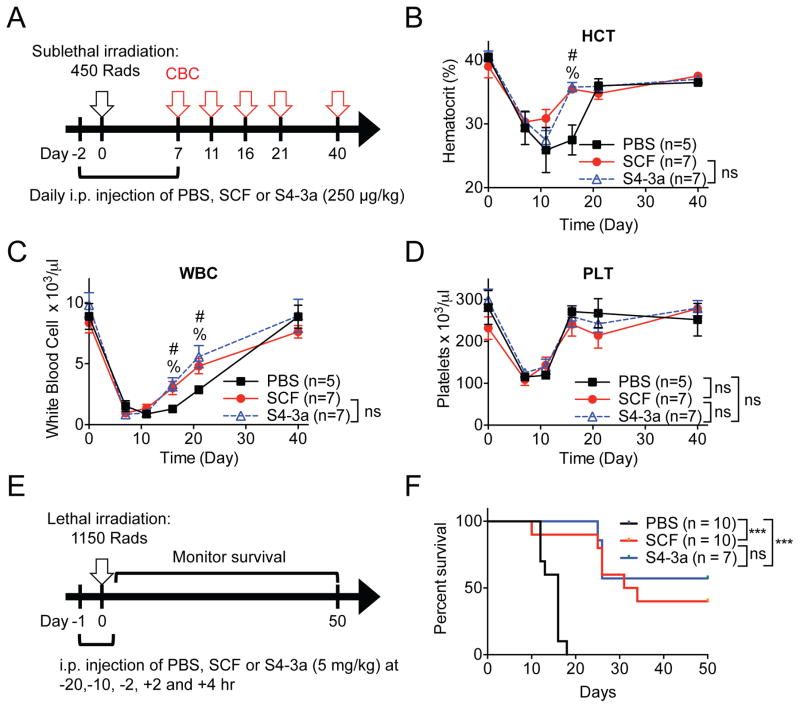
We asked whether the increased in vitro cell type-specific activity of S4-3a translated into in vivo functional biases. Two independent models were used to assess the in vivo therapeutic activity on HSPCs. We first evaluated the ability of SCF variants to stimulate hematopoietic expansion and recovery in mice exposed to sub-lethal irradiation as a pre-clinical model of radiation therapy for cancer (Figure 6A). A low daily intraperitoneal (i.p.) dose of 250 μg/kg was used based on published studies with a similar experimental setup (Molineux et al., 1991; Zsebo et al., 1992). Periodic analysis of complete blood counts (CBCs) revealed an expected sharp decline of hematocrit, white blood cells and platelets in the first 10 days post-irradiation (Figure 6B–6D). Although modestly, both SCF and S4-3a accelerated the recovery of hematocrit and white blood cells to a similar extent compared to PBS treatment (Figure 6B–6D). We suspect that more pronounced effects may be possible with a higher dose or additional modifications to increase serum half-life and bioavailability. Consequently, a higher dose of 5 mg/kg was used in the second model in attempt to increase bioavailability.
Figure 6. S4-3a and wild-type SCF induce similar proliferative and radioprotective effects in vivo.
(A) Experimental setup for analysis of hematopoietic recovery after sub-lethal irradiation. Mice received daily i.p. injections of SCF variants at 250 μg/kg or of PBS starting 2 days prior to sub-lethal TBI (450 Rads) and continued for a total of 10 days. Complete blood cell counts (CBC) were analyzed over a 40-day period as indicated. (B–D) Peripheral blood cell counts of (B) hematocrit (HCT), (C) white blood cells (WBC) and (D) platelets (PLT) over time for mice treated with indicated agents. Data represent mean ± SEM and are pooled from two independent experiments (n≥5). %p<0.05 (statistical significance between PBS and SCF) and #p<0.05 (statistical significance between PBS and S4-3a), and ns = not significant (i.e., p>0.05), by unpaired, two-tailed Student’s t test and corrected for multiple comparisons using the Holm-Sidak method. (E) Experimental setup for analyzing SCF prevention of radiation-induced mortality. Injections of 5 mg/kg SCF variants or PBS were administered i.p. at −20, −10, −2, +2 and +4 h of lethal total body irradiation (TBI; 1150 Rads). Survival was monitored daily for 50 days. (F) Survival of mice treated as described in (E). Data are pooled from two independent experiments (n≥7). ***p<0.001, and ns = not significant (i.e., p>0.05), by Log-rank Mantel-Cox test. See also Figure S5.
Treatment with recombinant PEGylated SCF pre- and post-lethal irradiation has been shown to produce a significant survival benefit in otherwise lethally irradiated mice by radioprotecting the HSPCs (Zsebo et al., 1992). Therefore, the radioprotective effect of SCF variants was compared using a similar treatment regimen prior to and post 1150 Rads of lethal irradiation (Figure 6E). Treatment with either SCF or S4-3a variants substantially, and comparably, increased the long-term survival of lethally irradiated mice whereas none of the saline-treated mice survived past day 18 (Figure 6F). While we cannot rule out the possibility that SCF variants have some radioprotective effects on intestinal stem cells (Leigh et al., 1995), it is more likely that the enhancement of overall survival post-exposure in the context of our experiment stems primarily from the effects of SCF variants on residual HSPCs. This is based on the known effects from the dosage of radiation we used and the time to death, both of which are characteristic of the hematopoietic compartment (Gudkov and Komarova, 2003; Leibowitz et al., 2014).
In rescuing the mice from lethal irradiation, a notable observation was made after the first i.p. injection of 5 mg/kg treatment: only the SCF-treated mice but not those receiving S4-3a experienced acute adverse reactions, accompanied by severe lethargy and shivering (data not shown). These symptoms are hallmarks of systemic anaphylaxis in mice. To quantify this observation, we administered a single dose of 5 mg/kg or 10 mg/kg of SCF, 10 mg/kg of S4-3a or PBS and measured their effects on body temperature over the course of an hour (Figure S5A). We chose to test a higher dose of 10 mg/kg to investigate if an adverse event could be induced in the S4-3a treated mice as no reaction was observed at 5 mg/kg. Consistent with previously observed lethargy and shivering, the SCF-treated mice experienced profound hypothermia, another signature of systemic anaphylaxis in mice, in a dose-dependent manner whereas even 10 mg/kg of S4-3a had no effect on body temperature (Figure S5B).
S4-3a has minimal capacity to induce mast cell-mediated anaphylaxis in vivo compared to SCF
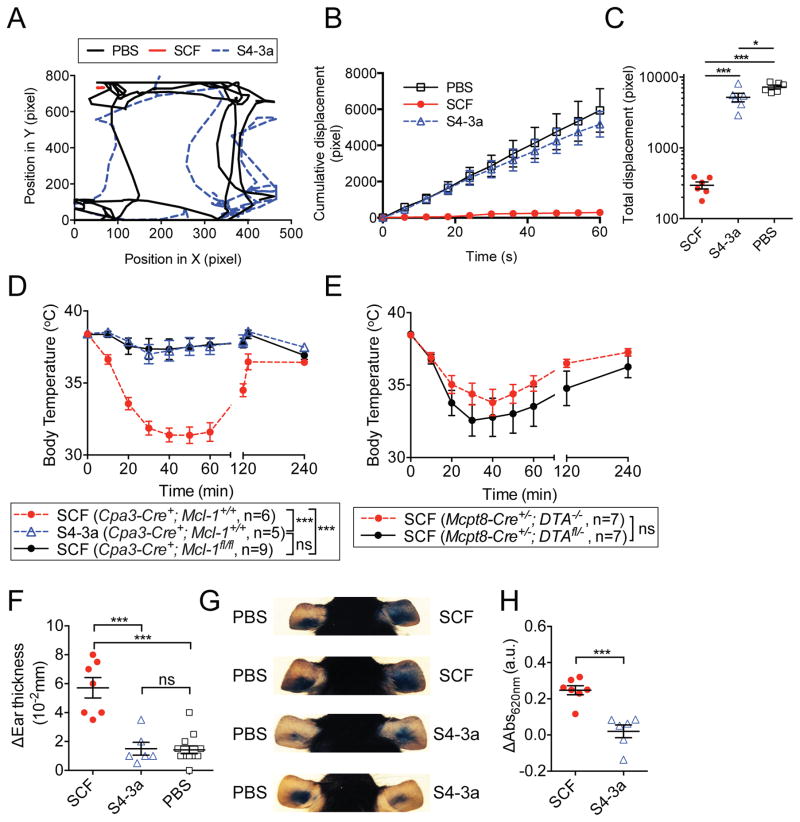
We assessed the effect of SCF variants on systemic anaphylaxis. For these experiments, a high dose of 10 mg/kg was used in order to enhance our ability to detect toxicities associated with the S4-3a variant. We filmed and tracked mouse movements in their cages for a one-minute time interval 20 minutes after i.p. administration of PBS or SCF variants (Movie S4–S6; Figure 7A–7C and S6A). Similar to previous observations, SCF treatment induced severe lethargy, which was visually apparent and quantifiable by minimal cumulative and total displacement during the time of tracking (Movie S5; Figure 7A–7C and S6A). This is in significant contrast to the S4-3a treated mice, which remained as active as the PBS treated mice (Movie S4 and S6; Figure 7A–7C and S6A).
Figure 7. S4-3a has reduced capacity compared to wild-type SCF to elicit mast cell activation and related anaphylaxis in vivo.
(A–E) Analysis of systemic anaphylaxis after i.p. injection of 10 mg/kg SCF variants. (A–C) Movement of mice treated with the indicated agents. (A) Trajectory tracking of a representative mouse from each treatment. (B) Cumulative displacement and (C) total displacement over a one-minute period. (D and E) Body temperature of (D) control C57BL/6;Cpa3-Cre+; Mcl-1+/+ or mast cell/basophil-deficient C57BL/6;Cpa3-Cre+; Mcl-1fl/fl mice and (E) control C57BL/6;Mcpt8-Cre+/−; DTA−/− or basophil-deficient C57BL/6;Mcpt8-Cre+/−; DTAfl/− mice was measured at the indicated time points post-injection. (F–H) Assessment of local cutaneous anaphylaxis after injection of 25 μl PBS alone or 50 μg/ml SCF variants in the left and right ear pinna, respectively. (F) Difference in ear pinnae thickness of individual mice determined 2 h after injection compared to baseline values. (G) Images of mouse ears showing Evans blue extravasation of four representative mice treated with the indicated agents 3 h earlier. (H) Quantification of Evans blue dye in ear pinnae (such as those shown in G), calculated by subtracting respective values obtained from PBS-injected control ear pinnae of the same mouse. (A–H) Data represent the mean ± SEM and are pooled from two independent experiments (n≥5). *p<0.05, ***p<0.001, and ns = not significant (i.e., p>0.05) by unpaired, two-tailed Student’s t test. See also Figure S6.
The peritoneal cells of these mice were then harvested and processed to search for signs of mast cell activation and degranulation. By May-Grünwald/Giemsa staining of the cytospun peritoneal cells, we found higher numbers of cells with macrophage-like morphology that had taken up granules released by mast cells in mice treated with SCF compared to those treated with saline or S4-3a (Figure S6B and S6C). This observation is common during anaphylaxis and suggests a higher degree of mast cell activation occurred with SCF vs. S4-3a challenge, since these granules were presumably released upon mast cell degranulation. In addition, peritoneal mast cells from SCF-treated mice displayed significantly decreased levels of cell surface c-Kit expression as compared to those isolated from saline or S4-3a-treated animals (Figure S6D), suggesting increased c-Kit-mediated mast cell activation in mice injected with SCF (Yee et al., 1994).
Consistent with these results, SCF treatment but not S4-3a treatment induced severe hypothermia in C57BL/6 Cpa3-Cre+; Mcl-1+/+ mast cell- and basophil-sufficient control mice (Figure 7D). In contrast, C57BL/6 Cpa3-Cre+; Mcl-1fl/fl mice, which retain endogenous expression of c-Kit but are deficient in mast cells and exhibit substantially decreased numbers of basophils (Lilla et al., 2011), did not develop hypothermia in response to SCF injection. Furthermore, specific basophil-deficient, mast cell-sufficient C57BL/6 Mcpt8-Cre+/−; DTAfl/− mice, as well as basophil- and mast cell-sufficient control C57BL/6 Mcpt8-Cre+/−; DTA−/− animals, each developed significant hypothermia upon SCF challenge (Figure 7E).
We subsequently evaluated the effect of SCF variants on local anaphylaxis because a wheal and flare reaction, caused by cutaneous anaphylaxis at the site of injection, was the most common side effect of SCF reported in human patients (Costa et al., 1996). Mediators released by activated mast cells increase vascular permeability and cause edema; therefore, tissue swelling and Evans blue extravasation are two conventional in vivo measures of local anaphylaxis (Taketomi et al., 2007). We found that intradermal injection of SCF, but not S4-3a, induced cutaneous anaphylaxis associated with significant ear swelling and vasodilation (assessed by Evans blue extravasation) (Figure 7F–7H).
Based on our findings in mouse models of systemic and local anaphylaxis, we conclude that the partial agonist variant of SCF, S4-3a, is significantly less potent in activating mast cells and producing mast cell-dependent pathology in vivo compared to SCF.
DICUSSION
We have engineered the SCF/c-Kit interaction using a mechanism-based strategy to modulate signaling amplitude. We report two principal findings: 1- Partial receptor agonism was sufficient to elicit full activation of some cell subsets (e.g. HSPCs), but not others (e.g. mast cells). 2- More broadly, partial agonism of an RTK (c-Kit) by an engineered ligand (SCF) resulted in biased activation of cell subsets in vivo, which resulted in reduced toxicity from mast cell activation. While our study is limited to a proof of concept in a model mouse system, the basic mechanistic insights into dimeric receptor signaling arising from these studies have clear translational implications. Growth factor and cytokine agonists present significant clinical challenges as a result of their functional pleiotropy, but hold promise for the treatment of a wide range of diseases, particularly in regenerative medicine and immunotherapy. The current study, together with previous studies demonstrating that the signaling amplitudes of dimeric cytokine receptors are ‘tunable’ through ligand engineering, begins to make a compelling case for revisiting GF and cytokine agonism clinically using partial agonistic variants.
Mechanistically, while our signaling data clearly indicate that S4-3a must dimerize c-Kit to some extent, the single molecule TIRF data shows that the degree of dimerization is minimal, yet still sufficient to activate partial signaling (Figure 2–4). One possibility is that the F63A mutation incompletely disrupts the SCF homodimerization interface. In a mass-action based mechanism, SCF F63A variants may be able to weakly self-associate at high local concentrations (e.g., those occurring on a cell surface) but elicit lower amplitude c-Kit signals than those variants with intact homodimerization interfaces. Another possible explanation is that the propensity of c-Kit to “zipper” together after SCF binding (Reshetnyak et al., 2015), may result in the disrupted S4-3a dimer interface being compensated to some degree by enforced proximity through interacting domains within the c-Kit homodimer, resulting in a quasi-stable receptor signaling dimer (Figure 1A) (Blechman et al., 1995; Yuzawa et al., 2007).
The two-step engineering process was essential to create a partial agonist to elicit biased in vivo cell activation in a controlled manner that would be difficult to mimic with low dose SCF. The affinity maturation of the partial agonist was necessary for it to act as a high-affinity ‘clamp’ on c-Kit that acted as an antagonist for the endogenous ligand, thereby controlling the signaling output (Figure 1 and 2). This dominant negative effect of the high-affinity partial agonist prevents, or at least mitigates the possibility of the endogenous ligand, in this case SCF, from overcoming the partial agonist. We demonstrated that 100 pM SCF induces roughly equivalent levels of receptor dimerization as that mediated by a saturating concentration of S4-3a (Figure 4C and 4D), but 100 pM SCF elicits lower levels of ERK and AKT signaling compared to S4-3a (Figure 2D and 2E). The qualitative difference in signaling suggests that S4-3a and 100 pM SCF would unlikely have similar biological outcomes. Indeed, 100 pM (~4 ng/ml) SCF induced significantly less ex vivo HSPC proliferation compared to S4-3a at a saturating dose (Figure 5B). Even if 100 pM SCF did elicit the same biological effects as S4-3a, controlled delivery of 100 pM SCF to elicit controlled efficacy would be difficult for two reasons. First, based on the sigmoidal signaling dose response curves (Figure 2D and 2E), 100 pM SCF lies on the unstable region of the response such that a slight variation in drug delivery engenders a large variation in the biological outcome. Second, it remains challenging to reliably and precisely deliver low dose biologics. For these reasons, low dose SCF is a less controllable and thus less ideal in vivo agent than S4-3a, which clamps partial receptor dimerization and partial signal activation at a stable level.
S4-3a appears to exhibit HSPC-biased activity compared to wild-type SCF in vitro. In particular, based on the two dose-response curves (Figure 5B and 5D), our data suggest: While HSPCs are intrinsically more sensitive to SCF than mast cells, as the EC50 for LSK cell proliferation is lower than that for BMCMC IL-6 release, S4-3a is more biased towards LSK cells with a 10-fold larger therapeutic window in vitro. This is calculated by comparing the difference between the EC50 values of S4-3a and SCF in HSPC proliferation (~30-fold) and BMCMC IL-6 release (~200-fold) (Figure 5B and 5D). Since mast cells have higher cell surface c-Kit expression (Figure S7), it is likely that the observed selectivity instead reflects different cell types possessing differential functional signaling thresholds.
Our data support evaluation of a human analog of S4-3a in clinical settings where wild-type SCF has been thought to have potential value, but such benefit may have been limited by mast cell-related toxicities of SCF. For instance, SCF has been shown to synergize with G-CSF in mobilizing bone marrow HSPCs into the blood to facilitate graft collection for hematopoietic cell transplantations (Briddell et al., 1993). However, because of the clear risk of toxicity related to the mast cell-activation properties of SCF (Costa et al., 1996), some subjects were excluded from clinical trials of SCF plus G-CSF either because they had a history of allergic disorders or because of adverse effects upon treatment with SCF that probably were related to effects of SCF on mast cells, which were observed even when such subjects had been pre-treated with pharmacological agents to counteract the effects of mast cell activation (Moskowitz et al., 1997). Moreover, da Silva et al. reported dose-limiting mast cell-related toxicities of SCF when it was administered in amounts beyond 20 μg/kg/day, which severely impeded the adequate evaluation of the full clinical potential of SCF as an agent to enhance the development and/or mobilization of hematopoietic progenitor cells alone or in combination with G-CSF (da Silva et al., 2004). A human analog of S4-3a would likely be free from these restrictions given its narrowed pleiotropy and selectivity for hematopoietic progenitor cells.
The intent of our study was to explore the concept that mechanism-based engineering of a receptor tyrosine kinase partial agonist can result in in vivo functional biases, thereby decoupling its pleiotropic actions. Our data suggest that this strategy may merit consideration for development of human therapeutics, and revisiting cytokine and GF systems where dose-limiting toxicity and off-target effects have limited clinical efficacy.
STAR METHODS TEXT
Contact for Reagent and Resource Sharing
Further information and requests for reagents may be directed to K.C. Garcia (kcgarcia@stanford.edu).
Experimental Model and Subjects
Animals
All animal care and experiments were carried out in accord with current National Institutes of Health guidelines and with the approval of the Stanford University Institutional Animal Care and Use Committee.
C57BL/6 mice were purchased from Jackson labs, maintained in the local animal facility and used for experiments at 8–10 weeks of age. C57BL/6 Cpa3-Cre+; Mcl-1+/+ mast cell/basophil-sufficient control or mast cell/basophil-deficient C57BL/6 Cpa3-Cre+; Mcl-1fl/fl mice (Lilla et al., 2011) were bred and maintained in the local animal facility and used for experiments at 8–10 weeks of age. C57BL/6 Mcpt8-Cre+/−; DTAfl/− basophil-deficient (Wada et al., 2010) or C57BL/6 Mcpt8-Cre+/−; DTA−/− littermate basophil-sufficient control mice were bred in the local animal facility and generated by cross-breeding of homozygous B6.129S4-Mcpt8tm1(cre)Lky/J with heterozygous (generated by crossing to C57BL/6J) B6.129P2-Gt(ROSA)26Sortm1(DTA)Lky/J originally obtained from Jackson labs. Male and female mice at 8–10 weeks of age were used for experiments. Basophil deficiency (or sufficiency) of mice was confirmed prior to experiments by flow cytometry analysis of basophils in peripheral blood using antibodies against FcεRIα (clone MAR-1; BioLegend) and CD49b (clone DX5; BioLegend). Mast cell deficiency (or sufficiency) was confirmed by using classical toluidine blue (Fisher Scientific) staining of skin biopsies.
Sample sizes were estimated based on previous experience with animal experiments of similar setup in our labs and on literature reports. All animal experiments were performed non-randomized and without blinding of the experimenters.
Isolation of primary bone marrow HSPCs and peritoneal mast cells
Bone marrow Lin−c-Kit+Sca-1+ LSK HSPCs
Lin− bone marrow cells were isolated from GFP+ or wild-type C57B/6J mice using a Lineage cell depletion kit (Miltenyi). Lin−Sca-1+c-Kit+ (LSK) cells were subsequently purified by FACS sorting on BD AriaIII SORP using PE conjugated antibodies against Lineage+ markers (CD3e (clone:145-2C11); CD4 (clone: RM4-5); CD8 (clone: 53-6.7); B220 (clone: RA3-6B2); Ter119 (clone: TER-119); Gr1 (clone: RB6-8C5); Mac1 (clone: M1/70)), Sca1 PE-Cy7 (clone: D7), c-Kit APC-Cy7 (clone: 2B8) (all from eBiosciences).
Peritoneal mast cells
Peritoneal lavage was used to extract mouse peritoneal cells. FcεRIα+c-Kit+ peritoneal mast cells were identified using anti-FcεRIα (clone: Mar-1) Pacific Blue (BioLegend) and anti-c-Kit APC-Cy7 (eBiosciences).
Tissue Culture Cells
MC/9 mast cells
The mouse mast cell line MC/9 was obtained from ATCC (ATCC designation CRL-8306) and was cultured in DMEM (Life Technologies) adjusted to contain 4.5 g/L glucose and 1.5 g/L sodium bicarbonate and supplemented with 2 mM L-glutamine, 0.05 mM 2-mercaptoethanol, 10% Rat T-STIM (Becton Dickenson) and 10% fetal bovine serum.
BMCMCs
Bone marrow-derived cultured mast cells (BMCMCs) were generated from bone marrow cells of female C57BL/6J mice by culture in Wehi-3-conditioned, IL-3-containing medium (DMEM containing 20% supernatant of Wehi-3 cells, 10% FCS, 50 μM β-mercaptoethanol, 2 mM L-glutamine, and 1% antibiotic-antimycotic solution) as previously described (Kalesnikoff and Galli, 2011). The cells were used after at least 4 weeks of culture. Flow cytometry was used to confirm c-Kit and FcεRIα expression (~95 % of cells were c-Kit- and FcεRIα-positive) using anti-c-Kit (clone 2B8: eBioscience) and anti-FcεRIα (clone Mar-1: Biolegend) antibodies; these cultures thus predominately consisted of mast cells.
PBCMCs
Human PBCMCs were obtained as previously described (Gaudenzio et al., 2016). Briefly, peripheral blood mononuclear cells were obtained from buffy coats of healthy blood donors at the Stanford Blood Center. CD34+ precursor cells were isolated from peripheral blood mononuclear cells (EasySep Human CD34 Positive Selection Kit, STEMCELL Technologies). CD34+ cells were maintained for 1 week under serum-free conditions using StemSpan medium (STEMCELL Technologies) supplemented with recombinant human IL-6 (50 ng/ml; Peprotech), human IL-3 (10 ng/ml; Peprotech) and 3% supernatant of CHO transfectants secreting murine SCF (a gift from Dr P. Dubreuil, Marseille, France, 3% correspond to ~50 ng/ml SCF). Thereafter, the cells were maintained in IMDM Glutamax I, sodium pyruvate, 2-mercaptoethanol, 0.5% BSA, insulin-transferrin selenium (all from Invitrogen), ciprofloxacin (10 mg/ml; Sigma-Aldrich), IL-6 (50 ng/ml) and 3% supernatant of CHO transfectants secreting mouse SCF. Before use in experiments, PBCMCs were tested for phenotype by flow cytometry (tryptase, chymase, CD117, FcεRI) and function (β-hexosaminidase release in response to FcεRI cross-linking) at 8–12 weeks. PBCMCs were ready for experiments after ~10 weeks in culture, at which point mast cells (CD117+FcεRI+) represented ~95–99% of all cells.
HeLa cells
HeLa cells were cultivated at 37°C and 5% CO2 in MEM Earle’s with stable glutamine supplemented with 10% fetal bovine serum (FBS), non-essential amino acids and HEPES buffer without addition of antibiotics. Cells were seeded in 60 mm cell-culture dishes to a density of ca. 60% confluence and transiently transfected via calcium-phosphate-precipitation according to standard protocols. For control transfection, model transmembrane proteins, comprised of an N-terminal of monomeric enhanced green fluorescent protein (mEGFP) or tandem-mEGFP linked to an artificial transmembrane domain (ALA)7KSSR were inserted into a modified version of pSems-26m (Covalys) including the N-terminal signal sequence of Igκ. For experimental transfection, a non-fluorescent (Y67F) variant of monomeric green fluorescent protein (mXFP) was N-terminally fused to mKit, and mXFP-mKit was inserted into the same vector as control transfection. Transfected HeLa cells were transferred onto 25 mm glass cover slides coated with a poly-L-lysine-graft-(polyethylene glycol) copolymer functionalized with RGD for minimizing non-specific binding of fluorescent NBs (You et al., 2014). Single molecule imaging experiments were conducted 48 h post transfection after mounting the cover slides into custom-designed microscopy chambers with a volume of 1 ml.
Method Details
Protein expression and purification
Mouse and human c-Kit extracellular domains 1–3 (mKitD1-3 and hKitD1-3; residues 26-314 and 26-311 according to Uniprot entries P05532 and P10721, respectively), were cloned into pAcGP67a (BD Biosciences) for secretion from Trichoplusia ni (High Five) cells (Invitrogen) using Baculogold baculovirus expression system (BD Biosciences). Both with a carboxy-terminal 8X histidine tag, mKitD1-3 and hKitD1-3 were purified by nickel-nitrilotriacetic acid (Ni-NTA) resin (Qiagen) and size exclusion chromatography with an AKTAPurifier (GE Healthcare) on a Superdex-75 column (GE Healthcare).
To produce recombinant proteins for yeast display selections and immobilization on streptavidin BIAcore chip (see below), mKitD1-3 and hKitD1-3 were expressed with a carboxy-terminal bio acceptor peptide tag (GLNDIFEAQKIEWHE) and purified as described above. The purified proteins were biotinylated in vitro with BirA ligase (produced in house) and then re-purified from the reaction mixture by size exclusion chromatography.
Mouse and human SCF (residues 26-166 and 26-166 according to Uniprot entries P20826 and P21583, respectively) and variants were cloned into pET28a (Novagen) with a C-terminal 8X histidine tag and a starting methionine residue. The SCF constructs were transformed into competent BL21(DE3) E. coli (Novagen). Bacterial culture was induced with 1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) at 37 °C for 4 h. Inclusion bodies were purified and dissolved in 8 M urea, 50 mM sodium acetate, pH 6.0, 0.1 mM ethylenediamine tetraacetic acid (EDTA, pH 8.0) and 1 mM dithiothreitol (DTT). Solubilized SCF was slowly injected into the vortex of stirring refolding buffer containing 2.5 M urea, 0.01 mM EDTA, 5 mM sodium acetate, 50 mM Tris-HCl (pH 8.5), 0.5 mM oxidized glutathione, 5.0 mM reduced glutathione and 0.2 mM phenylmethanesulfonylfluoride (PMSF) at 4°C. After 40 h, the refolding mixture was dialyzed thrice against 10 mM Tris-HCl (pH 8.0) and 150 mM sodium chloride. Thirty-percent followed by 70% ammonium sulfate precipitation were performed to remove misfolded aggregates and enrich for properly folded material. The 70% precipitates were resolubilized in 1 X HBS (hepes buffered saline) and purified by Ni-NTA and size exclusion chromatography with a Superdex-200 column (GE healthcare).
For in vitro and in vivo functional assays, endotoxin was removed using Triton X-114 as previously described (Weiskopf et al., 2013), and endotoxin removal was confirmed using the ToxinSensor Chromogenic LAL Endotoxin Assay Kit (Genscript).
For single-molecule TIRF experiments, the anti-GFP nanobody (NB) “enhancer” (Kirchhofer et al., 2010) was cloned into pET-21a with an additional cysteine at the C-terminus followed by a (PAS)5 sequence. Protein expression in E. coli Rosetta (DE3) and purification by immobilized metal ion affinity chromatography was carried out by standard protocols. Purified proteins were dialyzed and labeled with DY647 maleimide (Dyomics) and Rho11-maleimide (ATTO-TEC), respectively. Protein aggregates and free dye were subsequently removed by size exclusion chromatography. A labeling degree of > 90% was achieved as determined by UV/Vis spectrometry.
Size exclusion chromatography (molecular weight estimation)
Size exclusion chromatography of analytical amounts (~100 μg) of purified SCF or SCF F63A (SCFa) in PBS was performed using an AKTA purifier (GE Healthcare) on a Superdex-200 column (GE Healthcare). The elution flow rate was 0.5 ml/min. Absorbance reads at 280 nm were exported from UNICORN™ 5.11 software (GE Healthcare) and plotted using Prism 6 (GraphPad). The standard curve was constructed by using the elution volumes of Gel Filtration Standards (Bio-Rad) over the same Superdex-200 column and plotting log[expected molecular weights (MW) of the standards] (kDa) vs. elution volume (ml). The standard curve was fitted using linear regression with an R2 value of 0.981. The fitted equation was y = −0.219x + 5.08, where y is log[MW] in kDa, and x is the elution volume in ml. The estimated molecular weights of SCF and SCFa were calculated using the equation.
Yeast surface display and construction of first and second-generation SCF mutant libraries
Mouse and human SCF (residues 26-166) was displayed on the surface of S. cerevisiae strain EBY100 as an N-terminal fusion to Aga2 using the pYAL vector (Birnbaum et al., 2014).
Based on the crystal structure of the wild-type mouse c-Kit/SCF complex (PDB ID = 2O26) (Liu et al., 2007), we designed a first-generation ‘contact’ library that randomized the c-Kit-contact residues using degenerate codons. The contact residues and the respective degenerate codons used include, Gly5=RST (Gly, Ser, Thr, or Ala), Asn6=VAW (Asn, Asp, Gln, Glu, His, or Lys), Thr9=RMA (Thr, Asn, Asp, or Ala), Asp=VAW, Ile50=RKA (Ile, Arg, Val, or Gly), Leu54=NTT (Leu, Val, Ile or Phe), Thr57=RMA, Leu60=MKT (Leu, Ile, Arg or Ser), Asp61=VAW, Asp77=VAW, Lys81=ANA (Lys, Ile, Arg or Thr), Asp84=VAW, Asp85=VAW, Val87=NTT, Leu88=YWT (Leu, Phe, Tyr or His), Asn97=VAW, Ile98=MKT, Lys99=MAW (Lys, His, Asn, or Gln), Glu100=VAW, Arg104=CNT (Arg, His, Pro, or Leu). Electroporation, rescue and expansion of the yeast library were performed as described previously (Boder and Wittrup, 1997). Final library contained approximately 2 × 108 yeast transformants.
To obtain SCF variants with further enhanced affinity after the first-generation selections and evolution (see below), we designed a second-generation ‘affinity maturation’ library. In this library, the consensus mutations from the first-generation selections were retained using regular codons or codons with limited degeneracy: Asn6=GAT (Asp), Thr57=GAA (Glu), Asp77=CAT (His), Lys81=ATT (Ile), Val87=WTT (Phe or Ile), Leu88=TWT (Phe or Tyr). The positions that were diversified in the first-generation library but did not converge on a consensus amino acid residue were further diversified to all 20 amino acids using the NNK degenerate codon to interrogate a broader chemical space. These positions were Asp10, Leu54, Asp61, Asn97, Ile98, Lys99, Glu100, and Arg104.
Selections of the first-generation ‘contact’ library
Library selection was conducted as described previously with some modifications (Weiskopf et al., 2013). Briefly, the initial selections (rounds 1–3) were conducted using magnetic activated cell sorting (MACS). For round 1, 1.0 × 109 cells were selected with paramagnetic streptavidin microbeads (Miltenyi) that were pre-coated with 400 nM biotinylated mKitD1-3. For rounds 2 and 3, 1.0 × 108 yeast were stained and selected with 360 and 10 nM monomeric biotinylated mKitD1-3, respectively. To normalize apparent affinity by protein expression on the cell surface, library selection was performed using two-color fluorescence activated cell sorting (FACS) in the subsequent rounds. Yeast library was incubated with the target concentration of the bait (i.e. mKitD1-3) for each subsequent round, washed twice with PBE (PBS, pH 7.4 + 0.5% (w/v) BSA + 2 mM EDTA, pH 8.0) and co-labeled with streptavidin-Alexa fluor 647 (mKitD1-3 binding) and anti-c-Myc-Alexa Fluor 488 (SCF variant cell surface expression) for 10 min at 4 °C. Alexa647+Alexa488+ yeast were purified using a FACS Jazz cell (BD Biosciences) sorter using the gating strategy that has been previously described (Boder and Wittrup, 1997). For rounds 4 and 5, the yeast library was stained using 10 and 1 nM monomeric biotinylated mKitD1-3 for 1 h at 4 °C, respectively. At the conclusion of the selections, post-selection library at each round was simultaneously expanded and stained with 1 nM biotinylated mKitD1-3 followed by fluorescently labeled streptavidin to assess the enrichment of high-affinity clones via flow cytometry. In addition, 100 μl of post-round 5 ‘contact’ library was used to extract library DNA using the Zymoprep Yeast Plasmid Miniprep II Kit (Zymo Research), according to the manufacturer’s instructions. The extracted DNA was transformed into DH5α E. coli and plated to sequence the individual clones.
Selections of the second-generation affinity maturation library
The initial selection was performed exactly as the first round of selection of the first-generation contact library. For the second round of selection, yeast were selected with 100 nM monomeric biotinylated mKitD1-3 using the same procedure as in rounds 2 and 3 of the first-generation selections. For the third round of selection, the library was stained with 1 nM monomeric biotinylated mKitD1-3, and two-color FACS sorting was performed as described above. Kinetic selection was used for the final round of selection. Briefly, the library was stained with 100 nM monomeric biotinylated mKitD1-3 at room temperature (rt) for 1 h, washed with PBE, then incubated with 1 μM monomeric non-biotinylated mKitD1-3 at rt for 1 h. After washing twice with ice-cold PBE, the library was stained and FACS sorted as in round 3. All post-selection library at each round was expanded, stained with 100 nM biotinylated mKitD1-3 at rt for 1 h followed by competition with 1 μM monomeric non-biotinylated mKitD1-3 at rt for 1 h and assessed for binding via flow cytometry. Post-round 4 ‘affinity maturation’ library was sequenced as described above.
Surface plasmon resonance
Binding affinity and kinetics were measured with a BIAcore T100 (GE Healthcare). Protein concentrations were quantified by 280 nM absorbance with a Nanodrop2000 spectrometer (Thermo Scientific). A BIAcore SA sensor chip (GE Healthcare) was used to capture biotinylated mKitD1-3 and hKitD1-3 on different flow channels with a Rmax of ~ 1000 RU and ~100 RU, respectively. An unrelated biotinylated protein was immobilized with a similar RU values to control for nonspecific binding. Experiments were carried out at 25 °C, and measurements were made with serial dilutions of SCF variants in HBS-P+ buffer (GE Healthcare) supplemented with 0.1% (w/v) BSA. The SA sensor chip is regenerated with 2 mM MgCl2 after each binding cycle. All data were analyzed with the BIAcore T100 evaluation software version 2.0 with a 1:1 Langmuir binding model.
On-yeast competitive binding experiments
Biotinylated mKitD1-3 and hKitD1-3 was incubated with Alexa Fluor 647-conjugated streptavidin (produced in house) for 15 min at rt to form c-Kit tetramers. Labeled mouse and human c-Kit tetramers at 20 nM were combined with serial dilutions of soluble mSCF variants and simulatenously added to stain mSCF- and hSCF-expressing yeast, respectively. Yeast were incubated for 1 h at 4 °C then washed with PBE to remove unbound proteins. Samples were analyzed on an Accuri C6 flow cytometer (BD Biosciences). Data represent the mean fluorescence intensity normalized to maximal binding for each reagent, and points were fit to sigmoidal dose-response curves using Prism 6 (GraphPad).
Cell surface c-Kit internalization experiment
MC/9 mast cells were stimulated with 1 μM of SCF variants at 37 °C for 1, 5, 15, 30, 60 and 120 min. c-Kit internalization was terminated by placing the cells on ice. MC/9 cells were stained with a non-blocking anti-c-Kit antibody APC (clone: 2B8) (eBioscience) for 1 h at 4 °C and then washed twice to remove unbound antibodies. Cell surface c-Kit expression was analyzed using an Accuri C6 flow cytometer (BD Biosciences). Non-competitive binding of 2B8-APC to c-Kit was confirmed by staining unstimulated MC/9 cells with 2B8-APC in the presence and absence of 1 μM unlabeled wild-type SCF or high-affinity S4-3 dimers at 4 °C (Data not shown).
Phosphoflow signaling experiments
MC/9 mast cells were starved in serum free medium for 6–12 h at 37 °C prior to simulation. For signaling time course, MC/9 cells were stimulated with 1 μM of SCF variants at 37 °C for 1, 3, 5, 7.5, 10, 12.5, 15 and 30 min. For dose response, MC/9 cells were stimulated with titrating amounts of SCF variants for 5 min at 37 °C. Subsequently, the cells were fixed with 1:5 dilution of 16% paraformaldehyde (Electron Microscopy Sciences) with shaking for 15 min at rt, washed with PBE and permeabilized with cold 100% methanol overnight at −80 °C. Samples were washed again and stained with pERK-A647 (Cell Signaling, clone: 197G2) and p-AKT-A488 (BD Biosciences, clone: M89-61) in PBE for 1 h at 4 °C and then washed to remove unbound antibodies. An Accuri C6 flow cytometer (BD Biosciences) was used for analysis. Data represent the mean fluorescence intensity, and points were fit to sigmoidal dose-response curves using Prism 6 (GraphPad).
For Primity Bio Pathway, MC/9 cells were starved for 4 h and stimulated with 1 μM of either wild-type SCF or S4-3a for 5 and 15 min, then fixed and permeabilized as described above. The cells stained with a panel of phosphorylation specific antibodies from Primity Bio Pathway Phenotyping service and Table S1) (Krutzik and Nolan, 2003). Signaling profiles were analyzed with an LSRII cell analyzer with high throughput sampler (BD Biosciences). The Log2 ratio of the median fluorescence intensities (MFI) of the stimulated samples divided by the unstimulated control samples were calculated as a measure of response.
Analysis of cell surface c-Kit expression on primary mouse HSPCs and mast cells
Primary mouse Lin− bone marrow cells and peritoneal cells were obtained and separately stained for Lin−Sca-1+c-Kit+ bone marrow HSPCs and FcεRIα+c-Kit+ peritoneal mast cells using the antibodies described above. c-Kit expression was analyzed with an LSRII cell analyzer (BD Biosciences) at the Stanford Shared FACS Facility. Data was analyzed using FlowJo (Treestar).
Single molecule imaging and evaluation
Single molecule imaging experiments were conducted by total internal reflection fluorescence (TIRF) microscopy with an inverted microscope (Olympus IX71) equipped with a triple-line total internal reflection (TIR) illumination condenser (Olympus) and a back-illuminated electron multiplied (EM) CCD camera (iXon DU897D, Andor Technology) as described in more detail previously (Moraga et al., 2015b; Wilmes et al., 2015). A 150 × magnification objective with a numerical aperture of 1.45 (UAPO 150 ×/1.45 TIRFM, Olympus) was used for TIR illumination of the sample. All experiments were carried out at room temperature in medium without phenol red, supplemented with an oxygen scavenger and a redox-active photoprotectant to minimize photobleaching (Vogelsang et al., 2008). For cell surface labeling of mXFP-mKit, DY647- and Rho11-labeled NBs were added to the medium at equal concentrations (2 nM each) and incubated for at least 5 min. The labeled NBs were kept in the bulk solution during the whole experiment in order to ensure high equilibrium binding to mXFP-mKit. Under these conditions, an effective labeling efficiency with respect to single molecule co-localization of ~60% of mXGP-tagged cell surface proteins is achieved. Negative and positive control proteins for co-localization (mEGFP-TMD and (mEGFP)2-TMD) were labelled under the same conditions. Dimerization of mKit was probed before and after incubation with variable concentrations of SCF and S3-4a, respectively. Rho11 was excited by a 561 nm laser at 0.95 mW (~32 W/cm2) and DY647 by a 642 nm laser at 0.65 mW (~22 W/cm2). Fluorescence was detected using an spectral image splitter (DualView, Optical Insight) with a 640 DCXR dichroic beam splitter (Chroma) combined with the bandpass filter 585/40 (Semrock) for detection of RHO11 and 690/70 (Chroma) for detection of DY647 dividing each emission channel into 512×256 pixel. Image stacks of 150 frames were recorded at a time resolution of 32 ms/frame for each cell.
Single molecule localization was carried out using the multiple-target tracing (MTT) algorithm (Serge et al., 2008) and tracking was performed according to (Jaqaman et al., 2008). Immobile molecules were identified by spatiotemporal cluster analysis (Roder et al., 2014) and removed from the dataset (typically <15% of all localizations) prior to quantifying diffusion and dimerization because this fraction is biased by (i) labeled NBs non-specifically adsorbed onto the coverslide surface and (ii) endosomes located close to the plasma membrane. Receptor dimerization was quantified based on sequential co-localization and co-tracking analysis as described in detail recently (Wilmes et al., 2015). After aligning Rho11 and DY647 channels with sub-pixel precision by using a spatial transformation based on a calibration measurement with multicolor fluorescent beads (TetraSpeck microspheres 0.1 μm, Invitrogen), individual molecules detected in the both spectral channels of the same frame within a distance threshold of 100 nm were considered co-localized. For single molecule co-tracking analysis, the MTT algorithm was applied to this dataset of co-localized molecules to reconstruct co-locomotion trajectories (co-trajectories) from the identified population of co-localizations. For the co-tracking analysis, only trajectories with a minimum of 10 consecutive steps (~300 ms) were considered. The relative fraction of co-tracked molecules was determined with respect to the absolute number of trajectories from both channels and corrected for dimers stochastically double-labeled with the same fluorophore species as follows:
where A, B and AB are the numbers of trajectories observed for Rho11, DY647 and co-trajectories, respectively.
Proliferation of HSPCs ex vivo
5000 mouse LSK HSPCs from GFP+ C57B/6J mice were plated on irradiated (2000 Rads) OP9 stroma (ATCC) in Alpha MEM media, supplemented with 20% FBS, 1% Pen Strep Glut (Gibco), and either SCF or S4-3a at 5000, 500, 50 and 5 ng/ml. Total numbers of cells were counted using a Countess (Invirogen) after 6 days in culture.
Ex vivo colony formation assay
500 sorted LSK HSPC from C57B/6J mice were re-suspended in 100 μL PBS and subsequently mixed with 1 mL of Methocult basic medium (Stem cell technology) containing 100 ng/ml recombinant mouse IL-3 (Peptrotech) without or with the addition of 100 ng/ml or 5000 ng/ml of SCF or S4-3a. The mixture of cell and methylcellulose was subsequently vortexed for 30 sec, and plated on cell culture dishes (35 mm X 10 mm) using a 16G blunt needle and a 1-ml syringe 20–30 min after incubating at room temperature. Cells were cultured in a CO2 tissue culture incubator at 37 °C for 7 days in triplicates. Two independent experiments were performed. Colonies for granulocyte and macrophage progenitors (CFU-GM) were counted under the microscope (Nikon Eclipse TS100). The numbers of colonies were normalized to the highest colony count in 5000 ng/ml SCF group in each experimental replicate. Data from two experimental replicates were then pooled and plotted using Prism 6 (Graphpad).
In vitro IL-6 release by BMCMCs
C57BL/6J mouse BMCMCs were washed with DMEM, resuspended at 2 × 106 cells/ml and starved for 3 h in DMEM containing 10% FCS, 50 μM β-mercaptoethanol, 2 mM L-glutamine, and 1% antibiotic-antimycotic solution. BMCMCs were subsequently aliquoted and stimulated by addition of an equivalent volume of 2X concentrated stimulants in DMEM (resulting in a final density of 106 cells/ml). After 6 h of stimulation at 37 °C, cells were centrifuged and the supernatant was harvested and stored at −20 °C until analysis. IL-6 content in the supernatant was determined using the OptEIA mouse IL-6 ELISA Set (BD Biosciences) according to the manufacturer’s instructions.
In vitro β-hexosaminidase release by human PBCMCs
Human PBCMCs (105 cells/sample) were pre-sensitized with 1 μg/ml human IgE (Millipore) overnight, washed and stimulated with rabbit anti-human IgE (Bethyl), SCF variants or a combination of anti-IgE and SCF variants in 100 μl Tyrode’s buffer for 30 minutes. The concentrations of anti-IgE tested were 2000, 200, 20, or 2 ng/ml while those of SCF variants were 1000, 100, 10, or 1 ng/ml. For the combination stimulation, IgE pre-sensitized PBCMCs were treated with 1000, 100, 10 or 1 ng/ml of SCF variants in the presence of 2 ng/ml anti-IgE. The cells were then solubilized with 0.5% Triton X-100 in Tyrode’s buffer. The amount of β-hexosaminidase in supernatants and in the insoluble cell pellets were measured with p-nitrophenyl N-acetyl-β-D-glucosaminide in 0.1 M sodium citrate (pH 4.5). The reaction was stopped by adding 0.2 M glycine (pH 10.7). The release of the product 4-p-nitrophenol was detected at 405 nm. The extent of degranulation (i.e. β-hexosaminidase release) was calculated as the percentage of 4-p-nitrophenol absorbance in the supernatants, over the sum of absorbance in the supernatants and in cell pellets (total β-hexosaminidase) solubilized in detergent.
Acceleration of hematopoietic recovery after sublethal radiation damage
Eight- to 10-week old female C57BL/6J mice, weighing approximately 20 g, were injected intraperitoneally (i.p.) with PBS, SCF or S4-3a daily starting 2 days prior to sublethal total body irradiation (TBI) and continued until 7 days post-TBI, for a total of 10 days. The protein treatments were administered at 250 μg/kg. Sublethal dose of TBI at 450 Rads was given at Day 0, and complete blood count (CBC) was performed on Day 0, 7, 11, 16, 21 and 40 using Hematrue hematology analyzer (Heska). Peripheral blood samples was collected from the tail vein of mice.
Prevention of radiation-induced mortality
Eight- to 10-week old female C57BL/6J mice, weighing approximately 20 g, were given 1150 Rads of lethal TBI in split dose of equal intensity (i.e. 575 Rads x 2). Twenty, 10 and 2 h prior to the first dose, and 2 and 4 h after the second dose of TBI, mice were injected i.p. with PBS or 5 mg/kg of either SCF or S4-3a. A higher dose and slightly more frequent dosing of our non-PEGylated SCF variants were used to compensate for shorter serum half-life compared to the PEGylated SCF used in the previous study (Zsebo et al., 1992). Survival was monitored daily for 50 days.
Assessment of systemic anaphylaxis and characterization of peritoneal mast cells
C57BL/6 wild-type mice, mast cell/basophil-deficient C57BL/6 Cpa3-Cre+; Mcl-1fl/fl or C57BL/6 Cpa3-Cre+; Mcl-1+/+ mast cell/basophil-sufficient control mice, or C57BL/6 Mcpt8-Cre+/−; DTAfl/− basophil-deficient (Wada et al., 2010) or C57BL/6 Mcpt8-Cre+/−; DTA−/− littermate basophil-sufficient control mice (Lilla et al., 2011) were i.p. injected with either PBS, 5 or 10 mg/kg μg of SCF or S4-3a. Rectal temperature was measured at the indicated time points. One-minute movies of C57BL/6J mice were taken approximately 20 min after injection. The videos were sped up by 2-fold using iMovies. To track mouse movement and determine displacement, the videos were converted into a sequence of still images using MPEG Streamclip (downloaded from http://www.squared5.com) at 8 frames/s and analyzed using the TrackMate plugin for ImageJ. For analysis of c-Kit expression levels on peritoneal mast cells of C57BL/6J mice, the animals were sacrificed 1 h after injections and a peritoneal lavage with 5 ml of ice-cold PBS was performed to harvest the peritoneal cells as previously described (Gaudenzio et al., 2015). The peritoneal cell suspension was centrifuged, resuspended in 1 ml PBS 1% BSA and 200 μl were subsequently stained with 1:200 anti-c-Kit and anti-FcεRIα antibodies on ice for 15 min, analyzed by flow cytometry on an Accuri C6 flow cytometer (BD Biosciences).
Histological analysis of peritoneal mast cells was performed based on a published protocol (Gaudenzio et al., 2015). Briefly, 1 ml of peritoneal cell suspension was centrifuged and the pellet resuspended in 100 μl PBS 1% BSA and spun onto microscopy slides using a Cytospin system (Shandon). After air-drying for 30 min at rt, the slides were immersed 5 min in undiluted May-Grünwald stain (Sigma), briefly rinsed in PBS and immediately stained in 5% (diluted in tap water) modified Giemsa stain (Sigma) for 20 min. After a brief rinse in tap water, slides were allowed to air-dry and mounted using Permount media (Fisher Scientific). Images were captured with a Nikon E1000M microscope equipped with a Spotflex camera and Spot software version 5.1 (Diagnostic Instruments). For quantification of granule+ peritoneal cells (indicated by red arrowheads in Figure S6B) that were clearly distinguishable from non-degranulated mast cells (indicated by black arrows in Figure S6B), field of views were randomly selected and the fractions of granule+ peritoneal cells among at least 100 cells/sample was determined.
Assessment of local cutaneous anaphylaxis
Female and male C57BL/6J wild-type mice were anesthetized with isoflurane and injected with 250 μl sterile-filtered 1% Evans blue (Sigma) in PBS into the retro-orbital vein and allowed to recover. Fifteen min later, mice were anesthetized again and (after measurement of initial ear thickness using a gauge [Peacock]) intradermally injected into the ear pinnae with 25 μl of PBS alone (into the left ear pinna) or containing 50 μg/ml SCF or S4-3a (into the right ear pinna) using a 1 ml syringe equipped with a 30 G x 1/2 needle. Two h after injections, mice were anesthetized again and ear pinnae thickness was measured and photographs were taken. Three h after injections, mice were euthanized, ear pinnae were harvested, finely chopped and incubated in 300 μl Dimethyl sulfoxide (DMSO) in a 48-well tissue culture plate (Corning) for 6 h on a shaker at rt. Evans blue containing supernatant was collected, transferred into 96-well flat-bottom ELISA plate (Thermo Scientific) and absorbance at 620 nm wavelength was measured using a plate reader (Molecular Devices). Evans blue signal of supernatant extracted from PBS-injected ears was considered background and subtracted from signals obtained from the respective other ear injected with either SCF or S4-3a.
Quantification and Statistical Analysis
Statistical parameters including the number of n, definition of center and spread (mean ± SD or mean ± SEM), the type of statistical test and correction, and statistical significance are reported in the Figures and Figure Legends. Briefly, data are judged to be significant when p < 0.05 by unpaired, two-tailed Student’s t test or Log-rank Mantel-Cox test. We used the Holm-Sidak method to correct for multiple comparisons. We denote statistical significance as follows: ns, not significant (i.e. p > 0.05); *, p < 0.05; **, p <0.01; ***, p < 0.001. Graphs and statistical analyses except for Figure 4 and S3 were generated using Prism 6 (Graphpad). Figure 4 and S3 were generated using OriginPro 8G (OriginLab), and the box plots indicate the data distribution of the second and third quartile (box), median (line), mean (filled squares), and 1.5X interquartile range (whiskers).
KEY RESOURCES TABLE
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-mouse CD117 conjugated to APC-Cy7 (clone 2B8) | eBioscience | Cat#47-1171-82 |
| Anti-mouse CD117 conjugated to APC (clone 2B8) | eBioscience | Cat#17-1171-82 |
| Anti-mouse CD11b conjugated to PE (clone M1/70) | eBioscience | Cat#12-0112-83 |
| Anti-mouse CD3 conjugated to PE (clone 145-2C11) | eBioscience | Cat#12-0031-85 |
| Anti-mouse CD4 conjugated to PE (clone RM4-5) | eBioscience | Cat#12-0042-85 |
| Anti-human/mouse CD45R conjugated to PE (clone RA3-6B2) | eBioscience | Cat#12-0452-83 |
| Anti-mouse CD8a conjugated to PE (clone 53-6.7) | eBioscience | Cat#12-0081-85 |
| Anti-mouse Ly-6G conjugated to PE (clone RB6-8C5) | eBioscience | Cat#12-5931-83 |
| Anti-mouse TER-119 conjugated to PE (clone TER-119) | eBioscience | Cat#12-5921-83 |
| Anti-mouse Ly-6A/E (Sca-1) conjugated to PE-Cy7 (clone D7) | eBioscience | Cat#25-5981-82 |
| Anti-c-Myc-Alexa Fluor 488 | Cell Signaling Technology | Cat#2279S |
| Anti-AKT (pS473) conjugated to Alexa Fluor 488 (clone M89-61) | BD Biosciences | Cat#560404 |
| Anti-P-p38 (T180/Y182) conjugated to Alexa Fluor 647 (clone 28B10) | Cell Signaling Technology | Cat#4552S |
| Anti-pERK1/2 (T202/Y204) conjugated to PerCP-eFlour 710 (clone MILAN8R) | eBioscience | Cat#46-9109-42 |
| Anti-mouse FcεRIα conjugated to Pacific Blue (clone MAR-1) | BioLegend | Cat#134313 |
| Anti-mouse CD49b conjugated to FITC (clone DX5) | BioLegend | Cat#108906 |
| Anti-human IgE polyclonal | Bethyl Laboratories | Cat#A80-109A |
| Biological Samples | ||
| Human PBMCs | Stanford Blood Center | N/A |
| Mouse C57BL/6J bone marrow cells | This paper | N/A |
| Mouse C57BL/6J peritoneal cells | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Recombinant Mouse c-Kit extracellular domains 1-3 protein | This paper | N/A |
| Recombinant Human c-Kit extracellular domains 1-3 protein | This paper | N/A |
| Recombinant Mouse c-Kit extracellular domains 1-3- Biotin protein | This paper | N/A |
| Recombinant Human c-Kit extracellular domains 1-3-Biotin protein | This paper | N/A |
| Recombinant Mouse SCF: wild-type protein | This paper | N/A |
| Recombinant Human SCF: wild-type protein | This paper | N/A |
| Recombinant Mouse SCF: SCFa (with wild-type c-Kit interface and F63A monomer mutation) protein | This paper | N/A |
| Recombinant Mouse SCF: S4-3 (with high-affinity c-Kit interface) protein | This paper | N/A |
| Recombinant Mouse SCF: S4-3a (with high-affinity c-Kit interface and F63A monomer mutation) protein | This paper | N/A |
| Recombinant streptavidin conjugated to Alexa 647 | In house | N/A |
| Recombinant Human IL-6 | Peprotech | Cat#200-06 |
| Recombinant Human IL-3 | Peprotech | Cat#200-03 |
| Recombinant Murine IL-3 | Peprotech | Cat#213-13 |
| Recombinant Human IgE | Millipore | Cat#AG30P |
| Recombinant anti-GFP nanobody | (Kirchhofer et al., 2010) | N/A |
| DY647 maleimide | Dyomics | Cat#647-03 |
| Rho11-maleimide | ATTO-TEC | Cat#AD Rho11-41 |
| Isopropyl β-D-1-thiogalactopyranoside (IPTG) | Research Products International Corp. | Cat#I56000-100.0 |
| Urea | Fisher Scientific | Cat#BP169-212 |
| Sodium acetate | Fisher Scientific | Cat#BP333-1 |
| Ethylenediamine tetraacetic acid (EDTA) | Fisher Scientific | Cat#BP120-500 |
| Dithiothreitol (DTT) | Sigma-Aldrich | Cat#D0632-25G |
| Tris-HCl | Fisher Scientific | Cat#BP153-1 |
| Oxidized glutathione | Sigma-Aldrich | Cat#G4376-10G |
| Reduced glutathione | Sigma-Aldrich | Cat#G4251-50G |
| Phenylmethanesulfonylfluoride (PMSF) | Sigma-Aldrich | Cat#P7626-1G |
| Ammonium sulfate | Sigma-Aldrich | Cat#A4915-5KG |
| Sodium chloride | Fisher Scientific | Cat#S271-3 |
| Triton X-114 | Sigma_Aldrich | Cat#X114-100ML |
| Triton X-100 | Sigma-Aldrich | Cat#T9284-500ML |
| L-Glutamine | Invitrogen | Cat#25030081 |
| 2-mercaptoethanol | Invitrogen | Cat#21985023 |
| Rat T-STIM | BD Biosciences | Cat#354115 |
| Methanol | Fisher Scientific | Cat#A412P-4 |
| Paraformaldehyde | Electron Microscopy Sciences | Cat#15710 |
| Sodium pyruvate | Invitrogen | Cat#11360070 |
| Insulin-transferrin selenium | Invitrogen | Cat#41400045 |
| Ciprofloxacin | Sigma-Aldrich | Cat#17850-5G-F |
| P-nitrophenyl N-acetyl-β-D-glucosaminide | Sigma-Aldrich | Cat#N9376-100MG |
| May-Grünwald stain | Sigma-Aldrich | Cat#MG500-500ML |
| Modified Giemsa stain | Sigma-Aldrich | Cat#GS500-500ML |
| Evans blue dye | Sigma-Aldrich | Cat#E2129 |
| Toluidine Blue O | Fisher Scientific | Cat#T161-25 |
| Dimethyl sulfoxide (DMSO) | Sigma-Aldrich | Cat#D8418-50ML |
| Methocult basic medium | STEMCELL Technology | Cat#M3231 |
| Antibiotic-Antimycotic | Invitrogen | Cat#15240062 |
| Critical Commercial Assays | ||
| OptEIA™ mouse IL-6 ELISA Set | BD Biosciences | Cat#555240 |
| Zymoprep™ Yeast Plasmid Miniprep II Kit | Zymo Research | Cat#D2004 |
| Primity Bio Pathway Assay | Primity Bio | N/A |
| Lineage Cell Depletion Kit | Miltenyi Biotech | Cat#130-090-858 |
| ToxinSensor™ Chromogenic LAL Endotoxin Assay Kit | Genescript | Cat#L00350 |
| EasySep™ Human CD34 Positive Selection Kit | STEMCELL Technologies | Cat#18056 |
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Bacteria: E. coli BL21 (DE3) | Novagen | Cat#69450 |
| Bacteria: E. coli Rosetta | In house | |
| Bacteria: E. coli DH5α | In house | N/A |
| Yeast: S. cerevisiae EBY100 | ATCC | Cat#MYA-4941 |
| Yeast: S. cerevisiae EBY100 expressing mouse SCF | This paper | N/A |
| Yeast: S. cerevisiae EBY100 expressing human SCF | This paper | N/A |
| Insect: Trichoplusia ni (High Five) | Invitrogen | Cat#B85502 |
| Mouse: MC/9 | ATCC | Cat#CRL-8306 |
| Mouse: Bone marrow-cultured mast cells (BMCMC) | Kalesnikoff and Galli, 2011 | N/A |
| Mouse: OP9 | ATCC | Cat#CRL-2749 |
| Hamster: CHO transfectants secreting mouse SCF | Gift from Dr. P. Dubreuil, Marseille, France. | N/A |
| Human: Peripheral blood-cultured mast cells (PBCMC) | Gaudenzio et al., 2013; Joulia et al., 2015 | N/A |
| Human: HeLa | ATCC | Cat#CCL-2 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | Cat#000664 |
| Mouse: C57BL/6J Cpa3-Cre+;Mcl-l+/+ | Lilla et al., 2011 | N/A |
| Mouse: C57BL/6J Cpa3-Cre+;Mcl-lfl/fl | Lilla et al., 2011 | N/A |
| Mouse: C57BL/6J Mcpt8-Cre+/−;DTA−/− | Wada et al., 2010 | N/A |
| Mouse: C57BL/6J Mcpt8-Cre+/−;DTAfl/− | Wada et al., 2010 | N/A |
| Recombinant DNA | ||
| pAcGP67a | BD Biosciences | Cat#554756 |
| pAcGP67a_mKitD1-3 | This paper | N/A |
| pAcGP67a_hKitD1-3 | This paper | N/A |
| pAcGP67a_mKitD1-3-GLNDIFEAQKIEWHE | This paper | N/A |
| pAcGP67a_hKitD1-3-GLNDIFEAQKIEWHE | This paper | N/A |
| pET28a | Novagen | Cat#69864-3 |
| pET28a_mSCF-WT | This paper | N/A |
| pET28a_mSCF-F63A (SCFa) | This paper | N/A |
| pET28a_mSCF-S4-3 | This paper | N/A |
| pET28a_mSCF-S4-3-F63A (S4-3a) | This paper | N/A |
| pET21a_anti-GFP nanobody | Kirchhofer et al., 2010 | N/A |
| pYAL | Birnbaum et al., 2014 | N/A |
| pYAL_mSCF | This paper | N/A |
| pYAL_hSCF | This paper | N/A |
| pSems-26m | Covalys Biosciences | N/A |
| pSems_mXFP-mKit | This paper | N/A |
| pSems_mEGFP | This paper | N/A |
| pSems_tandem-mEGFP | This paper | N/A |
| Sequence-Based Reagents | ||
| Software and Algorithms | ||
| UNICORN™ 5.11 software | GE Healthcare | |
| Prism 6 | Graphpad | |
| BIAcore T100 evaluation software version 2.0 | GE Healthcare | |
| FlowJo v10 software | Tree Star Inc | |
| Spot software version 5.1 | Diagnostic Instruments | |
| iQ Live Cell Imaging Software | Andor Technologies | |
| MPEG Streamclip | http://www.squared5.com | |
| ImageJ | http://rsb.info.nih.gov/ij/index.html | |
| Other | ||
The table highlights the genetically modified organisms and strains, cell lines, reagents, software, and source data essential to reproduce results presented in the manuscript. Depending on the nature of the study, this may include standard laboratory materials (i.e., food chow for metabolism studies), but the Table is not meant to be comprehensive list of all materials and resources used (e.g., essential chemicals such as SDS, sucrose, or standard culture media don’t need to be listed in the Table). Items in the Table must also be reported in the Method Details section within the context of their use. The number of primers and RNA sequences that may be listed in the Table is restricted to no more than ten each. If there are more than ten primers or RNA sequences to report, please provide this information as a supplementary document and reference this file (e.g., See Table S1 for XX) in the Key Resources Table.
Please note that ALL references cited in the Key Resources Table must be included in the References list. Please report the information as follows:
REAGENT or RESOURCE: Provide full descriptive name of the item so that it can be identified and linked with its description in the manuscript (e.g., provide version number for software, host source for antibody, strain name). In the Experimental Models section, please include all models used in the paper and describe each line/strain as: model organism: name used for strain/line in paper: genotype. (i.e., Mouse: OXTRfl/fl: B6.129(SJL)-Oxtrtm1.1Wsy/J). In the Biological Samples section, please list all samples obtained from commercial sources or biological repositories. Please note that software mentioned in the Methods Details or Data and Software Availability section needs to be also included in the table. See the sample Table at the end of this document for examples of how to report reagents.
SOURCE: Report the company, manufacturer, or individual that provided the item or where the item can obtained (e.g., stock center or repository). For materials distributed by Addgene, please cite the article describing the plasmid and include “Addgene” as part of the identifier. If an item is from another lab, please include the name of the principal investigator and a citation if it has been previously published. If the material is being reported for the first time in the current paper, please indicate as “this paper.” For software, please provide the company name if it is commercially available or cite the paper in which it has been initially described.
-
IDENTIFIER: Include catalog numbers (entered in the column as “Cat#” followed by the number, e.g., Cat#3879S). Where available, please include unique entities such as RRIDs, Model Organism Database numbers, accession numbers, and PDB or CAS IDs. For antibodies, if applicable and available, please also include the lot number or clone identity. For software or data resources, please include the URL where the resource can be downloaded. Please ensure accuracy of the identifiers, as they are essential for generation of hyperlinks to external sources when available. Please see the Elsevier list of Data Repositories with automated bidirectional linking for details. When listing more than one identifier for the same item, use semicolons to separate them (e.g. Cat#3879S; RRID: AB_2255011). If an identifier is not available, please enter “N/A” in the column.
A NOTE ABOUT RRIDs: We highly recommend using RRIDs as the identifier (in particular for antibodies and organisms, but also for software tools and databases). For more details on how to obtain or generate an RRID for existing or newly generated resources, please visit the RII or search for RRIDs.
Please see the sample Table at the end of this document for examples of how reagents should be cited. To see how the typeset table will appear in the PDF and online, please refer to any of the research articles published in Cell in the August 25, 2016 issue and beyond.
Please use the empty table that follows to organize the information in the sections defined by the subheading, skipping sections not relevant to your study. Please do not add subheadings. To add a row, place the cursor at the end of the row above where you would like to add the row, just outside the right border of the table. Then press the ENTER key to add the row. You do not need to delete empty rows. Each entry must be on a separate row; do not list multiple items in a single table cell.
Supplementary Material
(A–C) First-generation library design and selection. (A) First-generation library design. Left: Co-crystal structure of mouse SCF/c-Kit is shown in cartoon representation (PDB: 2O26). Amino acid positions randomized in the first generation library are highlighted and shown in sphere representation (light blue). Right: Table of randomized positions of the ‘contact’ residue library with possible amino acid variants and the corresponding degenerate DNA codons (noted in the parentheses). (B) Histogram overlays assessing 1 nM mKitD1-3 staining of the library at each round via flow cytometry. MFI = mean fluorescence intensity. (C) Summary of SCF variant sequence analysis. The position of mutated residues and their corresponding sequences in wild-type SCF are denoted in the first two columns of the table. Each column represents a unique SCF clone. Red text color indicates the consensus mutations. (D) Second-generation library design. Left: Co-crystal structure of mouse SCF/c-Kit is shown in cartoon representation (PDB: 2O26). Amino acid positions highlighted in green indicate the set of consensus mutations obtained from the first-generation selections and were not randomized. Amino acid positions highlighted in orange are residues randomized in the second-generation ‘affinity maturation’ library. Right: Table of randomized positions, possible amino acid substitutions and the corresponding degenerate DNA codons (noted in the parentheses) for the second-generation library. (E) Chromatograms of purified SCF variants over a Superdex-75 size exclusion column with the retention time denoted on the top of each of the main peaks. (F) Purified SCF variants resolved on a 12% SDS-PAGE gel under reducing conditions.
(A) Representative SPR sensorgrams of indicated monomeric SCF variants binding to immobilized human c-Kit domains 1-3 (hKitD1-3). (B) On-yeast competitive blocking of mouse SCF/c-Kit and human SCF/c-Kit interactions by soluble mouse SCF variants. Yeast expressing wild-type mSCF or hSCF were stained with 20 nM fluorescently-labeled mouse or human c-KitD1-3 tetramers, respectively, in the presence of indicated unlabeled soluble mouse SCF variants. Data represent the mean ± SEM and are representative of two independent experiments. MFI = mean fluorescence intensity.
(A and B) Cell surface labeling of mXFP-mKit. (A) Density (Left) and ratio (Right) of single molecule localizations obtained after labeling cell surface mXFP-mKit by addition of anti-GPF NBs conjugated with Rho11 (red) and DY647 (blue), respectively. (B) Decay in the relative number of single molecule localizations due to photobleaching. (C and D) Diffusion properties of mXFP-mKit quantified from single molecule trajectories. (C) Step-length histogram (time-lapse: 160 ms) obtained for mXFP-mKit in absence of ligand and in presence of SCF and S4-3a, respectively. (D) Mean square displacement (MSD) analysis of mXFP-mKit diffusion properties in absence of ligand and in presence of SCF and S4-3a, respectively.
Dose response of β-hexosaminidase release by human PBCMCs treated with IgE, SCF or S4-3a at indicated concentrations (ng/ml) as single agents for 30 min ex vivo. Data represent mean ± SEM. ***p<0.001, and ns = not significant (i.e., p>0.05) by unpaired, two-tailed Student’s t test.
(A) Schematics of the experimental setup. C57BL/6 mice were injected i.p. with PBS, 5 or 10 mg/kg of SCF, or 10 mg/kg of S4-3a, and body temperatures were monitored at 10-min time intervals for 60 min. (B) Body temperature of mice treated as described in (A). Data represent mean ± SEM. *p<0.05, ***p<0.001, and ns = not significant (i.e., p>0.05) compared to the PBS-treated control group by unpaired, two-tailed Student’s t test.
(A–D) C57BL/6 mice were challenged by i.p. injection of PBS or 10 mg/kg of either SCF or S4-3a. (A) Mouse movements ~20 min after injection of PBS (left), SCF (middle) or S4-3a (right). The y- and x-axes indicate arbitrary limits of a mouse cage. Each color represents the trace of one mouse. (B–D) One h post-injection, peritoneal cells were harvested by peritoneal lavage. (B) Representative images of May-Grünwald/Giemsa-stained cytospin preparations of peritoneal cells from mice after the indicated treatments. Black arrows indicate examples of naïve (i.e., apparently non-degranulated) mast cells. Red arrowheads indicate cells with macrophage-like morphology that have taken up metachromatically-stained granules, which were presumably released upon mast cell activation and degranulation. (C) Quantification of granule+ peritoneal cells (that are not non-degranulated mast cells) from (B). (D) Flow cytometry analysis of surface expression of c-Kit on peritoneal FcεRIα+c-Kit+ mast cells. (C and D) Data are pooled from two independent experiments. ***p<0.001, and ns = not significant (i.e., p>0.05) by Student’s t test.
(A and B) Flow cytometry gating strategy to identify primary mouse (A) peritoneal FcεRIα+c-Kit+ mast cells and (B) bone marrow LSK HSPCs. (C) Flow cytometry analysis of cell surface c-Kit expression in the peritoneal mast cell and bone marrow LSK cell populations identified in (A and B).
Individual mXFP-mKit receptors expressed on HeLa cells and labelled by anti-GPF NBs conjugated with Rho11 (red channel) and DY647 (blue channel), respectively, in the absence of ligands. Frame rate: 30 Hz; playback: real time.
Dimerization of mXFP-mKit receptors expressed on HeLa cells and labelled by anti-GPF NBs conjugated with Rho11 (red channel) and DY647 (blue channel), respectively, in the presence of 100 nM SCF. Individual mKit dimers (magenta) were identified by co-tracking and the corresponding co-trajectories are depicted as yellow lines. Frame rate: 30 Hz; playback: real time.
Dimerization of mXFP-mKit receptors expressed on HeLa cells and labelled by anti-GPF NBs conjugated with Rho11 (red channel) and DY647 (blue channel), respectively, in the presence of 100 nM S4-3a. Co-trajectories are indicated as yellow lines. Frame rate: 30 Hz; playback: real time.
C57BL/6 mice were challenged by i.p. injection of saline. One-minute video was taken approximately 20 minutes post-injection and sped up 2-fold for the final 30-second clip.
C57BL/6 mice were challenged by i.p. injection of 10 mg/kg SCF. One-minute video was taken approximately 20 minutes post-injection and sped up 2-fold for the final 30-second clip.
C57BL/6 mice were challenged by i.p. injection of 10 mg/kg S4-3a. One-minute video was taken approximately 20 minutes post-injection and sped up 2-fold for the final 30-second clip.
CONTEXT and SIGNIFICANCE.
Stem cell factor (SCF) is a growth factor that acts through the c-Kit receptor tyrosine kinase to elicit hematopoietic progenitor expansion and radioprotection, but can be toxic when administered in vivo because it concurrently activates mast cells. We engineered a mechanism-based SCF partial agonist that impaired c-Kit dimerization to truncate downstream signaling amplitudes. This partial agonist retained therapeutic efficacy on the hematopoietic progenitors while exhibiting virtually no anaphylactic off-target effects on mast cells in vitro and in vivo, effectively decoupling SCF pleiotropy. The approach of biasing cell activation by tuning signaling outputs has applications to many dimeric receptor-ligand systems.
Acknowledgments
We thank D. Waghray, S. Fischer, N. Goriatcheva, T. Storm, L. Jerabek, J.P. Volkmer, J. Poyser, S. Jungers, and M. Tsai for technical assistance, discussions, and reagents. We thank Stanford Shared FACS facility and Stanford SIM1 FACS facility for access to high throughput FACS analyzers. We thank Xiaolin He for providing mouse c-Kit and SCF cDNAs. This work was supported by Siebel Stem Cell Institute (to C.C.M.H.); Max Kade Fellowship of the Max Kade Foundation and the Austrian Academy of Sciences and a Schroedinger Fellowship of the Austrian Science Fund (FWF: J3399-B21) (to P.S.); Stanford Molecular and Cellular Immunobiology NIH postdoctoral training grant (5T32 AI072905) (to J.T.S.); National Institutes of Health and National Cancer Institute (to S.J.G. and I.L.W.); California Institute for Regenerative Medicine (to J.A.S.); SFB 944 from the Deutsche Forschungsgemeinschaft (to J.P.); and Ludwig grant from Stanford University (to K.C.G.).
Footnotes
AUTHOR CONTRIBUTIONS
C.C.M.H. performed directed evolution of SCF and all on-yeast measurements. C.C.M.H. and M.G. prepared the recombinant proteins for all of the experiments. C.C.M.H. and J.T.S. measured the affinities of SCF variants via SPR. C.C.M.H., I.M., M.G. and T.S.W. conducted the signaling experiments. S.W. performed single-molecule TIRF microscopy. C.C.M.H., A.C., H.S.K. and P.J.S. conducted all experiments related to hematopoietic progenitors. C.C.M.H., P.S., N.G. and R.S. conducted all experiments related to mast cells. M.R.T. analyzed and quantified mouse movements. K.C.G. and C.C.M.H. wrote the manuscript with inputs from other authors. K.C.G., S.J.G., J.A.S., J.P., and I.L.W. supervised the research.
References
- Akahoshi M, Song CH, Piliponsky AM, Metz M, Guzzetta A, Abrink M, Schlenner SM, Feyerabend TB, Rodewald HR, Pejler G, et al. Mast cell chymase reduces the toxicity of Gila monster venom, scorpion venom, and vasoactive intestinal polypeptide in mice. The Journal of clinical investigation. 2011;121:4180–4191. doi: 10.1172/JCI46139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanasova M, Whitty A. Understanding cytokine and growth factor receptor activation mechanisms. Crit Rev Biochem Mol Biol. 2012;47:502–530. doi: 10.3109/10409238.2012.729561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldo BA. Side effects of cytokines approved for therapy. Drug Saf. 2014;37:921–943. doi: 10.1007/s40264-014-0226-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall LJ, Bradstock KF. G-CSF: From granulopoietic stimulant to bone marrow stem cell mobilizing agent. Cytokine & growth factor reviews. 2014;25:355–367. doi: 10.1016/j.cytogfr.2014.07.011. [DOI] [PubMed] [Google Scholar]
- Birnbaum ME, Mendoza JL, Sethi DK, Dong S, Glanville J, Dobbins J, Ozkan E, Davis MM, Wucherpfennig KW, Garcia KC. Deconstructing the peptide-MHC specificity of T cell recognition. Cell. 2014;157:1073–1087. doi: 10.1016/j.cell.2014.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blechman JM, Lev S, Barg J, Eisenstein M, Vaks B, Vogel Z, Givol D, Yarden Y. The fourth immunoglobulin domain of the stem cell factor receptor couples ligand binding to signal transduction. Cell. 1995;80:103–113. doi: 10.1016/0092-8674(95)90455-7. [DOI] [PubMed] [Google Scholar]
- Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- Briddell RA, Hartley CA, Smith KA, McNiece IK. Recombinant rat stem cell factor synergizes with recombinant human granulocyte colony-stimulating factor in vivo in mice to mobilize peripheral blood progenitor cells that have enhanced repopulating potential. Blood. 1993;82:1720–1723. [PubMed] [Google Scholar]
- Costa JJ, Demetri GD, Harrist TJ, Dvorak AM, Hayes DF, Merica EA, Menchaca DM, Gringeri AJ, Schwartz LB, Galli SJ. Recombinant human stem cell factor (kit ligand) promotes human mast cell and melanocyte hyperplasia and functional activation in vivo. The Journal of experimental medicine. 1996;183:2681–2686. doi: 10.1084/jem.183.6.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva MG, Pimentel P, Carvalhais A, Barbosa I, Machado A, Campilho F, Sousa SR, Miranda N, da Costa FL, Campos A, et al. Ancestim (recombinant human stem cell factor, SCF) in association with filgrastim does not enhance chemotherapy and/or growth factor-induced peripheral blood progenitor cell (PBPC) mobilization in patients with a prior insufficient PBPC collection. Bone Marrow Transplant. 2004;34:683–691. doi: 10.1038/sj.bmt.1704602. [DOI] [PubMed] [Google Scholar]
- Ding L, Saunders TL, Enikolopov G, Morrison SJ. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481:457–462. doi: 10.1038/nature10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floros T, Tarhini AA. Anticancer Cytokines: Biology and Clinical Effects of Interferon-alpha2, Interleukin (IL)-2, IL-15, IL-21, and IL-12. Seminars in oncology. 2015;42:539–548. doi: 10.1053/j.seminoncol.2015.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagari E, Tsai M, Lantz CS, Fox LG, Galli SJ. Differential release of mast cell interleukin-6 via c-kit. Blood. 1997;89:2654–2663. [PubMed] [Google Scholar]
- Galli SJ, Iemura A, Garlick DS, Gamba-Vitalo C, Zsebo KM, Andrews RG. Reversible expansion of primate mast cell populations in vivo by stem cell factor. The Journal of clinical investigation. 1993;91:148–152. doi: 10.1172/JCI116164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RV, Oliver P, Astle CM. Stem cell factor improves the repopulating ability of primitive hematopoietic stem cells after sublethal irradiation (and, to a lesser extent) after bone marrow transplantation in mice. Stem cells. 1998;16:112–119. doi: 10.1002/stem.160112. [DOI] [PubMed] [Google Scholar]
- Gaudenzio N, Sibilano R, Marichal T, Starkl P, Reber LL, Cenac N, McNeil BD, Dong X, Hernandez JD, Sagi-Eisenberg R, et al. Different activation signals induce distinct mast cell degranulation strategies. The Journal of clinical investigation. 2016;126:3981–3998. doi: 10.1172/JCI85538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudenzio N, Sibilano R, Starkl P, Tsai M, Galli SJ, Reber LL. Analyzing the Functions of Mast Cells In Vivo Using ‘Mast Cell Knock-in’ Mice. Journal of visualized experiments: JoVE. 2015 doi: 10.3791/52753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilfillan AM, Peavy RD, Metcalfe DD. Amplification mechanisms for the enhancement of antigen-mediated mast cell activation. Immunologic research. 2009;43:15–24. doi: 10.1007/s12026-008-8046-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudkov AV, Komarova EA. The role of p53 in determining sensitivity to radiotherapy. Nat Rev Cancer. 2003;3:117–129. doi: 10.1038/nrc992. [DOI] [PubMed] [Google Scholar]
- Hart Y, Reich-Zeliger S, Antebi YE, Zaretsky I, Mayo AE, Alon U, Friedman N. Paradoxical signaling by a secreted molecule leads to homeostasis of cell levels. Cell. 2014;158:1022–1032. doi: 10.1016/j.cell.2014.07.033. [DOI] [PubMed] [Google Scholar]
- Hill PB, MacDonald AJ, Thornton EM, Newlands GF, Galli SJ, Miller HR. Stem cell factor enhances immunoglobulin E-dependent mediator release from cultured rat bone marrow-derived mast cells: activation of previously unresponsive cells demonstrated by a novel ELISPOT assay. Immunology. 1996;87:326–333. doi: 10.1046/j.1365-2567.1996.455545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YR, Wu GM, Mendiaz EA, Syed R, Wypych J, Toso R, Mann MB, Boone TC, Narhi LO, Lu HS, et al. The majority of stem cell factor exists as monomer under physiological conditions. Implications for dimerization mediating biological activity. The Journal of biological chemistry. 1997;272:6406–6415. doi: 10.1074/jbc.272.10.6406. [DOI] [PubMed] [Google Scholar]
- Jaqaman K, Loerke D, Mettlen M, Kuwata H, Grinstein S, Schmid SL, Danuser G. Robust single-particle tracking in live-cell time-lapse sequences. Nature methods. 2008;5:695–702. doi: 10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Gurel O, Mendiaz EA, Stearns GW, Clogston CL, Lu HS, Osslund TD, Syed RS, Langley KE, Hendrickson WA. Structure of the active core of human stem cell factor and analysis of binding to its receptor kit. The EMBO journal. 2000;19:3192–3203. doi: 10.1093/emboj/19.13.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalesnikoff J, Galli SJ. Antiinflammatory and immunosuppressive functions of mast cells. Methods in molecular biology. 2011;677:207–220. doi: 10.1007/978-1-60761-869-0_15. [DOI] [PubMed] [Google Scholar]
- Kirchhofer A, Helma J, Schmidthals K, Frauer C, Cui S, Karcher A, Pellis M, Muyldermans S, Casas-Delucchi CS, Cardoso MC, et al. Modulation of protein properties in living cells using nanobodies. Nature structural & molecular biology. 2010;17:133–138. doi: 10.1038/nsmb.1727. [DOI] [PubMed] [Google Scholar]
- Klemm JD, Schreiber SL, Crabtree GR. Dimerization as a regulatory mechanism in signal transduction. Annu Rev Immunol. 1998;16:569–592. doi: 10.1146/annurev.immunol.16.1.569. [DOI] [PubMed] [Google Scholar]
- Krieg C, Letourneau S, Pantaleo G, Boyman O. Improved IL-2 immunotherapy by selective stimulation of IL-2 receptors on lymphocytes and endothelial cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11906–11911. doi: 10.1073/pnas.1002569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzik PO, Nolan GP. Intracellular phospho-protein staining techniques for flow cytometry: monitoring single cell signaling events. Cytometry A. 2003;55:61–70. doi: 10.1002/cyto.a.10072. [DOI] [PubMed] [Google Scholar]
- Leibowitz BJ, Wei L, Zhang L, Ping X, Epperly M, Greenberger J, Cheng T, Yu J. Ionizing irradiation induces acute haematopoietic syndrome and gastrointestinal syndrome independently in mice. Nature communications. 2014;5:3494. doi: 10.1038/ncomms4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh BR, Khan W, Hancock SL, Knox SJ. Stem cell factor enhances the survival of murine intestinal stem cells after photon irradiation. Radiat Res. 1995;142:12–15. [PubMed] [Google Scholar]
- Levin D, Schneider WM, Hoffmann HH, Yarden G, Busetto AG, Manor O, Sharma N, Rice CM, Schreiber G. Multifaceted activities of type I interferon are revealed by a receptor antagonist. Sci Signal. 2014;7:ra50. doi: 10.1126/scisignal.2004998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CL, Johnson GR. Stem cell factor enhances the survival but not the self-renewal of murine hematopoietic long-term repopulating cells. Blood. 1994;84:408–414. [PubMed] [Google Scholar]
- Lilla JN, Chen CC, Mukai K, BenBarak MJ, Franco CB, Kalesnikoff J, Yu M, Tsai M, Piliponsky AM, Galli SJ. Reduced mast cell and basophil numbers and function in Cpa3-Cre; Mcl-1fl/fl mice. Blood. 2011;118:6930–6938. doi: 10.1182/blood-2011-03-343962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JX, Migone TS, Tsang M, Friedmann M, Weatherbee JA, Zhou L, Yamauchi A, Bloom ET, Mietz J, John S, et al. The role of shared receptor motifs and common Stat proteins in the generation of cytokine pleiotropy and redundancy by IL-2, IL-4, IL-7, IL-13, and IL-15. Immunity. 1995;2:331–339. doi: 10.1016/1074-7613(95)90141-8. [DOI] [PubMed] [Google Scholar]
- Liu H, Chen X, Focia PJ, He X. Structural basis for stem cell factor-KIT signaling and activation of class III receptor tyrosine kinases. The EMBO journal. 2007;26:891–901. doi: 10.1038/sj.emboj.7601545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald-Obermann JL, Pike LJ. Different epidermal growth factor (EGF) receptor ligands show distinct kinetics and biased or partial agonism for homodimer and heterodimer formation. The Journal of biological chemistry. 2014;289:26178–26188. doi: 10.1074/jbc.M114.586826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S, Ring AM, Amarnath S, Spangler JB, Li P, Ju W, Fischer S, Oh J, Spolski R, Weiskopf K, et al. Interleukin-2 activity can be fine tuned with engineered receptor signaling clamps. Immunity. 2015;42:826–838. doi: 10.1016/j.immuni.2015.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molineux G, Migdalska A, Szmitkowski M, Zsebo K, Dexter TM. The effects on hematopoiesis of recombinant stem cell factor (ligand for c-kit) administered in vivo to mice either alone or in combination with granulocyte colony-stimulating factor. Blood. 1991;78:961–966. [PubMed] [Google Scholar]
- Moraga I, Richter D, Wilmes S, Winkelmann H, Jude K, Thomas C, Suhoski MM, Engleman EG, Piehler J, Garcia KC. Instructive roles for cytokine-receptor binding parameters in determining signaling and functional potency. Sci Signal. 2015a;8:ra114. doi: 10.1126/scisignal.aab2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraga I, Spangler J, Mendoza JL, Garcia KC. Multifarious determinants of cytokine receptor signaling specificity. Advances in immunology. 2014;121:1–39. doi: 10.1016/B978-0-12-800100-4.00001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraga I, Wernig G, Wilmes S, Gryshkova V, Richter CP, Hong WJ, Sinha R, Guo F, Fabionar H, Wehrman TS, et al. Tuning cytokine receptor signaling by re-orienting dimer geometry with surrogate ligands. Cell. 2015b;160:1196–1208. doi: 10.1016/j.cell.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskowitz CH, Stiff P, Gordon MS, McNiece I, Ho AD, Costa JJ, Broun ER, Bayer RA, Wyres M, Hill J, et al. Recombinant methionyl human stem cell factor and filgrastim for peripheral blood progenitor cell mobilization and transplantation in non-Hodgkin’s lymphoma patients--results of a phase I/II trial. Blood. 1997;89:3136–3147. [PubMed] [Google Scholar]
- Nepom GT, Ehlers M, Mandrup-Poulsen T. Anti-cytokine therapies in T1D: Concepts and strategies. Clinical immunology. 2013;149:279–285. doi: 10.1016/j.clim.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicola NA. Cytokine pleiotropy and redundancy: a view from the receptor. Stem cells. 1994;12(Suppl 1):3–12. discussion 12–14. [PubMed] [Google Scholar]
- Reshetnyak AV, Opatowsky Y, Boggon TJ, Folta-Stogniew E, Tome F, Lax I, Schlessinger J. The strength and cooperativity of KIT ectodomain contacts determine normal ligand-dependent stimulation or oncogenic activation in cancer. Mol Cell. 2015;57:191–201. doi: 10.1016/j.molcel.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riese DJ., 2nd Ligand-based receptor tyrosine kinase partial agonists: New paradigm for cancer drug discovery? Expert Opin Drug Discov. 2011;6:185–193. doi: 10.1517/17460441.2011.547468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roder F, Wilmes S, Richter CP, Piehler J. Rapid transfer of transmembrane proteins for single molecule dimerization assays in polymer-supported membranes. ACS chemical biology. 2014;9:2479–2484. doi: 10.1021/cb5005806. [DOI] [PubMed] [Google Scholar]
- Ronnstrand L. Signal transduction via the stem cell factor receptor/c-Kit. Cell Mol Life Sci. 2004;61:2535–2548. doi: 10.1007/s00018-004-4189-6. [DOI] [PubMed] [Google Scholar]
- Schlessinger J, Ullrich A. Growth factor signaling by receptor tyrosine kinases. Neuron. 1992;9:383–391. doi: 10.1016/0896-6273(92)90177-f. [DOI] [PubMed] [Google Scholar]
- Serge A, Bertaux N, Rigneault H, Marguet D. Dynamic multiple-target tracing to probe spatiotemporal cartography of cell membranes. Nature methods. 2008;5:687–694. doi: 10.1038/nmeth.1233. [DOI] [PubMed] [Google Scholar]
- Stroud RM, Wells JA. Mechanistic diversity of cytokine receptor signaling across cell membranes. Science’s STKE: signal transduction knowledge environment. 2004;2004:re7. doi: 10.1126/stke.2312004re7. [DOI] [PubMed] [Google Scholar]
- Taketomi Y, Sunaga K, Tanaka S, Nakamura M, Arata S, Okuda T, Moon TC, Chang HW, Sugimoto Y, Kokame K, et al. Impaired mast cell maturation and degranulation and attenuated allergic responses in Ndrg1-deficient mice. Journal of immunology. 2007;178:7042–7053. doi: 10.4049/jimmunol.178.11.7042. [DOI] [PubMed] [Google Scholar]
- Taylor AM, Galli SJ, Coleman JW. Stem-cell factor, the kit ligand, induces direct degranulation of rat peritoneal mast cells in vitro and in vivo: dependence of the in vitro effect on period of culture and comparisons of stem-cell factor with other mast cell-activating agents. Immunology. 1995;86:427–433. [PMC free article] [PubMed] [Google Scholar]
- Thomas D, Vadas M, Lopez A. Regulation of haematopoiesis by growth factors - emerging insights and therapies. Expert opinion on biological therapy. 2004;4:869–879. doi: 10.1517/14712598.4.6.869. [DOI] [PubMed] [Google Scholar]
- Tong J, Gordon MS, Srour EF, Cooper RJ, Orazi A, McNiece I, Hoffman R. In vivo administration of recombinant methionyl human stem cell factor expands the number of human marrow hematopoietic stem cells. Blood. 1993;82:784–791. [PubMed] [Google Scholar]
- Vogelsang J, Kasper R, Steinhauer C, Person B, Heilemann M, Sauer M, Tinnefeld P. A reducing and oxidizing system minimizes photobleaching and blinking of fluorescent dyes. Angewandte Chemie. 2008;47:5465–5469. doi: 10.1002/anie.200801518. [DOI] [PubMed] [Google Scholar]
- Wada T, Ishiwata K, Koseki H, Ishikura T, Ugajin T, Ohnuma N, Obata K, Ishikawa R, Yoshikawa S, Mukai K, et al. Selective ablation of basophils in mice reveals their nonredundant role in acquired immunity against ticks. The Journal of clinical investigation. 2010;120:2867–2875. doi: 10.1172/JCI42680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lupardus P, Laporte SL, Garcia KC. Structural biology of shared cytokine receptors. Annu Rev Immunol. 2009;27:29–60. doi: 10.1146/annurev.immunol.24.021605.090616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf K, Ring AM, Ho CC, Volkmer JP, Levin AM, Volkmer AK, Ozkan E, Fernhoff NB, van de Rijn M, Weissman IL, et al. Engineered SIRPalpha variants as immunotherapeutic adjuvants to anticancer antibodies. Science. 2013;341:88–91. doi: 10.1126/science.1238856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DE, Foxworthe D, Teepe M, Lyman SD, Anderson D, Eisenman J. Recombinant murine steel factor stimulates in vitro production of granulocyte-macrophage progenitor cells. J Cell Biochem. 1992;50:221–226. doi: 10.1002/jcb.240500302. [DOI] [PubMed] [Google Scholar]
- Wilmes S, Beutel O, Li Z, Francois-Newton V, Richter CP, Janning D, Kroll C, Hanhart P, Hotte K, You C, et al. Receptor dimerization dynamics as a regulatory valve for plasticity of type I interferon signaling. The Journal of cell biology. 2015;209:579–593. doi: 10.1083/jcb.201412049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yea K, Xie J, Zhang H, Zhang W, Lerner RA. Selection of multiple agonist antibodies from intracellular combinatorial libraries reveals that cellular receptors are functionally pleiotropic. Current opinion in chemical biology. 2015;26:1–7. doi: 10.1016/j.cbpa.2015.01.002. [DOI] [PubMed] [Google Scholar]
- Yee NS, Hsiau CW, Serve H, Vosseller K, Besmer P. Mechanism of down-regulation of c-kit receptor. Roles of receptor tyrosine kinase, phosphatidylinositol 3′-kinase, and protein kinase C. The Journal of biological chemistry. 1994;269:31991–31998. [PubMed] [Google Scholar]
- You C, Richter CP, Lochte S, Wilmes S, Piehler J. Dynamic submicroscopic signaling zones revealed by pair correlation tracking and localization microscopy. Analytical chemistry. 2014;86:8593–8602. doi: 10.1021/ac501127r. [DOI] [PubMed] [Google Scholar]
- Yuzawa S, Opatowsky Y, Zhang Z, Mandiyan V, Lax I, Schlessinger J. Structural basis for activation of the receptor tyrosine kinase KIT by stem cell factor. Cell. 2007;130:323–334. doi: 10.1016/j.cell.2007.05.055. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Zhang R, Joachimiak A, Schlessinger J, Kong XP. Crystal structure of human stem cell factor: implication for stem cell factor receptor dimerization and activation. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:7732–7737. doi: 10.1073/pnas.97.14.7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zsebo KM, Smith KA, Hartley CA, Greenblatt M, Cooke K, Rich W, McNiece IK. Radioprotection of mice by recombinant rat stem cell factor. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:9464–9468. doi: 10.1073/pnas.89.20.9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A–C) First-generation library design and selection. (A) First-generation library design. Left: Co-crystal structure of mouse SCF/c-Kit is shown in cartoon representation (PDB: 2O26). Amino acid positions randomized in the first generation library are highlighted and shown in sphere representation (light blue). Right: Table of randomized positions of the ‘contact’ residue library with possible amino acid variants and the corresponding degenerate DNA codons (noted in the parentheses). (B) Histogram overlays assessing 1 nM mKitD1-3 staining of the library at each round via flow cytometry. MFI = mean fluorescence intensity. (C) Summary of SCF variant sequence analysis. The position of mutated residues and their corresponding sequences in wild-type SCF are denoted in the first two columns of the table. Each column represents a unique SCF clone. Red text color indicates the consensus mutations. (D) Second-generation library design. Left: Co-crystal structure of mouse SCF/c-Kit is shown in cartoon representation (PDB: 2O26). Amino acid positions highlighted in green indicate the set of consensus mutations obtained from the first-generation selections and were not randomized. Amino acid positions highlighted in orange are residues randomized in the second-generation ‘affinity maturation’ library. Right: Table of randomized positions, possible amino acid substitutions and the corresponding degenerate DNA codons (noted in the parentheses) for the second-generation library. (E) Chromatograms of purified SCF variants over a Superdex-75 size exclusion column with the retention time denoted on the top of each of the main peaks. (F) Purified SCF variants resolved on a 12% SDS-PAGE gel under reducing conditions.
(A) Representative SPR sensorgrams of indicated monomeric SCF variants binding to immobilized human c-Kit domains 1-3 (hKitD1-3). (B) On-yeast competitive blocking of mouse SCF/c-Kit and human SCF/c-Kit interactions by soluble mouse SCF variants. Yeast expressing wild-type mSCF or hSCF were stained with 20 nM fluorescently-labeled mouse or human c-KitD1-3 tetramers, respectively, in the presence of indicated unlabeled soluble mouse SCF variants. Data represent the mean ± SEM and are representative of two independent experiments. MFI = mean fluorescence intensity.
(A and B) Cell surface labeling of mXFP-mKit. (A) Density (Left) and ratio (Right) of single molecule localizations obtained after labeling cell surface mXFP-mKit by addition of anti-GPF NBs conjugated with Rho11 (red) and DY647 (blue), respectively. (B) Decay in the relative number of single molecule localizations due to photobleaching. (C and D) Diffusion properties of mXFP-mKit quantified from single molecule trajectories. (C) Step-length histogram (time-lapse: 160 ms) obtained for mXFP-mKit in absence of ligand and in presence of SCF and S4-3a, respectively. (D) Mean square displacement (MSD) analysis of mXFP-mKit diffusion properties in absence of ligand and in presence of SCF and S4-3a, respectively.
Dose response of β-hexosaminidase release by human PBCMCs treated with IgE, SCF or S4-3a at indicated concentrations (ng/ml) as single agents for 30 min ex vivo. Data represent mean ± SEM. ***p<0.001, and ns = not significant (i.e., p>0.05) by unpaired, two-tailed Student’s t test.
(A) Schematics of the experimental setup. C57BL/6 mice were injected i.p. with PBS, 5 or 10 mg/kg of SCF, or 10 mg/kg of S4-3a, and body temperatures were monitored at 10-min time intervals for 60 min. (B) Body temperature of mice treated as described in (A). Data represent mean ± SEM. *p<0.05, ***p<0.001, and ns = not significant (i.e., p>0.05) compared to the PBS-treated control group by unpaired, two-tailed Student’s t test.
(A–D) C57BL/6 mice were challenged by i.p. injection of PBS or 10 mg/kg of either SCF or S4-3a. (A) Mouse movements ~20 min after injection of PBS (left), SCF (middle) or S4-3a (right). The y- and x-axes indicate arbitrary limits of a mouse cage. Each color represents the trace of one mouse. (B–D) One h post-injection, peritoneal cells were harvested by peritoneal lavage. (B) Representative images of May-Grünwald/Giemsa-stained cytospin preparations of peritoneal cells from mice after the indicated treatments. Black arrows indicate examples of naïve (i.e., apparently non-degranulated) mast cells. Red arrowheads indicate cells with macrophage-like morphology that have taken up metachromatically-stained granules, which were presumably released upon mast cell activation and degranulation. (C) Quantification of granule+ peritoneal cells (that are not non-degranulated mast cells) from (B). (D) Flow cytometry analysis of surface expression of c-Kit on peritoneal FcεRIα+c-Kit+ mast cells. (C and D) Data are pooled from two independent experiments. ***p<0.001, and ns = not significant (i.e., p>0.05) by Student’s t test.
(A and B) Flow cytometry gating strategy to identify primary mouse (A) peritoneal FcεRIα+c-Kit+ mast cells and (B) bone marrow LSK HSPCs. (C) Flow cytometry analysis of cell surface c-Kit expression in the peritoneal mast cell and bone marrow LSK cell populations identified in (A and B).
Individual mXFP-mKit receptors expressed on HeLa cells and labelled by anti-GPF NBs conjugated with Rho11 (red channel) and DY647 (blue channel), respectively, in the absence of ligands. Frame rate: 30 Hz; playback: real time.
Dimerization of mXFP-mKit receptors expressed on HeLa cells and labelled by anti-GPF NBs conjugated with Rho11 (red channel) and DY647 (blue channel), respectively, in the presence of 100 nM SCF. Individual mKit dimers (magenta) were identified by co-tracking and the corresponding co-trajectories are depicted as yellow lines. Frame rate: 30 Hz; playback: real time.
Dimerization of mXFP-mKit receptors expressed on HeLa cells and labelled by anti-GPF NBs conjugated with Rho11 (red channel) and DY647 (blue channel), respectively, in the presence of 100 nM S4-3a. Co-trajectories are indicated as yellow lines. Frame rate: 30 Hz; playback: real time.
C57BL/6 mice were challenged by i.p. injection of saline. One-minute video was taken approximately 20 minutes post-injection and sped up 2-fold for the final 30-second clip.
C57BL/6 mice were challenged by i.p. injection of 10 mg/kg SCF. One-minute video was taken approximately 20 minutes post-injection and sped up 2-fold for the final 30-second clip.
C57BL/6 mice were challenged by i.p. injection of 10 mg/kg S4-3a. One-minute video was taken approximately 20 minutes post-injection and sped up 2-fold for the final 30-second clip.